Abstract
In view of the rapidly progressing COVID-19 pandemic, our aim was to isolate and characterize SARS-CoV-2 from Indian patients. SARS-CoV-2 was isolated from nasopharyngeal swabs collected from the two members of a family without any history of (H/O) travel abroad. Both the virus isolates (8003 and 8004) showed CPE on day 3 post-inoculation, viral antigens by immunofluorescence assay and produced distinct, clear and uniform plaques. Infectious virus titers were 5 × 106 and 4 × 106 Pfu/ml by plaque assay and 107.5 and 107 by CPE-based TCID50/ml, respectively. Phylogenetic analysis grouped our isolates with the Italian strains. On comparison with Wuhan strain, 3 unique mutations were identified in nsp3 (A1812D), exonuclease (P1821S) of Orf1ab and spike protein (Q677H) regions, respectively. Both the viruses grouped with Italian strains of SARS-CoV-2 suggesting possible source being the virus imported from Italy. These fully characterized virus isolates will be useful in developing neutralization/virological assays for the evaluation of vaccines/antivirals.
Keywords: SARS-CoV-2, Plaque assay, TCID50, Phylogeny, Virus isolates, Mutation
Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the etiological agent of coronavirus disease 2019 (COVID-19) [1]. It has infected more than 49.1 million individuals and caused more than 1.2 million deaths globally (https://covid19.who.int/). First report of person-to-person transmission was established as an important mode of the virus transmission after the disease was documented in a family cluster of five patients [2].
SARS-CoV-2 is a betacoronavirus with single-stranded positive sense RNA genome of around 29.8 kb [1, 3]. The genome sequences of SARS-CoV-2 shared 96% similarity with bat coronavirus (RaTG13) suggesting possible Bat origin of SARS-CoV-2 [4]. The SARS-CoV-2 genome had unique polybasic cleavage motif PRRA in the S1-S2 junction region of spike protein. This cleavage site determines the infectivity and host range that may be responsible for increased human to human transmission by SARS-CoV-2.
Currently, several vaccines are under different phases of clinical trials / development. Being a RNA virus, SARS-CoV-2 is likely to mutate over time that may significantly impact transmission potential and response to vaccines / antivirals and utility of diagnostics developed based on earlier viruses. For this, it is essential to isolate and characterize SARS-CoV-2 at different times/different places. This study reports isolation and sequence analysis of SARS-CoV-2 at the beginning of emergence of COVID-19 in India. Importantly, the cases did not give history of (H/O) travel or contact with persons with H/O travel.
Results and discussion
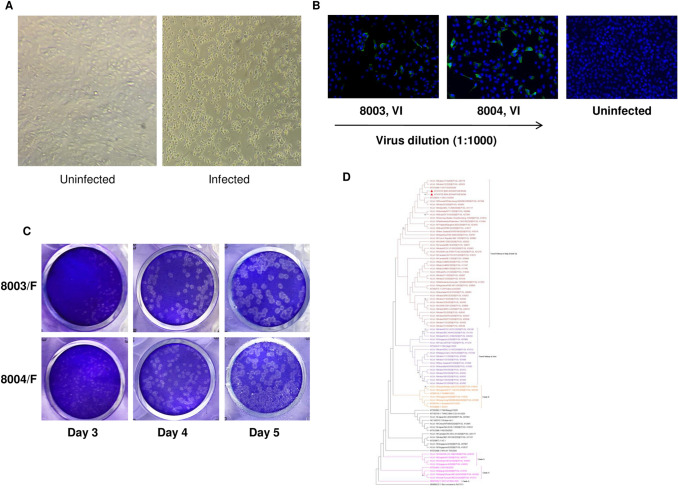
Nasopharyngeal swabs (NPS) from 5 COVID-19 patients (2 from the same family) were used to isolate the virus in Vero CCL-81 cells (ATCC, USA). Of these, 2 samples (8003 and 8004, IRSHA isolates) from a single family with Ct values < 15 in qRT-PCR showed distinct cytopathic effect (CPE, more than 90% cells detached) on day 3 post-inoculation (Fig. 1a). No cytopathic effects were observed for the remaining 3 samples even after one blind passage into T-25cm2 flask. Replication of SARS-CoV-2 was confirmed by immunofluorescence assay using serum sample from a convalescent-phase (27 days post-disease onset) COVID-19 patient. On day 2 post-infection, viral antigen(s) were detected only in infected cells (Fig. 1b). Successful virus isolation using nasopharyngeal swabs (NPS) was reported from Korea [5] and United States [6] (Vero CCL-81), Italy [7] (Vero E6) and Australia [8] (Vero cell line expressing human signalling lymphocytic activation molecule). Other countries used different sample types and cell lines for virus isolation. In the very first report of SARS-CoV-2 isolation from China, bronchoalveolar lavage sample was inoculated into primary culture of human airway epithelial cells [9]. Caco-2 cells were used for SARS-CoV-2 isolation in Germany [10]. Isolation of the virus from ocular fluid of COVID-19 patient suggests that human ocular cell types may be supporting SARS-CoV-2 replication [11].
Fig. 1.
Isolation and genomic characterization of SARS-CoV-2. a Phase-contrast microscopic image of uninfected control cells and cells infected with nasopharyngeal swab of SARS-CoV-2-infected patient exhibiting cytopathic effect, 3 days after inoculation. b Vero cells were infected with 1:1000 diluted 8003 and 8004 virus isolates. After 2 days post-infection, SARS-CoV-2 protein was detected with a convalescent patient’s serum and Alexa-fluor 488 conjugated goat anti-human-IgG. Nuclei were stained with DAPI. c Plaque morphology of two clinical isolates on Vero cells at day 3, day 4 and day 5 post-infection. Distinct plaques could be visualized on day 4 and day 5 post-infection. d Phylogenetic tree indicating complete genome sequences of SARS-CoV-2 isolates (n = 2) from Pune, India. Each strain is indicated by country, year of isolation, and GenBank accession number or GISAID number. The sequences obtained in this study are marked in filled colour triangles. Phylogenetic trees were constructed using neighbour joining method and distances were computed using maximum composite likelihood method in MEGA X software. Genetic distances were calculated using the p-distance model of nucleotide and amino acid substitution. The robustness of the resulting tree was assessed with 1000 bootstrap replicates
Plaque assay and CPE-based TCID50 were used to quantitate infectious viruses. Both the IRSHA isolates (8003-P0 and 8004-P0) formed clear and distinct plaques on day 4 post-infection (Fig. 1c). Plaque assay-based virus titers of 8003 and 8004 isolates were 3.0 × 106 and 7.5 × 106 PFU/mL, respectively. CPE-based virus titers of 8003 and 8004 were 10 4.34 and 10 5.6 TCID50 per mL, respectively, on day 3 post-infection and remained constant till day 5. Further passage in Vero CCL-81 cells did not lead to appreciable change in plaque assay-based titers; titers of 8003-P1 and 8004-P1 were 5 × 106 and 4.0 × 106 PFU/mL, respectively. However, the CPE-based titers showed significant increase on passaging and titers of 8003-P1 and 8004-P1 were 107.5 and 107 TCID50/mL, respectively.
For the genomic characterization, whole-genome sequencing of virus isolates from the first passage was performed using next-generation sequencing platform, Ion Proton system (Life technologies, USA) as reported earlier [12]. GenBank accession numbers for the generated sequences are MT416725 and MT416726. For phylogenetic analysis, 89 additional complete genome sequences were retrieved from GenBank and GISAID (https:www.gisaid.org) database. All 91 sequences were aligned using MAFFT v7.450, 2019 online program (https://mafft.cbrc.jp/alignment/software/).
The complete genome length of 8003 and 8004 viruses were 29,866 nucleotides (nt) with 5′untranslated region (UTR) of 265nt and 3′UTR of 188nt. These sequences were compared with reference sequence, NC045512 from Wuhan, China. At nucleotide level, IRSHA isolates were 99.96 ± 0.01% identical to the reference sequence. The distribution of the 11 mutations recorded was C241T (5′UTR), C313T, C3037T, C5700A, C14408T, C18928T (ORF1ab), A23403G, G23593C (spike protein) and G28881A, G28882A, G28883C (nucleocapsid phosphoprotein) (Table 1). Two mutations at positions, C313T and C3037T, were synonymous mutations with no change in amino acids, leucine (L) and phenylalanine (F). Eight mutations resulted in amino acid changes at positions 5700 (nsp3, A1812D), 14,408 (RdRp, P314L), 18,928 (3′ to 5′ exonuclease, exoN, P1821S), 23,403 (spike protein, D614G), and 23,593 (spike protein, Q677H). The mutations at G28881A, G28882A and G28883C resulted in amino acid changes, R9455K and G9456R in nucleocapsid (203 and 204) region.
Table 1.
Analysis of sequence variations and amino acid changes of Pune isolates in comparison to reference strain, NC045512 (Wuhan, China)
| Gene | 5′UTR | Orf1a | Orf1ab | Orf1ab | Orf1ab | Orf1ab | S | S | N | N | N |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Nucleotide variation | 241 | 313 | 3037 | 5700 | 14,408 | 18,298 | 23,403 | 23,593 | 28,881 | 28,882 | 28,883 |
| NC045512 | C | C | C | C | C | C | A | G | G | G | G |
| 8003-IRSHA (MT416725) | T | T | T | A | T | T | G | C | A | A | C |
| 8004-IRSHA (MT416726) | T | T | T | A | T | T | G | C | A | A | C |
| IND/c31 (EPI ISL 426,179) | T | T | T | C | T | C | G | G | A | A | C |
| IND/c32 (EPI ISL 420,555) | T | T | T | C | T | C | G | G | A | A | C |
| ISR IT0320 (MT276598) | T | T | T | C | T | C | G | G | A | A | C |
| Amino acid changes | Non-coding | CTC (L) to CTT (L) | TTC (F) to TTT (F) | GCT (A) to GAT (D) | CCT (P) to CTT (L) | CCT (P) to TCT (S) | GAT (D) to GGT (G) | CAG (Q) to CAC (H) | AGG (R) to AAA (K) | GGA (G) to CGA (R) | |
Nucleotide variation unique to 8003 and 8004 IRSHA isolates were highlighted in bold
Of these, 8 mutations were identical with c31/India, c32/India, and Israel sequences. A1812D, P1821S and Q677H mutations, respectively, in nsp3, exoN of ORF1ab and spike protein were unique to IRSHA isolates. A1812D mutation was in the protease domain of nsp3 region of orf1ab protein [13]. This mutation might modulate the function of nsp3 protein by affecting viral protein maturation process and innate immune signalling pathways. Mutation in exoN, P1821S might increase the fidelity of RNA synthesis by correcting nucleotide incorporation errors made by RdRp [14].
Phylogenetic analysis (Fig. 1d) revealed that IRSHA isolates were closely related to the Indian strains isolated from close contacts of infected patients with travel history to Italy (EPI ISL 420,555, EPI ISL 426,179). The nucleotide divergence within these sequences was found to be 0.02% while nucleotide identity with cluster of sequences with travel history to Italy was 99.97 ± 0.01%. The SARS-CoV-2 sequences of Italy origin were further segregated into two subgroups: IRSHA (Pune, western India) isolates clubbed with two other Indian sequences (India/c31/2020, India/c32/2020, Delhi, north India) from contacts of patients returned from Italy and viruses from Israel, Greece, Russia, Brazil, Germany and Netherlands (Fig. 1d). The other subgroup clustered with Indian isolates obtained from Italian tourists who visited India /their contacts and viruses from Italy and Spain [15]. Similar to Wuhan isolate, all these sequences belonged to the L strain of SARS-CoV-2, with C at 8782nd and T at 28114th position [16].
IRSHA sequences clustered together with sequences derived from Italy and other European countries are classified as clade G due to the presence of D614G mutation in the spike protein. Origin of D614G variant, located in the S1 domain of spike protein, was associated with accelerating the spread of infections in Europe and North America [17]. In our isolates, a unique mutation Q677H located near the S1-S2 junction region (681–684) might affect the SARS-CoV-2 fusion with cell membrane. Importantly, attenuated phenotype of SARS-CoV-2 variants was obtained following 15-30 bp deletion at S1-S2 junction region of the spike protein [18]. Detection of deletion at the flank sites of PRRAR in 3 clinical samples from Guangdong, China, is noteworthy [19]. In addition, circulation of other genetic variants such as 382 nucleotide deletion in orf8 region from eight hospitalized patients in Singapore and 81 nucleotide deletion in orf7a gene in a patient from Arizona, USA [20], has been reported and effect of such mutations on the transmissibility of the virus should be available soon.
In the initial phase of the pandemic, India reported cases in travellers from China (1st case), Italy and Iran [15, 21]. IRSHA sequences exhibited 99.94 ± 0.01% nucleotide identity with viral sequences obtained from patients having travel history to Iran. They formed separate cluster with four common nucleotide mutations at G1437A, G11083T (Orf1a), T28688C (Nucleocapsid) and G29742T (3′UTR).
Continuous evolution of SARS-CoV-2 was observed in viral sequences of Italy origin. The very first imported case of SARS-CoV-2 in Italy was in a Chinese traveller [22]. The sequence (GenBank accession No MT066156) was similar to Wuhan, China, except for a unique G251V mutation forming a separate clade V [22]. On several introductions / transmissions of SARS-CoV-2 in Europe, D614G mutation was identified in Italian patients with no travel history. These sequences formed a separate cluster, clade G [23] which spread to other countries including India as observed in this study.
Italy being a country of global choice for tourism, it is natural to identify COVID-19 cases among travellers from Italy and their close contacts. Though the patients from whom we isolated virus did not give H/O travel or contact with a traveller from abroad, sequence data suggest source of the virus to be related to the strain imported from Italy. Thus, sequence analysis can help in identifying origin and circulation of a particular virus strain in a population.
In summary, we report isolation and characterization of two Indian isolates from a single family without history of travel. Phylogenetic analysis revealed that source of the virus is related to the strain imported from Italy. The availability of fully characterized isolates along with well-standardized plaque and TCID50 assays will allow development of much required neutralization tests for understanding pathogenesis of the disease and evaluation of vaccines and other immunotherapeutics.
Acknowledgements
We are grateful for the valuable support from the health officials of Pune Municipal Corporation and Bharati Vidyapeeth University authorities to make this study feasible. We thank Tushar Bhosale, Shamburaje Pisal, Prajakta Rane, Mrunal Gosavi, Pravin Kore and Shweta Chelluboina for their technical assistance. We also thank Rahul Kadu for BSL-3 lab support.
Author’s contributions
VAA and ACM contributed to concept, study design and supervision; SS, HPP and STM performed the experiments, data analysis and interpretation; SP, SL and ACM involved in patient’s recruitment and clinical management; SS and HPP participated in writing of the manuscript; and VAA did review and editing of manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by Department of Biotechnology (DBT) – BIRAC (grant number: BT/NBM0095/02/18). The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Data availability
All data generated or analysed during this study are included in this article.
Declarations
Conflict of interest
None of the authors have any conflict of interest.
Ethical approval
This study was approved by Institutional Ethics Committee of Bharati Hospital & Research Centre at Bharati Vidyapeeth Deemed University (IEC/2020/25).
Consent to participate
Informed written consent was obtained from each participant before enrolment in this study.
Consent for publication
Not applicable.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Wu F, Zhao S, Yu B, Chen YM, Wang W, Song ZG, et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579(7798):265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chan JFW, Yuan S, Kok KH, To KKW, Chu H, Yang J, et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020;395(10223):514–523. doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gorbalenya AE, et al. The Species Severe Acute Respiratory Syndrome-Related Coronavirus: Classifying 2019-NCoV and Naming It SARS-CoV-2. Nat Microbiol. 2020;5(4):536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lau SKP, Luk HKH, Wong ACP, Li KSM, Zhu L, He Z, et al. Possible Bat Origin of Severe Acute Respiratory Syndrome Coronavirus 2. Emerg Infect Dis. 2020 doi: 10.3201/eid2607.200092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim J-M, Chung Y-S, Jo HJ, Lee N-J, Kim MS, Woo SH, et al. Identification of Coronavirus Isolated from a Patient in Korea with COVID-19. Osong Public Health Res Perspect. 2020;11(1):3–7. doi: 10.24171/j.phrp.2020.11.1.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harcourt J, Tamin A, Lu X, Kamili S, Sakthivel SK, Murray J, et al. Severe Acute Respiratory Syndrome Coronavirus 2 from Patient with 2019 Novel Coronavirus Disease. United States Emerg Infect Dis. 2020;26(6):1266–1273. doi: 10.3201/eid2606.200516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Licastro D, Rajasekharan S, Dal Monego S, Segat L, D’Agaro P, Marcello A. Isolation and full-length genome characterization of SARS-CoV-2 from COVID-19 cases in Northern Italy. J Virol. 2020;94(11):e00543–e620. doi: 10.1128/JVI.00543-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caly L, Druce J, Roberts J, Bond K, Tran T, Kostecki R, et al. Isolation and Rapid Sharing of the 2019 Novel Coronavirus (SARS-CoV-2) From the First Patient Diagnosed With COVID-19 in Australia. Med J Aust. 2020;212(10):459–462. doi: 10.5694/mja2.50569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382(8):727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoehl S, Rabenau H, Berger A, Kortenbusch M, Cinatl J, Bojkova D, et al. Evidence of SARS-CoV-2 infection in returning travelers from Wuhan. China N Engl J Med. 2020;382(13):1278–1280. doi: 10.1056/NEJMc2001899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Colavita F, Lapa D, Carletti F, Lalle E, Bordi L, Marsella P, et al. SARS-CoV-2 Isolation From Ocular Secretions of a Patient With COVID-19 in Italy With Prolonged Viral RNA Detection. Ann Intern Med. 2020 doi: 10.7326/M20-1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shrivastava S, Tiraki D, Diwan A, Lalwani SK, Modak M, Mishra AC, et al. Co-circulation of all the four dengue virus serotypes and detection of a novel clade of DENV-4 (genotype I) virus in Pune, India during 2016 season. PLoS ONE. 2018;13(2):e0192672. doi: 10.1371/journal.pone.0192672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rut W, Groborz K, Zhang L, Sun X, Zmudzinski M, Hilgenfeld R, et al (2020) Substrate specificity profiling of SARS-CoV-2 Mpro protease provides basis for anti-COVID-19 drug design. bioRxiv;2020.03.07.981928.
- 14.Pachetti M, Marini B, Benedetti F, Giudici F, Mauro E, Storici P, et al. (2020) Emerging SARS-CoV-2 mutation hot spots include a novel RNA-dependent-RNA polymerase variant. J Transl Med. 2020;18(1):179. doi: 10.1186/s12967-020-02344-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Potdar V, Cherian S, Deshpande G, Ullas P, Yadav P, Choudhary M, et al. Genomic analysis of SARS-CoV-2 strains among Indians returning from Italy, Iran & China, & Italian tourists in India. Indian J Med Res. 2020;151(2):255–260. doi: 10.4103/ijmr.IJMR_1058_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tang X, Wu C, Li X, Song Y, Yao X, Wu X, et al. On the origin and continuing evolution of SARS-CoV-2. Natl Sci Rev. 2020 doi: 10.1093/nsr/nwaa036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bhattacharyya C, Das C, Ghosh A, Singh AK (2020) Global Spread of SARS-CoV-2 Subtype with Spike Protein Mutation D614G is Shaped by Human Genomic Variations that Regulate Expression of TMPRSS2 and MX1 Genes. BioRxiv.
- 18.Lau S-Y, Wang P, Mok BW-Y, Zhang AJ, Chu H, Lee AC-Y, et al. Attenuated SARS-CoV-2 variants with deletions at the S1/S2 junction. Emerg Microbes Infect. 2020;9(1):837–842. doi: 10.1080/22221751.2020.1756700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu Z, Zheng H, Yuan R, Li M, Lin H, Peng J, et al. Identification of a common deletion in the spike protein of SARS-CoV-2. J Virol. 2020 doi: 10.1128/JVI.00790-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holland L, Kaelin E, Maqsood R, Estifanos B, LI W, Varsani A,, et al. An 81 nucleotide deletion in SARS-CoV-2 ORF7a identified from sentinel surveillance in Arizona (Jan-Mar 2020) J Virol. 2020 doi: 10.1128/JVI.00711-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yadav P, Potdar V, Choudhary M, Nyayanit D, Agrawal M, Jadhav S, et al. Full-genome sequences of the first two SARS-CoV-2 viruses from India. Indian J Med Res. 2020;151(2):200–209. doi: 10.4103/ijmr.IJMR_663_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Capobianchi MR, Rueca M, Messina F, Giombini E, Carletti F, Colavita F, et al. Molecular characterization of SARS-CoV-2 from the first case of COVID-19 in Italy. Clin Microbiol Infect. 2020 doi: 10.1016/j.cmi.2020.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stefanelli P, Faggioni G, Lo Presti A, Fiore S, Marchi A, Benedetti E, et al. Whole genome and phylogenetic analysis of two SARSCoV-2 strains isolated in Italy in January and February 2020: Additional clues on multiple introductions and further circulation in Europe. Euro Surveill. 2020 doi: 10.2807/1560-7917.ES.2020.25.13.2000305. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analysed during this study are included in this article.