Abstract
An outbreak of the COVID-19 pandemic is a major public health disease as well as a challenging task to people with comorbidity worldwide. According to a report, comorbidity enhances the risk factors with complications of COVID-19. Here, we propose and explore a mathematical framework to study the transmission dynamics of COVID-19 with comorbidity. Within this framework, the model is calibrated by using new daily confirmed COVID-19 cases in India. The qualitative properties of the model and the stability of feasible equilibrium are studied. The model experiences the scenario of backward bifurcation by parameter regime accounting for progress in susceptibility to acquire infection by comorbidity individuals. The endemic equilibrium is asymptotically stable if recruitment of comorbidity becomes higher without acquiring the infection. Moreover, a larger backward bifurcation regime indicates the possibility of more infection in susceptible individuals. A dynamics in the mean fluctuation of the force of infection is investigated with different parameter regimes. A significant correlation is established between the force of infection and corresponding Shannon entropy under the same parameters, which provides evidence that infection reaches a significant proportion of the susceptible.
Keywords: COVID-19, Comorbidity, Model calibration, Backward bifurcation, Sensitivity analysis, Shannon entropy
Introduction
COVID-19 was announced by the World health organization (WHO) as a major health hazard at the end of 2019. This was first seen in Wuhan province, China [1]. After that this infectious disease has been spread out in the whole world. To defeat the epidemic, the different countries of the world were in a lockdown scenario, the also successively unlock process due to socioeconomic and political impacts [2–6]. Moreover, the mortality and morbidity rate has varied across countries of the worlds [7–9]. COVID-19 is often present in human being as common symptoms, for example, cold-like illness including fever, muscle pain, fatigue, loss of taste and smell, sore throat, and dry cough [10–13].
COVID-19 can be transmitted from people to people through respiratory droplets from an infected human being or direct contact with contaminated surfaces or objects [14, 15]. As all vaccinations are in trial positions for the prevention of COVID-19. Various studies have pointed out the stage of comorbidity co-infection (like lung disease, heart disease, diabetes, etc) among the infected people. According to the Centers for Disease Control and Prevention, all people with comorbidity or medical condition have more risk to become infected than healthy or normal human being [16–18]. From the survey of clinical reports, this is further suggested that the confirmed COVID-19 patients are most having comorbidity co-infection which includes kidney disease, obesity those having basal metabolic rate (BMR) more than thirty, type-2 diabetes, COPD (chronic obstructive pulmonary disease) [19–21]. Moreover, sometimes this confirmed COVID-19 with comorbidity were admitted to the intensive care unit (ICU).
New daily cases of COVID-19 follow a very highly volatile pattern, i.e, deceasing-increasing arbitrarily [22, 23]. To implement fruitful public health measures in a scheduled time within a certain resource according to geographical state [24], it is utmost essential to study the diffusion or force of infection among the wider population. In order to explain this observation, entropy helps us to determine the heterogeneity of a daily number of cases and time of highest diffusion or force of infection [25]. Various methods of measuring entropy, like weighted entropy [26], maximum entropy [27], evolutionary Entropy [28], structural Entropy [29] are employed to forecast and study the transmission dynamics of pandemic like, COVID-19 [30, 31]. Here, the concept of Shannon entropy is indeed used and seemed to be appropriate to determine the spread of an epidemic as there is a significant analogy based on Boltzmann’s classical thermodynamical paradigm [32].
Various mathematical models have been formulated and studied the corresponding dynamics [33, 34]. Lie et al. proposed a data-driven COVID-19 model with distributed delay [35]. Global dynamics of a COVID-19 SEIR epidemic model has been investigated by Khyar et al. [36]. Further, different authors have suggested control strategies for disease transmission of COVID-19 [37–39]. The main aim of this paper is to study the qualitative behavior of COVID-19 transmission dynamics by bifurcation theory. Various qualitative dynamics have been studied in different mathematical model [40–42]. For this purpose, data-driven modeling is a powerful tool to investigate dynamical behavior in infectious disease [43]. Various mathematical models have been devolved as well as investigated the transmission dynamic of COVID-19 [44–48]. Indeed, considering comorbidity in modelling of COVID-19 is of noteworthy interest in ongoing studies and also has not been yet investigated according to literature.
The subsequent parts of the paper are as follows: In Sect. 2, we propose and explore a mathematical model of COVID-19 with comorbidity. The qualitative dynamics is investigated at feasible equilibrium by performing bifurcation analysis in Sect. 3. In Sect. 4, the model is calibrated and sensitivity analysis is also performed. Moreover, the transmission dynamics of mean fluctuation in force of infection is studied by measuring the corresponding Shannon entropy. Finally, in Sect. 5, we discuss and conclude the results from our proposed study.
Mathematical model
We develop here a SCEAIHR-type model by introducing threatened or comorbidity susceptible and hospitalized individuals in the dynamics of COVID-19 (SARS-Cov-2). The model consists of seven compartments, namely, susceptible individuals (S), comorbidity susceptible individuals (C), exposed individuals (E), asymptomatic individual (A), infected or infectious individuals with clinical symptoms (I), diagnosed and hospitalized individuals (H) and recovered individuals with no more infections (R). The total number of individuals is . Our main aim is to study the dynamics of COVID-19 with comorbidity co-infection. Based on a biological viewpoint, we formulate a nonlinear mathematical model to investigate the COVID-19 or SARS-Cov-2 pandemic in a certain time window:
| 1 |
The following initial values are considered in model (1) as follows:
| 2 |
Here, it is assumed that , where indicates the initial date of the outbreak of COVID-19 for the model (1). Susceptible individuals are generated by the recruitment of persons by birth or immigration to the community at a constant rate of . Population decreases after infection and can be transmitted at a rate , and by direct interaction between susceptible individuals with symptomatic infected, asymptomatic infected and hospitalized individuals respectively and at a rate , and by direct interaction between co-morbid susceptible individuals with symptomatic infected, asymptomatic individuals and hospitalized individuals respectively. Also, susceptible individuals often develop co-morbidity at a rate of . It is assumed that there is a recovery rate for asymptomatic, symptomatic, and hospitalized individuals, such as , , and respectively. The model incorporates certain demographic impacts by estimating the disease-induced mortality rate of hospitalized persons and natural death rate of each of the seven subpopulations. After the disease incubation period, , a fraction of the exposed individuals are symptomatically infected and the remaining fraction become asymptomatically infected. The exposed population also decreased at a rate of due to natural death. The pictorial diagram of our model is presented in Fig. 1. The description of model parameters is provided in Table 1.
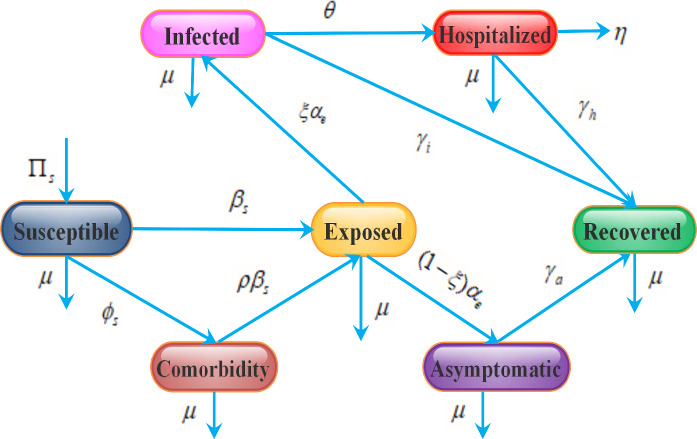
Fig. 1.
Schematic illustration of SCEAIHR model. The flow diagram exhibits the interaction of different stages of individuals in the model: susceptible (S), comorbidity (C), exposed (E), asymptomatic (A), infected (I), hospitalized (H) and recovered (R)
Table 1.
The values of the parameters used in the SCEAIHR model (1)
| Parameter | Description | Value | Reference |
|---|---|---|---|
| Average recruitment rate | 4.7387 | – | |
| Transmission rate | 1.6746 | Estimated | |
| Modification factor for asymptomatic | 0.4499 | Estimated | |
| Modification factor for hospitalized | 0.2918 | Estimated | |
| Rate of comorbidity development by susceptible | 0.0000017 | Estimated | |
| Average life expectancy at birth | 70.4 years | [50] | |
| Modification factor for comorbidity development | 0.1433 | Estimated | |
| COVID-19 incubation period | 5.2 days | [51] | |
| Fraction of exposed individuals to become infected | 0.62 | [52] | |
| Recovery rate of asymptomatic individuals | 0.73 | [52] | |
| Recovery rate of infected individuals | 0.79 | [52] | |
| Average on hospitalized rate of infected individuals | 0.1037 | Estimated | |
| Recovery rate of hospitalized individuals | 0.8368 | [53] | |
| Average case fatality rate | 0.0156 | [53] |
SCEAIHR model analysis
In this section, we study some basic properties of the SCEAIHR model (1) such as positivity and boundedness, basic reproduction number, stability analysis at biologically feasible equilibrium points with non-negative initial conditions .
Lemma 3.1
For initial values (2), the solutions of SCEAIHR model (1) remain positive through out the region in for all
Proof
To establish the positivity of the system (1), we take any solution emerging from non-negative region which remain positive for all In order to do this, we prove that the points of vector field on the each hyper plane are bounded by non-negative region . In the system (1), we see that Here, this assures the positivity of solutions in the region . is established as positive invariant set of SCEAIHR model.
Lemma 3.2
For initial values (2), the solutions of SCEAIHR model (1) are uniformly bounded in the region
Proof
In order to prove boundedness, we sum up all equations in the model (1), which provides Differentiating both sides, we get
which implies Without of loss of generality, we can express all equations in model (1) as . So, a bounded set can be defined
which is further a positive invariant set to the SCEAIHR model (1). Therefore, all solution trajectories initiating from interior of always remain within the domain . this assures that the growth of all individuals can’t be unbounded or exponential for time time window.
Infection free equilibrium and basic reproduction number
The basic reproduction number is a threshold value and characterizes the infectious disease. This is denoted by and defined by the number of secondarily infected individuals induced by single infected individuals as infectious during its incubation period within susceptible individuals. is a dimensionless number and quantifies the expectation of decreasing or increasing the disease outbreak. The system (1) has infection free equilibrium , given by 0, 0, 0, 0, 0, 0). The compartment E, A, I, H in system (1) are explicitly associated with disease outbreak. Thus we get matrix , for new infection and transition part respectively, given by
The variational matrix and , where can be calculated at The basic reproduction number is dominant eigenvalue of matrix So, we obtain
Stability of infection free equilibrium
Theorem 1
The infection-free equilibrium is locally asymptotically stable if and unstable
Proof
The variational matrix J of the system (1) is given by
| 3 |
For our convenience, we consider
| 4 |
The matrix at has unique and repeated eigenvalues as and respectively. The rest of eigenvalues are the roots of following equation as follow:
which can be expressed as follow:
Denote
Now substitute , if , where, represents real part of , then
Consequently, , which implies . So, eigenvalues corresponding characteristic equation have negative real parts for Thus, for , all eigenvalues are negative. According to Routh–Hurwitz stability criterion, infection free equilibrium is locally asymptotically stable for Again, if we consider , i.e, then
then, there exits so that . This means that there exits non-negative eigenvalue in matrix Hence, infection free equilibrium is unstable for
Existence of endemic equilibrium
Here, we study the existence of endemic equilibrium. Let be any endemic equilibrium of system (1). Moreover, The force of infection is
| 5 |
By solving the system (1) at equilibrium state, we get , , , , and .
Substituting the all expressions into (5), we get the non- zero equilibrium of (1) which satisfies quadratic equation in as follow:
| 6 |
where,
| 7 |
From (6), we can obtain endemic equilibrium of (1). Here, the coefficient A is always positive. The sign of coefficient C depends on lesser or greater value of than unity. It can be noted that multiple endemic equilibrium can be coexist for Thus, we can summarize as follows:
Theorem 2
The system (1) has
two endemic equilibrium if and and
at least one endemic equilibrium if iff
a unique endemic equilibrium if ( and ) or
no endemic equilibrium if iff and
From the above theorem, it can be noted that backward bifurcation is possible due to the coexistence of stable infection-free and endemic equilibrium for For this purpose, we find a critical value of for unique endemic equilibrium as follow:
Hence, backward bifurcation can occur with specific restriction . Now, we turn on the analysis for backward bifurcation.
Backward bifurcation
In order to investigate backward bifurcation at endemic equilibrium , we apply centre manifold theorem by considering bifurcation parameter , corresponded to
From variational matrix J in (3), we again consider few entries as follows:
| 8 |
Now, the variational matrix of system (1) at has right eigenvector corresponding to zero eigenvalue, given by , where
Similarly, at at , the variational matrix of system (1) has left eigenvector corresponding to zero eigenvalue, given by , where
We introduce a few notations for SCEAIHR model system as follows: and where Now, we compute second order partial of at infection-free equilibrium and obtain
The remaining partial derivatives at are zero. Now we evaluate the coefficient a and b of well-established Theorem 4.1 in Castillo-Chavez et al. [49] as follows:
Now, we substitute all above values to find the coefficient a and b at threshold , we get
and
Here, the value of b is always positive, the system (1) undergoes backward bifurcation at if , i.e
| 9 |
where,
Theorem 3
The SCEAIHR model system (1) exhibits a backward bifurcation at , if the parametric restriction in (9) holds.
Moreover, as comorbidity individual becomes always infected, backward bifurcation occurs in SCEAIHR model. Now for a case, it can be noted that comorbidity individuals do not get infection due to maintaining social distancing, wearing mask and proper sanitation, i.e, The bifurcation coefficient a is as follow :
As, , , by Theorem 4.1 in Castillo-Chavez et al., a transcritical bifurcation occurs at for and endemic equilibrium and the endemic equilibrium is locally asymptotically stable for Here, a backward bifurcation do not exist for .
Theorem 4
The SCEAIHR model system (1) experiences a transcritical bifurcation at with parametric restriction in (9) holds, i.e, .
Computer simulation and results
In this section, we perform numerical simulation and its biological interpretation to complement the analytical findings. In the previous section, local stability for infection-free equilibrium and the existence of endemic equilibrium are studied. Moreover, backward bifurcation and transcritical bifurcation are observed. In order to validate the analytical findings, we estimate parameter values of the SCEAIHR model (1).
Model calibration
For the duration from March, 2020 to October, 2020 is considered for model calibration. For this analysis, we have taken daily COVID-19 cases in India. Daily cases notified by COVID-19 were obtained for India from [53]. The model (1) is fitted for new hospitalized COVID cases in India on a regular basis. Due to the high infectivity, the notified patients become hospitalized quickly and it is also easy to confirm the hospitalized cases from the recorded results. We have enlisted the main model parameters, which are estimated from the data in Table 1. By adopting the model to the new regular cases reported, six parameters of the model have been estimated, such as (a) transmission rate (), (b) modification factor for asymptomatic () and symptomatic (), (c) rate of co-morbidity development by susceptible individuals (), (d) modification factor for comorbidity development () and (e) average hospitalized rate of infected individuals (). The data will also estimate some of the initial conditions of the model (1). In MATLAB, the nonlinear least-square solver has been used during the defined time span to fit the simulated new daily data of COVID-19 recorded by India. The confidence area is generated by using the Delayed Rejection Adaptive Metropolis (DRAM) algorithm. A description of this model fitting technique is given in [54]. Tables 1 and 2 provide the parameters and initial conditions respectively, which are estimated by the above technique. The fitting of India’s daily new hospitalized COVID-19 cases is seen in Fig. 2. The basic reproduction number is estimated at by using the fixed parameters from Table 1 and parameters that are estimated from the model.
Table 2.
Estimated initial population sizes for India
Fig. 2.
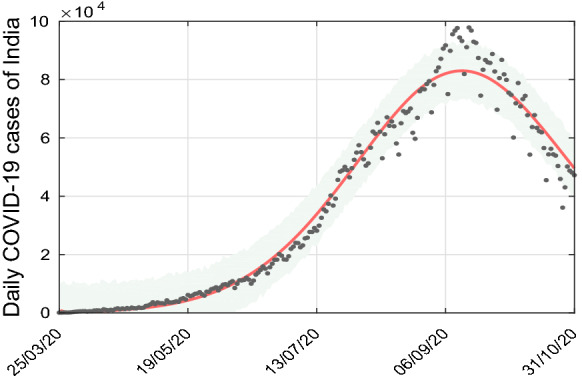
The SCEAIHR model fitted to daily new confirmed COVID-19 cases in India. Observed data points are shown in black dots and the solid red line depicts the model simulated curve
In order to recognize the most significant parameters to infected individual, we next perform sensitivity analysis followed by uncertainty analysis.
Sensitivity analysis
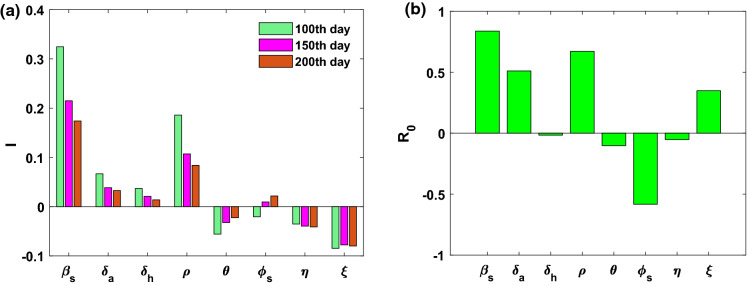
We apply PRCC(Partial rank correlation coefficient) method for sensitivity analysis and LHS (Latin hypercube sampling) method for uncertainty analysis. A detailed technique can obtained in Marino et. al, [55]. We assign uniform distribution the parameters namely, , , , , , and with confidence domain. We consider baseline values as parameter values. The outputs are given in Table 3 with bar diagram in Fig. 3a. From Fig. 3a, it can be seen that , and are influential parameter to infected individual. Moreover, we study the sensitivity indices of basic reproduction number . Here, only is negatively correlated, and , , , and are positively correlated to , given in Fig. 3b.
Table 3.
Sensitivity indices of the parameters of SCEAIHR model (1) to I and . i= 100, 150, 200 th day
| Description | ||||||||
|---|---|---|---|---|---|---|---|---|
| 0.3221 | 0.0622 | 0.0441 | 0.1918 | 0.0418 | 0.0235 | 0.0321 | 0.0914 | |
| 0.2214 | 0.0415 | 0.0350 | 0.1261 | 0.0358 | 0.0113 | 0.0372 | 0.0811 | |
| 0.1786 | 0.0321 | 0.0271 | 0.0938 | 0.0204 | 0.0225 | 0.0407 | 0.0830 | |
| 0.8400 | 0.4962 | 0.0035 | 0.7630 | 0.1093 | 0.5842 | 0.0636 | 0.3401 |
Fig. 3.
a and b PRCC indicating sensitivity indices to infected individual (I) and basic reproduction number . PRCC values of various parameters with the level of significance 0.05. Sample size = 500 for each parameters is taken based on LHS approach with uniform probability distribution
As basic reproduction number quantifies the expectation of decreasing or increasing the epidemic evolution, we next study the effects of parameter variation on .
Effects of parametric variation on basic reproduction number
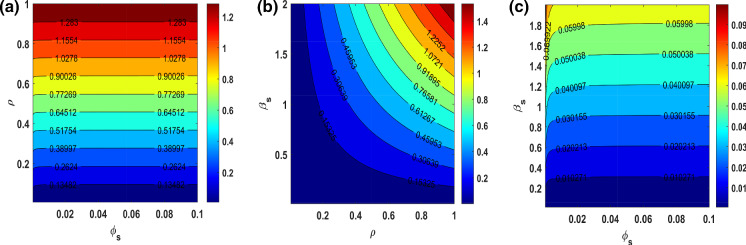
We further study the effects of parameter variations on basic reproduction number under , , in Fig. 4. We observe that only the increasing value of can change to in Fig. 4a. From Fig. 4b, the increasing value simultaneously changes the value of . Further, it can be also seen that the value of always remains less than one within the parametric plane of .
Fig. 4.
Contour plots indicating the nature of change in basic reproduction number() of SCEAIHR model under parametric planes. a versus . b versus . (c) versus
Bifurcation diagram
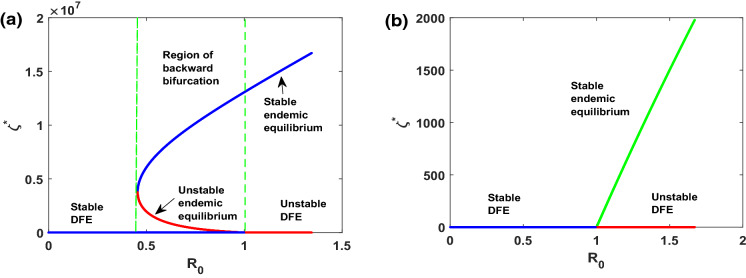
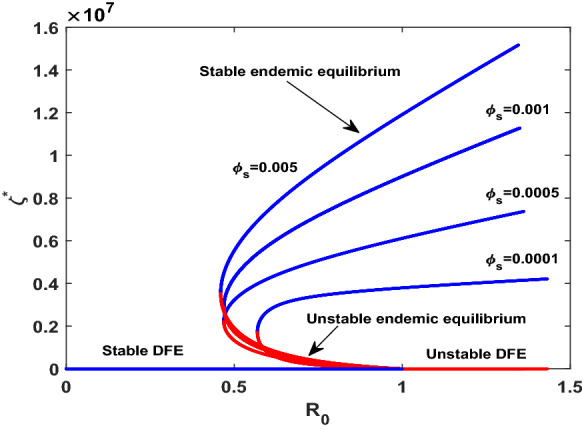
On investigation of the existence of endemic equilibrium, we derive the sub-threshold range of bistable equilibrium in the SCEAIHR model (1). Thus backward bifurcation can be seen within the domain with , given in Fig. 5a, where two positive endemic equilibrium coexist, i.e, asymptotically stable equilibrium (blue line) and unstable equilibrium (red line). From an epidemiological viewpoint, the scenario of backward bifurcation indicates that the model possesses two endemic equilibrium. However, infection-free equilibrium can exist only for . Indeed, this restriction is sufficient for the eradication of infection from the system. Moreover, it sometimes depends on the initial size of the sub-population of model.
Fig. 5.
a versus plot indicating backward bifurcation of SCEAIHR model in . b versus plot illustrating transcritical bifurcation of SCEAIHR model in with . All the remaining parameters values are reported in Table 1
On the other hand, if comorbidity individuals do not acquire infection, i.e, , the model (1) experiences transcritical with given in Fig. 5b. Here, only one endemic equilibrium (blue line) and infection-free equilibrium (red line) exist for It indicates that force of infection is not strong enough to spread out as all susceptible individuals take the precaution of disease spreading, like, wearing a mask, social distancing, maintaining sanitation.
Moreover, it can be also noted that increasing the value of becomes a challenging task for the eradication of infection. Numerically, it can be seen that a backward bifurcation regime increases gradually with increases of with , given in Fig. 6. Consequently, the value of is essential to be reduced to ensure the eradication of infection in such a way that and is close to one.
Fig. 6.
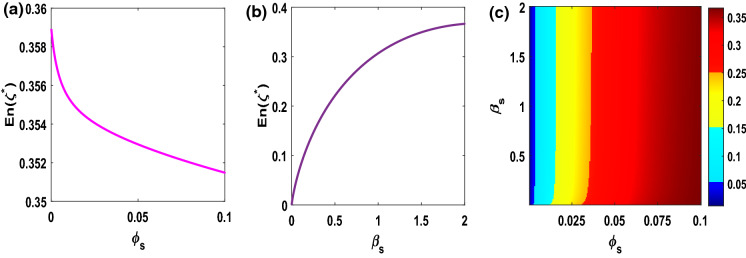
Impacts of variation in , on backward bifurcation with , keeping all parameters value remained same as in Table 1. The diagram exhibits that the extent of backward bifurcation regime increases gradually with the increasing of
Now we study the variation of infection intensity. For this purpose, we consider average of force of infection for daily basis cases.
Effects of parametric variation on force of infection
Now we investigate the fluctuation of force of infection by considering , where N being length of series . Figure 7a, b exhibit the fluctuations of average force of infection under and respectively. From Fig. 7a, b, increasing and decreasing trend can be observed in respectively. It indicates that average force of infection decreases with the increase of and average force of infection increases with the increase of in the model (1). Moreover, in order to understand the combined effect under on , a matrix plot is given in Fig. 7c. In Fig. 7c, it can be mentioned that the average force of infection is dependent on value of for any value of , defined in efferent color region of increased value of In order to quantitative measure the degree of disorder in force of infection. Shannon entropy [32] is employed for this purpose. Here, disorder means the non-uniform distribution of infection under distribution of susceptible.
Fig. 7.
a, b represent versus plot (with and versus plot (with . c represent over matrix plot, where . The corresponding color bar indicates values of . The values of the other parameters are taken as same, shown in Table 1
Entropy in force of infection
Shannon entropy is generally defined as
N being length of event and is probability of event , i.e, relative frequency of occurrence of non-recurrent event . The trend of on and are shown in Fig. 8a, b. Figure 8a shows that deceasing trend of with increase of . On the contrary, increases with in Fig. 8b. From Figs.7a, b and 8a, b, it can be noted that the trend of is highly correlated with the fluctuation in force of infection . Moreover, we also plot in Fig. 8c to study the dependence of on . Further comparing Figs. 7c and 8, we observe similar pattern of increasing entropy between and . This result reveals the additional evidence about the measure of dynamical disorder in force of infection in the model (1) for parameter regimes.
Fig. 8.
a, b represent versus plot (with and versus plot (with . (c) represent over matrix plot, where . The corresponding color bar indicates values of . The values of the other parameters are taken as same, shown in Table 1
Discussion and conclusion
In this paper, we have proposed and studied a mathematical model for disease dynamics of COVID-19 accounting of impacts of comorbidities on the complication of COVID-19. The proposed model has been calibrated by using new daily cases of India. The qualitative properties of the model have been studied and the basic reproduction number has been calculated by the method of the next-generation matrix. The model has asymptotically stable infection-free equilibrium for less than a unity of basic reproduction number. Using the center manifold theorem, the phenomena of backward bifurcation have been observed for increasing the modification factor of comorbidity development. This indicates that infection persists for less than a unity of basic reproduction number. The occurrence of backward bifurcation assures the rich dynamics of the model. From an epidemiological viewpoint, comorbidity individuals acquire more re-infection due to lack of surveillance and precautions like wearing masks, social distancing, proper sanitation, etc. Further, endemic equilibrium has become asymptotically stable if the modification factor for comorbidity is zero (), i,e. none of the comorbidity individual gets an infection and keeps maintained themselves with high surveillance. In these circumstances, more susceptible individuals have acquired infection and become exposed individuals as basic reproduction is more than unity, indeed, exposed individuals have a strong immunity to prevent COVID-19 infection in this context. Moreover, the increase of comorbidity development and modification factor simultaneously has increased the backward bifurcation regimes. Consequently, this can lead to a disaster situation.
Sensitivity analysis of the model reveals that transmission rate (), modification factor of comorbidity development (), comorbidity development () have a significant influence on infected individuals and basic reproduction number (). In order to control disease transmission, a policy might be implemented by enhancing surveillance with proper maintaining sanitation and keep safe from infected individuals.
On investigation of the pattern of transmission dynamics, the mean fluctuations of the force of infection have shown decreasing as well as increasing trend with comorbidity development () and transmission rate (). Consequently, the combined effect has shown an increasing trend to the average force of infection. From the dynamical viewpoint, the measure of disorder in force of infection, i.e, the Shannon entropy has provided evidence towards the higher entropy production indicating the more powerful force of infection, i.e, the infection reaches a significant proportion in susceptible individuals. This might lead to a devastating situation in society regarding disease transmission.
Finally, our study assures that the increase of force of infection builds up with the enhanced entropy production attributed to highly disorder in epidemic evolution. The production of entropy are concepts that can be helpful to guide computational biologists to elucidate the associations between mean fluctuation and changes in parameters to develop policy in disease control and novel vaccination strategies. Thus, it would be useful to include in future publications how vaccination policy can be fruitful.
Funding
Sk Shahid Nadim receives funding from Council of Scientific & Industrial Research as senior research fellowship (Grant No. 09/093(0172)/2016/EMR-I), Government of India, New Delhi.
Declarations
Conflict of interest
The authors have no conflict of interest concerning the publication of this manuscript.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Parthasakha Das, Email: parthasakha87das@gmail.com.
Sk Shahid Nadim, Email: nadimskshahid@gmail.com.
Samhita Das, Email: samhitamaths@yahoo.com.
Pritha Das, Email: prithadas01@yahoo.com.
References
- 1.Wang C, Horby PW, Hayden FG, Gao GF. A novel coronavirus outbreak of global health concern. The Lancet. 2020;395(10223):470–473. doi: 10.1016/S0140-6736(20)30185-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sun G-Q, Wang S-F, Li M-T, Li L, Zhang J, Zhang W, Jin Z, Feng G-L. Transmission dynamics of covid-19 in wuhan, china: effects of lockdown and medical resources. Nonlinear Dyn. 2020;101(3):1981–1993. doi: 10.1007/s11071-020-05770-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tian H, Liu Y, Li Y, Chieh-Hsi W. An investigation of transmission control measures during the first 50 days of the covid-19 epidemic in china. Science. 2020;368(6491):638–642. doi: 10.1126/science.abb6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Spelta A, Flori A, Pierri F, Bonaccorsi G, Pammolli F. After the lockdown: simulating mobility, public health and economic recovery scenarios. Sci. Rep. 2020;10(1):16950. doi: 10.1038/s41598-020-73949-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Saha, Jay, Chouhan, Pradip: Lockdown and unlock for covid-19 and its impact on residential mobility in india: an analysis of the covid-19 community mobility reports, 2020. Int. J. Infect. Dis. (2020) [DOI] [PubMed]
- 6.Acharya R, Porwal A. A vulnerability index for the management of and response to the covid-19 epidemic in india: an ecological study. Lancet Glob. Health. 2020;8(9):e1142–e1151. doi: 10.1016/S2214-109X(20)30300-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mekonen KG, Habtemicheal TG, Balcha SF. Modeling the effect of contaminated objects for the transmission dynamics of covid-19 pandemic with self protection behavior changes. Results Appl. Math. 2021;9:100134. doi: 10.1016/j.rinam.2020.100134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Walker PGT, Whittaker C, Watson OJ. The impact of covid-19 and strategies for mitigation and suppression in low- and middle-income countries. Science. 2020;369(6502):413–422. doi: 10.1126/science.abc0035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Singh AK, Misra A. Impact of covid-19 and comorbidities on health and economics: Focus on developing countries and india. Diabetes Metab. Syn. 2020;14(6):1625–1630. doi: 10.1016/j.dsx.2020.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brand SPC, Tildesley MJ, Keeling MJ. Rapid simulation of spatial epidemics: a spectral method. J. Theor. Biol. 2015;370:121–134. doi: 10.1016/j.jtbi.2015.01.027. [DOI] [PubMed] [Google Scholar]
- 11.Gralinski LE, Menachery VD. Return of the coronavirus: 2019-ncov. Viruses. 2020;12(2):135. doi: 10.3390/v12020135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang C, Wang Y, Li X, Ren L, Zhao J, Yi H, Zhang L, Fan G, Jiuyang X, Xiaoying G. Clinical features of patients infected with 2019 novel coronavirus in wuhan, china. The Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hogan AB, Jewell BL, Sherrard-Smith E, Vesga JF. Potential impact of the covid-19 pandemic on hiv, tuberculosis, and malaria in low-income and middle-income countries: a modelling study. Lancet Glob. Health. 2020;8(9):e1132–e1141. doi: 10.1016/S2214-109X(20)30288-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hui DS, Azhar EI, Madani TA, Ntoumi F, Kock R, Dar O, Ippolito G, Mchugh TD, Memish ZA, Drosten C. The continuing 2019-ncov epidemic threat of novel coronaviruses to global health-the latest 2019 novel coronavirus outbreak in wuhan, china. Int. J. Infect. Dis. 2019;91(264–266):2020. doi: 10.1016/j.ijid.2020.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thompson R. Pandemic potential of 2019-ncov. Lancet Infect Dis. 2020;20(3):280. doi: 10.1016/S1473-3099(20)30068-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang J, Zheng Y, Gou X, Ke P, Chen Z, Guo Q, Ji R, Wang H, Wang Y, Zhou Y. Prevalence of comorbidities and its effects in patients infected with sars-cov-2: a systematic review and meta-analysis. Int. J. Infect. Dis. 2020;94:91–95. doi: 10.1016/j.ijid.2020.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guan, W.-j., Liang, W.-h., et al.: Comorbidity and its impact on 1590 patients with covid-19 in china: A nationwide analysis. Eur. Respir. J. (2020) [DOI] [PMC free article] [PubMed]
- 18.Carreira, H., Strongman, H., Peppa, M., McDonald, H. I., dos Santos-Silva, I., Stanway, S., Smeeth, L., Bhaskaran, K.: Prevalence of covid-19-related risk factors and risk of severe influenza outcomes in cancer survivors: A matched cohort study using linked english electronic health records data. EClinicalMedicine, 29, (2020) [DOI] [PMC free article] [PubMed]
- 19.Gupta R, Hussain A, Misra A. Diabetes and covid-19: evidence, current status and unanswered research questions. Eur. J. Clin. Nutr. 2020;74(6):864–870. doi: 10.1038/s41430-020-0652-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee SC, Son KJ, Han CH, Jung JY, Park SC. Impact of comorbid asthma on severity of coronavirus disease (covid-19) Sci. Rep. 2020;10(1):21805. doi: 10.1038/s41598-020-77791-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Paramasivam A, Priyadharsini JV, Raghunandhakumar S, Elumalai P. A novel covid-19 and its effects on cardiovascular disease. Hypertens. Res. 2020;43(7):729–730. doi: 10.1038/s41440-020-0461-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guo Z-G, Sun G-Q, Wang Z, Jin Z, Li L, Li C. Spatial dynamics of an epidemic model with nonlocal infection. Appl. Math. Comput. 2020;377:125158. [Google Scholar]
- 23.Sun G-Q, Jusup M, Jin Z, Wang Y, Wang Z. Pattern transitions in spatial epidemics: mechanisms and emergent properties. Phys. Life Rev. 2016;19:43–73. doi: 10.1016/j.plrev.2016.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Núñez-López M, Ramos LA, Velasco-Hernández JX. Migration rate estimation in an epidemic network. Appl. Math. Model. 2021;89:1949–1964. doi: 10.1016/j.apm.2020.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lucia U, Deisboeck TS, Grisolia G. Entropy-based pandemics forecasting. Front. Phys. 2020;8:274. doi: 10.3389/fphy.2020.00274. [DOI] [Google Scholar]
- 26.Das P, Upadhyay RK, Das P, Ghosh D. Exploring dynamical complexity in a time-delayed tumor-immune model. Chaos. 2020;30(12):123118. doi: 10.1063/5.0025510. [DOI] [PubMed] [Google Scholar]
- 27.Tao Y. Maximum entropy method for estimating the reproduction number: An investigation for covid-19 in china and the united states. Phys. Rev. E. 2020;102:032136. doi: 10.1103/PhysRevE.102.032136. [DOI] [PubMed] [Google Scholar]
- 28.Rhodes CJ, Demetrius L. Evolutionary entropy determines invasion success in emergent epidemics. PLoS ONE. 2010;5(9):1–8. doi: 10.1371/journal.pone.0012951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Unlu, E.: Structural entropy of daily number of covid-19 related fatalities. (2020)
- 30.Wang Z, Broccardo M, Mignan A, Sornette D. The dynamics of entropy in the covid-19 outbreaks. Nonlinear Dyn. 2020;101(3):1847–1869. doi: 10.1007/s11071-020-05871-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bandt C. Entropy ratio and entropy concentration coefficient, with application to the covid-19 pandemic. Entropy. 2020;22:1315. doi: 10.3390/e22111315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shannon C. A mathematical theory of communication. Bell Syst. Tech. J. 1948;27(3):379–423. doi: 10.1002/j.1538-7305.1948.tb01338.x. [DOI] [Google Scholar]
- 33.Nazarimehr F, Pham V-T, Kapitaniak T. Prediction of bifurcations by varying critical parameters of covid-19. Nonlinear Dyn. 2020;101(3):1681–1692. doi: 10.1007/s11071-020-05749-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.He S, Peng Y, Sun K. Seir modeling of the covid-19 and its dynamics. Nonlinear Dyn. 2020;101(3):1667–1680. doi: 10.1007/s11071-020-05743-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu X, Zheng X, Balachandran B. Covid-19: data-driven dynamics, statistical and distributed delay models, and observations. Nonlinear Dyn. 2020;101(3):1527–1543. doi: 10.1007/s11071-020-05863-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Khyar O, Allali K. Global dynamics of a multi-strain seir epidemic model with general incidence rates: application to covid-19 pandemic. Nonlinear Dyn. 2020;102(1):489–509. doi: 10.1007/s11071-020-05929-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rohith G, Devika KB. Dynamics and control of covid-19 pandemic with nonlinear incidence rates. Nonlinear Dyn. 2020;101(3):2013–2026. doi: 10.1007/s11071-020-05774-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Das P, Das S, Upadhyay RK, Das P. Optimal treatment strategies for delayed cancer-immune system with multiple therapeutic approach. Chaos Solitons Fract. 2020;136:109806. doi: 10.1016/j.chaos.2020.109806. [DOI] [Google Scholar]
- 39.Huang J, Qi G. Effects of control measures on the dynamics of covid-19 and double-peak behavior in spain. Nonlinear Dyn. 2020;101(3):1889–1899. doi: 10.1007/s11071-020-05901-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Das P, Das P, Mukherjee S. Stochastic dynamics of michaelis-menten kinetics based tumor-immune interactions. Phys. A. 2020;541:123603. doi: 10.1016/j.physa.2019.123603. [DOI] [Google Scholar]
- 41.Das P, Mukherjee S, Das P. An investigation on michaelis - menten kinetics based complex dynamics of tumor - immune interaction. Chaos Solitons Fract. 2019;128:297–305. doi: 10.1016/j.chaos.2019.08.006. [DOI] [Google Scholar]
- 42.Das P, Das P, Das S. An investigation on monod-haldane immune response based tumor-effector-interleukin-2 interactions with treatments. Appl. Math. Comput. 2019;361:536–551. [Google Scholar]
- 43.Heesterbeek, H., Anderson, R.M., Andreasen, V., et al.: Modeling infectious disease dynamics in the complex landscape of global health. Science 347(6227), (2015) [DOI] [PMC free article] [PubMed]
- 44.Weitz JS, Beckett SJ, Coenen AR. Modeling shield immunity to reduce covid-19 epidemic spread. Nat. Med. 2020;26(6):849–854. doi: 10.1038/s41591-020-0895-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Giordano G, Blanchini F, Bruno R, et al. Modelling the covid-19 epidemic and implementation of population-wide interventions in italy. Nat. Med. 2020;26(6):855–860. doi: 10.1038/s41591-020-0883-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Das P, Mukherjee S, Das P, Banerjee S. Characterizing chaos and multifractality in noise-assisted tumor-immune interplay. Nonlinear Dyn. 2020;101:675–685. doi: 10.1007/s11071-020-05781-6. [DOI] [Google Scholar]
- 47.Nadim SKS, Chattopadhyay J. Occurrence of backward bifurcation and prediction of disease transmission with imperfect lockdown: A case study on covid-19. Chaos Solitons Fract. 2020;140:110163. doi: 10.1016/j.chaos.2020.110163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nadim, S.K.S., Ghosh, I., Chattopadhyay, J.: Short-term predictions and prevention strategies for covid-2019: A model based study. arXiv preprint arXiv:2003.08150, (2020) [DOI] [PMC free article] [PubMed]
- 49.Castillo-Chavez C, Song B. Dynamical models of tuberculosis and their applications. Math. Biosci. Eng. 2004;1(2):361. doi: 10.3934/mbe.2004.1.361. [DOI] [PubMed] [Google Scholar]
- 50.COVID-19 coronavirus outbreak. https://www.worldometers.info/coronavirus/#repro, (2020). Retrieved : 2020-12-15
- 51.Li, Q., Guan, X., Wu, P., Wang, X., Zhou, L., Tong, Y., Ren, R., Leung, K.S.M., Lau, E.H.Y., Wong, J.Y., et al.: Early transmission dynamics in wuhan, china, of novel coronavirus–infected pneumonia. New Engl. J. Med. (2020) [DOI] [PMC free article] [PubMed]
- 52.Sardar T, Nadim SKS, Rana S, Chattopadhyay J. Assessment of lockdown effect in some states and overall india: A predictive mathematical study on covid-19 outbreak. Chaos Solitons Fract. 2020;139:110078. doi: 10.1016/j.chaos.2020.110078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.India covid-19 tracker. https://www.covid19india.org/, 2020. Retrieved : 2020-04-03
- 54.Sardar T, Saha B. Mathematical analysis of a power-law form time dependent vector-borne disease transmission model. Math. Biosci. 2017;288:109–123. doi: 10.1016/j.mbs.2017.03.004. [DOI] [PubMed] [Google Scholar]
- 55.Marino S, Hogue IB, Ray CJ, Kirschner DE. A methodology for performing global uncertainty and sensitivity analysis in systems biology. J. Theor. Biol. 2008;254(1):178–196. doi: 10.1016/j.jtbi.2008.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]