Abstract
Rationale
Myocardial infarction (MI) is a leading cause of cardiovascular mortality globally. The improvement of microvascular function is critical for cardiac repair after MI. Evidence now points to long non-coding RNAs (lncRNAs) as key regulators of cardiac remodelling processes. The lncRNA Malat1 is involved in the development and progression of multiple cardiac diseases. Studies have shown that Malat1 is closely related to the regulation of endothelial cell regeneration. However, the potential molecular mechanisms of Malat1 in repairing cardiac microvascular dysfunction after MI remain unreported.
Methods and results
The present study found that Malat1 is upregulated in the border zone of infarction in mouse hearts, as well as in isolated cardiac microvascular endothelial cells (CMECs). Targeted knockdown of Malat1 in endothelial cells exacerbated oxidative stress, attenuated angiogenesis and microvascular perfusion, and as a result decreased cardiac function in MI mice. Further studies showed that silencing Malat1 obviously inhibited CMEC proliferation, migration and tube formation, which was at least in part attributed to disturbed mitochondrial dynamics and activation of the mitochondrial apoptosis pathway. Moreover, bioinformatic analyses, luciferase assays and pull-down assays indicated that Malat1 acted as a competing endogenous RNA (ceRNA) for miR-26b-5p and formed a signalling axis with Mfn1 to regulate mitochondrial dynamics and endothelial functions. Overexpression of Mfn1 markedly reversed the microvascular dysfunction and CMEC injuries that were aggravated by silencing Malat1 via inhibition of excessive mitochondrial fragments and mitochondria-dependent apoptosis.
Conclusions
The present study elucidated the functions and mechanisms of Malat1 in cardiac microcirculation repair after MI. The underlying mechanisms of the effects of Malat1 could be attributed to its blocking effects on miR-26b-5p/Mfn1 pathway-mediated mitochondrial dynamics and apoptosis.
Keywords: Myocardial infarction, Cardiac microvascular dysfunction, Malat1, miR-26b-5p, Mfn1
1. Introduction
Myocardial infarction (MI) is a common and severe cardiovascular disease that leads to heart failure and cardiac mortality [1]. The maintenance of cardiac function after MI is mainly dependent on angiogenesis and preserved microvascular function in the border zone of the infarcted myocardium. However, in addition to insufficient angiogenesis via self-healing in that region, microvascular dysfunction is the predominant secondary injury of the stenosed epicardial coronary artery. Increased microvascular flow resistance and compromised blood perfusion lead to an imbalance between nutritional supply and demand and in turn induce hypoxic injury and necrosis of cardiomyocytes, precipitating infarct expansion, cardiac insufficiency and irreversible end-stage heart diseases. Therefore, appropriate treatment for the improvement of microvascular dysfunction can slow down the process of cardiac pathologic remodelling and improve outcomes after MI.
Long non-coding RNAs (lncRNAs) are RNA molecules that do not possess protein-coding potential. Nevertheless, this RNA subgroup has been demonstrated to play crucial roles in the pathogenesis of many cardiovascular disorders, including MI, hypertension, and arrhythmia [2,3]. The lncRNA Malat1 is one of the most promising members involved in the angiogenic process of various diseases, such as diabetic foot ulcer, diabetic retinopathy, and central nervous system injury [[4], [5], [6]]. Moreover, Malat1 can reduce the apoptosis rate of brain microvascular endothelial cells (ECs) after ischaemic stroke [7], improve human umbilical vein endothelial cell (HUVEC) tube formation and endothelial barrier function [8], and promote high glucose-induced proliferation of human retinal vascular ECs [5]. However, the contribution of Malat1 to cardiac microvascular protection is currently a neglected topic. Therefore, the molecular mechanisms and signalling regulation of Malat1 in the cardiac microcirculation should be elucidated. Moreover, as accepted downstream targets of lncRNAs, microRNAs (miRNAs) have been regarded as molecular sponges that competitively bind lncRNAs and subsequently regulate the expression of target mRNAs [9]. Although several miRNAs have been reported to participate in regulating vascular functions after MI [10], the specific miRNA behind Malat1 in facilitating cardiovascular repair remains unknown.
Mitochondria are now considered arbiters of cell death and survival, facilitating endothelial repair or accelerating cell damage. By promoting oxidative stress and activating the mitochondrial apoptotic pathway, mitochondrial dysfunction can trigger and aggravate many cardiovascular diseases [11,12]. Conversely, enhancing mitochondrial functions via mitochondrial quality control measures showed preventive effects on microvascular collapse. Mitochondrial dynamics, including fission and fusion, are in responsible for clearing injured mitochondria and generating well-functioning mitochondria during stress. However, under severe injury states, overactive mitochondrial fission generates a mass of small and non-functioning mitochondria, which are the source of mitochondrial reactive oxygen species (ROS) that transduce apoptotic signals. Mitofusin-1 (Mfn1) promotes mitochondrial fusion and maintains mitochondrial functions in many pathologic conditions. Furthermore, Mfn1 is required for angiogenic potential and modulates signalling pathways in ECs [13]. Studies have demonstrated that non-coding RNAs are involved in modulating mitochondrial dynamics and mitochondrial functions via Mfn1 in many pathologic conditions. For example, downregulation of miR-140 could upregulate the expression of Mfn1, improve mitochondrial function and inhibit mitochondrial fission, contributing to the improvement of cell viability in cardiomyocytes and the reduction in infarct size in animal models [14]. It is now known that knocking down Malat1 in HepG2 cells aggravates mitochondrial functions and compromises mitochondrial metabolism [15]; however, whether Malat1 can affect mitochondrial dynamics in cardiac microvascular endothelial cells (CMECs) after ischaemic injury and thereby improve vascular functions remains unreported.
On the basis of the previous findings highlighted above, we hypothesized that Malat1 may have a role in protecting the cardiac microcirculation by modulating mitochondrial dynamics and functions. Our present study reported that silencing Malat1 exacerbated microvascular dysfunction by enhancing CMEC apoptosis partly through regulation of miR-26b-5p/Mfn1 pathway-associated mitochondrial dynamics. This study sheds light on a novel signalling axis in the cardiac microcirculation and provides a potential therapeutic strategy for MI treatment.
2. Methods
2.1. Animals
Four-week-old male C57BL/6 mice were purchased from Beijing Vital Charles River Laboratory (China). All mice were fed with standard chow and water. At 5 weeks of age, the mice were injected with His-tagged adeno-associated virus (AAV) 2/9 containing Malat1-specific small hairpin RNA (shMalat1) or scrambled shRNA via the caudal veins to establish an EC-specific Malat1 gene knockout model. Likewise, Flag-tagged AAV2/9 was used to overexpress Mfn1 in mouse ECs in vivo. Animal experiments were approved by the China Medical University Institutional Ethics Committee and followed the Guide for the Care and Use of Laboratory Animals (US National Institutes of Health publication, Doc. 2011–11490).
2.2. Mice MI model establishment
The MI mouse model was constructed by permanent ligation of the left anterior descending coronary artery (LAD). In short, mice were anaesthetized and subjected to thoracotomy to expose the hearts. Then, the LAD was ligated permanently. The sham group underwent the same operation but without LAD ligation. Echocardiography was performed to measure the cardiac function of each group, and experimental samples, including serum and cardiac tissues, were collected after euthanasia.
2.3. Serological tests
The activity of lactate dehydrogenase (LDH) was determined using an LDH activity test kit. The levels of cardiac troponin T (cTnT) and brain natriuretic peptide (BNP) were tested via enzyme-linked immunosorbent assay (ELISA) using a mouse cTnT ELISA kit and a mouse BNP ELISA kit. All of the reagents above were purchased from Solarbio (China), and the tests were performed according to the manufacturer's instructions.
2.4. Oxidative stress and nitric oxide (NO) content measurements
Protein samples from mouse hearts and CMECs were collected after lysis and centrifugation, followed by quantitation with a BCA Protein Assay Kit (Beyotime, China). Malondialdehyde (MDA) content was determined via a Lipid Peroxidation MDA Assay Kit (Beyotime, China). The enzyme activities of total superoxide dismutase (t-SOD) and Mn-SOD were assessed by the Cu/Zn-SOD and Mn-SOD assay kits. The contents of glutathione (GSH) and oxidized glutathione (GSSG) were detected using a GSH and GSSG Assay Kit (Beyotime, China). ROS generation and H2O2 content were detected by a Lucigenin chemiluminescence reagent kit (Genmed Scientifics, USA) and Amplex Red Hydrogen Peroxide/Peroxidase Assay Kit (Thermo Fisher, USA). The NO content was detected using a Micro NO Content Assay Kit (Solarbio, China). The assays above were carried out according to the manufacturer's instructions and were standardized by protein content.
2.5. Histochemistry and immunofluorescent staining
Tissue samples were fixed, dehydrated and paraffin-embedded for histological staining. Collagen deposition was stained by the Masson staining method, and quantitative analysis of collagen content was performed by ImageJ. Lycopersicon esculentum lectin (1 mg/ml) was injected via the tail vein and was allowed to circulate for 10 min to label perfused myocardial microvessels. Then, the heart was sliced into 8-μm frozen sections for immunofluorescent staining of CD31, eNOS and VEGFR2. All antibodies used for immunofluorescent staining were purchased from Abcam (USA).
2.6. Real-time PCR
Total RNA was extracted from mouse heart tissues or cells using TRIzol (Invitrogen, USA). The quantitative expression of Malat1, miR-26b-5p and mRNAs was detected by the SYBR Premix Ex Taq™ kit (TaKaRa, Japan). β-Actin was used as a reference control gene in each sample, and U6 was used as an internal control for miR-26b-5p. The relative gene expression data were calculated using the 2−ΔΔCT method. The primers used in this study are listed in Table S1.
2.7. Western blot assay
Total proteins from cardiac samples or CMECs were lysed and extracted by RIPA lysis buffer (Beyotime, China). After quantitation using a BCA Protein Assay Kit (Beyotime, China), protein samples were separated by SDS-PAGE, transferred to PVDF membranes, and blocked with 5% BSA. Following incubation with primary antibodies at 4 °C overnight, the membranes were incubated with HRP-labelled secondary antibodies for 1 h at room temperature. All antibodies were purchased from Abcam (UK) and diluted following the manufacturer's instructions. The immunoblots were developed using Pierce™ ECL Western Blotting Substrate (Thermo Scientific™, USA).
2.8. CMEC isolation and oxygen-glucose deprivation (OGD) treatment
CMECs were isolated from myocardial tissue using CD31-coupled microbeads as previously described and cultured in endothelial culture medium (ECM, ScienCell) [16]. To stimulate the hypoxic-ischaemic microenvironment in vitro, CMECs were cultured in Dynamic O2 and CO2 Subchamber Controller Oxycycler C42 (Biospherix, USA) at 37 °C for 24 h with a gas mixture consisting of 5% CO2 and 95% N2.
2.9. Cell transfection
Short interfering RNA for Malat1 (siMalat1), short interfering RNA for Mfn1 (siMfn1), miR-26b-5p mimic and inhibitor, and their negative controls (siNC, mimic-NC, inhibitor-NC), were purchased from RiboBio. The Mfn1 overexpression plasmid and its negative control (pcDNA3.1) were purchased from GenePharma (China). When the CMECs grew to 75% confluence, gene transfections were performed using Lipofectamine 3000 Transfection Reagent (Thermo Fisher, USA) according to the manufacturer's protocols.
2.10. Cell viability assay
Cell viability was determined by the Cell Counting Kit-8 assay (Biosharp). Following the manufacturer's instructions, a total of 110 μl working reagent was added to each well of a 96-well plate, and then the plate was incubated at 37 °C for 2 h. The spectrophotometric absorbance at 450 nm of each sample was detected by a Varioskan LUX Multimode Microplate Reader (Thermo Scientific™).
2.11. Cell migration assay
Cell migration ability was determined by the Transwell assay. For the Transwell assay, 5 × 105 cells were seeded in the upper chambers in ECM without FBS. ECM with 20% FBS was added to the lower chambers to induce cell penetration. After 24 h of incubation, the migrated CMECs were fixed with 4% paraformaldehyde solution (Solarbio, China) and stained with crystal violet. Random visual fields of each group were imaged by an optical microscope (OLYMPUS CKX53, Japan), and the number of migrated cells was counted manually.
2.12. Tube formation assay
The capacity of CMECs to form vascular rings in vitro was assessed by the tube formation assay. The Matrigel™ Basement Membrane Matrix (BD Biosciences, USA) was thawed in an ice bath for 12 h and then rapidly added to precooled 96-well plates (65 μl/well). After polymerization at 37 °C for 35 min, a 70 μl cell suspension was placed on the Matrigel coat at a density of 2 × 104 cells per well. The CMECs were incubated at 37 °C for 2 h. Images of each group were obtained by a DMIL LED microscope (Leica, Germany), and the cell tube formation capacity was quantified by calculating the tube length and branch points using Image-Pro Plus (Media Cybernetics, USA).
2.13. CMEC fluorescent staining
A total of 1.2 × 106 CMECs were plated in confocal dishes. Fluorescent images of each cell sample were obtained via TCS SP8 laser confocal microscopy (Leica, Germany) and quantitatively analysed using ImageJ. BeyoClick™ EdU-594 (Beyotime, China) was used to evaluate CMEC proliferation. After incubation with EdU for 2 h, the cells were fixed, permeabilized and stained with EdU colour-substrate solution. A TUNEL Apoptosis Assay Kit (Roche, Germany) was used to evaluate TUNEL-positive CMECs. According to the manufacturer's instructions, CMECs were fixed, permeabilized, and then stained with TUNEL detection solution in the dark for 60 min. An ROS assay kit (Beyotime, China) and a mitochondrial superoxide indicator (Invitrogen, USA) were utilized to label intracellular ROS and mitochondrial ROS (mtROS). The cells were rinsed and incubated with the two fluorescent probes in the dark at 37 °C for 20 min. The intracellular ROS and mtROS contents are expressed as the area of intensity (AOI). Mitochondrial membrane potential (MMP) depolarization was assessed by a JC-1 fluorescent probe and is represented as the ratio of red to green fluorescence intensity. Mitochondria and nuclei were labelled with MitoTracker Red solution and DAPI solution.
2.14. Luciferase reporter assay
The 3ʹ-UTRs of both Malat1 and Mfn1 contain conserved miR-26b-5p binding sites. Synthesis of the mutated 3ʹ'-UTR of Malat1 was accomplished using a QuikChange II XL Site-Directed Mutagenesis Kit (Stratagene), and the fragment was amplified by PCR. The PCR fragment was inserted into the XhoI and NotI downstream sites of the luciferase gene in the psiCHECK-2 vector. HEK 293 cells were co-transfected with the 3ʹ-UTR luciferase vector and miR-26b-5p mimics via Lipofectamine 3000 (Invitrogen). Renilla luciferase was used as internal reference. After 48 h of expression, luciferase activity was determined via the Dual-Luciferase Reporter Assay System (Promega). Similarly, wild-type and mutated forms of the Mfn1 3ʹ-UTR were used to investigate the relationship between Mfn1 and miR-26b-5p.
2.15. Pull-down assay with biotinylated miRNA
After 48 h of transfection with biotinylated miRNA (80 nM), CMECs were rinsed with PBS and incubated in ice-cold lysis buffer for 10 min with gentle vortexing. Then, the lysate was incubated with M − 280 streptavidin-coated magnetic beads (Sigma-Aldrich, USA). After 3 h of incubation at 4 °C, the magnetic beads were washed successively with lysis buffer, low-salt washing buffer and high-salt washing buffer. The bound RNAs were extracted by TRIzol and subjected to quantitative analysis by qRT-PCR.
2.16. RNA immunoprecipitation (RIP)
The RIP assay was performed using the Magna RIP RNA Binding Protein Immunoprecipitation Kit (Millipore, USA) according to the manufacturer's protocol. After transfection with miR-26b-5p mimics or negative control, CMECs were completely lysed in RIP lysis buffer. The cell lysates were incubated with anti-AGO2 or anti-IgG magnetic beads (Millipore, USA) at 4 °C for 7 h. After that, the protein components were extracted by incubating the magnetic beads with Proteinase K buffer, and RNAs were further extracted via TRIzol. The purified RNA was quantified by qRT-PCR.
3. Results
3.1. Silencing Malat1 in CMECs aggravated MI and microvascular dysfunction
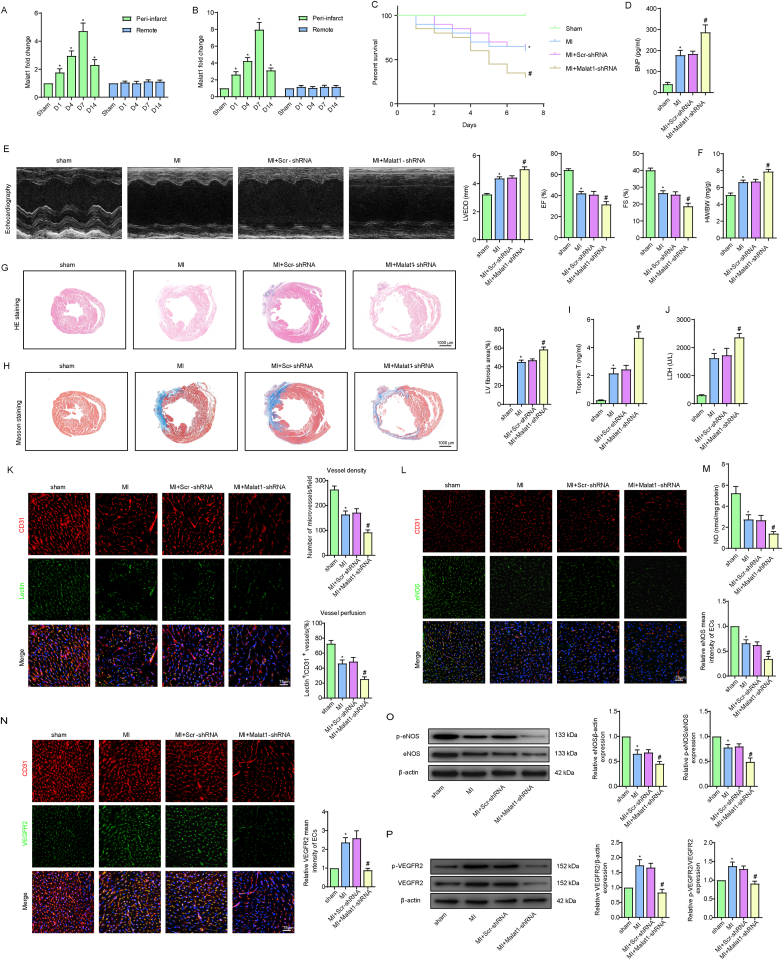
Malat1 has been reported to be upregulated in MI in rats, mice and patients [[17], [18], [19]]. To investigate the exact expression variation of Malat1 in cardiac tissues and microcirculation after myocardial infarction, the expression levels of Malat1 were determined in post-MI mouse hearts. The results showed that, in comparison to that in sham mice, Malat1 was significantly increased only in the border zone of infarction and reached a peak at day 7 (Fig. 1A). More evidence was acquired by isolating CMECs from sham and MI mouse hearts. Similarly, compared with the levels in the sham group counterparts, higher expression levels of Malat1 were found only in CMECs isolated from the border zone, with expression peaking at day 7 (Fig. 1B). Therefore, mice at 7 days after MI were selected as our study subjects.
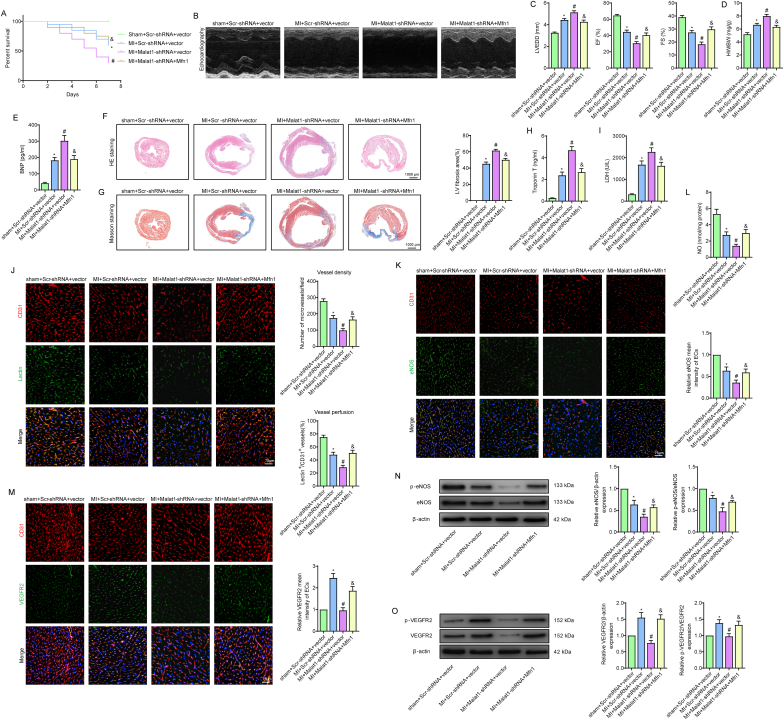
Fig. 1.
Silencing Malat1 in CMECs aggravated myocardial infarction (MI) and microvascular dysfunction. Time course of the relative expression of Malat1 in cardiac tissues and CMECs in the infarct border zone and remote areas after MI (A, B). One-week overall survival curve (C). Analysis of brain natriuretic peptide (BNP) levels in serum (D). The M-mode of echocardiography images and the data on left ventricular end-diastolic diameter (LVEDD), ejection fraction (EF) and fractional shortening (FS) for each group (E). Heart weight to body weight ratio (HW/BW) for each group (F). HE staining, Masson staining and quantification of cardiac fibrosis (G, H). Analyses of cardiac troponin T (cTnT) concentration and lactate dehydrogenase (LDH) activity in serum (I, J). Microvascular perfusion of the border zone was indicated by the ratio of lectin-perfused vessels (green) to CD31-positive ECs (red) (K). Immunofluorescence staining for eNOS (L). Nitric oxide (NO) content in the border zone of each group (M). Immunofluorescence staining for VEGFR2 (N). eNOS expression and phosphorylation at Ser1177 in the border zone were analysed by Western blotting (O). VEGFR2 expression and phosphorylation at Tyr1175 in the border zone were analysed by Western blotting (P). *p < 0.05 compared with the sham group, #p < 0.05 compared with the MI + Scr-shRNA group. n = 6 in each group.
To evaluate the potential effects of Malat1 on cardiac microvascular protection, we knocked down Malat1 in mouse ECs by AAV-shMalat1 (Fig. S1A-D). Our preliminary experiment showed that silencing Malat1 in normal mice had no effect on cardiac functions, microvascular density or perfusion (data not shown). However, Malat1 knockdown in MI mice significantly decreased the overall 7-day survival rate and severely exacerbated cardiac remodelling and dysfunction (Fig. 1C–F). The above changes could be attributed to expanded infarction, as indicated by the increased fibrosis deposition area and higher serum contents of cTnT and LDH (Fig. 1G–J). Additionally, knockdown of Malat1 aggravated oxidative damage after MI, as shown by the decreased GSH content, reduced t-SOD enzyme activity, and increased ROS, H2O2, MDA and GSSG levels in cardiac tissues (Fig. S3A).
Further experiments were performed to assess the function of Malat1 in coronary microcirculation after MI. In MI hearts, microvessel density (MVD) and lectin-perfused vessels were obviously decreased. More importantly, silencing Malat1 in ECs led to a further reduction in MVD and microvascular perfusion (Fig. 1K), indicating that Malat1 has a protective role in microvascular functions after MI. Moreover, knocking out Malat1 negatively affected the expression of eNOS, NO and VEGFR2 in the peri-infarct area, which was accompanied by reduced phosphorylation of eNOS at Ser1177 and VEGFR2 at Tyr1175 (Fig. 1L–P), suggesting that angiogenesis and endothelium-derived vasodilatation after MI were partly mediated by Malat1.
Mitochondrial dynamics and related apoptosis are topics in many myocardial microvascular diseases. In the present study, the mRNA expression levels of Fis1 and Drp1 were obviously increased in isolated CMECs after MI, along with decreased Mfn1, Mfn2 and Opa1 levels (Fig. S3B). This trend was only partially enhanced after Malat1 was inhibited because only DRP1, Fis1 and Mfn1 underwent greater changes in Malat1-silenced MI hearts than in MI hearts transfected with the scramble control (Fig. S3B). Additionally, the pro-apoptotic genes Bax and caspase 3 were increased to a much greater degree, and the anti-apoptotic genes Bcl-2, Mcl-1 and Bcl-xL were more greatly downregulated after Malat1 silencing (Fig. S3B).
Overall, these results suggested that knocking out Malat1 in CMECs accelerated infarction extension and exacerbated cardiac dysfunction, which was at least in part caused by the aggravated microvascular dysfunction induced by abnormal mitochondrial dynamics and related apoptosis.
3.2. Silencing Malat1 compromised CMEC functions by accelerating mitochondrial fragmentation and related apoptosis under hypoxia
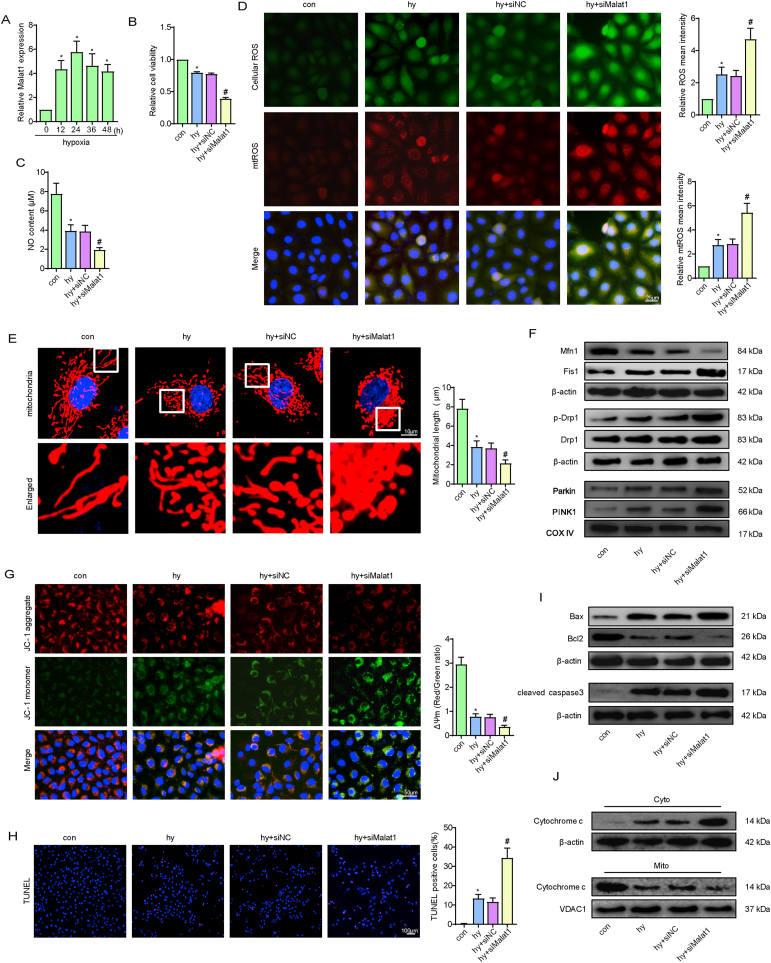
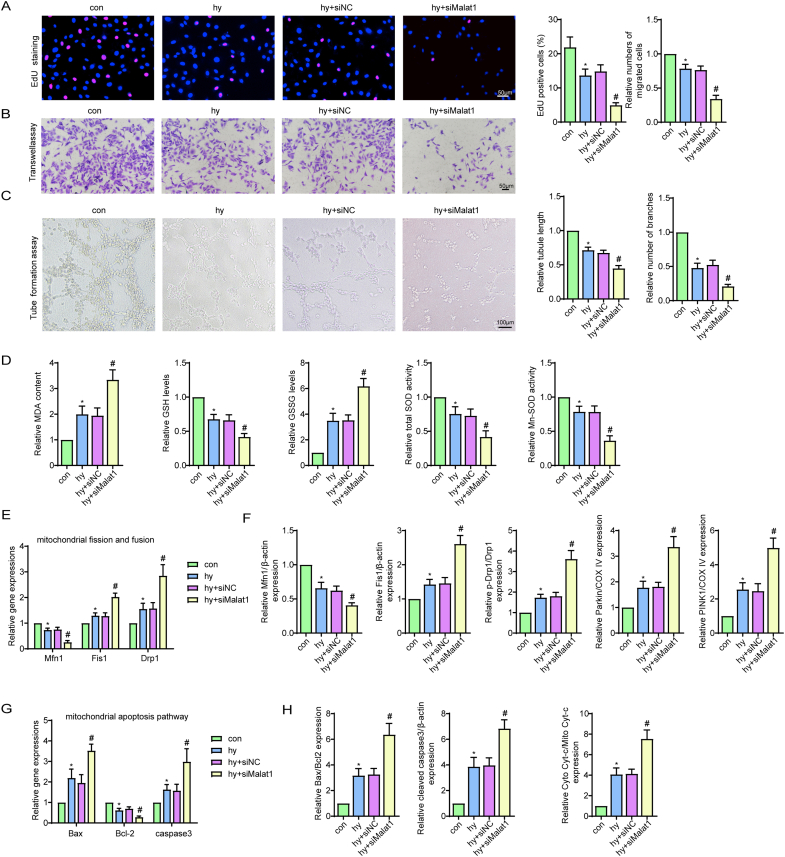
To provide more evidence for the role of Malat1 in regulating CMEC functions and repair, in vitro experiments were performed on CMECs exposed to hypoxia. Based on the finding that the expression of Malat1 was obviously increased in CMECs under hypoxia and peaked at 24 h (Fig. 2A), Malat1 was silenced via siRNA to explore its potential functions (Fig. S1E, F). Compared with the hypoxia group treated with siNC, the Malat1-knockdown hypoxia group had a more pronounced decrease in cell viability and proliferation ability, as detected by the CCK-8 and EdU staining methods (Fig. 2B, S4A). Moreover, Malat1 knockdown compromised cell migration and tube formation ability after hypoxia (Fig. S4B, C). Additionally, silencing Malat1 aggravated the dysregulation of NO synthesis in CMECs under hypoxia, suggesting compromised endothelium-mediated vasodilation (Fig. 2C).
Fig. 2.
Silencing Malat1 compromised CMEC functions by accelerating mitochondrial fragmentation and related apoptosis under hypoxia. Time course of the relative expression of Malat1 under hypoxia (A). Relative cell viability was measured by the CCK-8 assay (B). Nitric oxide (NO) content in CMECs (C). Cellular ROS (green) and mtROS (red) in CMECs were measured and quantified (D). Mitochondria were stained with MitoTracker (red), and mitochondrial morphology was quantified (E). The expression of mitochondrial fusion/fission-related proteins and mitophagy-related proteins in CMECs (F). Mitochondrial membrane potential (ΔΨm) was measured using the JC-1 assay (G). TUNEL staining of CMECs and the percentage of TUNEL-positive CMECs (red) were quantified (H). The expression of apoptosis-associated proteins in CMECs (I, J). *p < 0.05 compared with the control group, #p < 0.05 compared with the hy + siNC group. n = 5 in each group.
To examine the effects of Malat1 on hypoxia-induced oxidative damage in CMECs, several vital indices of oxidative stress were assessed. As expected, Malat1 knockdown plus hypoxic injury resulted in higher oxidative stress injury in CMECs than hypoxia alone, as demonstrated by the lower GSH content, less t-SOD and Mn-SOD enzyme activities, and more enhanced expression of intracellular ROS, mtROS, MDA and GSSG (Fig. 2D, S4D).
In the coronary microvascular field, the imbalance between mitochondrial fission and fusion has been highly associated with inferior mitochondrial function and activation of the mitochondrial apoptotic pathway. After hypoxia exposure, CMECs displayed partially fragmented mitochondrial networks, whereas more mitochondrial debris was found after siMalat1 transfection (Fig. 2E). Moreover, compared with cells in the siNC group, cells in the siMalat1 group showed higher expression of DRP1 and Fis1, a higher phosphorylation level of DRP1 at Ser616 and a greater reduction in Mfn1 (Fig. 2F, S4E, S4F). The above results indicated that the elevated Malat1 expression after hypoxia may be responsible to a certain extent for regulating mitochondrial dynamics. Mitophagy functions to clear up undesirable mitochondrial debris. The expression of Parkin and PINK1 was enhanced after hypoxia, indicating mitophagy occurred in response to hypoxia injury. Silencing Malat1 further increased mitophagy, as evidenced by more enhanced expression of Parkin and PINK1 (Fig. 2F, S4F). Enhanced mitochondrial fragmentation causes MMP depolarization, an early marker for mitochondrial apoptosis. In the present study, silencing Malat1 obviously worsened MMP depolarization in CMECs under hypoxic conditions (Fig. 2G). Accordingly, a much higher ratio of TUNEL-positive CMECs was observed after Malat1 deletion (Fig. 2H), which was coupled with more cytochrome C release from mitochondria to the cytoplasm and more transcriptional and translational modulation of mitochondrial apoptosis-related genes, including Bax, Bcl 2 and cleaved-caspase3 (Fig. 2I and J, S4G, S4H).
These results further confirmed that Malat1 played an important role in promoting CMEC functions by balancing mitochondrial dynamics and inhibiting mitochondria-dependent apoptosis.
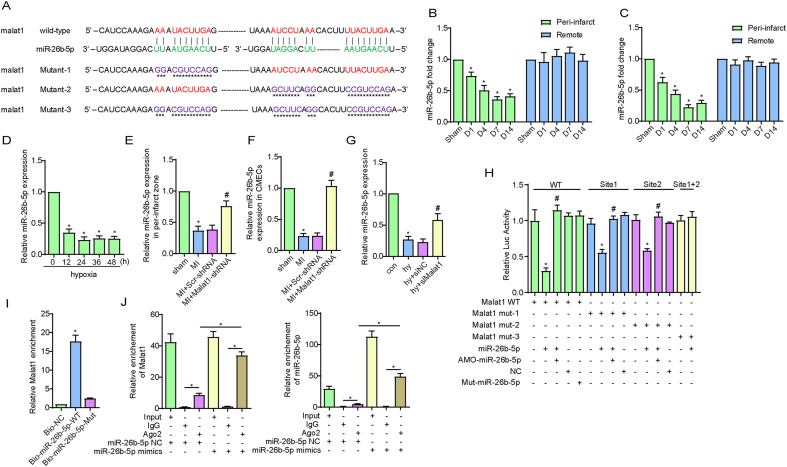
3.3. The lncRNA Malat1 targeted miR-26b-5p to act as a molecular sponge in CMECs
The mechanism by which lncRNAs function as miRNA molecular sponges to bind and sequester miRNAs and thereby indirectly regulate targeted mRNAs is known as the ceRNA network [9]. To determine the downstream miRNAs of Malat1 in ECs during hypoxia, we predicted the potential interaction of Malat1 with miR-26b-5p in starBase (http://starbase.sysu.edu.cn/) (Fig. 3A). Subsequently, a series of experiments were conducted to verify this prediction. We first determined that the expression of miR-26b-5p was decreased only in the border zone of infarction during the post-MI period and reached the lowest point at day 7 (Fig. 3B). Similar results were obtained in CMECs extracted from MI heart tissue (Fig. 3C). Additionally, in the CMEC hypoxia model, the miR-26b-5p expression level decline at 12 h and decreased to a minimum level at 24 h (Fig. 3D). Additionally, silencing Malat1 reversed the decrease in miR-26b-5p in CMECs both in vivo and in vitro (Fig. 3E–G). The above data suggested that Malat1 could suppress miR-26b-5p expression, whether directly or indirectly.
Fig. 3.
The lncRNA Malat1 targeted miR-26b-5p to act as a molecular sponge in CMECs. The predicted binding sites of Malat1 and miR-26b-5p and the mutated binding sites are indicated (A). Time course of the expression of miR-26b-5p in tissues and CMECs isolated from the border zone and remote areas of MI (B–C). Time course of the expression of miR-26b-5p in CMECs under hypoxia (D). Relative expression of miR-26b-5p in tissues and CMECs isolated from the border zone after silencing Malat1 (E, F). Relative expression of miR-26b-5p in CMECs under hypoxia after silencing Malat1 (G). Luciferase activities of reporter vectors containing luciferase genes and a fragment of Malat1 RNA containing wild-type or mutated miR-26b-5p binding sites (H). Malat1 was associated with miR-26b-5p. CMECs were transfected with biotinylated wild-type miR-26b-5p (Bio-miR-26b-5p-WT) or biotinylated mutant miR-26b-5p (Bio-miR-26b-5p-Mut) (I). The RIP assay for Malat1 was performed with an anti-AGO2 antibody in CMECs transfected with NC or mimics, and the expression of Malat1 and miR-26b-5p was detected (J). *p < 0.05 compared with the sham or NC group, #p < 0.05 compared with the adjacent group. n = 5 in each group.
Further work was performed to test the direct interaction between Malat1 and miR-26b-5p. A dual-luciferase reporter assay was performed to determine the binding potential between the two RNAs. The results showed that miR-26b-5p had inhibitory effects on the luciferase activities of the wild-type Malat1 and mutated Malat1 (one of the two miR-26b-5p binding sites was mutated) reporters, whereas the activity of the mutated Malat1 reporter with two mutated binding sites remained unaffected. Moreover, the mutated miR-26b-5p failed to exhibit the inhibitory effects described above (Fig. 3H). Next, an avidin-biotin pull-down assay was applied to analyse the binding between miR-26b-5p and Malat1. As shown in Fig. 3I, Malat1 was enriched by introducing miR-26b-5p, the effect of which was not observed with mutated miR-26b-5p. Argonaute 2 (Ago2) is a key component of the RNA-induced silencing complex (RISC), in which miRNAs silence genes and are regulated by lncRNAs. Compared with that in the IgG control, higher levels of miR-26b-5p and Malat1 were pulled down using the anti-AGO2 antibody. Moreover, the content of immunoprecipitated Malat1 was obviously higher in the miR-26b-5p overexpression group than in the NC group (Fig. 3J). The above results explicitly confirmed that Malat1 could directly bind miR-26b-5p in CMECs.
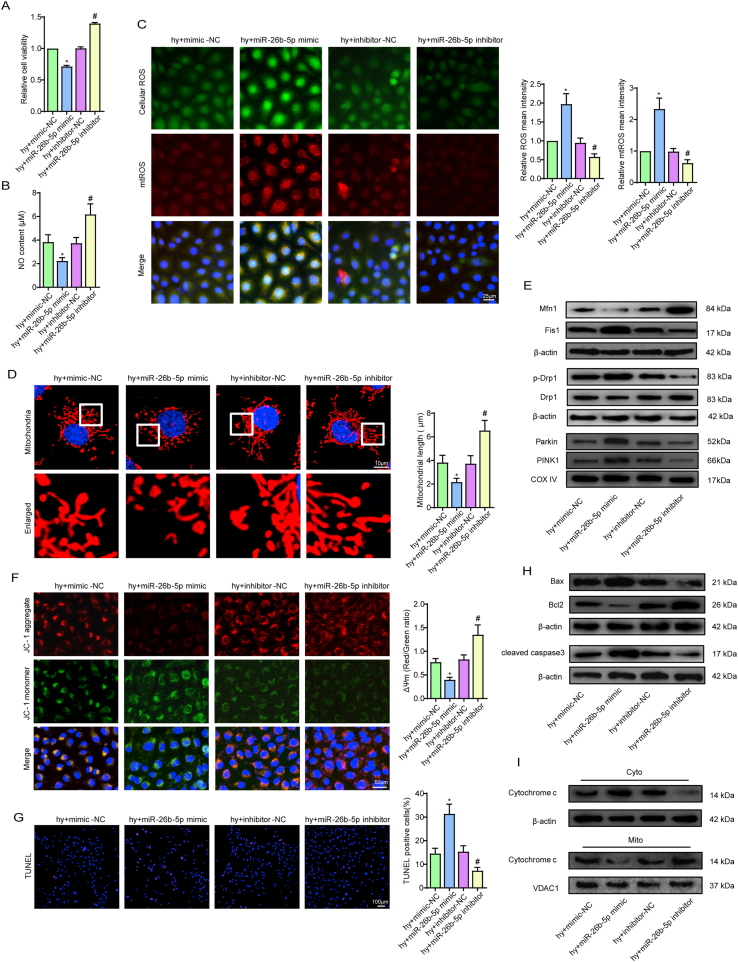
3.4. miR-26b-5p negatively regulated CMEC functions under hypoxic injury
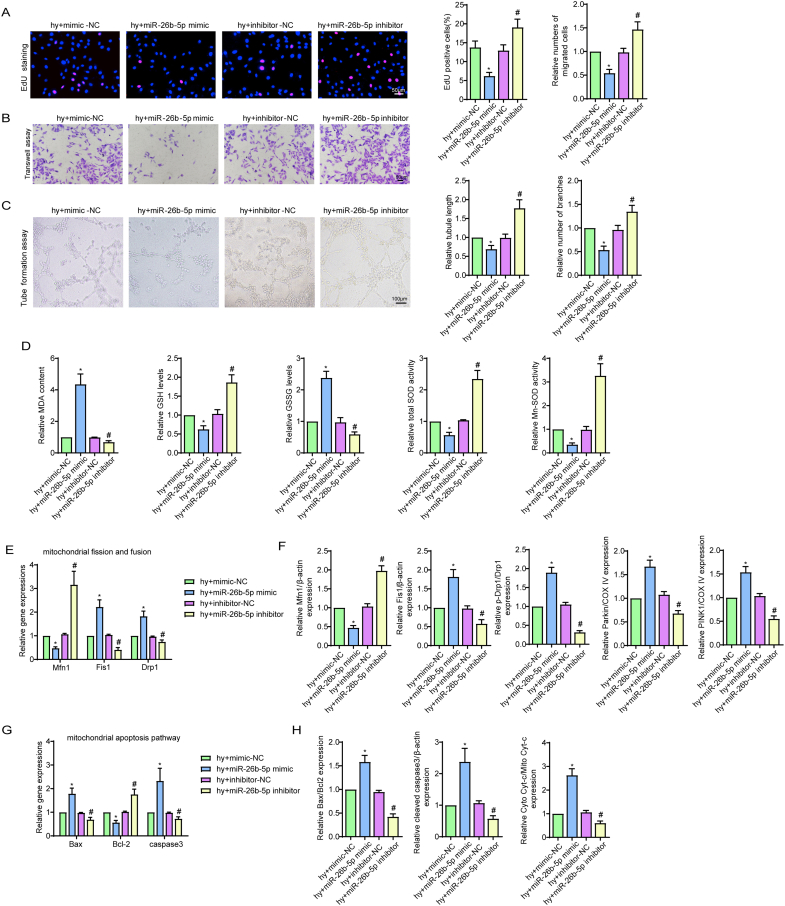
We next investigated the effects of miR-26b-5p on CMEC function by loss- and gain-of-function experiments (Fig. S1G, H). After hypoxic injury, overexpression of miR-26b-5p obviously reduced cell viability, proliferation ability, cell migration ability, tube formation ability and NO synthesis (Fig. 4A and B, S5A-C). In contrast, downregulating miR-26b-5p showed the completely opposite results (Fig. 4A and B, S5A-C). Additionally, miR-26b-5p overexpression increased hypoxia-mediated oxidative stress injury, as shown by the reduced GSH content, decreased t-SOD and Mn-SOD enzyme activities, and enhanced cellular ROS, mtROS, MDA and GSSG expression (Fig. S5D, 4C). The opposite trend was observed after transfection of the miR-26b-5p inhibitor (Fig. S5D, 4C). Next, based on our data that hypoxia alone or in combination with Malat1 inhibition caused fragmented mitochondria and cell apoptosis, we focused on the effects of miR-26b-5p on mitochondria. Transfection of hypoxic cells with the exogenous miR-26b-5p mimic caused more mitochondrial debris (Fig. 4D), reduced Mfn1 expression and increased DRP1 and Fis1 expression (Fig. 4E, S5E, S5F). Conversely, the mitochondrial length in miR-26b-5p inhibitor-transfected cells was longer than that in non-transfected hypoxic cells, and higher levels of Mfn1 and lower levels of DRP1 and Fis1 were found in this cell group (Fig. 4D and E, S5E, S5F). Under hypoxia condition, the expression of Parkin and PINK1was further increased after miR-26b-5p mimic transfection and was down-regulated by miR-26b-5p inhibition, indicating mitophagy was mediated by miR-26b-5p (Fig. 4E, S5F). Moreover, the miR-26b-5p mimic obviously enhanced cell apoptosis by activating the mitochondrial signalling pathway, as indicated by the markedly increased MMP depolarization and percentage of TUNEL-positive cells, enhanced cytochrome c release, upregulated expression of Bax and cleaved caspase 3, and decreased expression of Bcl-2 (Fig. 4F–I, S5G, S5H). In contrast, the completely opposite results were obtained after miR-26b-5p was inhibited (Fig. 4F–I, S5G, S5H).
Fig. 4.
miR-26b-5p negatively regulated CMEC functions under hypoxic injury. Relative cell viability was measured by the CCK-8 assay (A). Nitric oxide (NO) content in CMECs (B). Cellular ROS (green) and mtROS (red) in CMECs were measured and quantified (C). Mitochondria were stained with MitoTracker (red), and mitochondrial morphology was quantified (D). The expression of mitochondrial fusion/fission-related proteins and mitophagy-related proteins in CMECs (E). Mitochondrial membrane potential (ΔΨm) was measured by the JC-1 assay (F). TUNEL staining of CMECs and the percentage of TUNEL-positive CMECs (red) were quantified (G). The expression of apoptosis-associated proteins in CMECs (H, I). *p < 0.05 compared with the hy + mimic-NC group, #p < 0.05 compared with the hy + inhibitor-NC group. n = 5 in each group.Relative cell viability was measured by the CCK-8 assay (A). Nitric oxide (NO) content in CMECs (B). Cellular ROS (green) and mtROS (red) in CMECs were measured and quantified (C). Mitochondria were stained with MitoTracker (red), and mitochondrial morphology was quantified (D). The expression of mitochondrial fusion/fission-related proteins and mitophagy-related proteins in CMECs (E). Mitochondrial membrane potential (ΔΨm) was measured by the JC-1 assay (F). TUNEL staining of CMECs and the percentage of TUNEL-positive CMECs (red) were quantified (G). The expression of apoptosis-associated proteins in CMECs (H, I). *p < 0.05 compared with the hy + mimic-NC group, #p < 0.05 compared with the hy + inhibitor-NC group. n = 5 in each group.
In summary, the present data demonstrated that miR-26b-5p had negative effects on endothelial functions on account of destroying mitochondrial networks and providing a pro-apoptotic environment for CMECs under hypoxic injury.
3.5. Inhibiting miR-26b-5p alleviated Malat1 deletion-aggravated EC injuries under hypoxia
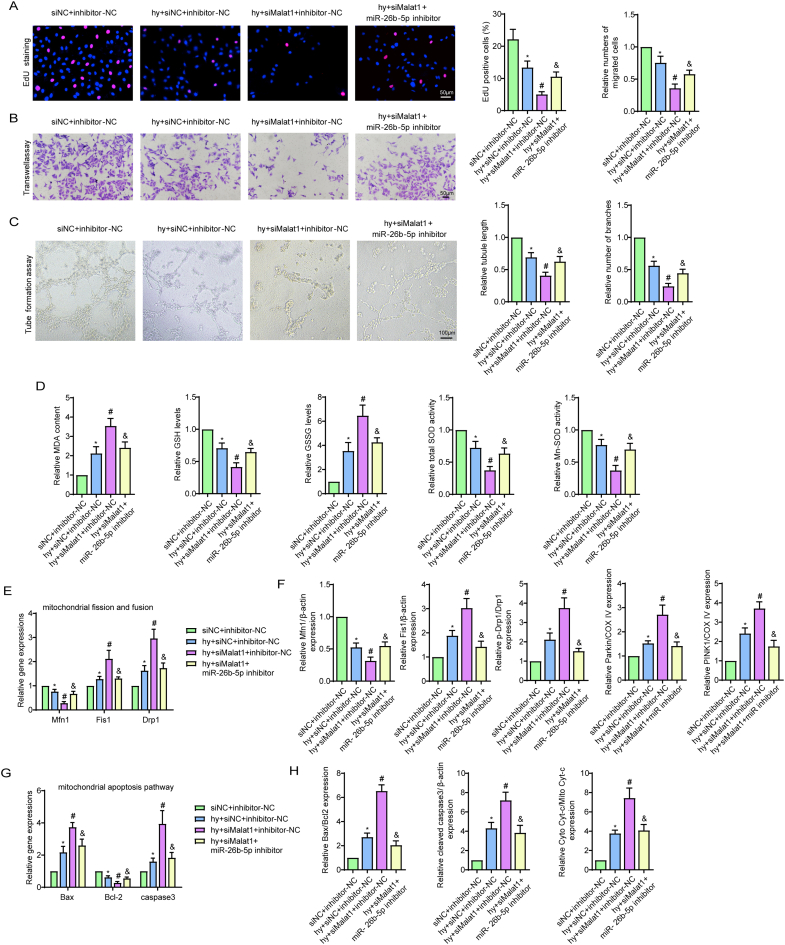
Considering that Malat1 was determined to be a molecular sponge for miR-26b-5p, further attention was given to the functional interactions between the two RNAs. As we have previously described, silencing Malat1 exacerbated CMEC dysfunction under hypoxia, including affecting cell viability, proliferation ability, cell migration, tube formation ability and NO synthesis (Fig. 5A and B, S6A-C). However, transfection with the miR-26b-5p inhibitor significantly reversed the adverse effects of Malat1 deletion, suggesting that miR-26b-5p is a downstream gene of Malat1 with antagonistic functions (Fig. 5A and B, S6A-C). Moreover, inhibiting miR-26b-5p also reversed the oxidative stress injury aggravated by Malat1 silencing, such as increased GSH content, increased t-SOD and Mn-SOD enzyme activities, and reduced cellular ROS, mtROS, MDA and GSSG expression (Fig. S6D, 5C).
Fig. 5.
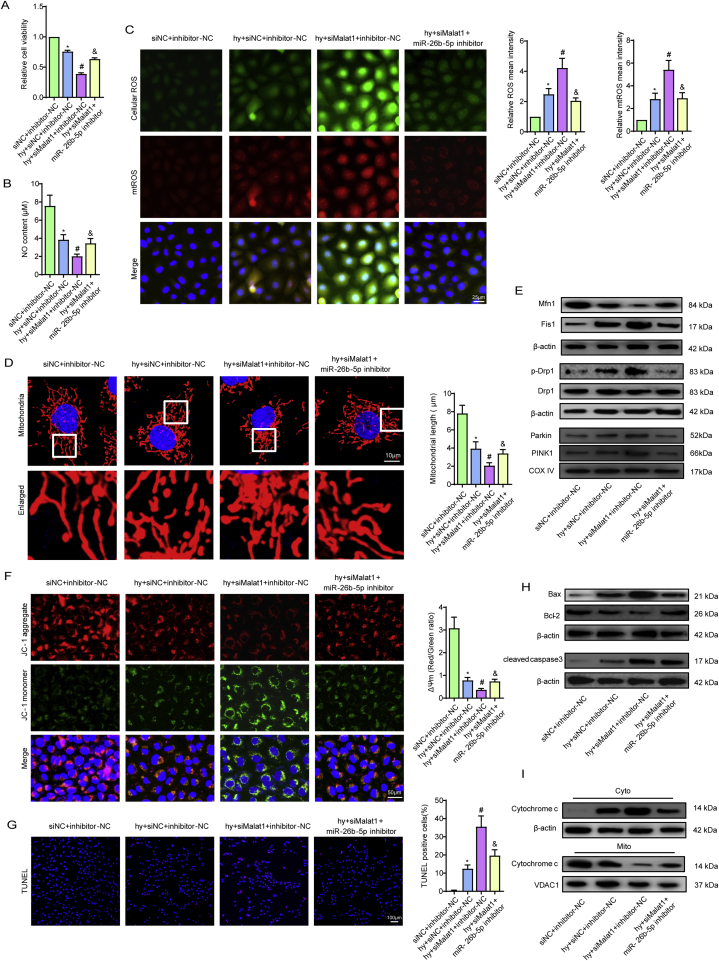
Inhibiting miR-26b-5p alleviated Malat1 deletion-aggravated EC injuries under hypoxia. Relative cell viability was measured by the CCK-8 assay (A). Nitric oxide (NO) content in CMECs (B). Cellular ROS (green) and mtROS (red) in CMECs were measured and quantified (C). Mitochondria were stained with MitoTracker (red), and mitochondrial morphology was quantified (D). The expression of mitochondrial fusion/fission-related proteins and mitophagy-related proteins in CMECs (E). Mitochondrial membrane potential (ΔΨm) was measured by the JC-1 assay (F). TUNEL staining of CMECs and the percentage of TUNEL-positive CMECs (red) were quantified (G). The expression of apoptosis-associated proteins in CMECs (H, I). *p < 0.05 compared with the siNC + inhibitor-NC group, #p < 0.05 compared with the hy + siNC + inhibitor-NC group, & p < 0.05 compared with the hy + siMalat1+inhibitor-NC group. n = 5 in each group.
Further experiments were performed to explore the influence of miR-26b-5p on Malat1-mediated mitochondrial functions. Compared to that in cells treated with siMalat1, less mitochondrial debris was found after miR-26b-5p inhibitor treatment (Fig. 5D), which was accompanied by reduced DRP1 and Fis1 expression and increased Mfn1 expression (Fig. 5E, S6E, S6F), indicating that silencing miR-26b-5p could reverse the imbalance between fission- and fusion-related proteins after Malat1 knockout. Moreover, miR-26b-5p inhibitor treatment suppressed the expression of Parkin and PINK1 in siMalat1 transfected cells (Fig. 5E, S6F). Silencing miR-26b-5p further inhibited anabatic MMP depolarization after Malat1 inhibition (Fig. 5F). In addition, co-transfection with the miR-26b-5p inhibitor diminished the ratio of TUNEL-labelled apoptotic cells (Fig. 5G), the leakage of cytochrome C, the increased expression of Bax and cleaved caspase3, and the reduction in Bcl-2 compared with transfection with Malat1 alone (Fig. 5H and I, S6G, S6H). The above data showed that miR-26b-5p, as a downstream signal of Malat1, exerts the opposite effects to Malat1 in regard to endothelial and mitochondrial protection.
3.6. Mfn1 is a direct downstream mRNA target of miR-26b-5p in CMECs
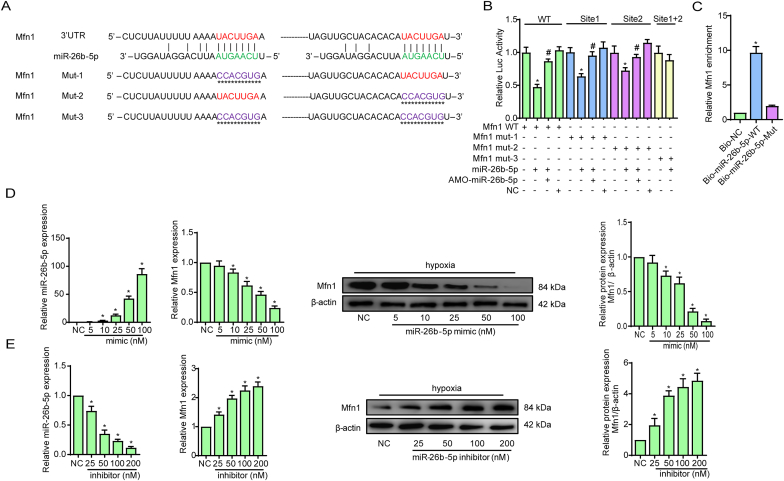
To further elucidate the ceRNA network associated with Malat1 and miR-26b-5p, attempts were made to identify the specific target mRNA of miR-26b-5p in regulating CMECs under hypoxia. Our main focus was mitochondrial dynamics-related genes with changed expression after exogenous Malat1 expression or miR-26b-5p treatment, including Drp1, Fis1 and Mfn1. TargetScan (http://www.targetscan.org/mamm_31/) predicted that Mfn1 possesses potential binding sites for miR-26b-5p (Fig. 6A). A dual-luciferase reporter assay showed that miR-26b-5p had an inhibitory effect on the luciferase activities of the wild-type and mutated Mfn1 (mutation in one of two miR-26b-5p binding sites) reporters, whereas the activity of the Mfn1 reporter remained unaffected when both binding sites were mutated (Fig. 6B). In addition, an avidin-biotin pull-down assay confirmed the specific interactions between miR-26b-5p and Mfn1. As shown in Fig. 6C, Mfn1 was enriched by introducing miR-26b-5p, while the mutated miR-26b-5p was unable to bind Mfn1. These results indicated the potential of miR-26b-5p to bind Mfn1 directly. Then, CMECs were transfected with different concentrations of the miR-26b-5p mimic or inhibitor to explore whether miR-26b-5p could modulate Mfn1 transcription and translation. As shown in Fig. 6D, both the gene and protein expression of Mfn1 was obviously reduced in a dose-dependent manner after miR-26b-5p mimic transfection. In contrast, transfecting increasing concentrations of the miR-26b-5p inhibitor gradually promoted Mfn1 expression (Fig. 6E). These results confirmed that miR-26b-5p could directly bind Mfn1 mRNA in CMECs and negatively control Mfn1 transcription.
Fig. 6.
Mfn1 is a direct downstream mRNA of miR-26b-5p in CMECs. The predicted binding sites of Malat1 and miR-26b-5p and the mutated binding sites are indicated (A). Luciferase activities of reporter vectors containing luciferase genes and a fragment of Mfn1 RNA containing wild-type or mutated miR-26b-5p binding sites (B). Mfn1 was associated with miR-26b-5p. CMECs were transfected with biotinylated wild-type miR-26b-5p (Bio-miR-26b-5p-WT) or biotinylated mutant miR-26b-5p (Bio-miR-26b-5p-Mut) (C). Concentration gradients of the miR-26b-5p mimic and inhibitor were transfected into CMECs, and quantitative analysis of the alteration of miR-26b-5p and the gene and protein expression of Mfn1 was performed (D-E). *p < 0.05 compared with the NC group, #p < 0.05 compared with the adjacent group. n = 5 in each group.
3.7. Mfn1 positively regulated CMEC functions after hypoxic injury
Mfn1 is of great importance to mitochondrial fusion and plays a pivotal role in a variety of cardiovascular disorders [20]. To further clarify the mechanism of action of Mfn1 in CMECs under hypoxia, we examined the effects of Mfn1 via knockout or overexpression methods in vitro. Fig. S1I-N shows that transfection was effective. Under hypoxia, CMEC viability, proliferation ability, migration ability, tube formation ability and NO synthesis were obviously decreased (Fig. 7A and B, S7A-C), the effects of which were further aggravated after Mfn1 silencing. In contrast, overexpression of Mfn1 clearly reversed this endothelial dysfunction (Fig. 7A and B, S7A-C). Subsequently, Mfn1 overexpression reduced the content of cellular ROS, mtROS, MDA and GSSG and increased the GSH content and enzyme activities of t-SOD and Mn-SOD, demonstrating the potential role of Mfn1 in alleviating cell oxidative stress (Fig. 7C, S7D).
Fig. 7.
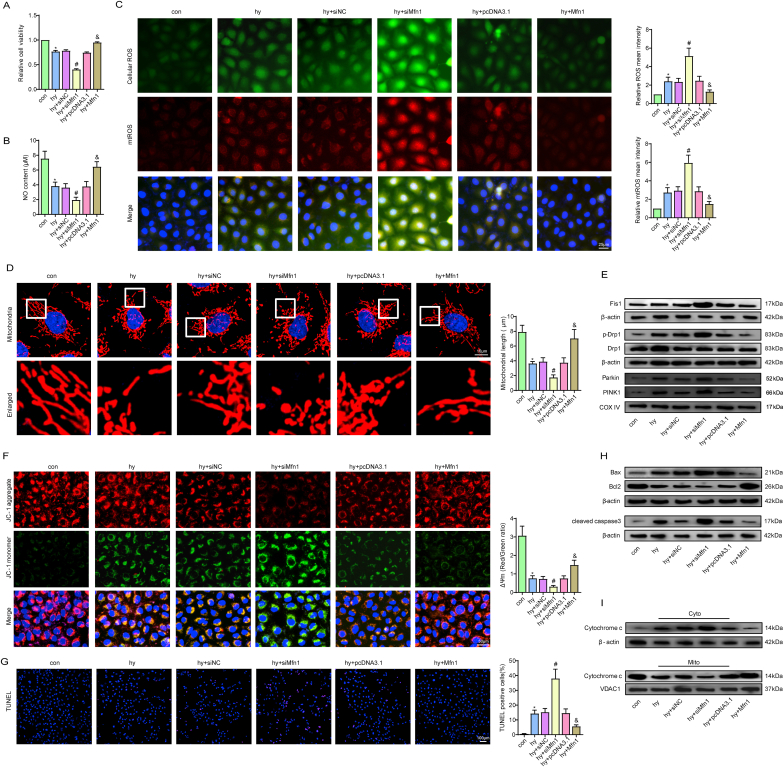
Mfn1 positively regulated CMEC functions after hypoxic injury. Relative cell viability was measured by the CCK-8 assay (A). Nitric oxide (NO) content in CMECs (B). Cellular ROS (green) and mtROS (red) in CMECs were measured and quantified (C). Mitochondria were stained with MitoTracker (red), and mitochondrial morphology was quantified (D). The expression of mitochondrial fusion/fission-related proteins and mitophagy-related proteins in CMECs (E). Mitochondrial membrane potential (ΔΨm) was measured by the JC-1 assay (F). TUNEL staining of CMECs and the percentage of TUNEL-positive CMECs (red) were quantified (G). The expression of apoptosis-associated proteins in CMECs (H, I). *p < 0.05 compared with the control group, #p < 0.05 compared with the hy + siNC group, & p < 0.05 compared with the hy + pcDNA3.1 group. n = 5 in each group.
Since mitochondrial fragmentation is highly associated with activation of mitochondrial apoptosis and myocardial microvascular dysfunction, the possible beneficial effects of Mfn1 on mitochondrial dynamics and mitochondrial apoptosis were investigated. Our data revealed that overexpression of Mfn1 significantly decreased mitochondrial debris, whereas the opposite trend was observed after silencing Mfn1 (Fig. 7D). Moreover, two other mitochondrial fission-related proteins, DRP1 and Fis1, were also strongly inhibited by Mfn1 overexpression (Fig. 7E, S7E, S7F). Since less mitochondrial debris occurred after Mfn1 overexpression, mitophagy was suppressed correspondingly, as evidenced by reduced expression of Parkin and PINK1 (Fig. 7E, S7F). Furthermore, Mfn1 markedly and inhibited MMP depolarization, decreased TUNEL-positive CMECs after hypoxic injury by preventing cytochrome c release from mitochondria, reducing Bax and cleaved caspase3 expression and increasing Bcl-2 expression, favouring Mfn1-mediated inhibition of endothelial apoptosis via a mitochondria-dependent pathway (Fig. 7F–I, S7G, S7H).
Altogether, our data confirmed that Mfn1 was a downstream target mRNA of miR-26b-5p and was to a great extent responsible for CMEC recovery under hypoxia by modulating mitochondrial dynamics and inhibiting mitochondria-dependent apoptosis.
3.8. Mfn1 reversed mitochondrial damage and apoptosis induced by silencing Malat1 in CMECs under hypoxia
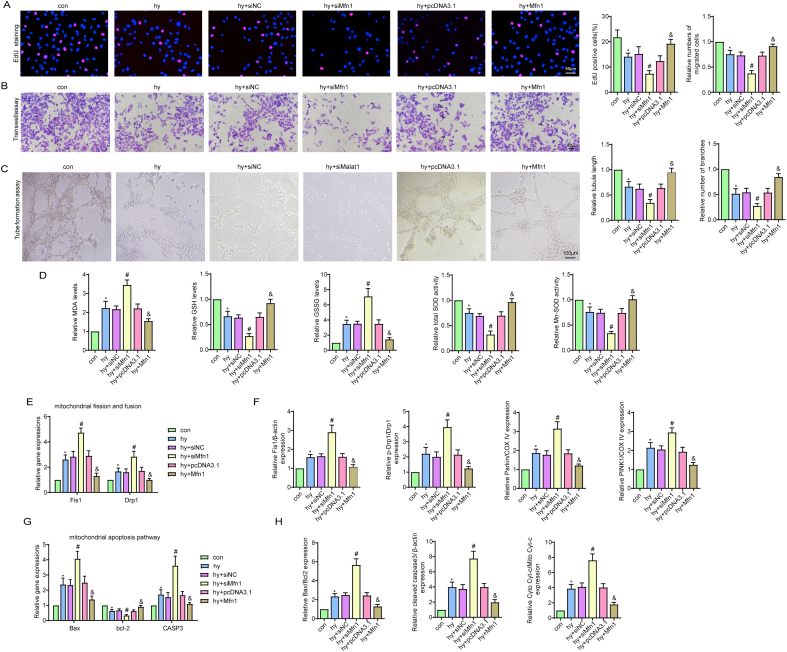
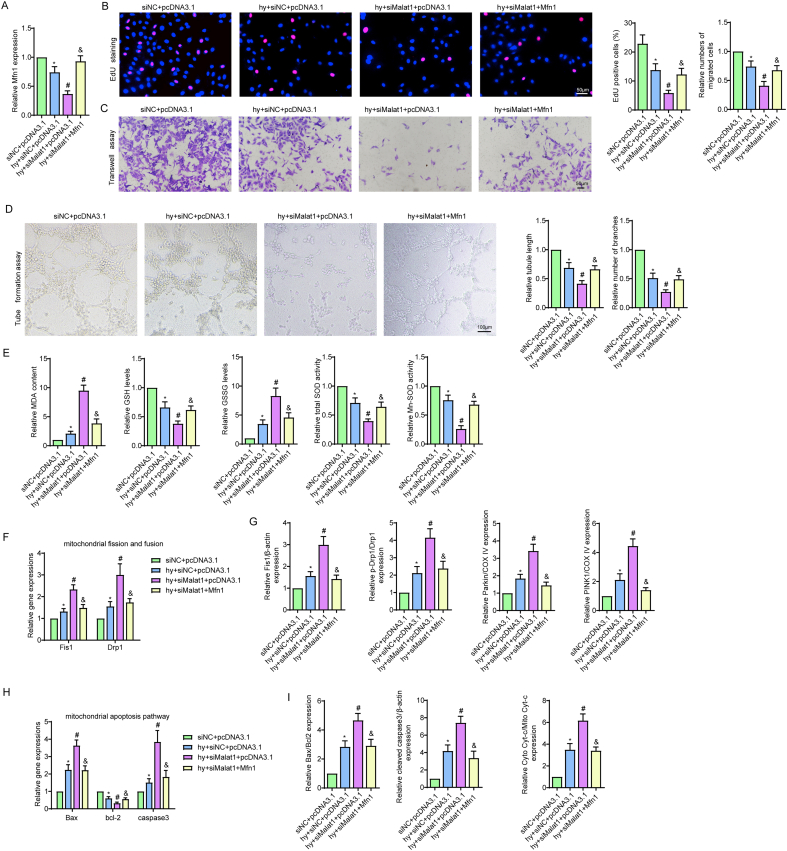
To further explore whether Mfn1 was involved in the beneficial effects of Malat1, we overexpressed Mfn1 in the siMalat1 group based on our previous finding that inhibiting Malat1 further reduced Mfn1 expression in hypoxic CMECs (Fig. 8A, S8A). Overexpression of Mfn1 reversed the siMalat1-induced reduction in cell viability, proliferation ability, cell migration ability, tube formation ability and NO synthesis in CMECs under hypoxia (Fig. 5, Fig. 8C, S8B-D). Additionally, exogenous overexpression of Mfn1 significantly diminished oxidative stress in cells transfected with siMalat1, as demonstrated by higher GSH content, higher t-SOD and Mn-SOD enzyme activities, and markedly suppressed expression of intracellular ROS, mtROS, MDA and GSSG (Fig. S8E, 8D).
Fig. 8.
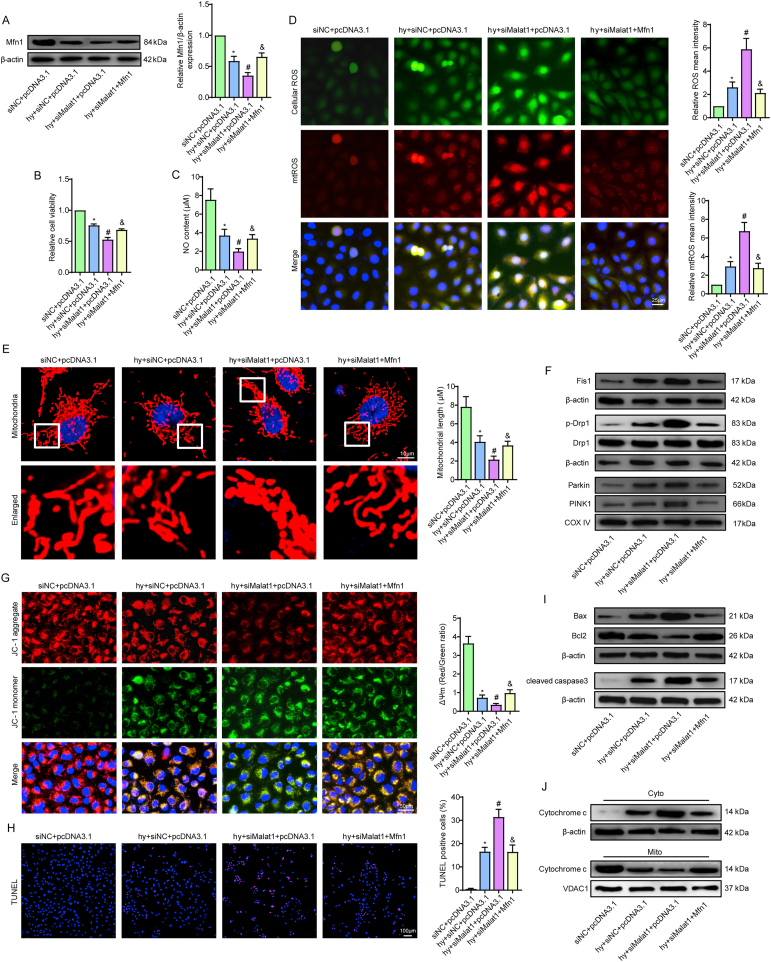
Mfn1 reversed mitochondrial damage and apoptosis induced by silencing Malat1 in CMECs under hypoxia. Relative protein expression of Mfn1 in CMECs (A). Relative cell viability was measured by the CCK-8 assay (B). Nitric oxide (NO) content in CMECs (C). Cellular ROS (green) and mtROS (red) in CMECs were measured and quantified (D). Mitochondria were stained with MitoTracker (red), and mitochondrial morphology was quantified (E). The expression of mitochondrial fusion/fission-related proteins and mitophagy-related proteins in CMECs (F). Mitochondrial membrane potential (ΔΨm) was measured by the JC-1 assay (G). TUNEL staining of CMECs and the percentage of TUNEL-positive CMECs (red) were quantified (H). The expression of apoptosis-associated proteins in CMECs (I, J). *p < 0.05 compared with the siNC + pcDNA3.1 group, #p < 0.05 compared with the hy + siNC + pcDNA3.1 group, & p < 0.05 compared with the hy + siMalat1+pcDNA3.1 group. n = 5 in each group.
Attempts were then made to illustrate the potential role of Mfn1 in mitochondrial injuries in cells treated with hypoxia and siMalat1 at the same time. As one of the major mitochondrial fusion-related proteins, Mfn1 functioned to fuse the debris into a filamentous structure, indicating that the mitochondrial network was properly re-established (Fig. 8E). Additionally, Mfn1 clearly offset the promoting effects of siMalat1 on DRP1, Fis1, Parkin and PINK1 expression (Fig. 8F, S8F, S8G). The anti-apoptotic effects of Mfn1 were demonstrated in our previous data. Considering that Mfn1 is a downstream gene of Malat1, overexpression of Mfn1 also reduced siMalat1-intensified endothelial apoptosis under hypoxia, which could be attributed to its inhibitory effects on the mitochondrial apoptosis pathway, as illustrated by the decreased cytochrome C release, reduced Bax and cleaved-caspase3 expression and increased Bcl-2 expression compared with those in hypoxic cells treated with siMalat1 alone (Fig. 8G–J, S8H, S8I).
Altogether, the above results suggested that Mfn1 could be regulated by Malat1 and had a role in restoring Malat1 deletion-induced CMEC dysfunction under hypoxia by ameliorating the mitochondrial dynamics and the mitochondrial apoptosis pathway.
3.9. Overexpressing Mfn1 reversed the aggravation of myocardial infarction and microvascular dysfunction after silencing Malat1 in MI hearts
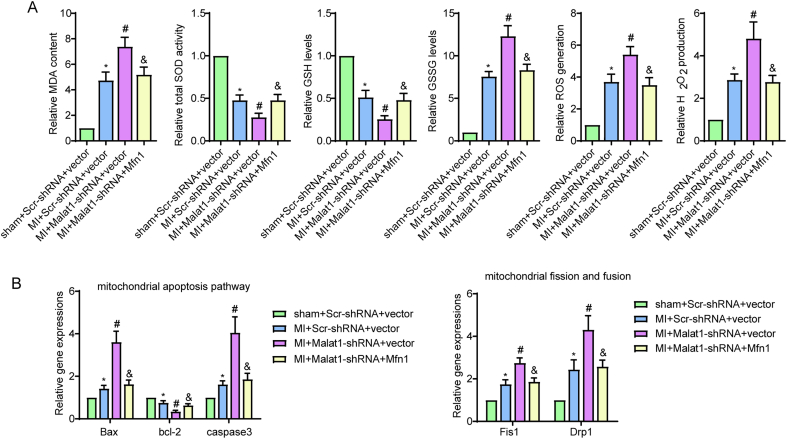
To gain more insight into the role of Mfn1 in cardiac microcirculation after MI and further demonstrate that Mfn1 is a functional downstream gene of Malat1 in vivo, AAV-shMalat1 and AAV-Mfn1 were co-transfected into ECs (Fig. S1O-S). Consistent with the data observed in our previous work, silencing Malat1 in mice caused significantly lower overall 7-day survival rates (Fig. 9A), more severe cardiac dysfunction and larger infarct size (Fig. 9B–I) compared to the control MI mice. However, reintroduction of Mfn1 in CMECs antagonized the negative effects of Malat1 deletion in terms of overall 7-day survival rates, cardiac dysfunction and infarct size (Fig. 9A–I). In addition, transfection of Mfn1 strongly suppressed the stimulating effects of Malat1 silencing on oxidative damage after MI, which was evidenced by the increased content of GSH, enhanced enzyme activity of t-SOD, and inhibited ROS, H2O2, MDA and GSSG expression in cardiac tissues (Fig. S9A). Moreover, after genetic ablation of Malat1 in MI mice, restoring Mfn1 expression effectively reversed microvascular deficiency and perfusion, which were accompanied by obviously increased total and phosphorylated VEGFR2 and eNOS levels (Fig. 9J–O).
Fig. 9.
Overexpressing Mfn1 reversed the aggravation of myocardial infarction and microvascular dysfunction induced by silencing Malat1 in MI hearts. One-week overall survival curve (A). The M-mode of echocardiography images and the data on left ventricular end-diastolic diameter (LVEDD), ejection fraction (EF) and fractional shortening (FS) for each group (B, C). Heart weight to body weight ratio (HW/BW) of each group (D). Analysis of serum brain natriuretic peptide (BNP) levels (E). HE staining, Masson staining and quantification of cardiac fibrosis (F, G). Analysis of serum cardiac troponin T (cTnT) concentration and lactate dehydrogenase (LDH) activity (H, I). Microvascular perfusion of the border zone was indicated by the ratio of lectin-perfused vessels (green) to CD31-positive ECs (red) (J). Immunofluorescence staining for eNOS (K). Nitric oxide (NO) content in the border zone of each group (L). Immunofluorescence staining for VEGFR2 (M). eNOS expression and phosphorylation at Ser1177 in the border zone were analysed by Western blotting (N). VEGFR2 expression and phosphorylation at Tyr1175 in the border zone were analysed by Western blotting (O).*p < 0.05 compared with the sham + Scr-shRNA + vector group, #p < 0.05 compared with the MI + Scr-shRNA + vector group, & p < 0.05 compared with the MI + Malat1-shRNA + vector group. n = 6 in each group.
Further experiments were performed via real-time PCR to determine the mechanisms by which Mfn1 reverses the adverse effects on microcirculation caused by Malat1 silencing. Our previous results indicated that mitochondrial fission progression was obviously enhanced to a greater degree after silencing Malat1 in vivo and in vitro. However, after re-expression of Mfn1, lower levels of DRP1 and Fis1 were detected in isolated CMECs (Fig. S9B). Moreover, the increase in Bax and caspase3 and decrease in Bcl-2 caused by Malat1 deletion were obviously reversed after restoring Mfn1 expression, indicating the great potential for Malat1 to inhibit apoptosis via the mitochondrial pathway that is inactivated by Mfn1 (Fig. S9B).
The above data directly supported the notion that Mfn1 is a downstream factor of Malat1 and exerts beneficial effects on cardiac function via the microvascular protection pathway partly by alleviating mitochondrial dynamics and apoptosis.
4. Discussion
The potential effects of Malat1 on cardiac microcirculation and heart function were identified in the present study. Malat1 was increased in cardiac tissues and isolated ECs in the peri-infarct region. CMEC-specific knockout of Malat1 aggravated endothelial dysfunction and thereby exacerbated infarction expansion, which led to poor prognosis after MI. The potential mechanisms could involve the decrease in the gene and protein expression of Mfn1 after Malat1 silencing through the regulation of miR-26b-5p, which in turn disturbed mitochondrial dynamics and activated the mitochondrial-dependent apoptosis signalling pathway in CMECs.
CMECs, as the main and essential components of the myocardial microcirculation, participate in nutrient exchange processes and energy metabolism regulation in the myocardial microenvironment [21]. MI leads to cardiac microcirculatory disturbance, mainly manifested as decreased CMEC functions and microvascular perfusion [22,23]. Previous studies have demonstrated that treatments or strategies aimed at protecting CMECs and microcirculation contribute to the improvement of cardiac function after MI [24]. As a well-known lncRNA enriched in ECs, Malat1 has been previously implicated in the proliferation and migration processes of multiple EC types in response to hypoxia. In addition, Malat1 possesses anti-apoptosis potential. Wang G et al. found that knockdown of Malat1 dramatically increased the apoptosis rate of human brain microvascular ECs under oxygen glucose deprivation/reoxygenation injury [25]. Several studies have shown the increased expression level of Malat1 in MI, suggesting that Malat1 has a role in the progression or repair of MI [17,26]. However, the underlying mechanisms are still not fully understood, and most studies so far have focused on cardiomyocytes and cardiac fibroblasts [17,18], and no study has focused on the influence of Malat1 on cardiac microcirculation after MI. The present study revealed that silencing Malat1 in CMECs led to higher 7-day overall cardiac mortality, infarction extension and more severe cardiac dysfunction after MI, the effects of which were largely caused by aggravated cardiac microvascular dysfunction in terms of reduced microvascular density and compromised blood perfusion. Moreover, knockdown of Malat1 in vitro decreased CMEC proliferation, migration, tube formation ability and NO synthesis abilities under hypoxia. The underlying mechanisms could be attributed to the possible role of Malat1 in regulating mitochondrial dynamics and the mitochondrial apoptotic pathway.
Mitochondria are vital organelles in ECs not only for their role in providing ATP but also for their function in transmitting extracellular and intracellular signals when ECs are subjected to environmental stresses or injuries. Persistent mitochondrial dysfunction contributes to hypoxia-induced EC damage, thus precipitating EC apoptosis, capillary destruction, oedema and cardiac microvascular injuries. Mitochondrial dynamics is one of the mitochondrial quality control (MQC) systems that eliminates injured mitochondria and generates new, healthy mitochondria. Mfn1 is an important protein that controls mitochondrial dynamics by regulating mitochondrial fusion. Knockdown of Mfn1 in ECs resulted in small and punctate mitochondria and accelerated the cellular apoptosis rate [13]. Mitophagy is a special autophagy flux that occurred to eliminate mitochondrial debris. Evidence noted Mfn1 overexpression inhibited mitochondrial fragmentation and thereby suppress mitophagy in skeletal muscle atrophy [27]. Mfn1 participates in different cardiovascular diseases, such as protecting myocardial cells against ischaemia-reperfusion injury and inhibiting the development of cardiac hypertrophy [28,29]. There are several lines of evidence that lncRNAs can modulate mitochondrial dynamics and, as a consequence, control mitochondrial functions and change disease prognosis. However, no study has demonstrated that Malat1 is an upstream regulatory signalling molecule of Mfn1, even outside of the cardiovascular research field. The present study explained for the first time the positive association between Malat1 and Mfn1. Silencing Malat1 reduced Mfn1 expression in CMECs; in contrast, overexpression of Mfn1 offset the negative effects of knocking out Malat1 on cardiac microvascular functions after MI and EC functions under hypoxia by regulating mitochondrial dynamics and its related mitochondrial apoptosis.
The ceRNA hypothesis is a basic mechanism to explain the possible functions of lncRNAs, by which lncRNAs affect the transcription process of target mRNA by directly sponging miRNA binding sites [30]. On the other hand, miRNAs repress target gene expression by binding the Ago-2 protein to form RISC. Importantly, it has been extensively indicated that miRNAs play essential roles in multiple cardiovascular diseases [9]. Herein, miR-26b-5p was predicted to be a specific target miRNA of Malat1 and acted as a connection between Malat1 and Mfn1. miR-26b-5p has been suggested to have a potential role in pathological cardiac hypertrophy, cardiac fibrosis and cerebral ischaemia-reperfusion injury, and its level is decreased in the plasma of patients who experience major cardiovascular events (MACEs) after MI [31,32]. Even though the latent mechanism is still unclear, we have reason to believe that miR-26b-5p may be associated with cardiac ischaemic injury. Notably, miR-26b-5p was implicated in the angiogenic process in liver fibrosis, hepatocellular carcinoma and high glucose-injured HUVECs [33], suggesting that miR-26b-5p is involved in the vascular development of certain diseases. By bioinformatics analysis, we predicted that miR-26b-5p was a direct target of Malat1 and confirmed their complementary regulatory relationship via luciferase reporter and pull-down assays. In the same way, Mfn1 was identified as its specific downstream signalling factor. Functionally, the inhibition of miR-26b-5p reversed the downregulation of Mfn1 under hypoxia and rebalanced mitochondrial dynamics and mitochondrial apoptosis signalling, consequently abrogating the harmful effects of silencing Malat1 on cardiac microvascular dysfunction.
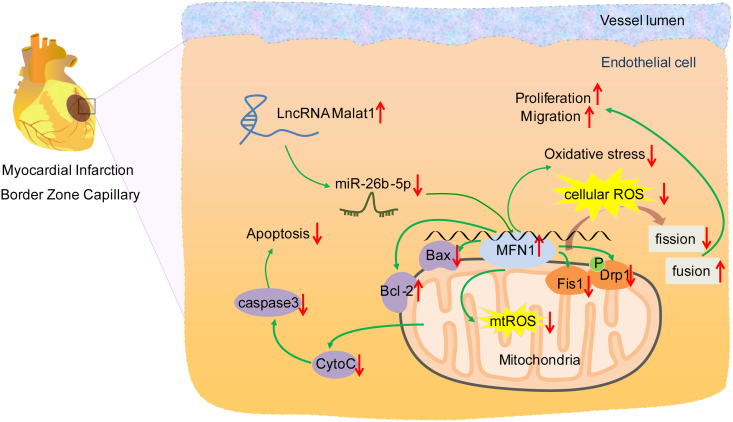
In conclusion, the present study confirmed a new ceRNA pathway in regulating mitochondrial homeostasis and cardiac microcirculation repair. Malat1 inactivated miR-26b-5p and subsequently increased Mfn1 expression, contributing to mitochondrial dynamics and inhibiting mitochondria-dependent apoptosis in CMECs. Malat1-mediated mitochondrial homeostasis enhances the resistance of cardiac microcirculation to hypoxic injury and thereby improves the prognosis of MI mice. These findings provide evidence that the Malat1/miR-26b-5p/Mfn1 axis is involved in mitochondrial dynamics in CMECs and may be a therapeutic alternative to facilitate microcirculation reconstruction after MI.
Funding
This work was supported by National Natural Science Foundation of China (grant nos. 82070267, 81800232, 81700078 and 81670320).
Availability of data and materials
The datasets generated or analysed in this study can be obtained upon reasonable request from the corresponding author.
Declaration of competing interest
The authors declare that they have no competing interests.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2021.101910.
Contributor Information
Yao Lu, Email: luyao19890122@163.com.
Dalin Jia, Email: jdl2001@126.com.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
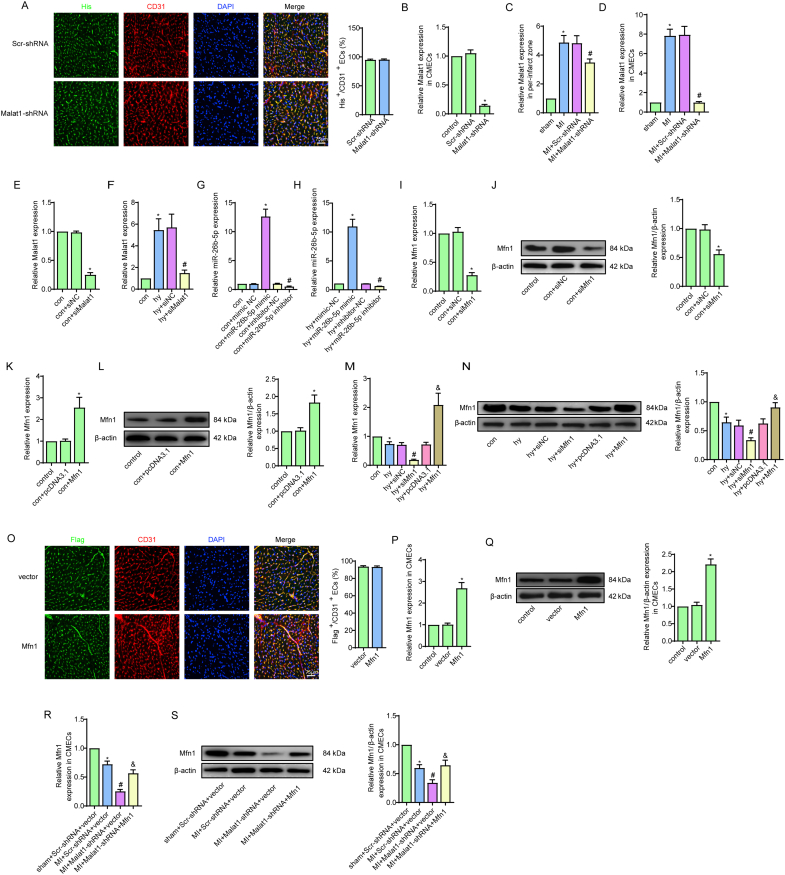
Fig. S1.
Transfection efficiency and expression of target genes.
Immunofluorescence staining of His-tagged AAV-shMalat1 (green) and CD31-labelled CMECs (red) and quantitative analysis of the percentage of His-positive CMECs (A). Transfection efficiency of AAV-shMalat1 in mouse CMECs (B). Relative expression of Malat1 in cardiac tissues and CMECs in the border zone 7 days after AAV-shMalat1 injection (C, D). The silencing efficiency of siMalat1 in CMECs under normoxia in vitro (E). Relative expression of Malat1 in CMECs under hypoxia after siMalat1 transfection (F). Transfection efficiency of the miR-26b-5p mimic and miR-26b-5p inhibitor in CMECs under normoxia in vitro (G). Relative expression of miR-26b-5p in CMECs under hypoxia after transfection of the miR-26b-5p mimic and miR-26b-5p inhibitor (H). PCR and Western blot results of the silencing efficiency of Mfn1 in CMECs under normoxia (I, J). PCR and Western blot results of the overexpression efficiency of Mfn1 in CMECs under normoxia (K, L). PCR and Western blot results of Mfn1 expression after knocking out or overexpressing Mfn1 under hypoxia (M, N). Immunofluorescence staining of Flag-tagged AAV-Mfn1 (green) and CD31-labelled CMECs (red) and quantitative analysis of the percentage of Flag-positive CMECs (O). Transfection efficiency of AAV-Mfn1 in mouse CMECs (P, Q). Gene and protein expression of Mfn1 in mouse CMECs (R–S). *p < 0.05 compared with sham, control or NC, #p < 0.05 compared with the adjacent group. n = 5 in each group
Fig. S2.
Malat1-mediated mitochondrial homeostasis enhanced cardiac microcirculation resistance after MI via the miR-26b-5p/Mfn1 axis.
The Malat1/miR-26b-5p/Mfn1 axis is involved in mitochondrial dynamics in CMECs, which may contribute to restoring mitochondrial dynamics and inhibiting mitochondria-dependent apoptosis in CMECs under hypoxia
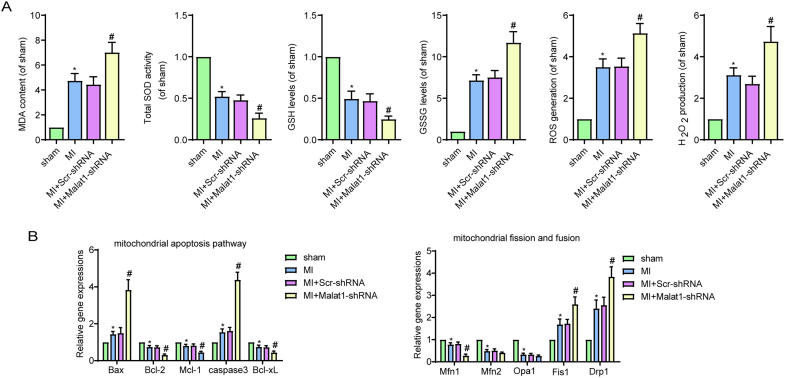
Fig. S3.
Silencing Malat1 in CMECs aggravated oxidative stress, apoptosis and mitochondrial dysfunction after myocardial infarction (MI).
MDA content, total SOD activities, GSH and GSSG levels, and ROS and H2O2 generation in the border zone (A). Relative expression of mitochondrial apoptosis-related genes and mitochondrial fusion- and fission-related genes (B). *p < 0.05 compared with the sham group, #p < 0.05 compared with the MI + Scr-shRNA group. n = 6 in each group
Fig. S4.
Silencing Malat1 compromised CMEC functions by inhibiting cellular proliferation, migration and promoting oxidative stress
Cell proliferation was measured by EdU staining, and the percentage of EdU-positive CMECs (red) was quantified (A). CMEC migration ability was measured by the Transwell assay (B). Images and quantitative analysis of the tube formation assay (C). MDA content, GSH and GSSG levels, and total SOD and Mn-SOD activities of CMECs (D). Relative expression of mitochondrial fusion/fission-related genes (E). Quantification of the expression of mitochondrial fusion/fission-related proteins and mitophagy-related proteins (F). Expression of mitochondrial apoptosis-related genes (G). Quantification of the expression of apoptosis-associated proteins (H). *p < 0.05 compared with the control group, #p < 0.05 compared with the hy + siNC group. n = 5 in each group
Fig. S5.
miR-26b-5p negatively regulated CMEC proliferation, migration and promoted oxidative stress
Cell proliferation was measured by EdU staining, and the percentage of EdU-positive CMECs (red) was quantified (A). CMEC migration ability was measured by the Transwell assay (B). Images and quantitative analysis of the tube formation assay (C). MDA content, GSH and GSSG levels, and total SOD and Mn-SOD activities of CMECs (D). Relative expression of mitochondrial fusion/fission-related genes (E). Quantification of the expression of mitochondrial fusion/fission-related proteins and mitophagy-related proteins (F). Expression of mitochondrial apoptosis-related genes (G). Quantification of the expression of apoptosis-associated proteins (H). *p < 0.05 compared with the hy + mimic-NC group, #p < 0.05 compared with the hy + inhibitor-NC group. n = 5 in each group
Fig. S6.
Inhibiting miR-26b-5p alleviated Malat1 deletion-aggravated EC dysfunction and oxidative stress under hypoxia.
Cell proliferation was measured by EdU staining, and the percentage of EdU-positive CMECs (red) was quantified (A). CMEC migration ability was measured by the Transwell assay (B). Images and quantitative analysis of the tube formation assay (C). MDA content, GSH and GSSG levels, and total SOD and Mn-SOD activities of CMECs (D). Relative expression of mitochondrial fusion/fission-related genes (E). Quantification of the expression of mitochondrial fusion/fission-related proteins and mitophagy-related proteins (F). Expression of mitochondrial apoptosis-related genes (G). Quantification of the expression of apoptosis-associated proteins (H). *p < 0.05 compared with the siNC + inhibitor-NC group, #p < 0.05 compared with the hy + siNC + inhibitor-NC group, & p < 0.05 compared with the hy + siMalat1+inhibitor-NC group. n = 5 in each group
Fig. S7.
Mfn1 positively regulated CMEC proliferation, migration and inhibited oxidative stress after hypoxic injury
Cell proliferation was measured by EdU staining, and the percentage of EdU-positive CMECs (red) was quantified (A). CMEC migration ability was measured by the Transwell assay (B). Images and quantitative analysis of the tube formation assay (C). MDA content, GSH and GSSG levels, and total SOD and Mn-SOD activities of CMECs (D). Relative expression of mitochondrial fusion/fission-related genes (E). Quantification of the expression of mitochondrial fusion/fission-related proteins and mitophagy-related proteins (F). Expression of mitochondrial apoptosis-related genes (G). Quantification of the expression of apoptosis-associated proteins (H). *p < 0.05 compared with the control group, #p < 0.05 compared with the hy + siNC group, & p < 0.05 compared with the hy + pcDNA3.1 group. n = 5 in each group
Fig. S8.
Mfn1 reversed EC dysfunction and oxidative stress induced by silencing Malat1 in CMECs under hypoxia.
Relative gene expression of Mfn1 in CMECs (A). Cell proliferation was measured by EdU staining, and the percentage of EdU-positive CMECs (red) was quantified (B). CMEC migration ability was measured by the Transwell assay (C). Images and quantitative analysis of the tube formation assay (D). MDA content, GSH and GSSG levels, and total SOD and Mn-SOD activities of CMECs (E). Relative expression of mitochondrial fusion/fission-related genes (F). Quantification of the expression of mitochondrial fusion/fission-related proteins and mitophagy-related proteins (G). Expression of mitochondrial apoptosis-related genes (H). Quantification of the expression of apoptosis-associated proteins (I). *p < 0.05 compared with the siNC + pcDNA3.1 group, #p < 0.05 compared with the hy + siNC + pcDNA3.1 group, & p < 0.05 compared with the hy + siMalat1+pcDNA3.1 group. n = 5 in each group
Fig. S9.
Overexpressing Mfn1 reversed the aggravation of oxidative stress, apoptosis and mitochondrial dysfunction induced by silencing Malat1 in MI hearts.
MDA content, total SOD activities, GSH and GSSG levels, and ROS and H2O2 generation in the border zone (A). Relative expression of mitochondrial apoptosis-related genes and mitochondrial fusion/fission-related genes (B). *p < 0.05 compared with the sham + Scr-shRNA + vector group, #p < 0.05 compared with the MI + Scr-shRNA + vector group, & p < 0.05 compared with the MI + Malat1-shRNA + vector group. n = 6 in each group
References
- 1.van Duijvenboden K. Conserved NPPB+ border zone switches from MEF2- to AP-1-driven gene program. Circulation. 2019;140(10):864–879. doi: 10.1161/CIRCULATIONAHA.118.038944. [DOI] [PubMed] [Google Scholar]
- 2.Tang R. A roadmap for fixing the heart: RNA regulatory networks in cardiac disease. Mol. Ther. Nucleic Acids. 2020;20:673–686. doi: 10.1016/j.omtn.2020.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Prestes P. A Guide to the short, long and circular RNAs in hypertension and cardiovascular disease. Int. J. Mol. Sci. 2020;21(10) doi: 10.3390/ijms21103666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jayasuriya R. Role of Nrf 2 in MALAT1/HIF-1α loop on the regulation of angiogenesis in diabetic foot ulcer. Free Radic. Biol. Med. 2020;156:168–175. doi: 10.1016/j.freeradbiomed.2020.05.018. [DOI] [PubMed] [Google Scholar]
- 5.Wang Y. Knockdown of MALAT1 attenuates high-glucose-induced angiogenesis and inflammation via endoplasmic reticulum stress in human retinal vascular endothelial cells. Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie. 2020;124:109699. doi: 10.1016/j.biopha.2019.109699. [DOI] [PubMed] [Google Scholar]
- 6.Zhang L., Wang H. Long non-coding RNA in CNS injuries: a new target for therapeutic intervention. Mol. Ther. Nucleic Acids. 2019;17:754–766. doi: 10.1016/j.omtn.2019.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gao Q., Wang Y. Long noncoding RNA MALAT1 regulates apoptosis in ischemic stroke by sponging miR-205-3p and modulating PTEN expression. Am. J. Tourism Res. 2020;12(6):2738–2748. [PMC free article] [PubMed] [Google Scholar]
- 8.Wu Q. Extracellular vesicles from human embryonic stem cell-derived cardiovascular progenitor cells promote cardiac infarct healing through reducing cardiomyocyte death and promoting angiogenesis. Cell Death Dis. 2020;11(5):354. doi: 10.1038/s41419-020-2508-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ala U. Competing endogenous RNAs, non-coding RNAs and diseases: an intertwined story. Cells. 2020;9(7) doi: 10.3390/cells9071574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peters M., Sampaio-Pinto V., da Costa Martins P. Non-coding RNAs in endothelial cell signalling and hypoxia during cardiac regeneration. Biochimica et biophysica acta. Molecular cell research. 2020;1867(3):118515. doi: 10.1016/j.bbamcr.2019.07.010. [DOI] [PubMed] [Google Scholar]
- 11.Zouein F., Booz G. Targeting mitochondria to protect the heart: a matter of balance? Clinical science (London, England. 1979;134(7):885–888. doi: 10.1042/CS20200236. 2020. [DOI] [PubMed] [Google Scholar]
- 12.Yan K. Mitochondrial miR-762 regulates apoptosis and myocardial infarction by impairing ND2. Cell Death Dis. 2019;10(7):500. doi: 10.1038/s41419-019-1734-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lugus J. Mitofusins are required for angiogenic function and modulate different signaling pathways in cultured endothelial cells. J. Mol. Cell. Cardiol. 2011;51(6):885–893. doi: 10.1016/j.yjmcc.2011.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li J. Mitofusin 1 is negatively regulated by microRNA 140 in cardiomyocyte apoptosis. Mol. Cell Biol. 2014;34(10):1788–1799. doi: 10.1128/MCB.00774-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao Y. Aberrant shuttling of long noncoding RNAs during the mitochondria-nuclear crosstalk in hepatocellular carcinoma cells. American journal of cancer research. 2019;9(5):999–1008. [PMC free article] [PubMed] [Google Scholar]
- 16.Chatterjee I. Endothelial lipid phosphate phosphatase-3 deficiency that disrupts the endothelial barrier function is a modifier of cardiovascular development. Cardiovasc. Res. 2016;111(1):105–118. doi: 10.1093/cvr/cvw090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guo X. LncRNA MALAT1 protects cardiomyocytes from isoproterenol-induced apoptosis through sponging miR-558 to enhance ULK1-mediated protective autophagy. J. Cell. Physiol. 2019;234(7):10842–10854. doi: 10.1002/jcp.27925. [DOI] [PubMed] [Google Scholar]
- 18.Huang S. Long noncoding RNA MALAT1 mediates cardiac fibrosis in experimental postinfarct myocardium mice model. J. Cell. Physiol. 2019;234(3):2997–3006. doi: 10.1002/jcp.27117. [DOI] [PubMed] [Google Scholar]
- 19.Vausort M., Wagner D., Devaux Y. Long noncoding RNAs in patients with acute myocardial infarction. Circ. Res. 2014;115(7):668–677. doi: 10.1161/CIRCRESAHA.115.303836. [DOI] [PubMed] [Google Scholar]
- 20.Su R. The mechanisms of PM and its main components penetrate into HUVEC cells and effects on cell organelles. Chemosphere. 2020;241:125127. doi: 10.1016/j.chemosphere.2019.125127. [DOI] [PubMed] [Google Scholar]
- 21.Pries A., Kuebler W., Habazettl H. Coronary microcirculation in ischemic heart disease. Curr. Pharmaceut. Des. 2018;24(25):2893–2899. doi: 10.2174/1381612824666180625142341. [DOI] [PubMed] [Google Scholar]
- 22.Wang J., Toan S., Zhou H. New insights into the role of mitochondria in cardiac microvascular ischemia/reperfusion injury. Angiogenesis. 2020;23(3):299–314. doi: 10.1007/s10456-020-09720-2. [DOI] [PubMed] [Google Scholar]
- 23.Wang J., Toan S., Zhou H. Mitochondrial quality control in cardiac microvascular ischemia-reperfusion injury: new insights into the mechanisms and therapeutic potentials. Pharmacol. Res. 2020;156:104771. doi: 10.1016/j.phrs.2020.104771. [DOI] [PubMed] [Google Scholar]
- 24.Yu H., Kalogeris T., Korthuis R. Reactive species-induced microvascular dysfunction in ischemia/reperfusion. Free Radic. Biol. Med. 2019;135:182–197. doi: 10.1016/j.freeradbiomed.2019.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang G., Wu Y., Zhu Y. Mechanism of MALAT1 preventing apoptosis of vascular endothelial cells induced by oxygen-glucose deficiency and reoxidation. Artificial cells, nanomedicine, and biotechnology. 2018;46:798–805. doi: 10.1080/21691401.2018.1436065. [DOI] [PubMed] [Google Scholar]
- 26.Wang X. Long non-coding RNAs H19, MALAT1 and MIAT as potential novel biomarkers for diagnosis of acute myocardial infarction. Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie. 2019;118:109208. doi: 10.1016/j.biopha.2019.109208. [DOI] [PubMed] [Google Scholar]
- 27.Yang X. Denervation drives skeletal muscle atrophy and induces mitochondrial dysfunction, mitophagy and apoptosis via miR-142a-5p/MFN1 axis. Theranostics. 2020;10(3):1415–1432. doi: 10.7150/thno.40857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dai S. Notch 1 protects against myocardial ischaemia-reperfusion injury via regulating mitochondrial fusion and function. J. Cell Mol. Med. 2020;24(5):3183–3191. doi: 10.1111/jcmm.14992. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 29.Wang T. NFATc3-dependent expression of miR-153-3p promotes mitochondrial fragmentation in cardiac hypertrophy by impairing mitofusin-1 expression. Theranostics. 2020;10(2):553–566. doi: 10.7150/thno.37181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lei L. Functions and regulatory mechanisms of metastasis-associated lung adenocarcinoma transcript 1. J. Cell. Physiol. 2018;234(1):134–151. doi: 10.1002/jcp.26759. [DOI] [PubMed] [Google Scholar]
- 31.Li H. Circular RNA circRNA_000203 aggravates cardiac hypertrophy via suppressing miR-26b-5p and miR-140-3p binding to Gata 4. Cardiovasc. Res. 2020;116(7):1323–1334. doi: 10.1093/cvr/cvz215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jakob P. Profiling and validation of circulating microRNAs for cardiovascular events in patients presenting with ST-segment elevation myocardial infarction. Eur. Heart J. 2017;38(7):511–515. doi: 10.1093/eurheartj/ehw563. [DOI] [PubMed] [Google Scholar]
- 33.Yang L. MicroRNA-26b-5p inhibits mouse liver Fibrogenesis and Angiogenesis by targeting PDGF receptor-beta. Molecular therapy. Nucleic acids. 2019;16:206–217. doi: 10.1016/j.omtn.2019.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated or analysed in this study can be obtained upon reasonable request from the corresponding author.