Abstract
Dysregulation of the balance between cell proliferation and cell death is a central feature of malignances. Death-associated protein kinase 3 (DAPK3) regulates programmed cell death including apoptosis and autophagy. Our previous study showed that DAPK3 downregulation was detected in more than half of gastric cancers (GCs), which was related to tumor invasion, metastasis, and poor prognosis. However, the precise molecular mechanism underlying DAPK3-mediated tumor suppression remains unclear. Here, we showed that the tumor suppressive function of DAPK3 was dependent on autophagy process. Mass spectrometry, in vitro kinase assay, and immunoprecipitation revealed that DAPK3 increased ULK1 activity by direct ULK1 phosphorylation at Ser556. ULK1 phosphorylation by DAPK3 facilitates the ULK1 complex formation, the VPS34 complex activation, and autophagy induction upon starvation. The kinase activity of DAPK3 and ULK1 Ser556 phosphorylation were required for DAPK3-modulated tumor suppression. The coordinate expression of DAPK3 with ULK1 Ser556 phosphorylation was confirmed in clinical GC samples, and this co-expression was correlated with favorable survival outcomes in patients. Collectively, these findings indicate that the tumor-suppressor roles of DAPK3 in GC are associated with autophagy and that DAPK3 is a novel autophagy regulator, which can directly phosphorylate ULK1 and activate ULK1. Thus, DAPK3 might be a promising prognostic autophagy-associated marker.
Subject terms: Tumour-suppressor proteins, Macroautophagy
Introduction
Dysregulation of cell death is a common feature of cancer cells and it leads to uncontrolled cell growth and drug resistance. Apoptosis and autophagy, two forms of programmed cell death, are critical in dictating cell fate and play a fundamental role in tissue and organism homeostasis. As apoptosis serves as a barrier that inhibits cancer cell growth and dissemination, restoring apoptotic activity, and targeting antiapoptotic activity in cancer cells are the current antitumor strategies [1, 2]. However, autophagy has been revealed as a “double-edged” sword, acting as both a tumor promoter and a tumor suppressor, depending on the tumor tissue of origin and the pattern of genetic mutations and epigenetic changes. Autophagy is a catabolic process that digests cellular contents within lysosomes, and excessive autophagy can cause cell death [3]. Autophagy induces tumor suppression by eliminating harmful cellular components and preventing DNA damage and genomic instability. On the other hand, autophagy can support tumor cell survival by maintaining a steady nutrient supply to facilitate cancer cell growth. The different impacts of autophagy on tumor development may be determined by the specific oncogene activation and/or tumor suppressor loss that drives malignant transformation and cancer progression [3–5]. Therefore, a better understanding of the context-specific role of autophagy in each tumor and the mechanisms involved will be crucial for effective autophagy-based cancer therapy.
The death-associated protein kinases (DAPKs) are a family of five Ser/Thr kinases with conserved catalytic domains and cell death-associated functions. DAPK1, DAPK2, and DAPK3 show more than 80% similarity in the amino acid sequences of their catalytic domains and are recognized as closely related homologs [6]. DAPK1 is the most well-studied member of the family, and DAPK1 downregulation by promoter methylation is a general phenomenon observed in most human cancers. DAPK1 suppresses tumor growth and metastasis by activating autophagy and apoptosis [7]. DAPK3 possesses tumor and metastasis suppressor properties in different types of cancers including prostate, colon, and ovarian cancer. Loss-of-function mutations, deletion and microRNA dysregulation lead to DAPK3 inactivation [8–11]. As a key protein kinase in prostate apoptosis response-4-induced apoptosis, DAPK3 suppresses tumorigenesis through enhancing apoptosis activity in prostate cancer [8, 12]. We previously showed that DAPK3 downregulation was associated with tumor invasion, metastasis, and poor prognosis in primary GC [10]. However, the precise molecular mechanism underlying the suppression of GC cell growth by DAPK3 remains unknown. Therefore, in this study, we investigated the tumor-suppressive roles of DAPK3 in GC cells with respect to autophagy induction. Our study reveals a new mechanistic insight into DAPK3 antitumor function and highlights the clinicopathological significance of DAPK3-modulated autophagy in GC.
Materials and methods
Clinical samples and cell lines
Primary GC specimens and their matched adjacent non-tumor tissues were obtained with informed consent from patients who underwent radical gastrectomy at the First Affiliated Hospital, Sun Yat-sen University. The clinical and pathological features of these 235 patients are provided in Table S1. This study has been approved by the Committees for Ethical Review at the First Affiliated Hospital, Sun Yat-sen University. GC cell lines MKN28 and GES-1 were provided by Jie Chen (the First Affiliated Hospital of Sun Yat-sen University). MKN45 and MGC803 cells were provided by Changhua Zhang (the First Affiliated Hospital of Sun Yat-sen University). All GC cell lines were maintained in Dulbecco’s modified Eagle’s medium (Gibco, USA) containing 1% penicillin and streptomycin (Gibco, USA), supplemented with 10% fetal bovine serum (Biological Industries, USA). All cell lines were checked by STR profiling and mycoplasma contamination was tested every 2 weeks.
In vitro functional assays
XTT proliferation assay, foci formation assay, colony formation in soft agar, invasion assay, and wound healing assay were used to test the in vitro functional roles of DAPK3. For XTT cell proliferation assay, cells in logarithmic growth phase were plated in 96-well plates at the appropriate density. Six duplicate wells were set for each group with a negative control. 10 μL of the XTT (Dojindo, Japan) substrate was then added into each well with a total volume of 100 μL medium, the plates were incubated at 37 °C for 3 h, and the OD values were measured with microplate reader at a wavelength of 490 nm. XTT assays were performed once a day until day 5. The growth curve was constructed by plotting absorbance against time. For foci formation assay, 2 × 102 cells were plated in 3.5 cm culture plates in triplicates with DMEM medium containing 10% FBS. Surviving colonies (>50 cells per colony) were stained with 0.05% crystal violet and counted after one week in culture. For soft agar assay, 5 × 103 cells were suspended in soft agar mixture (DMEM, 10% FBS and 0.4% Sea Plaque agarose) and were subsequently overlaid on the solidified 0.6% agar base. After 2 weeks, colonies (≥10 cells) were counted under the microscope in ten fields per well. For wound healing assay, the confluent monolayer of cells was scratched with a fine pipette tip, and cell migration into the wound was observed after 48 h by microscopy. Cell migration or invasion assays were performed using a 24-well transwell chamber (Coring, USA) according to the manufacturer’s instructions. Briefly, cells (MKN45, MGC803, and GES-1: 4 × 104; MKN28: 2 × 104) in DMEM medium with 2% FBS were layered in the upper chamber, and medium containing 15% FBS was applied to the lower chamber. The chambers were then incubated for 24 h for cell migration (without matrigel) or 48 h for invasion (with 250 μg/mL matrigel coating) at 37 °C. After removing the cells in the upper surface of filter with cotton swab, the invasive cells attached to the lower surface of the membrane were fixed with 4% paraformaldehyde solution, stained with 0.1% crystal violet and then quantified by counting the cell number at ten random fields under a microscope.
Animal experiments
All animal experiments were approved by and performed in accordance with the Committee of the Use of Live Animals in Teaching and Research at Sun Yat-sen University. For in vivo tumorigenicity assay, different numbers of GC cells (MKN45: 1 × 106; MGC803: 3 × 106) with DAPK3 overexpression or knockdown were subcutaneously injected into the left or right dorsal flank of each 4-week-old female BALB/c nude mouse (five mice per group), respectively. Tumor volume was measured weekly over a 4-week period. The tumor volume was measured weekly and calculated using the formula V = 0.5 × W2 × L (V, volume; L, length; W, width).
Fluorescence microscopy
GFP-LC3-expressing GC cells were generated by transfection with pEZ-M98-neo GFP-LC3, selection for neomycin resistance and cell sorting for GFP expression. Cells were grown on coverslips, washed in PBS and treated with EBSS for 4 h. Cells were rinsed three times with PBS and fixed for 20 min with 4% paraformaldehyde at room temperature. Images were obtained using Leica confocal laser scanning microscope with Ar-488 and DAPI laser excitation.
Transmission electron microscopy
Cells were harvested, immediately fixed in 2.5% glutaraldehyde, and treated with 1% osmium tetroxide. Samples were dehydrated in graded ethanol, embedded, and stained with uranyl acetate/lead citrate. The images were obtained using a transmission electron microscope (JEM-1400PLUS; JEOL Ltd, Japan).
Sample preparation and mass spectrometric analysis
Cells were collected and centrifuged at 300 × g for 5 min at 4 °C. Peptides were reconstituted in 0.5 M tetraethylammonium tetrahydroborate and processed using the TMT kit (Thermo Scientific, USA), according to the manufacturer’s protocol. The phospho-peptides were enriched with immobilized metal affinity chromatography and subjected to a nanospray ionization source followed by tandem MS (MS/MS) in the Q Exactive™ Plus mass spectrometer (Thermo Scientific, USA) coupled online to the UPLC. The resulting MS/MS data were processed using the MaxQuant search engine (v.1.5.2.8).
In vitro kinase assay and immunoprecipitation
For in vitro kinase assay, recombinant ULK1 (Catalog No. ab95322, Abcam, USA) or immunoprecipitated HA-ULK1 from transfected human embryonic kidney (HEK) 293T cells was incubated with 0.1 μg of recombinant human active DAPK3 (catalog No. D03-10G, Signal Chem, Canada) in a 1× kinase assay buffer (25 mM Tris-HCl, pH 7.5, 5 mM b-glycerophosphate, 2 mM dithiothreitol, 0.1 mM Na3VO4, 10 mM MgCl2) with cold ATP (10 mM, CST, USA) in a total volume of 25 μL for 30 min at 37 °C. The reaction was terminated by adding 12.5 μL 3× loading buffer and incubating at 100 °C for 10 min. The proteins were then analyzed by 12% SDS-PAGE and coomassie blue staining of SDS-PAGE gels was conducted to verify the protein purity [13].
For immunoprecipitation, cells expressing HA-tagged (wild type or S556A) or Flag-tagged (wild type or K42A) proteins were lysed in 50 mM Tris-HCl (pH7.5), 150 mM NaCl, 5 mM EDTA (pH8.0), 1% NP-40, 0.5% deoxycholate, 0.1%SDS, and Roche complete protease inhibitor mixture (Roche, UK), and incubated with indicated antibodies (anti-Flag, Abmart, Shanghai, China; anti-HA, Sigma, USA) at 4 °C for 4 h, and then mixed with 100 μL protein A/G agarose. This immune complex was added to the cell lysates and incubated overnight at 4 °C. The resulting beads were washed with lysis buffer five times before analysis.
IHC and evaluation
IHC staining was performed using the standard streptavidin-biotin-peroxidase complex method. The sections were incubated with anti-DAPK3 (Prosci, USA), anti-pULK1 Ser556 (Abcam, USA), and anti-LC3B (Novus, USA) primary antibodies. DAPK3 and pULK1 Ser556 immunostaining was evaluated using a semi-quantitative scoring system [10]. According to the evaluation criteria of LC3B staining [14, 15], LC3 positive dots in cells were regarded as autophagy-related in our study.
Statistical analysis
All statistical analyses were conducted using commercially available software (SPSS version 18.0; SPSS, Inc, Chicago, IL, USA). The associations of DAPK3, pULK1 Ser556, and LC3B expression with clinicopathologic characteristics was determined using the Chi-square test or Fisher exact test. Data from cell growth, foci formation, wound healing, Transwell, western blot, and qPCR assays were evaluated using Student’s t test. Survival analysis was performed using the Kaplan–Meier method for univariate analysis with a log-rank test for significance and Cox regression for multivariate analysis. P values < 0.05 were considered statistically significant.
Results
DAPK3 exhibits tumor suppressor activities in GC cells
Our previous immunohistochemistry (IHC) analysis revealed that DAPK3 expression correlated negatively with late clinical stages and poor prognosis in GC [10]. In order to gain insights into the tumor-suppressive function of DAPK3 in GC cells, we first examined DAPK3 protein levels in eight gastric cancer cell lines and a gastric epithelial cell line. MKN45 and MKN28 cells derived from the metastatic gastric cancers expressed low levels of DAPK3, whereas MGC803 cells derived from the primary gastric cancer showed high DAPK3 expression. The moderate level of DAPK3 was observed in GES-1 gastric epithelia cells (Supplementary Fig. S1a). Lentiviral-mediated human DAPK3 was stably transfected into MKN45 and MKN28 GC cells (Supplementary Fig. S1b, c). XTT proliferation assay showed that DAPK3 significantly inhibited tumor cell growth rate (Fig. 1a and Supplementary Fig. S2a). Foci formation and soft agar assays showed that DAPK3 significantly reduced colony frequency and size in both anchorage-dependent and independent cells (Fig. 1b, c and Supplementary Fig. S2b, c). Further, to test whether DAPK3 knockdown could enhance tumorigenicity, two short hairpin RNAs (shRNAs) were used for DAPK3 silencing in MGC803 and GES-1 cells (Supplementary Fig. S1d, e). DAPK3 knockdown in MGC803 and GES-1 cells strongly promoted cell growth and colony formation (Fig. 1a–c and Supplementary Fig. S2a–c). To validate the effect of DAPK3 on tumor growth in vivo, subcutaneous xenograft tumor mouse models were established. We found that the tumors derived from empty vector-transfected MKN45 cells became palpable within 7 days after injection in all five nude mice tested, whereas those derived from DAPK3-transfected MKN45 cells were not observed until 3 weeks after the injection. Tumor volumes were plotted against tumor development time as shown in Fig. 1d. The kinetic graph showed that the tumor size of vector-transfected MKN45 cells was always larger than that of DAPK3-transfected MKN45 cells. Conversely, tumors developed from DAPK3-silenced MGC803 cells were significantly larger than those from the control cells (Fig. 1d). Wound healing and transwell assay demonstrated that cell migration and invasion were markedly decreased after DAPK3 overexpression and increased after DAPK3 knockdown (Fig. 1e, f and Supplementary Fig. S2d,e). Next, we detected cell cycle profiles by flow cytometry. Compared with the control cells, DAPK3-overexpressed MKN45 and MKN28 cells showed a significant increase in the percentage of G0/G1-phase cells. Simultaneously, we observed a marked reduction in the percentage of G0/G1-phase cells in MGC803 and GES-1 cells, when DAPK3 was silenced (Fig. 1g and Supplementary Fig. S2f).
Fig. 1. Tumor-suppressive effects of DAPK3 in gastric cancer cells.
a Cell growth rates were evaluated by CCK-8 assay. The results were expressed as the mean ± SD of three independent experiments (***P < 0.001, Student’s t test). Representative images of foci formation (b) and colony formation in soft agar (c) from MKN45-Vec, MKN45-DAPK3, MGC803-shControl, and MGC803-shDAPK3 cells. The numbers of foci and colonies were calculated and showed in the bar chart. (***P < 0.001, Student’s t test). d Representative images of the xenograft tumors formed in nude mice. Growth curves of tumors derived from the indicated cell lines are shown (***P < 0.001, Student’s t test). e Cell migration was assessed by wound healing assay (original magnification, 200×, scale bars: 100 μm). f Transwell assay was performed to detect cell migration and invasion ability (***P < 0.001, Student’s t test; original magnification, 200×, scale bars: 100 μm). g Cell cycle profiles were examined by flow cytometry (***P < 0.001, Student’s t test).
DAPK3 is required for autophagy induction by amino acid deprivation
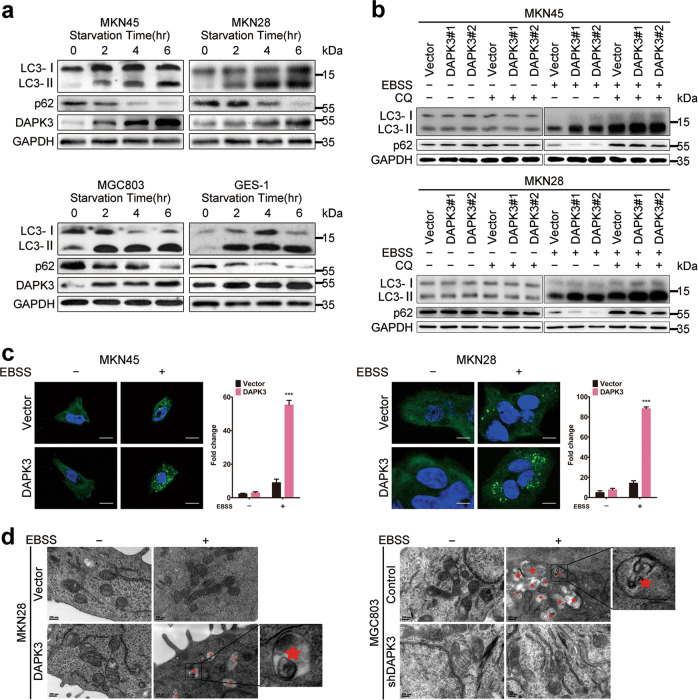
The above findings suggested that DAPK3 plays a pivotal inhibitory role in GC. We first presumed that the apoptosis induction by DAPK3 might be responsible for tumor growth suppression, because DAPK3 is involved in a wide array of apoptotic functions [16]. Therefore, we measured apoptosis in GC cells using flow cytometry and western blot. Intriguingly, DAPK3 upregulation did not affect apoptosis levels significantly (Supplementary Fig. S3). These data indicated that apoptosis is not the key mechanism underlying the DAPK3-mediated inhibition of tumor progression in these GC cell types. We next investigated whether DAPK3 regulates autophagy in GC cells. Endogenous DAPK3 expression was tested in MKN28, MKN45, MGC803, and GES-1 cells upon amino acid starvation using Earle’s balanced salt solution (EBSS). Western blot analysis showed significant increases in endogenous DAPK3 protein levels after autophagy promotion via EBSS treatment (Fig. 2a). Similarly, DAPK3 overexpression increased the microtubule-associated protein light chain 3 (LC3)-II/LC3-I ratio and LC3-II levels increased further after chloroquine (CQ)-mediated inhibition of autolysosome turnover. Meanwhile, SQSTM1/p62 levels decreased significantly in DAPK3-overexpressed cells under amino acid starvation. CQ treatment prevented SQSTM1/p62 degradation (Fig. 2b). Moreover, an increase in the number of GFP-LC3 puncta was found in DAPK3-overexpressed cells, suggesting that DAPK3 enhanced autophagosome formation (Fig. 2c). Transmission electron microscopy revealed a significant increase in autophagic vacuoles containing cytoplasmic structures and residual digested materials in DAPK3-overexpressed cells, compared with the control cells (Fig. 2d). DAPK3 knockdown in MGC803 and GES-1 cells markedly impaired the LC3-I to LC3-II conversion, SQSTM1/p62 degradation, GFP-LC3 puncta accumulation, and autophagic vacuole formation during amino acid starvation (Supplementary Fig. S4 and Fig. 2d). Thus, DAPK3 is required for the induction of amino acid starvation-mediated autophagy.
Fig. 2. DAPK3 regulated autophagy under amino acid starvation.
a Western blotting was performed to detect endogenous DAPK3 expression and autophagy in the indicated cells upon amino acid starvation (EBSS, 4 h). Cell lysates were immunoblotted with the indicated antibodies. GAPDH was used as a loading control. b Immunoblot analysis of autophagy fluxes in DAPK3-overexpressed and their respective control cells. The lysosomal inhibitor chloroquine was used to rescue LC3-II and SQSTM1/p62 breakdown. c GFP-LC3 punctate signals were assessed by confocal fluorescence microscopy (original magnification, 600×, scale bar:20 μm). d Autophagic vacuoles were analyzed by transmission electron microscopy. Red asterisk indicated double-membraned autophagic vacuoles containing cytoplasmic structures (scale bar: 200 nm).
Autophagy inhibition by ATG5 or ATG7 depletion impairs the tumor-suppressive function of DAPK3
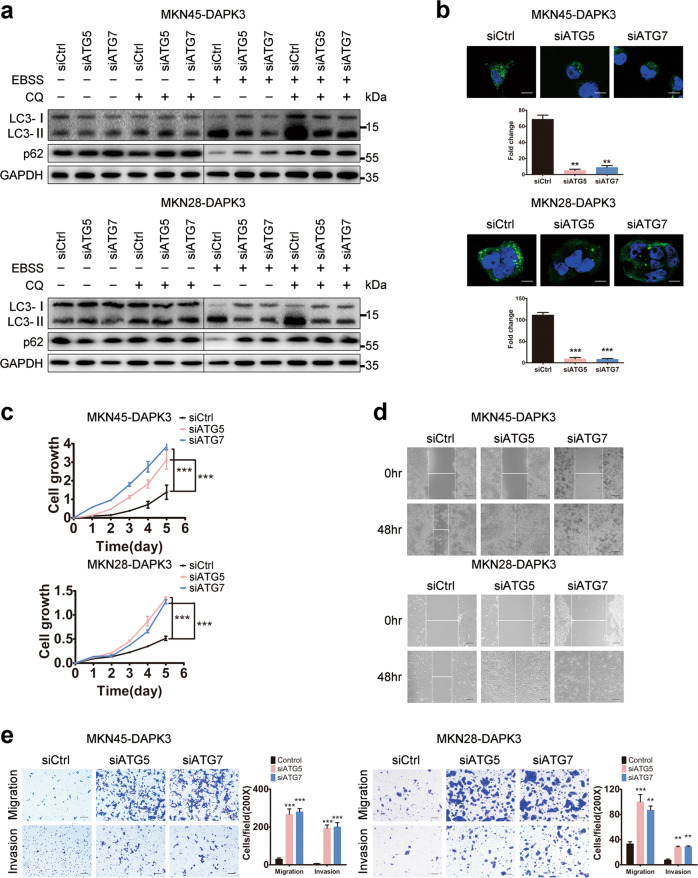
To ascertain whether DAPK3-mediated autophagy is associated with its tumor-suppressive function, autophagy activity was inhibited by siRNA-mediated autophagy-related 5 (ATG5) or autophagy-related 7 (ATG7) depletion in DAPK3-overexpressed cells and empty vector-transfected cells. The knockdown efficiency was verified by western blot and qPCR, and the results showed that ATG5 or ATG7 was significantly reduced after siATG5 or siATG7 transfection (Supplementary Fig. S5a–d). Knockdown of ATG5 or ATG7 markedly decreased the LC3-I to LC3-II conversion, SQSTM1/P62 degradation, and GFP-LC3 puncta formation in DAPK3-overexpressed cells, indicating that DAPK3-induced autophagy was impaired by ATG5 or ATG7 depletion (Fig. 3a, b and Supplementary Fig. S5e, f). We next examined the effect of autophagy inhibition on cell proliferation, migration and invasion in DAPK3-overexpressed cells, and empty vector-transfected cells. Cell growth assay showed that cell growth rates in DAPK3-overexpressed cells with ATG5 or ATG7 depletion were significantly higher than those control GC cells. Meanwhile, ATG5 or ATG7 knockdown promoted cell migration and invasion (Fig. 3c–e and Supplementary Fig. S6). These results showed that ATG5 or ATG7 knockdown could rescue GC cells from DAPK3-induced growth and metastasis arrest, suggesting that autophagy activity is required for the tumor-suppressive function of DAPK3.
Fig. 3. Autophagy inhibition promoted cell growth, migration, and invasion.
DAPK3-overexpressed MKN45 cells were transfected with siRNAs against ATG5, ATG7, or control siRNA, respectively. Autophagy activity was examined by western blot assay (a) and GFP-LC3 fluorescence assay (b) (original magnification, 600×, scale bar: 20 μm). Cell growth rates, migration and invasion were measured by XTT assay (c), wound healing assay (d) and transwell assay (e) respectively (***P < 0.001, Student’s t test) (original magnification, 200×, scale bars: 100 μm).
DAPK3 regulates autophagy through ULK1 activation
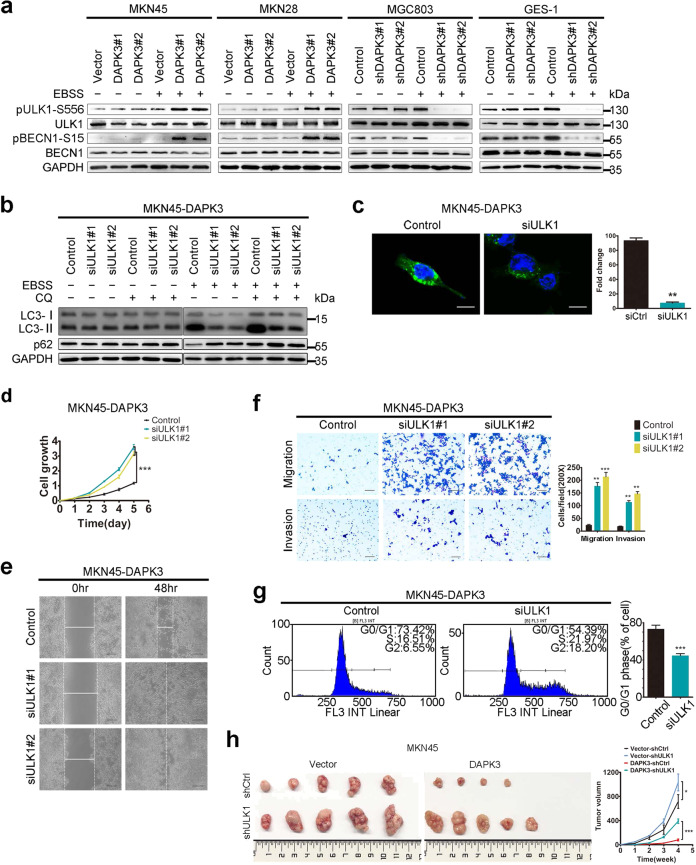
To elucidate the molecular mechanisms underlying DAPK3-mediated autophagy, we used mass spectrometry (MS)-based quantitative phosphoproteome analysis to profile the differentially expressed proteins in DAPK3-overexpressed MKN28 cells and the control cells under amino acid starvation. We observed an approximate twofold increase in ULK1 phosphorylation at serine 556 (Ser556) in DAPK3-overexpressed cells, compared with the control cells. We used the immunoblot assay to validate the significant upregulations of phosphorylation levels of ULK1 at Ser556 in DAPK3-overexpressed cells after amino acid starvation (Fig. 4a). We also detected the phosphorylation status of Beclin-1, a substrate of the ULK1, which is required for VPS34 complex activation and full autophagic induction [17]. As anticipated, DAPK3 increased Beclin-1 phosphorylation at Ser15, revealing that DAPK3 certainly regulated ULK1 activation (Fig. 4a). To further determine the roles of ULK1 in DAPK3 function, ULK1 was blocked by siRNA in DAPK3-overexpressed cells and empty vector control cells. ULK1 knockdown strongly inhibited autophagic fluxes, autophagosome formation and enhanced the tumorigenicity of DAPK3-overexpressed cells in vitro (Fig. 4b–g, Supplementary Figs. S7, S8). Next, we confirmed the effect of ULK1 on tumor growth in nude mice by subcutaneous injection of DAPK3-overexpressed or empty vector control cells in which ULK1 was depleted by shRNA. Tumors size was significantly increased in the nude mice inoculated with ULK1-silenced MKN45-DAPK3 cells, suggesting that DAPK3-mediated tumor suppression depends on ULK1 activation (Fig. 4h).
Fig. 4. DAPK3 increased amino acid starvation-induced autophagy via ULK1 activation.
a Western blotting for ULK1, ULK1 Ser556 phosphorylation, Beclin1 and Beclin1 Ser15 phosphorylation in DAPK3-overexpressed, DAPK3-silenced, and their respective control cells. DAPK3-overexpressed MKN45 cells were transfected with siRNAs against ULK1 or control siRNA. Autophagy activity was examined by western blot assay (b) and GFP-LC3 fluorescence assay (c) (original magnification, 600×, scale bar: 20 μm). Cell growth rates, cell migration, invasion and cell cycle profile were measured by XTT assay (d), wound healing assay (e), transwell assay (f) and flow cytometry (g) respectively (***P < 0.001, Student’s t test). (original magnification, 200×, scale bars: 100 μm). h Representative images of the xenograft tumors formed in nude mice. Growth curves of tumors derived from the indicated cell lines were shown (***P < 0.001, Student’s t test).
ULK1 activation is directly regulated by mTOR and AMPK. AMPK positively regulates ULK1 to induce mitophagy through phosphorylating ULK1 at Ser556 [18, 19]. High mTOR activity suppresses ULK1 activation by ULK1 phosphorylation at Ser758 when nutrients are in plentiful supply. Upon nutrient starvation, mTOR activity is inhibited and ULK1 Ser758 dephosphorylation leads to ULK1 activation [20]. It has been reported that DAPK3 downregulates the AKT activity in prostate cancer cells [8]. In this study, we examined the effect of DAPK3 on AKT/mTOR signaling pathway during autophagy induction. DAPK3 strongly reduced AKT/mTOR activity, as indicated by decreased phosphorylation of AKT at Thr308, mTOR at Ser2448, P70S6K at Thr389, 4E-BP1 at Thr37/46, and ULK1 at Ser758 (Supplementary Fig. S9). Our study ascertained that DAPK3 negatively regulated AKT and mTORC1 activity during autophagy induction, suggesting that inhibition of AKT/mTOR signaling pathway is an alternative mechanism of DAPK3-modulated autophagy. In addition, we further investigated whether DAPK3 activates ULK1 dependent on AMPK. AMPK inhibition by compound C did not affect DAPK3-induced ULK1 activation in GC cells (Supplementary Fig. S10). These observations indicated that DAPK3 could enhance ULK1 Ser556 phosphorylation in an AMPK-independent manner.
DAPK3 activates ULK1 through direct phosphorylation of Ser556
To investigate the possibility that DAPK3 directly phosphorylates ULK1, we used the GPS software (http://gps.biocuckoo.org) to predict the ULK1 phosphorylation sites for DAPK3 and found that ULK1 might be phosphorylated at Ser556 by DAPK3 (Fig. 5a).We initially performed in vitro kinase assay involving the incubation of recombinant active DAPK3 with recombinant ULK1 in the presence of cold ATP. DAPK3 phosphorylated recombinant ULK1 at Ser556, suggesting that DAPK3 could directly phosphorylate ULK1 at Ser556 (Fig. 5b). It was worth noting that DAPK3 did not phosphorylate ULK1 at Ser317 and Ser777 which are major sites for AMPK-induced ULK1 activation (Supplementary Fig. S11a). To confirm ULK1 Ser556 phosphorylation by DAPK3, we transfected wild-type (WT) or DAPK3 non-phosphorylatable (S556A) ULK1 cDNA into HEK 293T cells in which endogenous ULK1 was abolished by lentiviral hairpin shRNA, and performed in vitro DAPK3 kinase assay with immunoprecipitated HA-UKL1 WT and HA-UKL1 556A mutant as substrates (Supplementary Fig. S11b). DAPK3 also strongly phosphorylated ULK1 at Ser556 in vitro kinase assay and the phosphorylation level of Ser556 ULK1 was increased further under amino acid starvation (Fig. 5c). However, Ser556 mutation significantly impeded phosphorylation at this site (Fig. 5c). Next, we investigated the interaction between DAPK3 and ULK1 in vivo. HEK293T cells were transfected with Flag-tagged DAPK3 or HA-tagged ULK1. Endogenous ULK1 bound Flag-tagged DAPK3 and HA-tagged ULK1 immunoprecipitated endogenous DAPK3 from HEK293T cells (Supplementary Fig. S11c, d).To further verify the DAPK3 phosphorylation site in ULK1 in vivo, we detected Ser556 ULK1 phosphorylation by immunoprecipitation of HA-tagged ULK1 from DAPK3-overexpressed and empty vector-transfected HEK293T cells in which wild-type or mutant S556AULK1 cDNA was transfected after endogenous ULK1 depletion. As expected, high level of ULK1 Ser556 phosphorylation was observed in DAPK3-overexpressed cells with WT ULK1 (Fig. 5d and Supplementary Fig. S12a). Additionally, we tested whether the DAPK3 kinase activity was required for DAPK3-mediated phosphorylation of ULK1 at Ser556. WT DAPK3 or K42A, a kinase-inactivated mutant of DAPK3, was transfected into DAPK3-silenced HEK 293T cells (Supplementary Fig. S11e). The increase in ULK1 phosphorylation at Ser556 due to WT DAPK3 overexpression was markedly higher than that due to DAPK3 K42A overexpression, indicating that DAPK3 kinase activity was necessary for ULK1 Ser556 phosphorylation (Fig. 5e). We also observed that ULK1 with mutated Ser556 and catalytically inactive DAPK3 did not impair the interaction between ULK1 and DAPK3, suggesting that the phosphorylation status of ULK1 Ser556 did not affect the binding between ULK1 and DAPK3.
Fig. 5. DAPK3 directly activated ULK1 by phosphorylating Ser556.
a GPS software was used for predicting the DAPK3 phosphorylation sites in ULK1 (http://gps.biocuckoo.org/). b In vitro kinase assay. Recombinant ULK1 was incubated with active recombinant DAPK3 in the presence of cold ATP. The levels of ULK1-S556 phosphorylation were detected by western blotting. Coomassie blue staining of SDS-PAGE gels was used to assess the protein purity and sizes. c In vitro DAPK3 kinase assay with immunoprecipitated HA-ULK1 WT and HA-ULK1556A mutant as substrates. d HA-ULK1 WT or S556A mutant was transfected together with Flag-DAPK3 or Flag-empty vector into HEK293T cells in which endogenous ULK1 was abolished by shRNA. HA-ULK1 proteins were immunoprecipitated. p-ULK1(Ser556), FIP200, ATG13 and ATG101 were examined by western blotting. e Flag-DAPK3 proteins were immunoprecipitated from the HEK293T cells in which endogenous DAPK3 was depleted by shRNA and Flag-DAPK3 WT or K42A (a kinase-inactive mutant) was transfected. The amount of p-ULK1(Ser556), FIP200, ATG13, and ATG101 coimmunoprecipitated with Flag antibody was analyzed by western blotting. f The amount of Vps34 complex member Beclin1, ATG14, and Vps34 was detected by western blotting in DAPK3-overexpressed and empty vector-transfected HEK293T cells in which wild type ULK1 or ULK1-S556A mutant was transfected. The phosphorylation levels of BECN1 at Ser15 and ATG14 at Ser29 were evaluated by western blotting as well. g The expression of Vps34 complex members and the phosphorylation levels of BECN1 at Ser15 and ATG14 at Ser29 were evaluated by western blotting in the HEK293T cells expressing DAPK3 WT or K42A.
The ULK1 complex regulates initiation and formation of an autophagosome through activation of VPS34 complex [21]. We immunoprecipitated HA-ULK1 as a key molecule and analyzed the interaction between ULK1 and the other ULK1 complex components in DAPK3-overexpressed cells and empty vector control cells. DAPK3 expression markedly strengthened the interaction between WT ULK1 and its binding partners, FIP200, ATG13, and ATG101 under starvation, whereas loss of the Ser556 phosphorylation site in ULK1 partially impaired these interactions (Fig. 5d, Supplementary Figs. S11f and S12a, b). Next, we further explored the effect of DAPK3 kinase activity on ULK1 complex formation. ATG101 was found in the anti-Flag immunoprecipitates and the amounts of ATG101 was increased in WT DAPK3 cells upon starvation. FIP200 and ATG13 were co-immunoprecipitated with Flag-tagged WT DAPK3 upon starvation (Fig. 5e and Fig. S11g). Knockdown of ULK1 attenuated the interaction between DAPK3 and ULK1 complex components, suggesting that the role of DAPK3 in ULK1 complex is dependent on ULK1 activation (Fig. S11h). However, the VPS34 complex components were absent in the anti-Flag immunoprecipitates from HEK 293T cells overexpressing WT DAPK3 or K42A. ULK1 are conducive to activation of the VPS34 complex through direct phosphorylation of Beclin1 and ATG14L [17, 22]. Then, we explored the roles of DAPK3 in the VPS34 complex by western blot assay. Our study showed that the phosphorylation of ULK1 Ser556 and DAPK3 kinase activity were required for activation of the VPS34 complex in HEK293T cells under starvation (Fig. 5f, g, Supplementary Figs. S11i and S12c–e). Collectively, these data implied that ULK1 phosphorylation by DAPK3 enhances the ULK1 complex formation and the VPS34 complex activation upon starvation.
Ser556 phosphorylation in ULK1 facilitates DAPK3-mediated autophagy and tumor suppression
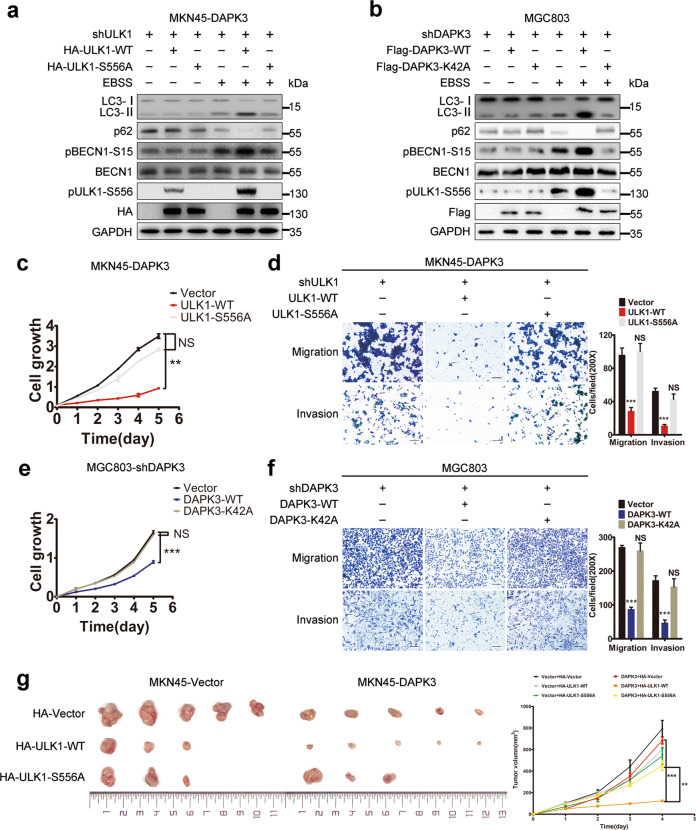
To determine the functional significance of ULK1 phosphorylation in DAPK3-regulated autophagy and tumor suppression, WT ULK1 or S556A ULK1 cDNA was transfected into DAPK3-overexpressed and empty vector control MKN45 cells (Supplementary Fig. S13a). DAPK3-overexpressed MKN45 cells with shRNA-induced endogenous ULK1 downregulation exhibited defective autophagy and increased tumorigenicity. Reconstitution of WT ULK1 cDNA, but not phosphorylation defective S556A mutant, restored autophagy and tumor suppression (Fig. 6a, c, d and Fig. S13b–d). To further assess the effect of UKL1 phosphorylation on tumor growth in vivo, we performed tumor xenograft experiments. The xenograft from DAPK3-overexpressed MKN45 cells with WT ULK1 grew at a slower rate than those derived from S556A mutant-expressed cells. WT ULK1 expression showed a more remarkable suppression of tumor growth in DAPK3-overexpressed MKN45 cells, compared with empty vector control cells (Fig. 6g). Our findings indicated that DAPK3 kinase activity was required for ULK1 Ser556 phosphorylation. We then analyzed the importance of DAPK3 kinase activity for autophagy induction and tumor inhibition. As expected, MGC803 cells with DAPK3 WT showed higher autophagy levels and decreased cell growth rate, motility and invasion than those cells with DAPK3 K42A (Fig. 6b, e, f). Taken together, these results denoted that the kinase activity of DAPK3 and ULK1 phosphorylation at Ser556 were required for DAPK3 function.
Fig. 6. Ser556 phosphorylation in ULK1 facilitates DAPK3-mediated autophagy and tumor suppression in GC cells.
a HA-ULK1 WT or S556A mutant were transfected into DAPK3-overexpressed MKN45 cells with endogenous ULK1 depletion by shRNA. Cell lysates were immunoblotted as indicated. b DAPK3-silenced MGC803 cells transfected with Flag-DAPK3-WT or Flag-DAPK3-K42A were placed in EBSS starvation media or full media for 4h and immunoblotted as indicated. Cell growth rate, migration, and invasion ability of MKN45-DAPK3 cells were measured by XTT assay (c) and transwell assay (d) (***P < 0.001, student t test) (original magnification, 200×, scale bars:100 μm). Cell growth rate, migration, and invasion were examined by XTT assay (e) and transwell assay (f) in MGC803-shDAPK3 cells (***P < 0.001, student t test, original magnification, 200×, scale bars: 100 μm). g 1×107 DAPK3-overexpressed and empty vector-transfected MKN45 cells expressing wild type ULK1 or ULK1-S556A mutant were subcutaneously injected into nude mice. Representative images of the xenograft tumors formed in the nude mice. Growth curves of tumors derived from the indicated cell lines were shown (***P < 0.001, Student’s t test).
DAPK3 expression is positively correlated with ULK1 phosphorylation at Ser556 and LC3B in surgical GC specimens
Our in vitro studies showed that ULK1 Ser556 phosphorylation by DAPK3 was crucial for autophagy induction and tumor suppression. Therefore, we assessed the clinical relevance between DAPK3 and autophagy activation in clinical GC specimens. The expression of DAPK3, pULK1 Ser556, and LC3B was measured by IHC in 235 primary GCs (Supplementary Table S1). The cytoplasmic expression of DAPK3, pULK1 Ser556, and LC3B in non-neoplastic tissues was higher than that in tumor tissues (Supplementary Fig. S14a–c). High expression of DAPK3, pULK1 Ser556, and LC3B was observed in 41.7%, 40%, and 34% of GCs, respectively. Correlation analysis demonstrated that DAPK3 expression was positively related to immunostaining of pULK1 Ser556 and LC3B in GC tissues (Fig. 7a, b and Supplementary Table S2). The association of DAPK3, pULK1 Ser556, and LC3B levels with clinicopathological parameters is summarized in Table 1 and Supplementary Table S3. Intriguingly, co-expression of DAPK3 and pULK1 Ser556 was inversely associated with poor tumor differentiation, tumor invasion, and more advanced clinical GC stages. Similarly, DAPK3 and LC3B co-expression was frequently detected in early stage tumors and negatively correlated with cancer progression. Kaplan–Meier analysis showed that patients with DAPK3 and pULK1 Ser556 co-expression had longer overall and cancer-specific survival time. Likewise, co-expression of DAPK3 and LC3B predicted a good prognosis in patients (Fig. 7c, d and Supplementary Fig. S14d–g). Further multivariate Cox regression analysis also indicated that co-expression of DAPK3 and pULK1 Ser556 as well as co-expression of DAPK3 and LC3B were independent prognostic factors for favorable overall and cancer-specific survival of GC patients. Of the other parameters, TNM stage was also identified as an independent prognostic factor (Supplementary Table S4). Collectively, these data illustrate that DAPK3-associated autophagy exerts distinct inhibitory effects on progression of human GC.
Fig. 7. The co-expression of DAPK3 with ULK1 Ser556 phosphorylation or LC3B predicted favorable survival in gastric cancer patients.
Immunohistochemical staining of DAPK3, phospho-ULK1 (Ser556), and LC3B in human GC specimens. Representative images showed DAPK3, pULK1-Ser556, and LC3B expression patterns in different GC cases. GC case 1 showed negative expression of DAPK3 and pULK1 Ser556; GC case 2 showed high expression of DAPK3 and pULK1 Ser556 (a). GC case 3 showed negative expression of DAPK3 and LC3B. GC case 4 showed high expression of DAPK3 and LC3B. The arrows indicated LC3 positive puncta (b) (original magnification, 200×, scale bars: 100 μm). Kaplan–Meier analysis of overall and cancer-specific survival in GC patients with co-expression of DAPK3 and pULK1 Ser556 (c) or DAPK3 and LC3B (d). P values were calculated by the log-rank test.
Table 1.
The association of DAPK3, pULK1 Ser556, and LC3B with clinicopathological features in 235 gastric cancers.
| Clinicopathological features | Total cases | DAPK3 and pULK1-Ser556 high expression | P value | DAPK3 and LC3B high expression | P value |
|---|---|---|---|---|---|
| Chi2 | Chi2 | ||||
| No. of patients | 235 | 69 (29.4%) | 59 (25.1%) | ||
| Age (mean 56 years) | 0.735 | 0.0.271 | |||
| ≤56 | 122 | 37 (30.3%) | 27 (22.1%) | ||
| >56 | 113 | 32 (28.3%) | 32 (28.3%) | ||
| Sex | 0.271 | 0.915 | |||
| Male | 158 | 50 (31.6%) | 40 (25.3%) | ||
| Female | 77 | 19 (24.7%) | 19 (24.7%) | ||
| Histological typea | 0.002 | 0.011 | |||
| WA | 72 | 33 (45.8%) | 29 (40.3%) | ||
| PA | 126 | 27 (21.4%) | 23 (18.3%) | ||
| MA | 14 | 2 (14.3%) | 2 (14.3%) | ||
| SRC | 15 | 3 (20%) | 3 (20.0%) | ||
| UC | 8 | 4 (50.0%) | 2 (25.0%) | ||
| Tumor stage | <0.0001 | 0.033 | |||
| pT1 | 29 | 18 (65.5%) | 10 (34.5%) | ||
| pT2 | 14 | 7 (50%) | 6 (42.9%) | ||
| pT3 | 81 | 25 (30.9%) | 25 (30.9%) | ||
| pT4 | 111 | 19 (17.1%) | 18 (16.2%) | ||
| Lymph-node metastasis | <0.0001 | 0.003 | |||
| pN0 | 112 | 50 (44.6%) | 39 (34.8%) | ||
| pN1 | 57 | 11 (19.3%) | 13 (22.8%) | ||
| pN2 | 56 | 8 (14.3%) | 7 (122.5%) | ||
| pN3 | 10 | 0 (0.0%) | 0 (0.0%) | ||
| Organ metastasis | <0.0001 | <0.0001 | |||
| M0 | 174 | 62 (35.6%) | 54 (31.0%) | ||
| M1 | 61 | 7 (11.5%) | 5 (8.2%) | ||
| TNM stage | <0.0001 | <0.0001 | |||
| I | 36 | 21 (58.3%) | 12 (33.3%) | ||
| II | 89 | 36 (40.4%) | 35 (39.3%) | ||
| III | 49 | 5 (10.2%) | 7 (14.3%) | ||
| IV | 61 | 7 (11.5%) | 5 (8.2%) | ||
aHistological type: WA well/moderately differentiated adenocarcinoma, PA poorly differentiated adenocarcinoma, MA mucinous adenocarcinoma, SRC signet ring cell carcinoma, UC undifferentiated.
Discussion
Our present study showed that the tumor suppressive function of DAPK3 depends on its pro-autophagic activity. DAPK3 is a novel regulator of ULK1, which can directly phosphorylate ULK1. DAPK3-associated autophagy is correlated with favorable survival outcome in GC patients. Thus, DAPK3 might be a promising prognostic autophagy-associated marker.
The inhibitory roles of DAPK3 in carcinoma was previously proved to be associated with its pro-apoptotic capacity. Our present study revealed the impact of DAPK3-mediated autophagy on tumor suppression. DAPK3 can enhance autophagy induction through activating ULK1 and suppressing AKT/mTOR signaling pathway upon nutrient deprivation. Several reports had showed that DAPK3 participates in the autophagy regulatory process via directly phosphorylating Beclin-1 Ser-90 and promotes autophagy initiation [13, 23]. Here, we demonstrated, for the first time, that DAPK3 was a novel regulator of ULK1 and that it increased ULK1 activity by direct phosphorylation of ULK1 at Ser556. ULK1 protein kinase exerts its function in a complex with FIP200, ATG13, and ATG101 which can stabilize and enhance ULK1 kinase activity. Upon autophagy induction, the ULK1 complex is a crucial node, transducing upstream signals to the downstream of the autophagy pathway [24]. ULK1 activates the downstream substrate VPS34 complex through direct phosphorylation of the complex members such as Beclin1 and ATG14L. The Vps34 complex is responsible for the production of the phospholipid phosphatidylinositol 3-phosphate at the site of forming autophagosome precursor, termed as phagophore [25]. Ser556 in ULK1 is the crucial AMPK phosphorylation site. Ser556 phosphorylation by AMPK is required for mitophagy during amino acid starvation or hypoxia [18, 19]. Phosphorylation of the site by AMPK is also important for the interaction of ULK1 with the adapter protein YWHAB/14-3-3 and localization of ATG9 to peri-nuclear [26]. Our study identified DAPK3 as a new direct upstream kinase of ULK1. The catalytically inactive DAPK3 and inhibition of Ser556 phosphorylation in ULK1 by mutation of this residue to alanine functionally impaired ULK1 complex formation, VPS34 complex activation and autophagic response during starvation, implying that Ser556 phosphorylation of ULK1 by DAPK3 is indispensable for efficient autophagy induction. We noticed that ULK1 activation by DAPK3 increased the protein levels of ATG14L and Vps34. Recent studies have reported that the degradations of ATG14L and Vsp34 by ubiquitination are inhibited during autophagy [27, 28]. Whether ULK1 controls the stability of the Vsp34 complex components is needed to investigate. The kinase activity of DAPK3 and Ser556 phosphorylation were also important for tumor suppression by DAPK3. The physical interaction between DAPK3 and ULK1 in drosophila and mammalian cells has been reported previously. DAPK3 is shown to be a substrate of ULK1 and to subsequently promote myosin II activation and ATG9 trafficking during autophagy [29]. In addition, several lines of evidence indicate the involvement of ULK1 in glucose and lipid metabolism, pentose phosphate pathway and innate immunity via non-autophagy pathway [30, 31]. Of note, we observed that expression of phospho-ULK1 alone was not a predicting factor for patient survival, whereas co-expression of DAPK3 and phospho-ULK1 predicted good prognosis of GC patients, implying that the function of ULK1 in cancer development is complicated. Although a serine/proline-rich region of ULK1 is taken as the site for many posttranslational modification events [25], further investigation is still needed to determine the DAPK3 binding site on ULK1 and to clarify the effect of DAPK3 on other functions of ULK1. Taken together, ours and other studies highlight that ULK1 and DAPK3 can activate each other through phosphorylation to exert their pro-autophagic roles in different stages of autophagy.
The first link between autophagy and tumor suppression was revealed from the findings of allelic deletion of the autophagy gene BECN1 in human breast and ovarian cancers [32]. In addition, mice deficient for essential autophagy genes such as ATG5, ATG7, or ATG4C are susceptible to tumorigenesis [33]. Several mechanisms seem to contribute to the tumor suppressive functions of autophagy. Autophagy responses decrease DNA damage and genomic instability that ultimately suppress malignant transformation [34]. Clearance of autophagy adapter protein p62/SQSTM1 by autophagy in tumor cells can attenuate the DNA damage response, pro-tumorigenic signaling pathways and tumor growth [35]. Moreover, autophagy may suppress cell proliferation and tumorigenesis by facilitating oncogene-induced senescence [36]. Recent studies show that autophagy restricts epithelial–mesenchymal transition (EMT) and aberrant cell cycle progression in cancer cells via lysosome-dependent degradation of EMT inducers and cell cycle proteins [37–39]. Our study demonstrated that the DAPK3-mediated tumor suppression depended on autophagy, because DAPK3-overexpressed cells exhibited increased tumorigenicity in vivo and in vitro when ATG7, ATG5, or ULK1 was depleted. DAPK3 expression was well correlated with autophagic capacity in clinical gastric cancer samples, and thus predicted favorable survival of patients. Consistent with our results, knockdown of DAPK3 causes enhanced tumor growth rate and reduced autophagy activity in prostate cancer [40]. However, the function of autophagy in cancer is complicated; depending on the context, autophagy can suppress or enhance tumorigenesis. The outcome may be determined by the status of key oncogenes and tumor suppressors during cancer initiation and progression [33, 41]. Oncogenic RAS activation induces autophagy to overcome metabolic stress and promote tumor cell survival [42, 43]. Nevertheless, autophagy defects also promote tumorigenesis of K-RASG12D-driven lung cancer [44]. P53 status probably influences the effects of autophagy on cancer development. Autophagy inhibition accelerates tumor formation in K-Ras-driven pancreatic tumor mouse models with P53 loss, whereas autophagy deficiency delayed Palb2-associated mammary tumorigenesis in P53 wild type mice [45, 46]. A previous study showed that DAPK3 phosphorylates P21 and MDM2, and may therefore regulate the P53 pathway indirectly by targeting P53-inducible gene products [47]. Whether the regulatory role of DAPK3 in P53 pathway influences the function of autophagy in gastric cancer needed to be investigated further.
Availability of supplementary data
Detailed methods and materials are attached.
Supplementary information
Acknowledgements
This work was supported by grants from The National Natural Science Foundation of China (81072047 and 81302079) and Science and Technology Planning Project of Guangdong Province, China (2016A030303005 and 2019A030317005). The authors thank Professor Jie Chen and Chang-hua Zhang (the First Affiliated Hospital of Sun Yat-sen University) for providing GC cells MGC803, MKN45, MKN28, and normal gastric epithelial cells GES-1. The authors also thank Run-jun He (the First Affiliated Hospital of Sun Yat-sen University) for providing technical assistance in flow cytometry.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Edited by E. Baehrecke
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Yu-Long He, Email: heyulong@mail.sysu.edu.cn.
Xin-Yuan Guan, Email: xyguan@hku.hk.
Jiong Bi, Email: bijiong@mail.sysu.edu.cn.
Supplementary information
The online version of this article (10.1038/s41418-020-00627-5) contains supplementary material, which is available to authorized users.
References
- 1.Fuchs Y, Steller H. Programmed cell death in animal development and disease. Cell. 2011;147:742–58. doi: 10.1016/j.cell.2011.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wong RS. Apoptosis in cancer: from pathogenesis to treatment. J Exp Clin Cancer Res. 2011;30:87. doi: 10.1186/1756-9966-30-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang Z, Klionsky DJ. Mammalian autophagy: core molecular machinery and signaling regulation. Curr Opin Cell Biol. 2010;22:124–31.. doi: 10.1016/j.ceb.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.White E. The role for autophagy in cancer. J Clin Investig. 2015;125:42–6. doi: 10.1172/JCI73941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zappavigna AL S, Vitale G, Merola N, Facchini S, Caraglia M. Autophagic cell death: a new frontier in cancer research. Adv Biosci Biotechnol. 2013;4:250–62. [Google Scholar]
- 6.Shiloh R, Bialik S, Kimchi A. The DAPK family: a structure-function analysis. Apoptosis: Int J Program Cell death. 2014;19:286–97. doi: 10.1007/s10495-013-0924-5. [DOI] [PubMed] [Google Scholar]
- 7.Chen HY, Lee YR, Chen RH. The functions and regulations of DAPK in cancer metastasis. Apoptosis. 2014;19:364–70. doi: 10.1007/s10495-013-0923-6. [DOI] [PubMed] [Google Scholar]
- 8.Das TP, Suman S, Papu John AM, Pal D, Edwards A, Alatassi H, et al. Activation of AKT negatively regulates the pro-apoptotic function of death-associated protein kinase 3 (DAPK3) in prostate cancer. Cancer Lett. 2016;377:134–9. doi: 10.1016/j.canlet.2016.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brognard J, Zhang YW, Puto LA, Hunter T. Cancer-associated loss-of-function mutations implicate DAPK3 as a tumor-suppressing kinase. Cancer Res. 2011;71:3152–61. doi: 10.1158/0008-5472.CAN-10-3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bi J, Lau SH, Hu L, Rao HL, Liu HB, Zhan WH, et al. Downregulation of ZIP kinase is associated with tumor invasion, metastasis and poor prognosis in gastric cancer. Int J Cancer. 2009;124:1587–93. doi: 10.1002/ijc.24164. [DOI] [PubMed] [Google Scholar]
- 11.Cai Z, Cao R, Zhang K, Xue Y, Zhang C, Zhou Y, et al. Oncogenic miR-17/20a forms a positive feed-forward loop with the p53 kinase DAPK3 to promote tumorigenesis. J Biol Chem. 2015;290:19967–75. doi: 10.1074/jbc.M115.661504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boosen M, Vetterkind S, Kubicek J, Scheidtmann KH, Illenberger S, Preuss U. Par-4 is an essential downstream target of DAP-like kinase (Dlk) in Dlk/Par-4-mediated apoptosis. Mol Biol Cell. 2009;20:4010–20. doi: 10.1091/mbc.E09-02-0173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fujiwara N, Usui T, Ohama T, Sato K. Regulation of beclin 1 protein phosphorylation and autophagy by protein phosphatase 2A (PP2A) and death-associated protein kinase 3 (DAPK3) J Biol Chem. 2016;291:10858–66. doi: 10.1074/jbc.M115.704908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schlafli AM, Berezowska S, Adams O, Langer R, Tschan MP. Reliable LC3 and p62 autophagy marker detection in formalin fixed paraffin embedded human tissue by immunohistochemistry. Eur J Histochem. 2015;59:2481. doi: 10.4081/ejh.2015.2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu Z, Baquero MT, Yang H, Yang M, Reger AS, Kim C, et al. DIRAS3 regulates the autophagosome initiation complex in dormant ovarian cancer cells. Autophagy. 2014;10:1071–92. doi: 10.4161/auto.28577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Usui T, Okada M, Yamawaki H. Zipper interacting protein kinase (ZIPK): function and signaling. Apoptosis. 2014;19:387–91. doi: 10.1007/s10495-013-0934-3. [DOI] [PubMed] [Google Scholar]
- 17.Russell RC, Tian Y, Yuan H, Park HW, Chang YY, Kim J, et al. ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nat cell Biol. 2013;15:741–50. doi: 10.1038/ncb2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tian W, Li W, Chen Y, Yan Z, Huang X, Zhuang H, et al. Phosphorylation of ULK1 by AMPK regulates translocation of ULK1 to mitochondria and mitophagy. FEBS Lett. 2015;589:1847–54. doi: 10.1016/j.febslet.2015.05.020. [DOI] [PubMed] [Google Scholar]
- 19.Egan DF, Shackelford DB, Mihaylova MM, Gelino S, Kohnz RA, Mair W, et al. Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science. 2011;331:456–61. doi: 10.1126/science.1196371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13:132–41. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zachari M, Ganley IG. The mammalian ULK1 complex and autophagy initiation. Essays Biochem. 2017;61:585–96. doi: 10.1042/EBC20170021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Park JM, Jung CH, Seo M, Otto NM, Grunwald D, Kim KH, et al. The ULK1 complex mediates MTORC1 signaling to the autophagy initiation machinery via binding and phosphorylating ATG14. Autophagy. 2016;12:547–64. doi: 10.1080/15548627.2016.1140293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shani G, Marash L, Gozuacik D, Bialik S, Teitelbaum L, Shohat G, et al. Death-associated protein kinase phosphorylates ZIP kinase, forming a unique kinase hierarchy to activate its cell death functions. Mol Cell Biol. 2004;24:8611–26. doi: 10.1128/MCB.24.19.8611-8626.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xie Y, Kang R, Sun X, Zhong M, Huang J, Klionsky DJ, et al. Posttranslational modification of autophagy-related proteins in macroautophagy. Autophagy. 2015;11:28–45. doi: 10.4161/15548627.2014.984267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wong PM, Puente C, Ganley IG, Jiang X. The ULK1 complex: sensing nutrient signals for autophagy activation. Autophagy. 2013;9:124–37. doi: 10.4161/auto.23323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mack HI, Zheng B, Asara JM, Thomas SM. AMPK-dependent phosphorylation of ULK1 regulates ATG9 localization. Autophagy. 2012;8:1197–214. doi: 10.4161/auto.20586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang T, Dong K, Liang W, Xu D, Xia H, Geng J, et al. G-protein-coupled receptors regulate autophagy by ZBTB16-mediated ubiquitination and proteasomal degradation of Atg14L. Elife. 2015;4:e06734. doi: 10.7554/eLife.06734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xiao J, Zhang T, Xu D, Wang H, Cai Y, Jin T, et al. FBXL20-mediated Vps34 ubiquitination as a p53 controlled checkpoint in regulating autophagy and receptor degradation. Genes Dev. 2015;29:184–96. doi: 10.1101/gad.252528.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tang HW, Wang YB, Wang SL, Wu MH, Lin SY, Chen GC. Atg1-mediated myosin II activation regulates autophagosome formation during starvation-induced autophagy. EMBO J. 2011;30:636–51. doi: 10.1038/emboj.2010.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ro SH, Jung CH, Hahn WS, Xu X, Kim YM, Yun YS, et al. Distinct functions of Ulk1 and Ulk2 in the regulation of lipid metabolism in adipocytes. Autophagy. 2013;9:2103–14. doi: 10.4161/auto.26563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Konno H, Konno K, Barber GN. Cyclic dinucleotides trigger ULK1 (ATG1) phosphorylation of STING to prevent sustained innate immune signaling. Cell. 2013;155:688–98. doi: 10.1016/j.cell.2013.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, et al. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402:672–6. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- 33.Galluzzi L, Pietrocola F, Bravo-San Pedro JM, Amaravadi RK, Baehrecke EH, Cecconi F, et al. Autophagy in malignant transformation and cancer progression. EMBO J. 2015;34:856–80. doi: 10.15252/embj.201490784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lorin S, Hamai A, Mehrpour M, Codogno P. Autophagy regulation and its role in cancer. Semin Cancer Biol. 2013;23:361–79. doi: 10.1016/j.semcancer.2013.06.007. [DOI] [PubMed] [Google Scholar]
- 35.Mathew R, Karp CM, Beaudoin B, Vuong N, Chen G, Chen HY, et al. Autophagy suppresses tumorigenesis through elimination of p62. Cell. 2009;137:1062–75. doi: 10.1016/j.cell.2009.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu H, He Z, Simon HU. Autophagy suppresses melanoma tumorigenesis by inducing senescence. Autophagy. 2014;10:372–3. doi: 10.4161/auto.27163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lv Q, Hua F, Hu ZW. DEDD, a novel tumor repressor, reverses epithelial-mesenchymal transition by activating selective autophagy. Autophagy. 2012;8:1675–6. doi: 10.4161/auto.21438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang Y, Xiong H, Liu D, Hill C, Ertay A, Li J, et al. Autophagy inhibition specifically promotes epithelial-mesenchymal transition and invasion in RAS-mutated cancer cells. Autophagy. 2019;15:886–99.. doi: 10.1080/15548627.2019.1569912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zheng K, He Z, Kitazato K, Wang Y. Selective autophagy regulates cell cycle in cancer therapy. Theranostics. 2019;9:104–25.. doi: 10.7150/thno.30308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Puto LA, Brognard J, Hunter T. Transcriptional repressor DAXX promotes prostate cancer tumorigenicity via suppression of autophagy. J Biol Chem. 2015;290:15406–20. doi: 10.1074/jbc.M115.658765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cicchini M, Karantza V, Xia B. Molecular pathways: autophagy in cancer—a matter of timing and context. Clin Cancer Res. 2015;21:498–504. doi: 10.1158/1078-0432.CCR-13-2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guo JY, Chen HY, Mathew R, Fan J, Strohecker AM, Karsli-Uzunbas G, et al. Activated Ras requires autophagy to maintain oxidative metabolism and tumorigenesis. Genes Dev. 2011;25:460–70. doi: 10.1101/gad.2016311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lock R, Roy S, Kenific CM, Su JS, Salas E, Ronen SM, et al. Autophagy facilitates glycolysis during Ras-mediated oncogenic transformation. Mol Biol Cell. 2011;22:165–78. doi: 10.1091/mbc.E10-06-0500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guo JY, Karsli-Uzunbas G, Mathew R, Aisner SC, Kamphorst JJ, Strohecker AM, et al. Autophagy suppresses progression of K-ras-induced lung tumors to oncocytomas and maintains lipid homeostasis. Genes Dev. 2013;27:1447–61. doi: 10.1101/gad.219642.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rosenfeldt MT, O’Prey J, Morton JP, Nixon C, MacKay G, Mrowinska A, et al. p53 status determines the role of autophagy in pancreatic tumour development. Nature. 2013;504:296–300. doi: 10.1038/nature12865. [DOI] [PubMed] [Google Scholar]
- 46.Huo Y, Cai H, Teplova I, Bowman-Colin C, Chen G, Price S, et al. Autophagy opposes p53-mediated tumor barrier to facilitate tumorigenesis in a model of PALB2-associated hereditary breast cancer. Cancer Discov. 2013;3:894–907. doi: 10.1158/2159-8290.CD-13-0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Burch LR, Scott M, Pohler E, Meek D, Hupp T. Phage-peptide display identifies the interferon-responsive, death-activated protein kinase family as a novel modifier of MDM2 and p21WAF1. J Mol Biol. 2004;337:115–28. doi: 10.1016/j.jmb.2003.10.081. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.