Abstract
The emergence of a new severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic has become a significant health concern worldwide. Undoubtedly, a better understanding of the innate and adaptive immune responses against SARS-CoV-2 and its relationship with the coronavirus disease 2019 (COVID-19) pathogenesis will be the sole basis for developing and applying therapeutics. This review will summarize the published results that relate to innate immune responses against infections with human coronaviruses including SARS-CoV-1 and SARS-CoV-2 in both humans and animal models. The topics encompass the innate immune sensing of the virus to the dysregulation of various innate immune cells during infection and disease progression.
Keywords: Coronavirus, COVID-19, SARS-CoV-2, Innate immunity
INTRODUCTION
The outbreak of novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and the resulting coronavirus disease 2019 (COVID-19) began in Wuhan, Hubei Province, China in December 2019. In March 2020, the World Health Organization (WHO) declared COVID-19 a global pandemic. As of December 30, 2020, COVID-19 has resulted in more than 81 million cases and over 1.7 million mortalities worldwide. Because COVID-19 emerges unpredictably and spreads rapidly, it poses a continuous threat to human health. Patients with COVID-19 have been reported to experience multiple symptoms ranging from mild symptoms such as cough, fever, fatigue, dyspnea, and occasional diarrhea to severe symptoms characterized by respiratory failure, acute respiratory distress syndrome (ARDS), and multi-organ failure (1). Especially, in COVID-19 ARDS patients, cytokine storm is a common characteristic which is defined as increased plasma levels of inflammatory cytokines, including IL-6, IL-1β, and TNF-α. And this cytokine storm induces immune cell infiltration into inflamed lungs to cause alveolar damage and diminish lung function (2). Emerging studies imply that innate immune systems act as first responders for the detection and clearance of SARS-CoV-2 infection and also play a role in COVID-19 severity. Therefore, understanding the role of innate immune responses in SARS-CoV-2 is important to uncover mechanisms of host responses and improve our knowledge for disease pathogenesis and developing vaccines and immunotherapeutics. In this review, we primarily focus on the recent findings related to the host's innate immune responses and immune evasion mechanisms against SARS-CoV-2 infection and pathophysiology.
INNATE SENSING OF SARS-CoV-2
Pattern recognition of SARS-CoV-2 by TLRs & retinoic acid-inducible gene I (RIG-I)
Against most RNA viruses including coronaviruses, innate immune signals are triggered through the detection of RNA by the pattern recognition receptors (PRRs) such as endosomal TLRs and cytoplasmic RIG-I like receptors (RLRs) (Fig. 1). Among the TLR family, TLR7 and TLR8 recognize ssRNA, and TLR3 recognizes dsRNA. After ssRNA binding, TLR7 and TLR8 induce the recruitment of adaptor protein MYD88, IL-1 receptor-associated kinase (IRAK)-4, TNF receptor-associated factor (TRAF) 6, and the transcription factor IFN regulatory factor (IRF)-7. Phosphorylation of IRF-7 finally results in increased expression of type I IFNs genes. Recognition of dsRNA via TLR3 leads to recruitment of adaptor protein TIR-domain-containing adaptor protein-inducing IFN-β (TRIF) and induces pro-inflammatory cytokines containing type I IFNs. RLRs, including RIG-I and MDA5, are known as sensors for cytosolic dsRNA. Recognition of cytoplasmic dsRNA through RLRs stimulates TRAF-family-member-associated NF-κB activator binding kinase 1 (TBK1), inducible IκB kinase (IKK-i), and IRF-7 downstream signals through IFN-β promoter stimulator I (IPS-1, also called mitochondrial antiviral-signaling protein [MAVS]), an adaptor protein localized in the mitochondria membrane and eventually induces pro-inflammatory cytokines and type I IFNs (3). Although these PRRs are vital for inducing anti-viral responses, little is known about the effects of SARS-CoV-2 infection. However, considering that SARS-CoV-2 shows a high level of conservation with other coronaviruses, it is worth referring to the previous studies on SARS-CoV-1 and Middle East respiratory syndrome coronavirus (MERS-CoV). In the case of MERS-CoV, a study revealed that TLR7 was essential for sensing of MERS-CoV by plasmacytoid dendritic cells (pDCs), the primary producers of type I IFNs (4). Moreover, MERS-CoV evades immune responses by suppressing TLR signaling. For example, MERS-CoV limits the TLR signaling by their spike glycoprotein which induced the IRAK-M expression, a negative regulator of TLR signaling, and peroxisomal proliferator-activated receptor (PPAR)-γ, a transcriptional repressor (5). Likewise, after the infection with SARS-CoV-1, while the expression level of TLR7 of infected monocytes was upregulated, MYD88 and TRIF, the adaptor proteins of the TLR pathway, were all downregulated and IRAK-M was upregulated. It indicates that after SARS-CoV-1 infection, even though TLR7 was upregulated, the transduction of TLR7 downstream signals may be impaired in the infected monocytes (6). In the case of RIG-I, it has been reported that nucleocapsid protein of SARS-CoV-1 inhibits the RIG-I ubiquitination and activation by interacting with tripartite motif protein 25 (TRIM25) E3 ubiquitin ligase, resulting in suppression of type I IFNs production (7). However, further studies are needed to demonstrate the impact of SARS-CoV-2 in the PRRs and PRRs-relating downstream signals.
Figure 1. Innate sensing of viral infection through TLR and RIG-I in coronaviruses.
The viral entry into the cell is largely mediated through cell surface receptors such as ACE2 for SARS-CoV-1 and DPP4 for MERS-CoV. After the viral entrance, the innate immune signaling cascades are triggered through the detection of cytosolic or endosomal nucleic acids. ssRNA in the endosome is recognized by TLR7/8. The TLR7/8 recruits and activates MYD88, IRAK-4, and TRAF-6, which results in phosphorylation of IRF-7. The dsRNA in the endosome is recognized by TLR3 and through TRIF, the IKK-i/TBK1 complex is activated, phosphorylating IRF-7. Likewise, the cytosolic dsRNA is recognized by RIG-I, which activates IKK-i/TBK1 complex through IPS-1 on mitochondria, which results in phosphorylation of IRF-7. The phosphorylated IRF-7 finally results in increased expression of type I IFN genes. In the meantime, binding of MERS-CoV's spike glycoprotein to DPP4 induces the expression of IRAK-M, a negative regulator of TLR signaling, and PPAR-γ, a transcriptional repressor for TLR7/8. The IRAK-M is also reported to be upregulated with the infection of SARS-CoV-1. The N protein in SARS-CoV-1, interacting with TRIM25 in the cytosol, inhibits the ubiquitination and activation of RIG-I.
DPP4, dipeptidyl peptidase 4.
Production of cytokines and chemokines on SARS-CoV-2 infection
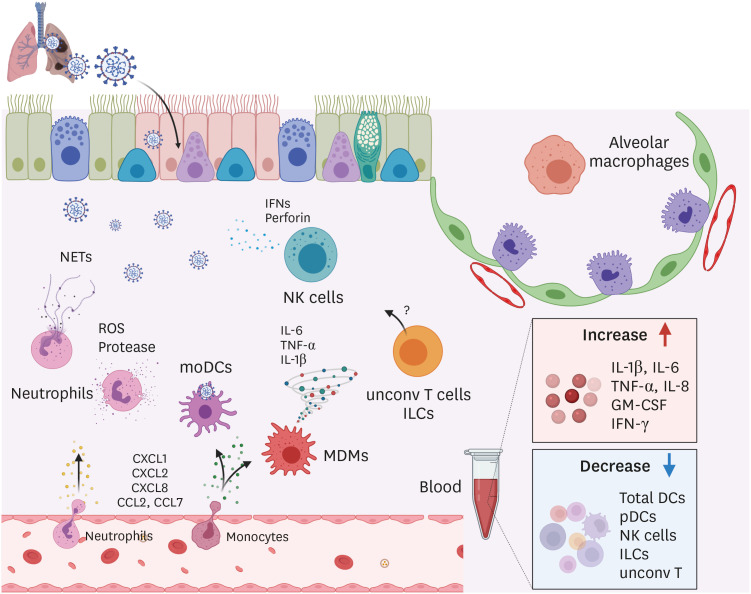
Several studies have tried to link various immunological parameters to disease severity in COVID-19 patients. Compared to healthy people, elevated levels of multiple pro-inflammatory cytokines such as IL-1β, TNF-α, IL-8, GM-CSF, and IFN-γ were detected in the serum from COVID-19 patients (Fig. 2) (8). Furthermore, a study identified that even in the COIVD-19 patients' group, circulating macrophage-related pro-inflammatory cytokines such as IL-6 and TNF-α were significantly increased in severe cases compared to mild cases. However, this study could not detect the IL-1β in the serum in most of the COVID-19 patients regardless of disease course (9). Similarly, a study performing the RNA analysis extracted from blood found that the expression of pro-inflammatory genes, including IL-2, IL-6, TNF, and IFNA1/13, was higher in severe cases compared to mild cases (10). Besides, the increased concentration of IL-6 in serum is consistently observed in critically ill patients (11). Taken together, it indicates that systemically elevated pro-inflammatory cytokines are commonly observed in COVID-19 patients and especially IL-6 and TNF-α are also positively correlated with disease severity (12).
Figure 2. Innate immune responses against SARS-CoV-2 in human.
Upon SARS-CoV-2 infection, chemokines such as CXCLl, CXCL2, CXCL8, CCL2, and CCL7 were increased in the lung and recruit neutrophils and monocytes. Activated neutrophils release reactive oxygen species (ROS), proteases, and NETs which are involved in tissue damage and pathology in infected lungs. Monocytes differentiate into macrophages and DCs in infected lungs, functioning as MDMs and moDCs, respectively. Activated MDMs produce pro-inflammatory cytokines including IL-6, TNF-α, and IL-1β. Excessive production of those cytokines results in cytokine storm and mortality in SARS-CoV-2 patients. moDCs uptake viral Ags and migrate to the draining lymph nodes. NK cells secrete cytotoxic granzymes and perforins, and IFNs. Exhaustion phenotypes of NK cells were found in SARS-CoV-2 patients. The exact role of ILCs and unconv T cells in SARS-CoV-2 infection remains to be addressed. In the blood of SARS-CoV-2 patients, levels of IL-1β, IL-6, TNF-α, IL-8, GM-CSF, and IFN-γ were significantly increased while the frequency of total DCs, pDCs, NK cells, ILCs, and unconv T cells was decreased.
In addition to systemic inflammatory conditions, local inflammation in the lungs is also a phenomenon commonly observed in COVID-19 patients. RNA-sequencing analysis of bronchoalveolar lavage fluid (BALF) from patients with COVID-19 revealed that neutrophil chemoattractant CXCL1, CXCL2, and CXCL8, and monocyte-derived macrophages (MDMs) chemoattractant CCL2 were highly expressed compared to healthy control (Fig. 2) (13,14). It suggests that elevated levels of chemokines in the lungs facilitate the migration and accumulation of those innate immune cells to the lungs. And these recruited immune cells contribute to inducing tissue damage and a massive amount of pro-inflammatory cytokines which may lead to cytokine storm.
Since the pathology observed in COVID-19 patients is driven by an increased level of circulating pro-inflammatory cytokines and recruitment of a large number of neutrophils and monocytes, those cytokines and chemokines are the good targets for the treatment of COVID-19. For example, IL-6, a critical cytokine associated with disease severity in COVID-19 patients, is a promising target for the COVID-19 therapy. IL-6 blocking agents, including tocilizumab, sarilumab (anti-IL-6R Abs), and siltuximab (anti-IL-6 Ab) were developed and tested for their efficacy in alleviating the COVID-19 disease in clinical trials (15). In addition to IL-6, GM-CSF contributing to the recruitment and activation of macrophages and monocytes has been proposed as potential targets for blockade (Gimsilumab) (16).
INNATE IMMUNE CELLS IN HUMANS
Neutrophils
Neutrophils are the first cells to be recruited to the inflamed tissue following stimulation by chemotactic factors released from infected epithelial cells (17). However, neutrophils are considered to play a major role in causing ARDS and acute lung injury (ALI), as activation and recruitment of neutrophils is a hallmark event in the progression of ARDS and ALI (18,19). Although neutrophil activation is vital for the host defense, activated neutrophils result in lung damage by releasing ROS and proteases by degranulation, and generating neutrophil extracellular traps (NETs) (2,20). Multiple studies have reported on the relationship between neutrophil infiltration and pathogenesis even in the COVID-19 patients because of these hallmarks of neutrophils. Significant neutrophilic infiltrate was identified in autopsied COVID-19 patients (21). Moreover, higher counts of peripheral neutrophils are observed and represent a predictor of poor outcome in COVID-19 ARDS patients. And a study observed that patients with severe symptoms had significantly higher peripheral neutrophil counts (22,23). Increased transcriptional levels of neutrophil chemoattractant CXCL2 and CXCL8 in BALF and PBMCs of COVID-19 patients support the relevance between enhanced neutrophil recruitment and disease severity in COVID-19 patients (Fig. 2) (14). Moreover, the neutrophil-to-lymphocyte ratio (NLR) was proposed as the most meaningful marker for predicting COVID pathogenesis (22).
Infiltration may not be the only mechanism by which neutrophils cause pathology in COVID-19 patients. ROS, proteases, and NETs released by activated neutrophils can also contribute to pathology (Fig. 2). SARS-CoV-2 infections cause redox imbalance and induce ROS accumulation, which induces tissue damage, thrombosis, and red blood cell dysfunction, contributing to COVID-19 disease severity (24,25). NETs, which comprise extracellular DNA, fibers and granular proteins, such as neutrophil elastase (NE) or myeloperoxidase, are released by neutrophils in response to many infectious agents (26,27). A recent study revealed that NETs released by neutrophils contribute to organ damage and mortality in COVID-19 patients (28,29). Furthermore, NETs play a significant role in inducing a cytokine storm (28). Because of their deleterious role in COVID-19 patients, strategies that inhibit the synthesis or promote the fragmentation of NETs are proposed to alleviate the COVID-19 disease. For example, inhibitors of NE, peptidyl arginine deiminase type 4 (PAD4), and gasdermin D which are the key enzymes required for NETs formation have been developed for the treatment of COVID-19 and undergone clinical trials (28,30). Since pro-inflammatory cytokines and molecules produced by activated neutrophils could be associated with the disease severity, targeting those molecules may be good strategies for alleviating the COVID-19 disease.
Macrophages
Macrophages provide effective host defense against pathogens, by phagocytosis, processing, and presenting foreign Ags that enter the airways. After infected with a respiratory virus, the lung harbors at least two distinct macrophage populations, including alveolar macrophages (AMs) and MDMs. AMs reside in the airspaces near placing with alveolar epithelial cells and are important for sustaining tissue-homeostasis (31). But during influenza infection, AMs are pro-inflammatory and play a critical role in viral clearance (32). Following the resolution of influenza infection, AMs are differentiated as immunosuppressive phenotype expressing high levels of negative regulators such as CD200 receptor (31), which is critical for maintaining airway immune homeostasis. In contrast to tissue-resident AMs, CC-chemokine receptor 2+ monocytes are recruited to the inflamed lungs enriched in CCL2 and CCL7 (Fig. 2) (13) and differentiate into pro-inflammatory macrophages through activation of JAK-STAT pathways (33). In general, during viral infection, these activated MDMs secrete pro-inflammatory cytokines, including IL-6, TNF-α, and IL-1β, which are the main players of inducing cytokine storm (Fig. 2) (34,35). And significantly, these macrophage populations are related to the progression of clinical disease. Single-cell RNA sequencing (scRNA-seq) on BALF cells from COVID-19 patients revealed that BALF consisted of a distinct macrophage population. In contrast to healthy donors (HD) and mild patients whose lungs are dominated by FABP4+ AMs, severe patients showed an increase in inflammatory MDMs and pro-fibrotic SPP1+ macrophages and depletion of AMs (35). Collectively, this observation suggests that recruitment of peripheral monocytes to the inflamed lung and their differentiation into MDMs can accelerate COVID-19 disease severity, and excessive inflammatory lung environment may delay the conversion of pro-inflammatory AMs into the anti-inflammatory phenotype, leading to exacerbation of the disease. Therefore, it is critical to developing therapeutic strategies toward inhibiting inflammatory MDMs accumulation in the lungs and properly modulating AMs activation status.
Dendritic cells (DCs)
DCs belong to the Ag-presenting cells (APCs) and they efficiently process and present Ag to T cells to induce priming of Ag-specific T cell responses. DCs can mainly be divided into 2 populations: conventional dendritic cells (cDCs) and pDCs. cDCs are further subdivided into CD103 or CD8α expressing type 1 cDCs (cDC1) and CD11b+ type 2 cDCs (cDC2), and they are functionally specialized in cross-presentation and induction of CD4+ T cell responses, respectively (36). Especially in the lungs, there are 2 main subsets, including CD103+ and CD11bhi respiratory DCs, which are tissue-resident and critical for the induction of T cell-mediated responses. Besides, upon inflammation, MHC IIlowCD11clow monocyte-derived DCs (moDCs) are recruited to the inflamed tissue (37). Because of their critical role in immunity to respiratory viruses, DCs with impaired function could eventually lead to immunopathology in respiratory diseases, including COVID-19 (23,38).
During acute SARS-CoV-2 infection, the frequency of total DCs in PBMC from patients was significantly decreased and they did not upregulate co-stimulatory molecules like CD80 after maturation stimulation compared to those of HD (Fig. 2). Furthermore, in contrast to cDCs derived from HD, cDCs from acute patients could not induce proliferation of T cells and production of antiviral molecules, indicating that acute patient-derived cDCs are functionally impaired for maturation and T cell activation (38). This demonstrates that they cannot lead to an appropriate anti-viral response during SARS-CoV-2 infection. In addition to cDCs, pDCs, major producers of type I IFNs, were decreased in the blood, and they were functionally impaired upon COVID-19 infection (Fig. 2) (39). However, it is necessary to study further whether these dysfunctional DCs are associated with disease severity in COVID-19 patients.
Cytotoxic innate lymphoid cells (ILCs) – NK cells
Unlike T and B cells, ILCs lack expression of somatically rearranged Ag receptors, thereby absent of any degree of Ag specificity. The ILC family is divided into two main groups – “cytotoxic” NK cells and “non-cytotoxic” ILC populations. And non-cytotoxic ILCs consist of 3 distinct groups: group 1 ILCs (ILC1s), group 2 ILCs (ILC2s), and group 3 ILCs (ILC3s) (40).
NK cells are cytotoxic lymphocytes that serve as a first-line defense against viral infections. NK cell activation can result in cytotoxic degranulation and inflammatory cytokines production for killing target cells (41). Due to their cytotoxic functions against viruses, several studies found the features of NK cells in COVID-19 patients. The number of NK cells in the peripheral blood of patients was decreased compared to healthy people, and even in the COVID-19 patients' group, severe cases had a further decrease in NK cell counts compared to mild cases (Fig. 2) (23,42,43,44,45). The NK cell population comprises cytotoxic CD56dimCD16+ NK cells and cytokine-producing CD56brightCD16− NK cells conferred with immunoregulatory functions. Decreased percentage of cytotoxic CD56dimCD16+ NK cells in severe cases compared to mild cases was observed in COVID-19 patients. It suggests that the decrease of cytotoxic CD56dimCD16+ NK cells may be one reason that severe patients could not correctly control against SARS-CoV-2 infection (44). Additionally, lower NK cell numbers correlated with higher plasma concentrations of IL-6 (46). And there was a report showing that in in vitro system, IL-6 and soluble IL-6 receptors impair perforin and granzyme B (GZMB) production by NK cells from HD, which could be restored by treatment with the IL-6R inhibitor tocilizumab (47). These results suggest that enhanced IL-6 may downregulate the number and function of NK cells in COVID-19 patients.
Consistently, multiple studies have reported that NK cells become functionally exhausted in COVID-19 patients. Expression of inhibitory receptor NK group 2 member A (NKG2A) on NK cells results in the functional exhaustion of NK cells in chronic viral infection and cancer (48,49). The frequency of NKG2A+ NK cells in the COVID-19 patients was significantly increased and other exhaustion markers, LAG-3 and TIM-3, were also upregulated (23,50). Importantly, the frequency of NKG2A+ NK cells was decreased in convalescing patients (42). Besides, the production of IFN-γ, TNF-α, IL-2, and CD107a was also reduced, indicating impaired cytotoxicity (23,42). These results suggest the functional exhaustion of cytotoxic NK cells in COVID-19 patients. However, another paper showed that the expression of GZMA and perforin on NK cells was increased in patients compared with HD, which indicates high cytotoxic potential. Moreover, perforin expression in NK cells was significantly increased according to the severity of the disease (45). Collectively, the effect of SARS-CoV-2 infection on functional exhaustion of NK cells is controversial, so further studies are required to understand NK cell exhaustion status according to the COVID-19 disease course.
Non-cytotoxic ILCs
Non-cytotoxic ILC populations are further divided into three subsets, including ILC1, ILC2, and ILC3 based on their ability to produce Th1, Th2, and Th17 cell-associated cytokines, respectively. Those ILCs are present in both lymphoid and non-lymphoid tissues and are primarily tissue-resident cells and are particularly abundant at the mucosal tissues, including intestine and lung. Although the role of ILCs has not yet been investigated in human respiratory viral diseases, it has been demonstrated through mouse studies. Among the ILC populations, ILC2s accumulate in the lung and play a critical role in restoring airway epithelial integrity and tissue homeostasis after influenza virus infection (51,52). Recently, the characteristics of the ILC population observed in COVID-19 patients have been reported. Circulating ILCs from COVID-19 patients were depleted compared with HD (Fig. 2). Among the circulating ILC population, the frequency of ILC2s was decreased in severe cases but not in mild cases, indicating that ILC2s may be differentially modulated depending on the disease severity. This is supported by the negative correlation between the frequency of ILC2s and the coagulation factor D-dimer and organ/muscle damage markers that reflect the disease severity in the COVID-19 patients (53). Therefore, it suggests that low ILC2 levels in COVID-19 patients could indicate a more severe disease outcome. However, further research is necessary to clarify the role of other ILC population in the context of SARS-CoV-2 infection.
Unconventional T (unconv T) cells
Besides conventional CD4+ and CD8+ T cells, the T cell compartment comprises several lineages of cells that share functional profiles of innate and adaptive immunity. They are called unconv T cells. Critical subsets of these lineages include invariant NK T (iNKT) cells, mucosal-associated invariant T (MAIT) cells, and γδT cells. In contrast to conventional T cells which express αβ TCR recognizing peptide Ags presented by MHC molecules, unconv T cells are not restricted to classic MHC molecules and recognize non-peptide Ags (54). Considering that these cells rapidly respond to non-cognate stimulation by releasing large amounts of cytokines, they may serve an important role in the initial response and defense against various pathogens. And in recent years, many groups have elucidated that NKT cells (55,56), MAIT cells (57,58), and γδT cells (59,60) contribute to anti-viral influenza immunity and lung tissue repair. Several studies have analyzed unconv T cells in the blood of severe COVID-19 patients, showing decreases in number and frequency of MAIT, iNKT cells, and Vδ2+ subset of γδT cells population in severe COVID-19 patients (Fig. 2) (61,62). And this is paralleled by a high frequency of MAIT cells in the airway which may indicate the possibility of recruitment from the blood. Furthermore, all types of unconv T cells in the blood of COVID-19 patients showed increased expression of the activation markers such as CD69 and PD-1, or CD25 (61,63). Simultaneously, the frequency of CD69 and PD-1 expressing MAIT and γδT cells was significantly increased in the airway compared to blood in matched COVID-19 patients. Collectively, these data suggest that activated unconv T cells can be recruited into the inflamed lung. In this study, the activated phenotypes of unconv T cells are likely due to the increased level of IL-18, a cytokine associated with unconv T cell activation during viral infection (58,64). Importantly, unconv T cells can be used as biomarkers, for example, CD69 expressing blood MAIT and iNKT cells may serve as a biomarker for the predictive of the clinical course (61). Therefore, considering the role of unconv T cells in anti-viral immune responses, in-depth studies are required to clarify their functions in COVID-19 patients further.
INNATE IMMUNE RESPONSES IN ANIMAL MODELS FOR SARS-CoV-2 INFECTION
From mouse to non-human primate, various animal models have been utilized to investigate therapeutics and vaccines against COVID-19 (Table 1). The mouse is the most utilized laboratory animal for the investigation of various human diseases. However, since the absence of a proper receptor for viral entrance to host cells, there was a limitation in utilizing mouse models for investigating viral infection including SARS-CoV-1 and SARS-CoV-2 (65,66). Researchers introduced various techniques that enable viral entrance and infection to the mouse system to solve this problem. One of the conventional methods is using mouse-adapted viruses. Serial passaging of the human virus in the mice allows the virus to be adapted (67). These approaches have been applied for SARS-CoV-1 infection in mice (68,69,70). Infection of the adapted virus in BALB/c mice induced accumulation of inflammatory monocyte and macrophages to the lung, which is associated with pulmonary pathogenesis with increased levels of cytokines and chemokines (69). Delayed type I IFN signaling after virus infection caused the pathogenesis since IFN-I orchestrates inflammatory response on SARS-CoV-1 infection. The AMs from the infected mice inhibited airway DCs' activation and T cell proliferation in vitro (71). More recently, mouse adaptation of human SARS-CoV-2 has been tried to assess the efficacy of vaccines and therapeutics (71,72,73,74). Significant infiltration of monocytes and other inflammatory cells was identified in mice infected with mouse-adapted SARS-CoV-2 as in mice with SARS-CoV-1 (74). However, further studies remain to address the impact of mouse-adapted virus infection on various innate immune cells. Another method is introducing the human angiotensin-converting enzyme (ACE) 2 gene to the mouse. K18-human angiotensin-converting enzyme 2 (hACE2) transgenic mice are the most frequently used strain, where human ACE2 is expressed on epithelial cells by human keratin 18 promoter, allowing the entrance of SARS-CoVs to mouse epithelial cells (75). This model can utilize patient-derived virus directly without the adaptation process in mice. The infection of SARS-CoV-2 in K18-hACE2 mice showed increased infiltration of inflammatory cells, including lymphocytes, macrophages, and neutrophils (76,77,78). Other cell populations including DCs, NK cells, and γδT cells were also accumulated in the lung. Monocyte accumulation was observed in the BALF, which coincided with the decreased number of AMs in the lung tissue (79).
Table 1. Innate immune response against SARS-CoV-1 and SARS-CoV-2 using various animal & organoid models.
| Model | Innate immune response | Reference | ||
|---|---|---|---|---|
| Small laboratory animals | ||||
| Mouse | ||||
| Viral adaptation | SARS-CoV-1 | |||
| Delayed IFN-I signaling | Channappanavar et al. (69) | |||
| Inflammatory monocytes accumulation to lung | Page et al. (68) | |||
| SARS-CoV-2 | ||||
| Infiltration of monocytes and other inflammatory cells | Dinnon et al. (74) | |||
| K18-hACE2 transgenic mice | SARS-CoV-2 | |||
| Infiltration of inflammatory cells; macrophages, neutrophils, and lymphocytes | Bao et al. (76); Israelow et al. (77) | |||
| Increased DCs, NK cells, and γδT cells | Hassan et al. (78) | |||
| Syrian hamster | SARS-CoV-2 | |||
| Prominent infiltration of inflammatory cells; neutrophils and monocytes | Sia et al. (80) | |||
| Ferret | SARS-CoV-2 | |||
| Increase of pneumocytes, macrophages, and neutrophils | Shi et al. (82) | |||
| Infiltration of macrophages, lymphocytes, and plasma cells in liver | Ryan et al. (83) | |||
| Non-human primate | ||||
| Rhesus macaque | SARS-CoV-1 | |||
| Infiltration of inflammatory macrophages and monocytes in lungs | Liu et al. (89) | |||
| Increased activation of lung macrophages | Clay et al. (90) | |||
| SARS-CoV-2 | ||||
| Infiltration of lymphocytes and monocytes in lungs | Shan et al. (87); Zheng et al. (88) | |||
| Age-related increased viral replication and pneumonia severity after infection | Yu et al. (93) | |||
| Organoid | ||||
| Airway epithelial organoid | SARS-CoV-2 | |||
| Responses related to innate immunity can be induced in the absence of immune cells | Katsura et al. (94); Elbadawi and Efferth. (95) | |||
| Alveolar type II cells mediate inflammatory state, secreting various cytokines and IFNs | ||||
The sequence similarity in the ACE2 receptor in hamsters and humans enables the Syrian hamster to be infected by patient-derived SARS-CoV-2 without the adaptation process (80). The infection of SARS-CoV-2 to the Syrian hamster resulted in prominent infiltration of inflammatory cells like neutrophils and monocytes to the lung. Ferrets were also available for the SARS-CoV-2 infection study, showing rapid transmission after infection (81). The increase of inflammatory cells including pneumocytes, macrophages, and neutrophils was identified through a histological assessment on the lung from the infected ferret (82). Another research on ferret identified cellular infiltration, including neutrophils and macrophages in bronchiolar lumina and macrophages, lymphocytes, and plasma cells in the liver, respectively (83). Non-human primate animal models have been utilized by many researchers to test therapeutics and vaccines against SARS-CoV-2 (84,85,86). Rhesus macaque model has been widely used in SARS-CoV-2 study, showing virulence and pathogenesis including pneumonia (87,88). Consistent with small animal models, SARS-CoV-1 infection to rhesus macaques results in significant infiltration of inflammatory macrophages and monocytes in the lungs (89). Another study in rhesus macaques also revealed that lung macrophages but not monocyte-derived DCs after the virus infection showed activated phenotype, indicating a higher frequency of activation marker CD86 (90). Likewise, many studies in rhesus macaques infected with SARS-CoV-2 revealed moderate infiltration of lymphocytes and monocytes to the lungs (85,91,92,93).
Organoids are miniaturized organs with 3D structures established from multipotent adult tissue stem cells or induced pluripotent stem cells. The maintenance of organ structure and organ-specific cell types allows many researchers to utilize the organoid system to assess pathogenesis in humans. The infection of SARS-CoV-2 to the organoid system could reveal many factors for inflammatory induction mediated by epithelial cells. Transcriptomic profiling revealed the inflammatory state mediated by alveolar type II cells which express numerous IFN, cytokines, and genes related to cell death after SARS-CoV-2 infection (94). Other researches with organoids also revealed the ability of the infection inducing innate immune response without recruitment of immune cells. However, most studies with the organoid system are restricted to their landscape for innate response in epithelial cells without the recruitment of immune cells. Therefore, the co-culture model of immune cells and organoids may provide a useful tool to investigate the interplay between epithelial cells and innate immune cells upon SARS-CoV-2 infection (95).
CONCLUSION
This review summarized the scientific knowledge from the general understanding of the innate anti-viral responses against various human coronaviruses, including SARS-CoV-2 in both human and animal models. In essence, the early anti-viral responses of epithelial cells and tissue-resident innate immune cells, such as macrophages and DCs, are crucial to the disease progression. Paradoxically, one major cause of severe disease by COVID-19 is associated with excessive responses by the recruited innate immune cells such as neutrophils and monocytes. The increase of innate immune cells is often followed by the severe loss of T cells, particularly CD8+ T cell subsets in patients. Those dysregulated immune cell composition represented with the neutrophil to lymphocyte ratio is closely correlated with disease pathogenesis including ARDS. Therefore, the critical question is how innate immunity is dysregulated, and if so, how to regulate the imbalanced host innate immune responses against SARS-CoV-2. Finding the answers to these questions will be important not only for a better understanding of basic knowledge but also for developing and applying therapeutics against COVID-19.
ACKNOWLEDGEMENTS
This study was supported by the BK21 funded by the Ministry of Education, Republic of Korea (4120200313623), by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (NRF-2020R1A6A3A01099978), by the grants from Genexine, Inc., Korea, and NeoImmuneTech, Inc.
Abbreviations
- ACE2
angiotensin-converting enzyme
- ALI
acute lung injury
- AM
alveolar macrophage
- APC
Ag-presenting cell
- ARDS
acute respiratory distress syndrome
- BALF
bronchoalveolar lavage fluid
- cDC
conventional dendritic cell
- COVID-19
coronavirus disease 2019
- DC
dendritic cell
- hACE2
human angiotensin-converting enzyme 2
- HD
healthy donors
- IKK-i
inducible IκB kinase
- ILC
innate lymphoid cell
- iNKT
invariant NK T
- IPS-1
IFN-β promoter stimulator I
- IRAK
IL-1 receptor associated kinase
- IRF
IFN regulatory factor
- GZMB
granzyme B
- MAIT
mucosal-associated invariant T
- MAVS
mitochondrial antiviral-signaling protein
- MDM
monocyte-derived macrophage
- MERS-CoV
Middle East respiratory syndrome coronavirus
- moDC
monocyte-derived dendritic cell
- NE
neutrophil elastase
- NET
neutrophil extracellular trap
- NKG2A
NK group 2 member A
- NLR
neutrophil-to-lymphocyte ratio
- PAD4
peptidyl arginine deiminase type 4
- pDC
plasmacytoid dendritic cell
- PPAR
peroxisomal proliferator-activated receptor
- PRR
pattern recognition receptor
- RIG-I
retinoic acid-inducible gene I
- RLR
RIG-I like receptor
- SARS-CoV
severe acute respiratory syndrome coronavirus
- scRNA-seq
single-cell RNA sequencing
- TBK1
TRAF-family-member-associated NF-κB activator binding kinase 1
- TRAF
TNF receptor-associated factor
- TRIF
TIR-domain-containing adaptor protein-inducing IFN-β
- TRIM25
tripartite motif protein 25
- unconv T
unconventional T
- WHO
World Health Organization
Footnotes
Conflict of Interest: The authors declare no potential conflicts of interest.
- Conceptualization: Lee SW, Kang YW.
- Investigation: Kang YW, Park S, Lee KJ, Moon D.
- Supervision: Lee SW.
- Visualization: Park S, Kim YM.
- Writing - original draft: Kang YW, Park S.
- Writing - review & editing: Lee SW, Kang YW, Park S, Lee KJ, Moon D, Kim YM.
References
- 1.Wu C, Chen X, Cai Y, Xia J, Zhou X, Xu S, Huang H, Zhang L, Zhou X, Du C, et al. Risk factors associated with acute respiratory distress syndrome and death in patients with coronavirus disease 2019 pneumonia in Wuhan, China. JAMA Intern Med. 2020;180:934–943. doi: 10.1001/jamainternmed.2020.0994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang SC, Tsai YF, Pan YL, Hwang TL. Understanding the role of neutrophils in acute respiratory distress syndrome. Biomed J. 2020 doi: 10.1016/j.bj.2020.09.001. Forthcoming. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 4.Scheuplein VA, Seifried J, Malczyk AH, Miller L, Höcker L, Vergara-Alert J, Dolnik O, Zielecki F, Becker B, Spreitzer I, et al. High secretion of interferons by human plasmacytoid dendritic cells upon recognition of Middle East respiratory syndrome coronavirus. J Virol. 2015;89:3859–3869. doi: 10.1128/JVI.03607-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Al-Qahtani AA, Lyroni K, Aznaourova M, Tseliou M, Al-Anazi MR, Al-Ahdal MN, Alkahtani S, Sourvinos G, Tsatsanis C. Middle east respiratory syndrome corona virus spike glycoprotein suppresses macrophage responses via DPP4-mediated induction of IRAK-M and PPARγ. Oncotarget. 2017;8:9053–9066. doi: 10.18632/oncotarget.14754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hu W, Yen YT, Singh S, Kao CL, Wu-Hsieh BA. SARS-CoV regulates immune function-related gene expression in human monocytic cells. Viral Immunol. 2012;25:277–288. doi: 10.1089/vim.2011.0099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hu Y, Li W, Gao T, Cui Y, Jin Y, Li P, Ma Q, Liu X, Cao C. The severe acute respiratory syndrome coronavirus nucleocapsid inhibits type I interferon production by interfering with TRIM25-mediated RIG-I ubiquitination. J Virol. 2017;91:e02143-16. doi: 10.1128/JVI.02143-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, Zhang L, Fan G, Xu J, Gu X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen G, Wu D, Guo W, Cao Y, Huang D, Wang H, Wang T, Zhang X, Chen H, Yu H, et al. Clinical and immunological features of severe and moderate coronavirus disease 2019. J Clin Invest. 2020;130:2620–2629. doi: 10.1172/JCI137244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ong EZ, Chan YF, Leong WY, Lee NM, Kalimuddin S, Haja Mohideen SM, Chan KS, Tan AT, Bertoletti A, Ooi EE, et al. A dynamic immune response shapes COVID-19 progression. Cell Host Microbe. 2020;27:879–882.e2. doi: 10.1016/j.chom.2020.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen X, Zhao B, Qu Y, Chen Y, Xiong J, Feng Y, Men D, Huang Q, Liu Y, Yang B, et al. Detectable serum severe acute respiratory syndrome coronavirus 2 viral load (RNAemia) is closely correlated with drastically elevated interleukin 6 level in critically ill patients with coronavirus disease 2019. Clin Infect Dis. 2020;71:1937–1942. doi: 10.1093/cid/ciaa449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gong J, Dong H, Xia QS, Huang ZY, Wang DK, Zhao Y, Liu WH, Tu SH, Zhang MM, Wang Q, et al. Correlation analysis between disease severity and inflammation-related parameters in patients with COVID-19: a retrospective study. BMC Infect Dis. 2020;20:963. doi: 10.1186/s12879-020-05681-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou Z, Ren L, Zhang L, Zhong J, Xiao Y, Jia Z, Guo L, Yang J, Wang C, Jiang S, et al. Heightened innate immune responses in the respiratory tract of COVID-19 patients. Cell Host Microbe. 2020;27:883–890.e2. doi: 10.1016/j.chom.2020.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xiong Y, Liu Y, Cao L, Wang D, Guo M, Jiang A, Guo D, Hu W, Yang J, Tang Z, et al. Transcriptomic characteristics of bronchoalveolar lavage fluid and peripheral blood mononuclear cells in COVID-19 patients. Emerg Microbes Infect. 2020;9:761–770. doi: 10.1080/22221751.2020.1747363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vabret N, Britton GJ, Gruber C, Hegde S, Kim J, Kuksin M, Levantovsky R, Malle L, Moreira A, Park MD, et al. Immunology of COVID-19: current state of the science. Immunity. 2020;52:910–941. doi: 10.1016/j.immuni.2020.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mehta P, Porter JC, Manson JJ, Isaacs JD, Openshaw PJ, McInnes IB, Summers C, Chambers RC. Therapeutic blockade of granulocyte macrophage colony-stimulating factor in COVID-19-associated hyperinflammation: challenges and opportunities. Lancet Respir Med. 2020;8:822–830. doi: 10.1016/S2213-2600(20)30267-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Williams AE, José RJ, Mercer PF, Brealey D, Parekh D, Thickett DR, O’Kane C, McAuley DF, Chambers RC. Evidence for chemokine synergy during neutrophil migration in ARDS. Thorax. 2017;72:66–73. doi: 10.1136/thoraxjnl-2016-208597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grommes J, Soehnlein O. Contribution of neutrophils to acute lung injury. Mol Med. 2011;17:293–307. doi: 10.2119/molmed.2010.00138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.ARDS Definition Task Force. Ranieri VM, Rubenfeld GD, Thompson BT, Ferguson ND, Caldwell E, Fan E, Camporota L, Slutsky AS. Acute respiratory distress syndrome: the Berlin Definition. JAMA. 2012;307:2526–2533. doi: 10.1001/jama.2012.5669. [DOI] [PubMed] [Google Scholar]
- 20.Camp JV, Jonsson CB. A role for neutrophils in viral respiratory disease. Front Immunol. 2017;8:550. doi: 10.3389/fimmu.2017.00550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fox SE, Akmatbekov A, Harbert JL, Li G, Quincy Brown J, Vander Heide RS. Pulmonary and cardiac pathology in African American patients with COVID-19: an autopsy series from New Orleans. Lancet Respir Med. 2020;8:681–686. doi: 10.1016/S2213-2600(20)30243-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu J, Liu Y, Xiang P, Pu L, Xiong H, Li C, Zhang M, Tan J, Xu Y, Song R, et al. Neutrophil-to-lymphocyte ratio predicts critical illness patients with 2019 coronavirus disease in the early stage. J Transl Med. 2020;18:206. doi: 10.1186/s12967-020-02374-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wilk AJ, Rustagi A, Zhao NQ, Roque J, Martínez-Colón GJ, McKechnie JL, Ivison GT, Ranganath T, Vergara R, Hollis T, et al. A single-cell atlas of the peripheral immune response in patients with severe COVID-19. Nat Med. 2020;26:1070–1076. doi: 10.1038/s41591-020-0944-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laforge M, Elbim C, Frère C, Hémadi M, Massaad C, Nuss P, Benoliel JJ, Becker C. Tissue damage from neutrophil-induced oxidative stress in COVID-19. Nat Rev Immunol. 2020;20:515–516. doi: 10.1038/s41577-020-0407-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klok FA, Kruip MJ, van der Meer NJ, Arbous MS, Gommers D, Kant KM, Kaptein FH, van Paassen J, Stals MA, Huisman MV, et al. Confirmation of the high cumulative incidence of thrombotic complications in critically ill ICU patients with COVID-19: an updated analysis. Thromb Res. 2020;191:148–150. doi: 10.1016/j.thromres.2020.04.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brinkmann V, Reichard U, Goosmann C, Fauler B, Uhlemann Y, Weiss DS, Weinrauch Y, Zychlinsky A. Neutrophil extracellular traps kill bacteria. Science. 2004;303:1532–1535. doi: 10.1126/science.1092385. [DOI] [PubMed] [Google Scholar]
- 27.Van Avondt K, Hartl D. Mechanisms and disease relevance of neutrophil extracellular trap formation. Eur J Clin Invest. 2018;48(Suppl 2):e12919. doi: 10.1111/eci.12919. [DOI] [PubMed] [Google Scholar]
- 28.Barnes BJ, Adrover JM, Baxter-Stoltzfus A, Borczuk A, Cools-Lartigue J, Crawford JM, Daßler-Plenker J, Guerci P, Huynh C, Knight JS, et al. Targeting potential drivers of COVID-19: neutrophil extracellular traps. J Exp Med. 2020;217:e20200652. doi: 10.1084/jem.20200652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zuo Y, Yalavarthi S, Shi H, Gockman K, Zuo M, Madison JA, Blair C, Weber A, Barnes BJ, Egeblad M, et al. Neutrophil extracellular traps in COVID-19. JCI Insight. 2020;5:e138999. doi: 10.1172/jci.insight.138999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Veras FP, Pontelli MC, Silva CM, Toller-Kawahisa JE, de Lima M, Nascimento DC, Schneider AH, Caetité D, Tavares LA, Paiva IM, et al. SARS-CoV-2-triggered neutrophil extracellular traps mediate COVID-19 pathology. J Exp Med. 2020;217:e20201129. doi: 10.1084/jem.20201129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hussell T, Bell TJ. Alveolar macrophages: plasticity in a tissue-specific context. Nat Rev Immunol. 2014;14:81–93. doi: 10.1038/nri3600. [DOI] [PubMed] [Google Scholar]
- 32.Tate MD, Pickett DL, van Rooijen N, Brooks AG, Reading PC. Critical role of airway macrophages in modulating disease severity during influenza virus infection of mice. J Virol. 2010;84:7569–7580. doi: 10.1128/JVI.00291-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Merad M, Martin JC. Pathological inflammation in patients with COVID-19: a key role for monocytes and macrophages. Nat Rev Immunol. 2020;20:355–362. doi: 10.1038/s41577-020-0331-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schulert GS, Grom AA. Pathogenesis of macrophage activation syndrome and potential for cytokine-directed therapies. Annu Rev Med. 2015;66:145–159. doi: 10.1146/annurev-med-061813-012806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liao M, Liu Y, Yuan J, Wen Y, Xu G, Zhao J, Cheng L, Li J, Wang X, Wang F, et al. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat Med. 2020;26:842–844. doi: 10.1038/s41591-020-0901-9. [DOI] [PubMed] [Google Scholar]
- 36.Eisenbarth SC. Dendritic cell subsets in T cell programming: location dictates function. Nat Rev Immunol. 2019;19:89–103. doi: 10.1038/s41577-018-0088-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Braciale TJ, Sun J, Kim TS. Regulating the adaptive immune response to respiratory virus infection. Nat Rev Immunol. 2012;12:295–305. doi: 10.1038/nri3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou R, To KK, Wong YC, Liu L, Zhou B, Li X, Huang H, Mo Y, Luk TY, Lau TT, et al. Acute SARS-CoV-2 infection impairs dendritic cell and T cell responses. Immunity. 2020;53:864–877.e5. doi: 10.1016/j.immuni.2020.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Arunachalam PS, Wimmers F, Mok CK, Perera RA, Scott M, Hagan T, Sigal N, Feng Y, Bristow L, Tak-Yin Tsang O, et al. Systems biological assessment of immunity to mild versus severe COVID-19 infection in humans. Science. 2020;369:1210–1220. doi: 10.1126/science.abc6261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Artis D, Spits H. The biology of innate lymphoid cells. Nature. 2015;517:293–301. doi: 10.1038/nature14189. [DOI] [PubMed] [Google Scholar]
- 41.van Eeden C, Khan L, Osman MS, Cohen Tervaert JW. Natural killer cell dysfunction and its role in COVID-19. Int J Mol Sci. 2020;21:6351. doi: 10.3390/ijms21176351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zheng M, Gao Y, Wang G, Song G, Liu S, Sun D, Xu Y, Tian Z. Functional exhaustion of antiviral lymphocytes in COVID-19 patients. Cell Mol Immunol. 2020;17:533–535. doi: 10.1038/s41423-020-0402-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Song CY, Xu J, He JQ, Lu YQ. Immune dysfunction following COVID-19, especially in severe patients. Sci Rep. 2020;10:15838. doi: 10.1038/s41598-020-72718-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li M, Guo W, Dong Y, Wang X, Dai D, Liu X, Wu Y, Li M, Zhang W, Zhou H, et al. Elevated exhaustion levels of NK and CD8+ T cells as indicators for progression and prognosis of COVID-19 disease. Front Immunol. 2020;11:580237. doi: 10.3389/fimmu.2020.580237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jiang Y, Wei X, Guan J, Qin S, Wang Z, Lu H, Qian J, Wu L, Chen Y, Chen Y, et al. COVID-19 pneumonia: CD8+ T and NK cells are decreased in number but compensatory increased in cytotoxic potential. Clin Immunol. 2020;218:108516. doi: 10.1016/j.clim.2020.108516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang W, Liu X, Wu S, Chen S, Li Y, Nong L, Lie P, Huang L, Cheng L, Lin Y, et al. Definition and risks of cytokine release syndrome in 11 critically ill COVID-19 patients with pneumonia: analysis of disease characteristics. J Infect Dis. 2020;222:1444–1451. doi: 10.1093/infdis/jiaa387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cifaldi L, Prencipe G, Caiello I, Bracaglia C, Locatelli F, De Benedetti F, Strippoli R. Inhibition of natural killer cell cytotoxicity by interleukin-6: implications for the pathogenesis of macrophage activation syndrome. Arthritis Rheumatol. 2015;67:3037–3046. doi: 10.1002/art.39295. [DOI] [PubMed] [Google Scholar]
- 48.André P, Denis C, Soulas C, Bourbon-Caillet C, Lopez J, Arnoux T, Bléry M, Bonnafous C, Gauthier L, Morel A, et al. Anti-NKG2A mAb is a checkpoint inhibitor that promotes anti-tumor immunity by unleashing both T and NK cells. Cell. 2018;175:1731–1743.e13. doi: 10.1016/j.cell.2018.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li F, Wei H, Wei H, Gao Y, Xu L, Yin W, Sun R, Tian Z. Blocking the natural killer cell inhibitory receptor NKG2A increases activity of human natural killer cells and clears hepatitis B virus infection in mice. Gastroenterology. 2013;144:392–401. doi: 10.1053/j.gastro.2012.10.039. [DOI] [PubMed] [Google Scholar]
- 50.Hadjadj J, Yatim N, Barnabei L, Corneau A, Boussier J, Smith N, Péré H, Charbit B, Bondet V, Chenevier-Gobeaux C, et al. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science. 2020;369:718–724. doi: 10.1126/science.abc6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Monticelli LA, Sonnenberg GF, Abt MC, Alenghat T, Ziegler CG, Doering TA, Angelosanto JM, Laidlaw BJ, Yang CY, Sathaliyawala T, et al. Innate lymphoid cells promote lung-tissue homeostasis after infection with influenza virus. Nat Immunol. 2011;12:1045–1054. doi: 10.1031/ni.2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Califano D, Furuya Y, Roberts S, Avram D, McKenzie AN, Metzger DW. IFN-γ increases susceptibility to influenza A infection through suppression of group II innate lymphoid cells. Mucosal Immunol. 2018;11:209–219. doi: 10.1038/mi.2017.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.García M, Kokkinou E, Carrasco García A, Parrot T, Palma Medina LM, Maleki KT, Christ W, Varnaitė R, Filipovic I, Ljunggren HG, et al. Innate lymphoid cell composition associates with COVID-19 disease severity. Clin Transl Immunology. 2020;9:e1224. doi: 10.1002/cti2.1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pellicci DG, Koay HF, Berzins SP. Thymic development of unconventional T cells: how NKT cells, MAIT cells and γδ T cells emerge. Nat Rev Immunol. 2020;20:756–770. doi: 10.1038/s41577-020-0345-y. [DOI] [PubMed] [Google Scholar]
- 55.Gottschalk C, Mettke E, Kurts C. The role of invariant natural killer T cells in dendritic cell licensing, cross-priming, and memory CD8+ T cell generation. Front Immunol. 2015;6:379. doi: 10.3389/fimmu.2015.00379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kok WL, Denney L, Benam K, Cole S, Clelland C, McMichael AJ, Ho LP. Pivotal advance: invariant NKT cells reduce accumulation of inflammatory monocytes in the lungs and decrease immune-pathology during severe influenza A virus infection. J Leukoc Biol. 2012;91:357–368. doi: 10.1189/jlb.0411184. [DOI] [PubMed] [Google Scholar]
- 57.van Wilgenburg B, Loh L, Chen Z, Pediongco TJ, Wang H, Shi M, Zhao Z, Koutsakos M, Nüssing S, Sant S, et al. MAIT cells contribute to protection against lethal influenza infection in vivo . Nat Commun. 2018;9:4706. doi: 10.1038/s41467-018-07207-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Loh L, Wang Z, Sant S, Koutsakos M, Jegaskanda S, Corbett AJ, Liu L, Fairlie DP, Crowe J, Rossjohn J, et al. Human mucosal-associated invariant T cells contribute to antiviral influenza immunity via IL-18-dependent activation. Proc Natl Acad Sci U S A. 2016;113:10133–10138. doi: 10.1073/pnas.1610750113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Palomino-Segura M, Latino I, Farsakoglu Y, Gonzalez SF. Early production of IL-17A by γδ T cells in the trachea promotes viral clearance during influenza infection in mice. Eur J Immunol. 2020;50:97–109. doi: 10.1002/eji.201948157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Poccia F, Agrati C, Castilletti C, Bordi L, Gioia C, Horejsh D, Ippolito G, Chan PK, Hui DS, Sung JJ, et al. Anti-severe acute respiratory syndrome coronavirus immune responses: the role played by Vγ9Vδ2 T cells. J Infect Dis. 2006;193:1244–1249. doi: 10.1086/502975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jouan Y, Guillon A, Gonzalez L, Perez Y, Boisseau C, Ehrmann S, Ferreira M, Daix T, Jeannet R, François B, et al. Phenotypical and functional alteration of unconventional T cells in severe COVID-19 patients. J Exp Med. 2020;217:e20200872. doi: 10.1084/jem.20200872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Carissimo G, Xu W, Kwok I, Abdad MY, Chan YH, Fong SW, Puan KJ, Lee CY, Yeo NK, Amrun SN, et al. Whole blood immunophenotyping uncovers immature neutrophil-to-VD2 T-cell ratio as an early marker for severe COVID-19. Nat Commun. 2020;11:5243. doi: 10.1038/s41467-020-19080-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lei L, Qian H, Yang X, Zhang X, Zhang D, Dai T, Guo R, Shi L, Cheng Y, Zhang B, et al. The phenotypic changes of γδ T cells in COVID-19 patients. J Cell Mol Med. 2020;24:11603–11606. doi: 10.1111/jcmm.15620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tyznik AJ, Verma S, Wang Q, Kronenberg M, Benedict CA. Distinct requirements for activation of NKT and NK cells during viral infection. J Immunol. 2014;192:3676–3685. doi: 10.4049/jimmunol.1300837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liu Z, Xiao X, Wei X, Li J, Yang J, Tan H, Zhu J, Zhang Q, Wu J, Liu L. Composition and divergence of coronavirus spike proteins and host ACE2 receptors predict potential intermediate hosts of SARS-CoV-2. J Med Virol. 2020;92:595–601. doi: 10.1002/jmv.25726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li F, Li W, Farzan M, Harrison SC. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science. 2005;309:1864–1868. doi: 10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]
- 67.Roberts A, Deming D, Paddock CD, Cheng A, Yount B, Vogel L, Herman BD, Sheahan T, Heise M, Genrich GL, et al. A mouse-adapted SARS-coronavirus causes disease and mortality in BALB/c mice. PLoS Pathog. 2007;3:e5. doi: 10.1371/journal.ppat.0030005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Page C, Goicochea L, Matthews K, Zhang Y, Klover P, Holtzman MJ, Hennighausen L, Frieman M. Induction of alternatively activated macrophages enhances pathogenesis during severe acute respiratory syndrome coronavirus infection. J Virol. 2012;86:13334–13349. doi: 10.1128/JVI.01689-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Channappanavar R, Fehr AR, Vijay R, Mack M, Zhao J, Meyerholz DK, Perlman S. Dysregulated type I Interferon and inflammatory monocyte-macrophage responses cause lethal pneumonia in SARS-CoV-infected mice. Cell Host Microbe. 2016;19:181–193. doi: 10.1016/j.chom.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhao J, Zhao J, Van Rooijen N, Perlman S. Evasion by stealth: inefficient immune activation underlies poor T cell response and severe disease in SARS-CoV-infected mice. PLoS Pathog. 2009;5:e1000636. doi: 10.1371/journal.ppat.1000636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang J, Shuai L, Wang C, Liu R, He X, Zhang X, Sun Z, Shan D, Ge J, Wang X, et al. Mouse-adapted SARS-CoV-2 replicates efficiently in the upper and lower respiratory tract of BALB/c and C57BL/6J mice. Protein Cell. 2020;11:776–782. doi: 10.1007/s13238-020-00767-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gu H, Chen Q, Yang G, He L, Fan H, Deng YQ, Wang Y, Teng Y, Zhao Z, Cui Y, et al. Adaptation of SARS-CoV-2 in BALB/c mice for testing vaccine efficacy. Science. 2020;369:1603–1607. doi: 10.1126/science.abc4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Leist SR, Dinnon KH, 3rd, Schäfer A, Tse LV, Okuda K, Hou YJ, West A, Edwards CE, Sanders W, Fritch EJ, et al. A mouse-adapted SARS-CoV-2 induces acute lung injury and mortality in standard laboratory mice. Cell. 2020;183:1070–1085.e12. doi: 10.1016/j.cell.2020.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dinnon KH, 3rd, Leist SR, Schäfer A, Edwards CE, Martinez DR, Montgomery SA, West A, Yount BL, Jr, Hou YJ, Adams LE, et al. A mouse-adapted model of SARS-CoV-2 to test COVID-19 countermeasures. Nature. 2020;586:560–566. doi: 10.1038/s41586-020-2708-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zheng J, Wong LR, Li K, Verma AK, Ortiz ME, Wohlford-Lenane C, Leidinger MR, Knudson CM, Meyerholz DK, McCray PB, Jr, et al. COVID-19 treatments and pathogenesis including anosmia in K18-hACE2 mice. Nature. 2021;589:603–607. doi: 10.1038/s41586-020-2943-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bao L, Deng W, Huang B, Gao H, Liu J, Ren L, Wei Q, Yu P, Xu Y, Qi F, et al. The pathogenicity of SARS-CoV-2 in hACE2 transgenic mice. Nature. 2020;583:830–833. doi: 10.1038/s41586-020-2312-y. [DOI] [PubMed] [Google Scholar]
- 77.Israelow B, Song E, Mao T, Lu P, Meir A, Liu F, Alfajaro MM, Wei J, Dong H, Homer RJ, et al. Mouse model of SARS-CoV-2 reveals inflammatory role of type I interferon signaling. J Exp Med. 2020;217:e20201241. doi: 10.1084/jem.20201241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hassan AO, Case JB, Winkler ES, Thackray LB, Kafai NM, Bailey AL, McCune BT, Fox JM, Chen RE, Alsoussi WB, et al. A SARS-CoV-2 infection model in mice demonstrates protection by neutralizing antibodies. Cell. 2020;182:744–753.e4. doi: 10.1016/j.cell.2020.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Winkler ES, Bailey AL, Kafai NM, Nair S, McCune BT, Yu J, Fox JM, Chen RE, Earnest JT, Keeler SP, et al. SARS-CoV-2 infection of human ACE2-transgenic mice causes severe lung inflammation and impaired function. Nat Immunol. 2020;21:1327–1335. doi: 10.1038/s41590-020-0778-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sia SF, Yan LM, Chin AW, Fung K, Choy KT, Wong AY, Kaewpreedee P, Perera RA, Poon LL, Nicholls JM, et al. Pathogenesis and transmission of SARS-CoV-2 in golden hamsters. Nature. 2020;583:834–838. doi: 10.1038/s41586-020-2342-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kim YI, Kim SG, Kim SM, Kim EH, Park SJ, Yu KM, Chang JH, Kim EJ, Lee S, Casel MA, et al. Infection and rapid transmission of SARS-CoV-2 in ferrets. Cell Host Microbe. 2020;27:704–709.e2. doi: 10.1016/j.chom.2020.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Shi J, Wen Z, Zhong G, Yang H, Wang C, Huang B, Liu R, He X, Shuai L, Sun Z, et al. Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS-coronavirus 2. Science. 2020;368:1016–1020. doi: 10.1126/science.abb7015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ryan KA, Bewley KR, Fotheringham SA, Slack GS, Brown P, Hall Y, Wand NI, Marriott AC, Cavell BE, Tree JA, et al. Dose-dependent response to infection with SARS-CoV-2 in the ferret model and evidence of protective immunity. Nat Commun. 2021;12:81. doi: 10.1038/s41467-020-20439-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lu S, Zhao J, Dong J, Liu H, Zhu Y, Li H, Liu L, Yang Y, Sun S, Song Y, et al. Effective treatment of SARS-CoV-2-infected rhesus macaques by attenuating inflammation. Cell Res. 2021;31:229–232. doi: 10.1038/s41422-020-00414-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chandrashekar A, Liu J, Martinot AJ, McMahan K, Mercado NB, Peter L, Tostanoski LH, Yu J, Maliga Z, Nekorchuk M, et al. SARS-CoV-2 infection protects against rechallenge in rhesus macaques. Science. 2020;369:812–817. doi: 10.1126/science.abc4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mercado NB, Zahn R, Wegmann F, Loos C, Chandrashekar A, Yu J, Liu J, Peter L, McMahan K, Tostanoski LH, et al. Single-shot Ad26 vaccine protects against SARS-CoV-2 in rhesus macaques. Nature. 2020;586:583–588. doi: 10.1038/s41586-020-2607-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Shan C, Yao YF, Yang XL, Zhou YW, Gao G, Peng Y, Yang L, Hu X, Xiong J, Jiang RD, et al. Infection with novel coronavirus (SARS-CoV-2) causes pneumonia in Rhesus macaques. Cell Res. 2020;30:670–677. doi: 10.1038/s41422-020-0364-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zheng H, Li H, Guo L, Liang Y, Li J, Wang X, Hu Y, Wang L, Liao Y, Yang F, et al. Virulence and pathogenesis of SARS-CoV-2 infection in rhesus macaques: a nonhuman primate model of COVID-19 progression. PLoS Pathog. 2020;16:e1008949. doi: 10.1371/journal.ppat.1008949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Liu L, Wei Q, Lin Q, Fang J, Wang H, Kwok H, Tang H, Nishiura K, Peng J, Tan Z, et al. Anti-spike IgG causes severe acute lung injury by skewing macrophage responses during acute SARS-CoV infection. JCI Insight. 2019;4:e123158. doi: 10.1172/jci.insight.123158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Clay C, Donart N, Fomukong N, Knight JB, Lei W, Price L, Hahn F, Van Westrienen J, Harrod KS. Primary severe acute respiratory syndrome coronavirus infection limits replication but not lung inflammation upon homologous rechallenge. J Virol. 2012;86:4234–4244. doi: 10.1128/JVI.06791-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Rockx B, Kuiken T, Herfst S, Bestebroer T, Lamers MM, Oude Munnink BB, de Meulder D, van Amerongen G, van den Brand J, Okba NM, et al. Comparative pathogenesis of COVID-19, MERS, and SARS in a nonhuman primate model. Science. 2020;368:1012–1015. doi: 10.1126/science.abb7314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Munster VJ, Feldmann F, Williamson BN, van Doremalen N, Pérez-Pérez L, Schulz J, Meade-White K, Okumura A, Callison J, Brumbaugh B, et al. Respiratory disease in rhesus macaques inoculated with SARS-CoV-2. Nature. 2020;585:268–272. doi: 10.1038/s41586-020-2324-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yu P, Qi F, Xu Y, Li F, Liu P, Liu J, Bao L, Deng W, Gao H, Xiang Z, et al. Age-related rhesus macaque models of COVID-19. Animal Model Exp Med. 2020;3:93–97. doi: 10.1002/ame2.12108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Katsura H, Sontake V, Tata A, Kobayashi Y, Edwards CE, Heaton BE, Konkimalla A, Asakura T, Mikami Y, Fritch EJ, et al. Human lung stem cell-based alveolospheres provide insights into SARS-CoV-2-mediated interferon responses and pneumocyte dysfunction. Cell Stem Cell. 2020;27:890–904.e8. doi: 10.1016/j.stem.2020.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Elbadawi M, Efferth T. Organoids of human airways to study infectivity and cytopathy of SARS-CoV-2. Lancet Respir Med. 2020;8:e55–e56. doi: 10.1016/S2213-2600(20)30238-1. [DOI] [PMC free article] [PubMed] [Google Scholar]