Abstract
POLD1, the catalytic subunit of DNA polymerase δ, plays a critical role in DNA synthesis and DNA repair processes. Moreover, POLD1 is downregulated in replicative senescence to mediate aging. In any case, the components of age-related downregulation of POLD1 expression have not been fully explained. In this article, we elucidate the mechanism of the regulation of POLD1 at the transcription level and found that the transcription factor CCCTC-binding factor (CTCF) was bound to the POLD1 promoter area in two sites. The binding level of CTCF for the POLD1 promoter appeared to be related to aging and was confirmed to be positively controlled by the CTCF level. Additionally, cell senescence characteristics were detected within the cells transfected with short hairpin RNA (shRNA)-CTCF, pLenti-CMV-CTCF, shRNA-POLD1, and pLenti-CMV-POLD1, and the results showed that the CTCF may contribute to the altered expression of POLD1 in aging. In conclusion, the binding level of CTCF for the POLD1 promoter intervened by an age-related decrease in CTCF and downregulated the POLD1 expression in aging. Moreover, the decrease in CTCF-mediated POLD1 transcription accelerates the progression of cell aging.
Keywords: POLD1, transcription factor, CTCF, aging, transcriptional regulation
Introduction
Aging is the major risk factor for most chronic diseases that account for the bulk of morbidity, mortality, and health costs in developed and developing countries (Harper, 2014; Childs et al., 2015; Kirkland and Tchkonia, 2017). There are various theories on the occurrence of aging (Da et al., 2016), and one of the theories is genomic instability and decreased proliferation ability, which is considered a major driving force of aging (Sun et al., 2018; Hou et al., 2019). POLD1, as the catalytic subunit of DNA polymerase delta and endowed with polymerase and exonuclease activities, plays a critical role in DNA synthesis and DNA repair processes (Cui et al., 2019). Our previous study found that POLD1 expression was downregulated as cell aging in human lymphocytes and preliminarily confirmed that the expression of POLD1 was essential in cell cycle regulation and DNA damage repair processes (Wang et al., 2012; Song et al., 2015; Maierhofer et al., 2019). However, the mechanism of age-related downregulation of POLD1 expression has not been fully explained.
Clearly, transcriptional regulation is the main regulatory mechanism of gene expression (Tepper et al., 2013), and several defined transcription factors (TFs) can bind to the POLD1 promoter to regulate cell cycle and aging (Li and Lee, 2001; Song et al., 2009). Identifying the POLD1 associated with other TFs is important to understand the mechanism of the regulation of POLD1 in aging. The CCCTC-binding factor (CTCF) TF, which can bind to DNA sequences or proteins through its special 11 zinc finger structure that forms CTCF–DNA and CTCF–protein complexes to regulate gene expression, plays an essential role in cell growth and differentiation (Chang et al., 2010; Song and Kim, 2017; Peng et al., 2019). Besides, many studies have shown that CTCF can bind to other key aging genes to regulate cell aging progress (Wendt et al., 2008). Therefore, the correlation between CTCF and POLD1 is worth exploring.
Previous studies have indicated that POLD1 and CTCF are closely related to cell growth and differentiation. In this study, CTCF was found to be one of the key TFs of POLD1, and it was hypothesized that the attenuation of the binding level of CTCF to the POLD1 promoter, mediated by an age-related decline in CTCF, downregulated POLD1 expression and accelerated replicative senescence.
Results
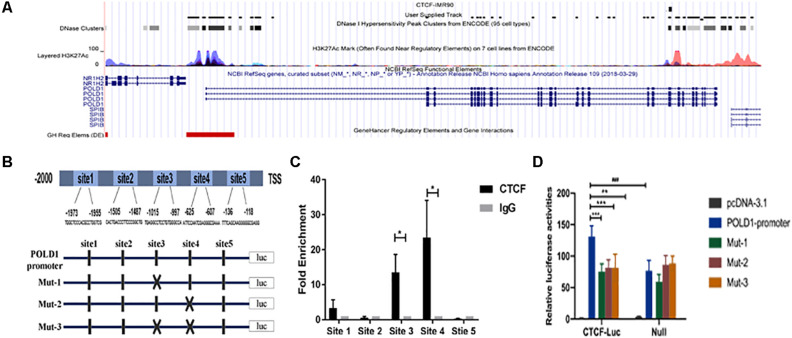
CTCF Mainly Binds to the POLD1 Promoter in Two Sites
We analyzed the Encyclopedia of DNA Elements (ENCODE) database and found several transcription factor binding sites (TFBSs) of CTCF in the POLD1 promoter (Figure 1A). Potential specific CTCF-binding sites were analyzed with the assistance of the JASPAR database, and the results showed five potential CTCF-binding sites in the promoter of POLD1 with a higher score (Figure 1B). To directly determine which site CTCF binds to the POLD1 promoter, chromatin immunoprecipitation (ChIP)-qPCR assays were performed in lymphocyte cells. The ChIP results demonstrated that CTCF binds mainly to the POLD1 promoter in site 3 (−1015 to −997) and site 4 (−625 to −607) regions (Figure 1C). Evidence for transcriptional activation in a dual-luciferase reporter system showed that the region from the POLD1 promoter containing all sites displayed increased reporter activity relative to the control vector. Interestingly, mutation of site 3 or site 4 greatly reduced promoter activity, demonstrating that these are functional CTCF-binding sites in the human POLD1 promoter (Figure 1D).
FIGURE 1.
CCCTC-binding factor (CTCF) mainly binds to the DNA polymerase delta 1, catalytic subunit (POLD1) promoter in two sites. (A) ENCODE database analysis showing potential CTCF-binding sites in the POLD1 promoter. (B) The JASPAR database predicted five higher score binding sites of CTCF to the POLD1 promoter, and the mutations of the two CTCF-binding sites were designed. (C) The binding sites of CTCF to the POLD1 promoter were analyzed by chromatin immunoprecipitation (ChIP) assay in lymphocyte cells (n = 40). *P < 0.05 vs. IgG-negative control group. (D) Luciferase activity assay demonstrated that the CTCF led to an increase in POLD1 promoter activity and that mutation of site 3 or 4 greatly reduced promoter activity in HEK293T cells (n = 3). **P < 0.01, ***P < 0.005 vs. POLD1 promoter wild type (WT) + CTCF. ###P < 0.001 vs. POLD1 promoter WT. Data were analyzed using Student’s t-test, and data are shown as mean ± SEM, with three independent experiments in each group.
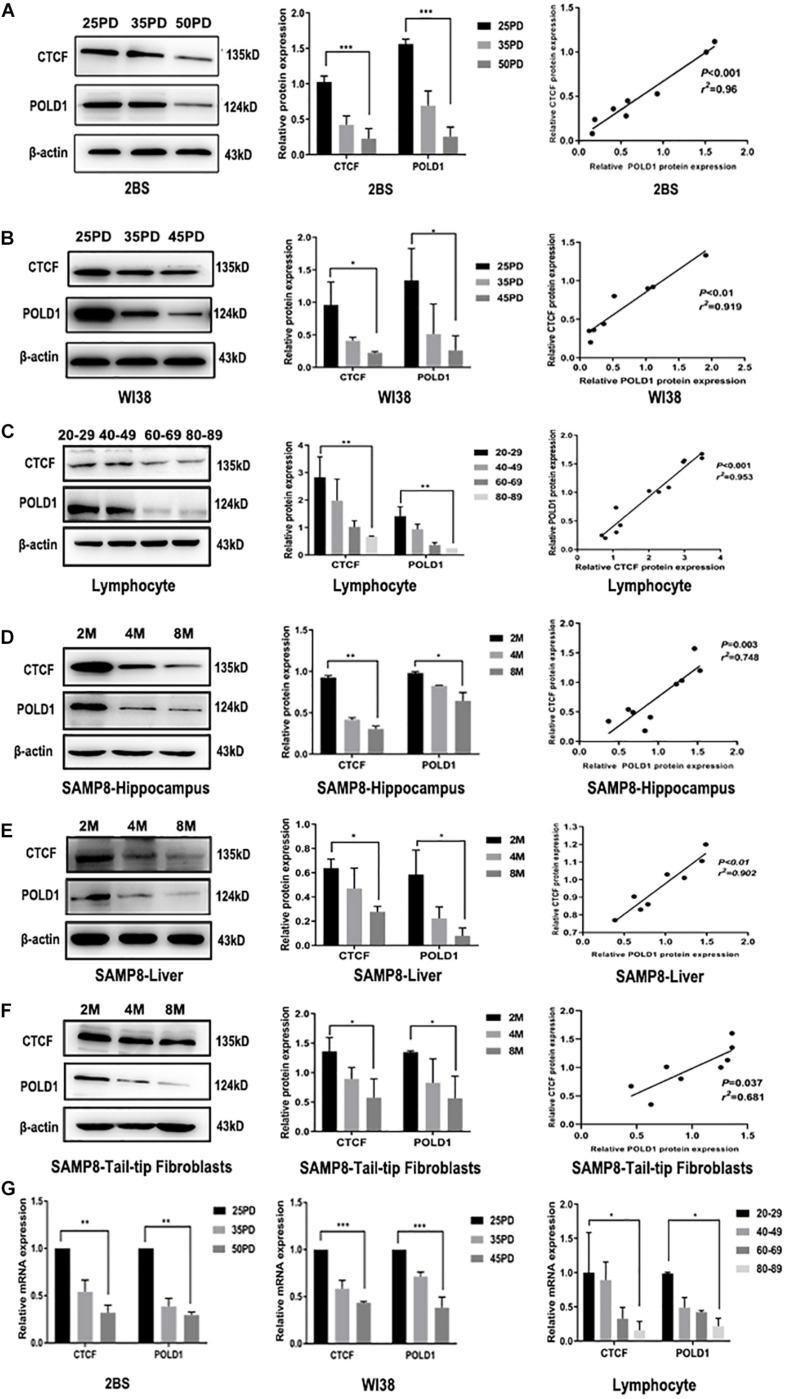
Both CTCF and POLD1 Expression Levels Were Reduced With Aging
The expression levels of POLD1 and CTCF were detected in different population doublings (PDs) of human embryonic lung diploid fibroblasts (2BS) (25, 35, and 50 PDs) and human fetal lung fibroblast (WI38) (25, 35, and 45 PDs), lymphocytes of healthy people of different ages (20–29, 40–49, 60–69, and 80–89 years), and senescence-accelerated mouse prone (SAMP8) mice (Chen et al., 2019) at different ages in months (2, 4, and 8 months). The results showed that the protein expression levels of both CTCF and POLD1 were reduced consistently with aging, and there was a positive correlation between CTCF and POLD1 expression (Figures 2A–F). A similar result was found at the mRNA level (Figure 2G). These results indicate that reduced CTCF level could be responsible for the downregulation of POLD1.
FIGURE 2.
Both CCCTC-binding factor (CTCF) and DNA polymerase delta 1, catalytic subunit (POLD1) expression levels were reduced with aging. (A) Protein expression levels and relationship of CTCF and POLD1 in different population doublings (PDs) of 2BS cells (n = 3). (B) Protein expression levels and relationship of CTCF and POLD1 in different PDs of WI38 cells (n = 3). (C) Protein expression levels of CTCF and POLD1 in the lymphocytes of healthy people of different ages (n = 20 in every group). (D) Protein expression levels of CTCF and POLD1 in the hippocampus of SAMP8 mice at different ages in months (n = 18). (E) Protein expression levels of CTCF and POLD1 in livers of SAMP8 mice at different ages in months (n = 18). (F) Protein expression levels of CTCF and POLD1 in the 3PD mouse tail-tip fibroblasts of SAMP8 mice at different age in months (n = 18). (G) mRNA expression levels of CTCF and POLD1 in different PDs of 2BS, WI38 cells, and lymphocytes of healthy people of different ages. β-actin was used for the normalization of data. Data were analyzed using one-way ANOVA, and data are shown as mean ± SEM, with three independent experiments in each group. *P < 0.05, **P < 0.01, ***P < 0.005. The correlation was determined using Pearson’s correlation coefficient.
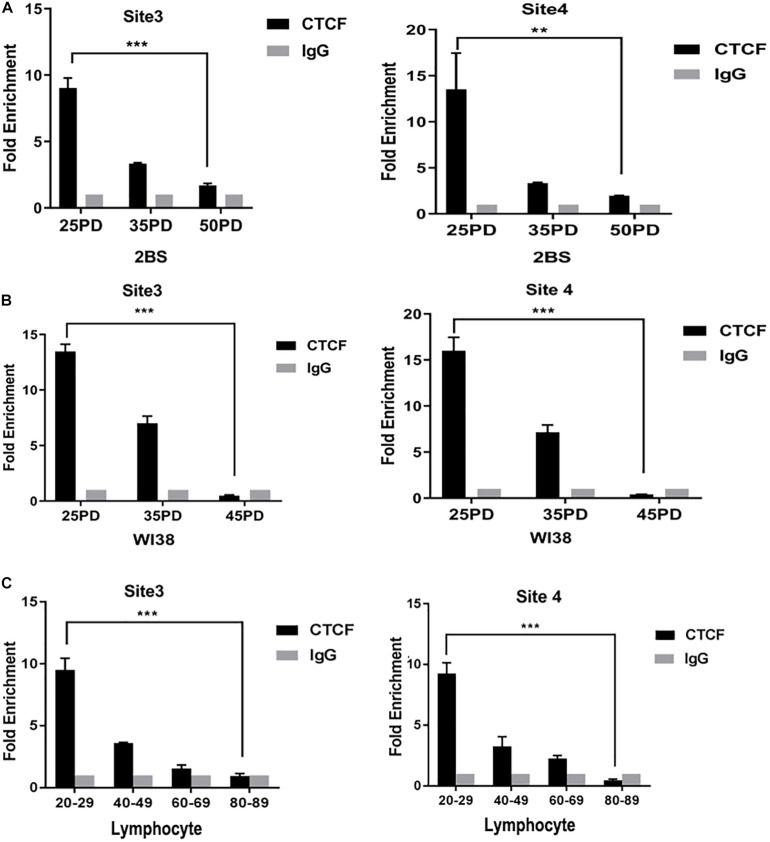
Binding Level of CTCF in the POLD1 Promoter Decreased With Aging
To further explore the change in the binding level of CTCF to POLD1 promoter, the binding level of CTCF in POLD1 promoter was detected in different PDs of 2BS and WI38 cells and in the lymphocytes of healthy people of different ages. The results showed that the binding levels of CTCF to the POLD1 promoter at sites 3 and 4 were both markedly decreased with aging in 2BS cell, WI38 cell, and lymphocytes (Figures 3A–C). These results suggested that the reduced level of CTCF could be related to the decreased binding level of CTCF to the POLD1 promoter to downregulate the expression of POLD1.
FIGURE 3.
The binding level of CCCTC-binding factor (CTCF) in the DNA polymerase delta 1, catalytic subunit (POLD1) promoter decreased with aging. (A) Binding level of CTCF at the POLD1 promoter in 2BS cells from different population doublings (PDs). (B) Binding level of CTCF at the POLD1 promoter in WI38 cells from different PDs. (C) Binding level of CTCF at the POLD1 promoter in lymphocytes from healthy people of different ages. Densitometric analysis of the band intensity in different groups, normalized to the input. Data were compared using one-way ANOVA, and values represent the mean ± SEM with three independent experiments in each group. **P < 0.01, ***P < 0.005.
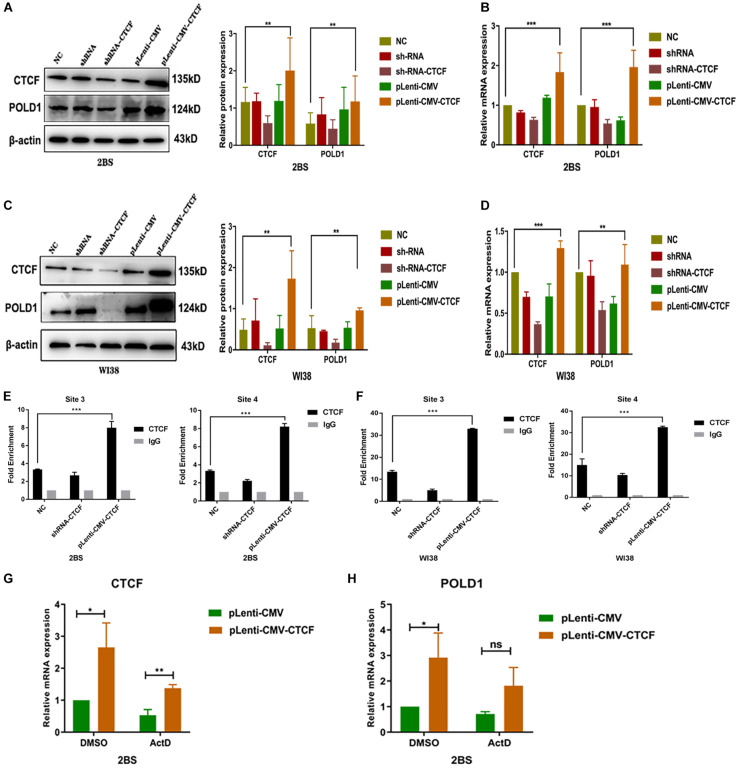
CTCF Upregulated POLD1 Expression by Elevating the Binding Level of CTCF to the POLD1 Promoter
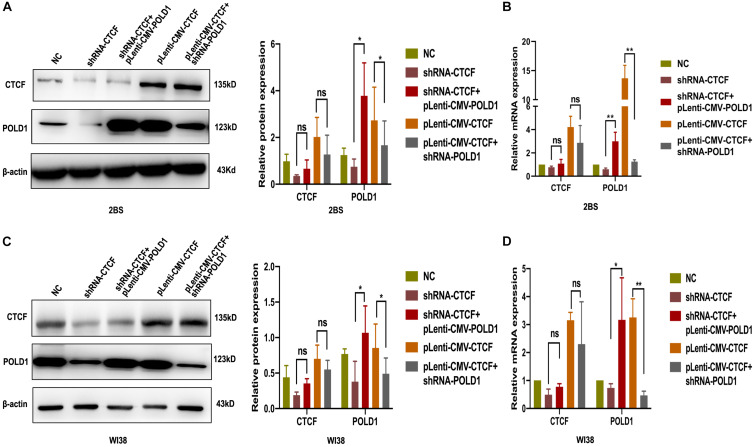
To illustrate the biological function of CTCF in POLD1 gene regulation, RNA interference (RNAi) and overexpression methods were used to regulate CTCF expression in 30 PD 2BS and 28 PD WI38 cells. The protein and mRNA expression levels of POLD1 and CTCF were determined by Western blot and quantitative reverse transcription-polymerase chain reaction (RT-qPCR), respectively. The results showed that the expression of CTCF and POLD1 was dramatically downregulated after transfection with short hairpin RNA (shRNA)-CTCF and upregulated after transfection with the CTCF eukaryotic expression lentivirus vector (Figures 4A–D). Furthermore, the binding level of CTCF to the POLD1 promoter at sites 3 and 4 in cells transfected with shRNA-CTCF was slightly lower than that of the control cells, and the binding level was higher in cells transfected with pLenti-CMV-CTCF than in the control cells (Figures 4E,F).
FIGURE 4.
CCCTC-binding factor (CTCF) upregulated DNA polymerase delta 1, catalytic subunit (POLD1) expression by elevating the binding level of CTCF to the POLD1 promoter. (A,C) Protein expression levels of CTCF and POLD1 in 2BS (A) and WI38 cells (C). (B,D) mRNA expression levels of CTCF and POLD1 in 2BS (B) and WI38 cells (D). (E,F) Site 3 and 4 binding levels of CTCF at the POLD1 promoter in 2BS (E) and WI38 cells (F) transfected with short hairpin RNA (shRNA) and CTCF overexpression lentivirus. (G,H) CTCF (G) and POLD1 (H) mRNA levels in 2BS cells transfected with pLenti-CMV-CTCF lentivirus. Data were compared by one-way ANOVA and Student’s t-test, and data are shown as the mean ± SEM, with three independent experiments in each group. *P < 0.05, **P < 0.01, ***P < 0.005.
These results indicate that CTCF regulates POLD1 gene expression through transcription. To further verify this speculation, actinomycin D, a transcriptional inhibitor, was used. The results showed that the POLD1 upregulation induced by CTCF overexpression was blocked by actinomycin D (10 μg/ml) (Figures 4G,H).
POLD1 Could Rescue the Effect of CTCF on POLD1 Expression
Given that CTCF could affect the expression of POLD1, we tested whether overexpression or knockdown of POLD1 could interfere with the effect of CTCF on the POLD1 gene. shRNA-CTCF and pLenti-CMV-POLD1 were cotransfected into 2BS and WI38 cells, and pLenti-CMV-CTCF and shRNA-POLD1 were cotransfected into 2BS and WI38 cells. The protein expression and mRNA levels of CTCF and POLD1 were confirmed by Western blot and RT-qPCR, respectively. Upregulated expression of POLD1 was found in cells with shRNA-CTCF + pLenti-CMV-POLD1 than with shRNA-CTCF, and decreased POLD1 expression was observed in cells transfected with pLenti-CMV-CTCF + shRNA-POLD1 than with pLenti-CMV-CTCF (Figures 5A–D). These results showed that the POLD1 gene could rescue the effect of CTCF on the expression of POLD1.
FIGURE 5.
DNA polymerase delta 1, catalytic subunit (POLD1) rescued the effect of CCCTC-binding factor (CTCF) on POLD1 expression. (A,C) Protein expression of POLD1 and CTCF in 2BS (A) and WI38 cells (C). (B,D) mRNA expression of POLD1 and CTCF in 2BS (B) and WI38 (D) cells. Data were compared by Student’s t-test, and data are shown as the mean ± SEM, with three independent experiments in each group. *P < 0.05, **P < 0.01.
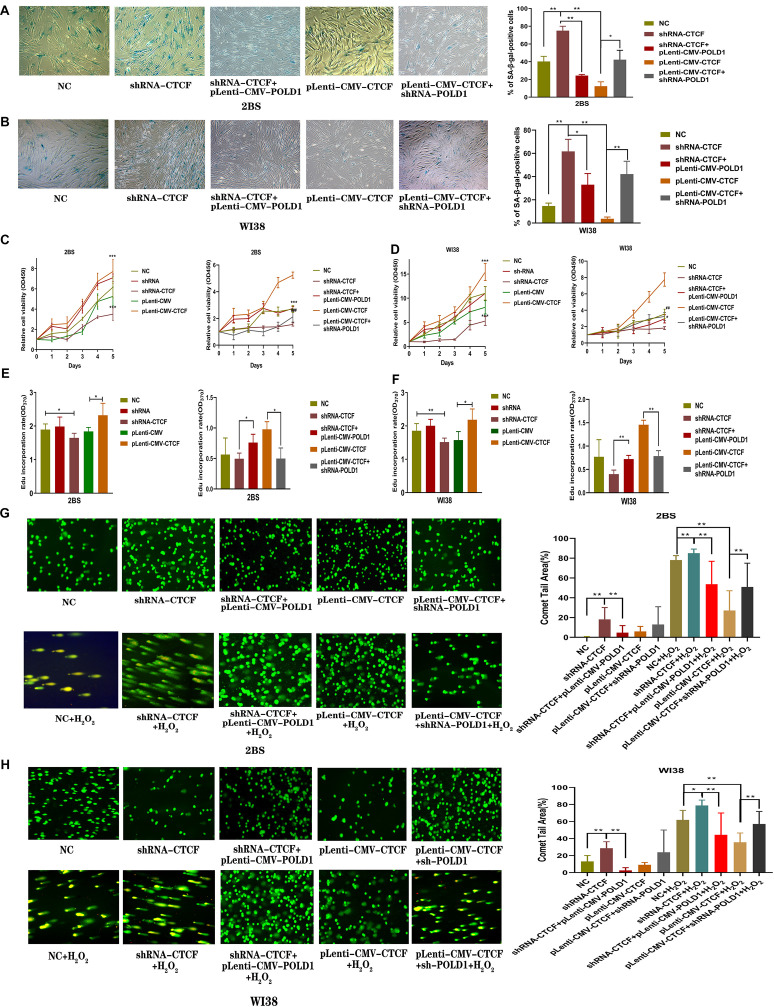
CTCF Was Involved in the Progression of Cellular Senescence, and POLD1 Could Rescue the Effect of Downregulated CTCF on Aging
To demonstrate the potential role of CTCF in cell senescence, the levels of senescence-associated β-galactosidase (SA-β-gal) staining, cell proliferation, DNA synthesis, and DNA damage were tested in 2BS and WI38 cells transfected with shRNA-CTCF, shRNA-CTCF + pLenti-CMV-POLD, pLenti-CMV-CTCF, or pLenti-CMV-CTCF + shRNA-POLD1. The results showed that the percentage of SA-β-gal-positive cells in 2BS and WI38 cells with CTCF knockdown was significantly increased relative to those of other groups, while the overexpression of POLD1 significantly attenuated the effects of shRNA-CTCF on aging. Accordingly, the percentage of SA-β-gal-positive cells in CTCF-overexpressing 2BS and WI38 cells was significantly decreased relative to that in other groups, and the percentage of positive cells was upregulated with the knockdown of POLD1 (Figures 6A,B).
FIGURE 6.
CCCTC-binding factor (CTCF) was involved in the progression of cellular senescence, and DNA polymerase delta 1, catalytic subunit (POLD1) rescued the effect of the downregulated CTCF on aging. (A,B) Senescence-associated β-galactosidase (SA-β-gal) staining of 2BS (A) and WI38 cells (B) transfected with the indicated lentivirus. The percentage of SA-β-gal-positive cells counted in each condition was shown. The experiments shown are representative of three biological replicates. Data were compared by Student’s t-test; *P < 0.05, **P < 0.01. (C,D) Proliferation of 2BS and WI38 cells transfected with the indicated lentivirus measured by Cell Counting Kit-8 (CCK-8) assay. The absorbance at 450 nm was measured at 0–5 days. Data are shown as the mean ± SEM of the ratio for the absorbance. Data were compared by Student’s t-test, and data are shown as the mean ± SEM, with three independent experiments in each group; ***P < 0.005 vs. the NC group; ##P < 0.01, ###P < 0.005, Lenti-CMV-CTCF vs. pLenti-CMV-CTCF + shRNA-POLD1. (E,F) An 5-ethynyl-2′-deoxyuridine (EdU) incorporation assay was used to detect the rate of DNA synthesis. EdU-positive cells were quantified after 72 h of treatment by the ratio of the absorbance at 370 nm. Data were compared by one-way ANOVA and Student’s t-test, and data are shown as the mean ± SEM, with three independent experiments in each group; *P < 0.05, **P < 0.01. (G,H) Effects of altered CTCF expression on DNA damage in 2BS and WI38 cells transfected with the indicated lentivirus (magnification, × 100). The percentage of the tail area determined by CAPS software (n = 50). Data were compared by one-way ANOVA and Student’s t-test, and data are shown as the mean ± SEM. *P < 0.05, **P < 0.01.
The results of the Cell Counting Kit-8 (CCK8) assay showed that the proliferation of 2BS and WI38 cells transfected with shRNA-CTCF decreased significantly compared with that of the other groups of cells, and the proliferative capacity was restored with POLD1 overexpression. Cells transfected with pLenti-CMV-CTCF showed a significantly increased proliferative capacity compared with other groups of cells, and a decrease in proliferative capacity was observed in cells with POLD1 downregulation (Figures 6C,D).
The 5-ethynyl-2′-deoxyuridine (EdU) incorporation rate was quantified. The rate of EdU incorporation was lower in cells with CTCF knockdown than that in other groups of cells, and the rate was raised in cells with coinfected pLenti-CMV-POLD1. The rate of EdU incorporation was higher in cells overexpressing CTCF than in other groups of cells, and the rate decreased in cells coinfected with shRNA-POLD1 (Figures 6E,F). These results were consistent with the outcome of the CCK-8 assay, which indicated that CTCF promoted cell growth and proliferation through the POLD1 gene. DNA lesions were assessed using a single-cell gel electrophoretic assay (comet assay) under alkaline conditions. H2O2 can damage DNA (Zivkovic et al., 2019). The percentage of the comet tail area represents the level of DNA breakage. DNA damage levels were higher in the normal control, shRNA-CTCF, shRNA-CTCF + pLenti-CMV-POLD1, pLenti-CMV-CTCF, and pLenti-CMV-CTCF + shRNA-POLD1 cells when 2BS and WI38 cells were treated with H2O2. Notably, the DNA damage level in 2BS and WI38 cells transfected with shRNA-CTCF increased significantly, and this effect was reversed by POLD1 overexpression. We also found that CTCF overexpression in 2BS and WI38 cells significantly decreased the DNA damage level when cells were treated with H2O2, while the DNA damage level increased with the knockdown of POLD1 (Figures 6G,H). These results suggest that CTCF elevates DNA damage repairability, which could be attributed to POLD1 upregulation.
Discussion
Aging refers to the phenomenon that the body’s physiological and psychological adaptability to the environment gradually decreases and tends to die, which is characterized by a series of specific changes in cellular morphology and senescence-associated genes (Pazolli and Stewart, 2008). Aging in cells is defined as a loss of replicative capacity, an arrest of the cell cycle, a shortened telomeric length, evident alterations of aging-related genes, and the appearance of aging phenotypes (McHugh and Gil, 2018). A decline in DNA synthesis and an increase in DNA breakage represent the most damaging DNA injuries that can compromise genomic integrity and viability, which are the main features of senescent cells (Gea et al., 2020).
POLD1, which is a central mediator of DNA replication and repair, plays an essential role in senescence to regulate the cell cycle and DNA damage repair. Some studies have shown that POLD1 gene mutation and expression were associated with the pathogenesis of Werner syndrome and MDPL (mandibular hypoplasia, deafness, progeroid features, and lipodystrophy) syndrome (Reinier et al., 2015; Fiorillo et al., 2018). CTCF is a TF that functions in many nuclear processes, including genomic organization, transcriptional regulation, insulator activity, and homologous recombination (HR)-mediated repair (Shan et al., 2019). CTCF mutations in humans are linked to microcephaly and intellectual disability (Gregor et al., 2013). Importantly, some findings identify the critical role of CTCF in selecting the homologous recombination repair pathway and in maintaining proper telomere replication and chromosome stability to participate in the progression of aging (Beishline et al., 2017; Hwang et al., 2019). In addition, CTCF could collaborate with Cockayne syndrome group B protein (CSB) to protect cells from oxidative stress and therefore delay aging (Lake et al., 2016). CTCF could also bind with the human longevity gene forkhead box O3 (FOXO3) and bind close to the peak of age-associated DNA changes and thereby participate in aging progression (Badarinarayan et al., 2017; Flachsbart et al., 2017; Han et al., 2020). In the present study, we successfully showed that CTCF, when binding to the POLD1 promoter, regulates the aging process.
First, we used the ENCODE database to predict whether CTCF binds to POLD1 in IMR90 cells. As shown in Figure 1, the more activated chromatin open region labeled with H3K27Ac, the denser the binding site of CTCF on the POLD1 promoter, which is consistent with the traditional view that TFs usually bind to DNA motifs in open chromatin regions (Zhu et al., 2016). The JASPAR database was also used to predict specific binding sites of CTCF in the POLD1 promoter, and five higher score sites were chosen to be verified by ChIP-qPCR. The results showed that CTCF bound mainly in the regions from −1,015 to −997 (site 3) and −625 to −607 (site 4) in the POLD1 promoter (Figure 1C). Furthermore, the activity of the POLD1 promoter was enhanced with the transfection of CTCF, and sites 3 and 4 played critical roles in the regulation of POLD1 by CTCF. These results indicated that CTCF could play a positive role in the transcriptional regulation of POLD1.
The age-related decrease in CTCF and POLD1 expression, as well as in the positive relationship between CTCF and POLD1 expression levels, were also observed in 2BS and WI38 cells, human lymphocytes, and SAMP8 mice (Figure 2). Moreover, the binding level of CTCF to the POLD1 promoter was verified to be attenuated with aging in 2BS cells, WI38 cells, and human lymphocytes (Figure 3). The results above suggested that the decrease in CTCF expression could be responsible for POLD1 downregulation by attenuating the binding level in senescence.
To test this hypothesis, CTCF expression was knocked down or upregulated in this study. Overexpression of CTCF in cells increased POLD1 transcription, and this CTCF-induced POLD1 upregulation was blocked by actinomycin D, an inhibitor of transcription, a further proof that CTCF regulates POLD1 at the transcriptional level (Figure 4). Compared with those in the normal control, both the expression of POLD1 and the CTCF-binding level to the POLD1 promoter were confirmed to be increased with higher CTCF expression and decreased with lower CTCF expression (Figure 4). In addition, the expression of POLD1 was altered by cotransfection with shRNA-POLD1 or pLenti-CMV-POLD1 (Figure 5). These results illustrated that the CTCF-binding level to the POLD1 promoter was positively regulated by CTCF expression and fully proved that the decrease in CTCF-mediated transcription was attributed to POLD1 downregulation in aging by regulating the binding level of CTCF to the POLD1 promoter.
To investigate the effect of CTCF expression level on cellular senescence, SA-β-gal staining, cell proliferation, DNA synthesis, and DNA damage levels were measured in cells transfected with shRNA-CTCF, shRNA-CTCF + pLenti-CMV-POLD1, pLenti-CMV-CTCF, and pLenti-CMV-CTCF + shRNA-POLD1. A higher percentage of SA-β-gal-positive cells, a striking decrease in cell proliferation, a lower rate of EdU incorporation, and a notable increase in DNA damage under oxidative stress were observed in cells transfected with shRNA-CTCF compared with control cells. Moreover, the opposite change was observed in 2BS and WI38 cells transfected with pLenti-CMV-CTCF compared with the control cells. In addition, the effect of CTCF on aging was reversed by exogenously altered POLD1 expression (Figure 6). These results indicated that CTCF participates in the aging process by regulating POLD1 transcription, which is responsible for DNA synthesis and DNA damage repair. Further investigation found that metformin, as an antiaging drug (Glossmann and Lutz, 2019), can promote the expression of CTCF and POLD1. Moreover, the decreased level of p16INK4a, elevated DNA synthesis, and reduced SA-β-Gal-positive rates were observed in 2BS cells cultured with 4 mM metformin. These results demonstrated that metformin could delay replicative senescence as CTCF agonist, and CTCF delays replicative senescence as a key TF of POLD1 (Supplementary Figure S1).
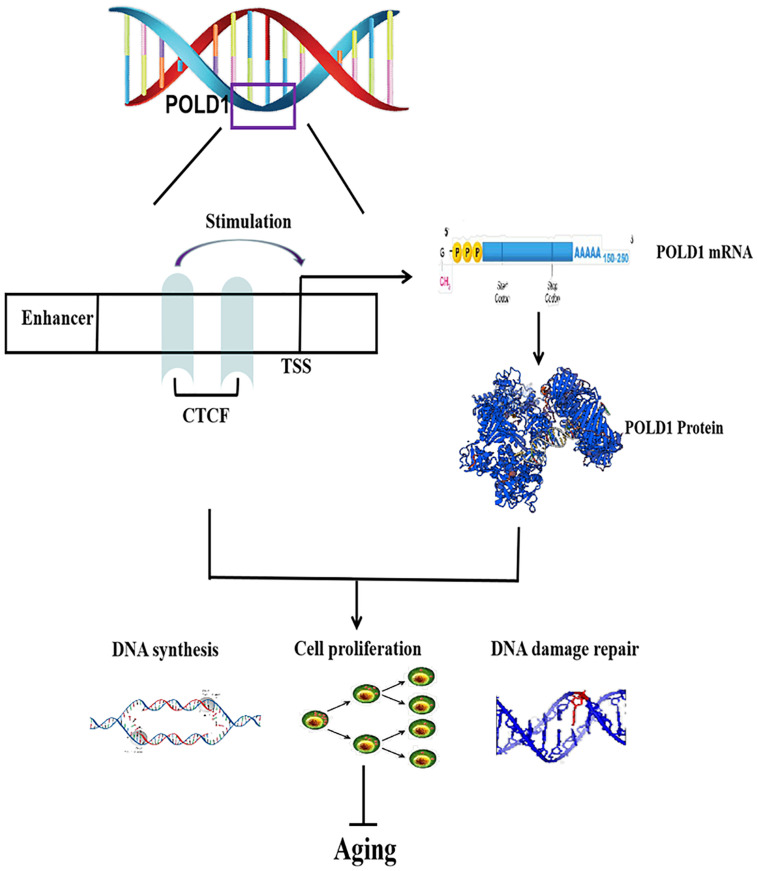
The role of POLD1 in aging may be regulated by multiple TFs or other factors (Gao et al., 2019), but our results propose the following POLD1 mechanism of aging: A decrease in POLD1 expression regulated by the decreased expression of the TF CTCF can accelerate aging by reducing cell proliferation, DNA synthesis, and DNA damage repair ability (Figure 7). As we first proved that CTCF expression levels reduced with aging and focused on the level of CTCF expression that affects the binding level of CTCF with the POLD1 promoter, it is worth to further explore if there are other factors that influence the binding affinity of CTCF with the POLD1 promoter.
FIGURE 7.
Regulatory mechanism of CCCTC-binding factor (CTCF) on aging. CTCF binds to the DNA polymerase delta 1, catalytic subunit (POLD1) promoter, and stimulates POLD1 expression to inhibit the progression of aging. TSS, transcription start site.
Materials and Methods
Cell Lines and Culture Conditions
All cell lines, including 2BS and WI38, were purchased from the National Infrastructure of Cell Line Resource (Beijing, China). All cell lines were recently authenticated and tested for mycoplasma contamination when we purchased them. 2BS and WI38 cells were cultured in minimum essential medium (MEM; Gibco, Gaithersburg, MD, United States). All culture media were supplemented with 10% fetal bovine serum (FBS, Gibco), 100 U/ml penicillin, and 100 μg/ml streptomycin (Gibco), and cells were then cultured in a humidified incubator with 5% CO2 at 37°C. Mouse tail-tip fibroblasts (TTFs) were isolated from SAMP8 mice at different months of age and cultured at 37°C in Dulbecco’s modified Eagle’s medium (DMEM; Gibco) containing non-essential amino acids (Gibco) and 10% FBS (Gibco). Cell cultures were expanded through sequential subculturing using trypsin–EDTA (Gibco) to achieve a higher PD level.
Animals
A total of 18 male SAMP8 mice were purchased from the First Affiliated Hospital of Tianjin University of Traditional Chinese Medicine (Tianjin, China). They were divided into three groups according to age: 2 months (2M, n = 6), 4 months (4M, n = 6), and 8 months (8M, n = 6). All animal experiments were approved by the Bioethics Committee of Xuanwu Hospital of Capital Medical University and complied with the National Institutes of Health Guide for the Care and Use of Laboratory Animals.
Lymphocyte Isolation
Blood with anticoagulant EDTA-K2 was collected from volunteers who signed informed consent forms. The volunteers were recruited from the hospital staff, medical students, and healthy people from the health screening center of Xuanwu Hospital. Donors with cancer, blood disease, or recent inflammation were excluded. Eighty participants were enrolled in this study and divided into four age groups (in years): ages 20–29 (n = 20; mean age, 25), 40–49 (n = 20; mean age, 46), 60–69 (n = 20; mean age, 65), and 80–89 (n = 20; mean age, 86). Peripheral blood mononuclear cells were obtained by Ficoll-Paque (Solarbio, Beijing, China) density gradient centrifugation. The use of human specimens was approved by the appropriate institutional review boards.
Bioinformatic Analysis
The 2,000-bp sequence upstream of the POLD1 transcription start site was obtained from the Ensembl genome browser1, and databases of ENCODE2 and JASPAR3 were used to predict the TFBS of CTCF on the POLD1 promoter. The binding sites predicted by the JASPAR database are shown in Table 1.
TABLE 1.
Binding site prediction of CTCF in the POLD1 promoter region.
| Model ID | Relative score | Start | End | Strand | Predicted site sequence |
| Site 1 | 0.78 | −1,973 | −1955 | −1 | CGACCAGGCGTGGGAGCCA |
| Site 2 | 0.77 | −1,505 | −1,487 | −1 | CAGCCGGGAAGGGTCAGTG |
| Site 3 | 0.77 | −1,015 | −997 | −1 | TGGCCCACAGGAGGCCTCA |
| Site 4 | 0.77 | −625 | −607 | 1 | ATTCCAATCGAGGGCGAAA |
| Site 5 | 0.77 | −136 | −118 | 1 | TTTCAGCAAGGGGGCGAGG |
Western Blotting
Briefly, total cellular and tissue proteins were extracted using radioimmunoprecipitation (Solarbio) assay buffer containing protease inhibitors (Solarbio). The protein concentration was determined using a BCA Protein Assay kit (Thermo Fisher Scientific, Massachusetts, United States). Then, 15 μg of cell or tissue lysate from each sample was separated by 8 or 12% sodium dodecyl sulfate (SDS) polyacrylamide gel electrophoresis. Electrophoretic transfer of proteins from gels onto nitrocellulose membranes was carried out in a trans-blotting chamber. Membranes were blocked by immersing in 5% non-fat milk (w/v) in TBST for 1 h to inhibit non-specific binding before being incubated with primary antibodies against CTCF (1:1,000; Abcam, Cambridge, United Kingdom), POLD1 (1:1,000; Abcam), and β-actin (1:5,000; Zhongshan Boil Tech Co., Beijing, China) at 4°C overnight. After rinsing with phosphate-buffered saline (PBS)/0.1% Tween-20, membranes were incubated with horseradish peroxidase-conjugated secondary antibodies (1:5,000; Zhongshan Boil Tech Co.). Immunocomplexes were visualized by incubation using an Enhanced Chemiluminescence Kit (Millipore, Massachusetts, United States). The bands were quantified with ImageJ software (National Institutes of Health, NIH, United States), and the target protein level was normalized to the β-actin level.
RT-qPCR
Total RNA was isolated from cells and tissues with TRIzoL reagent (Thermo Fisher Scientific) and then reverse transcribed. RT-qPCR was performed using SYBR (Takara, Japan) on a Roche 480 machine (Basel, Switzerland). Quantification was performed by 2–Δ Δ CT to calculate relative expression levels by subtracting the cycle threshold (CT) value of the control gene from the CT value of CTCF and POLD1. The primer sequences for PCR amplification of CTCF, POLD1, and β-actin are listed in Table 2.
TABLE 2.
Antibodies and sequences used for Western blotting and RT-qPCR.
| Protein | Mfrs./Made in | Cat no. |
| Primary antibodies | ||
| CTCF | Abcam/UK | Ab188408 |
| POLD1 | Abcam/UK | Ab186407 |
| β-actin | Zhongshan Boil Tech Co./China | TA-09 |
| Goat anti-rabbit IgG | Abcam/UK | ab6721 |
| Secondary antibodies | ||
| HRP-conjugated anti-mouse IgG | Zhongshan Boil Tech Co./China | ZB2305 |
| HRP-conjugated anti-rabbit IgG | Zhongshan Boil Tech Co./China | Zb5301 |
| Gene | Primer sequences | |
| β-actin | Forward | ACAGAGCCTCGCCTTTGC |
| Reverse | CCACCATCACGCCCTGG | |
| CTCF | Forward | TTCAGGTGGTTAAAGTGGGGGCCAA TGGAG |
| Reverse | TCCTCTGTATAACGCAGTTTGCTCTTTTTG | |
| POLD1 | Forward | GCTCCGCTCCTACACGCTCAA |
| Reverse | GGTCTGGTCGTTCCCATTCTGC | |
| ChIP assay promoter-specific qPCR | ||
| Site 1 | Forward | AGCCAGGTCTAGTCCAAGCACAGAG |
| Reverse | CGCAGGCTCACCAGCTTCATGA | |
| Site 2 | Forward | AGACCTATCGGCTCTCATCCCTTGG |
| Reverse | GCGTGCTGGGCTTGAGGTGTAA | |
| Site 3 | Forward | GGACCTGGACTTCTGTCCGAACAAC |
| Reverse | CCCGAGTCTTGGTAGGAGGATGGTT | |
| Site 4 | Forward | CGAAGCGACGAAGTTCCTCCAATCC |
| Reverse | GCGCCCGGTACACTACTCTTAGGAT | |
| Site 5 | Forward | AGACCGCACGAGGTCGTGAAGGTA |
| Reverse | CAAACAGCGTTTCCCGCCACAG | |
Chromatin Immunoprecipitation Assay
ChIP assays were carried out using the ChIP assay kit (Thermo Fisher Scientific) according to the manufacturer’s protocol. 2BS and WI38 cells were fixed and processed for ChIP. Briefly, cells were rinsed with PBS before being cross-linked in a 1% formaldehyde solution for 10 min, and the cross-linking reaction was stopped by the addition of 2.5 M glycine to a final concentration of 0.125 M followed by an additional 5 min of gentle swirling. Cells were washed once with 4°C sterile PBS and then collected by adding 1 ml of 4°C sterile PBS containing protease inhibitors. Cells were scraped from the dish with a razor blade and transferred into a tube, which was centrifuged at 3,000 × g for 5 min at 4°C. Then, 1.0 μl of Micrococcal Nuclease (ChIP Grade) (10 U/μl) was added, and the tube was vortexed and incubated in a 37°C water bath for 15 min, mixing by inversion every 5 min. For immunoprecipitation, 10 μg of CTCF antibody (Abcam) and 2 μl of normal rabbit IgG (Abcam) were added to the sample, which was then incubated at 4°C with rotation for 12–16 h and washed according to the ChIP assay protocol. Cross-links were reversed on all samples, including 10% input according to the protocol. DNA was extracted from the digested samples using PCR purification columns following the manufacturer’s instructions (QIAGEN, Maryland). Then, 5 μl of the ChIP DNA sample was used in the subsequent 20 μl real-time PCR mixture. The primers used in the qPCR assessment are listed in Table 2. Normal rabbit IgG was used as negative control, and input was used as the positive amplification control, indicating 10% input DNA.
Luciferase Reporter Assay
The 3′ untranslated region (UTR) of the POLD1 luciferase reporter plasmid (pHS-AVC-LW2301) was obtained from SyngenTech (Beijing, China). For the assessment of CTCF function on POLD1 promoter activity, genomic fragments harboring putative CTCF-binding sites in the human POLD1 promoter (∼1.8 kb upstream of the translation start site) were subcloned into the pGL4-Luc reporter vector (Promega) using Infusion 2.0 Dry-Down PCR cloning kit (Clontech, Shanghai, China). Promoter activity was further validated by mutation of the putative CTCF-binding site on the promoter at −1,015 to −997 or −625 to −607 by selecting these sites. For the luciferase reporter assay, 293T cells were seeded in a 24-well plate and incubated for 24 h before transfection. Subsequently, luciferase constructs, POLD1, and POLD1 + CTCF were co-transfected into 293T cells using Lipofectamine 3000. Cells were collected at 48 h after transfection and measured using the Dual-Luciferase Reporter System (Promega, WI, United States), according to manufacturer’s protocols. Three independent experiments were performed, and data were presented as mean ± SD.
Lentivirus Transfection
To investigate the function of CTCF, we transduced pLenti-CMV-CTCF vector lentiviral constructs to overexpress CTCF in 2BS and WI38 cells. The empty pLenti-CMV vector served as a negative control. shRNA targeting CTCF (shRNA-CTCF) was used to knock down CTCF expression, and the negative control shRNA was purchased from GeneChem (Shanghai, China). We also purchased shRNA-POLD1 to knock down the expression of POLD1 and pLenti-CMV-POLD1 to overexpress POLD1 from GeneChem. 2BS and WI38 cells were infected with shRNA, shRNA-CTCF, empty pLenti-CMV, shRNA-POLD1, pLenti-CMV-CTCF, shRNA-POLD1, and pLenti-CMV-POLD1. The supernatant was removed after 16 h and replaced with fresh culture medium.
Senescence-Associated (β-Galactosidase Staining
SA-β-gal (Beyotime, Beijing, China) activity was determined 72 h after transfection with lentivirus. Briefly, cells were washed with PBS and fixed for 15 min. Then, cells were washed three times with PBS and stained with X-gal solution for 6–24 h at 37°C. The population of SA-β-gal-positive cells was determined by counting 100 cells per dish, and images were taken using a phase-contrast microscope at 100× magnification (Olympus, Japan). The proportions of cells positive for SA-β-gal activity are presented as a ratio of the number of positive cells to the total number of cells counted in each dish. The results are expressed as the mean of triplicates ± SD.
Cell Counting Kit-8 Assay
The proliferation of 2BS and WI38 cells was tested using CCK-8 kit (Beyotime). Briefly, cells were seeded in 96-well plates at 5 × 103 cells/well with 100 μl of complete culture medium. At 0, 1, 2, 3, 4, and 5 days after adhesion, 10 μl of CCK-8 solution was added to each well, and the plates were incubated at 37°C for 2 h. The absorbance was measured at 450 nm using a microplate reader (Thermo Fisher Scientific).
5-Ethynyl-2′-Deoxyuridine Incorporation Assay
The DNA synthesis rate was assessed using the Beyo-ClickTM EdU kit (Beyotime). The cells were incubated with 10 μM EdU for 2 h, followed by staining according to the manufacturer’s instructions. The absorbance values of all wells at 370 nm were determined with an enzyme immunoassay instrument (Thermo Fisher Scientific).
Comet Assay
Comet assays were used to examine 2BS and WI38 cells subjected to oxidative DNA damage under different conditions. At 72 h after transfection with CTCF overexpression lentivirus/shRNA-CTCF/shRNA-POLD1/pLenti-CMV-POLD1, the cells were treated with 100 μM H2O2 for 5 min at 4°C in the dark. Alkaline neutral comet assays were performed using a comet assay kit (Trevigen, Gaithersburg, MD, United States) according to manufacturer’s protocol. The mixture was loaded on the slide. The slide was immersed in an ice-cold lysis solution at 4°C for 60 min, and then alkaline unwinding solution was added for 60 min at 4°C. Electrophoresis was performed at 21 volts for 30 min in alkaline electrophoresis solution. After electrophoresis, the slides were washed with deionized H2O and then stained with SYBR Gold (1:10,000 dilution) for 30 min. A fluorescence microscope (OLYMPUS, Japan) was then used to observe the state of the cells. The percentage of the tail area of each cell was analyzed using CASP software (version 1.2.3, download in http://casplab.com/).
Statistical Analysis
Statistical examination was carried out using GraphPad Prism version 8 (GraphPad Software, La Jolla, CA, United States). All data are displayed as mean ± SD (standard deviation). The difference between two groups was analyzed with a two-tailed t-test with variance equality. Differences among more than two groups were analyzed by one-way analysis of variance (ANOVA). CCK-8 data were analyzed by two-way ANOVA with repeated measures. The relationship between CTCF and POLD1 expression was calculated by Spearman’s rho test. P < 0.05 was considered to indicate a statistically significant result.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Ethics Statement
The studies involving human participants were reviewed and approved by the Bioethics Committee of Xuanwu Hospital of Capital Medical University. The patients/participants provided their written informed consent to participate in this study. The animal study was reviewed and approved by the Bioethics Committee of Xuanwu Hospital of Capital Medical University.
Author Contributions
YH and PW conceived the study. YH conducted the experiments. QS and SG performed the bioinformatics analysis. XZ conducted the data analysis. YW drafted the report. LJ and MC provided statistical support. JF and PW provided funding support. All authors interpreted the data and contributed to the final version of this report.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abbreviations
- 2BS
human embryonic lung diploid fibroblasts
- ChIP
chromatin immunoprecipitation
- CCK-8
Cell Counting Kit-8
- EdU
5-ethynyl-2 ′ -deoxyuridine
- Pol δ
DNA polymerases delta
- PD
population doubling
- RT-qPCR
quantitative reverse transcription-polymerase chain reaction
- SA- β-gal
senescence-associated β-galactosidase
- SAMP8
senescence-accelerated mouse prone 8
- shRNA
short hairpin RNA
- TFBS
transcription factor binding site
- WI38
human fetal lung fibroblasts
- TTFs
mouse tail-tip fibroblasts.
Funding. This work was supported by grants from the National Natural Science Foundation of China (Codes: 81871714, 81901406, and 81472007), Beijing Municipal Administration of Hospital’s Ascent Plan (Code: DFL20180803), and Research and Development Foundation of Capital Medical University (Code: PYZ19131).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2021.618586/full#supplementary-material
Metformin could act as agonist of CTCF to delay cell senescence. (A) Protein expression of CTCF, POLD1, and p16INK4a in 2BS cells treated with 4 mM metformin for 24 h. (B) SA-β-gal staining of 2BS cells treated with 4 mM metformin for 24 h. (C) EdU-positive cells were quantified after 24 h of treatment by the ratio of the absorbance at 370 nm. Data were compared by Student’s t-test, and data are shown as the mean ± SEM, with three independent experiments in each group. *P < 0.05, ***P < 0.001.
References
- Badarinarayan N., ElSharawy A., May S., Luzius A., Torres G. G., Jentzsch M. (2017). Identification and characterization of two functional variants in the human longevity gene FOXO3. Nat. Commun. 8:2063. 10.1038/s41467-017-02183-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beishline K., Vladimirova O., Tutton S., Wang Z., Deng Z., Lieberman P. M. (2017). CTCF driven TERRA transcription facilitates completion of telomere DNA replication. Nat. Commun. 8:2114. 10.1038/s41467-017-02212-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang J., Zhang B., Heath H., Galjart N., Wang X., Milbrandt J. (2010). Nicotinamide adenine dinucleotide (n.d.)-regulated DNA methylation alters CCCTC-binding factor (CTCF)/cohesin binding and transcription at the BDNF locus. Proc. Natl. Acad. Sci. U S A. 107 21836–21841. 10.1073/pnas.1002130107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F. J., Liu B., Wu Q., Liu J., Xu Y. Y., Zhou S. Y., et al. (2019). Icariin delays brain aging in senescence-accelerated mouse prone 8 (SAMP8) model via inhibiting autophagy. J. Pharmacol. Exp. Ther. 369 121–128. 10.1124/jpet.118.253310 [DOI] [PubMed] [Google Scholar]
- Childs B. G., Durik M., Baker D. J., van Deursen J. M. (2015). Cellular senescence in aging and age-related disease: from mechanisms to therapy. Nat. Med. 21 1424–1435. 10.1038/nm.4000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y., Keles S., Charbonnier L. M., Jule A. M., Henderson L., Celik S. C., et al. (2019). Combined immunodeficiency caused by a loss-of-function mutation in DNA polymerase delta 1. J. Allergy Clin. Immunol. 145 391–401. 10.1016/j.jaci.2019.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da C. J., Vitorino R., Silva G. M., Vogel C., Duarte A. C., Rocha-Santos T. (2016). A synopsis on aging-Theories, mechanisms and future prospects. Ageing Res. Rev. 29 90–112. 10.1016/j.arr.2016.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiorillo C., D’Apice M. R., Trucco F., Murdocca M., Spitalieri P., Assereto S., et al. (2018). Characterization of MDPL fibroblasts carrying the recurrent p.Ser605del mutation in POLD1 gene. DNA Cell Biol. 10.1089/dna.2018.4335 Online ahead of print. [DOI] [PubMed] [Google Scholar]
- Flachsbart F., Dose J., Gentschew L., Geismann C., Caliebe A., Knecht C., et al. (2017). Identification and characterization of two functional variants in the human longevity gene FOXO3. Nat. Commun. 8:2063. 10.1038/s41467-017-02183-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao S., Song Q., Liu J., Zhang X., Ji X., Wang P. (2019). E2F1 mediates the downregulation of POLD1 in replicative senescence. Cell Mol. Life Sci. 76 2833–2850. 10.1007/s00018-019-03070-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gea J., Ausin P., Martinez-Llorens J. M., Barreiro E. (2020). Respiratory muscle senescence in ageing and chronic lung diseases. Eur. Respir. Rev. 29:200087. 10.1183/16000617.0087-2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glossmann H. H., Lutz O. M. D. (2019). Metformin and aging: a review. Gerontology 65 581–590. 10.1159/000502257 [DOI] [PubMed] [Google Scholar]
- Gregor A., Oti M., Kouwenhoven E. N., Hoyer J., Sticht H., Ekici A. B., et al. (2013). De novo mutations in the genome organizer CTCF cause intellectual disability. Am. J. Hum. Genet. 93 124–131. 10.1016/j.ajhg.2013.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y., Franzen J., Stiehl T., Gobs M., Kuo C. C., Nikoliæ M., et al. (2020). New targeted approaches for epigenetic age predictions. BMC Biol. 18:71. 10.1186/s12915-020-00807-802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper S. (2014). Economic and social implications of aging societies. Science 346 587–591. 10.1126/science.1254405 [DOI] [PubMed] [Google Scholar]
- Hou Y., Dan X., Babbar M., Wei Y., Hasselbalch S. G., Croteau D. L., et al. (2019). Ageing as a risk factor for neurodegenerative disease. Nat. Rev. Neurol. 15 565–581. 10.1038/s41582-019-0244-247 [DOI] [PubMed] [Google Scholar]
- Hwang S. Y., Kang M. A., Baik C. J., Lee Y., Hang N. T., Kim B. G., et al. (2019). CTCF cooperates with CtIP to drive homologous recombination repair of double-strand breaks. Nucleic Acids Res. 47 9160–9179. 10.1093/nar/gkz639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkland J. L., Tchkonia T. (2017). Cellular senescence: a translational perspective. EBioMedicine 21 21–28. 10.1016/j.ebiom.2017.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lake R. J., Boetefuer E. L., Won K. J., Fan H. Y. (2016). The CSB chromatin remodeler and CTCF architectural protein cooperate in response to oxidative stress. Nucleic Acids Res. 44 2125–2135. 10.1093/nar/gkv1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Lee M. Y. (2001). Transcriptional regulation of the human DNA polymerase delta catalytic subunit gene POLD1 by p53 tumor suppressor and Sp1. J. Biol. Chem. 276 29729–29739. 10.1074/jbc.M101167200 [DOI] [PubMed] [Google Scholar]
- Maierhofer A., Flunkert J., Oshima J., Martin G. M., Poot M., Nanda I., et al. (2019). Epigenetic signatures of werner syndrome occur early in life and are distinct from normal epigenetic aging processes. Aging Cell 18:e12995. 10.1111/acel.12995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHugh D., Gil J. (2018). Senescence and aging: causes, consequences, and therapeutic avenues. J. Cell Biol. 217 65–77. 10.1083/jcb.201708092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazolli E., Stewart S. A. (2008). Senescence: the good the bad and the dysfunctional. Curr. Opin. Genet. Dev. 18 42–47. [DOI] [PubMed] [Google Scholar]
- Peng W. X., He R. Z., Zhang Z., Yang L., Mo Y. Y. (2019). LINC00346 promotes pancreatic cancer progression through the CTCF-mediated Myc transcription. Oncogene 38 6770–6780. 10.1038/s41388-019-0918-z [DOI] [PubMed] [Google Scholar]
- Reinier F., Zoledziewska M., Hanna D., Smith J. D., Valentini M., Zara I., et al. (2015). Mandibular hypoplasia, deafness, progeroid features and lipodystrophy (MDPL) syndrome in the context of inherited lipodystrophies. Metabolism 64 1530–1540. 10.1016/j.metabol.2015.07.022 [DOI] [PubMed] [Google Scholar]
- Shan Z., Li Y., Yu S., Wu J., Zhang C., Ma Y., et al. (2019). CTCF regulates the FoxO signaling pathway to affect the progression of prostate cancer. J. Cell Mol. Med. 23 3130–3139. 10.1111/jcmm.14138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J., Hong P., Liu C., Zhang Y., Wang J., Wang P. (2015). Human POLD1 modulates cell cycle progression and DNA damage repair. BMC Biochem. 16:14. 10.1186/s12858-015-0044-47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song N., Zhu X., Shi L., An J., Wu Y., Sang J. (2009). Identification and functional analysis of a CDE/CHR element in the POLD1 promoter. Sci. China C Life Sci. 52 551–559. 10.1007/s11427-009-0077-75 [DOI] [PubMed] [Google Scholar]
- Song S. H., Kim T. Y. (2017). CTCF, cohesin, and chromatin in human cancer. Genom. Inform. 15 114–122. 10.5808/GI.2017.15.4.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L., Yu R., Dang W. (2018). Chromatin architectural changes during cellular senescence and aging. Genes (Basel) 9:211. 10.3390/genes9040211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tepper R. G., Ashraf J., Kaletsky R., Kleemann G., Murphy C. T., Bussemaker H. J. (2013). PQM-1 complements DAF-16 as a key transcriptional regulator of DAF-2-mediated development and longevity. Cell 154 676–690. 10.1016/j.cell.2013.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. L., Guo H. L., Wang P. C., Liu C. G. (2012). Age-dependent down-regulation of DNA polymerase delta1 in human lymphocytes. Mol. Cell Biochem. 371 157–163. 10.1007/s11010-012-1432-1436 [DOI] [PubMed] [Google Scholar]
- Wendt K. S., Yoshida K., Itoh T., Bando M., Koch B., Schirghuber E., et al. (2008). Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature 451 796–801. 10.1038/nature06634 [DOI] [PubMed] [Google Scholar]
- Zhu H., Wang G., Qian J. (2016). Transcription factors as readers and effectors of DNA methylation. Nat. Rev. Genet. 17 551–565. 10.1038/nrg.2016.83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zivkovic L., Bajic V., Topalovic D., Bruic M., Spremo-Potparevic B. (2019). Antigenotoxic effects of biochaga and dihydroquercetin (Taxifolin) on H2O2-induced DNA damage in human whole blood cells. Oxid. Med. Cell Longev. 2019:5039372. 10.1155/2019/5039372 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Metformin could act as agonist of CTCF to delay cell senescence. (A) Protein expression of CTCF, POLD1, and p16INK4a in 2BS cells treated with 4 mM metformin for 24 h. (B) SA-β-gal staining of 2BS cells treated with 4 mM metformin for 24 h. (C) EdU-positive cells were quantified after 24 h of treatment by the ratio of the absorbance at 370 nm. Data were compared by Student’s t-test, and data are shown as the mean ± SEM, with three independent experiments in each group. *P < 0.05, ***P < 0.001.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.