Abstract
While the capacity to regenerate tissues or limbs is limited in mammals, including humans, axolotls are able to regrow entire limbs and major organs after incurring a wound. The wound blastema has been extensively studied in limb regeneration. However, due to the inadequate characterization of ECM and cell subpopulations involved in the regeneration process, the discovery of the key drivers for human limb regeneration remains unknown. In this study, we applied large-scale single-cell RNA sequencing to classify cells throughout the adult axolotl limb regeneration process, uncovering a novel regeneration-specific mitochondria-related cluster supporting regeneration through energy providing and the ECM secretion (COL2+) cluster contributing to regeneration through cell–cell interactions signals. We also discovered the dedifferentiation and re-differentiation of the COL1+/COL2+ cellular subpopulation and exposed a COL2-mitochondria subcluster supporting the musculoskeletal system regeneration. On the basis of these findings, we reconstructed the dynamic single-cell transcriptome of adult axolotl limb regenerative process, and identified the novel regenerative mitochondria-related musculoskeletal populations, which yielded deeper insights into the crucial interactions between cell clusters within the regenerative microenvironment.
Subject terms: Development, Gene expression
Introduction
The ability to fully regenerate damaged and lost tissues would be of tremendous benefit for clinical medicine; however, the limited regeneration capacity of many human tissues, including limbs, presents a formidable clinical hurdle [1]. The axolotl is a widely used model organism due to its powerful ability to reconstitute a fully functional limb after amputation throughout adulthood. After amputation, a blastema forms which is defined as a progenitor cell pool that is capable of regenerating limbs by autonomously patterning itself to precisely replace the lost structures [2]. Lineage tracing studies illustrated that blastema cells originate from the mesodermal tissues including dermal fibroblasts, Schwann cells, and myogenic cells [3]. Meanwhile, many studies based on transcriptomes and proteomic analysis provided much information concerning the key molecular mechanisms in blastema formation [4–6]. The oncogenes were reported to burst in the early stage [7] and two crucial genes cirbp and kazald1 were discovered upregulated in blastemas [8]. Despite this accumulated knowledge, the cell populations and their interactions that contribute to blastema constitution still remain unclear.
Traditional sequencing methods using whole tissues, mask potential cell heterogeneity within the tissue and may add bias to be understanding of the cellular hierarchy. Single-cell RNA-sequencing (scRNA-seq) analysis allows unbiased and high-throughput analysis of gene expression profiles from an individual cell level in order to investigate cell population heterogeneity [9]. ScRNA-seq reveals novel cell types and reconstructs lineage hierarchies of tissue types such as human pluripotent stem cells, blood dendritic cells, neurons, and uterus epithelium [10–14]. Recently, the scRNA-seq was performed on axolotl to uncover the connective tissue (CT) subpopulations with their characters and mesenchymal cellular diversity during regeneration [15–17]. However, the mechanism of regeneration-specific cell populations still need further exploration.
In this study, scRNA-seq was performed on blastema cells collected limb tissues at four regenerative stages of adult axolotls. The transcriptome profiling displayed a comprehensive axolotl regeneration cell atlas with the reconstructed cell connectivity map and regenerative pseudo-temporal trajectory. We found the dynamics of the regenerative musculoskeletal system cells and identified a novel mitochondria-related cell population which plays a vital energy supporting character in regeneration. All of these provide a greater understanding of the limb regeneration process.
Results
Dynamic patterns of gene expression along axolotl limbs regeneration time course
To evaluate the heterogeneity of the regenerating limb at an individual cellular level, we performed single-cell RNA-seq over a time course of adult axolotl limb regeneration. Samples were collected from the adult axolotl regenerated distal limb tips at 3, 7, and 21 days post amputation (3, 7, 21 dpa) which represent the early, middle, and late stage of limb regeneration respectively, as well as the unamputated forelimbs (0 dpa) (Figs. 1A and S1A). These samples were isolated as single cells for sequencing using Fluidigm C1 system with HT IFCs (Fig. 1A) which could detect an average of 5000 genes per cell. Our sequencing data were mapped with the axolotl transcriptome assemblies [7, 8, 18]. After data quality control and removal of multiple cell samples, 938 cells were selected for further analysis (Fig. S1B, C). In order to understand the transcriptome changes in our data during regeneration, differentially expressed (DE) genes and gene set enrichment analysis (GSEA) analysis were performed. The results revealed the marker genes of four time points (Fig. S1D–F; Table S1), with GSEA results revealing that 0 dpa cells were related to cartilage morphogenesis, 3 dpa cells were related to functions involving cell proliferation, and cellular immune response, while at 21 dpa, the blastema displayed the functions of endoderm development and limb morphogenesis (Fig. S2A, B). These results demonstrated the integral dynamic gene patterns along regeneration and proved the reliability of our data.
Fig. 1. Single-cell profiling revealed cell heterogeneity in axolotl limb tissues from different regeneration stage.
A Schematic diagram of the blastemal time course experiment. The red box on limbs indicated the amputation location on axolotls’ limbs (upper left). The pictures show that the amputated tissues were trypsinized to yield a single-cell suspension (upper middle) and trapped using the Fluidigm C1 auto prepare system (C1 high-throughput integrated fluidics circuits (HT IFCs) and HiSeq systems) for the downstream single-cell mRNA-seq experiment. B T-distributed stochastic neighbor embedding (t-SNE) visualizations of all cell clusters identified using the computational pipeline. C T- SNE visualizations of 4 days post amputation identified using the computational pipeline. D Heatmap of top 20 differentiated gene of seven clusters. E T-SNE distribution of cells from each sampling stage. Red dots represent the cells from the corresponding time point, and gray dots represent all the cells from four stages. F The proportion of cells at each time point in each cluster. G Violin plots of marker genes from seven cluster.
Cell heterogeneity in blastemas of different amputation time points by single-cell profiling
General DE analysis probably masks the cell heterogeneity within each time point. To identify cell types of axolotl limb regeneration, we used Seurat package [19] implemented in R to perform t-distributed stochastic neighbor embedding (t-SNE) analysis, which led to consistent clustering of different cell types into distinct regions (Fig. 1B, C) with each cluster possessing a unique set of signature genes (Fig. 1D, G; Table S2). Each group has a different cell quantity proportion at different time points (Figs. 1E, F and S3A–C). GO analysis was performed on the upregulated genes of each cluster (Fig. S3D). Based on these, the seven clusters (C0–C6) were identified as the general cluster, mitochondria cluster (e.g., CYTB, ATP6), CT cluster (e.g., FBN2, MDK), chondrocyte cluster (e.g., COL2A1, MATN1), inflammation cluster (e.g., CCL5, PTPRC), cycling cluster (e.g., SLC2A8, PIF1), and apical epithelium cap (AEC) cluster (e.g., KRT12, KRT8) (Fig. 1B, F). The mitochondria cluster was shown in high proportion in cells from regenerating stages (3, 7, and 21 dpa) when compared to cells from 0 dpa (Figs. 1D and S3A–C). In order to gain more knowledge on this particular cluster, we removed mitochondria-associated genes and re-evaluated the differences, because mitochondrial genes were significantly expressed, possibly masking other highly expressed genes. Interestingly, the GO functions were mainly related to the regeneration and development of neuron and stem cells as well as regulation of programmed cell death (Fig. S3d). These results were consistent with recent studies demonstrating that mitochondrial activity influences the cell differentiation and response to stress [20].
Immediately after amputation, a wound epithelium was formed by migrating epithelial cells [21]; it would interact with the stump tissues to form an AEC which is essential in creating a regenerative environment for blastema [22, 23]. The AEC cluster of cells highly expressed epidermis marker genes KRT12 and KRT8 (Fig. 1F) which represented the AEC formed by the migrating epithelial cells. Moreover, the GO analysis revealed that this cluster was related to not only epidermis development but also Schwann cell and stem cell proliferation (Fig. S3F, Table S9), which indicates the traits of the AEC cluster including inducing the differentiation in the underlying stump tissue, attracting cells accumulating below and being in response to nerve signals [24].
To confirm that the t-SNE results were capable of corroborating the gene expression sites in axolotl regenerative limbs, we detected the marker genes of each cluster by immunofluorescence staining over a time course of limb regeneration. MDK+ cells, representing the CT cluster, mainly distributed at the basal layers at all four time points (Fig. S4A). PTPRC+ cells mainly distributed at the stroma under the AEC at 3, 7, and 21 dpa, as well as subcutaneous tissue at 0 dpa, which represented the location of the inflammation cluster (Fig. S4B). PIF1, as the marker of the cycling cluster, was mainly detected at the stroma in close proximity to the AEC at 3, 7, and 21 dpa, and the subcutaneous tissue at 0 dpa (Fig. S4C). KRT8+ cells mainly located at AEC at 3, 7, and 21 dpa, and the epidermis at 0 dpa (Fig. S4D).
The microenvironment regulation among seven cell clusters during the regeneration process
Different cell clusters that co-existed within the same environment, physically surrounded each other. The interactions among these cell clusters may also play a part in regulating cell state and determining cell fate. Thus, we next constructed a connectivity map of the seven clusters for each corresponding time point respectively in order to reveal the potential interactions of different clusters by using known ligand–receptor pairs [25, 26]. 928 ligand–receptor pairs were detected among seven clusters in our connectivity map (Fig. 2A).
Fig. 2. Connectivity maps revealed the microenvironment regulation of seven cell clusters during regeneration process.
A Circle plot depicts the total number of ligand–receptor interactions between each subcluster in each time point post amputation. B Top receptor–ligand expression between chondrocyte cluster and AEC cluster. C GO terms of top receptor–ligand genes between chondrocyte cluster and AEC cluster.
The connectivity map revealed that the C3 (chondrocyte cluster) had the most ligands to C6 (AEC cluster) when compared to other clusters in all four temporal groups (Figs. 2A and S5C). Further analysis of the ligand–receptor pairs between C3 and C6 showed abundant extracellular matrix (COL1A1, COL3A1, MATN1), and growth factor (ITGB1, EGFR, BMPR2, VEGF) interactions (Fig. 2B) with the GO terms positive regulation of cell migration and adhesion, tissue morphogenesis, epithelial cell proliferation, and skeletal system development (Figs. 2C and S5A). These results imply that through releasing ECM to generate cell-to-cell interaction, cartilage promotes the migration and morphogenesis of AEC which contributes to the formation of blastema. The C4 (inflammation cluster) had the strongest connectivity at 3 dpa which indicated the vital role that early inflammation has in releasing cytokines to induce cell differentiation and triggering the blastema formation which consistent with the previous finding [27]. Overall, the connectivity map revealed abundant interaction information among cell clusters which can help provided orientation for further researches.
The mitochondria cluster supported cell division in regenerative limbs
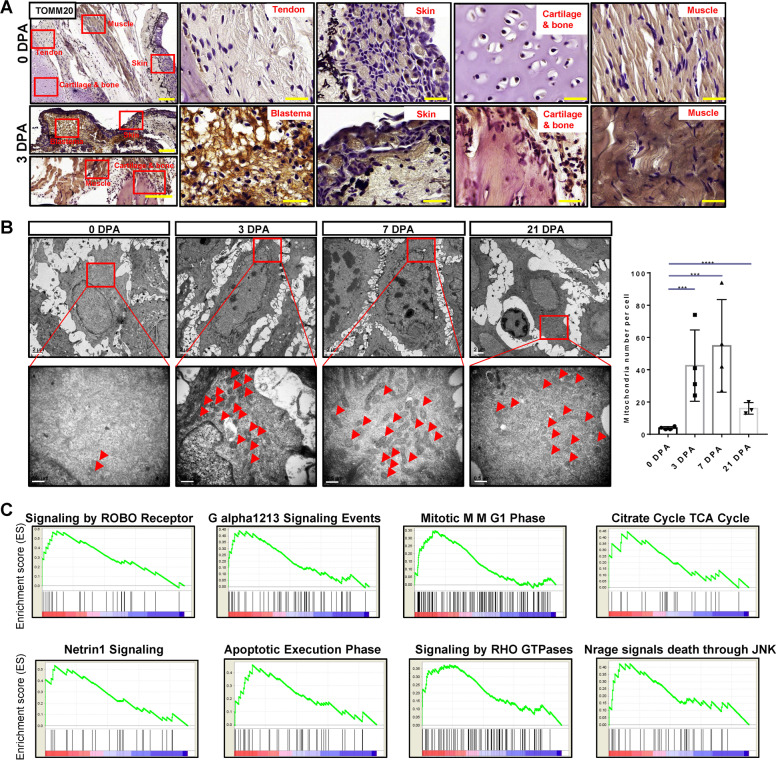
To confirm the mitochondria activity in the regeneration process, we detected the mitochondrion-marker TOMM20 using immunochemistry staining. The TOMM20 had the highest expression in blastema of the 3 dpa tissues. We also found that TOMM20 had a higher expression level in the skin and muscle of the 3 dpa tissues compared to the 0 dpa tissues (Fig. 3A). Additionally, TEM results of the epidermis (Fig. S5D) indicated that the average number of mitochondria per cell in regenerative tissues was significantly higher than in normal limb tissues (Fig. 3B). These results indicated that the higher mitochondria activities in the mitochondria cluster play a key role in the regeneration process. To recognize the functions of the mitochondria cluster, GSEA analysis was performed and revealed that the citrate cycle TCA cycle pathway was upregulated in the mitochondria cluster, which points at the reliability of the identity of the mitochondria cluster. The cell division-associated signaling pathways including signaling by ROBO receptor, G alpha1213 signaling events, apoptotic execution phase, the energy metabolism-associated signalings such as signaling by RHO GTPases, and cell death regulation-associated pathways including Nrage signals death through JNK were upregulated in the mitochondria cluster (Fig. 3C). These indicated that the mitochondria cluster supported cell division and regulated cell death in regenerative tissues.
Fig. 3. The mitochondria cluster specifically existed in regenerative limbs.
A Immunostaining of TOMM20 in tendon, skin, cartilage, and muscle of 0 dpa tissues, and blastema, skin, cartilage and muscle of 3 dpa tissues. Scale bar, 50 μm. B TEM of regenerative samples at each time points. Scale bar, upper 2 μm, lower 0.5 μm. C The top 8 signaling pathways in mitochondria cluster compared with the rest clusters by GSEA signaling pathway analysis.
The pseudo-temporal ordering of individual cells from all clusters
Most cell-state transitions, including development and regeneration, are characterized by flows of gene expression changes. However, a heterogeneous tissue, at the same time point of regeneration, also contained cells at different stages during development progression. To uncover the differentiation relationship of cells during the regeneration process, we used MONOCLE algorithm that applies machine learning to single-cell transcriptome sequencing data [28, 29]. This method makes use of gene expression variations to define a pseudo-temporal biological process using an unsupervised procedure, and order cells along a reconstructed ‘trajectory’ of regeneration called ‘pseudo-temporal ordering’.
Monocle compresses all single-cell transcription datasets in a multidimensional space into a two-dimension space for projection. In our data, cells within this two-dimension space (represented by dots) are ordered in the “pseudo-temporal ordering”, with a line through all dots/cells showing a path of the developmental trajectory/timeline. The trajectory of seven clusters displayed a clear large central branch, with few minor branches emanating from it (Fig. 4A–C). In this trajectory, the cycling cluster showed an inchoate state while the chondrocyte cluster was at the end of the central branch. The minor branch 1 and 4 mainly containing AEC cells; while the minor branch 2 and 3 mainly contained the mitochondria cluster cells (Figs. 4A and S6A). The arrangement of cells on the pseudotime trajectory was almost consistent with the true time except a fraction of cells from 0 dpa located in the primitive stage of the trajectory, suggesting that some original stem cells may still exist in unamputated limb tissues (Figs. 4B and S6B). To investigate the cell alterations in the musculoskeletal system in regeneration, we termed the marker genes, including COL2A1 for chondrocytes, ACAN (Aggrecan) for extracellular matrix in cartilaginous tissue, COL1A1/A2 for CTs including bone, dermis and tendon, THBS4 for tendon cells and POSTN (Periostin) for osteoblasts or periosteum (Figs. 4D and S6C). Results showed that they all slightly fluctuated in the early period and then increased remarkably during the middle and later periods. We then investigated the expressional changes of the BMP, FGF and, TGF-β signaling pathway members along the pseudotime (Figs. 4D and S6C); results showed that members of FGF and BMP families were expressed with a rising trend. In contrast, TGF-β signaling pathway members FOSB and ZFYVE16 increased at the later stage, while JUNB was stably expressed with small fluctuations.
Fig. 4. Pseudo-temporal ordering of individual cells.
A Pseudotime ordering of single cells using 2D PCA. Each data point represents a single cell colored by Fig. 3A clusters age collected. B Pseudotime ordering of single cells using 2D PCA. Each data point represents a single cell colored by postamputation time point collected. C Pseudotime ordering of single cells using 2D PCA. Each data point represents a single cell colored Pseudotime. D Expression profiles of Tendon and skeleton system marker genes, different cell signaling pathway components, and blastema marker genes along pseudotime ordered by seven clusters. E Heatmap shows the gene expression dynamics during limb regeneration (left). Genes (row) are clustered and cells (column) are ordered according to the pseudotime development. Gene clusters A, B, and C were selected for GO analysis (right).
To determine cellular states within regeneration, we performed k-means clustering on all genes, revealed three gene clusters according to the expressing trend of genes, hereafter labeled cluster A to C with GO analysis, representing pseudo-temporal early, late, and mid-late stages, respectively (Fig. 4E). The results revealed that cell proliferation was mainly at the early stage with immune response at mid-late stage, while the maturation of cell differentiation and determination occurred during the late stage.
A continuous transcriptional transition of the musculoskeletal system population in response to the stimuli
The clustering results showed that the COL2A1+ cells were included not only in C3 but also in C2 (with a lower expression compared to C3) (Fig. S2F), which indicated that the COL2A1+ cells with different phenotypes persisted throughout the whole regeneration process. To determine the variances of gene expression in the COL2A1+ cells and other musculoskeletal system cells in different regeneration stage, we performed t-SNE on each time-point cells (Fig. 5A). We attempted to identify the marker genes, COL2A1, ACAN, COL1A1, COL1A2, THBS4, and POSTN (Figs. 5B–E and S7D). COL1A1/2+ (including COL1A1 and COL1A2) and COL2A1+ cells had different expression patterns during the regeneration process as reflected by t-SNE plots (Fig. 5B–E). COL1A1/2+ cells were in a high proportion (COL1A1 58%, COL1A2 72%) at 0 dpa, then the number decreased at 3 dpa (COL1A1 20%, COL1A2 21%), and re-increased (COL1A1 47%, COL1A2 58%) at 7 dpa; the fluctuation then stopped and the COL1A1/2+ cell number was maintained (COL1A1 55%, COL1A2 70%) at 21 dpa (Figs. 5B–E and S7B, C). By contrast, the proportion of COL2A1+ cells was high (72%) at 0 dpa, and then sharply declined (9%) at 3 dpa and (2%) at 7 dpa, and re-increased at (43%) 21 dpa (Figs. 5B–E and S7B, C). Immunostaining analysis also showed that COL1 had a higher expression level at 21 dpa in the skeleton, muscle, skin, and blastema (Fig. S8), further supporting a mechanism whereby dedifferentiation of the cells.
Fig. 5. Identification of different lineage cells within 0–21 dpa axolotl limbs singe-cell RNA-seq data.
A t-SNE visualizations of cells at each postamputation stage. Colors and associated numbers represent individual clusters. t-SNE visualization of 0 dpa (B), 3 dpa (C), 7 dpa (D), and 21 dpa (E) scRNA-seq data overlaid with expression of the cell-type-specific markers COL2A1, COL1A2, COL1A1, THBS4, POSTN, and ACAN. Cells are color-coded according to expression level, ranging from low detected to the highest detected levels (gray, yellow, orange, and red). Cluster numbers are as shown in (A).
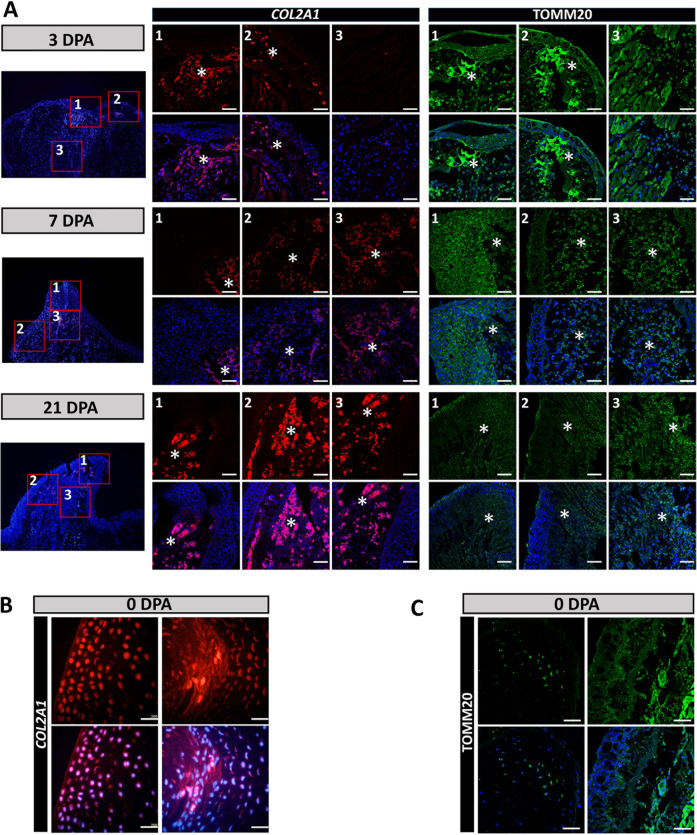
We next sought to confirm the existence of the COL2A1 cell population (Fig. 5). We performed RNA FISH of COL2A1 (Fig. 7, A middle, and B). The COL2A1+ cells mainly located at the top of blastema under the AEC and close to AEC at all regeneration time points. The COL2A1+ cells increased in 21 dpa regeneration, which confirmed the dynamic COL2A1 expression profiles generated from the single-cell RNA-seq data (Fig. 5).
Fig. 7. Location of COL2-mito subcluster during the regeneration process.
A Location of COL2-mito subcluster at 3, 7, and 21 dpa. The left part is the entire sample staining by DAPI. The middle part is representative RNA in situ hybridization probing for COL2A1 of blastemas from three locations at 3, 7, and 21 dpa. The right part is the immunofluorescence of TOMM20 antibody of blastemas from three locations at 3, 7, and 21 dpa. The middle and right parts labeled by number 1, 2, 3 are corresponding to the red boxes labeled by 1, 2, 3 at the left part, and 1, 2, 3 respectively represent the top of blastema, AEC with subcutaneous tissues, and blastema center. The positive positions are labeled by asterisks. B Representative RNA in situ hybridization probing for COL2A1 in 0 dpa samples. C The immunofluorescence of TOMM20 on 0 dpa samples. Scale bars, 100 μm (A, B, and C).
Meanwhile, the proportion of POSTN+ cells (44, 10, 22, and 57%) and THBS4+ cells (46, 13, 12, 23%) were consistent with the expression trend of COL1A1/2+ cells (Figs. 5B–E and S7B, C), indicating that the musculoskeletal cells may regenerate from one cell source. Conversely, ACAN, representing the cartilaginous extracellular matrix, showed a different expression profile in that it was highly expressed at 0 dpa (34%) then gradually decreased during the regeneration process until 21 dpa (4% at 3 dpa, 1% at 7 dpa, 1% at 21 dpa) (Fig. 5B–E and S7B, C).
A novel subcluster in COL2+ cells
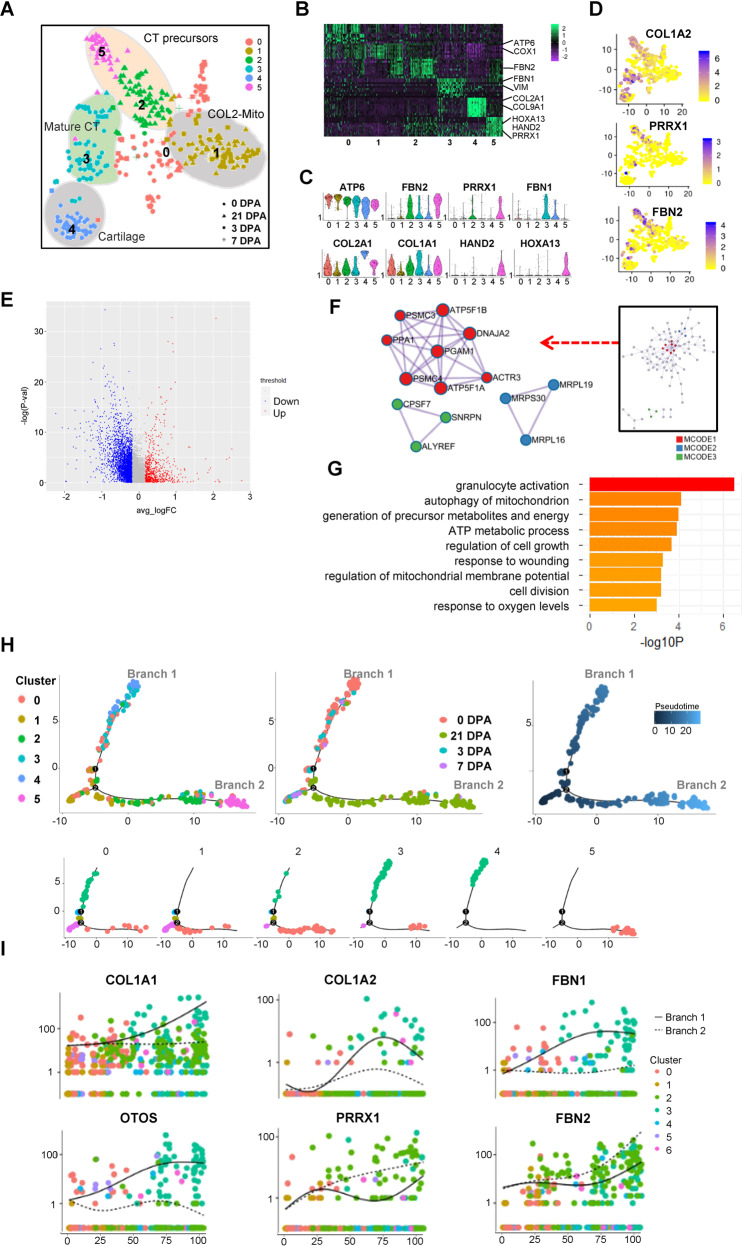
Given these results, we then extracted the COL2+ cells and divided them into six subclusters with unsupervised Seurat clustering methods (Fig. 6A). The subclusters 0 and 1 both highly expressed mitochondrial genes, such as ATP6 and COX1 (Fig. 6B, C). Notably, the subcluster 1 specifically expressed mitochondrion genes and was fully contained in the mitochondrion cluster in Fig. 2. The subclusters 2, 3, and 5 were CT lineage cells with highly expressed COL1A1. Distinctly, the subcluster 3 was labeled mature CT with the marker gene FBN1, while the subclusters 2 and 5 were labeled with the marker genes PPRX1 and FBN2 (Fig. 6B–D), representing the CT precursors. Meanwhile, in the COL2-CT precursors subpopulation, the subcluster 5 highly expressed axolotl upper limb development key gene HOXA13 and HAND2 (Fig. 6B, C) and showed a strong cell cycling process (Fig. S9A–C), which represented an actively proliferating COL2+ CT regenerative key subcluster.
Fig. 6. ScRNA-seq analysis revealed COL2-mito subcluster in COL2A1+ cells during the regeneration process.
A T-distributed stochastic neighbor embedding (t-SNE) visualizations of COL2A1+ cells subclusters identified using the computational pipeline. B Heatmap of top 10 differentiated gene of six subclusters. C Violin plots of subclusters’ marker genes. D Feature Plot of CT marker genes’ expression in individual cells. E Volcano plot of the differentiation gene expression of COL2-mito subcluster and the other subclusters. F The protein–protein interaction network analyzed by the molecular complex detection (MCODE) algorithm. G −Log p values for each GO term are shown as histograms. Bars color-coded according to −Log p value, ranging from low detected to the highest detected levels (yellow and red). H Pseudotime ordering of single cells using 2D PCA. Each data point represents a single cell colored by cluster in A (left), ampution time points (middle), and pseudotime (right). I Expression profiles of skeleton system marker genes, different cell signaling pathway components, and blastema marker genes along pseudotime ordered byseven clusters in (A).
As mitochondria showed the regenerative specificity, we then examined whether the COL2-mito subcluster had a regenerative supporting function for the musculoskeletal system. The molecular complex detection (MCODE) algorithm was applied on the marker genes to identify densely connected network components in the protein–protein interaction network (Fig. 6E, F). In the MCODE network, mitochondrial ATP synthase subunits proteins encoded genes (ATP5F1A, ATP5F1B, PSMC3 et al.) were strongly connected with each other. Meanwhile, the mitochondrial ribosomal proteins encoded genes (MRPL16/19/30) and mRNA splicing pathway-associated genes (CPSF7, SNRPN, and ALYREF) were separated connected together, which demonstrated increased mitochondria and high translation activity. GO analysis indicated that this subcluster performed the energy metabolite associated functions such as response to oxygen levels, and ATP metabolic process (Fig. 6G). Moreover, this subcluster showed a response to wounding, including granulocyte activation and cell division. The pseudo-temporal trajectory of COL2+ cells originated by the mitochondria subcluster and bifurcated into two big branches (Fig. 6H); The CT precursors subclusters (subcluster 2 and 5) were mainly located in branch 1, and the mature CT subclusters and cartilage subclusters (subcluster 3 and 4) mainly composed branch 2 (Fig. 6H). In this trajectory, COL1A1/2, FBN1, and OTOS were only increasingly expressed in branch 1, while FBN2 expression was higher in branch 2 than branch 1 (Fig. 6I).
To validate the COL2-mito subcluster in the blastema, we simultaneously performed RNA FISH probing for COL2A1 and immunofluorescence of TOMM20 to identify their location within the regeneration (Fig. 7). TOMM20 has a highest expression in 3 dpa blastema and mainly located at AEC, which is corresponding to previous results (Figs. 1B, 3A, B and 7A, right). At the top of the blastema, the tissue under AEC, and the blastema center, COL2A1+ cells and TOMM20+ cells all have overlapping parts, which indicated the COL2-mito subcluster location in blastema (Fig. 7A). COL2A1+ cells are also detected in the cartilage of 0 dpa samples overlapping a spot of TOMM20+ cells, signifying the location of the 0 dpa part in the COL2-mito subcluster (Fig. 7B, C).
Discussion
In this study, we applied scRNA-seq analysis and provided single-cell transcriptomics that examined the gene expression level of individual cells in axolotl limbs from the early to late stages of regeneration. We computationally identified seven cell clusters in unamputated and regenerated limbs, including a chondrocyte cluster contributing to blastema forming by ECM, and a novel mitochondria cluster supporting the regeneration process. We defined the dedifferentiation and re-appearing process of COL1A1/2+ and COL2A1+ populations after amputation, and described how the COL2-mito subcluster supported the musculoskeletal system regeneration by providing energy.
Chondrocytes as the origin of bone cells play an important role in skeletal system development [30]. Previous studies on axolotl limb regeneration model showed that the majority cartilagenous skeleton contributes to cartilage regeneration after amputation [3]. In our research, the chondrocyte cluster, through secreting ECM, displayed a strong ligand–receptor interaction with the AEC clusters, contributing to regeneration. The ECM proteins are known to contribute to the structural formation of the matrix during tissue repair and development in mammal models [31]. Meanwhile, ECM proteins also were displayed to regulate heart regeneration in zebrafish model [32]. In view that the role of ECM during axolotl limb regeneration was comparatively rarely explored, our results demonstrated that the ECM act as a novel medium for cartilage contributing to limb regeneration.
The importance of COL2 in regeneration is now more evident. In our results, COL1+/COL2+ cells (0 dpa) expressed decreased COL1A1/A2 and COL2A1 at the early regeneration stage, and re-increased COL1A1/A2 at 7 dpa with a delayed re-increased COL2A1 expression. Previous studies have demonstrated that chondrocyte dedifferentiation is characterized by the upregulation of type I collagen and downregulation of type II collagen, changing chondrocytes into fibroblast-like cells in vitro [33]. Combining the previous studies that the expression of the COL1A1/2 is earlier than COL2A1 during limb regeneration [34, 35] and the chondrocytes would dedifferentiated during digits regeneration [36], the downregulation of COL2A1 in COL1+/COL2+ cells may represent the dedifferentiation and trans-differentiation during regeneration that transformed into a primitive state, as the origin of the musculoskeletal system. Our data provided novel evidence for the dedifferentiation and re-differentiation process of the musculoskeletal system from a single-cell level, and may provide valuable clues in the understanding of musculoskeletal system regeneration.
Indeed, previous studies on regeneration have revealed the links between energy metabolism and transcription rate [37, 38], in addition to clarifying the role of the mitochondria in as well as their roles on the modulation of stem cell differentiation, niche adhesion, and teratoma formation [20, 39]. Meanwhile, the distinct mitochondrial profile subpopulation contained in tumor cells was termed as the cancer stem cells [40, 41]. So that, the mitochondrial population maybe not only a cell differentiation state, but also an independent cell population that exists during regeneration. In regeneration, some types of cells act as niche support for stem cells such as nerves [42]. In our results, the mitochondria cluster as a specific regenerative cell cluster demonstrated a high translation rate and stem cell functions. This indicated that axolotl limb regeneration relied on a particular cell cluster with a quantity of mitochondria to offer enough energy intermediated for stress reaction, cell differentiation, and tissue remodeling and the COL2-mitochondria subpopulation perform such functions in musculoskeletal system regeneration. Furthermore, the chondrocyte dedifferentiation is known to respond to aspects of the microenvironment, such as oxidative stress [43]. These data demonstrated that axolotl limbs could generate population cells with the large amount of mitochondria after amputation, not only to deliver energy for a plethora of cellular activities but also to mitigate the oxidative stress which arises in successfully regenerated limbs. It is possible that these dynamics may be relevant for the clinical treatment of tissue regeneration.
The recent scRNA-seq researches on axolotl exposed the CT clusters with their characters, mesenchymal cellular diversity, and epithelial to mesenchymal transition during regeneration [15–17]. The identification and dynamic analysis results of the CT cluster and epidermis cluster from Gerber et al. and Leigh et al. were consistent with our analysis of CT cluster and AEC cluster in our research, thus further supporting the reproducibility of our experimental procedure.
In addition to the axolotl limb model for regenerative researches, several mammalian regeneration models have also been studied [44, 45]. In axolotl regeneration, the immune responses to amputation and some signaling pathways are similar to the mouse digit tip model [46, 47]. In contrast, the axolotl relies on the cartilaginous skeleton for regeneration rather than direct ossification as in mouse digit tip regeneration [48]. Additionally, the importance of mitochondria in stem cells and liver regeneration in mammals [49, 50] is similar to the function of our mitochondria cluster in axolotl limb regeneration—an until-now undiscovered parallel which may offer further elucidation into energy metabolism in regeneration. Our results revealed novel insights into the regeneration biology which occurs in the axolotl and other species, hopefully directing future functional studies.
Material and methods
Axolotl husbandry, surgery, and cell isolation
All animal experiments were performed with the approval of the Zhejiang University Ethics Committee (ZJU11602). We established an axolotl colony (Ambystoma mexicanum) with the axolotls obtained from the Guilin Salamander Breeding Base. The adult axolotls were randomly selected for experiments. For all surgical procedures, the animals were anesthetized with 0.03% ethyl 3-aminobenzoate until they were unresponsive to a tail pinch stimulus. We amputated juvenile axolotl right forelimbs at the upper limb level. The 0 dpa cells were harvested from 1 mm of adult upper forelimbs. The 3 and 7 dpa cells were harvested from 1 mm proximal to the regenerative distal tissues (n = 4). After dissection, tissues were finely chopped in DMEM and digested in dissociation solution(C/T, 0.5 ml collagenase 2% + 10 ml, trypsin) at 37 °C then centrifuged at 1600 rpm. The precipitation was re-suspended to obtain a concentration of approximately one million cells per milliliter.
C1 single-cell mRNA sequencing
Blastema and non-regenerating limb stump cells at 5 × 105 cells/ml were loaded onto two 17–25 μm C1 Single-Cell Auto Prep integrated fluidic circuits (Fluidigm) and cell capture was performed according to the manufacturer’s instructions. Individual capture sites were inspected under a light microscope to confirm the presence of single cells. Empty capture wells and wells containing multiple cells or cell debris were discarded for quality control. A SMARTer Ultra Low RNA kit (Clontech) and an Advantage 2 PCR Kit (Clontech) were used for cDNA generation. An ArrayControl RNA Spots and Spikes kit (with spike numbers 1, 4, and 7) (Ambion) was used to monitor technical variability, and the dilutions used were as recommended by the manufacturer. The concentration of cDNA for every single well was determined by Qubit™ dsDNA HS Assay Kit. Multiplex sequencing libraries were generated using the TruePrepTM DNA Library Prep Kit V2 and the Nextera XT Index Kit (Illumina). Libraries were pooled and subjected to sequencing on an Illumina HiSeq 2000 (Illumina).
Bulk RNA-seq library construction
We used mRNA Capture Beads (VAHTS mRNA-seq v2 Library Prep Kit for Illumina, Vazyme) to extract mRNA from total RNA. PrimeScript™ Double Strand cDNA Synthesis Kit (TaKaRa) was used to synthesize double-stranded cDNA from purified polyadenylated mRNA templates. We used TruePrep DNA Library Prep Kit V2 for Illumina (TaKaRa) to prepare cDNA libraries for Illumina sequencing.
Data analysis pipeline
We mainly referred to the transcriptome from Bryant’s research [8] and used the other two transcriptomes and one genome as an aid. The clean reads were obtained from the high quality region (10–60 nt) of the sequenced Illumina raw reads, and were mapped with three de novo transcriptome assemblies from previous studies containing both axolotl and Chinese giant salamander, in addition to the genome of Xenopus laevis [7, 8, 18] (609,180 transcripts) through Bowtie. The methods for functional annotation were performed according to the methods published by Bryant [8] and we got 83,296 transcripts were ultimately annotated. The functional annotation of de novo assembled transcriptomes was performed by Trinotate (http://trinotate.github.io). After data quality control and removal of 72 multiple cell samples, 938 cells with >212,000 transcripts (15,650 genes) were selected for further analysis.
Sequencing data analysis
The differential expression genes (DE genes) were calculated by DEseq2 R package. ScRNA-seq expression data were analyzed with Seurat v2.0.1 (PCA, Cluster, t-SNE and cluster) [19]. In brief, the Seurat object was generated from digital gene expression matrices. In the standard preprocessing workflow of Seurat, we selected 15,608 variable genes for PCA, whereupon we performed cell cluster and t-SNE.
Marker genes of each cell cluster were outputted for GO analysis. GOEAST (http://omicslab.genetics.ac.cn/GOEAST/) was used to perform GO analysis. We used GSEA for GO analysis and signaling pathway analysis. Cell clusters were annotated with the information of marker genes and GO analysis results. Digital gene expression matrices with annotations from Seurat were analyzed by Monocle v2.3.6 (pseudotime analysis). The pseudo-temporal gene trend heatmap was performed by placing all single cells in their pseudotime order by compressing small branches into a larger central branch and performed k-means clustering on all genes (p value < 0.05). The cell–cell interactions visualized by the connectivity map were constructed by R package Circlize 0.4.3 [51].
The count of cell–cell interactions was based on the ligand–receptor pairings. GSEA was performed using GSEA software. The signaling pathway analysis used the gene sets KEGG, REACTOME and BIOCARTA (www.broadinstitute.org/gsea/doc/GSEAUserGuideFrame.html). The protein–protein interaction enrichment analysis was carried out with the databases BioGrid, InWeb_IM, and OmniPath.
Histological examination
The harvested specimens were immediately fixed in 4% (w/v) paraformaldehyde in phosphate-buffered saline for 24 h (n = 3). The samples were then dehydrated through an alcohol gradient, cleared, and embedded in paraffin blocks. Histological sections (7 mm) were prepared using a microtome and subsequently stained with hematoxylin and eosin (H&E), and Safranin O.
Immunostaining
Paraffin sections for immunohistochemistry were treated with 0.05% Trypsin-EDTA (Gibco), 3% (v/v) hydrogen peroxide in methanol, and 1% (w/v) BSA. Sections were incubated overnight at 4 °C with primary antibodies for TOMM20 (Abcam, ab78547), COL1 (Santa Cruz, sc-8784), MDK (Affinity, DF6054), PTPRC (Affinity, DF6839), PIF1 (Affinity, DF9256), and KRT8 (Affinity, AF6097). For immunohistochemical assays, sections were then incubated with secondary antibodies (Beyotime Biotechnology, China) for 2 h at room temperature. The DAB substrate system (ZSGB-bio, Beijing, China) was used for color development. Hematoxylin staining was utilized to reveal the cell nuclei. For immunofluorescence assays, sections were then incubated with secondary antibodies (Invitrogen) for 2 h at room temperature. DAPI staining was utilized to reveal the cell nuclei (n = 3, 3 times the experiment shown was replicated in the laboratory).
Transmission electron microscopy
The samples (tissues from 0, 3, 7, and 21 dpa axolotl limbs) were fixed and handled by standard procedures as described previously [52]. Mitochondria were counted per 300,00× picture and each sample was averaged from ten pictures and analyzed at least three times. Image-Pro Plus (IPP 6.0, Media Cybernetics, Rockville, MD, http://www.mediacy.com.cn/cn/index/index.asp) software was used to counts the mitochondria numbers.
RNA FISH assay
Fluorescence-conjugated Col2a1 probes for RNA FISH were generated in line with the protocols of Biosearch Technologies. All RNA hybridization experiments were performed as described in manuals of Biosearch Technologies. Treated samples were visualized by confocal microscopy (FV1000, Olympus) (n = 3, 3 times the experiment shown was replicated in the laboratory).
Statistical analysis
Assuming a onefold change in mitochondria number, with an alpha error of 0.05%, two-tailed, the data shown are mean ± SEM of the number of independent experiments indicated (n). The represent experiments performed on at least three separate occasions with similar outcomes. The comparisons between groups were performed using a t test using GraphPad Prism software. Statistical differences were determined based on a P value (*p < 0.05, **p < 0.01, and ***p < 0.001). The mitochondria were compared in quantitative blinded analyses.
Supplementary information
Figure. S1 Single-Cell RNA-Seq data features and changes during axolotl limb regeneration the time course.
Figure. S2 Cell heterogeneity in axolotl limb regeneration tissues.
Figure S3. Connectivity map reveals interactions among clusters.
Figure S4. Pseudo-temporal ordering of cells from all time points.
Figure S5. Identification of different lineage cells within 0dpa–21dpa axolotl limbs Singe-cell RNA-seq data.
Figure S6. Validation of COL1 variance in regeneration.
Figure S7. Proliferation analysis of COL2+ sub-clusters.
Figure S8. Validation of COL1 variance in regeneration.
Figure S9. Proliferation analysis of COL2+ sub-clusters.
S1: Upregulated gene by DE analysis in 4 time points
Acknowledgements
This work was supported by the National key research and development program of China [2017YFA0104902]; and the National Natural Science Foundation of China (NSFC) grants [81972099, 81871764, 82072463, 81522029, 81772418, 31570987, 81330041, 81401781, 81572157]. Zhejiang Provincial Natural Science Foundation of China (LR20H060001). Fundamental Research Funds for the Central Universities. Written on behalf of the AME Translational Medicine Collaborative Group. We are grateful to the Core Facilities of Zhejiang University School of Medicine, and Bio-Ultrastructure Analysis Lab. of Analysis Center of Agrobiology and Environmental Sciences of the Zhejiang University for technical assistance.
Data availability
All data needed to evaluate the conclusions in the paper are available in the GSA (CRA002291) or present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
Code availability
All code follows the official tutorial. Relative information may be requested from the authors.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no competing interests.
Footnotes
Edited by I. Amelio
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Tian Qin, Chun-mei Fan, Ting-zhang Wang
Contributor Information
Zi Yin, Email: yinzi@zju.edu.cn.
Xiao Chen, Email: chenxiao-610@zju.edu.cn.
Supplementary information
The online version of this article (10.1038/s41418-020-00640-8) contains supplementary material, which is available to authorized users.
References
- 1.Shieh SJ, Cheng TC. Regeneration and repair of human digits and limbs: fact and fiction. Regeneration. 2015;2:149–68. doi: 10.1002/reg2.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brockes JP. Amphibian limb regeneration: rebuilding a complex structure. Science. 1997;276:81–7. doi: 10.1126/science.276.5309.81. [DOI] [PubMed] [Google Scholar]
- 3.Kragl M, Knapp D, Nacu E, Khattak S, Maden M, Epperlein HH, et al. Cells keep a memory of their tissue origin during axolotl limb regeneration. Nature. 2009;460:60–5. doi: 10.1038/nature08152. [DOI] [PubMed] [Google Scholar]
- 4.Looso M, Preussner J, Sousounis K, Bruckskotten M, Michel CS, Lignelli E, et al. A de novo assembly of the newt transcriptome combined with proteomic validation identifies new protein families expressed during tissue regeneration. Genome Biol. 2013;14:R16. doi: 10.1186/gb-2013-14-2-r16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nacu E, Gromberg E, Oliveira CR, Drechsel D, Tanaka EM. FGF8 and SHH substitute for anterior-posterior tissue interactions to induce limb regeneration. Nature. 2016;533:407–10. doi: 10.1038/nature17972. [DOI] [PubMed] [Google Scholar]
- 6.Haas BJ, Whited JL. Advances in decoding axolotl limb regeneration. Trends Genet. 2017;33:553–65. doi: 10.1016/j.tig.2017.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stewart R, Rascón CA, Tian S, Nie J, Barry C, Chu LF, et al. Comparative RNA-seq analysis in the unsequenced axolotl: the oncogene burst highlights early gene expression in the blastema. PLoS Comput Biol. 2013;9:e1002936. doi: 10.1371/journal.pcbi.1002936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bryant DM, Johnson K, DiTommaso T, et al. A tissue-mapped axolotl de novo transcriptome enables identification of limb regeneration factors. Cell Rep. 2017;18:762–76. doi: 10.1016/j.celrep.2016.12.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lanctôt C. Single cell analysis reveals concomitant transcription of pluripotent and lineage markers during the early steps of differentiation of embryonic stem cells. Stem Cells. 2015;33:2949–60. doi: 10.1002/stem.2108. [DOI] [PubMed] [Google Scholar]
- 10.Han X, Chen H, Huang D, Chen H, Fei L, Cheng C, et al. Mapping human pluripotent stem cell differentiation pathways using high throughput single-cell RNA-sequencing. Genome Biol. 2018;19:47. doi: 10.1186/s13059-018-1426-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Villani AC, Satija R, Reynolds G, Sarkizova S, Shekhar K, Fletcher J, et al. Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science. 2017;356:1955–6. doi: 10.1126/science.aah4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li H, Horns F, Wu B, Xie Q, Li J, Li T, et al. Classifying drosophila olfactory projection neuron subtypes by single-cell RNA sequencing. Cell. 2017;171:1206–20.e22. doi: 10.1016/j.cell.2017.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu B, An C, Li Y, Yin Z, Gong L, Li Z, et al. Reconstructing lineage hierarchies of mouse uterus epithelial development using single-cell analysis. Stem Cell Rep. 2017;9:381–96. doi: 10.1016/j.stemcr.2017.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Han X, Wang R, Zhou Y, Fei L, Sun H, Lai S, et al. Mapping the Mouse. Cell Atlas Microwell-Seq Cell. 2018;172:1091–107.e17. doi: 10.1016/j.cell.2018.02.001. [DOI] [PubMed] [Google Scholar]
- 15.Gerber T, Murawala P, Knapp D, et al. Single-cell analysis uncovers convergence of cell identities during axolotl limb regeneration. Science. 2018;362:eaaq0681. doi: 10.1126/science.aaq0681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leigh ND, Dunlap GS, Johnson K, Mariano R, Oshiro R, Wong AY, et al. Transcriptomic landscape of the blastema niche in regenerating adult axolotl limbs at single-cell resolution. Nat Commun. 2018;9:5153. doi: 10.1038/s41467-018-07604-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li H, Wei X, Zhou L, Zhang W, Wang C, Guo Y et al. Dynamic cell transition and immune response landscapes of axolotl limb regeneration revealed by single-cell analysis. Protein Cell. 2020 10.1007/s13238-020-00763-1. [DOI] [PMC free article] [PubMed]
- 18.Geng X, Li W, Shang H, Gou Q, Zhang F, Zang X, et al. A reference gene set construction using RNA-seq of multiple tissues of Chinese giant salamander, Andrias davidianus. Gigascience. 2017;6:1–7. doi: 10.1093/gigascience/gix006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Satija R, Farrell JA, Gennert D, Schier AF, Regev A. Spatial reconstruction of single-cell gene expression data. Nat Biotechnol. 2015;33:495–502. doi: 10.1038/nbt.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schieke SM, Ma M, Cao L, McCoy JP, Liu C, Hensel NF, et al. Mitochondrial metabolism modulates differentiation and teratoma formation capacity in mouse embryonic stem cells. J Biol Chem. 2008;283:28506–12. doi: 10.1074/jbc.M802763200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roensch K, Tazaki A, Chara O, Tanaka EM. Progressive specification rather than intercalation of segments during limb regeneration. Science. 2013;342:1375–9. doi: 10.1126/science.1241796. [DOI] [PubMed] [Google Scholar]
- 22.Christensen RN, Tassava RA. Apical epithelial cap morphology and fibronectin gene expression in regenerating axolotl limbs. Dev Dyn. 2000;217:216–24. doi: 10.1002/(SICI)1097-0177(200002)217:2<216::AID-DVDY8>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 23.Endo T, Bryant SV, Gardiner DM. A stepwise model system for limb regeneration. Dev Biol. 2004;270:135–45. doi: 10.1016/j.ydbio.2004.02.016. [DOI] [PubMed] [Google Scholar]
- 24.SINGER M, INOUE S. The nerve and the epidermal apical cap in regeneration of the forelimb of adult triturus. J Exp Zool. 1964;155:105–16.. doi: 10.1002/jez.1401550108. [DOI] [PubMed] [Google Scholar]
- 25.Camp JG, Sekine K, Gerber T, Loeffler-Wirth H, Binder H, Gac M, et al. Multilineage communication regulates human liver bud development from pluripotency. Nature. 2017;546:533–8. doi: 10.1038/nature22796. [DOI] [PubMed] [Google Scholar]
- 26.Zepp JA, Zacharias WJ, Frank DB, Cavanaugh CA, Zhou S, Morley MP, et al. Distinct mesenchymal lineages and niches promote epithelial self-renewal and myofibrogenesis in the lung. Cell. 2017;170:1134–48.e10. doi: 10.1016/j.cell.2017.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Godwin JW, Pinto AR, Rosenthal NA. Macrophages are required for adult salamander limb regeneration. Proc Natl Acad Sci USA. 2013;110:9415–20. doi: 10.1073/pnas.1300290110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trapnell C, Cacchiarelli D, Grimsby J, Pokharel P, Li S, Morse M, et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol. 2014;32:381–6. doi: 10.1038/nbt.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qiu X, Mao Q, Tang Y, Wang L, Chawla R, Pliner HA, et al. Reversed graph embedding resolves complex single-cell trajectories. Nat Methods. 2017;14:979–82. doi: 10.1038/nmeth.4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hinton RJ, Jing Y, Jing J, Feng JQ. Roles of chondrocytes in endochondral bone formation and fracture repair. J Dent Res. 2017;96:23–30. doi: 10.1177/0022034516668321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Midwood KS, Williams LV, Schwarzbauer JE. Tissue repair and the dynamics of the extracellular matrix. Int J Biochem Cell Biol. 2004;36:1031–7. doi: 10.1016/j.biocel.2003.12.003. [DOI] [PubMed] [Google Scholar]
- 32.Garcia-Puig A, Mosquera JL, Jiménez-Delgado S, García-Pastor C, Jorba I, Navajas D, et al. Proteomics analysis of extracellular matrix remodeling during zebrafish heart regeneration. Mol Cell Proteom. 2019;18:1745–55. doi: 10.1074/mcp.RA118.001193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cheng T, Maddox NC, Wong AW, Rahnama R, Kuo AC. Comparison of gene expression patterns in articular cartilage and dedifferentiated articular chondrocytes. J Orthop Res. 2012;30:234–45. doi: 10.1002/jor.21503. [DOI] [PubMed] [Google Scholar]
- 34.Asahina K, Obara M, Yoshizato K. Expression of genes of type I and type II collagen in the formation and development of the blastema of regenerating newt limb. Dev Dyn. 1999;216:59–71. doi: 10.1002/(SICI)1097-0177(199909)216:1<59::AID-DVDY8>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 35.Mitogawa K, Makanae A, Satoh A, Satoh A. Comparative analysis of cartilage marker gene expression patterns during axolotl and xenopus limb regeneration. PLoS ONE. 2015;10:e0133375. doi: 10.1371/journal.pone.0133375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tanaka HV, Ng NC, Yang YZ, Casco-Robles MM, Maruo F, Tsonis PA, et al. A developmentally regulated switch from stem cells to dedifferentiation for limb muscle regeneration in newts. Nat Commun. 2016;7:11069. doi: 10.1038/ncomms11069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.das NRP, Jones NS, Andreu L, Gupta R, Enver T, Iborra FJ. Connecting variability in global transcription rate to mitochondrial variability. PLoS Biol. 2010;8:e1000560. doi: 10.1371/journal.pbio.1000560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kiryu-Seo S, Kiyama H. Mitochondrial behavior during axon regeneration/degeneration in vivo. Neurosci Res. 2019;139:42–7. doi: 10.1016/j.neures.2018.08.014. [DOI] [PubMed] [Google Scholar]
- 39.Wang ZA, Huang J, Kalderon D. Drosophila follicle stem cells are regulated by proliferation and niche adhesion as well as mitochondria and ROS. Nat Commun. 2012;3:769. doi: 10.1038/ncomms1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen H, Chan DC. Mitochondrial dynamics in regulating the unique phenotypes of cancer and stem cells. Cell Metab. 2017;26:39–48. doi: 10.1016/j.cmet.2017.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xie Q, Wu Q, Horbinski CM, Flavahan WA, Yang K, Zhou W, et al. Mitochondrial control by DRP1 in brain tumor initiating cells. Nat Neurosci. 2015;18:501–10. doi: 10.1038/nn.3960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kumar A, Brockes JP. Nerve dependence in tissue, organ, and appendage regeneration. Trends Neurosci. 2012;35:691–9. doi: 10.1016/j.tins.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 43.Ashraf S, Cha BH, Kim JS, Ahn J, Han I, Park H, et al. Regulation of senescence associated signaling mechanisms in chondrocytes for cartilage tissue regeneration. Osteoarthr Cartil. 2016;24:196–205. doi: 10.1016/j.joca.2015.07.008. [DOI] [PubMed] [Google Scholar]
- 44.Fernando WA, Leininger E, Simkin J, Li N, Malcom CA, Sathyamoorthi S, et al. Wound healing and blastema formation in regenerating digit tips of adult mice. Dev Biol. 2011;350:301–10. doi: 10.1016/j.ydbio.2010.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kierdorf U, Kierdorf H. Antler regrowth as a form of epimorphic regeneration in vertebrates—a comparative view. Front Biosci. 2012;4:1606–24. doi: 10.2741/483. [DOI] [PubMed] [Google Scholar]
- 46.Takeo M, Chou WC, Sun Q, Lee W, Rabbani P, Loomis C, et al. Wnt activation in nail epithelium couples nail growth to digit regeneration. Nature. 2013;499:228–32. doi: 10.1038/nature12214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Johnson GL, Masias EJ, Lehoczky JA. Cellular heterogeneity and lineage restriction during mouse digit tip regeneration at single-cell resolution. Dev Cell. 2020;52:525–40. doi: 10.1016/j.devcel.2020.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Han M, Yang X, Lee J, Allan CH, Muneoka K. Development and regeneration of the neonatal digit tip in mice. Dev Biol. 2008;315:125–35. doi: 10.1016/j.ydbio.2007.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mastrodonato M, Portincasa P, Mentino D, Rossi R, Resta L, Ferri D, et al. Caveolin-1 and mitochondrial alterations in regenerating rat liver. Microsc Res Tech. 2012;75:1026–32. doi: 10.1002/jemt.22027. [DOI] [PubMed] [Google Scholar]
- 50.Khacho M, Clark A, Svoboda DS, Azzi J, MacLaurin JG, Meghaizel C, et al. Mitochondrial dynamics impacts stem cell identity and fate decisions by regulating a nuclear transcriptional program. Cell Stem Cell. 2016;19:232–47. doi: 10.1016/j.stem.2016.04.015. [DOI] [PubMed] [Google Scholar]
- 51.Gu Z, Gu L, Eils R, Schlesner M, Brors B. circlize Implements and enhances circular visualization in R. Bioinformatics. 2014;30:2811–2. doi: 10.1093/bioinformatics/btu393. [DOI] [PubMed] [Google Scholar]
- 52.Chen X, Yin Z, Chen JL, Shen WL, Liu HH, Tang QM, et al. Force and scleraxis synergistically promote the commitment of human ES cells derived MSCs to tenocytes. Sci Rep. 2012;2:977. doi: 10.1038/srep00977. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure. S1 Single-Cell RNA-Seq data features and changes during axolotl limb regeneration the time course.
Figure. S2 Cell heterogeneity in axolotl limb regeneration tissues.
Figure S3. Connectivity map reveals interactions among clusters.
Figure S4. Pseudo-temporal ordering of cells from all time points.
Figure S5. Identification of different lineage cells within 0dpa–21dpa axolotl limbs Singe-cell RNA-seq data.
Figure S6. Validation of COL1 variance in regeneration.
Figure S7. Proliferation analysis of COL2+ sub-clusters.
Figure S8. Validation of COL1 variance in regeneration.
Figure S9. Proliferation analysis of COL2+ sub-clusters.
S1: Upregulated gene by DE analysis in 4 time points
Data Availability Statement
All data needed to evaluate the conclusions in the paper are available in the GSA (CRA002291) or present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
All code follows the official tutorial. Relative information may be requested from the authors.