Abstract
Objective
Chromovert® Technology is presented as a new cell engineering technology to detect and purify living cells based on gene expression.
Methods
The technology utilizes fluorogenic oligonucleotide signaling probes and flow cytometry to detect and isolate individual living cells expressing one or more transfected or endogenously-expressed genes.
Results
Results for production of cell lines expressing a diversity of ion channel and membrane proteins are presented, including heteromultimeric epithelial sodium channel (αβγ-ENaC), sodium voltage-gated ion channel 1.7 (NaV1.7-αβ1β2), four unique γ-aminobutyric acid A (GABAA) receptor ion channel subunit combinations α1β3γ2s, α2β3γ2s, α3β3γ2s and α5β3γ2s, cystic fibrosis conductance regulator (CFTR), CFTR-Δ508 and two G-protein coupled receptors (GPCRs) without reliance on leader sequences and/or chaperones. In addition, three novel plasmid-encoded sequences used to introduce 3′ untranslated RNA sequence tags in mRNA expression products and differentially-detectable fluorogenic probes directed to each are described. The tags and corresponding fluorogenic signaling probes streamline the process by enabling the multiplexed detection and isolation of cells expressing one or more genes without the need for gene-specific probes.
Conclusions
Chromovert technology is provided as a research tool for use to enrich and isolate cells engineered to express one or more desired genes.
Keywords: Cell engineering, Genetic engineering, Flow cytometry, Fluorescence-activated cell sorting (FACS), Cell line, GPCR, Ion channel
Introduction
Mammalian cell lines, especially those produced using immortalized lines like human embryonic kidney 293 (HEK 293) and Chinese hamster ovary (CHO) cells, are widely used in biological research, including in drug discovery and to produce biologic drugs. In general, cell line production begins with the introduction of one or more DNA sequences of interest into a cell culture resulting in a diversity of genetically distinguished cells based on the site and copy number of integration events. The goal is to produce, in a reasonable time frame, clonal cell lines that meet desired criteria for an application of interest.
Despite multiple advances in cell engineering including selection of HEK 293 and CHO host cell subtypes with improved functionality, e.g., HEK 293T cells (Pear et al. 1993) and serum-free, suspension adapted DG44 CHO cells (Urlaub et al. 1983), co-transfection and/or tagging of DNA sequences of interest with markers including autofluorescent proteins such as green fluorescent protein, GFP (Chalfie et al. 1994), N-terminal tagging of protein expression products with leader sequences (e.g., using rhodopsin leader sequences) to enhance folding, assembly and/or membrane integration of protein expression products (Krautwurst et al. 1998), the addition of chaperones (Mohan et al. 2007) and strategies to amplify the copy number of genetic recombination events (Cockett et al. 1990), the engineering of cells treated to comprise one or more added genes of interest remains challenging even given precision genetic engineering tools such as clustered regularly interspaced short palindromic repeats, or CRISPR (Nowogrodzki 2019). For instance, the use of cell lines to produce recombinant adeno-associated virus (rAAV) as required for numerous approved gene therapies has been limited due to the unpredictable and overly time-consuming nature of the process even though cell lines are highly sought-after to replace current rate-limiting rAAV production methods that involve transient transfection of multiple plasmids into HEK 293T cells (Wang et al. 2019).
Here, we present Chromovert technology as a research tool for production of mammalian cell lines comprising one or more sequences of interest. The method is based upon the broadly-applicable principles of fluorescence-resonance energy transfer (FRET) and nucleic acid hybridization using fluorogenic oligonucleotide signaling probes originally reported for in tubo qRT-PCR applications (Marras et al. 1999) transfected into living cells. The termini of the signaling probes are covalently linked to a fluorophore or quencher paired to absorb its emission. The termini are designed to form a 4–7 base-pair stem juxtaposing the fluorophore and quencher pair. In the presence of target sequence, hybridization of the sequence-specific probe results in a fluorogenic conformational change. Flow cytometry is then used to detect and isolate positive cells that fluoresce above background. Thousands of individual clones can then be isolated and expanded using automated cell culture methods. Functional testing over time in the absence of selective pressure is used to select final clones.
Results
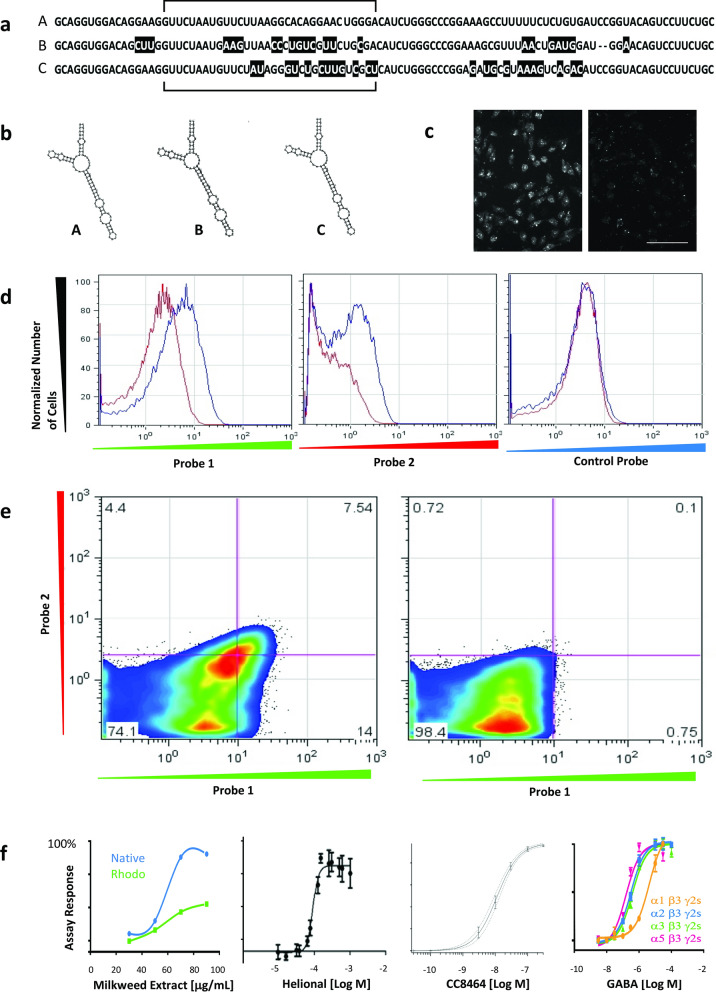
Data are presented for the production of 11 unique cell lines expressing ion channel or GPCR proteins. The cell lines were produced to express a total of 26 transfected cDNAs. All cell lines were prepared using the same standardized protocol without any gene-specific signaling probes. Generic signaling probes directed to three plasmid-encoded sequence tags, termed Chromo-Tags™, were used to detect and isolate cells expressing any cDNAs required to produce all the cell line described in this report. Three Chromo-Tags that vary by at least 50% in pairwise comparisons over the 24-nucleotide region indicated and that share similar predicted mRNA folding based on the mFold algorithm (Zuker 2003) were designed, as shown in Fig. 1a and b, respectively. Representative data for the multiplexed detection of the three Chromo-Tag sequences expressed in cells co-transfected with expression plasmids each comprising an expression cassette encoding one of the Chromo-Tags using differentially-labeled signaling probes directed to the 24-nucleotide region of the Chromo-Tags are shown in Fig. 1c–e.
Fig. 1.
Detection of gene expression in individual living cells using Chromovert technology. a Three artificial RNA sequences, termed Chromo-Tags or C-Tags A, B and C, used to tag mRNAs of interest for detection using signaling probes are shown. Color-coding is used to indicate sequence variations between Chromo-Tag A and either Chromo-Tag B or C. The Chromo-Tags vary from each other by at least 50% within the 24-nucleotide region indicated. b Predicted RNA folding for Chromo-Tags A, B and C using the mFold algorithm is shown. c Detection of a target RNA by a signaling probe in transfected cells vs. control-transfected cells using fluorescence microscopy is shown on the left and right panels, respectively. Scale bar = 100 µm. d Cell cytometric analysis for the detection of Chromo-Tags A and B using differentially-labeled signaling probes to Chromo-Tags A, B and C in transfected cells (blue traces) versus control-transfected cells (red traces) is shown. Fluorescence above background corresponding to signaling probes directed to Chromo-Tags A and B is shown in the left and center panels, respectively. Background fluorescence corresponding to control signaling probe directed to Chromo-Tag C is shown in the right panel. X-axis: fluorescence signal corresponding to the indicated probe, Y-axis: normalized cell count. e 2D dot plots for the simultaneous detection of Chromo-Tags A and B in transfected cells (left panel) versus control-transfected cells (right panel) are shown. Percentages of cells falling in each quadrant are indicated. X-axis and Y-axis: fluorescence signal corresponding to the indicated probe. f Far left panel shows single-point HTS screening data for cell lines expressing the human bitter taste receptor GPCR T2R16 (“Native”) or the T2R16 comprising amino acids 1–39 of rhodopsin as an N-terminal tag (“Rhodo”) in response to milkweed extract using a fluorescent calcium flux cell-based assay, as indicated. Center left panel shows a dose-response cure for Helional in the native human odorant receptor OR3A1 cell line (“hOR3A1”) using a calcium flux cell-based assay, as indicated. Center right panel shows a dose-response curve for the inhibition of current in the NaV1.7-αβ1β2 stable cell line by a small molecule compound CC8464 (n = 7). X-axis: CC8464 concentration, Y-axis: fraction of normalized current blocked using a whole-cell patch-clamp assay. Far right panel shows the response of the GABAA α1β3γ2s, α2β3γ2s, α3β3γ2s and α5β3γ2s ion channel cell lines to GABA using a fluorescent membrane potential cell-based assay, as indicated
Chromovert technology was used to produce cell lines expressing GPCRs. Figure 1f, far left and center left panels show calcium flux cell-based assay results for three GPCR cell lines produced using the Chromovert method. Functional cell-based responses for the cell line comprising the native human bitter taste receptor T2R16 and a second cell line comprising a modified version of the same GPCR in response to milkweed extract are shown in Fig. 1f, far left panel. The modified T2R26 comprises an N-terminal amino acid extension corresponding to amino acids 1–39 of rhodopsin previously reported to be required for the functional expression of human bitter GPCRs (Chandrashekar et al. 2000). Both cell lines were co-transfected to express the promiscuous murine G-alpha protein, Gα15 (Offermanns and simon 1995). Figure 1f, center left panel shows the response of the Chromovert technology-enabled cell line expressing the native human odorant GPCR hOR31A and Gα15 to α-methyl-3,4-methylendioxy-hydrocinnamal-dehyde, or Helional. The hOR3A1 cell line was also produced without reliance on any chaperones or N-terminal amino acid leader sequences.
Chromovert technology was used to produce cell lines expressing ion channels. Figure 1f, center right and far right panels show cell-based assay results for five heteromultimeric ion channel cell lines produced using Chromovert technology and Chromo-Tags. The functional cell-based response of the NaV1.7 cell line to a NaV1.7 inhibitor, Chromocell compound CC8464 (Babich et al. 2016), is shown in Fig. 1f, center right panel. A combinatorial panel of cell lines and cell-based assays for four GABAA receptor ion channel subunit combinations α1β3γ2s, α2β3γ2s, α3β3γ2s or α5β3γ2s was also generated, as shown in Fig. 1f, far right panel. High-throughput screening (HTS) of a library of pharmaceutically active compounds (LOPAC, Sigma 1280) was performed using the panel. We identified 34 LOPAC compounds that activated at least one of the cell lines at least as much as GABA itself, including all known GABAA agonists included in LOPAC: GABA, Propofol, Isoguvacine hydrochloride, Muscimol hydrobromide, Piperidine-4-sulphonic acid, 3-alpha, 21-dihydroxy-5-alpha-pregnan-20-one, 5-alpha-pregnan-3alpha-ol-11,20-dione, 5-alpha-pegnan-3alpha-ol-20-one and Tracazolate (Johnston 2005). In addition, at least four compounds not previously described as GABAA agonists but known to have other activities associated with GABAA ion channels were identified, including: etazolate, androsterone, chlormezanone and ivermectin. The screen further identified compounds which, until now, were not reported to interact with GABAA, including: dipyridamole, niclosamide, tyrphostin A9 and I-OMe-Tyrphostin AG 538.
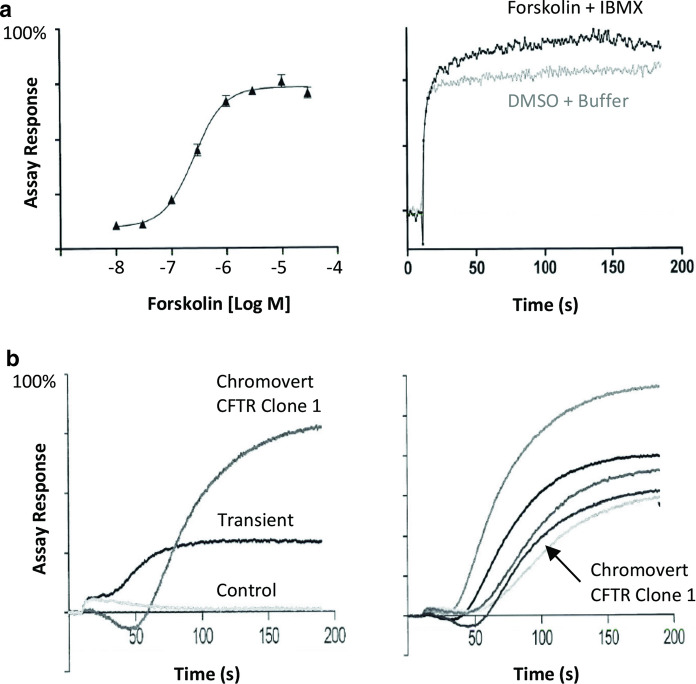
Chromovert technology was also used to produce CFTR and CFTR-Δ508 cell lines and cell-based assays. Figure 2a, left panel shows a dose–response curve for the CFTR cell line to Forskolin. Figure 2a, right panel shows that the Chromovert technology-enabled CFTR-Δ508 cell line enables a functional cell-based assay for CFTR-Δ508 in the absence of any added trafficking correctors. Figure 2b shows the functional cell-based responses of multiple Chromovert technology-enabled CFTR cell lines compared to transiently-transfected cells.
Fig. 2.
Comparison of Chromovert technology-enabled CFTR cell line to traditional results using a fluorescent membrane potential assay. a Left panel shows the dose response curve of the CFTR cell line to forskolin, resulting in an EC50 value of 256 nM. Right panel shows the response of the CFTR-Δ508 cell line to 30 µM Forskolin + 100 µM IBMX (3-isobutyl-1-methyl xanthine) versus DMSO + Buffer in the absence of any trafficking correctors. b Left panel shows the response of the one Chromovert CFTR cell line (Clone 1), cells transiently-transfected with CFTR and control-transfected cells to an EC50 concentration of Forskolin. Right panel shows the response of Clone 1 compared to additional Chromovert CFTR clonal cell line isolates
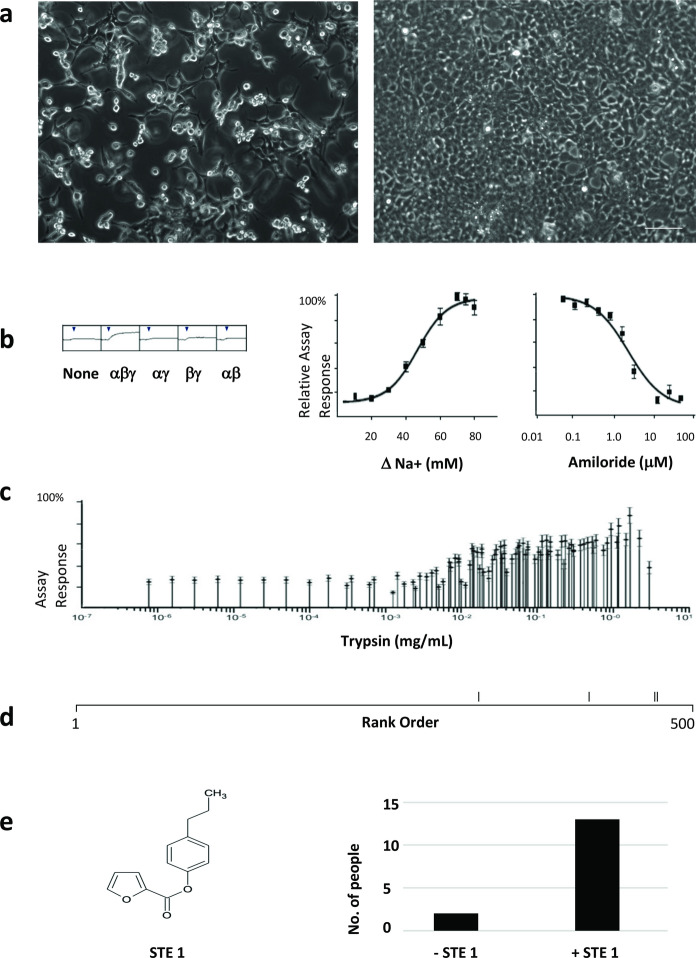
Finally, Chromovert technology was used to create a cell line for ENaC. ENaC causes cell swelling and is cytotoxic when expressed in transfected cell cultures, as shown in Fig. 3a. Chromovert technology enabled an αβγ-ENaC cell line as shown in Fig. 3b using a cell-based assay selective for αβγ-ENaC activity. Limited proteolysis of the cell line was used to achieve functionally distinct proteolyzed variants, as shown in Fig. 3c. HTS of a compound library against non-proteolyzed versus proteolyzed ENaC variants resulted in largely non-overlapping hits, as shown in Fig. 3d. One potentiator identified during HTS of proteolyzed ENaC and confirmed to enhance the salty taste of sodium chloride using human sensory studies is shown in Fig. 3e.
Fig. 3.
Creation and testing of a stable αβγ-ENaC cell line. a Expression of αβγ-ENaC is cytotoxic in transiently-transfected cultured cells (left) versus control-transfected cells (right) as visualized by phase-contrast microscopy. Scale bar = 100 µm. b Left panel shows a functional cell-based assay selective for αβγ-ENaC using cells transiently-transfected cells. X-axis: time, Y-axis, assay response. Arrows: addition of solution to increase sodium by 75 mM. Right panel shows functional characterization of the Chromovert technology-enabled ENaC cell line using the assay. Responses to increasing changes in sodium concentration and inhibition of the response to a 75 mM increase in sodium concentration by amiloride are shown. c Multiple functionally-distinct proteolyzed variants of αβγ-ENaC created using limited proteolysis in the context of the cell-based assay are shown (n = 9). d The only four compounds among the top 500 hits identified in HTS of a compound library screened using proteolyzed ENaC that were also identified in HTS of the same library screened using non-proteolyzed ENaC are rank-ordered for their activity on non-proteolyzed ENaC. e The chemical structure and human sensory taste testing results for 4-propylphenyl 2-furoate, a potentiator of proteolyzed αβγ-ENaC identified using HTS termed salt-taste enhancing compound 1 (STE1), are shown. In pairwise comparisons of 45 mM NaCl ± 15 µg STE1/mL using a human sensory taste testing protocol approved by an institutional review board, 13 out of 15 participants selected the sample containing STE1 as more salty
Discussion
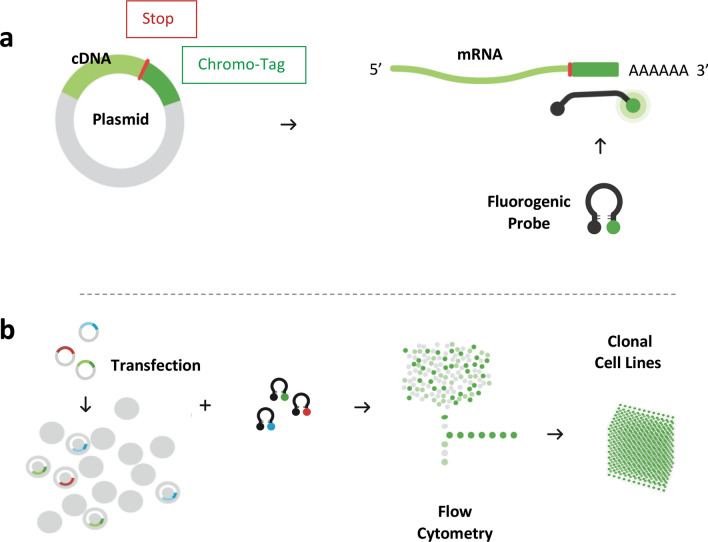
Chromovert technology utilizes fluorogenic oligonucleotide signaling probes to detect and isolate individual cells expressing one or more genes of interest, see technology flowchart in Fig. 4. The technology was used to create cell lines for a diversity of proteins. All the cell lines in this report were created using only signaling probes directed to plasmid-encoded RNA tags, termed Chromo-Tags. Chromo-Tags are 3´ untranslated RNA sequence tags subcloned for expression downstream of the stop codon of cDNAs of interest. Unlike epitope tags, Chromo-Tags are not translated into protein such that the amino acid sequences of expression products are not altered. Chromo-Tags streamline the cell engineering method by obviating the need to design and develop gene-specific probes. The NaV1.7, GABAA, CFTR, ENaC and GPCR example cell lines were all produced using the same Chromo-Tags and protocols. Whereas numerous cell-based approaches have been reported to access each of these proteins, the Chromovert technology-enabled cell lines and cell-based assays contributed to research and development efforts relating to each of these proteins, as described below.
Fig. 4.
Chromovert technology flowchart. a cDNAs are subcloned for expression of mRNAs comprising 3′ untranslated plasmid-encoded Chromo-Tag sequences for detection using fluorogenic oligonucleotide signaling probes. Protein expression products remain untagged. b To create cell lines, one or more Chromo-Tagged cDNAs are transfected into cells, the transfected cells are exposed to differentially-labeled signaling probes and individual positive cells are isolated using flow cytometry. Downstream testing is used to select final cell lines
First, whereas GPCRs are commonly reported to require N-terminal amino acid extensions (e.g., leader sequences derived from rhodopsin) and/or chaperones for functional expression in cultured cells (Krautwurst et al. 1998; Mohan et al. 2007), the native and full-length functional GPCR cell lines in this report were produced using neither. In the case of human odorant receptor hOR3A1, the Chromovert technology-enabled cell line for the native receptor matched literature results (Wetzel et al. 1999). In the case of human bitter taste receptor T2R16, our results indicate that N-terminal rhodopsin leader sequences may in some cases affect the physiological form and function of the native receptors.
Second, NaV1.7 inhibitors are highly sought as non-addictive pain blockers for use to help address the opioid epidemic however stable cell lines comprising multiple accessory subunits have not been reported. The NaV ion channel family is comprised of large pore-forming α-subunits and up to four accessory β-subunits, β1, β2, β3 and β4, which modulate their activity (O’Malley and Isom 2015). HTS of the Chromovert technology-enabled NaV1.7-αβ1β2 cell line resulted in the discovery and development of Chromocell compound CC8464 (Babich et al. 2016), a clinical lead compound that has been advanced through human Phase I studies with potential to address the opioid epidemic.
Third, results using the Chromovert technology-enabled panel of four GABAA cell lines provide proof-of-concept data for the production of more comprehensive combinatorial panels of GABAA subunits for use to increase understanding of subunit combinations that underlie in vivo function and drug pharmacology. The GABAA ion channel family includes at least 19 different subunits (6 alpha, 3 beta, 3 gamma, 1 delta, 1 epsilon, 1 theta, 1 pi, and 3 rho subunits) thought to form pentameric ion channels comprising 2 alpha subunits, 2 beta subunits and a third subunit (Olsen and Sieghart 2008). Drugs, including FDA-approved drugs and failed candidate molecules, may be profiled against such panels to identify in vitro correlates for desired pharmacology and unwanted side-effects.
Fourth, comparisons of Chromovert technology-enabled CFTR and CFTR-Δ508 cell lines suggest certain advantages compared to traditional cell-based assays. For instance, whereas transient transfection is generally regarded to produce more robust cell-based assay results as compared to cell lines, Chromovert technology enabled cell lines with 2 to 6 fold or more functionality. Previously, no CFTR-Δ508 cell lines have been reported to result in cell-based assay for the detection of CFTR-Δ508 in the absence of trafficking correctors used to release the mutant CFTR trapped in intracellular compartments. In contrast, the Chromovert technology-enabled CFTR-Δ508 cell line resulted in a detectable cell-based assay signal in the absence of any trafficking correctors. This CFTR mutant is the most prevalent in cystic fibrosis (Lemna et al. 1990). The creation of a cell-based assay for CFTR-Δ508 that does not necessitate the use of trafficking correctors may provide for more physiologically relevant drug discovery.
Finally, ENaC is a highly-sought heteromultimeric ion channel drug target comprising α, β and γ subunits expressed on the apical surface of epithelial cells where it conducts sodium and plays a role in fluid balance (Fornius 2013). In the lung, ENaC is a validated drug target in pulmonary edema (PE), chronic obstructive pulmonary disease (COPD) and cystic fibrosis (CF), with potential to treat coronavirus disease 2019, COVID-19, (Matalon et al. 2015; Eisenhut and Shin 2020). ENaC has also been postulated as the putative human salt taste receptor (Roper 2015), where salt taste enhancers could be used to reduce dietary sodium. However, cell-based drug discovery against this validated drug target has been hampered due to its cytotoxicity in cultured cells. Only Chromovert technology enabled the creation of an αβγ-ENaC cell line. The creation of the ENaC cell line in turn enabled limiting proteolysis to achieve functionally distinct proteolyzed variants, resulting in the discovery of an ENaC potentiator and human sensory confirmation of ENaC’s role as a human salt taste receptor.
Many cultured cells exhibit vast cell-to-cell genetic diversity. For example, according to the product specifications at ATCC at https://www.atcc.org/, the HEK 293 cell line is characterized as follows: “This is a hypotriploid human cell line. The modal chromosome number was 64, occurring in 30% of cells. The rate of cells with higher ploidies was 4.2%.” Compared to other cell line production methods, it is possible that the signaling probes used in Chromovert technology may exploit the heterogeneity of transfected cell cultures by serving as biomarkers to detect and isolate those cells that are compatible with the viable and functional expression of biological targets including proteins that are otherwise cytotoxic in cell cultures.
Materials and methods
Production of cell lines
Each gene of interest was subcloned into an independent mammalian expression vector containing a Chromo-Tag and drug selectable marker and transfected into HEK 293T and CHO cells cultured using DMEM supplemented with 10% fetal bovine serum, a 4 mM l-Glutamine, 10 mM Hepes pH 7.4. After 1–3 days cells were transferred to fresh media containing mild selective pressure for 7–14 days. On the day of the experiment, each well of four to six 24-well tissue culture plates was plated with 200–400 µL of transfected cells at 1.25–2 × 106 cells/mL in serum-free media and exposed to differentially-labeled signaling probes directed to the expression products. For each well, 1.7–2.5 µL of each probe at a concentration of 2.5–5 µM was added to 50 µL of serum free media in tube A and 0.3–0.75 µL of transfection reagent, using Lipofectamine for HEK cells (Thermo Fisher 18324) and jetPEI for CHO (Polyplus Transfection 101), added to tube B. The contents of tubes A and B were mixed and incubated at room temperature for 25–30 min prior to addition to the well. Following a 1–2 h incubation at 37 °C in a tissue culture incubator, serum-free media was added to the well up to a total volume of 1 mL. The plates were spun at 2000 RPM for 4 min. 750 µL of media was aspirated and replaced with 250 µL of fresh serum-free media. The cells were incubated for an additional 2–2.5 h and then used to isolate individual positive cells using Beckman Coulter Epic Altra or Becton Dickinson FACS Aria flow cytometers.
Fluorogenic probes directed to plasmid-encoded sequences downstream of the 3´ stop codon of the cDNAs of interest were used. A first Chromo-Tag comprising portions of the reverse-complement of the mRNA sequence of the human Vav gene and two derivate Chromo-Tag sequences were generated and used. Signaling probes directed to the Chromo-Tags are listed below including fluorophores and quenchers. BHQ2 was also used. 5-Methyl-dC and 2-Amino dA chemical modifications were included for the underlined bases except in probes used for the ENaC cell line.
Signaling probes directed to Chromo-Tags
Chromo-Tag A:5´–Cy5 GCCAGTCCCAGTTCCTGTGCCTTAAGAACCTCGC BHQ3–3´
Chromo-Tag B:5´–CY5.5 GCGAGTCGCAGAACGACAGGGTTAACTTCCTCGC BHQ3–3´
Chromo-Tag C:5´–Fam GCGAGAGCGACAAGCAGACCCTATAGAACCTCGC BHQ1–3´
Individual positive cells were expanded in 96-well multiter plates. Cell lines were binned based on growth rate, maintained using automated cell culture methods and functional cell-based assays were used to select final clones. The ENaC cell line was created without robotic cell culture methods and a custom low-sodium F-12 HAM media (Sigma N4888) formulation was used for cell culture (Shekdar and Langer 2017).
Cell-based assays
Functional testing using fluorescent cell-based assays of cells cultured in the absence of any antibiotic-mediated selectable pressure was performed over time (typically for at least 4–6 weeks) to identify inherently stable clones. Commonly available fluorescent membrane potential and calcium flux kits and reagents were used for fluorescent cell-based assays in 96-well and 384-well plates using the Molecular Devices FLIPR 3 and Hamamatsu FDSS 6000 plate readers as described (Shekdar and Langer 2011, 2017; Shekdar et al. 2012a, b). Electrophysiology of ion channel cell lines was obtained using the Nanion Technologies Patchliner patch clamp system. The patch clamp recordings were conducted according to Nanion’s standard procedure (Obergrussberger et al. 2014). Currents were measured using the whole-cell configuration. The extracellular solution contained the following: 140 mM NaCl, 4 mM KCl, 1 mM MgCl2, 2 mM CaCl2, 5 mM glucose, 10 mM Hepes, pH 7.4 (adjusted with NaOH). The intracellular solution for sodium current recordings contained the following: 50 mM CsCl, 10 mM NaCl, 60 mM CsF, 20 mM EGTA, 10 mM Hepes, pH 7.2 (adjusted with CsOH). For transient transfection studies, Fugene 6 (Promega E2691) was used to transfect expression plasmid into cells cultured as above and cells were assayed three days post-transfection.
Acknowledgements
In loving memory of Chromocell co-founder and Rockefeller University Professor Günter Blobel, doctoral advisor to Kambiz Shekdar, and with deep gratitude to Chromocell co-founder and CEO Christian Köpfli, Research Foundation to Cure AIDS Chairwoman Karen Hagberg, Columbia University Medical Center/New York-Presbyterian Hospital Chairman of Medicine Don Landry, and all colleagues and advisors.
Author contributions
KS conceptualized and performed the first proof-of-concept experiments for Chromovert technology and Chromo-Tags and conceptualized cell-based proteolysis of ENaC. DS optimized Chromovert technology. JL performed flow cytometry and experiments. JA, AL, LS, PS, OB, OD, and SV performed experiments. All authors contributed to data analysis. KS wrote the manuscript.
Funding
This work was privately funded by Chromocell.
Data availability
Data generated and analyzed in this study are included in this publication. Materials are available from the corresponding author.
Compliance with ethical standards
Conflict of interest
KS is CSO, co-founder and director of Chromocell Corporation with equity in the company, sole member of Secondcell Bio, LLC established to make Chromovert Technology available to the research community and president of Research Foundation to Cure AIDS, with a license to the technology in the field of curing HIV infection. DS is founder of D3 Insight Consulting, LLC. All authors performed their work as employees of Chromocell.
Ethical approval
Human sensory studies were performed according to protocols approved by an independent institutional review board.
Informed consent
Human sensory studies were performed with written informed consent obtained from all participants.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Babich O, Garyantes T, Luo R, Palling D, Venkatachalan S, Wang-Fischer Y (2016) Sodium channel modulators for the treatment of pain and diabetes. US Patent 9458118
- Chalfie M, Tu Y, Euskirchen G, Ward WW, Prasher DC. Green fluorescent protein as a marker for gene expression. Science. 1994;263:802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- Chandrashekar J, Mueller KL, Hoon MA, Adler E, Feng L, Guo W, Zuker CS, Ryba NJ. T2Rs function as bitter taste receptors. Cell. 2000;100:703–711. doi: 10.1016/S0092-8674(00)80706-0. [DOI] [PubMed] [Google Scholar]
- Cockett M, Bebbington CR, Yarranton GT. High level expression of tissue inhibitor of metalloproteinases in Chinese hamster ovary cells using glutamine synthetase gene amplification. Biotechnology. 1990;8:662–667. doi: 10.1038/nbt0790-662. [DOI] [PubMed] [Google Scholar]
- Eisenhut M, Shin J. Pathways in the pathophysiology of coronavirus 19 lung disease accessible to prevention and treatment. Front Physiol. 2020;11:872. doi: 10.3389/fphys.2020.00872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornius M. Treatment of pulmonary edema by ENaC activators/stimulators. Curr Mol Pharmacol. 2013;6:13–27. doi: 10.2174/1874467211306010003. [DOI] [PubMed] [Google Scholar]
- Johnston G. GABA(A) receptor channel pharmacology. Curr Pharm Des. 2005;11:1867–1885. doi: 10.2174/1381612054021024. [DOI] [PubMed] [Google Scholar]
- Krautwurst D, Yau KW, Reer RR. Identification of ligands for olfactory receptors by functional expression of a receptor library. Cell. 1998;95:917–926. doi: 10.1016/S0092-8674(00)81716-X. [DOI] [PubMed] [Google Scholar]
- Lemna, et al. Mutation analysis for heterozygote detection and the prenatal diagnosis of cystic fibrosis. N Engl J Med. 1990;322:291–296. doi: 10.1056/NEJM199002013220503. [DOI] [PubMed] [Google Scholar]
- Marras SA, Kramer FR, Tyagi S. Multiplex detection of single-nucleotide variations using molecular beacons. Genet Anal. 1999;14:151–156. doi: 10.1016/S1050-3862(98)00018-7. [DOI] [PubMed] [Google Scholar]
- Matalon S, Bartoszewski R, Collawn JF. Role of epithelial sodium channels in the regulation of lung fluid homeostasis. Am J Physiol Lung Cell Mol Physiol. 2015;309:L1229–1238. doi: 10.1152/ajplung.00319.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohan C, Park SH, Chung JY, Lee GM. Effect of doxycycline-regulated protein disulfide isomerase expression on the specific productivity of recombinant CHO cells: thrombopoietin and antibody. Biotechnol Bioeng. 2007;98:611–615. doi: 10.1002/bit.21453. [DOI] [PubMed] [Google Scholar]
- Nowogrodzki A (2019) The challenge of using CRISPR to knock in genes The Scientist. https://www.the-scientist.com/lab-tools/jacking-up-gene-knock-ins-65504
- Obergrussberger A, Haarmann C, Rinke I, Becker N, Guinot D, Brueggemann A, Stoelzle-Feix S, George M, Fertig N. Automated patch clamp analysis of nAChα7 and Na(V)1.7 channels. Curr Protoc Pharmacol. 2014;65(1):1–48. doi: 10.1002/0471141755.ph1113s65. [DOI] [PubMed] [Google Scholar]
- Offermanns S, Simon M. Gα15 and Gα16 couple a wide variety of receptors to phospholipase C. J Biol Chem. 1995;270:15175–15180. doi: 10.1074/jbc.270.25.15175. [DOI] [PubMed] [Google Scholar]
- Olsen RW, Sieghart W. International union of pharmacology LXX subtypes of γ aminobutyric acid A receptors: classification on the basis of subunit composition, pharmacology, and function. Update Pharmacol Rev. 2008;60:243–260. doi: 10.1124/pr.108.00505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Malley HA, Isom LL. Sodium channel β subunits: emerging targets in channelopathies. Annu Rev Physiol. 2015;77:481–504. doi: 10.1146/annurev-physiol-021014-071846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pear WS, Nolan GP, Scott ML, Baltimore D. Production of high-titer helper-free retroviruses by transient transfection. PNAS. 1993;90:8392–8396. doi: 10.1073/pnas.90.18.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roper SD. The taste of table salt. Pflugers Arch. 2015;467:457–463. doi: 10.1007/s00424-014-1683-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shekdar K, Langer J (2011) Cell lines expressing GABA receptor and methods of using them US Pat Publ 2011/0003711
- Shekdar K, Langer J (2017) Cell lines expressing ENaC and methods of using them. US Patent 9534035
- Shekdar K, Sawchuk D, Shah P (2012) Novel cell lines and methods. US Patent Publ 2012/0015841
- Shekdar K, Venkatachalan S, Langer J, Sawchuk D (2012) Cell lines expressing CFTR and methods of using them. US Patent Publ 2012/0058918
- Urlaub G, Emmanuel K, Carothers AM, Chasin LA. Deletion of the diploid dihydrofolate reductase locus from cultured mammalian cells. Cell. 1983;33:405–412. doi: 10.1016/0092-8674(83)90422-1. [DOI] [PubMed] [Google Scholar]
- Wang D, Tai PWL, Gao G. Adeno associated virus vector as a platform for gene therapy delivery. Nat Rev Drug Discov. 2019;18:358–378. doi: 10.1038/s41573-019-0012-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wetzel H, Oles M, Wellerdieck C, Kuczkowiak M, Gisselmann G, Hatt H. Specificity and sensitivity of a human olfactory receptor functionally expressed in human embryonic kidney 293 cells and xenopus laevis oocytes. J Neurisci. 1999;19:7426–7433. doi: 10.1523/JNEUROSCI.19-17-07426.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data generated and analyzed in this study are included in this publication. Materials are available from the corresponding author.