Abstract
Coronavirus disease 2019 caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has been spreading worldwide since its outbreak in December 2019, and World Health Organization declared it as a pandemic on March 11, 2020. SARS-CoV-2 is highly contagious and is transmitted through airway epithelial cells as the first gateway. SARS-CoV-2 is detected by nasopharyngeal or oropharyngeal swab samples, and the viral load is significantly high in the upper respiratory tract. The host cellular receptors in airway epithelial cells, including angiotensin-converting enzyme 2 and transmembrane serine protease 2, have been identified by single-cell RNA sequencing or immunostaining. The expression levels of these molecules vary by type, function, and location of airway epithelial cells, such as ciliated cells, secretory cells, olfactory epithelial cells, and alveolar epithelial cells, as well as differ from host to host depending on age, sex, or comorbid diseases. Infected airway epithelial cells by SARS-CoV-2 in ex vivo experiments produce chemokines and cytokines to recruit inflammatory cells to target organs. Same as other viral infections, IFN signaling is a critical pathway for host defense. Various studies are underway to confirm the pathophysiological mechanisms of SARS-CoV-2 infection. Herein, we review cellular entry, host-viral interactions, immune responses to SARS-CoV-2 in airway epithelial cells. We also discuss therapeutic options related to epithelial immune reactions to SARS-CoV-2.
Keywords: COVID-19, Coronavirus, SARS-CoV-2, Respiratory system, Epithelial cells
INTRODUCTION
In the ongoing coronavirus disease 2019 (COVID-19) pandemic caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), almost all countries are struggling with the global economic, social, and health crisis. The respiratory virus SARS-CoV-2 was first recognized as an outbreak of pneumonia in December 2019 (1). It is transmitted through droplets to the airways and then mainly attacks the respiratory system (2,3,4). COVID-19 patients present various clinical symptoms, including fever, cough, sore throat, myalgia, headache, loss of smell or taste, nausea or vomiting, and diarrhea (5). Fever and respiratory symptoms are the main symptoms, and clinical manifestations of COVID-19 patients range from asymptomatic to systemic cytokine storm (5,6).
SARS-CoV-2 is highly contagious and transmitted through direct contact, respiratory droplets, or possibly aerosols (7,8). Diagnostic test using polymerase chain reaction detects the nucleic acids of SARS-CoV-2 with nasopharyngeal or oropharyngeal swab samples (9). Nasopharyngeal or nasal swabs have shown higher viral loads compared with oropharyngeal or oral swabs (10,11). This finding may suggest that the nasal or nasopharyngeal epithelium is the first gateway for viral invasion and transmission (12). Some cellular interactions between host cells and coronavirus have been discovered through the previous pandemic of SARS-CoV in 2003 and the epidemic of Middle East respiratory syndrome (MERS)-CoV in 2013 (13,14). The viral envelop spike (S) protein trimer of SARS-CoV and SARS-CoV-2 binds to the human angiotensin-converting enzyme 2 (ACE2), and S protein is subsequently primed by host cellular protease, transmembrane serine protease 2 (TMPRSS2) (15,16). Clinical manifestations and complications in specific organs have been associated with the cellular expression of ACE2 and TMPRSS2 (17,18). S protein of SARS-CoV-2 binds human ACE2 with 10 to 20-fold higher affinity than SARS-CoV, which may potentiate the infectivity of the virus (19). The nasal epithelium is the gateway and reservoir for SARS-CoV-2, but the epithelial barrier function and subsequent immune reaction can play a key role in protecting the host from infection (20). It is necessary to understand the host's immune reaction to SARS-CoV-2 and to develop therapeutic strategies to control COVID-19.
Global researches on COVID-19 has been proceeding very rapidly and urgently, and much has been revealed within a short period of time. However, until now, extensive studies on the pathophysiological mechanism of SARS-CoV-2 and host epithelial cells are warranted, and further studies on downstream of immune reaction are also needed. Many experts and researchers have emphasized the importance of therapeutics and vaccination from the beginning of the pandemic (21). Unfortunately, there are no definite curable antiviral drugs for SARS-CoV-2, and various clinical trials are underway (22). In this review, we focused on the cell tropism, host-viral interaction, and immune response of SARS-CoV-2 in airway epithelial cells, based on the latest findings published so far. Also, we reviewed therapeutic targets for the epithelial immune response.
CELLULAR ENTRY OF SARS-COV-2 VIA AIRWAY EPITHELIAL CELLS
The primary role of ACE2 is the maturation of angiotensin of the renin-angiotensin system, which controls blood pressure and vasoconstriction (23). ACE2 is expressed in the heart, blood vessels, kidney, esophagus, ileum, colon, upper and lower airways, cornea, liver, gallbladder, and testis (12). However, compared to other organs, the amount of gene or protein expression of ACE2 in the airways is low (12,24). Still, entry of SARS-CoV-2 depends on the expression of receptors (ACE2, TMPRSS2, or cathepsin B and cathepsin L [CatB/L]) of the airways as the first gateway for the respiratory virus to initiate infection, and the distribution of receptors in the upper airway increases the infectivity of the virus (12). Originally, ACE2 plays a protective role in the acute lung injury in respiratory viral infections, such as SARS-CoV and influenza virus (25,26,27).
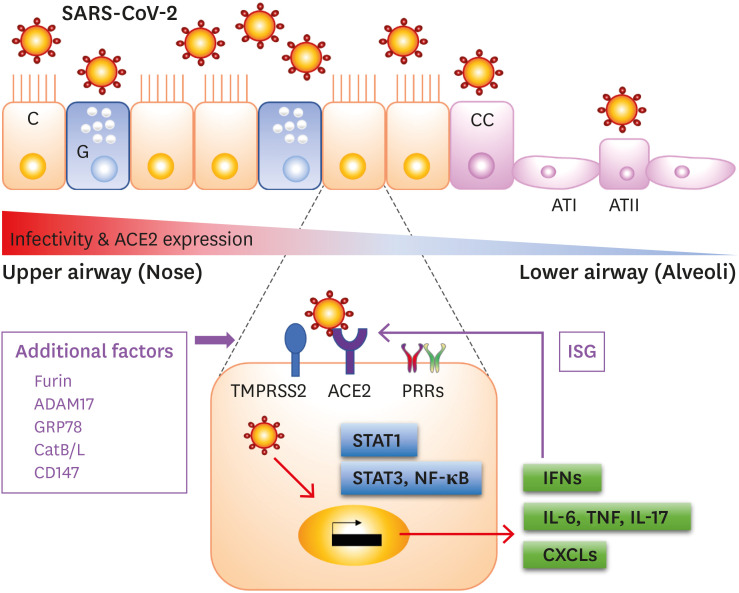
The trimeric S protein of SARS-CoV-2 is cleaved into S1 and S2, and S1 directly binds to the ACE2 (28). Another cleavage site on S2 is subsequently cleaved by host proteases. To understand the host infection of SARS-CoV-2, we reviewed recent studies of the processes involved in the epithelial cell entry (Fig. 1).
Figure 1. An overview of SARS-CoV-2 infection via airway epithelial cells. ACE2 is expressed in various types of airway epithelial cells, such as C, G, CC, and type II pneumocytes (ATII). The infectivity and ACE2 expression are gradually decreased from the upper airway (red color) to lower airway (blue color). Cellular protease, TMPRSS2, is promoting the cellular entry of the virus with other co-factors. SARS-CoV-2 can activate transcription factors and can stimulate to produce proinflammatory cytokines, chemokines, and IFNs. ACE2 is upregulated by IFNs, so it acts as an ISG.
C, ciliated cells; G, goblet cells; CC, club cells; ATI, alveolar type I cell; ATII, alveolar type II cell; ADAM17, a disintegrin and metalloprotease domain metallopeptidase domain 17.
Single-cell RNA-sequencing (scRNA-seq) and immunostaining analyses
ACE2 is a well-known cellular receptor of SARS-CoV and SARS-CoV-2 in humans, while MERS-CoV binds dipeptidyl peptidase 4 (29). Many researchers have identified the cellular expression of ACE2 using scRNA-seq data from non-SARS-CoV-2-infected samples. In the upper airway, 1.3% of all secretory cells and 4% of goblet cells expressing ACE2 in the ethmoid sinus and inferior turbinate from healthy human donors (30). Another study confirmed the SARS-CoV-2 tropism of host cells and genetic expression of viral entry-related genes in the nasal epithelium (12). The authors found that ACE2 was highly expressed in nasal ciliated and secretory cells, and ACE2 was also detected in nasal epithelial cells cultured in vitro under the air-liquid interface (ALI) condition, a method of culturing cells that expose the apical surface in the air to resemble the airway (31). ACE2 was identified using immunofluorescence staining on the motile cilia of the nasal turbinate, ethmoid sinus, uncinate process, trachea, and bronchus (24). However, another study using immunofluorescence staining showed that ACE2 abundantly observed in the apical surface of sustentacular cells in the olfactory mucosa harvested from patients with chronic rhinosinusitis, and it was expressed at low levels in the respiratory epithelium (32). The authors suggested that SARS-CoV-2 replicates actively in the nasal and olfactory mucosa, and the expression of ACE2 at specific sites is associate with nasal symptoms, such as anosmia.
TMPRSS2, a host cellular protease promoting the cellular entry of the virus, was broadly expressed in the nasal epithelial cells (4% of goblet cells express ACE2 and 28% express TMPRSS2) (30). This finding suggests that ACE2 is a limiting factor of the transmissibility of SARS-CoV-2 to host epithelial cells (12). The expression of ACE2 on nasopharyngeal swab was higher in patients with COVID-19 than healthy control, and ACE2-positive cells were not increased after SARS-CoV-2 infection, suggesting that ACE2 expression in the upper airway is related to susceptibility for infection (33). In COVID-19 patients, secretory cells were ACE2+/TMPRSS2+/Furin+ cells on scRNA-seq data (33). Furin is a pro-protein convertase and can cleave S protein of SARS-CoV-2, but not SARS-CoV (34). In particular, Furin was abundant in the glandular cells of the olfactory epithelium (30).
In the lower respiratory tract, ACE2 was mainly expressed by type II pneumocyte (alveolar epithelial type II cell) and ciliated cells. Lung ACE2 protein expression was detected using immunohistochemistry of human tissue microarrays, and mucin 1-positive type II pneumocytes exhibited double staining with ACE2, consistent with scRNA-seq data (24). Co-expression of ACE2 and TMPRSS2 was identified in 3.8% of type II pneumocytes, while 6.7% express ACE2 and 29.5% express TMPRSS2, respectively (30). TMPRSS2 was abundantly expressed in type II pneumocytes, club cells, ciliated cells, and type I pneumocytes, and ACE2+ TMPRSS2+ cells included type II pneumocytes, nasal secretory cells, and absorptive enterocytes (30). Lukassen et al. (35) reported that a cluster of ACE2+ cells was identified in the subsegmental bronchial branches and characterized between secretory and ciliated cells, which are referred to as transient secretory cells. These cells co-expressed TMPRSS2 and Furin and highly expressed Rho GTPase, associated with viral replication in the host cells (35).
Additional host proteases or cellular receptors have been known to facilitate cellular entry of SARS-CoV-2 into the host cells, including a disintegrin and metalloprotease domain metallopeptidase domain 17, CatB/L, 78-kDa glucose-regulated protein (GRP78), and CD147 (36). Another host factor known to bind furin-cleaved substrates, neuropilin-1 (NRP1), is one that participates in the cellular entry and infectivity of SARS-CoV-2, and NRP1+ cells were identified in the olfactory epithelium from human COVID-19 autopsies (37,38). These molecules have been studied for their potential as therapeutic targets, which will be discussed later in this review (39).
Airway epithelial cell culture
Respiratory epithelial cell culture systems have been utilized for researches in a variety of fields. Compared with immortalized cell lines, such as Vero E6 or Huh7 cells, fully differentiated human bronchial epithelial cells (HBECs) were effectively infected with SARS-CoV-2, reflecting the biological phenomena occurring in humans (2,40). Submerged or ALI culture with airway epithelial cell lines (A549, bronchial epithelium transformed with Ad12-SV40 2B [BEAS-2B], and Calu-3 cells) or primary cells were analyzed using scRNA-seq or immunostaining (Table 1). Researchers have uncovered the same patterns of expression of cellular entry-related molecules as in human tissues (41). Normal HBECs in ALI were analyzed scRNA-seq and expressed ACE2 and TMPRSS2 (42). The type II pneumocyte cell line, A549 cells, exhibited strong expression of ACE2 in immunohistochemistry (43). Contrarily, some in vitro studies using A549 cell lines showed a low rate of infection and low expression of ACE2 (16,44). Therefore, researchers supplemented A549 cells with an exogenous vector expressing ACE2 for SARS-CoV-2 in vitro infection (45). Primary HBECs were expressing ACE2 and TMPRSS2, while TMPRSS2 was broadly expressed than ACE2 (35). The SARS-CoV-2 infection also occurred in primary human nasal epithelial cells (HNECs) which were obtained from the inferior turbinate of healthy donors (46). Cultured HNECs expressed ACE2 in the apical side and TMPRSS2 throughout the layer, respectively (46).
Table 1. Summary of airway epithelial cell types used in cell culture experiments with SARS-CoV-2.
| Cell types | Characteristics | Findings | |
|---|---|---|---|
| Primary HNEC | Primary cell | Express ACE2 and TMPRSS2 (46) | |
| SARS-CoV-2 infection on the apical surface | |||
| - Secretion of CXCL10 (46) | |||
| Primary HBEC | Primary cell | Express ACE2 and TMPRSS2 (35,42) | |
| SARS-CoV-2 infection on the apical surface | |||
| - Downregulation of tight junction molecules and loss of cilia (48) | |||
| - Production of IL-6, CXCL9, CXCL10, and CXCL11 (50) | |||
| - Induction of CCL20, CXCL1, IL-1β, IL-6, CXCL3, CXCL5, CXCL6, CXCL2, CXCL16, and TNF (45) | |||
| Therapeutic effect: remdesivir (42), camostat mesylate and CatB/L inhibitor (16) | |||
| A549 cell | Type II pneumocyte cell line | Strong expression (43) or low expression (16,44) of ACE2 | |
| BEAS-2B cell | Normal bronchial epithelial cell line | Therapeutic effect: meplazumab (102) | |
| Calu-3 cell | Lung cancer cell line | Therapeutic effect: camostat mesylate and CatB/L inhibitor (16), IFN-α and IFN-λ (106) | |
| Vero E6 cell | Kidney epithelial cell line | Therapeutic effect: meplazumab (102), human soluble ACE2 (99), IFN-α and IFN-λ (106) | |
Primary HBECs differentiated in ALI culture were infected by SARS-CoV-2, and viral particles were found on the apical surface of ciliated and secretory cells on transmission electron microscopy (47). During the long-term period of ALI culture (up to 51 days), the recurrent replication peak of SARS-CoV-2 was 7 to 10 days, and primary HBECs exhibited decreased tight junction molecule, zonula occludens-1, and loss of cilia (48). As SARS-CoV-2 is attached to the cilia of HBECs in a scanning electron microscopy (49), the virus did not infect HBECs on the basolateral side of the ALI culture system (48). In addition, no viral particles of SARS-CoV-2 were detected in the basolateral chamber (46). Longitudinal analysis of scRNA-seq from HBECs cultured in ALI condition showed that cellular tropism of the virus expands from ciliated cells to basal and club cells, suggesting that basal stem cells are secondary targets (50). Accordingly, ACE2 expression was increased during productive infection in both infected and bystander cells (50). Recombinant SARS-CoV-2, the infectious-clone-derived SARS-CoV-2 mNeonGreen (icSARS-CoV-2-mNG) (51), infected 12.5% of normal HBECs at ALI. Most of the infected cell types were ciliated cells, which was confirmed by flow cytometry and scRNA-seq, but basal cells were not infected by icSARS-CoV-2-mNG (42).
Several studies have been reported the data of SARS-CoV-2 infection of proximal and distal lung organoids cultures (52,53). About 10% of type II pneumocytes organoids or KRT5+ basal cell organoids revealed SARS-CoV-2 nucleocapsid protein expression, and ACE2 was found in the apical cell membranes (54). Human pluripotent stem cell-derived lung organoid also expressed type II pneumocyte-like cells rich in ACE2, TMPRSS2, and Furin, and infected organoids were induced robust chemokines, such as transcripts of TNF and IL-17 signaling (55). Researchers suggest that airway organoids can provide more physiologic responses to the viral infection compared to conventional cell cultures and can be used for drug screening (56).
Upregulation or downregulation of ACE2 in the airway
ACE2 is a transmembrane protein that is highly expressed in vascular endothelial cells of the lung, and the level of ACE2 expression in patients with COVID-19 varies from host to host (57). Viral receptor gene expression of host cells might be influenced by age, sex, smoking, medications, and comorbid diseases (58).
In an analysis using scRNA-seq of lung tissue and HBECs, there was no difference in ACE2 expression between males and females (35). The expression levels of ACE2 and TMPRSS2 in children were lower than in adults in nasal and bronchial epithelial tissue assessed using transcriptomic datasets (59). RNA analysis using a cytology brush sample of nasal epithelium, the expression of ACE2 gene in nasal cytology samples increased significantly with age after adjusted for sex and asthma (60). However, ACE2 expression of bronchioalveolar lavage fluid samples or overall scRNA-seq of the lung had no dependency on age (35,61). Protein levels of ACE2 in lung tissue scored using immunohistochemistry did not show significant differences among 29 subjects with different demographics, such as age, sex, and comorbidities (62). However, aging itself is the most decisive factor in serious illness for SARS-CoV-2 infection (63). In aged COVID-19 patients, the ability to eliminate the virus is impaired and proinflammatory response is augmented (64). The weakened epithelial barrier function and excessive reactive oxygen species production might exaggerate host damage by SARS-CoV-2 in the elderly (65,66).
ACE2 plays an important role in the renin-angiotensin system as a negative regulator by inactivating angiotensin II, which mediates vasoconstriction (67,68). Angiotensin II induces bronchoconstriction, vasoconstriction, fibrosis, cytokine release, and eventually, lung tissue injury (69). Of note, the history of taking ACE inhibitor or angiotensin II receptor blockers did not affect ACE2 expression in the upper airways (24). However, further investigation is needed to identify the effect of ACE inhibitor or angiotensin II receptor blockers in COVID-19.
Gene expression of ACE2 was the highest in male smokers compared to female smokers or non-smokers (70). Immunohistochemistry of TMPRSS2 and CD147 displayed higher positive cell counts in smokers with chronic obstructive pulmonary disease (COPD) than healthy control, whereas those of ACE2 and GRP78 revealed no differences in 98 human lung samples (36). Transcriptomic analysis of adult COPD patients showed that ACE2 and TMPRSS2 were upregulated compared with healthy controls (59). Furthermore, the level of ACE2 gene expression was negatively correlated with forced expiratory volume in 1 second and was overexpressed in COPD patients and current smokers (71). These results were consistent with clinical risk factors for COVID-19, including older age, smoking, and COPD (1). Purkayastha et al. (72) performed in vitro study to determine the direct effects of cigarette smoke exposure on airway basal stem cells cultured in ALI and infecting SARS-CoV-2. Cigarette smoke exposure increased the number of SARS-CoV-2 infected cells and induced apoptosis.
In contrast, allergy and asthma do not seem to increase the risk of COVID-19 (58). Gene expression data from asthma patients were used to evaluate the association with type 2 inflammation. Type 2 cytokines, IL13, but not IL4 and IL5, was inversely correlated with ACE2 expression, while IL4 expression was positively correlated with TMPRSS2 (73). In scRNA-seq data from asthmatic children and healthy controls, the expression of TMPRSS2 was related to mucus goblet cell markers and type 2 inflammation markers, whereas that of ACE2 was negatively correlated with type 2 inflammation (74). In the children's cohort, ACE2 expression in the nasal epithelium was lower in allergic sensitized subjects than non-atopic controls, regardless of asthma status (75). ALI cultured primary HBECs treated with 10 ng/ml (48 h) of IL-13 exhibited downregulation of ACE2 and upregulation of TMPRSS2 in both asthmatic and non-asthmatic atopic groups (73). In addition, chronic stimulation with IL-13 for 10 days on tracheal epithelial culture showed the same results that decreased ACE2 and increased TMPRSS2 expression (74). TMPRSS2 was highly expressed in secretory cells by IL-13 stimulation, suggesting that TMPRSS2 is associated with a mucus secretory network. However, the role and mechanism of ACE2 and TMPRSS2 in type 2 inflammatory disease is not yet clearly elucidated. In chronic rhinosinusitis, ACE2 and TMPRSS2 expressions seem to be regulated by the inflammatory milieu (76). Initially, it was known that the expression level of ACE2 was low in mucosal tissues of patients with chronic rhinosinusitis (30,77). Same as asthma, ACE2 expression was reduced in eosinophilic nasal polyps characterized by strong type 2 inflammation. However, non-eosinophilic nasal polyps with increased IFN-γ showed higher expression of ACE2 than eosinophilic polyps and control tissues (78).
INTERACTION OF EPITHELIAL CELLS AND IMMUNE CELLS IN SARS-COV-2 INFECTION
Generally, viral infection in airway epithelial cells induces virus-linked pyroptosis, epithelial disruption, loss of cilia, vascular leakage, and triggers local immune responses (79,80,81). Using pattern recognition receptors (PRRs), airway epithelial cells can detect the pathogen-associated molecular patterns. Single-stranded RNA virus such as SARS-CoV-2 was sensed by TLR7 and TLR8. After PRR activation, transcription of antiviral genes and secretion of cytokines/chemokines are subsequently processed (82). However, the role of PRRs and the innate immune response of SARS-CoV-2 are under investigation.
After intracellular PRRs sense viral RNAs, transcription factors are activated to initiate cellular antiviral responses, including induction of type I and III IFNs and secretion of chemokines (83,84,85). During infection and replication of the SARS-CoV-2, airway epithelial cells secrete inflammatory mediators and play a role in triggering host immunologic reactions (41). Unlike other respiratory viruses, cells infected with SARS-CoV-2 induce a low level of IFNs but drive strong expression of chemokines and cytokines (45). Type I IFN response impairment was associated with severe cases among COVID-19 patients (86). According to the gene set enrichment analysis of peripheral white blood cells from COVID-19 patients, innate and inflammatory pathways were increased in severe and critical patients, whereas type I IFN responses and IFN-stimulated genes (ISGs) were downregulated in those groups (86). Additionally, patients with severe COVID-19 had impaired IFN-α production when stimulated with TLR ligands in dendritic cells (87). Some patients with severe COVID-19 presented multisystemic inflammation due to cytokine storm, with a prolonged elevation of cytokines and chemokines (88,89). These imbalances in the immune reaction conferring low IFNs and high cytokines might cause serious and fatal disease in COVID-19 patients (90). Bronchioalveolar lavage fluids of COVID-19 patients were analyzed using scRNA-seq, and the results showed different immunologic profiles according to the disease severity (91,92). Severe cases had more vital interaction between epithelial cells and immune cells and more potent inflammatory macrophages and cytotoxic T cells (33).
Epithelial cell-derived cytokines and chemokines
Proinflammatory cytokines were elevated in the serum of COVID-19 patients, with higher levels of these cytokines associated with severe cases (93). Especially, serum IL-6 levels correlated with acute respiratory distress syndrome, respiratory failure, and increased mortality (94). Innate immune response in ALI culture with scRNA-seq resulted that infected cells (ciliated, basal, and club cells) produced IL-6, CXCL9, CXCL10, and CXCL11 (50). Bronchial secretory cells infected with SARS-CoV-2 expressed IL-6 at 24 h post-infection, which was blocked by remdesivir pretreatment (42). Another SARS-CoV-2 infection study showed that CXCL10 was detected in the basolateral chamber of ALI culture system of HNECs (46). Normal HBECs infected by SARS-CoV-2 induced CCL20, CXCL1, IL-1β, IL-6, CXCL3, CXCL5, CXCL6, CXCL2, CXCL16, and TNF (45). In COVID-19 patients, ciliated cells expressed CCL15, and secretory cells highly expressed CXCL1, CXCL3, CXCL6, CXCL16, and CXCL17, resulting in recruitment of monocytes, macrophages, neutrophils, T cells, and mast cells (33). SARS-CoV-2 infection can activate NF-κB via PRRs and angiotensin II can co-activate NF-κB and IL6-STAT3 axis (95). IL-6-mediated proinflammatory responses are associated with cytokine release syndrome in COVID-19 (89).
IFN signaling
Several studies confirmed that ACE2 is behavior as an ISG in human airway epithelial cells (30,33). ACE2 was directly enhanced by IFN-α stimulation in primary HNECs (30), same as ACE2 was upregulated by IFN-β in HBECs (96). Upper and lower airway epithelial cells, especially ACE2+ TMPRSS2+ cells, have been found to be upregulated with genes involved in IFN signaling, including ADAR, GBP2, OAS1, JAK1, and DUOX2 (12,30). Induction of IFN in normal HBECs was strongly correlated with levels of SARS-CoV-2 replication, and ISGs were robustly induced in ciliated cells (42). SARS-CoV-2 in vitro infection did not elicit type I or III IFN expression but exhibit moderate levels of ISGs and proinflammatory cytokines in normal HBECs and unmodified A549 cells (45). Conversely, human parainfluenza virus 3 and respiratory syncytial virus induced high levels of IFNs and ISGs. Recently, Onabajo et al. (97) have identified the role of truncated isotype of ACE2, dACE2, as an ISG. IFNs and respiratory viruses, including SARS-CoV-2 induced dACE2, but not full-length ACE2, in various human cell lines, while overexpressed dACE2 does not bind to SARS-CoV-2. STAT1 was one of the best predictors for ACE2 expression in the airway epithelium (33). Interestingly, phosphorylation of STAT1 was blocked in infected lower airway cells, while uninfected bystander cells showed STAT1 phosphorylation and nuclear translocation, resulting in ISGs induction (52).
EPITHELIAL CELL-RELATED THERAPEUTIC OPTIONS
Blocking the cellular entry of SARS-CoV-2
ACE2 acts as a receptor of viral entry of SARS-CoV-2 and SARS-CoV, while protecting from lung injury (98). Human recombinant soluble ACE2 (hrsACE2) was treated with SARS-CoV-2 infected Vero E6 cells, and hrsACE2 inhibited the viral infection in a dose-dependent manner (99). The protective role of hrsACE2 has been proved in acute respiratory distress syndrome (100,101). TMPRSS2 inhibitor (camostat mesylate) and endosomal cysteine proteases CatB/L inhibitor (ammonium chloride or E-64d) block SARS-CoV-2 priming and entry in primary HBECs and Calu-3 cells (16). An anti-CD147 monoclonal Ab, meplazumab, can block SARS-CoV-2 amplification in Vero E6 and BEAS-2B cell lines (102). COVID-19 patients treated with meplazumab had a shorter recovery time and lower severity of the disease (103).
Therapeutic role of IFNs
To overcome viral IFN evasion strategies, antiviral activity of type I and III IFNs have been studied in SARS-CoV and MERS-CoV (104,105). In human Calu-3 and Vero E6 cell line, IFN-α and IFN-λ inhibited SARS-CoV-2 replication, while only IFN-α blocked SARS-CoV (106). Another study on Calu-3 and Vero E6 cells pretreated with IFN-α demonstrated that only SARS-CoV-2 was attenuated and failed to phosphorylate of STAT1 and express ISGs (107). Overall, SARS-CoV-2 is more sensitive to IFN treatment than SARS-CoV (108). Type III IFN, also known as IFN-λ, plays an essential role as a first defense line at the mucosal barrier surface, such as respiratory, gastrointestinal, and genitourinary tracts (109). Unlikely type I IFNs, IFN-λ receptor, IFNLR, is restricted to epithelial cells and neutrophils (110). Therefore, treatment with IFN-λ at an early stage of COVID-19 would elicit ISG and antiviral response in epithelial cells (111). However, Broggi et al. (112) recently reported that IFN-λ produced by dendritic cells in the lower airway in SARS-CoV-2 infection, and chronic exposure of IFN-λ on lung epithelial cells compromise barrier function, predisposing secondary bacterial infection. Taken together, further meticulous studies are warranted to evaluate the therapeutic roles of IFNs in COVID-19.
CONCLUSION
Airway epithelial cells are crucial for cell entry and viral replication of SARS-CoV-2. Expression of ACE2, the host cellular receptor for SARS-CoV-2, is regulated by a variety of host factors, such as age, sex, smoking, and comorbidities. Once exposed to SARS-CoV-2, the amount of virus entering the host tissues depends on the expression level of principal receptors in the airway epithelial cells. The subsequent immune response and inflammatory substances secreted by infected epithelial cells play an essential role in initiating infection and triggering inflammation. Therefore, ACE2 expression and epithelial cell-derived inflammatory markers are associated with the clinical course and prognosis of COVID-19. Reduced or blocked type I IFN response leads to an imbalance of the immune system in COVID-19 patients, and it can be a biomarker of impairment of innate immunity to SARS-CoV-2 infection. Further studies on the interaction between SARS-CoV-2 and airway epithelial cells and their potential as therapeutic targets are needed.
ACKNOWLEDGEMENTS
This work was supported by a grant from the National Research Foundation of Korea (2020R1A4A2002903 to H.W.S. and 2020R1I1A3075353 to G.R.) and a grant of the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (grant No. HI17C1669). This work was supported by the Soonchunhyang University Research Fund.
Abbreviations
- ACE2
Angiotensin-converting enzyme 2
- ALI
air-liquid interface
- BEAS-2B
bronchial epithelium transformed with Ad12-SV40 2B
- CatB/L
cathepsin B and cathepsin L
- COPD
chronic obstructive pulmonary disease
- COVID-19
coronavirus disease 2019
- GRP78
78-kDa glucose-regulated protein
- HBEC
human bronchial epithelial cell
- HNEC
human nasal epithelial cell
- hrsACE2
human recombinant soluble ACE2
- icSARS-CoV-2-mNG
infectious-clone-derived SARS-CoV-2 mNeonGreen
- ISG
IFN-stimulated gene
- MERS
Middle East respiratory syndrome
- NRP1
neuropilin-1
- PRR
pattern recognition receptors
- SARS-CoV-2
severe acute respiratory syndrome coronavirus 2
- scRNA-seq
single-cell RNA-sequencing
- S
spike
- TMPRSS2
transmembrane serine protease 2
Footnotes
Conflict of Interest: The authors declare no potential conflicts of interest.
- Conceptualization: Ryu G, Shin HW.
- Data curation: Ryu G, Shin HW.
- Formal analysis: Ryu G, Shin HW.
- Funding acquisition: Ryu G, Shin HW.
- Visualization: Ryu G, Shin HW.
- Writing - original draft: Ryu G, Shin HW.
- Writing - review & editing: Ryu G, Shin HW.
References
- 1.Wang D, Hu B, Hu C, Zhu F, Liu X, Zhang J, Wang B, Xiang H, Cheng Z, Xiong Y, et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA. 2020;323:1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu F, Zhao S, Yu B, Chen YM, Wang W, Song ZG, Hu Y, Tao ZW, Tian JH, Pei YY, et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, Si HR, Zhu Y, Li B, Huang CL, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Larsen JR, Martin MR, Martin JD, Kuhn P, Hicks JB. Modeling the onset of symptoms of COVID-19. Front Public Health. 2020;8:473. doi: 10.3389/fpubh.2020.00473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Riphagen S, Gomez X, Gonzalez-Martinez C, Wilkinson N, Theocharis P. Hyperinflammatory shock in children during COVID-19 pandemic. Lancet. 2020;395:1607–1608. doi: 10.1016/S0140-6736(20)31094-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Richard M, Kok A, de Meulder D, Bestebroer TM, Lamers MM, Okba NMA, Fentener van Vlissingen M, Rockx B, Haagmans BL, Koopmans MPG, et al. SARS-CoV-2 is transmitted via contact and via the air between ferrets. Nat Commun. 2020;11:3496. doi: 10.1038/s41467-020-17367-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang R, Li Y, Zhang AL, Wang Y, Molina MJ. Identifying airborne transmission as the dominant route for the spread of COVID-19. Proc Natl Acad Sci U S A. 2020;117:14857–14863. doi: 10.1073/pnas.2009637117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zou L, Ruan F, Huang M, Liang L, Huang H, Hong Z, Yu J, Kang M, Song Y, Xia J, et al. SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N Engl J Med. 2020;382:1177–1179. doi: 10.1056/NEJMc2001737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vlek ALM, Wesselius TS, Achterberg R, Thijsen SFT. Combined throat/nasal swab sampling for SARS-CoV-2 is equivalent to nasopharyngeal sampling. Eur J Clin Microbiol Infect Dis. 2021;40:193–195. doi: 10.1007/s10096-020-03972-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang H, Liu Q, Hu J, Zhou M, Yu MQ, Li KY, Xu D, Xiao Y, Yang JY, Lu YJ, et al. Nasopharyngeal swabs are more sensitive than oropharyngeal swabs for COVID-19 diagnosis and monitoring the SARS-CoV-2 load. Front Med (Lausanne) 2020;7:334. doi: 10.3389/fmed.2020.00334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sungnak W, Huang N, Bécavin C, Berg M, Queen R, Litvinukova M, Talavera-López C, Maatz H, Reichart D, Sampaziotis F, et al. SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat Med. 2020;26:681–687. doi: 10.1038/s41591-020-0868-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fung M, Otani I, Pham M, Babik J. Zoonotic coronavirus epidemics: SARS, MERS, and COVID-19. Ann Allergy Asthma Immunol. 2020 doi: 10.1016/j.anai.2020.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peiris JS, Yuen KY, Osterhaus AD, Stöhr K. The severe acute respiratory syndrome. N Engl J Med. 2003;349:2431–2441. doi: 10.1056/NEJMra032498. [DOI] [PubMed] [Google Scholar]
- 15.Li W, Moore MJ, Vasilieva N, Sui J, Wong SK, Berne MA, Somasundaran M, Sullivan JL, Luzuriaga K, Greenough TC, et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu NH, Nitsche A, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bertram S, Heurich A, Lavender H, Gierer S, Danisch S, Perin P, Lucas JM, Nelson PS, Pöhlmann S, Soilleux EJ. Influenza and SARS-coronavirus activating proteases TMPRSS2 and HAT are expressed at multiple sites in human respiratory and gastrointestinal tracts. PLoS One. 2012;7:e35876. doi: 10.1371/journal.pone.0035876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Du M, Cai G, Chen F, Christiani DC, Zhang Z, Wang M. Multiomics evaluation of gastrointestinal and other clinical characteristics of COVID-19. Gastroenterology. 2020;158:2298–2301.e7. doi: 10.1053/j.gastro.2020.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wrapp D, Wang N, Corbett KS, Goldsmith JA, Hsieh CL, Abiona O, Graham BS, McLellan JS. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gallo O, Locatello LG, Mazzoni A, Novelli L, Annunziato F. The central role of the nasal microenvironment in the transmission, modulation, and clinical progression of SARS-CoV-2 infection. Mucosal Immunol. 2020 doi: 10.1038/s41385-020-00359-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bedford J, Enria D, Giesecke J, Heymann DL, Ihekweazu C, Kobinger G, Lane HC, Memish Z, Oh MD, Sall AA, et al. COVID-19: towards controlling of a pandemic. Lancet. 2020;395:1015–1018. doi: 10.1016/S0140-6736(20)30673-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gordon DE, Jang GM, Bouhaddou M, Xu J, Obernier K, White KM, O'Meara MJ, Rezelj VV, Guo JZ, Swaney DL, et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583:459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Patel VB, Zhong JC, Grant MB, Oudit GY. Role of the ACE2/angiotensin 1–7 axis of the renin-angiotensin system in heart failure. Circ Res. 2016;118:1313–1326. doi: 10.1161/CIRCRESAHA.116.307708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee IT, Nakayama T, Wu CT, Goltsev Y, Jiang S, Gall PA, Liao CK, Shih LC, Schürch CM, McIlwain DR, et al. ACE2 localizes to the respiratory cilia and is not increased by ACE inhibitors or ARBs. Nat Commun. 2020;11:5453. doi: 10.1038/s41467-020-19145-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Imai Y, Kuba K, Rao S, Huan Y, Guo F, Guan B, Yang P, Sarao R, Wada T, Leong-Poi H, et al. Angiotensin-converting enzyme 2 protects from severe acute lung failure. Nature. 2005;436:112–116. doi: 10.1038/nature03712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuba K, Imai Y, Rao S, Gao H, Guo F, Guan B, Huan Y, Yang P, Zhang Y, Deng W, et al. A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus-induced lung injury. Nat Med. 2005;11:875–879. doi: 10.1038/nm1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang F, Guo J, Zou Z, Liu J, Cao B, Zhang S, Li H, Wang W, Sheng M, Liu S, et al. Angiotensin II plasma levels are linked to disease severity and predict fatal outcomes in H7N9-infected patients. Nat Commun. 2014;5:3595. doi: 10.1038/ncomms4595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yan R, Zhang Y, Li Y, Xia L, Guo Y, Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367:1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Raj VS, Mou H, Smits SL, Dekkers DH, Müller MA, Dijkman R, Muth D, Demmers JA, Zaki A, Fouchier RA, et al. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature. 2013;495:251–254. doi: 10.1038/nature12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ziegler CGK, Allon SJ, Nyquist SK, Mbano IM, Miao VN, Tzouanas CN, Cao Y, Yousif AS, Bals J, Hauser BM, et al. SARS-CoV-2 receptor ACE2 is an interferon-stimulated gene in human airway epithelial cells and is detected in specific cell subsets across tissues. Cell. 2020;181:1016–1035.e19. doi: 10.1016/j.cell.2020.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tata PR, Mou H, Pardo-Saganta A, Zhao R, Prabhu M, Law BM, Vinarsky V, Cho JL, Breton S, Sahay A, et al. Dedifferentiation of committed epithelial cells into stem cells in vivo . Nature. 2013;503:218–223. doi: 10.1038/nature12777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen M, Shen W, Rowan NR, Kulaga H, Hillel A, Ramanathan M, Jr, Lane AP. Elevated ACE-2 expression in the olfactory neuroepithelium: implications for anosmia and upper respiratory SARS-CoV-2 entry and replication. Eur Respir J. 2020;56:2001948. doi: 10.1183/13993003.01948-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chua RL, Lukassen S, Trump S, Hennig BP, Wendisch D, Pott F, Debnath O, Thürmann L, Kurth F, Völker MT, et al. COVID-19 severity correlates with airway epithelium-immune cell interactions identified by single-cell analysis. Nat Biotechnol. 2020;38:970–979. doi: 10.1038/s41587-020-0602-4. [DOI] [PubMed] [Google Scholar]
- 34.Coutard B, Valle C, de Lamballerie X, Canard B, Seidah NG, Decroly E. The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antiviral Res. 2020;176:104742. doi: 10.1016/j.antiviral.2020.104742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lukassen S, Chua RL, Trefzer T, Kahn NC, Schneider MA, Muley T, Winter H, Meister M, Veith C, Boots AW, et al. SARS-CoV-2 receptor ACE2 and TMPRSS2 are primarily expressed in bronchial transient secretory cells. EMBO J. 2020;39:e105114. doi: 10.15252/embj.20105114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aguiar JA, Tremblay BJ, Mansfield MJ, Woody O, Lobb B, Banerjee A, Chandiramohan A, Tiessen N, Cao Q, Dvorkin-Gheva A, et al. Gene expression and in situ protein profiling of candidate SARS-CoV-2 receptors in human airway epithelial cells and lung tissue. Eur Respir J. 2020;56:2001123. doi: 10.1183/13993003.01123-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cantuti-Castelvetri L, Ojha R, Pedro LD, Djannatian M, Franz J, Kuivanen S, van der Meer F, Kallio K, Kaya T, Anastasina M, et al. Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science. 2020;370:856–860. doi: 10.1126/science.abd2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Daly JL, Simonetti B, Klein K, Chen KE, Williamson MK, Antón-Plágaro C, Shoemark DK, Simón-Gracia L, Bauer M, Hollandi R, et al. Neuropilin-1 is a host factor for SARS-CoV-2 infection. Science. 2020;370:861–865. doi: 10.1126/science.abd3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Drożdżal S, Rosik J, Lechowicz K, Machaj F, Kotfis K, Ghavami S, Łos MJ. FDA approved drugs with pharmacotherapeutic potential for SARS-CoV-2 (COVID-19) therapy. Drug Resist Updat. 2020;53:100719. doi: 10.1016/j.drup.2020.100719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Milewska A, Kula-Pacurar A, Wadas J, Suder A, Szczepanski A, Dabrowska A, Owczarek K, Marcello A, Ochman M, Stacel T, et al. Replication of severe acute respiratory syndrome coronavirus 2 in human respiratory epithelium. J Virol. 2020;94:e00957-20. doi: 10.1128/JVI.00957-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sallenave JM, Guillot L. Innate immune signaling and proteolytic pathways in the resolution or exacerbation of SARS-CoV-2 in COVID-19: key therapeutic targets? Front Immunol. 2020;11:1229. doi: 10.3389/fimmu.2020.01229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fiege JK, Thiede JM, Nanda HA, Matchett WE, Moore PJ, Montanari NR, Thielen BK, Daniel J, Stanley E, Hunter RC, et al. Single cell resolution of SARS-CoV-2 tropism, antiviral responses, and susceptibility to therapies in primary human airway epithelium. PLoS Pathog. 2021;17:e1009292. doi: 10.1371/journal.ppat.1009292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hamming I, Timens W, Bulthuis ML, Lely AT, Navis G, van Goor H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J Pathol. 2004;203:631–637. doi: 10.1002/path.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Harcourt J, Tamin A, Lu X, Kamili S, Sakthivel SK, Murray J, Queen K, Tao Y, Paden CR, Zhang J, et al. Severe acute respiratory syndrome coronavirus 2 from patient with coronavirus disease, united states. Emerg Infect Dis. 2020;26:1266–1273. doi: 10.3201/eid2606.200516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Blanco-Melo D, Nilsson-Payant BE, Liu WC, Uhl S, Hoagland D, Møller R, Jordan TX, Oishi K, Panis M, Sachs D, et al. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell. 2020;181:1036–1045.e9. doi: 10.1016/j.cell.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gamage AM, Tan KS, Chan WO, Liu J, Tan CW, Ong YK, Thong M, Andiappan AK, Anderson DE, Wang Y, et al. Infection of human nasal epithelial cells with SARS-CoV-2 and a 382-nt deletion isolate lacking ORF8 reveals similar viral kinetics and host transcriptional profiles. PLoS Pathog. 2020;16:e1009130. doi: 10.1371/journal.ppat.1009130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhu N, Wang W, Liu Z, Liang C, Wang W, Ye F, Huang B, Zhao L, Wang H, Zhou W, et al. Morphogenesis and cytopathic effect of SARS-CoV-2 infection in human airway epithelial cells. Nat Commun. 2020;11:3910. doi: 10.1038/s41467-020-17796-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hao S, Ning K, Kuz CA, Vorhies K, Yan Z, Qiu J. Long-term modeling of SARS-CoV-2 infection of in vitro cultured polarized human airway epithelium. MBio. 2020;11:e02852-20. doi: 10.1128/mBio.02852-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ehre C. SARS-CoV-2 infection of airway cells. N Engl J Med. 2020;383:969. doi: 10.1056/NEJMicm2023328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ravindra NG, Alfajaro MM, Gasque V, Wei J, Filler RB, Huston NC, Wan H, Szigeti-Buck K, Wang B, Montgomery RR, et al. Single-cell longitudinal analysis of SARS-CoV-2 infection in human bronchial epithelial cells. bioRxiv. 2020 [Google Scholar]
- 51.Xie X, Muruato A, Lokugamage KG, Narayanan K, Zhang X, Zou J, Liu J, Schindewolf C, Bopp NE, Aguilar PV, et al. An infectious cDNA clone of SARS-CoV-2. Cell Host Microbe. 2020;27:841–848.e3. doi: 10.1016/j.chom.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lamers MM, van der Vaart J, Knoops K, Riesebosch S, Breugem TI, Mykytyn AZ, Beumer J, Schipper D, Bezstarosti K, Koopman CD, et al. An organoid-derived bronchioalveolar model for SARS-CoV-2 infection of human alveolar type II-like cells. EMBO J. 2020 doi: 10.15252/embj.2020105912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lamers MM, Beumer J, van der Vaart J, Knoops K, Puschhof J, Breugem TI, Ravelli RB, Paul van Schayck J, Mykytyn AZ, Duimel HQ, et al. SARS-CoV-2 productively infects human gut enterocytes. Science. 2020;369:50–54. doi: 10.1126/science.abc1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Salahudeen AA, Choi SS, Rustagi A, Zhu J, van Unen V, de la O SM, Flynn RA, Margalef-Català M, Santos AJ, Ju J, et al. Progenitor identification and SARS-CoV-2 infection in human distal lung organoids. Nature. 2020;588:670–675. doi: 10.1038/s41586-020-3014-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Han Y, Duan X, Yang L, Nilsson-Payant BE, Wang P, Duan F, Tang X, Yaron TM, Zhang T, Uhl S, et al. Identification of SARS-CoV-2 inhibitors using lung and colonic organoids. Nature. 2021;589:270–275. doi: 10.1038/s41586-020-2901-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mulay A, Konda B, Garcia G, Yao C, Beil S, Sen C, Purkayastha A, Kolls JK, Pociask DA, Pessina P, et al. SARS-CoV-2 infection of primary human lung epithelium for COVID-19 modeling and drug discovery. bioRxiv. 2020 doi: 10.1101/2020.06.29.174623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Matheson NJ, Lehner PJ. How does SARS-CoV-2 cause COVID-19? Science. 2020;369:510–511. doi: 10.1126/science.abc6156. [DOI] [PubMed] [Google Scholar]
- 58.Yao Y, Wang H, Liu Z. Expression of ACE2 in airways: Implication for COVID-19 risk and disease management in patients with chronic inflammatory respiratory diseases. Clin Exp Allergy. 2020;50:1313–1324. doi: 10.1111/cea.13746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Saheb Sharif-Askari N, Saheb Sharif-Askari F, Alabed M, Temsah MH, Al Heialy S, Hamid Q, Halwani R. Airways expression of SARS-CoV-2 receptor, ACE2, and TMPRSS2 is lower in children than adults and increases with smoking and COPD. Mol Ther Methods Clin Dev. 2020;18:1–6. doi: 10.1016/j.omtm.2020.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bunyavanich S, Do A, Vicencio A. Nasal gene expression of angiotensin-converting enzyme 2 in children and adults. JAMA. 2020;323:2427–2429. doi: 10.1001/jama.2020.8707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schouten LR, van Kaam AH, Kohse F, Veltkamp F, Bos LD, de Beer FM, van Hooijdonk RT, Horn J, Straat M, Witteveen E, et al. Age-dependent differences in pulmonary host responses in ARDS: a prospective observational cohort study. Ann Intensive Care. 2019;9:55. doi: 10.1186/s13613-019-0529-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ortiz ME, Thurman A, Pezzulo AA, Leidinger MR, Klesney-Tait JA, Karp PH, Tan P, Wohlford-Lenane C, McCray PB, Jr, Meyerholz DK. Heterogeneous expression of the SARS-coronavirus-2 receptor ACE2 in the human respiratory tract. EBioMedicine. 2020;60:102976. doi: 10.1016/j.ebiom.2020.102976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Brodin P. Immune determinants of COVID-19 disease presentation and severity. Nat Med. 2021;27:28–33. doi: 10.1038/s41591-020-01202-8. [DOI] [PubMed] [Google Scholar]
- 64.Bajaj V, Gadi N, Spihlman AP, Wu SC, Choi CH, Moulton VR. Aging, immunity, and COVID-19: how age influences the host immune response to coronavirus infections? Front Physiol. 2021;11:571416. doi: 10.3389/fphys.2020.571416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Inde Z, Yapp C, Joshi GN, Spetz J, Fraser C, Deskin B, Ghelfi E, Sodhi C, Hackam D, Kobzik L, et al. Age-dependent regulation of SARS-CoV-2 cell entry genes and cell death programs correlates with COVID-19 disease severity. bioRxiv. 2020 doi: 10.1101/2020.09.13.276923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Meftahi GH, Jangravi Z, Sahraei H, Bahari Z. The possible pathophysiology mechanism of cytokine storm in elderly adults with COVID-19 infection: the contribution of “inflame-aging”. Inflamm Res. 2020;69:825–839. doi: 10.1007/s00011-020-01372-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Donoghue M, Hsieh F, Baronas E, Godbout K, Gosselin M, Stagliano N, Donovan M, Woolf B, Robison K, Jeyaseelan R, et al. A novel angiotensin-converting enzyme-related carboxypeptidase (ACE2) converts angiotensin I to angiotensin 1-9. Circ Res. 2000;87:E1–E9. doi: 10.1161/01.res.87.5.e1. [DOI] [PubMed] [Google Scholar]
- 68.Crackower MA, Sarao R, Oudit GY, Yagil C, Kozieradzki I, Scanga SE, Oliveira-dos-Santos AJ, da Costa J, Zhang L, Pei Y, et al. Angiotensin-converting enzyme 2 is an essential regulator of heart function. Nature. 2002;417:822–828. doi: 10.1038/nature00786. [DOI] [PubMed] [Google Scholar]
- 69.Hamming I, Cooper ME, Haagmans BL, Hooper NM, Korstanje R, Osterhaus AD, Timens W, Turner AJ, Navis G, van Goor H. The emerging role of ACE2 in physiology and disease. J Pathol. 2007;212:1–11. doi: 10.1002/path.2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang H, Rostami MR, Leopold PL, Mezey JG, O'Beirne SL, Strulovici-Barel Y, Crystal RG. Expression of the SARS-CoV-2 ACE2 receptor in the human airway epithelium. Am J Respir Crit Care Med. 2020;202:219–229. doi: 10.1164/rccm.202003-0541OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Leung JM, Yang CX, Tam A, Shaipanich T, Hackett TL, Singhera GK, Dorscheid DR, Sin DD. ACE-2 expression in the small airway epithelia of smokers and COPD patients: implications for COVID-19. Eur Respir J. 2020;55:55. doi: 10.1183/13993003.00688-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Purkayastha A, Sen C, Garcia G, Jr, Langerman J, Shia DW, Meneses LK, Vijayaraj P, Durra A, Koloff CR, Freund DR, et al. Direct exposure to SARS-CoV-2 and cigarette smoke increases infection severity and alters the stem cell-derived airway repair response. Cell Stem Cell. 2020;27:869–875.e4. doi: 10.1016/j.stem.2020.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kimura H, Francisco D, Conway M, Martinez FD, Vercelli D, Polverino F, Billheimer D, Kraft M. Type 2 inflammation modulates ACE2 and TMPRSS2 in airway epithelial cells. J Allergy Clin Immunol. 2020;146:80–88.e8. doi: 10.1016/j.jaci.2020.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sajuthi SP, DeFord P, Li Y, Jackson ND, Montgomery MT, Everman JL, Rios CL, Pruesse E, Nolin JD, Plender EG, et al. Type 2 and interferon inflammation regulate SARS-CoV-2 entry factor expression in the airway epithelium. Nat Commun. 2020;11:5139. doi: 10.1038/s41467-020-18781-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jackson DJ, Busse WW, Bacharier LB, Kattan M, O'Connor GT, Wood RA, Visness CM, Durham SR, Larson D, Esnault S, et al. Association of respiratory allergy, asthma, and expression of the SARS-CoV-2 receptor ACE2. J Allergy Clin Immunol. 2020;146:203–206.e3. doi: 10.1016/j.jaci.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang M, Wang C, Zhang L. Inflammatory endotypes of CRSwNP and responses to COVID-19. Curr Opin Allergy Clin Immunol. 2021;21:8–15. doi: 10.1097/ACI.0000000000000700. [DOI] [PubMed] [Google Scholar]
- 77.Wang H, Song J, Pan L, Deng YK, Yao Y, Wang ZC, Liao B, Ma J, Zeng M, Liu Z. Regional differences in ACE2 expression in the sinonasal mucosa of adult Chinese patients with chronic rhinosinusitis. Allergy. 2020 doi: 10.1111/all.14623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang M, Bu X, Fang G, Luan G, Huang Y, Akdis CA, Wang C, Zhang L. Distinct expression of SARS-CoV-2 receptor ACE2 correlates with endotypes of chronic rhinosinusitis with nasal polyps. Allergy. 2020 doi: 10.1111/all.14665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Narasaraju T. Histopathologic changes and SARS-CoV-2 immunostaining in the lung of a patient with COVID-19. Ann Intern Med. 2020;173:323–324. doi: 10.7326/L20-0894. [DOI] [PubMed] [Google Scholar]
- 80.Chen IY, Moriyama M, Chang MF, Ichinohe T. Severe acute respiratory syndrome coronavirus viroporin 3a activates the nlrp3 inflammasome. Front Microbiol. 2019;10:50. doi: 10.3389/fmicb.2019.00050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Fink SL, Cookson BT. Apoptosis, pyroptosis, and necrosis: mechanistic description of dead and dying eukaryotic cells. Infect Immun. 2005;73:1907–1916. doi: 10.1128/IAI.73.4.1907-1916.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tay MZ, Poh CM, Rénia L, MacAry PA, Ng LFP. The trinity of COVID-19: immunity, inflammation and intervention. Nat Rev Immunol. 2020;20:363–374. doi: 10.1038/s41577-020-0311-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lazear HM, Schoggins JW, Diamond MS. Shared and distinct functions of type i and type iii interferons. Immunity. 2019;50:907–923. doi: 10.1016/j.immuni.2019.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hur S. Double-stranded RNA sensors and modulators in innate immunity. Annu Rev Immunol. 2019;37:349–375. doi: 10.1146/annurev-immunol-042718-041356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sokol CL, Luster AD. The chemokine system in innate immunity. Cold Spring Harb Perspect Biol. 2015;7:a016303. doi: 10.1101/cshperspect.a016303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hadjadj J, Yatim N, Barnabei L, Corneau A, Boussier J, Smith N, Péré H, Charbit B, Bondet V, Chenevier-Gobeaux C, et al. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science. 2020;369:718–724. doi: 10.1126/science.abc6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Arunachalam PS, Wimmers F, Mok CKP, Perera RA, Scott M, Hagan T, Sigal N, Feng Y, Bristow L, Tak-Yin Tsang O, et al. Systems biological assessment of immunity to mild versus severe COVID-19 infection in humans. Science. 2020;369:1210–1220. doi: 10.1126/science.abc6261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lucas C, Wong P, Klein J, Castro TBR, Silva J, Sundaram M, Ellingson MK, Mao T, Oh JE, Israelow B, et al. Longitudinal analyses reveal immunological misfiring in severe COVID-19. Nature. 2020;584:463–469. doi: 10.1038/s41586-020-2588-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Moore JB, June CH. Cytokine release syndrome in severe COVID-19. Science. 2020;368:473–474. doi: 10.1126/science.abb8925. [DOI] [PubMed] [Google Scholar]
- 90.Sa Ribero M, Jouvenet N, Dreux M, Nisole S. Interplay between SARS-CoV-2 and the type I interferon response. PLoS Pathog. 2020;16:e1008737. doi: 10.1371/journal.ppat.1008737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bost P, Giladi A, Liu Y, Bendjelal Y, Xu G, David E, Blecher-Gonen R, Cohen M, Medaglia C, Li H, et al. Host-viral infection maps reveal signatures of severe COVID-19 patients. Cell. 2020;181:1475–1488.e12. doi: 10.1016/j.cell.2020.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Liao M, Liu Y, Yuan J, Wen Y, Xu G, Zhao J, Cheng L, Li J, Wang X, Wang F, et al. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat Med. 2020;26:842–844. doi: 10.1038/s41591-020-0901-9. [DOI] [PubMed] [Google Scholar]
- 93.Lee JS, Park S, Jeong HW, Ahn JY, Choi SJ, Lee H, Choi B, Nam SK, Sa M, Kwon JS, et al. Immunophenotyping of COVID-19 and influenza highlights the role of type I interferons in development of severe COVID-19. Sci Immunol. 2020;5:eabd1554. doi: 10.1126/sciimmunol.abd1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chen G, Wu D, Guo W, Cao Y, Huang D, Wang H, Wang T, Zhang X, Chen H, Yu H, et al. Clinical and immunological features of severe and moderate coronavirus disease 2019. J Clin Invest. 2020;130:2620–2629. doi: 10.1172/JCI137244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hirano T, Murakami M. COVID-19: a new virus, but a familiar receptor and cytokine release syndrome. Immunity. 2020;52:731–733. doi: 10.1016/j.immuni.2020.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zhuang MW, Cheng Y, Zhang J, Jiang XM, Wang L, Deng J, Wang PH. Increasing host cellular receptor-angiotensin-converting enzyme 2 expression by coronavirus may facilitate 2019-nCoV (or SARS-CoV-2) infection. J Med Virol. 2020;92:2693–2701. doi: 10.1002/jmv.26139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Onabajo OO, Banday AR, Stanifer ML, Yan W, Obajemu A, Santer DM, Florez-Vargas O, Piontkivska H, Vargas JM, Ring TJ, et al. Interferons and viruses induce a novel truncated ACE2 isoform and not the full-length SARS-CoV-2 receptor. Nat Genet. 2020;52:1283–1293. doi: 10.1038/s41588-020-00731-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhang H, Penninger JM, Li Y, Zhong N, Slutsky AS. Angiotensin-converting enzyme 2 (ACE2) as a SARS-CoV-2 receptor: molecular mechanisms and potential therapeutic target. Intensive Care Med. 2020;46:586–590. doi: 10.1007/s00134-020-05985-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Monteil V, Kwon H, Prado P, Hagelkrüys A, Wimmer RA, Stahl M, Leopoldi A, Garreta E, Hurtado Del Pozo C, Prosper F, et al. Inhibition of SARS-CoV-2 infections in engineered human tissues using clinical-grade soluble human ACE2. Cell. 2020;181:905–913.e7. doi: 10.1016/j.cell.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Khan A, Benthin C, Zeno B, Albertson TE, Boyd J, Christie JD, Hall R, Poirier G, Ronco JJ, Tidswell M, et al. A pilot clinical trial of recombinant human angiotensin-converting enzyme 2 in acute respiratory distress syndrome. Crit Care. 2017;21:234. doi: 10.1186/s13054-017-1823-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhang H, Baker A. Recombinant human ACE2: acing out angiotensin II in ARDS therapy. Crit Care. 2017;21:305. doi: 10.1186/s13054-017-1882-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wang K, Chen W, Zhang Z, Deng Y, Lian JQ, Du P, Wei D, Zhang Y, Sun XX, Gong L, et al. CD147-spike protein is a novel route for SARS-CoV-2 infection to host cells. Signal Transduct Target Ther. 2020;5:283. doi: 10.1038/s41392-020-00426-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bian H, Zheng ZH, Wei D, Zhang Z, Kang WZ, Hao CQ, Dong K, Kang W, Xia JL, Miao JL, et al. Meplazumab treats COVID-19 pneumonia: an open-labelled, concurrent controlled add-on clinical trial. medRxiv. 2020 [Google Scholar]
- 104.Lau SKP, Lau CCY, Chan KH, Li CPY, Chen H, Jin DY, Chan JFW, Woo PCY, Yuen KY. Delayed induction of proinflammatory cytokines and suppression of innate antiviral response by the novel Middle East respiratory syndrome coronavirus: implications for pathogenesis and treatment. J Gen Virol. 2013;94:2679–2690. doi: 10.1099/vir.0.055533-0. [DOI] [PubMed] [Google Scholar]
- 105.Cameron MJ, Ran L, Xu L, Danesh A, Bermejo-Martin JF, Cameron CM, Muller MP, Gold WL, Richardson SE, Poutanen SM, et al. Interferon-mediated immunopathological events are associated with atypical innate and adaptive immune responses in patients with severe acute respiratory syndrome. J Virol. 2007;81:8692–8706. doi: 10.1128/JVI.00527-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Felgenhauer U, Schoen A, Gad HH, Hartmann R, Schaubmar AR, Failing K, Drosten C, Weber F. Inhibition of SARS-CoV-2 by type I and type III interferons. J Biol Chem. 2020;295:13958–13964. doi: 10.1074/jbc.AC120.013788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lokugamage KG, Hage A, de Vries M, Valero-Jimenez AM, Schindewolf C, Dittmann M, Rajsbaum R, Menachery VD. Type I interferon susceptibility distinguishes SARS-CoV-2 from SARS-CoV. J Virol. 2020;94:e01410-20. doi: 10.1128/JVI.01410-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Mantlo E, Bukreyeva N, Maruyama J, Paessler S, Huang C. Antiviral activities of type I interferons to SARS-CoV-2 infection. Antiviral Res. 2020;179:104811. doi: 10.1016/j.antiviral.2020.104811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Andreakos E, Salagianni M, Galani IE, Koltsida O. Interferon-λs: front-line guardians of immunity and homeostasis in the respiratory tract. Front Immunol. 2017;8:1232. doi: 10.3389/fimmu.2017.01232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Broggi A, Granucci F, Zanoni I. Type III interferons: balancing tissue tolerance and resistance to pathogen invasion. J Exp Med. 2020;217:217. doi: 10.1084/jem.20190295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Prokunina-Olsson L, Alphonse N, Dickenson RE, Durbin JE, Glenn JS, Hartmann R, Kotenko SV, Lazear HM, O'Brien TR, Odendall C, et al. COVID-19 and emerging viral infections: the case for interferon lambda. J Exp Med. 2020;217:217. doi: 10.1084/jem.20200653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Broggi A, Ghosh S, Sposito B, Spreafico R, Balzarini F, Lo Cascio A, Clementi N, De Santis M, Mancini N, Granucci F, et al. Type III interferons disrupt the lung epithelial barrier upon viral recognition. Science. 2020;369:706–712. doi: 10.1126/science.abc3545. [DOI] [PMC free article] [PubMed] [Google Scholar]