Abstract
Oxidative stress is ubiquitously involved in disease etiology or progression. While the damaging effects have been well characterized, how cells deal with oxidative stress for prevention or removal of damage remains to be fully elucidated. Works from our laboratory have revealed de novo Nrf2 protein translation when cells are encountering low to mild levels of oxidative stress. Nrf2 encodes a transcription factor controlling a myriad of genes important for antioxidation, detoxification, wound repair and tissue remodeling. Here we report a role of FUBP1 in regulating de novo Nrf2 protein translation. An increase of FUBP1 binding to Nrf2 5′UTR due to H2O2 treatment has been found by LC-MS/MS, Far Western blot and ribonucleoprotein immunoprecipitation assays. Blocking FUBP1 expression using siRNA abolished H2O2 from inducing Nrf2 protein elevation or Nrf2 5′UTR activity. While no nuclear to cytoplasmic translocation was detected, cytosolic redistribution to the ribosomal fractions was observed due to oxidant treatment. The presence of FUBP1 in 40/43S ribosomal fractions confirm its involvement in translation initiation of Nrf2 protein. When tested by co-immunoprecipitation with eIF4E, eIF2a, eIF3η and eIF1, only eIF3η was found to gain physical interaction with FUBP1 due to H2O2 treatment. Our data support a role of FUBP1 for promoting the attachment of 40S ribosomal subunit to Nrf2 mRNA and formation of 43S pre-initiation complex for translation initiation of Nrf2 protein under oxidative stress.
Keywords: Oxidative stress, Protein translation, 5′UTR, RNA binding Protein, Initiation factors
1. Introduction
Oxidative stress plays a role in a vast variety of diseases, from etiology to progression. The endogenous sources of oxidative stress consist of reactive oxygen species (ROS) generated from damaged mitochondria, inflammatory reaction or activity of oxidases. Deviation of electron transport in the mitochondrial respiratory chain causes formation of superoxide, which is converted to hydrogen peroxide (H2O2) by superoxidase dismutases. H2O2 forms highly reactive hydroxyl radicals (•OH) via the Fenton reaction for attacking macromolecules inside cells, and high doses of H2O2 can cause cell death. Necrotic cell death triggers inflammatory reaction, with activated neutrophils and macrophages releasing ROS via the plasma membrane associated NADPH oxidases. Like NADPH oxidases that are present in a variety of cell types, the cytoplasmic xanthine oxidase consists another line of ROS generating enzyme. Activation of NADPH oxidases or xanthine oxidase cause further increases of ROS at the cellular level. Despite of the well-established knowledge for damaging effects of ROS, clinical trials have not provided clear evidence of cytoprotection with antioxidant supplements, supporting the importance of understanding the paradox of oxidative stress.
The Nrf2 gene encodes a transcription factor best known as the master controller for a cluster of antioxidant and detoxification genes. These downstream genes contain the Antioxidant Response Element (ARE) in their promoters, allowing Nrf2 binding for transcriptional activation. Typical examples of Nrf2 downstream genes include NAD(P)H: quinone oxidoreductase 1 (NQO1), glutamate-cysteine ligase catalytic subunit (Gclc) and regulatory subunit (Gclm), glutathione S-transferases (GSTs), heme oxygenase-1 (HO-1), and UDP-glucuronosyltransferase [1,2]. Microarray and chromatin immunoprecipitation sequencing studies have revealed that Nrf2 status affects the genes beyond antioxidant and detoxication response, from anabolic metabolism to cell proliferation or extracellular matrix remodeling [[3], [4], [5], [6], [7], [8]]. Genome-wide CRISPR screening revealed Nrf2 as an upstream regulator of autophagy genes [9]. These downstream events depict Nrf2 as a key regular for cellular defense as well as damage removal and tissue repair.
We have found that low to mild doses of oxidants cause rapid elevation of Nrf2 protein in vitro and in vivo by de novo protein translation [[10], [11], [12], [13]]. It is long known that physical or chemical stress causes an inhibition of protein synthesis in general. Increasing evidence supports that selective protein translation occurs to meet the demand of the cellular state at stress. For example, during viral infection, while general protein synthesis is inhibited, a specific set of proteins are synthesized by a 5′-methyl cap independent but the internal ribosomal entry site (IRES) dependent mechanism [[14], [15], [16]]. IRES mediated protein translation has been reported during nutrient deprivation [[17], [18], [19]]. Similar 5′ cap independent mechanisms have been adopted by mammalian cells for selective protein translation under stress conditions [20,21]. De novo translation of selective proteins presents an evolutionary advantage for cells to deal with oxidative stress.
Works from our laboratory have demonstrated increased Nrf2 protein translation involving the interaction of RNA binding proteins with 5′ untranslated region (5′UTR) of Nrf2 mRNA [[11], [12], [13]]. We have used LC-MS/MS based proteomics to reveal the proteins gaining the binding to Nrf2 5′UTR under oxidative stress. We have discovered that La antigen and EF1a as two RNA binding proteins important for oxidant induced de novo Nrf2 protein synthesis [12,13]. Here we report the discovery of 1Far Upstream Binding Protein 1 (FUBP1) playing a role in oxidant induced Nrf2 protein translation.
2. Experimental procedures
2.1. Cell culture
Hela cells were obtained from ATCC and cultured in DMEM with 10% FBS. Cells were subcultured weekly and seeded in 6-well, 24-well or 96-well plates or 100 mm dishes. When the culture reached 80% confluency, the cells were placed in 0.5% FBS/DMEM for 16–20 hours serum starvation before H2O2 treatment for 10 min. Culture medium was replaced with fresh DMEM containing 0.5% FBS for additional culture period before harvesting for measurements at designated time.
2.2. Cell viability
Cells seeded in 24- or 96-well plates were treated with various doses of H2O2 for 10 minutes before changing medium to fresh DMEM with 0.5% FBS for additional 4 hours of culture. Cells in 24 wells plate were fixed with 2% paraformaldehyde and stained with 0.1% Coomassie blue for recording cell morphology under a microscope with 20x lens (Rebel, Echo, San Diego). For metabolic activity, Cell Counting Kit-8 (CCK-8, APExBIO, Boston) was added to cells in 96- well plate for 2-hour incubation under cell culture condition at 37 °C in 5% CO2 incubator. The conversion of water-soluble tetrazolium salt WST-8 to orange colored formazan by cellular dehydrogenase was quantified by measuring the absorbance at 450 nm using a microplate reader (ClarioStarPlus, BMG Labtech, Offenburg, Germany).
2.3. In vitro transcription
Construction of the template plasmid pJet hsa Nrf2 5′UTR has been described previously [12]. To generate Nrf2 5′UTR RNA for protein binding, 10 μg of the plasmid DNA was linearized with XbaI (Fermentas) and extracted with phenol/chloroform/isoamyl alcohol (25:24:1), followed by isopropanol precipitation and 70% ethanol wash. After the pellet was air-dried, 1 μg of the linearized DNA was used for in vitro transcription with a MEGAscript T7 kit (Ambion) in the presence of Biotin-11-UTP (Invitrogen) following the manufacturer's instruction. After incubation for 2 hrs at 37 °C, the reaction was terminated and RNA was extracted with phenol/chloroform/isoamyl alcohol, followed by isopropanol precipitation and 70% ethanol wash. The RNA probe was resuspended in nuclease free water for affinity chromatography.
2.4. RNA affinity chromatography for identification of RNA binding proteins
Cells were harvested in nucleic acid binding buffer [10 mM HEPES (pH 7.6), 5 mM MgCl2, 40 mM KCl, 1 mM DTT, 5% glycerol, 5 mg/ml heparin] containing 1x protease inhibitors in a cocktail (Sigma Aldrich) and lysed by sonication 3 times for 5 seconds each on ice. To remove cell debris and nuclei, the lysate was centrifuged at 18,000×g at 4°C for collecting supernatants to perform RNA binding as described [12,22]. The supernatant containing 500 μg of proteins was incubated with 5 μg of the biotinylated Nrf2 RNA probe on ice for 1 hour, followed by addition of 0.2 ml of Streptavidin Sepharose beads (GE Healthcare) that were pre-equilibrated with the binding buffer. After incubation overnight at 4 °C with rotation, the beads were loaded on a 2 ml column (Pierce) and washed three times with 2 ml of 1 M NaCl in the nucleic acids binding buffer by gravity. The captured proteins were released by boiling in SDS-PAGE loading buffer and resolved on a 10% SDS-PAGE gel. The gel was silver-stained with a mass spectrometry compatible kit (BioRad) for picking unique bands for LC-MS/MS analyses at the Proteomics Core Service at the University of Arizona as described [12]. Alternatively, the proteins on the gel were transferred to PVDF membrane (BioRad) for Western blot analyses as described [13].
2.5. RNA- protein complex immunoprecipitation and qPCR analysis
HeLa cells were lysed on ice with nucleic acid binding buffer containing RNase inhibitor (1U/ml, Fermentas) and protease inhibitors as described above. Anti-FUBP1 antibodies or control IgG (2 μg) were first incubated with 100 μl of Protein A/G Plus beads (Santa Cruz Biotech) for 1 hour at room temperature, followed by three washes with nucleic acid binding buffer to remove unbound antibodies or IgG. The beads were then incubated with 500 μg cell lysates for 4 hours at 4 °C with rotation. After 5 washes with nucleic acid binding buffer, bound RNA was extracted with Trizol (Invitrogen) from the beads and precipitated with 10 μg glycogen.
Alternatively, HeLa cells were lysed in HEPES buffer (10 mM HEPES, pH 7.6, 5 mM MgCl2, 100 mM KCl, 0.5% NP-40, with freshly added 2 mM DTT, 100 U/ml RNase inhibitor and 1x protease inhibitors). Cell lysates were pre-incubated with protein A-agarose for 1 hour at 4 °C to remove non-specific binding material. The pre-cleaned cell lysates were incubated with anti-FUBP1 antibodies or control IgG (2 μg) with rotation at 4 °C overnight and additional 4 hours in the presence of protein A agarose at 4 °C. The immunocomplexes were washed 3 times with the HEPES buffer, before digestion with 3 μg of proteinase K at 50 °C for 30 min and RNA extraction using Trizol. The RNA was precipitated by isopropanol with 10 μg glycogen as a carrier.
The precipitated RNA was converted to cDNA with a Maxima First Strand Synthesis kit (Fermentas) for subsequent qPCR analysis. The design and synthesis of qPCR primer sets were described previously [12]: 5′- CAGGTTGCCCACATTCCCAAATCA-3′ and 5′- AGCAATGAAGACTGGGCTCTCGAT-3′ for human Nrf2 (NM_006164). qPCR was performed with SYBR Green Master Mix (Fermentas) on a BioRad CFX96 thermal cycler and Nrf2 mRNA abundance was calculated with BioRad CFX Manager Software. The cycle threshold (CT) values of control IgG precipitates were averaged for calculation of ΔCT of Nrf2 mRNA from anti- FUBP1 antibody or IgG precipitates of H2O2 treated groups.
2.6. Production of recombinant FUBP1 and EF1α proteins
Human EF1α (NM_001402.5, 1.4 kb) and FUBP1 (NM_003902, 1.9 kb) open reading frames (ORFs) were PCR amplified with Phusion II high fidelity DNA polymerase (Fermentas) and the following primer sets: CTCTGTCGACGGAAAGGAAAAGACTCATATC (EF1α Forward, ATG eliminated, underline indicating restriction site) and CGCAAGCTTTCATTTAGCCTTCTGAGCTTTCT (EF1α Reverse); CTGTCGACGCAGACTATTCAACAGTGC (FUBP1 Forward, ATG eliminated) and GCAAGCTTTTATTGGCCCTGAGGTGCTGG (FUBP1 Reverse). After verification of the presence of a single PCR band at the expected size (1.4 kb for EF1α or 1.9 kb for FUBP1), the PCR products were digested with SalI and HindIII (Fermentas), followed by extraction with phenol/chloroform/isoamyl alcohol and ethanol precipitation. The digested products were then ligated with SalI and HindIII linearized pEcoli-Nterm 6xHN (Clontech) to generate 6xHN-EF1α or 6xHN-FUBP1 plasmids. The sequence of the inserts was confirmed by the Sequencing Core service at the University of Arizona. The plasmids were then used to transform BL21 competent E.coli. Production of recombinant proteins were induced with 0.1 mM of IPTG (Sigma-Aldrich) incubated for 6 hours at room temperature after the cells grew to 0.6 at OD600. The recombinant proteins were purified with Ni-NTA agarose beads (Qiagen) according to the manufacturer's instruction. The eluted proteins were resolved with a 10% SDS-PAGE gel to check the homogeneity with BioSafe Coomassie Staining (BioRad).
2.7. Electrophoretic mobility shift assay (EMSA)
The RNA probe was generated with in vitro transcription of the XbaI linearized pJet hsa Nrf2 5′UTR incorporating [α-32P] ATP (50 μCi) into the sense RNA strand using a MEGAscript T7 kit (Ambion). Unlabeled RNA transcripts were used for cold competition controls. Recombinant proteins (1 μg) was incubated with the 32P-RNA probe in binding buffer (20 mM HEPES pH 7.5, 3 mM MgCl2, 30 mM KCl, 1 mM DTT, 5% glycerol, 5 mg/ml heparin, 1U RNase inhibitor) in a total volume of 20 μl for 30 min on ice. For cold probe competition, 10 μg of the unlabeled RNA probe were added to the reactions and incubated for an additional 30 minutes on ice. The samples were resolved on 4% polyacrylamide gels (60:1 acrylamide/bisacrylamide) in 0.5X TBE buffer. The gel was dried and exposed to a phosphor screen and scanned with a Storm Phosphorimager (GE Healthcare Life Sciences).
2.8. siRNA transfection and luciferase assay
FUBP1 siRNA and control were purchased from Santa Cruz Biotechnology, Inc (sc-43760 and sc-37007) or were designed and synthesized as described [23]. The siRNA was transfected to HeLa cells using Lipofectamine 3000. Cells were treated with H2O2 at 72 hours after transfection for measurements of FUBP1 and Nrf2 protein levels. Cotransfection with pRF bicistronic vector of Nrf2 5′UTR and H2O2 treatment of HeLa cells were based on the protocol reported previously [12]. Luciferase activity was measured using a Dual-Luciferase Assay Kit (Promega) and luminometer.
2.9. Immunocytochemistry
Cells were seeded on 0.13–0.17 mm thick circular cover glasses in 24-well plates and treated with H2O2 for 10 min when the confluence reached 50%. At one hour after H2O2 treatment, cells were fixed and permeabilized by incubating in 100% methanol at −20 °C for 10 minutes. After blocking with 2% BSA in phosphate saline containing 1% Tween 20, the cells were incubated overnight at 4 °C with FUBP1 antibody (1: 50 dilution, sc-271241; Santa Cruz Biotechnology). Alexa Fluor 488 conjugated Chicken anti-Mouse IgG (Invitrogen, A-21200) was used as secondary antibody at 1:2000 dilution. ProLong™ Gold Antifade Mountant with DAPI (Invitrogen, P36931) was added to the cells before mounting and observation under a fluorescence microscope. Images were captured under a 63× objective with a Zeiss Axio Observer Microscope. NIH Image J (v1.52r) was utilized to quantify total fluorescence per cell versus nuclear green fluorescence. Cytosolic FUBP1 fluorescence intensity was calculated by subtracting total cellular fluorescence with nuclear FUBP1 fluorescence signal.
2.10. Isolation of ribosomes
Hela cells were treated with 100 μg/ml cycloheximide (CHX) for 5 minutes at 37 °C before harvesting. After removing the medium, cells were washed twice and scrapped off in ice cold PBS containing 10 μg/ml CHX. Cell pellets were collected by spinning down at 500×g for 5 minutes and resuspended in 425 μl hypotonic buffer [5 mM Tris-HCl (pH 7.5), 1.5 mM KCl, 2.5 mM MgCl2 and 1x protease inhibitor cocktail (EDTA-free)] containing freshly added 10 μg/ml CHX, 1 mM DTT and 100U RNase Inhibitor. After adding 10% Triton X-100 and 10% sodium deoxycholate to a final concentration of 0.5%, the cell lysates were vortexed for 5 seconds then sit on ice for 5 minutes before collection of cytoplasmic fractions. Cell debris, nuclei and mitochondria were removed by two times centrifugation at 16000×g for 5 minutes. The supernatant was transferred onto the top of 10%+35% sucrose cushion and centrifuged at 240,000×g with SW41 rotor for 4 hours at 4 °C. Ribosomal pellets were collected at the bottom and resuspended in Trizol or 1×Laemmli sample buffer (2% SDS, 10% glycerol, 62.5 mM Tris-Cl, pH 6.8, 0.002% bromphenol blue, 5% 2-mercaptoethanol) for RT-PCR or Western blot respectively.
For separation of ribosomal subunits, HeLa cells were treated with 5 μg/ml CHX 10 minutes before harvesting. The cytoplasm was extracted with low salt lysis buffer (20 mM Tris, pH 7.5, 140 mM KCl, 3 mM MgCl2, 1 mM DTT) after cells being washed twice with ice-cold PBS containing 0.5 μg/ml CHX and centrifuged at 400×g for 5 minutes to remove nuclei and cell debris. The supernatant was transferred an Eppendorf tube and centrifuged at 18000×g for 10 minutes two times to remove mitochondrial fractions. The supernatant was loaded onto a linear sucrose gradient (15–50% w/v) and centrifuged at 240,000×g with a SW41 rotor for 1.5 hours at 4 °C. The gradient was displaced upright with 60% sucrose and the distribution of ribosome was recorded at absorbance of OD 254nm with a BioLogic LC System (BioRad) for collecting fractions as described [12,13].
2.11. Immunoprecipitation
Hela cells were seeded and treated in 100 mm dishes. When harvesting, cells were washed twice with ice-cold PBS, lysed in radioimmunoprecipitation (RIPA) buffer (1% Triton X-100, 140 mM NaCl, 0.1% SDS, 0.1% sodium deoxycholate, 10 mM Tris, pH 8.0) containing 1 mM DTT, 1 mM PMSF and 1x protease inhibitors (Pierce Protease Inhibitor Tablet, ThermoFisher Scientific), sonicated on ice for 10 seconds and centrifuged to remove insoluble debris. Cell lysate was pre-cleared with 10 μl Pierce Protein A/G Agarose beads for 1 hour and then incubated with 1 μg FUBP1 antibody (sc-271241; Santa Cruz) or normal mouse IgG (sc-2025; Santa Cruz) for 16 hours at 4 °C. After mixing with 30 μl Pierce Protein A/G Agarose beads for 2 hours with rotation at 4 °C, antibody-protein complex was captured by centrifugation. After three washes with RIPA buffer, captured proteins were eluted by 1×Laemmli sample buffer and analyzed by Western blot.
2.12. Western blot
For total cell lysates, Hela cells in culture dishes were harvested in 1×Laemmli sample buffer. For collecting cytosol versus nuclear enriched fractions, Hela cells were scrapped in ice-cold PBS and span down by centrifugation. The cell pellets were triturated by pipetting in PBS containing 0.5% IGEPAL CA-630 and protease inhibitors. Upon centrifugation, the supernatant was collected as cytosolic fraction and the pellet was collected as nuclear fraction, for denaturing using the Laemmli sample buffer. After boiling and sonicating, samples from equal cell number or equal protein concentration were loaded onto the SDS-PAGE gel. After resolving, proteins were transferred to a PVDF membrane using Bio-Rad Trans-Blot Turbo Transfer System and blotted with following antibodies: Nrf2 antibody (sc-13032), FUBP1 antibody (sc-271241), GAPDH antibody (sc-32233), Lamin B1 antibody (sc-377000), S6 antibody (sc-74459), L36a antibody (sc-100831), La antibody (sc-33593), EF1α antibody (sc-377439), eIF4E antibody (sc-9976), eIF3ŋ antibody (sc-16377), eIF2α antibody (sc-133132), or eIF1 antibody (sc-390122). The bound antibodies were recognized by anti-mouse IgG-HRP (A9044-2ML), anti-rabbit IgG-HRP (A9169-2ML), or anti-goat IgG-HRP (sc-2354). All antibodies were obtained from Santa Cruz Biotechnology, except anti-mouse or anti-rabbit secondary antibodies, which were purchased from Mlillipore Sigma-Aldrich.
2.13. Statistics
Data are presented as means ± SD. Means were compared by 2-tailed Student's t-test or one-way ANOVA with both S–N–K and LSD tests when comparing multiple groups. p ≤ 0.05 was considered significant and is labeled in the figures with *.
3. Results
3.1. Proteomic identification of Nrf2 5′UTR binding proteins
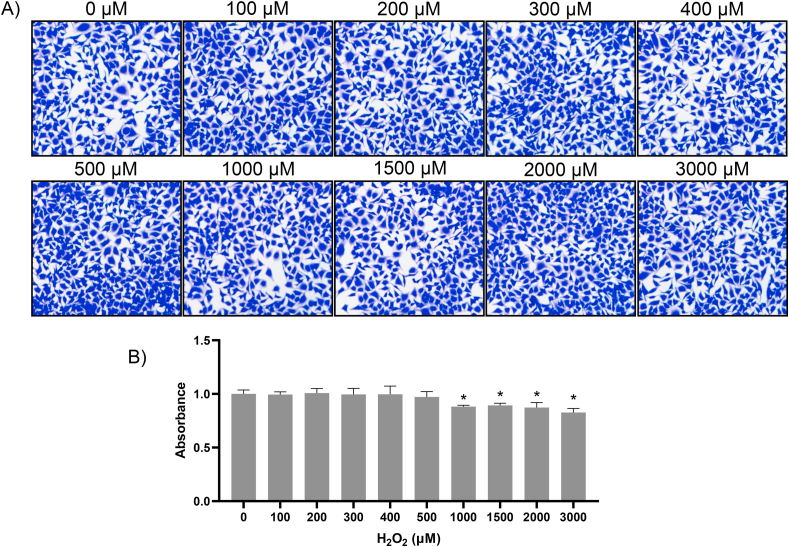
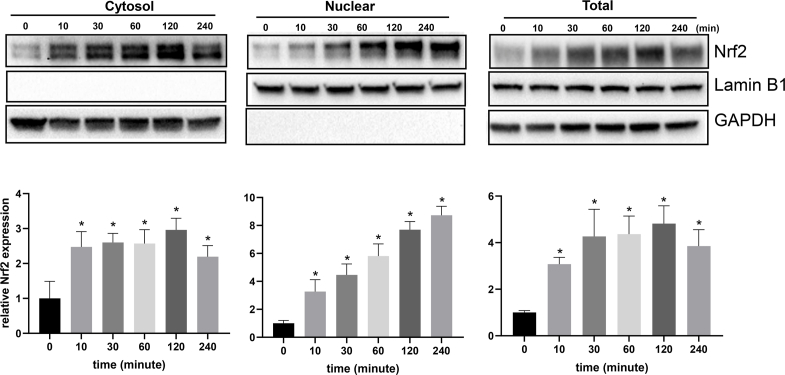
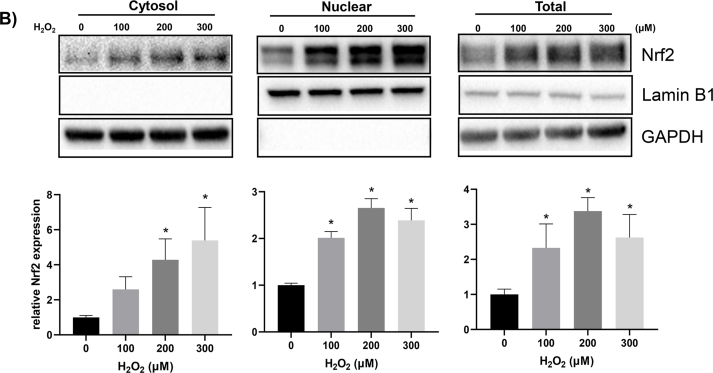
Previous works from our laboratory showed that H2O2 at the non-lethal doses, 50–300 μM caused rapid increase of Nrf2 via de novo protein synthesis in HEK293 cells and cardiomyocytes in culture [10,13]. Hela cells were able to tolerate H2O2 up to 3000 μM H2O2 without morphological sign of toxicity with our treatment protocol (Fig. 1A). H2O2 at 1000 μM or above caused inhibition of cellular metabolism as shown by CCK-8 assay (Fig. 2B). The time course and dose response reconfirmed that HeLa cells were able to increase Nrf2 protein, with significant elevation of Nrf2 detectable within 10 min of non-lethal H2O2 treatment in total cell lysates and cytosolic fractions, preceding the increase of Nrf2 in the nuclear fraction, consistent with de novo Nrf2 protein translation occurring in the cytosol (Fig. 2). Increased nuclear localization of Nrf2 is in agreement with the literature for Nrf2 activation as a transcription factor. H2O2 at 100–300 μM remained the ideal dose range for Nrf2 protein induction in HeLa cells (Fig. 3).
Fig. 1.
Define Sublethal Doses of H2O2. HeLa cells were treated with the dose of H2O2 as indicated for 10 min before changing medium to fresh DMEM with 0.5% FBS. After 4 hours of culture, cell morphology was recorded by fixing cells and staining with Coomassie blue (A). Cell viability or metabolic activity was measure by CCK-8 as described in the Methods. The bar graph represents the average ± SD from three independent experiments. * indicates statistically significant difference between control versus the H2O2 treated group (p < 0.05). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Fig. 2.
H2O2 Treatment Causes Rapid Increases of Nrf2 Protein Levels. HeLa cells were treated with non-lethal 100 μM H2O2 for 10 min before harvesting at the indicated time points for subcellular fractionation and Western blotting to measure the Nrf2 protein level. GAPDH was used as a loading control for the cytosolic fraction and total cell lysate, whereas Lamin B1 was used as a loading control for the nuclear fraction. The bar graphs show the summary of three independent experiments as the average ± SD of the intensity of Nrf2 band over GAPDH or Lamin B1 band. The image is from one Western blot result representative of three independent experiments. * indicates statistically significant difference between control versus the H2O2 treated group (p < 0.05).
Fig. 3.
H2O2 Dose Dependent Induction of Nrf2 Protein. HeLa cells were treated with non-lethal 100–300 μM of H2O2 for 10 min before harvesting at 1 h later for subcellular fractionation and Western blot to measure the Nrf2 protein level as described in Fig. 2.
To understand the mechanism of de novo Nrf2 protein translation under oxidative stress, LC-MS/MS based proteomic approach was employed to identify the proteins binding to Nrf2 5′UTR [12]. The method involved RNA affinity chromatography with biotinylated Nrf2 5′UTR as a bait to pull down binding proteins from the lysates of control and 100 μM H2O2 treated Hela cells. The proteins were separated by SDS gel electrophoresis for identification of differentially displayed bands between control and 100 μM H2O2 treated cells with gel image shown previously [12]. Bands above 37kDa appeared in H2O2 treated cell lysates but were absent in control were excised for LC-MS/MS analysis carried out by a linear quadrupole ion trap ThermoFinnigan LTQ mass spectrometer as described [12]. The instrument detected 10 exclusive unique peptides matching human isoform 1 of Far Upstream Element Binding Protein 1 (FUBP1) (NM_003902, Table 1). These peptides cover 116 out of 644 amino acids, or 18% of the FUBP1 protein sequence.
Table 1.
FUBP1 peptides detected by LC-MS/MS.
| Peptide | Charge | Xcorr | dCN |
|---|---|---|---|
| (K)IGGDAGTSLNSNDYGYGGQK(R) | 2+ | 4.08 | 0.43 |
| 2+ | 3.37 | 0.21 | |
| (K)IQIAPDSGGLPER(S) | 2+ | 4.32 | 0.44 |
| (R)SCMLTGTPESVQSAK(R) | 2+ | 5.25 | 0.46 |
| (K)RLLDQIVEK (G) | 2+ | 2.76 | 0.21 |
| (R)LLDQIVEK(G) | 2+ | 2.91 | 0.16 |
| (R)IGGNEGIDVPIPR(F) | 2+ | 3.77 | 0.42 |
| (R)IQFKPDDGTTPER(I) | 2+ | 3.59 | 0.25 |
| 2+ | 3.60 | 0.17 | |
| 2+ | 3.17 | 0.10 | |
| (F)NFIVPTGK(T) | 2+ | 2.13 | 0.22 |
| (K)AWEEYYK(K) | 2+ | 1.99 | 0.37 |
| (Y)AQTSPQGMPQHPPAPQGQ (−) | 2+ | 3.55 | 0.38 |
| 2+ | 3.46 | 0.31 | |
| 2+ | 3.51 | 0.28 | |
| 2+ | 3.32 | 0.13 |
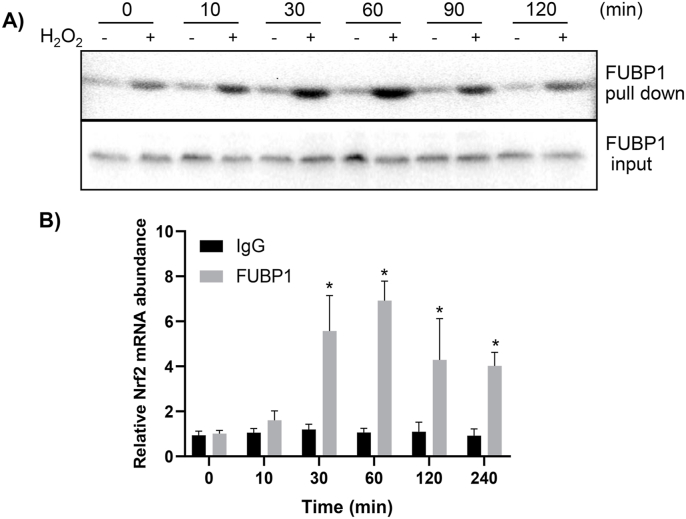
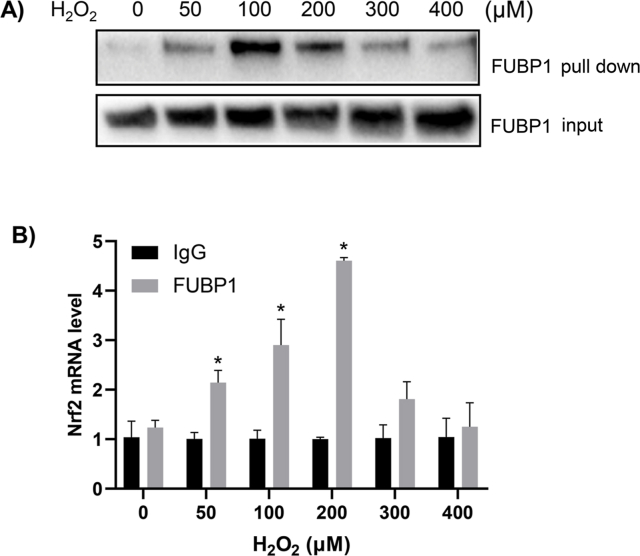
Far Western Blot analysis was used to reveal that oxidative stress caused an increased binding of endogenous FUBP1 to Nrf2 5′UTR. The binding proteins isolated by RNA affinity chromatography made of biotinylated Nrf2 5′UTR were used for detection of FUBP1 protein by Western Blot. A time dependent increase of FUBP1 protein was found for binding to Nrf2 5′UTR in H2O2 treated cells (Fig. 4A). To confirm the finding and to demonstrate such interaction indeed occurring in vivo, we used ribonucleoprotein immunoprecipitation (RNP-IP) to examine FUBP1 interaction with Nrf2 mRNA at the cellular level. In RNP-IP experiments, anti-FUBP1 antibody was used to pull down endogenous FUBP1 together with any bound RNA species. The presence of Nrf2 mRNA in the complex was quantitated by RT-PCR. The results showed that H2O2 treatment indeed caused a time dependent increase of FUBP1 binding to Nrf2 5′UTR in HeLa cells (Fig. 4B). Both types of assays showed increased FUBP1 binding to Nrf2 mRNA started at 10 min and reached the highest level at 30–60 min (Fig. 4), consistent with the time course of Nrf2 protein elevation. Both Far Western Blot and RNP-IP experiments also showed a H2O2 dose dependent FUBP1 interaction with Nrf2 mRNA (Fig. 5A&B). These results demonstrated that FUBP1 indeed increases its interaction with Nrf2 mRNA during oxidative stress.
Fig. 4.
Time Dependent FUBP1 Binding to Nrf2 mRNA. HeLa cells were treated with 100 μM H2O2 for 10 min and harvested at indicated time point for in vitro binding using Nrf2 5′UTR as a probe (A), or for immunoprecipitation using antibodies against FUBP1 for detection of associated Nrf2 mRNA (B). The proteins bound to Nrf2 5′UTR probe were pull down for Western blot to detect FUBP1, with FUBP1 levels in total cell lysates serving as an input (A). The immunoprecipitates of FUBP1 were used for real time RT-PCR to detect Nrf2 mRNA, and the abundance of Nrf2 mRNA was calculated with ΔCT by comparing the copy number from the immunoprecipitates of FUBP1 group or IgG group to that of the average of IgG group at 0 time point. The results are from one experiment representative of three independent experiments. * indicates statistically significant difference between control versus the H2O2 treated group (p < 0.05).
Fig. 5.
H2O2 Dose Dependent FUBP1 Binding to Nrf2 mRNA. HeLa cells were treated with H2O2 for 10 mins at the indicated dose and harvested at 1 hour after for in vitro binding using Nrf2 5′UTR as a probe (A) or for immunoprecipitation using antibodies against FUBP1 for detection of associated Nrf2 mRNA (B), as described in Fig. 4.
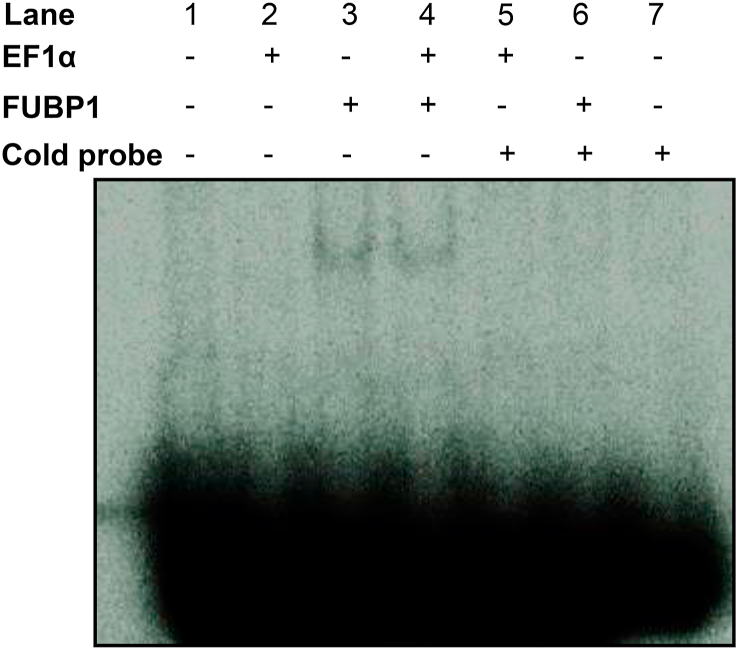
To further demonstrate the interaction of FUBP1 with Nrf2 5′UTR, we performed EMSA assay to reveal the direct binding in vitro. We have reported that the eukaryotic elongation factor 1a (EF1α) participated in de novo Nrf2 protein translation by binding to the G-quadruplex region of Nrf2 5′UTR [13], therefore EF1α was included to address whether or not EF1α enhance FUBP1 binding to Nrf2 mRNA. As shown in Fig. 6, the recombinant FUBP1 was able to bind to the probe made of Nrf2 5′UTR (Lane 3), whereas recombinant EF1α did not affect such binding (Lane 4). The binding of recombinant FUBP1 to Nrf2 5′UTR is specific since addition of excessive cold probe abolished such binding (Lane 6). This provides an additional piece of evidence for FUBP1 binding to Nrf2 5′UTR.
Fig. 6.
Recombinant FUBP1 Is Capable of Nrf2 5′UTR Binding. EMSA was performed using in vitro transcribed Nrf2 5′UTR probe and recombinant FUBP1 protein as described in the Methods.
3.2. FUBP1 mediates Nrf2 protein translation
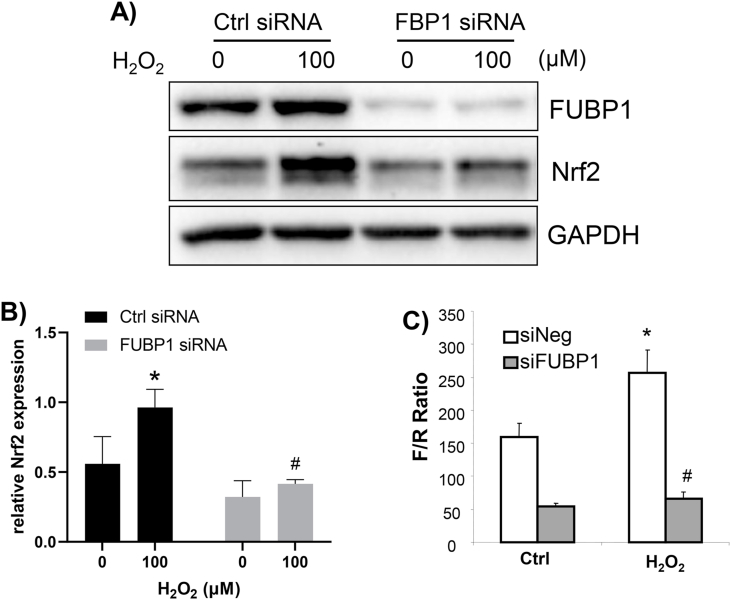
We addressed whether FUBP1 regulated de novo Nrf2 protein translation upon oxidative stress by blocking FUBP1 expression using siRNA. When cells were transfected with FUBP1 specific siRNA, we observed reduced level of FUBP1 protein and inhibition of H2O2 from inducing Nrf2 protein (Fig. 7A and B). To demonstrate that FUBP1 indeed regulated Nrf2 protein translation through Nrf2 5′ UTR, we used a dicistronic reporter containing Nrf2 5′UTR to assess the activity of IRES [12,13]. The dicistronic reporter contains a SV40 promoter for transcription of both Renilla and Firefly luciferases. The construct allows 5′ 7-methyl guanine cap-dependent translation of Renilla luciferase and Nrf2 5′UTR-mediated translation of Firefly luciferase. The ratio of Firefly luciferase over Renilla luciferase reflects Nrf2 5′UTR mediated protein translation. As expected, H2O2 treatment caused an induction of Nrf2 5′UTR activity (Fig. 7C). Blocking FUBP1 expression using siRNA resulted in a reduction of H2O2 induced Nrf2 5′UTR activity (Fig. 7C). These results support that FUBP1 was required for IRES-mediated Nrf2 translation upon oxidative stress.
Fig. 7.
FUBP1 Is Required for Nrf2 Protein Induction and Nrf2 5′UTR Activation. FUBP1 siRNA or negative control was introduced to HeLa cells at 72 hours prior to H2O2 treatment. Cells were harvested at 1 hour after 10 min of 100 μM H2O2 treatment, for Western blot to detect FUBP1 or Nrf2 protein from total cell lysates using GAPDH as a loading control (A, B). FUBP1 siRNA or negative control was cotransfected with pRF bicistronic vector of Nrf2 5′UTR in HeLa cells. At 72 hours after, cells were treated with H2O2 for measurements of luciferases at 1 hour after (C). The results of one experiment representative of three are shown (A, C), or are summarized as average ± SD of Nrf2 band intensity over that of GAPDH from three experiments (B). * indicates significant difference between control versus the H2O2 treated group (p < 0.05), whereas # indicates significant difference between two H2O2 treated groups with FUBP1 siRNA versus with control siRNA (p < 0.05).
3.3. Ribosomal association of FUBP1 and interaction with eIFs under oxidative stress
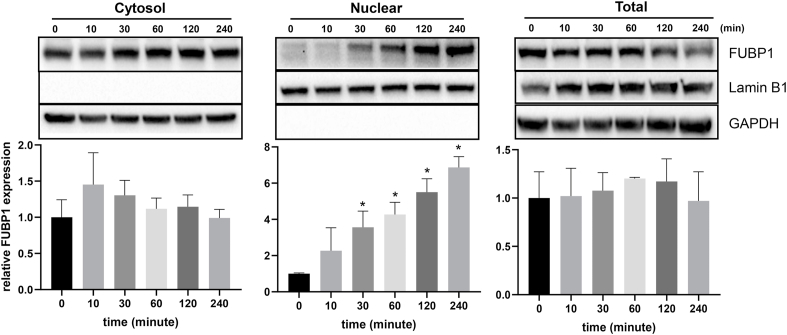
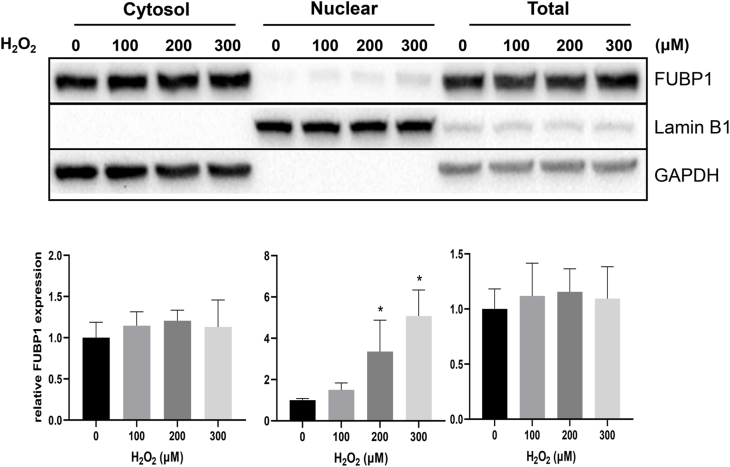
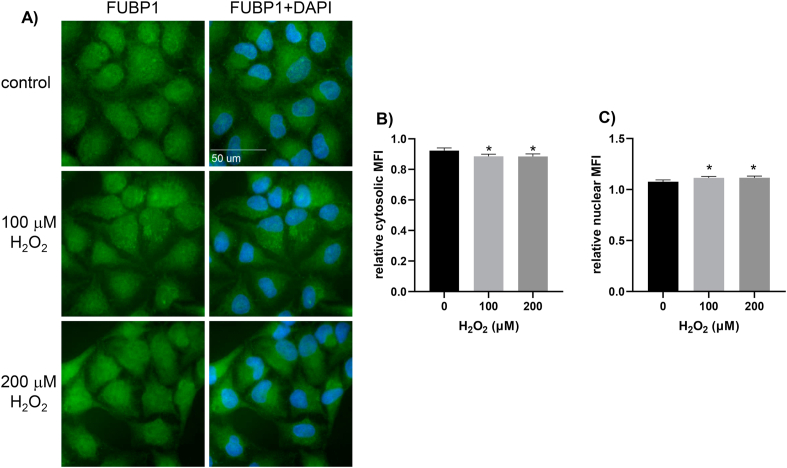
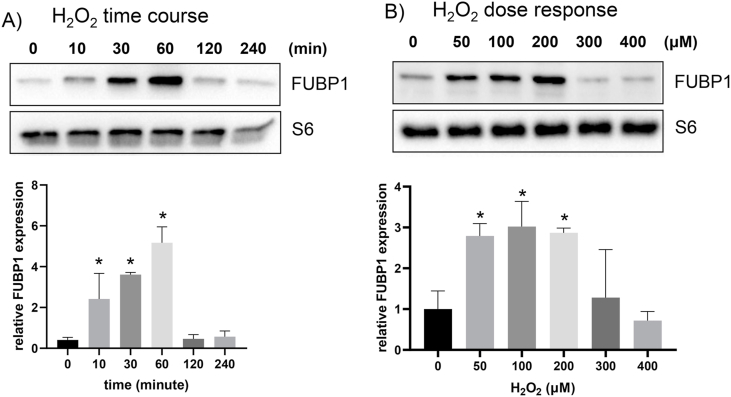
In an effort to understand how FUBP1 increases its binding to Nrf2 5′UTR upon oxidative stress, we tested whether or not FUBP1 protein abundance or subcellular localization changed. Total FUBP1 protein level did not show clear difference in H2O2 treated versus control samples in time course or dose response experiments (Fig. 8, Fig. 9). Lack of increases of FUBP1 in the cytosolic fractions does not support that H2O2 causes cytoplasmic translocation of FUBP1 (Fig. 8, Fig. 9). An observed increase in nuclear FUBP1 does not explain its involvement in Nrf2 protein translation, since protein translation occurs in the cytoplasm. Consistently with subcellular fractionation data, immunofluorescence staining showed no increased signal in cytoplasmic FUBP1 due to H2O2 treatment (Fig. 10).
Fig. 8.
Lack of Time Dependent Nuclear to Cytoplasmic Translocation of FUBP1. Hela cells were treated with 100 μM H2O2 for 10 minutes and subsequently cultured in fresh DMEM containing 0.5% FBS for indicated time point before harvesting for total cell lysates or fractionation into cytosolic and nuclear extracts. The level of FUBP1 protein was determined by Western Blot. Lamin B1 was used as a loading control for nuclear fraction, whereas GAPDH was used as a loading control for cytosolic or total proteins. The bar graphs represent average ± SD of FUBP1 band intensity over that of corresponding loading control from three independent experiments. * indicates statistically significant difference between control versus the H2O2 treated group (p < 0.05).
Fig. 9.
Lack of Dose Dependent Nuclear to Cytoplasmic Translocation of FUBP1. Hela cells were treated with 0, 100, 200 or 300 μM H2O2 for 10 minutes and cultured in freshly changed DMEM containing 0.5% FBS for another 1 hour. Cells were harvested for total cell lysates or fractionated into cytosolic and nuclear extracts for Western blot as described in Fig. 8.
Fig. 10.
Immunocytochemistry Confirmation of the Lack of Nuclear to Cytoplasmic Translocation of FUBP1. Hela cells were treated with 0, 100 or 200 μM H2O2 for 10 minutes and incubated 1 hour in fresh DMEM containing 0.5% FBS. Cells were then processed for immunocytochemistry to stain for FUBP1 protein using Alexa Fluor 488 conjugated secondary antibody for visualization with a 63× lens (A). The bar graphs showed the mean fluorescence intensity (MFI) of cytosolic versus whole cell area (B), or nuclear versus whole cell area (C) as average ± SD from 12 different fields chosen randomly for quantification using Image J. * indicates significant different between Control versus H2O2 treated group by student's t-test (p < 0.05).
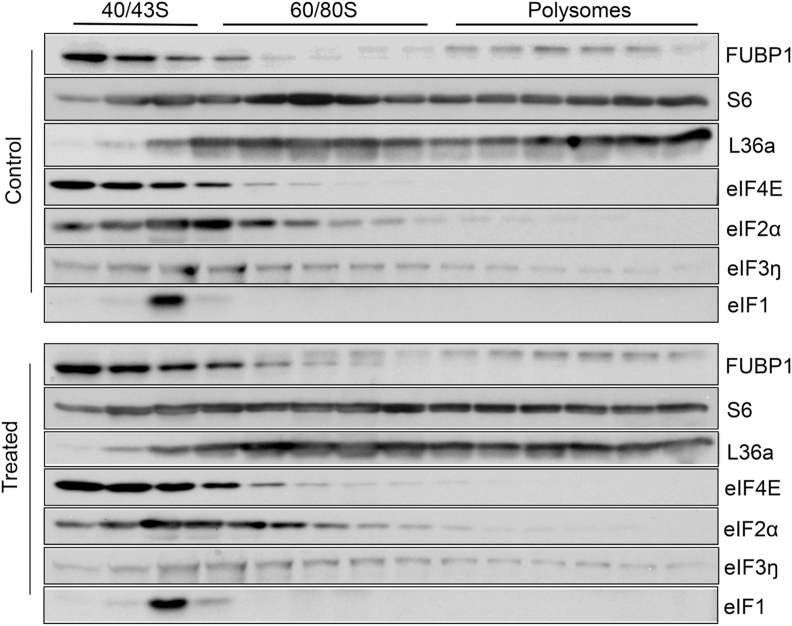
The process of protein translation is divided into three stages: initiation, elongation and termination, with initiation as the rate limiting step. In the initiation stage, the target mRNA strand is occupied by 43S ribosomes, whereas during the elongation stage, the strand of mRNA is bound with multiple ribosomes, i.e. polysomes. We first isolated total ribosomal fractions to address the presence of FUBP1. A time and dose dependent increase of FUBP1 was observed in the total ribosomal fractions (Fig. 11). When ribosomes were collected to separate the subunit of 40/43S, 60/80S, and polysomes, we found the presence of FUBP1 in 40/43S ribosomal fraction, indicating a role of FUBP1 in translation initiation (Fig. 12).
Fig. 11.
Time and Dose Dependent Increase of FUBP1 Association with Ribosomes. Hela cells were treated with 100 μM H2O2 for 10 min before harvesting at indicate time points (A) or were treated with various doses of H2O2 for 10 min before harvesting at 60 min after (B). Cells were lysed for total ribosome isolation using sucrose cushion and ultracentrifugation as described in the methods. Ribosomal proteins were resolved by SDS-PAGE for Western blot to detect FUBP1 protein using S6 as a loading control. The bar graphs showed average ± SD of FUBP1 band intensity over that of corresponding loading control from three independent experiments. * indicates significant different between control versus the H2O2 treated group (p < 0.05).
Fig. 12.
Presence of FUBP1 in the 40/43S Ribosomal Fraction. Hela cells were treated with 100 μM H2O2 for 10 min and harvested 1 hour after for isolation of ribosomes and separation of ribosomal subunits using a fractionator as described in the Methods. The proteins collected in the fractions between 8 and 21 min were used for Western blot to detect indicated proteins.
Translation initiation requires the assembly of 43S pre-initiation complex that contains the mRNA strands, 40S ribosomal subunit and a set of translation initiation factors (eIFs). The eIF4E is known for recognizing 5’ methyl cap of the mRNA strands for translation initiation during general protein translation. The eIF2a binds to GTP/Met-tRNA for interaction with 40S ribosomal subunit. eIF3 binds to eIF1, eIF4G, eIF5 and 40S ribosomal subunit, and promotes the attachment of the ribosomal complex to the mRNA strands and subsequent codon scanning. The eIF1 facilitates eIF2a/GTP/Met-tRNA binding to 40S ribosomes, ensures the fidelity of initiation codon selection and promotes ribosomal scanning. The presence of these eIFs in the initiation complex has been verified as shown in 40/43S ribosomal complex (Fig. 12).
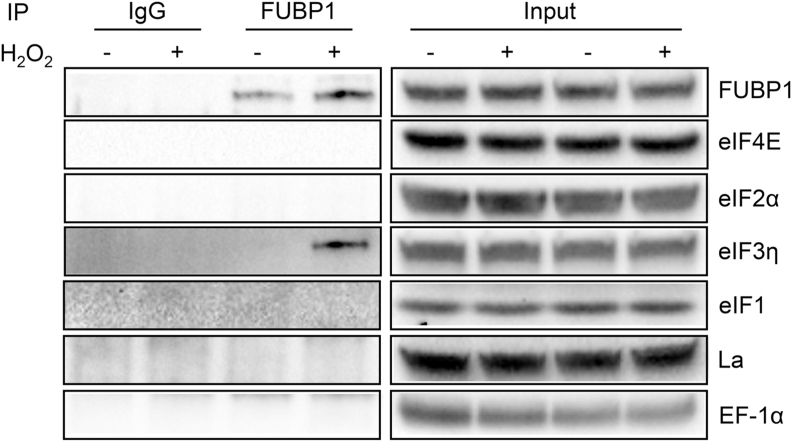
We reasoned that FUBP1 might bind to Nrf2 mRNA and interact with eIFs important for translation initiation. Given the availabilities of the antibodies from commercial sources, we examined the interaction of FUBP1 with eIFs using immunoprecipitation. We found that FUBP1 bound to eIF3η but not eIF4E, eIF2a or eIF1 when cells were treated with H2O2 (Fig. 13). This supports a role of FUBP1 in promoting 40S ribosomal attachment to Nrf2 mRNA and formation of 43S pre-initiation complex for translation initiation.
Fig. 13.
Immunoprecipitation of FUBP1 for Detection of eIFs. Hela cells were treated with 100 μM H2O2 for 10 minutes and lysed in RIPA buffer 1 hour after for immunoprecipitation. FUBP1 was captured and pulled down from cell lysate by anti-FUBP1 antibody and agarose-protein A/G beads. Mouse IgG was used as a control for immunoprecipitation. Protein aliquots from total cell lysates were used as input or loading control. Immunoprecipitated proteins and input were resolved by SDS-PAGE gel for detection of binding proteins as indicated.
In order to understand the mechanism underlying the gain of FUBP1 interaction with Nrf2 mRNA, eIF3η and ribosomes, we tested whether or not any posttranslational modifications occurred as a result of oxidative stress. With 2-D Western blot, we found that H2O2 treatment decreased the FUBP1 isoelectric point, indicating an increase of positive charges in these samples (data not shown). Antibodies against phospho Serine, Threonine or Tyrosine failed to detect any increase in FUBP1 due to H2O2 treatment (data not shown), consistent with the lack of negative charged posttranslational modification such as phosphorylation. How oxidative stress causes FUBP1 to increase its binding to Nrf2 mRNA and to trigger the onset of Nrf2 protein translation remains to be determined.
4. Discussion
In this study, we have found that FUBP1 protein increases its binding to Nrf2 mRNA and is required for de novo Nrf2 protein translation under oxidative stress. Mechanistically, FUBP1 did not show cytoplasmic translocation, but is present in 40/43S ribosomal fraction and shows a H2O2 dose dependent increase in total ribosomal fractions. Interestingly, FUBP1 interacts with eIF3η specifically, suggesting its role in attaching 43S pre-initiation complex to Nrf2 mRNA for translation initiation. Our data have revealed a novel mechanism of oxidative stress induced de novo Nrf2 protein translation.
FUBP1 protein was first discovered as a co-activator for maximizing c-myc transcription due to its binding to Far Upstream Element (FUSE), in an AT-rich region −1.5 kb upstream of the c-Myc promoter [24]. The core of FUSE in c-myc gene is 5′-TATATTCCCTCGGGATTTTTTATTTTGTG-3’ [24]. Through binding to FUSE, FUBP1 coordinates with additional cis-elements and their corresponding transcription factors to maximize the rate of transcription of the c-Myc gene. Aberrant expression of FUBP1 has been associated with various types of cancers, therefore emerging as a novel oncogene.
FUBP1 can bind to RNA at pyrimidine rich sequences and has been reported to bind several cellular mRNA species and viral RNA strands [24]. This protein contains four tandem K-homology (KH) motifs, similar to those in Heterogeneous Nuclear Ribonucleoprotein K (hnRNP-K) [25]. KH motifs mediate protein binding to single stranded DNA or RNA [26]. In fact, KH3 or KH4 of hnRNP is sufficient for binding to single stranded TTTT or ATTC sequence, respectively [27]. Nrf2 5′UTR contains one UUUU sequence and one AUUAC, and 7 patches of pyrimidine rich sequences, providing the potential binding sites for FUBP1. Importantly FUBP1 has been reported to play a role in IRES mediated translation of p27Kip mRNA [24]. Our data showing FUBP1 binding to Nrf2 mRNA are consistent with a role of FUBP1 in IRES mediated Nrf2 protein translation under oxidative stress.
FUBP1 is also known as an RNA splicing factor. The observed nuclear localization of FUBP1 supports its potential role as a splicing factor. FUBP1 promotes or facilitates splicing of Duchenne Muscular Dystrophy (DMD) gene and MDM2 oncogene [28,29]. Splicing regulatory elements are present in the exons and introns either as an enhancers or silencers. FUBP1 can bind to an exonic splicing silencer of cardiac triadin gene to repress its alternative splicing [30]. Recently, NCBI and Ensembl Genomic databases indicate that Nrf2 gene encodes 8 or 14 transcripts respectively, suggesting that Nrf2 gene undergoes alternative splicing. The interplay between alternative splicing and protein translation remains to be studied.
FUBP1 joins La/SSB and EF1a in the list of proteins found to increase binding to Nrf2 mRNA when cells are experiencing oxidative stress. We did not find FUBP1 interaction with La/SSB or EF1a, suggesting that each of these proteins bind to a distinct area of Nrf2 mRNA. Additionally, FUBP1 appears to respond to oxidative stress in a manner different from La/SSB or EF1a. While EF1a binds to the G-quadruplex in Nrf2 5′UTR [13], La/SSB translocates from the nuclei to the cytoplasm upon oxidative stress [12]. Cytoplasmic translocation of FUBP1 was reported in early phase of Japanese Encephalitis Virus (JEV) or Enterovirus 71 (EV71) infection [31,32]. Unlike these viral proteins, Nrf2 protein translation via FUBP1 participation does not appear to involve its nuclear to cytoplasmic translocation. The fact that FUBP1 was detected in the cytosol and increased its presence in the ribosomal fractions suggests that cytosolic redistribution of FUBP1 to be associated with ribosomes is important for de novo Nrf2 protein translation.
The interaction between FUBP1 and eIF3η was enhanced upon H2O2 treatment (Fig. 13). The initiation factor eIF3η is one of 13 subunits for 800 kD eIF3 complex that promotes assembly of the 43S pre-initiation complex and its association with the mRNA pre-occupied with eIF4F complex (eIF4E, eIF4G and eIF4A). With IRES mediated viral protein translation, involvement of eIF3 has been long established [33,34]. For cellular IRES, eIF3 has been shown to mediate 5′UTR dependent translation initiation of XIAP and c-Jun [[35], [36], [37]]. The physical interaction of FUBP1 with eIF3η discovered here supports a role of FUBP1 in promoting the attachment of 43S pre-initiation ribosomal complex to Nrf2 mRNA for translation initiation.
A plausible explanation for the observed increase of FUBP1 binding to Nrf2 mRNA is that FUBP1 undergoes posttranslational modifications upon oxidative stress. FUBP1 was reported to be ubiquitinated by p38/JTV-1 at the C-terminus, leading to its proteolysis [38]. Interestingly, FUBP1 can also be de-ubiquitinated by Ubiquitin-specific protease 22 (USP22), resulting in a reduction of FUBP1 binding to FUSE [39]. Posttranslational modifications, such as ubiquitination or sumoylation cause changes the molecular weights and/or charges of proteins, which can be detected by 2-dimensional electrophoresis. Lack of molecular weight change in 2-D Western blot argues against ubiquitination or sumoylation in FUBP1 due to H2O2 treatment. While 2-D electrophoresis showed a lowered PI of FUBP1 in cells treated with H2O2 treatment, antibodies detecting phosphorylation failed to indicate such posttranslational modification. Initial detection of FUBP1 using LC-MS/MS did not reveal phosphorylation or ubiquitination. Therefore, the signaling pathway leading to increased FUBP1 binding to Nrf2 mRNA remains to be determined.
Acknowledgement
The research programs under the direction of Dr. Qin M. Chen have been supported by NIH R01 HL089958, R01 GM111337, R01 GM125212, R01 GM126165, Holsclaw Endowment, and the University of Arizona College of Pharmacy start-up fund.
References
- 1.Chen Q.M., Maltagliati A.J. Nrf2 at the heart of oxidative stress and cardiac protection. Physiol. Genom. 2018;50(2):77–97. doi: 10.1152/physiolgenomics.00041.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cuadrado A., Rojo A.I., Wells G., Hayes J.D., Cousin S.P., Rumsey W.L., Attucks O.C., Franklin S., Levonen A.L., Kensler T.W., Dinkova-Kostova A.T. Therapeutic targeting of the NRF2 and KEAP1 partnership in chronic diseases. Nat. Rev. Drug Discov. 2019;18(4):295–317. doi: 10.1038/s41573-018-0008-x. [DOI] [PubMed] [Google Scholar]
- 3.Thimmulappa R.K., Mai K.H., Srisuma S., Kensler T.W., Yamamoto M., Biswal S. Identification of Nrf2-regulated genes induced by the chemopreventive agent sulforaphane by oligonucleotide microarray. Canc. Res. 2002;62(18):5196–5203. [PubMed] [Google Scholar]
- 4.Lee J.M., Calkins M.J., Chan K., Kan Y.W., Johnson J.A. Identification of the NF-E2-related factor-2-dependent genes conferring protection against oxidative stress in primary cortical astrocytes using oligonucleotide microarray analysis. J. Biol. Chem. 2003;278(14):12029–12038. doi: 10.1074/jbc.M211558200. [DOI] [PubMed] [Google Scholar]
- 5.Malhotra D., Portales-Casamar E., Singh A., Srivastava S., Arenillas D., Happel C., Shyr C., Wakabayashi N., Kensler T.W., Wasserman W.W., Biswal S. Global mapping of binding sites for Nrf2 identifies novel targets in cell survival response through ChIP-Seq profiling and network analysis. Nucleic Acids Res. 2010;38(17):5718–5734. doi: 10.1093/nar/gkq212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mitsuishi Y., Motohashi H., Yamamoto M. The Keap1-Nrf2 system in cancers: stress response and anabolic metabolism. Front. Oncol. 2012;2:200. doi: 10.3389/fonc.2012.00200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sayin V.I., LeBoeuf S.E., Singh S.X., Davidson S.M., Biancur D., Guzelhan B.S., Alvarez S.W., Wu W.L., Karakousi T.R., Zavitsanou A.M., Ubriaco J., Muir A., Karagiannis D., Morris P.J., Thomas C.J., Possemato R., Vander Heiden M.G., Papagiannakopoulos T. Activation of the NRF2 antioxidant program generates an imbalance in central carbon metabolism in cancer. elife. 2017;6 doi: 10.7554/eLife.28083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee S.B., Sellers B.N., DeNicola G.M. The regulation of NRF2 by nutrient-responsive signaling and its role in anabolic cancer metabolism. Antioxidants Redox Signal. 2018;29(17):1774–1791. doi: 10.1089/ars.2017.7356. [DOI] [PubMed] [Google Scholar]
- 9.Kerins M.J., Liu P., Tian W., Mannheim W., Zhang D.D., Ooi A. Genome-wide CRISPR screen reveals autophagy disruption as the convergence mechanism that regulates the NRF2 transcription factor. Mol. Cell Biol. 2019;39(13) doi: 10.1128/MCB.00037-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Purdom-Dickinson S.E., Sheveleva E.V., Sun H., Chen Q.M. Translational control of Nrf2 protein in activation of antioxidant response element by oxidants. Mol. Pharm. 2007;72:1074–1081. doi: 10.1124/mol.107.035360. [DOI] [PubMed] [Google Scholar]
- 11.Xu B., Zhang J., Strom J., Lee S., Chen Q.M. Myocardial ischemic reperfusion induces de novo Nrf2 protein translation. Biochim. Biophys. Acta. 2014;1842(9):1638–1647. doi: 10.1016/j.bbadis.2014.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang J., Dinh T.N., Kappeler K., Tsaprailis G., Chen Q.M. La autoantigen mediates oxidant induced de novo Nrf2 protein translation. Mol. Cell. Proteomics. 2012;11(6) doi: 10.1074/mcp.M111.015032. M111 015032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee S.C., Zhang J., Strom J., Yang D., Dinh T.N., Kappeler K., Chen Q.M. G-quadruplex in the NRF2 mRNA 5' untranslated region regulates de novo NRF2 protein translation under oxidative stress. Mol. Cell Biol. 2017;37(1) doi: 10.1128/MCB.00122-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee K.M., Chen C.J., Shih S.R. Regulation mechanisms of viral IRES-driven translation. Trends Microbiol. 2017;25(7):546–561. doi: 10.1016/j.tim.2017.01.010. [DOI] [PubMed] [Google Scholar]
- 15.Walsh D., Mathews M.B., Mohr I. Tinkering with translation: protein synthesis in virus-infected cells. Cold Spring Harb. Perspect. Biol. 2013;5(1):a012351. doi: 10.1101/cshperspect.a012351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fraser C.S., Doudna J.A. Structural and mechanistic insights into hepatitis C viral translation initiation. Nat. Rev. Microbiol. 2007;5(1):29–38. doi: 10.1038/nrmicro1558. [DOI] [PubMed] [Google Scholar]
- 17.Wu C.A., Huang D.Y., Lin W.W. Beclin-1-independent autophagy positively regulates internal ribosomal entry site-dependent translation of hypoxia-inducible factor 1alpha under nutrient deprivation. Oncotarget. 2014;5(17):7525–7539. doi: 10.18632/oncotarget.2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gilbert W.V., Zhou K., Butler T.K., Doudna J.A. Cap-independent translation is required for starvation-induced differentiation in yeast. Science. 2007;317(5842):1224–1227. doi: 10.1126/science.1144467. [DOI] [PubMed] [Google Scholar]
- 19.Garcia-Jimenez C., Goding C.R. Starvation and pseudo-starvation as drivers of cancer metastasis through translation reprogramming. Cell Metabol. 2019;29(2):254–267. doi: 10.1016/j.cmet.2018.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yamasaki S., Anderson P. Reprogramming mRNA translation during stress. Curr. Opin. Cell Biol. 2008;20(2):222–226. doi: 10.1016/j.ceb.2008.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Spriggs K.A., Stoneley M., Bushell M., Willis A.E. Re-programming of translation following cell stress allows IRES-mediated translation to predominate. Biol. Cell. 2008;100(1):27–38. doi: 10.1042/BC20070098. [DOI] [PubMed] [Google Scholar]
- 22.Cok S.J., Acton S.J., Sexton A.E., Morrison A.R. Identification of RNA-binding proteins in RAW 264.7 cells that recognize a lipopolysaccharide-responsive element in the 3-untranslated region of the murine cyclooxygenase-2 mRNA. J. Biol. Chem. 2004;279(9):8196–8205. doi: 10.1074/jbc.M308475200. [DOI] [PubMed] [Google Scholar]
- 23.Olanich M.E., Moss B.L., Piwnica-Worms D., Townsend R.R., Weber J.D. Identification of FUSE-binding protein 1 as a regulatory mRNA-binding protein that represses nucleophosmin translation. Oncogene. 2011;30(1):77–86. doi: 10.1038/onc.2010.404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang J., Chen Q.M. Far upstream element binding protein 1: a commander of transcription, translation and beyond. Oncogene. 2013;32(24):2907–2916. doi: 10.1038/onc.2012.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Braddock D.T., Louis J.M., Baber J.L., Levens D., Clore G.M. Structure and dynamics of KH domains from FBP bound to single-stranded DNA. Nature. 2002;415(6875):1051–1056. doi: 10.1038/4151051a. [DOI] [PubMed] [Google Scholar]
- 26.Valverde R., Edwards L., Regan L. Structure and function of KH domains. FEBS J. 2008;275(11):2712–2726. doi: 10.1111/j.1742-4658.2008.06411.x. [DOI] [PubMed] [Google Scholar]
- 27.Braddock D.T., Baber J.L., Levens D., Clore G.M. Molecular basis of sequence-specific single-stranded DNA recognition by KH domains: solution structure of a complex between hnRNP K KH3 and single-stranded DNA. EMBO J. 2002;21(13):3476–3485. doi: 10.1093/emboj/cdf352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miro J., Laaref A.M., Rofidal V., Lagrafeuille R., Hem S., Thorel D., Mechin D., Mamchaoui K., Mouly V., Claustres M., Tuffery-Giraud S. FUBP1: a new protagonist in splicing regulation of the DMD gene. Nucleic Acids Res. 2015;43(4):2378–2389. doi: 10.1093/nar/gkv086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jacob A.G., Singh R.K., Mohammad F., Bebee T.W., Chandler D.S. The splicing factor FUBP1 is required for the efficient splicing of oncogene MDM2 pre-mRNA. J. Biol. Chem. 2014;289(25):17350–17364. doi: 10.1074/jbc.M114.554717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li H., Wang Z., Zhou X., Cheng Y., Xie Z., Manley J.L., Feng Y. Far upstream element-binding protein 1 and RNA secondary structure both mediate second-step splicing repression. Proc. Natl. Acad. Sci. U.S.A. 2013;110(29):E2687–E2695. doi: 10.1073/pnas.1310607110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chien H.L., Liao C.L., Lin Y.L. FUSE binding protein 1 interacts with untranslated regions of Japanese encephalitis virus RNA and negatively regulates viral replication. J. Virol. 2011;85(10):4698–4706. doi: 10.1128/JVI.01950-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang P.N., Lin J.Y., Locker N., Kung Y.A., Hung C.T., Huang H.I., Li M.L., Shih S.R. Far upstream element binding protein 1 binds the internal ribosomal entry site of enterovirus 71 and enhances viral translation and viral growth. Nucleic Acids Res. 2011;39(22):16. doi: 10.1093/nar/gkr682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Khan D., Bhat P., Das S. HCV-like IRESs sequester eIF3: advantage virus. Trends Microbiol. 2014;22(2):57–58. doi: 10.1016/j.tim.2013.12.009. [DOI] [PubMed] [Google Scholar]
- 34.Aitken C.E. Long-lost cousins? eIF3 recognition of the HCV IRES and cellular mRNAs. J. Mol. Biol. 2020;432(7):1856–1860. doi: 10.1016/j.jmb.2020.03.006. [DOI] [PubMed] [Google Scholar]
- 35.Thakor N., Smith M.D., Roberts L., Faye M.D., Patel H., Wieden H.J., Cate J.H.D., Holcik M. Cellular mRNA recruits the ribosome via eIF3-PABP bridge to initiate internal translation. RNA Biol. 2017;14(5):553–567. doi: 10.1080/15476286.2015.1137419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee A.S., Kranzusch P.J., Cate J.H. eIF3 targets cell-proliferation messenger RNAs for translational activation or repression. Nature. 2015;522(7554):111–114. doi: 10.1038/nature14267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Walker M.J., Shortridge M.D., Albin D.D., Cominsky L.Y., Varani G. Structure of the RNA specialized translation initiation element that recruits eIF3 to the 5'-UTR of c-jun. J. Mol. Biol. 2020;432(7):1841–1855. doi: 10.1016/j.jmb.2020.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim M.J., Park B.J., Kang Y.S., Kim H.J., Park J.H., Kang J.W., Lee S.W., Han J.M., Lee H.W., Kim S. Downregulation of FUSE-binding protein and c-myc by tRNA synthetase cofactor p38 is required for lung cell differentiation. Nat. Genet. 2003;34(3):330–336. doi: 10.1038/ng1182. [DOI] [PubMed] [Google Scholar]
- 39.Atanassov B.S., Dent S.Y. USP22 regulates cell proliferation by deubiquitinating the transcriptional regulator FBP1. EMBO Rep. 2011;12(9):924–930. doi: 10.1038/embor.2011.140. [DOI] [PMC free article] [PubMed] [Google Scholar]