Abstract
Introduction
Diagnosing renal pathologies is important for performing treatments. However, classifying every glomerulus is difficult for clinicians; thus, a support system, such as a computer, is required. This paper describes the automatic classification of glomerular images using a convolutional neural network (CNN).
Method
To generate appropriate labeled data, annotation criteria including 12 features (e.g., “fibrous crescent”) were defined. The concordance among 5 clinicians was evaluated for 100 images using the kappa (κ) coefficient for each feature. Using the annotation criteria, 1 clinician annotated 10,102 images. We trained the CNNs to classify the features with an average κ ≥0.4 and evaluated their performance using the receiver operating characteristic–area under the curve (ROC–AUC). An error analysis was conducted and the gradient-weighted class activation mapping (Grad-CAM) was also applied; it expresses the CNN’s focusing point with a heat map when the CNN classifies the glomerular image for a feature.
Results
The average κ coefficient of the features ranged from 0.28 to 0.50. The ROC–AUC of the CNNs for test data varied from 0.65 to 0.98. Among the features, “capillary collapse” and “fibrous crescent” had high ROC–AUC values of 0.98 and 0.91, respectively. The error analysis and the Grad-CAM visually showed that the CNN could not distinguish between 2 different features that had similar visual structures or that occurred simultaneously.
Conclusion
The differences in the texture or frequency of the co-occurrence between the different features affected the CNN performance; thus, to improve the classification accuracy, methods such as segmentation are required.
Keywords: artificial intelligence, convolutional neural network, deep learning, glomerular image, renal pathology
Understanding renal pathology is important when making diagnoses and decisions for the course(s) of treatment.1,2 When interpreting renal pathology, all the glomeruli must be carefully observed individually. This is a time-consuming task for clinicians and requires the support of a computer.3
In recent years, there have been advancements in technologies related to artificial intelligence known as deep learning.4 Through deep learning, general image recognition is often more accurate than by humans. In particular, the convolutional neural network (CNN), which is based on the neocognitron,5 has played a major role. The CNN has demonstrated high performance in image recognition and object extraction even in the medical field. For example, Saha et al. reported that they detected mitosis from pathological glass slides of breast cancer using a CNN with a 92% accuracy and an 88% recall.6
Research on pathological glomerular images using machine learning, including the CNN, involves the detection of glomeruli from a whole slide image (WSI) and the classification of glomeruli. As an example of glomerular detection from a WSI, Gallero et al. reported that their CNN detected glomeruli from a periodic acid−Schiff (PAS) −stained WSI with an F1 score of 0.937.7 Kawazoe et al. reported that they detected glomeruli from PAS-, periodic acid-methenamine-silver (PAM)−, Masson trichrome−, and Azan-stained WSIs, with F1 scores ranging from 0.876 to 0.925 using a modified CNN.8 Bukowy et al. reported that they constructed a CNN to detect glomeruli from trichrome-stained WSIs with an average precision and recall of 96.94% and 96.79%.9 Hermsen et al. constructed a CNN to segment WSIs into 11 classes such as “glomeruli” or “interstitium” from the PAS-stained WSI, and it detected 92.7% of all glomeruli.10
Regarding glomerular classification, George et al. reported that they used the k−nearest neighbor algorithm and succeeded in classifying whether the glomerular images contained proliferative lesions with 92.3% precision.11 In another study, Shruti et al. constructed a CNN to determine whether the image obtained from trichrome-stained WSI is a “non-global sclerosis, global sclerosis, or non-glomerular” with 89.66% to 95.06% accuracy.12 Similarly, John et al. used deep learning to diagnose whether the glomeruli from hematoxylin and eosin−stained WSIs of the transplanted kidney are sclerotic, with an F1 score of 0.865 to 0.879.13 Chagas et al. combined a CNN and a support vector machine to classify the hypercellularity for glomerular images from hematoxylin and eosin− and PAS-stained WSIs, with 98.8% to 99.6% accuracy.14
To automatically classify the glomerular images using CNN, the ground truth for features is essential for all the images. However, in the routine diagnosis of kidney disease, scores are not always recorded for all of the features for all glomeruli, and they are often recorded only for representative glomeruli. Therefore, the pathological reports recorded in daily medical care often do not have complete ground truth for all glomeruli. Furthermore, the existing annotation criterion for a specific disease, such as the Oxford classification,15 or the annotation criterion for only particular diseases, such as the multicenter Nephrotic Syndrome Study Network (NEPTUNE) scoring system,16 cannot be applied to various diseases in their current format. Therefore, annotation criteria that can be applied to various kidney diseases should be newly developed. First, to create the ground truth of glomerular images, this study established the annotation criteria for glomerular images and evaluated the degree of concordance among clinicians based on the criteria. Next, CNNs that classified the various features of the glomerular images were developed, and their performance was evaluated. Finally, the concordance between the CNN score and the clinicians’ score was assessed, and an error analysis was performed to determine computer-related issues for the renal pathology images.
Materials and Methods
Dataset Collection
This study collected 293 PAS-stained WSIs from 3 hospitals: 99 WSIs from the University of Tokyo Hospital (UTH) from 2010 to 2017; 88 WSIs from the Tazuke Kofukai Medical Research Institute, Kitano Hospital (KH), from 2014 to 2017; and 106 WSIs from the University of Tsukuba Hospital (UTSH) from 2009 to 2016. From the 293 WSIs, the glomerular image was manually extracted in a rectangular shape so that 1 major glomerulus was included, which yielded 10,202 images. Supplementary Table S1 lists the major diagnoses of the 293 WSIs for the 10,202 images. The extracted glomerular images were converted into the png format using Openslide17 and were saved as a single image file.
Development of Annotation Criteria
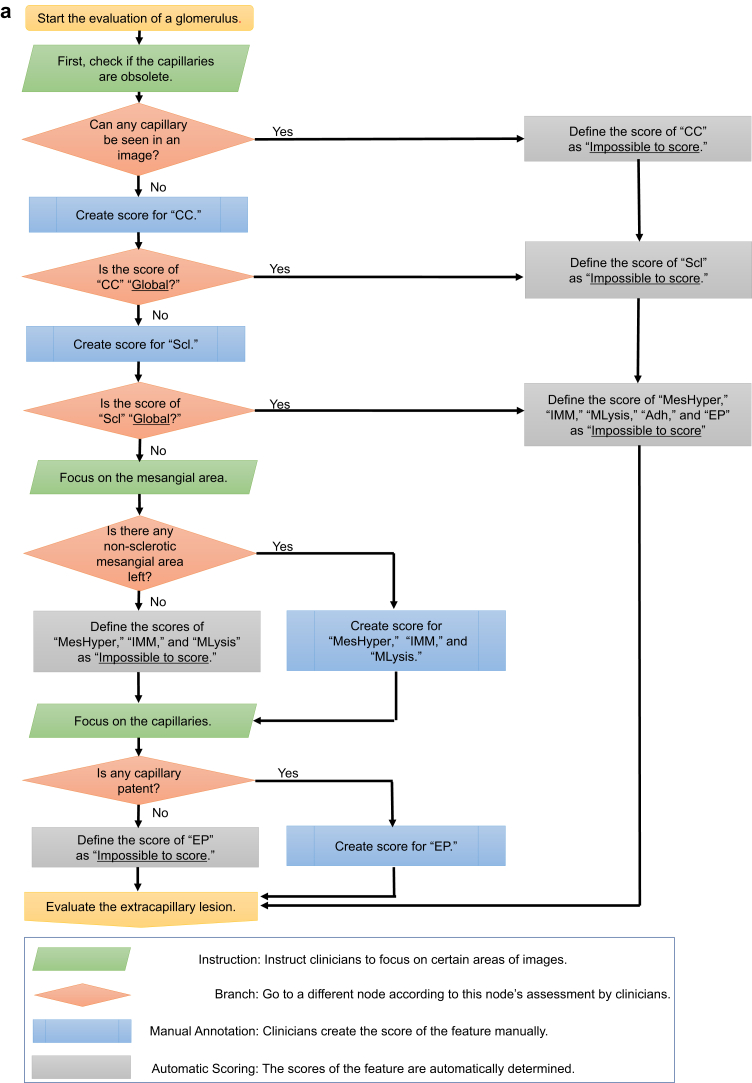
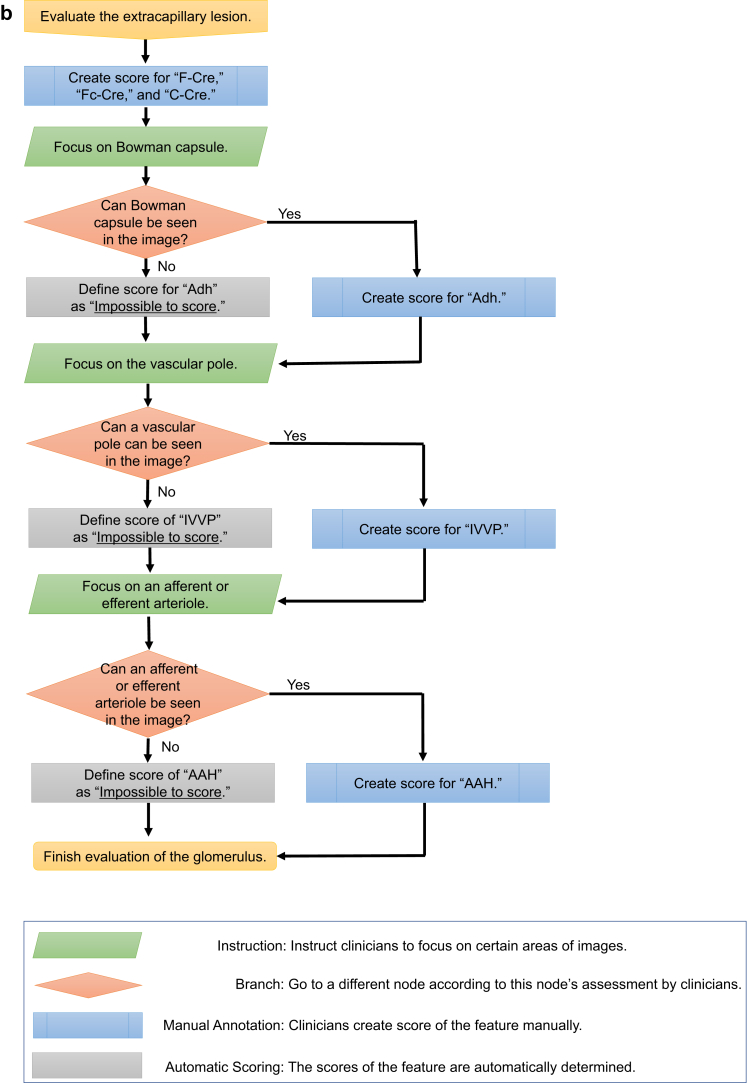
Five co-authored clinicians (1 physician and 4 pathologists) developed the annotation criteria for various kidney diseases based on the Delphi method,18 because we were to guarantee the quality of the annotation criteria and annotated labels; Krause et al.19 showed that the quality of the annotated label affected the validity of its performance in the machine learning model for diabetic retinopathy. Consequently, the annotation criteria consisted of the following 3 elements: (i) “feature”: finding the name (e.g., “mesangial hypercellularity”); (ii) “score”: the possible value of the feature (e.g., “normal,” “mild,” “moderate,” “severe”); and (iii) “regulation”: definitions for determining the score (e.g., “count the number of mesangial cells …”). The annotation criteria also included 12 features, which are important to diagnose the PAS-stained images. The 12 features are as follows: (i) capillary collapse (CC); (ii) sclerosis (Scl); (iii) mesangial hypercellularity (MesHyper); (iv) increased mesangial matrix (IMM); (v) mesangiolysis (MLysis); (vi) endocapillary proliferation (EP); (vii) fibrous crescent (F-Cre); (viii) fibrocellular crescent (Fc-Cre); (ix) cellular crescent (C-Cre); (x) adhesion (Adh); (xi) afferent/efferent arteriolar hyalinosis (AAH); and (xii) increased vasculature around the vascular pole (IVVP). The granularity of the score and the regulation were also discussed and agreed upon by the 5 clinicians. In some images, the score of the particular features could not be obtained; these were classified as “impossible to score.” Supplementary Table S2 lists the annotation criteria that were developed for this study. Supplementary Figure S1 depicts the example images of annotation for each feature. In addition to those criteria, an annotation flowchart was developed to clarify the dependency between the features, which sometimes made the annotation complicated. Figure 1a and b show the annotation flowcharts.
Figure 1.
(a, b) Flowcharts of the annotation for an image of a glomerulus. These 2 flowcharts represent the order followed to create a feature score. AAH, afferent/efferent arteriolar hyalinosis; Adh, adhesion; CC, capillary collapse; C-Cre, cellular crescent; EP, endocapillary proliferation; F-Cre, fibrous crescent; Fc-Cre, fibrocellular crescent; IMM, increased mesangial matrix; IVVP, increased vasculature around the vascular pole; MesHyper, mesangial hypercellularity; MLysis, mesangiolysis; Scl, sclerosis.
Concordance Evaluation of Annotated Features Between Clinicians
Out of 10,202 images, the first author selected 100 images for concordance evaluation. These 100 images included relatively “easy-to-score” images but also “hard-to-score” images in which different clinicians might assign different scores. Five clinicians annotated all 12 features for the 100 images except for 2 images in which 2 clinicians considered them to be ineligible to be scored. Supplementary Figure S2 shows the 98 images used for concordance evaluation. After the 5 clinicians finished annotating the images, the Cohen kappa (κ) coefficients20 of the 5 clinicians were calculated.
Development and Evaluation of CNN
One physician annotated all 12 features that were included in the developed annotation criteria for the remaining 10,102 glomerular images. The breakdown list of the 10,102 images for the features and scores is presented in Table 1.
Table 1.
Breakdown of the annotated images
| Feature | Score | ||||
|---|---|---|---|---|---|
| CC | (−) | Segmental | Global | Impossible to score | |
| 8837 | 187 | 1022 | 56 | ||
| Scl | (−) | Segmental | Global | Impossible to score | |
| 8499 | 448 | 82 | 1083 | ||
| MesHyper | Normal | Mild | Moderate | Severe | Impossible to score |
| 5411 | 1874 | 726 | 878 | 1213 | |
| IMM | (−) | (+) | Impossible to score | ||
| 1569 | 7320 | 1213 | |||
| MLysis | (−) | (+) | Impossible to score | ||
| 8777 | 112 | 1213 | |||
| EP | (−) | Segmental | Global | Impossible to score | |
| 8409 | 496 | 34 | 1163 | ||
| F-Cre | (−) | (+) | |||
| 8611 | 1491 | ||||
| Fc-Cre | (−) | (+) | |||
| 9413 | 689 | ||||
| C-Cre | (−) | (+) | |||
| 9733 | 369 | ||||
| Adh | (−) | (+) | Impossible to score | ||
| 6826 | 1925 | 1351 | |||
| IVVP | (−) | (+) | Impossible to score | ||
| 2160 | 144 | 7798 | |||
| AAH | (−) | (+) | Impossible to score | ||
| 798 | 235 | 9069 | |||
For each of the 12 features that were made, the distribution of the scores for all of the 10,102 images are listed. Each number represents the number of images scored for each score. AAH, afferent/efferent arteriolar hyalinosis; Adh, adhesion; C-Cre, cellular crescent; CC, capillary collapse; EP, endocapillary proliferation; F-Cre, fibrous crescent; Fc-Cre, fibrocellular crescent; IMM, increased mesangial matrix; IVVP, increased vasculature around the vascular pole; MesHyper, mesangial hypercellularity; MLysis, mesangiolysis; Scl, sclerosis.
The CNNs were developed for the features for which the κ coefficients were ≥0.40 to classify the features as positive or negative. The binary class breakdown list of the 10,102 images of the 5 features for the CNN is presented in Table 2. The strength of the correlation between the features was calculated using the phi coefficient (Table 3). There was a strong correlation between CC and F-Cre (phi coefficient = 0.67).
Table 2.
Breakdown of data used for the convolutional neural network (CNN)
| Score Feature |
Negative | Positive | Ratio of negative to positive |
|---|---|---|---|
| CC | None, Impossible to score | Segmental, Global | |
| 8893 | 1209 | 7.36 | |
| IMM | (−), Impossible to score | (+) | |
| 2782 | 7320 | 0.38 | |
| MLysis | (−), Impossible to score | (+) | |
| 9990 | 112 | 89.20 | |
| F-Cre | (−) | (+) | |
| 8611 | 1491 | 5.78 | |
| IVVP | (−), Impossible to score | (+) | |
| 9958 | 144 | 69.15 |
Each number shows the number of images contained in the score. CC, capillary collapse; F-Cre, fibrous crescent; IMM, increased mesangial matrix; IVVP, increased vasculature around the vascular pole; MLysis, mesangiolysis.
Table 3.
Phi coefficient among the 5 features
| CC | 1.00 | ||||
|---|---|---|---|---|---|
| IMM | –0.50 | 1.00 | |||
| MLysis | –0.04 | 0.04 | 1.00 | ||
| F-Cre | 0.67 | –0.36 | –0.02 | 1.00 | |
| IVVP | –0.03 | 0.03 | 0.00 | –0.03 | 1.00 |
| CC | IMM | MLysis | F-Cre | IVVP |
This table shows the phi coefficient between the 2 features among the 5 features. Each number shows the phi coefficient between the 2 features. The phi coefficient ranges from −1 to 1, where 1 represents a perfect positive correlation, −1 represents a perfect negative correlation, and 0 represents no correlation at all. CC, capillary collapse; F-Cre, fibrous crescent; IMM, increased mesangial matrix; IVVP, increased vasculature around the vascular pole; MLysis, mesangiolysis.
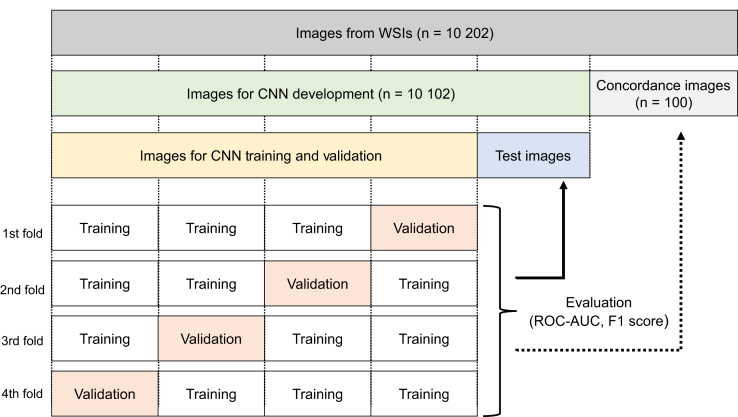
The performance of the CNN was evaluated on the validation, test, and concordance data, respectively, over the 4-fold cross-validation (Figure 2). The performance was measured using the average ROC-AUC and F1 score via 4-fold stratified cross-validation for the 10,102 images, and also for the 98 images used for the concordance evaluation.
Figure 2.
Schematic representation of the cross-validation. This figure shows how all images were divided for training, validation, testing, and concordance. First, 10,202 glomerular images were collected from the whole slide images (WSIs). Next, 100 glomerular images were chosen for the concordance data. Finally, the remaining 10,102 images were divided into 5 groups, 1 for the test data and the other 4 groups as the training data and validation data that were used for convolutional neural network training. ROC−AUC, receiver operating characteristic−area under the curve.
This study used a 50-layer residual network (ResNet50) as the structure of the CNN, which demonstrates the high performance in general image recognition and is unlikely to fall into poor local minima because of its structure21; hence, it would achieve high classification performance. This investigation also adopted transfer learning,22 which uses the parameters that were trained by the ImageNet dataset.23 The weighted softmax cross entropy loss was used as a loss function because the dataset for this study was unbalanced between positive and negative. For the weighted softmax cross entropy, the reciprocal of the ratio of the number of images in the class to the total number of images was set for each class. As the optimizing method, adaptive moment estimation (Adam), which has a fast convergence speed and can easily obtain good accuracy without parameter tuning, was used with an empirically determined hyperparameter (α = 1.0 × 10−7). All original images were resized to 224 × 224 pixels for the CNN input.
At the time of CNN training, data augmentation was applied to randomly rotate (0°, 90°, 180°, and 270°) and invert the images. The losses for the training and validation data were calculated for each epoch. The training was stopped at the point at which the loss of the validation data did not decrease for at least 50 continuous epochs.
Python3 was used as the programming language, and Chainer (version 4.0.0) was used as the deep learning library. The calculations were performed using NVIDIA DGX-1 (OS: Ubuntu 16.04, main memory: 515 GB, CPU: Intel Xeon CPU E5-2698 v4, GPU: 8 Tesla V100-SXM).
Concordance Evaluation Between the CNN Score and Clinicians’ Score
For the features for which the ROC–AUC for the test data exceeded 0.9, this study compared the CNN’s predictive score with the 5 clinicians’ scores for the concordance data and performed error analysis. Moreover, to perform the error analysis visually and to help understand the CNN’s properties, gradient-weighted class activation mapping24 (Grad-CAM) was applied for the concordance data; it is a technique that expresses the CNN’s focusing point using a heat map calculated by summing up the intermediate gradient of the CNN for each channel.
Results
Concordance Between Clinicians
Table 4 lists the results of the Cohen κ coefficient of the 5 clinicians for the 98 images. The average κ coefficient ranged from 0.28 to 0.50, and was ≥0.40 (moderate agreement) for the 5 features (CC, IMM, MLysis, F-Cre, and IVVP).
Table 4.
Results of Cohen κ coefficient by the 5 clinicians for the concordance images
| A/B | A/C | A/D | A/E | B/C | B/D | B/E | C/D | C/E | D/E | Average | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| CC | 0.33 | 0.30 | 0.50 | 0.35 | 0.51 | 0.51 | 0.40 | 0.44 | 0.35 | 0.41 | 0.41 |
| Scl | 0.18 | 0.36 | 0.46 | 0.44 | 0.46 | 0.31 | 0.26 | 0.52 | 0.38 | 0.44 | 0.38 |
| MesHyper | 0.44 | 0.37 | 0.35 | 0.37 | 0.42 | 0.27 | 0.28 | 0.29 | 0.36 | 0.35 | 0.35 |
| IMM | 0.45 | 0.52 | 0.62 | 0.45 | 0.43 | 0.42 | 0.45 | 0.53 | 0.45 | 0.42 | 0.47 |
| MLysis | 0.54 | 0.48 | 0.68 | 0.53 | 0.25 | 0.39 | 0.41 | 0.45 | 0.56 | 0.56 | 0.49 |
| EP | 0.19 | 0.38 | 0.54 | 0.35 | 0.18 | 0.30 | 0.41 | 0.34 | 0.35 | 0.51 | 0.36 |
| F-Cre | 0.59 | 0.50 | 0.20 | 0.64 | 0.72 | 0.04 | 0.59 | 0.02 | 0.50 | 0.28 | 0.41 |
| Fc-Cre | 0.34 | 0.20 | 0.30 | 0.23 | 0.72 | 0.39 | 0.56 | 0.12 | 0.44 | 0.33 | 0.36 |
| C-Cre | 0.47 | 0.57 | 0.24 | 0.33 | 0.44 | 0.26 | 0.45 | 0.22 | 0.53 | 0.40 | 0.39 |
| Adh | 0.16 | 0.58 | 0.26 | 0.37 | 0.18 | 0.29 | 0.23 | 0.08 | 0.28 | 0.33 | 0.28 |
| IVVP | 0.40 | 0.40 | 0.45 | 0.56 | 0.41 | 0.55 | 0.56 | 0.56 | 0.46 | 0.63 | 0.50 |
| AAH | 0.25 | 0.16 | 0.41 | 0.47 | 0.35 | 0.46 | 0.37 | 0.39 | 0.26 | 0.60 | 0.37 |
The rows indicate the features, and the column indicate the pairs of clinicians (e.g., A/B implies clinicians A and B) and their average value. Each number shows the Cohen κ coefficient. The Cohen κ coefficient ranges from −1 to 1, where 1 represents perfect concordance, −1 represents perfect discordance, and 0 represents completely no concordance. Adh, adhesion; AAH, afferent/efferent arteriolar hyalinosis; C-Cre, cellular crescent; CC, capillary collapse; EP, endocapillary proliferation; F-Cre, fibrous crescent; Fc-Cre, fibrocellular crescent; IMM, increased mesangial matrix; IVVP, increased vasculature around the vascular pole; MesHyper, mesangial hypercellularity; MLysis, mesangiolysis; Scl, sclerosis.
CNN Performance
Table 5 summarizes the ROC–AUC and the F1 score for the validation data, test data, and concordance data. The ROC–AUC for the test data varied from 0.65 to 0.98. Among the 5 features, CC and F-Cre demonstrated high ROC–AUCs of 0.98 and 0.91, respectively.
Table 5.
Overall results of the convolutional neural network (CNN) performance
| Feature | Validation data |
Test data |
Concordance data |
|||
|---|---|---|---|---|---|---|
| ROC–AUC | F1 score | ROC–AUC | F1 score | ROC–AUC | F1 score | |
| CC | 0.97 | 0.79 | 0.98 | 0.79 | 0.84 | 0.59 |
| IMM | 0.82 | 0.82 | 0.79 | 0.79 | 0.76 | 0.72 |
| MLysis | 0.87 | 0.12 | 0.76 | 0.08 | 0.37 | 0.04 |
| F-Cre | 0.90 | 0.61 | 0.91 | 0.63 | 0.69 | 0.53 |
| IVVP | 0.62 | 0.04 | 0.65 | 0.05 | 0.47 | 0.04 |
Each number shows the average receiver operating characteristic–area under the curve (ROC–AUC) or the F1 score over the 4 folds by each feature. CC, capillary collapse; F-Cre, fibrous crescent; IMM, increased mesangial matrix; IVVP, increased vasculature around the vascular pole; MLysis, mesangiolysis.
Concordance Between the CNN Score and Clinicians’ Score
As shown in Table 5, there were 2 features (CC and F-Cre) with ROC–AUCs for the test data that exceeded 0.9 (CC and F-Cre). Error analysis was conducted for these 2 features. Tables 6 and 7 present the comparisons for CC and F-Cre, respectively. As shown in Table 6, for CC, there are 13 completely true-positive (cTP) images that the CNN predicted to be positive, and all 5 clinicians scored them positively. In addition, there were no completely false-positive (cFP) images that the CNN predicted to be negative; however, no clinician scored these images positively. There were 33 completely true-negative (cTN) images that the CNN predicted to be negative, and no clinician scored them as positive. There was 1 completely false-negative (cFN) image that the CNN predicted to be negative; however, all of the clinicians scored it as positive. Table 7 demonstrates that for the F-Cre, there are no cTP images but there are 20 cFP images, 37 cTN images, and 1 cFN image.
Table 6.
Comparison between the convolutional neural network (CNN) predictive score and the clinicians’ feature for the capillary collapse (CC)
| No. of clinicians out of 5 who scored positive |
|||||||
|---|---|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | 4 | 5 | ||
| CNN’s predictive score over 4 folds | 0.75–1.00 | 0 a | 2 | 2 | 5 | 7 | 10b |
| 0.50–0.75 | 0a | 2 | 2 | 2 | 2 | 3b | |
| 0.25–0.50 | 9c | 5 | 0 | 0 | 1 | 1d | |
| 0.00–0.25 | 24d | 11 | 4 | 2 | 4 | 0d | |
This confusion matrix shows the comparison between the CNN’s predictive score and the clinicians’ feature for the CC. Rows show the CNN’s predictive score for an image, calculated as an average of the CNN’s softmax probability over the 4 folds. Columns show the number of clinicians out of 5 who scored it as positive. Each number represents the number of images.
Images in which the CNN predicted it to be positive but no clinician scored it as positive (the CNN’s completely false-positive result).
Images in which the CNN predicted it to be positive and all of the clinicians scored it as positive (the CNN’s completely true-positive result).
Images in which the CNN predicted it to be negative and no clinician scored it as positive (the CNN’s completely true-negative result).
Images in which the CNN predicted it to be negative but all of the clinicians scored it as positive (the CNN’s completely false-negative result).
Table 7.
Comparison between the convolutional neural network (CNN) predictive score and the clinicians’ feature for the fibrous crescent (F-Cre)
| No. of clinicians out of 5 who scored positive |
|||||||
|---|---|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | 4 | 5 | ||
| CNN’s predictive score over 4 folds | 0.75–1.00 | 9a | 4 | 2 | 7 | 6 | 0b |
| 0.50–0.75 | 11a | 5 | 1 | 2 | 1 | 0b | |
| 0.25–0.50 | 20c | 2 | 1 | 3 | 0 | 1d | |
| 0.00–0.25 | 17c | 4 | 1 | 0 | 1 | 0d | |
This confusion matrix shows the comparison between the CNN predictive score and the clinicians’ feature for the F-Cre. Rows show the CNN’s predictive score for an image, which was calculated as an average of the CNN’s softmax probability over the 4 folds. Columns show the number of clinicians out of 5 who scored it as positive. Each number shows the number of images.
Images in which the CNN predicted it to be positive, but no clinician scored it as positive (CNN’s completely false-positive result).
Images in which the CNN predicted it to be positive and all of the clinicians scored it as positive (the CNN’s completely true-positive result).
Images in which the CNN predicted it to be negative and no clinician scored it as positive (the CNN’s completely true-negative result).
Images in which the CNN predicted it to be negative, but all clinicians scored it as positive (the CNN’s completely false-negative result).
Visualization of the CNN’s Focusing Point
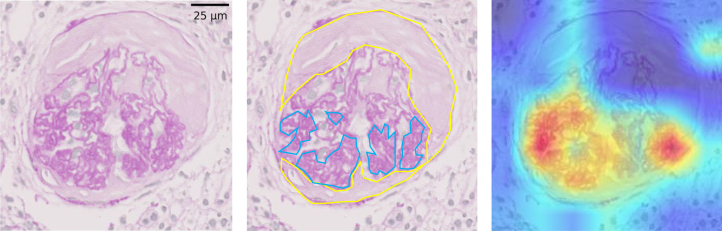
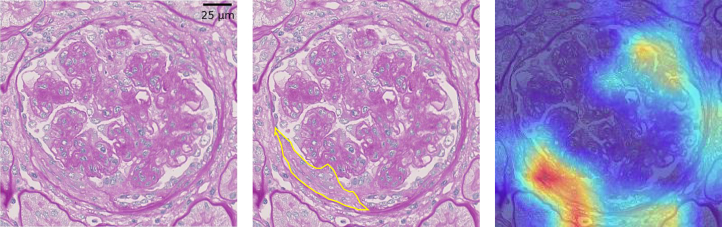
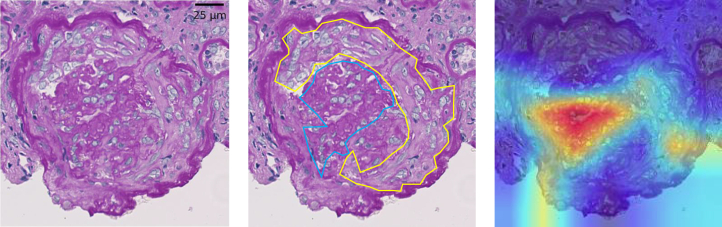
We show the 3 images of Grad-CAM to understand the properties of the CNN qualitatively. Figure 3 depicts a cTP image for CC where the CNN and all 5 clinicians scored it as positive. In this figure, the focus of the CNN was on the collapsed capillary, which is an actual pathological lesion. On the other hand, Figures 4 and 5 show the cFP images for the F-Cre where the CNN scored it as positive but none of the 5 clinicians scored it as positive. In Figure 4, the focus of the CNN was the extracapillary lesion, which is an actual pathological lesion of the Fc-Cre; however, the CNN misclassified it as F-Cre. In Figure 5, the focus of the CNN was not the extracapillary lesion but the collapsed capillary lesion.
Figure 3.
Example of the true positive score of the convolutional neural network (CNN) for the capillary collapse (CC). (Left) One of the periodic acid−Schiff (PAS)−stained images from the concordance data. The CNN diagnosed this image as “capillary collapse is positive,” and all 5 clinicians scored it as “capillary collapse is positive.” (Center) The blue lines depict the collapsed capillary, and the yellow lines show the fibrous crescents. (Right) Visualization of the CNN’s focusing points using the Gradient-weighted Class Activation Mapping when the CNN diagnosed the capillary collapse as positive or negative. Red lesions are the CNN’s focusing points. This shows that the CNN was focused correctly on the capillary area.
Figure 4.
Example of the false-positive case of the convolutional neural network (CNN) for the fibrous crescent (F-Cre). (Left) One of the periodic acid−Schiff (PAS)−stained images from the concordance data. The CNN diagnosed this image as “F-Cre is positive,” but none of the 5 clinicians scored it as “F-Cre is positive,” whereas 4 of the 5 clinicians scored it as “Fibrocellular Crescent (Fc-Cre) is positive.” (Center) The yellow line shows the area of Fc-Cre. (Right) Visualization of the CNN’s focusing points using the gradient-weighted class activation mapping when the CNN diagnosed the F-Cre as positive or negative. The red lesions are the CNN’s focusing points. This shows that the CNN focused correctly on the extracapillary area but “misdiagnosed” it as “F-Cre is positive.”
Figure 5.
Another example of the false-positive case of the convolutional neural network (CNN) for the fibrous crescent (F-Cre). (Left) One of the periodic acid−Schiff (PAS)−stained images from the concordance data. The CNN diagnosed this image as “F-Cre is positive” but none of the 5 clinicians scored it as “F-Cre is positive.” (Center) The blue line shows the area where the capillary was collapsed and obsoleted, and the yellow line depicts the fibrocellular crescent area. (Right) Visualization of the CNN’s focusing points using the gradient-weighted class activation mapping when the CNN diagnosed the F-Cre as positive or negative. The red lesions are the CNN’s focusing points. This reveals that the CNN accidentally focused on the capillary area although it needs to focus on the extracapillary lesion.
Discussion
Clinicians’ Concordance
As listed in Table 4, the κ coefficient ranged from 0.28 to 0.50, which indicates a fair-to-moderate concordance. The study about concordance in the Oxford classification for IgA nephropathy15 measured the concordance by using intraclass correlation coefficients (ICCs), and their ICCs ranged from 0.03 to 0.90. The same variety of ICCs were also reported in the Japanese cohort study using the Oxford classification for IgA nephropathy where the ICCs ranged from 0.26 to 0.89.25 Because the ICCs are analogous with the κ coefficient,26 the discordance was also reported in those studies.
As for the feature of MesHyper, the κ coefficient is 0.35, whereas the κ coefficient for the mesangial hypercellularity reported in the NEPTUNE scoring system16 was 0.54 to 0.64. Even though a direct comparison is difficult because of differences in the 2 study designs, the difference in the number of classes (5 for this study and 2 for NEPTUNE) might have led to the difference in the κ score.
One possible reason that may have caused discordance is that single-stained WSIs were used for scoring the images. In some cases, clinicians see the WSIs for several stains, and they try to assign a score for an image. Combining other stained WSIs, such as PAM or Azan, might improve the concordance between clinicians.
CNN Performance
As shown in Table 5, the ROC–AUC for the CNN for the test data exceeded 0.9 for the CC and the F-Cre. The 2 features may have demonstrated a high performance due to the positive-to-negative ratio (described in Table 2), and the width of the pathological lesions were not so narrow. In contrast, features such as IVVP with a low performance demonstrated a high ratio of negative to positive, and a small pathological lesion may have caused the low performance. To improve the accuracy, the data should be collected to improve the class balance with the resampling methods.
As displayed in Table 5, there is a difference in the performance between the concordance data and the test data for some features. This seems to be due to the sampling method. Specifically, the test data were selected to have a good representation of the whole dataset by the stratified folding method; however, the concordance data were intentionally selected to contain “hard-to-score” images.
Concordance Between the CNN and Clinicians
As shown in Tables 6 and 7, for the CC, there was no cFP case and only 1 cFN case out of 98 images. Meanwhile, for the F-Cre, there were 20 cFP cases and 1 cFN case. F-Cre has more cFP images compared with CC. The reason why there are so many cFP images for the F-Cre is discussed in the next section.
Visualization of CNN’s Focusing Point
A visualization of the cTP case for the CC is illustrated in Figure 3, and the cFP cases for F-Cre are presented in Figures 4 and 5. In Figure 4, the CNN cannot distinguish between the 2 different features, which are F-Cre and Fc-Cre. It is hypothesized that F-Cre and Fc-Cre have a similar crescentic structure but different textures. On the other hand, in Figure 5, the CNN cannot distinguish between CC and F-Cre, which tended to occur simultaneously in this dataset (as depicted in Table 3). This is because during the training process, the CNN merely focuses on the frequently occurring lesion, which is not always a true pathological lesion. This phenomenon has been reported as an inherent bias derived from the dataset.24 By correcting the balance of co-occurrence between the 2 different features, this may enable the CNN to focus on the appropriate pathological lesion and to make the correct classification.
Except for the CC and F-Cre, there are several other strong correlations between the features of the renal pathology in terms of pathogenesis. For example, in most cases of IgA nephropathy, endocapillary proliferation tends to accompany mesangial hypercellularity.27 To avoid the inherent bias caused by this correlation, it is suggested that instead of classifying the features directly, the glomerular image should be divided into different lesions according to its structures, such as the capillary lesion, the mesangial lesion, and the extracapillary lesion, and semantic segmentation28 can be applied to extract the lesions from the glomerular image.
In conclusion, this study established the annotation criteria and an annotation flowchart to automatically classify the glomerular images using CNNs. The annotation criteria included 12 features and the degree of concordance. This was indicated by the average κ coefficient among the 5 clinicians for each feature that ranged from 0.28 to 0.50. One clinician annotated 10,102 images and the performance of the CNN for the 5 features that were evaluated. The ROC–AUC values for the test data ranged from 0.65 to 0.98. In particular, CC and F-Cre demonstrated high ROC–AUC values of 0.98 and 0.91, respectively. The error analysis indicated that the CNN cannot distinguish features that have a similar visual structure or are likely to occur simultaneously. To address this problem, labeling every pixel with the structure class such as mesangial lesions or capillary lesions and applying semantic segmentation might improve the performance of the glomerular classification.
One of the limitations of this study is that the annotation criteria should be validated by other clinicians, and more images for concordance evaluation are desirable. Another limitation is that the images for the CNN development were annotated only by 1 clinician. Thus, there should be unintended bias in the dataset, and the results of the CNN performance might not be extrapolated to the other datasets.
This study has 3 important implications for future research on automatic glomerular classification. The first implication is that the discordance among the clinicians is similar to what has been reported in the literature. The second implication is that the width of the pathological lesion seems to affect the performance of the CNN. The third implication is that providing the predictive score and visualizing the focus of the CNN would support clinicians in diagnosing renal pathology, which would improve the quality of the clinicians’ assessment.
Disclosure
All the authors declared no competing interests.
Acknowledgments
This research was supported by the Health Labour Sciences Research Grants (grant number 28030401), Japan, and the Japan Science and Technology Agency, Promoting Individual Research to Nurture the Seeds of Future Innovation and Organizing Unique, Innovative Network (grant number JPMJPR1654), Japan. The Department of Artificial Intelligence in Healthcare, Graduate School of Medicine, the University of Tokyo is an endowment department that is supported with an unrestricted grant from I&H Co., Ltd. and EM SYSTEMS company; however, these sponsors had no control over the interpretation, writing, or publication of this work.
Author Contributions
RY and YK designed the experiments. RY, YS-D, HU, AS, and MN established the annotation criteria and flowchart and performed the concordance experiment. RY conducted the deep learning and analyzed the data. RY, YK, KS, ES, and KO wrote the article. TT, HN, HU, TU, MN, NK, AS, and MN collected the dataset. KO obtained the funding for this research.
Data Access
The dataset used in this study was collected by UTH, KH, and UTSH. The experiments and the data collection were approved by the institutional review board of each hospital (UTH:11455, KH:P17-05-004, UTSH:H29-160). The datasets are not available to the public, and restrictions apply to their use. The code is available at https://github.com/ryama-tokyo/glomerular_classification.
Translational Statement
When diagnosing kidney diseases, scoring every glomerulus is important but time-consuming; hence, scoring requires the support of a computer. This study demonstrates the performance of a convolutional neural network when classifying the glomerular features. This study visually demonstrated that there are cases where the convolutional neural network cannot distinguish 2 different features that had a similar visual structure, or they occurred simultaneously. This result suggests the need to divide the glomerular image into different lesions according to its structures for computers to perform a proper assessment.
Footnotes
Table S1. Annotation criteria for the 12 features that are important for diagnostics
Table S2. Breakdown list of the whole slide images (WSIs)
Figure S1. Annotation example images for the 12 features.
Figure S2. Images used for concordance evaluation.
Supplementary Material
Supplementary File (PDF)
Table S1. Annotation criteria for the 12 features that are important for diagnostics
Table S2. Breakdown list of the whole slide images (WSIs)
Figure S1. Annotation example images for the 12 features.
Figure S2. Images used for concordance evaluation.
References
- 1.Pfister M., Jakob S., Frey F.J. Judgment analysis in clinical nephrology. Am J Kidney Dis. 1999;34:569–575. doi: 10.1016/s0272-6386(99)70088-0. [DOI] [PubMed] [Google Scholar]
- 2.Kitterer D., Gürzing K., Segerer S. Diagnostic impact of percutaneous renal biopsy. Clin Nephrol. 2015;84:311–322. doi: 10.5414/CN108591. [DOI] [PubMed] [Google Scholar]
- 3.He L., Long L.R., Antani S. Histology image analysis for carcinoma detection and grading. Comput Methods Programs Biomed. 2012;107:538–556. doi: 10.1016/j.cmpb.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.LeCun Y., Bengio Y., Hinton G. Deep learning. Nature. 2015;521:436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 5.Fukushima K., Miyake S. Neocognitron: a new algorithm for pattern recognition tolerant of deformations and shifts in position. Pattern Recognit. 1982;15:455–469. [Google Scholar]
- 6.Saha M., Chakraborty C., Racoceanu D. Efficient deep learning model for mitosis detection using breast histopathology images. Comput Med Imaging Graph. 2018;64:29–40. doi: 10.1016/j.compmedimag.2017.12.001. [DOI] [PubMed] [Google Scholar]
- 7.Gallego J., Pedraza A., Lopez S. Glomerulus classification and detection based on convolutional neural networks. J Imaging. 2018;4:20. [Google Scholar]
- 8.Kawazoe Y., Shimamoto K., Yamaguchi R. Faster R-CNN-based glomerular detection in multistained human whole slide images. J Imaging. 2018;4:91. [Google Scholar]
- 9.Bukowy J.D., Dayton A., Cloutier D. Region-based convolutional neural nets for localization of glomeruli in trichrome-stained whole kidney sections. J Am Soc Nephrol. 2018;29:2081–2088. doi: 10.1681/ASN.2017111210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hermsen M., Bel T., den Boer M. Deep learning-based histopathologic assessment of kidney tissue. J Am Soc Nephrol. 2019;30:1968–1979. doi: 10.1681/ASN.2019020144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barros G.O., Navarro B., Duarte A. PathoSpotter-K: a computational tool for the automatic identification of glomerular lesions in histological images of kidneys. Sci Rep. 2017;7:1–8. doi: 10.1038/srep46769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kannan S., Morgan L.A., Liang B. Segmentation of glomeruli within trichrome images using deep learning. Kidney Int Reports. 2019;4:955–962. doi: 10.1016/j.ekir.2019.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marsh J.N., Matlock M.K., Kudose S. Deep learning global glomerulosclerosis in transplant kidney frozen sections. IEEE Trans Med Imaging. 2018;37:2718–2728. doi: 10.1109/TMI.2018.2851150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chagas P., Souza L., Araújo I. Classification of glomerular hypercellularity using convolutional features and support vector machine. Artif Intell Med. 2020;103:101808. doi: 10.1016/j.artmed.2020.101808. [DOI] [PubMed] [Google Scholar]
- 15.Roberts I.S.D., Cook H.T., Troyanov S. The Oxford classification of IgA nephropathy: pathology definitions, correlations, and reproducibility. Kidney Int. 2009;76:546–556. doi: 10.1038/ki.2009.168. [DOI] [PubMed] [Google Scholar]
- 16.Barisoni L., Troost J.P., Nast C. Reproducibility of the NEPTUNE descriptor-based scoring system on whole-slide images and histologic and ultrastructural digital images. Mod Pathol. 2016;29:671–684. doi: 10.1038/modpathol.2016.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goode A., Gilbert B., Harkes J. OpenSlide: a vendor-neutral software foundation for digital pathology. J Pathol Inform. 2013;4:27. doi: 10.4103/2153-3539.119005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sackman H. Delphi assessment: expert opinion, forecasting and group process. United States Air Force Proj RAND. 1974 R-1283-PR:130. [Google Scholar]
- 19.Krause J., Gulshan V., Rahimy E. Grader variability and the importance of reference standards for evaluating machine learning models for diabetic retinopathy. Ophthalmology. 2018;125:1264–1272. doi: 10.1016/j.ophtha.2018.01.034. [DOI] [PubMed] [Google Scholar]
- 20.Cohen J. Weighted kappa: nominal scale agreement provision for scaled disagreement or partial credit. Psychol Bull. 1968;70:213–220. doi: 10.1037/h0026256. [DOI] [PubMed] [Google Scholar]
- 21.Kawaguchi K., Bengio Y. Depth with nonlinearity creates no bad local minima in ResNets. Neural Networks. 2019;118:167–174. doi: 10.1016/j.neunet.2019.06.009. [DOI] [PubMed] [Google Scholar]
- 22.Pan S.J., Yang Q. A survey on transfer learning. IEEE Trans Knowl Data Eng. 2010;22:1345–1359. [Google Scholar]
- 23.Deng J., Dong W., Socher R. ImageNet: a large-scale hierarchical image database. In: 2009 IEEE Conference on Computer Vision and Pattern Recognition. 2009:248–255. https://ieeexplore.ieee.org/document/5206848 Available at: Accessed March 26, 2019. [Google Scholar]
- 24.Selvaraju R.R., Cogswell M., Das A. Grad-CAM: visual explanations from deep networks via gradient-based localization. 2016. https://arxiv.org/abs/1610.02391 Available at:
- 25.Hisano S., Joh K., Katafuchi R. Reproducibility for pathological prognostic parameters of the Oxford classification of IgA nephropathy: a Japanese cohort study of the Ministry of Health, Labor and Welfare. Clin Exp Nephrol. 2017;21:92–96. doi: 10.1007/s10157-016-1258-8. [DOI] [PubMed] [Google Scholar]
- 26.Landis J.R., Koch G.G. The measurement of observer agreement for categorical data. Biometrics. 1977;33:159. [PubMed] [Google Scholar]
- 27.Zhou X.J., Laszik Z.G., Nadasdy T., editors. Silva’s Diagnostic Renal Pathology. 2nd ed. Cambridge University Press; Cambridge: 2017. [Google Scholar]
- 28.Lateef F., Ruichek Y. Survey on semantic segmentation using deep learning techniques. Neurocomputing. 2019;338:321–348. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary File (PDF)