Abstract
Introduction
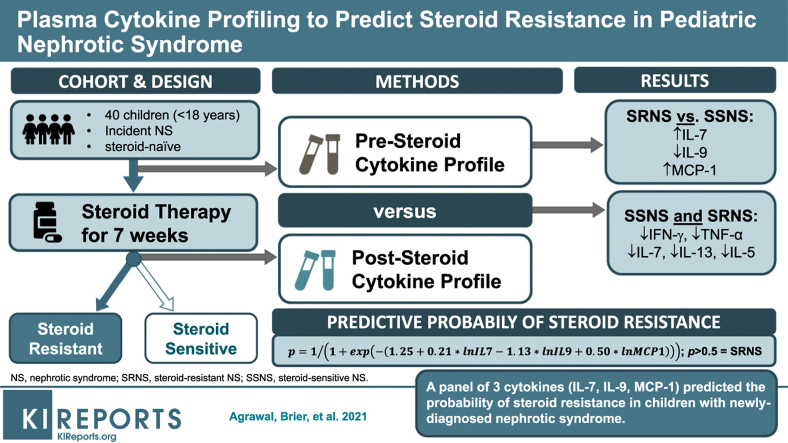
Glucocorticoids (GCs) are the primary treatment for nephrotic syndrome (NS), although ∼10% to 20% of children develop steroid-resistant NS (SRNS). Unfortunately, there are no validated biomarkers able to predict SRNS at initial disease presentation. We hypothesized that a plasma cytokine panel could predict SRNS at disease presentation, and identify potential pathways regulating SRNS pathogenesis.
Methods
Paired plasma samples were collected from 26 children with steroid-sensitive NS (SSNS) and 14 with SRNS at NS presentation and after ∼7 weeks of GC therapy, when SSNS versus SRNS was clinically determined. Plasma cytokine profiling was performed with a panel of 27 cytokines.
Results
We identified 13 cytokines significantly different in Pretreatment SSNS versus SRNS samples. Statistical modeling identified a cytokine panel (interleukin [IL]-7, IL-9, monocyte chemoattractant protein–1 [MCP-1]) able to discriminate between SSNS and SRNS at disease presentation (receiver operating characteristic [ROC] value = 0.846; sensitivity = 0.643; specificity = 0.846). Furthermore, GC treatment resulted in significant decreases in plasma interferon-γ (IFN-γ), tumor necrosis factor–α (TNF-α), IL-7, IL-13, and IL-5 in both SSNS and SRNS patients.
Conclusions
These studies suggest that initial GC treatment of NS reduces the plasma cytokines secreted by both CD4+ TH1 cells and TH2 cells, as well as CD8+ T cells. Importantly, a panel of 3 cytokines (IL-7, IL-9, and MCP-1) was able to predict SRNS prior to GC treatment at disease presentation. Although these findings will benefit from validation in a larger cohort, the ability to identify SRNS at disease presentation could greatly benefit patients by enabling both avoidance of unnecessary GC-induced toxicity and earlier transition to more effective alternative treatments.
Keywords: biomarkers, cytokines, glucocorticoids, Steroids, steroid-resistant nephrotic syndrome, steroid-sensitive nephrotic syndrome
Graphical abstract
Nephrotic syndrome (NS) is among the most common forms of kidney disease seen in children. Often referred to as the Shalhoub hypothesis, the role of T cells in idiopathic NS was first reported in 1974.1 Although both localized and systemic forms of immune-mediated mechanisms are well established and studied in immunologic forms of glomerular disease, such as glomerulonephritis and membranous nephropathy,2 reversible immune dysregulation is also a key feature of idiopathic NS, such as in minimal change disease and focal segmental glomerulosclerosis.3, 4, 5, 6, 7 Several clinical and experimental observations suggest that T-cell functional abnormalities and dysregulated cytokines are also features of idiopathic NS, which continues to be treated with GCs and other immunosuppressive medications.3,8,9 However, GCs have many side effects, and ∼50% of adults and ∼10% to 20% of children with NS present with or develop steroid resistance.10, 11, 12 Unfortunately, there are no validated biomarkers able to predict steroid resistance in NS, and the molecular mechanisms regulating GC resistance remain unclear. Thus, although most NS patients receive GC as initial therapy, a notable percentage will subsequently develop SRNS but will by then already have had toxic side effects from prolonged GC exposure, as well as potential disease progression while receiving an ineffective therapy.
To try to better identify those patients highly unlikely to respond to GC, and thus avoid its unnecessary toxicity, a few studies have tried to identify urinary, plasma, and salivary biomarkers for SRNS in children using cytokine profiling and omics approaches such as proteomics, metabolomics, and transcriptomics.8,13, 14, 15, 16, 17, 18, 19, 20 As a result, recently T lymphocytes expressing inflammatory cytokines and macrophage migration inhibitory factor in plasma, and MCP-1 in urine have been implicated in persistent proteinuria and SRNS in childhood NS.8,19,20 However, despite these efforts, we still lack understanding of the molecular mechanisms regulating GC resistance in NS, and there remain no plasma or urine biomarkers that have been validated to reliably predict GC resistance in NS prior to initiation of GC therapy.
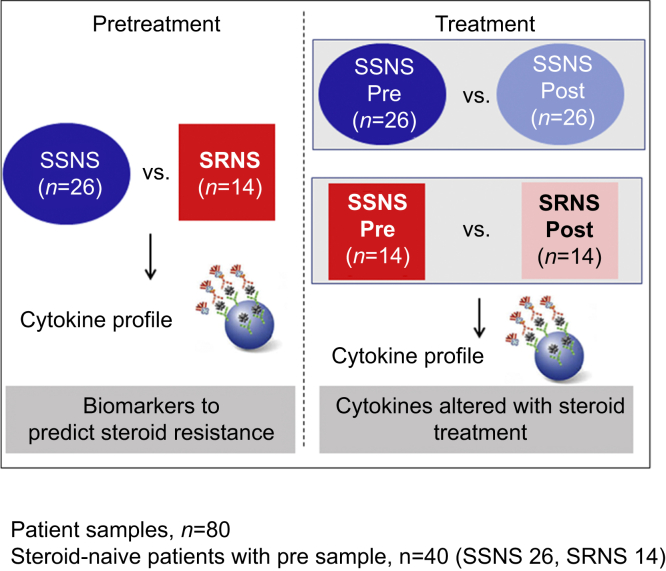
In this context, the present study was designed to test the hypothesis that a plasma cytokine panel could predict SRNS at clinical presentation and identify the cytokines altered with steroid treatment in childhood NS. To test this hypothesis, we quantified 27 cytokines (Table 1) on paired plasma samples obtained from children with NS at clinical presentation (prior to GC initiation) and again after ∼7 weeks of therapy when they were determined to have either SSNS or SRNS (Figure 1). Based on the prior literature, we specifically studied cytokines known to be released from CD4+ TH1 cells (e.g., IFN-γ), CD4+ TH2 cells (e.g., IL-4, IL-5, IL-10, and IL-13), TH17 cells (IL-17), and CD8+ cytotoxic T cells, as well as those produced prominently by macrophages, dendritic cells, and lymphoblasts (e.g., TNF-α, IL-12, and IL-15). We also studied chemotactic cytokines known to be released by a variety of immune cells (MCP-1/CCL2, MIP-1α/CCL3, and MIP-1β/CCL4) (Table 1),21 as well as growth factors such as platelet-derived growth factor, fibroblast growth factor, and vascular endothelial growth factor.
Table 1.
Description of cytokines analyzed in this study
| Cytokine | Description |
|---|---|
| IL-1β | Interleukin 1β |
| IL-1ra | Interleukin 1 receptor antagonist |
| IL-2 | Interleukin 2 |
| IL-4 | Interleukin 4 |
| IL-5 | Interleukin 5 |
| IL-6 | Interleukin 6 |
| IL-7 | Interleukin 7 |
| IL-8 | Interleukin 8 / CXCL8 (C-X-C motif chemokine ligand 8) |
| IL-9 | Interleukin 9 |
| IL-10 | Interleukin 10 |
| IL-12(p70) | Interleukin 12(p70) [heterodimer of IL-12A(p35) and IL12B(p40)] |
| IL-13 | Interleukin 13 |
| IL-15 | Interleukin 15 |
| IL-17A | Interleukin 17A |
| Eotaxin | CCL11 (C-C motif chemokine ligand 11) |
| FGF basic | Fibroblast growth factor basic / FGF2 |
| G-CSF | Granulocyte colony-stimulating factor / CSF3 (colony-stimulating factor 3) |
| GM-CSF | Granulocyte macrophage colony-stimulating factor / CSF2 (colony-stimulating factor 2) |
| IFN-γ | Interferon-γ |
| IP-10 | Interferon (IFN)-γ inducible protein / CXCL10 (C-X-C motif chemokine ligand 10) |
| MCP-1 (MCAF) | Monocyte chemoattractant protein–1 (monocyte chemotactic and activating factor) / CCL2 (C-C motif chemokine ligand 2) |
| MIP-1α | Macrophage inflammatory protein 1α / CCL3 (C-C motif chemokine ligand 3) |
| PDGF-BB | Platelet-derived growth factor B subunit homodimer |
| MIP-1β | Macrophage inflammatory protein 1β / CCL4 (C-C motif chemokine ligand 4) |
| RANTES | Regulated upon activation, normal T cell expressed and presumably secreted / CCL5 (C-C motif chemokine ligand 5) |
| TNF-α | Tumor necrosis factor α |
| VEGF | Vascular endothelial growth factor |
Figure 1.
Hypothesis and study model. Cytokine profile analysis of paired (before [Pre] and after [Post] ∼7 weeks of GC therapy) steroid-sensitive nephrotic syndrome (SSNS) and steroid-resistant nephrotic syndrome (SRNS) plasma samples can be used to identify novel predictive biomarkers and molecular pathways of steroid resistance.
Materials and Methods
Study Approval and Ethics Statement
All research protocols and consent documents were approved by the Institutional Review Board of Nationwide Children's Hospital as the coordinating center (approval numbers IRB07-00400, IRB12-00039, and IRB05-00544), as well as by each of the other participating centers of the PNRC. Informed written consent (and assent, where appropriate) was obtained from the parents of all participants before samples were collected, in accordance with the Declaration of Helsinki.
Pediatric Nephrotic Syndrome Patients and Plasma Collection
Pediatric NS patients aged between 18 months and 18 years were included in this study if they exhibited 3+ proteinuria and edema. The clinical response of each patient to GC (i.e., SRNS or SSNS) was assessed ∼7 weeks after initial presentation as is also shown in Table 2. Paired plasma samples were collected from 26 SSNS and 14 SRNS patients, obtained both at initial disease presentation and after ∼7 weeks of GC therapy, when SSNS versus SRNS was clinically determined. All the pretreatment samples were collected before even a single dose of steroids and were termed as steroid naïve. A total of 80 samples were used in this study from 14 SRNS and 26 SSNS patients. SSNS was defined as disease remission after an average of 7 weeks of GC therapy, and SRNS was defined as a failure to enter complete remission in response to GC treatment as measured by proteinuria or dipstick reading.
Table 2.
Patient demographics for the SSNS and SRNS cohorts in this study
| Total (n = 40) | SSNS (n = 26) | SRNS (n = 14) | P value | |
|---|---|---|---|---|
| Weeks between pre- and post-treatment, mean ± SD | 7.0 ± 0.4 | 6.9 ± 0.5 | 7.0 ± 0.6 | ns |
| Steroid-naïve disease onset patients, n (%) | 40 (100) | 26 (100) | 14 (100) | |
| Age, mean ± SD | 7.0 ± 0.4 (n = 38) | 5.7 ± 0.7 (n = 24) | 9.5 ± 1.0 (n = 14) | 0.0041a |
| Sex, n (%) | ||||
| Female | 22 (55) | 13 (50) | 9 (64) | |
| Male | 17 (42) | 12 (46) | 5 (36) | |
| Not reported | 1 | 1 | ||
| Height, mean ± SD | 124.2 ± 4.1 (n = 39) | 114.5 ± 4.2 (n = 25) | 141.4 ± 6.5 (n = 14) | 0.0010a |
| Weight, mean ± SD | 35.2 ± 3.6 (n = 39) | 26.2 ± 2.4 (n = 25) | 51.4 ± 7.7 (n = 14) | 0.0003a |
| Race, n (%) | ns | 0.0011b | ||
| White | 17 (42.5) | 11 (42.3) | 6 (42.8) | |
| Asian | 4 (10) | 4 (15.4) | 0 (0) | |
| African American | 12 (30) | 6 (23) | 6 (42.8) | |
| Biracial | 2 (5) | 1 (3.8) | 1 (7) | |
| Native American | 1 (2.5) | 1 (3.8) | 0 (0) | |
| Not reported | 4 (10) | 3 (11.5) | 1 (7) |
SRNS, steroid-resistant nephrotic syndrome; SSNS, steroid-sensitive nephrotic syndrome.
Significance determined by t test.
Significance determined by χ2 test.
Cytokine Profiling Assay
The cytokine profiling was performed using a 27-cytokine panel on a Luminex Technology platform using a bead-based fluorescence assay. The Bio-Plex Pro Human Cytokine 27-Plex Immunoassay (Bio-Rad, Hercules, CA) panel consisted of 27 major cytokines, including IL-1β, IL-1ra, IL-2, IL-4, IL-5, IL-6, IL-7, IL-8, IL-9, IL-10, IL-12(p70), IL-13, IL-15, IL-17A, eotaxin, fibroblast growth factor basic, granulocyte colony–stimulating factor (G-CSF), granulocyte macrophage colony-stimulating factor, IFN-γ, IP-10, MCP-1 (monocyte chemotactic and activating factor), MIP-1α, platelet-derived growth factor B subunit homodimer (PDGF-BB), MIP-1β, RANTES (regulated upon activation, normal T cell expressed and presumably secreted), TNF-α, and vascular endothelial growth factor. Similar methodology using cytokine multiplex bead array assay has recently been successfully used to measure circulating cytokines in patients with albuminuric and nonalbuminuric diabetic kidney disease.22 Corresponding human recombinant proteins provided with the kits were used to generate standard curves, and the values in patient plasma samples used in duplicates were measured and interpolated from the standard curves. Plasma samples were prepared by drawing 8 ml of peripheral blood into a CPT tube that contained 1 mL of 100-mM sodium citrate solution, polyester gel, polysaccharide/sodium diatrizoate solution (FICOLL Hypaque solution), and a silicone coating (Becton & Dickenson, Franklin Lakes, NJ). These were processed immediately to isolate plasma or shipped overnight at ambient temperature before processing. The isolated plasma was maintained in NCH Biopathology core biorepository at –80 °C until use.
Statistical Analyses
To test the hypothesis (Figure 1) that cytokine concentrations can be used to predict GC resistance, both univariate and multivariate analyses were performed. All statistical analyses were performed in SPSS, version 26 (International Business Machine Corp). The cytokines that fell below the lower limit of detection in the assay were recoded as one-half the value of that lower limit.23 The univariate analyses consisted of t tests, determining the ROC curve for the prediction of SRNS and binary logistic regression. Values of the ROC curve >0.5 were predictive of GC resistance, whereas values <0.5 were predictive of steroid sensitivity.
The multivariate analyses of the cytokine panel for the prediction of SRNS were performed by binary logistic regression using backward conditional elimination of the individual cytokine values. Regression using the complete cytokine panel failed to converge. A selection process was thus employed where only those cytokines that had a model summary statistic (–2 log likelihood) <50.0 were used. This resulted in 13 cytokines for analysis. Default logistic regression options were chosen, including a probability of 0.05 for stepwise entry and 0.10 for stepwise removal of a cytokine during model development.
To identify the altered plasma cytokine levels between SRNS and SSNS patients with GC treatment, analysis of variance with repeated measures was performed (Figure 1).
Results
Patients
Forty children with initial presentation of NS that had received no GC prior to the first sample collection (pretreatment) were included in the cytokine profile analyses, and detailed demographics were obtained from all patients (Table 2). Of these, paired samples were used from 26 SSNS and 14 SRNS patients. Patients clinically phenotyped as SSNS had entered complete remission of proteinuria within an average of ∼7 weeks of GC therapy, whereas patients who did not achieve remission during this time frame were phenotyped as SRNS. Because all patients were newly diagnosed with NS, no additional steroid sparing immunosuppressive therapies were administered to these pediatric patients between collection of the pre- and post-treatment samples. As has been reported previously, children with SRNS presented at a later age than from those with SSNS (9.5 vs. 5.7 years; P = 0.004).14,17,24 As expected, this difference in age was also reflected in significant differences in height and weight between the 2 cohorts (P < 0.001, see Table 2). Moreover, although the SSNS cohort comprised 23% African Americans and 42% Caucasians, the SRNS cohort comprised 43% of each, thus reflecting a significantly greater incidence of SRNS in African American children (P = 0.001).
Cytokines Predictive of Steroid Resistance in Pretreatment Samples
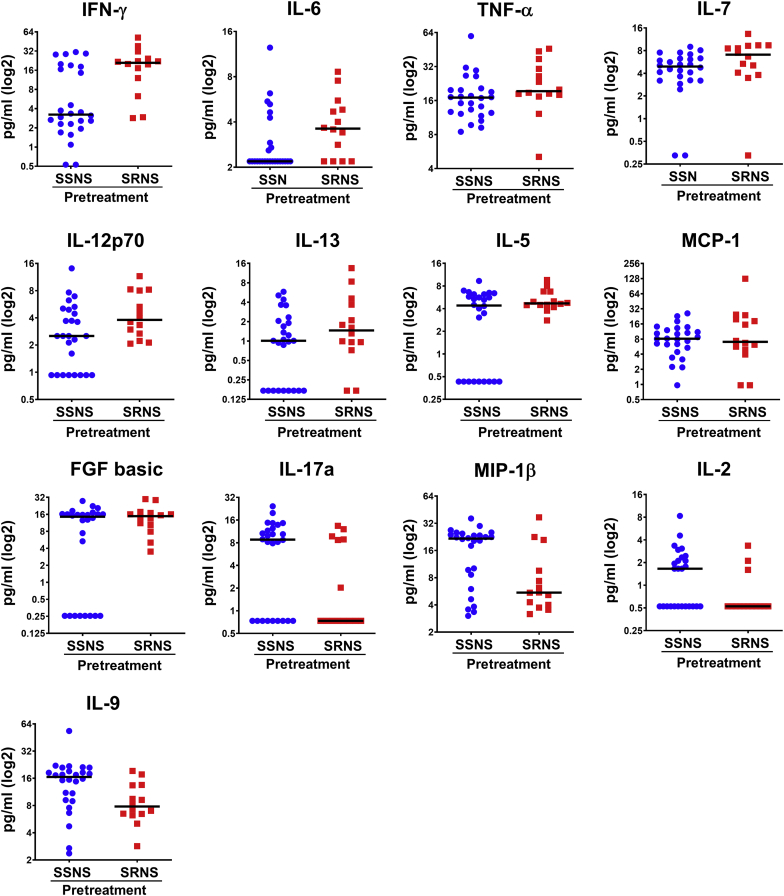
In order to test the hypothesis (Figure 1) that cytokine concentrations can be used to predict GC resistance, both univariate and multivariate analyses were performed between SSNS and SRNS on pretreatment samples. The results of the univariate analyses for the prediction of SRNS are shown in Table 3. ROC values ranged from 0.792 to 0.250. In addition, levels of INF-γ, MIP-1α, IL-6, IL-1b, MIP-1β, IL-2, and IL-9 all exhibited ROC values significantly different from 0.5 (P < 0.05) (Table 3, column I). Univariate logistic regression further identified IFN-γ, IL-7, MIP-1β, and IL-9 as cytokines that were statistically significant (Table 3, column II). These results are also somewhat consistent with the ROC analyses. Cytokine concentrations were also compared between SRNS and SSNS following log2 transformation (Table 3, column III), which resulted in statistically significant differences for INF-γ, IL-6, IL-12, IL-5, fibroblast growth factor basic, MIP-1β, IL-2, and IL-9. A total of 27 cytokines were explored for their ability to predict SRNS. However, because a significant proportion of IL-15 (38/40) and granulocyte macrophage colony-stimulating factor (36/40) results were below the lower limit of detection, these were not included in the final analysis, thus restricting our analyses to 25 cytokines.
Table 3.
Summary of univariate AUC and logistic regression analyses for predicting SRNS (pretreatment) and comparison of SRNS versus SSNS (pre- vs. post-treatment)
| Cytokine | I. |
II |
III |
IV |
|
|---|---|---|---|---|---|
| Univariate predicting SRNS |
Univariate logistic regression, P value |
t test pre-SRNS versus SSNS, P value |
–2 log likelihood | ||
| ROC | P value | ||||
| IFN-γ | 0.792 | 0.004 | 0.013 | 0.003 | 44.0 |
| MIP-1α | 0.774 | 0.007 | 0.831 | 0.095 | 51.7 |
| IL-6 | 0.740 | 0.017 | 0.110 | 0.039 | 49.5 |
| IL-1b | 0.723 | 0.027 | 0.591 | 0.081 | 51.5 |
| TNF-α | 0.679 | 0.075 | 0.158 | 0.160 | 49.6 |
| IL-7 | 0.673 | 0.086 | 0.046 | 0.287 | 47.0 |
| IL-12p70 | 0.662 | 0.108 | 0.151 | 0.040 | 49.6 |
| IL-8 | 0.638 | 0.171 | 0.688 | 0.860 | 51.6 |
| VEGF | 0.638 | 0.171 | 0.998 | 0.164 | 51.8 |
| IL-13 | 0.606 | 0.294 | 0.153 | 0.154 | 49.3 |
| IL-5 | 0.569 | 0.494 | 0.083 | 0.022 | 48.3 |
| G-CSF | 0.561 | 0.546 | 0.673 | 0.222 | 51.6 |
| MCP-1 | 0.551 | 0.611 | 0.311 | 0.641 | 49.4 |
| FGF basic | 0.545 | 0.656 | 0.179 | 0.047 | 49.9 |
| IP-10 | 0.538 | 0.703 | 0.730 | 0.457 | 51.8 |
| IL-10 | 0.524 | 0.811 | 0.944 | 0.629 | 51.8 |
| Eotaxin | 0.452 | 0.633 | 0.284 | 0.322 | 50.5 |
| IL-4 | 0.426 | 0.464 | 0.463 | 0.745 | 51.2 |
| PDGF-BB | 0.385 | 0.252 | 0.297 | 0.174 | 50.2 |
| IL-17A | 0.324 | 0.080 | 0.065 | 0.084 | 47.8 |
| RANTES | 0.314 | 0.065 | 0.181 | 0.854 | 50.4 |
| IL-1ra | 0.309 | 0.058 | 0.370 | 0.144 | 50.9 |
| MIP-1β | 0.269 | 0.022 | 0.027 | 0.012 | 46.0 |
| IL-2 | 0.260 | 0.017 | 0.098 | 0.048 | 47.7 |
| IL-9 | 0.250 | 0.013 | 0.021 | 0.037 | 44.7 |
AUC, area under the ROC curve; ROC, receiver operating characteristic; SRNS, steroid-resistant nephrotic syndrome; SSNS, steroid-sensitive nephrotic syndrome.
The 13 cytokines selected for multivariate analysis are shown in bold. For descriptions of the cytokines, refer to Table 1.
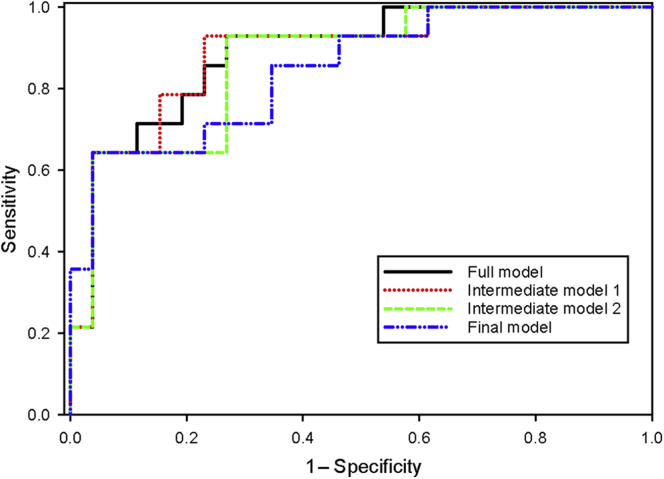
Furthermore, 13 cytokines (Table 3, column IV, and Figure 2) were combined into a panel to predict SRNS using logistic regression with backwards elimination and are shown in Table 4 and Figure 3. Only those cytokines that were found to have some ability to discriminate between SRNS and SSNS using the results shown in Table 3 (column IV) were analyzed. This resulted in a selection of 13 cytokines as the starting point for the backward elimination logistic regression. The results of the progression of the modeling are shown in 4 steps, where changes in the ROC values occurred. ROC values ranged between 0.887 and 0.846 for the full and reduced models, respectively. Sensitivity ranged from 0.714 to 0.643, and specificity ranged from 0.885 to 0.846. The final, parsimonious, statistically robust model consisted of 3 cytokines, IL-7, IL-9, and MCP-1, which yielded an ROC value of 0.846, with sensitivity of 0.643 and specificity of 0.846. The equation for this model was as follows, where lnIL7, lnIL9, and lnMCP1 are the log base 2 values for those cytokines:
Figure 2.
Cytokines predictive of steroid resistance in pretreatment samples. Cytokines found to be significantly different in pretreatment plasma samples between SSNS versus SRNS are shown. These 13 cytokines were identified using univariate analysis and t tests between the pretreatment samples of 26 children with SSNS and 14 children with SRNS, as depicted in Table 3. FGF, fibroblast growth factor; IFN-γ, interferon-γ; IL, interleukin; MCP-1, monocyte chemoattractant protein–1; MIP-1β, macrophage inflammatory protein 1β; SRNS, steroid-resistant nephrotic syndrome; SSNS, steroid-sensitive nephrotic syndrome; TNF-α, tumor necrosis factor α.
Table 4.
Sequential multivariate panels of cytokines to optimize prediction of SRNS
| Cytokine panel | ROC | P value | Sensitivity | Specificity |
|---|---|---|---|---|
| Full model | ||||
| IL-2, IL-5, IL-6, IL-7, IL-9, IL-12, IL-13, IL-17A, FGF basic, IFN-γ, MCP-1, MIP-1β, TNF-α | 0.887 (0.783–0.992) | <0.001 | 0.714 | 0.885 |
| Intermediate model 1 | ↓ | |||
| IL-2, IL-7, IL-9, IL-12, IL-13, IL-17A, FGF basic, MCP-1 | 0.885 (0.775–0.994) | <0.001 | 0.643 | 0.923 |
| Intermediate model 2 | ↓ | |||
| IL-7, IL-9, IL-12, IL-13, FGF basic, MCP-1 | 0.865 (0.750–0.981) | <0.001 | 0.643 | 0.923 |
| Final model | ↓ | |||
| IL-7, IL-9, MCP-1 | 0.846 (0.720–0.972) | <0.001 | 0.643 | 0.846 |
ROC, receiver operating characteristic; SRNS, steroid-resistant nephrotic syndrome.
A final statistically robust panel of 3 cytokines is bolded. For descriptions of the cytokines, refer to Table 1.
Figure 3.
ROC curves for sequential models of cytokines to optimize prediction of SRNS. Changes in the ROC values with the progression of the modeling starting with 13 cytokines are shown at 4 steps. ROC values ranged between 0.887 and 0.846 for the full and final models, respectively. Sensitivity ranged from 0.714 to 0.643, and specificity ranged from 0.885 to 0.846. The final, parsimonious, statistically robust model consisted of 3 cytokines, IL-7, IL-9, and MCP-1, which yielded an ROC value of 0.846, with a sensitivity of 0.643 and a specificity of 0.846. IL, interleukin; MCP-1, monocyte chemoattractant protein–1; ROC, receiver operating characteristic; SRNS, steroid-resistant nephrotic syndrome.
There would thus be different combinations of cytokine concentrations that could result in a P >0.5 and we might conclude that the patient has SRNS.
Identification of Cytokines Different Between Pre- and Post-treatment Samples in SSNS versus SRNS
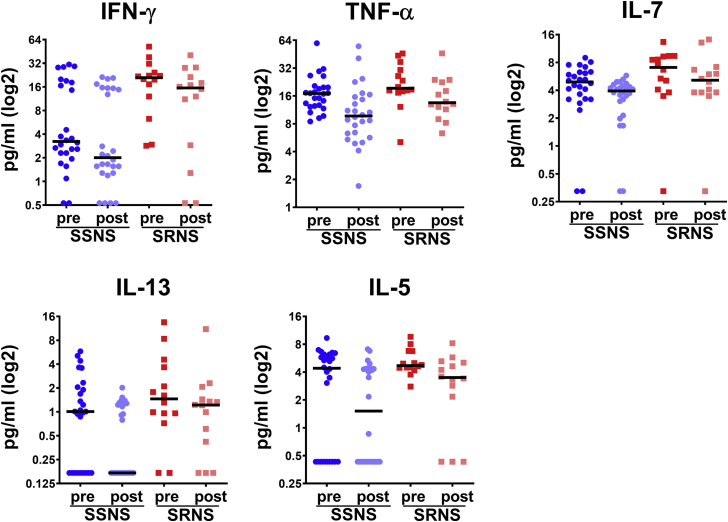
To identify the altered plasma cytokine levels between SRNS and SSNS patients with GC treatment, analysis of variance with repeated measures was performed, with the results shown in Table 5. GC treatment resulted in statistically significantly decreased concentrations of IFN-γ, TNF-α, IL-7, IL-13, and IL-5 (Figure 4). However, there were no statistically significant interactions between treatment (pretreatment, post-treatment) and group (SRNS, SSNS). Furthermore, Table 5 also shows the results of a comparison of cytokine concentrations between SSNS and SRNS patients, both in the pre- and post-treatment samples. The concentrations of INF-γ, IL-5, IL-7, IL-17A, and MIP-1β were all significantly higher in patients with SRNS than in those with SSNS, in both the pre- and post-treatment samples, as evidenced by a lack of statistical significance in the interaction term shown in Table 5.
Table 5.
Repeated measures analysis of group (SSNS vs. SRNS) and treatment (pre- vs. post-treatment with glucocorticoid)
| Steroid treatment (pre vs. post) | Group (SSNS vs. SRNS) | Interaction | |
|---|---|---|---|
| IFN-γ | 0.006 | 0.004 | 0.267 |
| MIP-1α | 0.292 | 0.742 | 0.994 |
| IL-6 | 0.274 | 0.072 | 0.687 |
| IL-1β | 0.743 | 0.880 | 0.694 |
| TNF-α | <0.001 | 0.227 | 0.584 |
| IL-7 | 0.012 | 0.006 | 0.609 |
| IL-12 | 0.410 | 0.673 | 0.156 |
| IL-8 | 0.648 | 0.562 | 0.489 |
| VEGF | 0.830 | 0.991 | 0.736 |
| IL-13 | 0.001 | 0.074 | 0.612 |
| IL-5 | 0.003 | 0.048 | 0.616 |
| G-CSF | 0.469 | 0.810 | 0.409 |
| MCP-1 | 0.058 | 0.124 | 0.213 |
| FGF basic | 0.056 | 0.418 | 0.426 |
| IP-10 | 0.063 | 0.914 | 0.627 |
| IL-10 | 0.654 | 0.604 | 0.313 |
| Eotaxin | 0.141 | 0.280 | 0.836 |
| IL-4 | 0.465 | 0.357 | 0.868 |
| PDGF-BB | 0.755 | 0.222 | 0.617 |
| IL-17A | 0.191 | 0.044 | 0.679 |
| RANTES | 0.551 | 0.206 | 0.441 |
| IL-1ra | 0.529 | 0.268 | 0.727 |
| MIP-1β | 0.651 | 0.020 | 0.760 |
| IL-2 | 0.398 | 0.138 | 0.871 |
| IL-9 | 0.776 | 0.101 | 0.730 |
SRNS, steroid-resistant nephrotic syndrome; SSNS, steroid-sensitive nephrotic syndrome.
Cytokines that were significantly altered with glucocorticoid treatment are bolded. For descriptions of the cytokines, refer to Table 1.
Figure 4.
Identification of cytokines different between pre- versus post-treatment samples in SSNS versus SRNS. Glucocorticoid treatment resulted in statistically significantly decreased levels of IFN-γ, TNF-α, IL-7, IL-13, and IL-5 among children with both SSNS and SRNS. The plasma levels of these cytokines are shown for 26 children with SSNS (pre- and post-treatment) and 14 children with SRNS (pre- and post-treatment). Notably, the relative differences in pre- and post-treatment levels were statistically similar between those children with SSNS and SRNS as depicted in Table 5. Furthermore, levels of IFN-γ, IL-7, IL-13, and IL-5 were different in both Pre and post-treatment samples in SSNS versus SRNS (Table 5). IFN-γ, interferon-γ; IL, interleukin; SRNS, steroid-resistant nephrotic syndrome; SSNS, steroid-sensitive nephrotic syndrome; TNF-α, tumor necrosis factor α.
Discussion
NS is among the most common forms of kidney disease seen in children, and reversible immune dysregulation is a key feature of idiopathic NS.1,4, 5, 6, 7 Although GCs remain the primary treatment for NS, ∼50% of adults and ∼10% to 20% of children with NS present with or develop SRNS, which results in prolonged exposure to a drug with significant toxicity and little or no clinical benefit.10, 11, 12 In the present study, we performed plasma cytokine profiling using a 27-cytokine panel in 40 children with new-onset NS (26 SSNS and 14 SRNS) using their pre- and post-treatment plasma samples to identify biomarkers able to predict SRNS at the time of initial disease presentation. We identified 13 cytokines in pretreatment samples that were further analyzed for their ability to predict SRNS. Modeling using backward elimination logistic regression analysis identified a panel of 3 cytokines (IL-7, IL-9, and MCP-1) that was able to discriminate children with SRNS versus SSNS at the time of initial disease presentation (prior to initiation of GC treatment) with an ROC value of 0.846, sensitivity of 0.643, and specificity of 0.846. Although these findings will benefit from validation in a larger cohort, the ability to identify children with SRNS at disease presentation could be highly beneficial to patients, as it would obviate the need to expose them to ineffective but toxic GC therapy and also enable earlier transition to more effective alternative treatments.
The present study is in alignment with recent efforts to identify and develop predictive biomarkers for SRNS and to understand the molecular mechanisms underlying steroid resistance, we hope, to guide both diagnostic and therapeutic decisions related to NS. Although we utilized statistical analyses of the cytokine profiles generated from plasma samples of children with SSNS and SRNS to answer these questions in the present study, we and others have also made other efforts toward the same goal using proteomic, metabolomic, and transcriptomic approaches to identify additional interesting candidates that are either predictive of SRNS or molecularly associated with SRNS.14, 15, 16, 17, 18 Over the past few years, serum concentrations of a few cytokines have been associated with relapse in NS. For example, elevated serum levels of IL-8 have been associated with NS relapse, and anti-IL8 antibodies have been shown to neutralize the ability of peripheral blood mononuclear cell culture supernatant to induce albuminuria in rats.25,26 Moreover, elevated levels of IL-1β, IL-6, and IL-8 have been reported in the urine samples of 37 children with idiopathic NS during relapse versus remission or healthy controls.27 Another study of 150 children with idiopathic NS and 569 healthy controls reported significant associations of IL-4, IL-6, and TNF-α polymorphisms between children with NS versus controls, and between SRNS versus SSNS, suggesting that steroid responsiveness may be determined by these polymorphisms.28 A few other reports have identified cytokines in plasma, urine, and saliva as markers of persistence of proteinuria or SRNS in childhood NS.13 A Brazilian study compared peripheral blood leukocytes and their intracellular cytokines in post-therapy samples from 44 pediatric patients, classified as having persistent proteinuria (partial remission) versus low proteinuria (complete remission). These investigators identified higher levels of inflammatory markers with persistent proteinuria. Moreover, another study reported increased expression of TNF-α in CD4 lymphocytes, as well as reduced expression of IFN-γ in CD8 lymphocytes, in patients with NS versus healthy controls, regardless of the level of proteinuria.8 In addition, a recent Italian study identified macrophage migration inhibitory factor as a good plasma predictor of steroid resistance in 21 children with NS (7 with SRNS, 7 with SSNS, and 7 with steroid-dependent NS) in pretreatment samples using 21-plex and 29-plex panels to analyze 48 cytokines, and this finding was extrapolated in 41 additional patients.20 Furthermore, a recent Japanese investigation studied cytokines in plasma and urine samples from 18 patients with NS, at onset and after therapy, and identified MCP-1 to be elevated in SRNS after therapy.19 These recent studies and our own findings suggest that although commercially available, clinically relevant cytokine biomarkers are not yet available, such approaches do have significant potential to be able to predict SRNS in childhood NS across varied demographics, despite the need for confirmatory validation studies in larger cohorts.
The combination of IL-7, IL-9, and MCP-1 in our binary logistic regression model allowed for the prediction of SRNS with an ROC of 0.846. This is similar to that obtained with a urine proteomics approach using a panel of 5 or 10 biomarkers (ROC = 0.85–0.92) in childhood NS.15 With regard to the individual cytokines comprising this panel, IL-7 is a pleiotropic cytokine secreted by stromal cells in the bone marrow and thymus and has central roles in modulating T- and B-cell development and T-cell homeostasis.29 In a paired sample study of 18 children with idiopathic NS, serum IL-7 levels were found to be elevated both before and after GC treatment, as well as in the absence of treatment versus healthy controls.30 IL-9 is produced mainly by TH9, CD4+ T cells, in addition to a variety of other cells such as mast cells, natural killer cells, TH2, TH17, and TH9 cells in different amounts. A recent study reported that IL-9 knockout resulted in increased and accelerated proteinuria and glomerulosclerosis and deteriorated kidney function in an adriamycin-induced nephropathy model in mice.31,32 Moreover, IL-9 treatment protected wild-type mice from glomerulosclerosis and kidney failure in the same model of nephropathy. These findings suggested that IL-9 is protective in experimental glomerulosclerosis, and thus it could be a potential therapeutic pathway for the treatment of chronic kidney disease. In accordance with these findings, our studies showed that IL-9 plasma levels were elevated in SSNS compared to SRNS patients. MCP-1 or CCl2 belongs to a family of chemoattractant cytokines that are produced in response to proinflammatory cytokines, and plays a major role in selectively recruiting monocytes, neutrophils, and lymphocytes, as well as in inducing chemotaxis through the activation of G-protein–coupled receptors.33 MCP-1 is a key chemokine regulating migration and infiltration of monocytes and macrophages and is known to be induced and involved in various diseases.33 A recent study investigated the effect of the MCP-1 2518 A/G polymorphism on the incidence and clinical course of biopsy-proven focal segmental glomerulosclerosis in children and found that the AA genotype might be a risk factor for disease progression.34 Moreover, these authors found that urinary levels of MCP-1 were significantly higher in focal segmental glomerulosclerosis versus SSNS and healthy controls, although serum levels of MCP-1 were comparable between these groups.34 Another study also similarly reported elevated urinary levels of MCP-1 in SRNS after therapy.19
A strength of this study, in comparison to prior studies of cytokines in NS, is that it used a very carefully phenotyped cohort of children collected across a diverse area in North America, with all children documented to have had no GC exposure prior to the collection of all pretreatment samples. In addition, multiple statistical methodologies and modeling approaches were used to arrive at a predictive biomarker panel of 3 cytokines that individually would not have been able to discriminate between the 2 groups. However, because this study was limited to a cohort of 26 SSNS and 14 SRNS patients, future validation studies with a larger cohort will certainly be needed to validate the ability of the identified 3-cytokine panel to predict SRNS at disease presentation. Another potential limitation could be that our findings of both age and race associations among our patients with SSNS versus SRNS implies some selection bias in our cohort. Nonetheless, it is well known that SRNS typically presents at older ages, associated with higher average heights and weights, compared to children presenting with SSNS.14,17,24 In addition, our finding of an increased incidence of SRNS among African Americans is consistent with prior published literature on this disease.35 These findings, in conjunction with the fact that this study included children recruited over several years through the PNRC at multiple sites across North America, suggest instead that our sample was likely representative of the general population of children with NS in North America. Of note, one school of thought suggests that the release of cytokines is influenced by age in children, and that this needs to be taken into consideration for cytokine-based diagnostic assays.36 However, in meta-analysis studies this age-association has been shown consistently primarily only for increasing IFN-γ and TNF-α levels, and decreasing IL-13 levels, with age.37 Although we observed decreases in these cytokines following GC treatment in both SSNS and SRNS patients, and higher IFN-γ, TNF-α, and IL-13 levels prior to GC treatment in both SSNS and SRNS patients (see Figures 2 and 4), none of these were identified as a member of our 3-cytokine panel predictive of SRNS. Thus, we elected not to perform age-related corrections in our statistical analyses. Lastly, although we now understand that up to one-third of children with SRNS may have a monogenic cause of disease, when these studies were initiated there was neither institutional review board approval nor resources available to perform genetic screening of the enrolled patients. Despite this limitation, however, these studies were still able to identify a 3-cytokine biomarker panel able to predict SRNS at disease presentation regardless of any underlying genetic mutations, which is likely to be clinically more useful at the time of initial NS presentation. In contrast to these limitations, the major strength of this study was the evaluation of carefully phenotyped patients from whom paired plasma samples were obtained, both before any GC treatment (i.e., completely steroid naïve) and after several weeks of GC treatment when they were clinically determined to have SRNS or SSNS. Indeed, our requirement to only include steroid-naïve patient samples as pretreatment samples in our analysis resulted in the exclusion of 50% of the patients enrolled in this study. This level of stringency in excluding any possible bias due to early GC exposure for the pretreatment samples ensured that all pretreatment samples reflected only the disease state at presentation, whereas the post-treatment samples reflected the cumulative effects to GC treatment on disease.
In summary, our studies suggest that a small panel of 2 interleukins (IL-7 and IL-9) and a chemokine (MCP-1) can discriminate between children with SRNS versus SSNS at initial disease presentation, and thus predict SRNS prior to the initiation of GC therapy (ROC value = 0.846). Moreover, we identified significant decreases in the plasma levels of the cytokines IFN-γ, TNF-α, IL-5, IL-7, and IL-13 in response to GC treatment in both patients with SSNS and those with SRNS. These findings thus suggest that GC treatment effectively reduces the plasma levels of cytokines secreted by CD4+ TH1 cells, TH2 cells, as well as CD8+ cells in children with new-onset NS. This study lays a strong foundation for a future validation study with a larger cohort to confirm the ability of the identified 3-cytokine biomarker panel to predict SRNS at initial disease presentation, and thus benefit patients by enabling both the avoidance of unnecessary GC-induced toxicity and earlier transition to more effective alternative treatments.
Acknowledgments
We thank the Pediatric Nephrology Research Consortium (PNRC; formerly the Midwest Nephrology Consortium [MWPNC]), its participating centers, physicians, and study and nurse coordinators for their contributions toward the collection of the plasma samples used in this study. These include Drs. John Mahan, Hiren Patel and Richard F. Ransom (NCH, Columbus, OH, USA), Cynthia Pan (Medical College of Wisconsin, Milwaukee, WI, USA), Denis F. Geary (The Hospital for Sick Children, Toronto, ON, Canada), Myra L . Chang (West Virginia University, Charleston, WV, USA), Keisha L. Gibson (University of North Carolina, NC, USA), Franca M. Iorember (Louisiana State University, New Orleans, LA, USA), Patrick D. Brophy (Children’s Hospital, University of Iowa, Iowa City, IA), Tarak Srivastava (Children’s Mercy Hospital, Kansas City, MO, USA), and Larry A. Greenbaum (Emory University School of Medicine, Atlanta, GA, USA). We also thank the Biopathology Core at NCH for storing and maintaining the sample biorepository. We also thank Jyotsna Nateri and Dr. Mark Hall for their valuable contributions to this work in acquiring the cytokine assay data. This work was supported in part by an NIDDK (NIH, USA) award to WES (R01 DK 095059), BAK (K08DK103982), and the American Heart Association (USA) Career Development Award to SA (CDA34110287).
Author Contributions
SA conceptualized and designed the studies, performed experiments, analyzed and interpreted the data, prepared the figures and tables, and drafted and edited the manuscript. MEB designed and performed statistical analyses and model predictions, interpreted the data, prepared the tables, and drafted and edited the manuscript. BAK interpreted the data and edited the manuscript. WES conceptualized and designed the studies, interpreted the data, and edited the manuscript. Pediatric nephrologists and author members of the PNRC collected patient samples and prepared their medical records. All the authors approve of the final version of the manuscript.
Footnotes
Table S1. Cytokines measured in this study and their concentration ranges in the nephrotic syndrome patient plasma samples.
Contributor Information
William E. Smoyer, Email: William.Smoyer@Nationwidechildrens.org.
Pediatric Nephrology Research Consortium:
John Mahan, Hiren Patel, Richard F. Ransom, Cynthia Pan, Denis F. Geary, Myra L. Chang, Keisha L. Gibson, Franca M. Iorember, Patrick D. Brophy, Tarak Srivastava, and Larry A. Greenbaum
Appendix
Contributing Members of the Pediatric Nephrology Research Consortium: Drs. John Mahan, Hiren Patel, and Richard F. Ransom (NCH, Columbus, Ohio, USA); Cynthia Pan (Medical College of Wisconsin, Milwaukee, Wisconsin, USA); Denis F. Geary (The Hospital for Sick Children, Toronto, Ontario, Canada); Myra L. Chang (West Virginia University, Charleston, West Virginia, USA); Keisha L. Gibson (University of North Carolina, Chapel Hill, North Carolina, USA); Franca M. Iorember (Louisiana State University, New Orleans, Louisiana, USA); Patrick D. Brophy (Children’s Hospital, University of Iowa, Iowa City, Iowa, USA); Tarak Srivastava (Children’s Mercy Hospital, Kansas City, Missouri, USA); and Larry A. Greenbaum (Emory University School of Medicine, Atlanta, Georgia, USA).
Disclosure
All the authors declared no competing interests.
Supplementary Material
Table S1. Cytokines measured in this study and their concentration ranges in the nephrotic syndrome patient plasma samples.
References
- 1.Shalhoub R.J. Pathogenesis of lipoid nephrosis: a disorder of T-cell function. Lancet. 1974;2:556–560. doi: 10.1016/s0140-6736(74)91880-7. [DOI] [PubMed] [Google Scholar]
- 2.Kurts C., Panzer U., Anders H.J. The immune system and kidney disease: basic concepts and clinical implications. Nat Rev Immunol. 2013;13:738–753. doi: 10.1038/nri3523. [DOI] [PubMed] [Google Scholar]
- 3.Pereira Wde F., Brito-Melo G.E., Guimaraes F.T. The role of the immune system in idiopathic nephrotic syndrome: a review of clinical and experimental studies. Inflamm Res. 2014;63:1–12. doi: 10.1007/s00011-013-0672-6. [DOI] [PubMed] [Google Scholar]
- 4.Cho M.H., Lee H.S., Choe B.H. Interleukin-8 and tumor necrosis factor-alpha are increased in minimal change disease but do not alter albumin permeability. Am J Nephrol. 2003;23:260–266. doi: 10.1159/000072065. [DOI] [PubMed] [Google Scholar]
- 5.Araya C., Diaz L., Wasserfall C. T regulatory cell function in idiopathic minimal lesion nephrotic syndrome. Pediatr Nephrol. 2009;24:1691–1698. doi: 10.1007/s00467-009-1214-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Araya C.E., Wasserfall C.H., Brusko T.M. A case of unfulfilled expectations. Cytokines in idiopathic minimal lesion nephrotic syndrome. Pediatr Nephrol. 2006;21:603–610. doi: 10.1007/s00467-006-0026-5. [DOI] [PubMed] [Google Scholar]
- 7.Cara-Fuentes G., Clapp W.L., Johnson R.J. Pathogenesis of proteinuria in idiopathic minimal change disease: molecular mechanisms. Pediatr Nephrol. 2016;31:2179–2189. doi: 10.1007/s00467-016-3379-4. [DOI] [PubMed] [Google Scholar]
- 8.Guimaraes F.T.L., Melo G., Cordeiro T.M. T-lymphocyte-expressing inflammatory cytokines underlie persistence of proteinuria in children with idiopathic nephrotic syndrome. J Pediatr (Rio J) 2018;94:546–553. doi: 10.1016/j.jped.2017.08.005. [DOI] [PubMed] [Google Scholar]
- 9.Gipson D.S., Trachtman H., Kaskel F.J. Clinical trial of focal segmental glomerulosclerosis in children and young adults. Kidney Int. 2011;80:868–878. doi: 10.1038/ki.2011.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nourbakhsh N., Mak R.H. Steroid-resistant nephrotic syndrome: past and current perspectives. Pediatric Health Med Ther. 2017;8:29–37. doi: 10.2147/PHMT.S100803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Canetta P.A., Radhakrishnan J. The Evidence-Based Approach to Adult-Onset Idiopathic Nephrotic Syndrome. Front Pediatr. 2015;3:78. doi: 10.3389/fped.2015.00078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eddy A.A., Symons J.M. Nephrotic syndrome in childhood. Lancet. 2003;362:629–639. doi: 10.1016/S0140-6736(03)14184-0. [DOI] [PubMed] [Google Scholar]
- 13.Polak D., Borovitz Y., Clyman-Levy D. Salivary cytokines in children with nephrotic syndrome versus healthy children: a comparative study. J Clin Med. 2020;9 doi: 10.3390/jcm9092691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Agrawal S., Merchant M.L., Kino J. Predicting and defining steroid resistance in pediatric nephrotic syndrome using plasma proteomics. Kidney Int Rep. 2020;5:66–80. doi: 10.1016/j.ekir.2019.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bennett M.R., Pleasant L., Haffner C. A novel biomarker panel to identify steroid resistance in childhood idiopathic nephrotic syndrome. Biomark Insights. 2017;12 doi: 10.1177/1177271917695832. 1177271917695832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bennett M.R. Biomarkers of therapeutic response in primary nephrotic syndrome: response. Pediatr Nephrol. 2013;28:161–162. doi: 10.1007/s00467-012-2256-z. [DOI] [PubMed] [Google Scholar]
- 17.Gooding J.R., Agrawal S., McRitchie S. Predicting and defining steroid resistance in pediatric nephrotic syndrome using plasma metabolomics. Kidney Int Rep. 2020;5:81–93. doi: 10.1016/j.ekir.2019.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kang H.G., Seo H., Lim J.H. Markers of disease and steroid responsiveness in paediatric idiopathic nephrotic syndrome: whole-transcriptome sequencing of peripheral blood mononuclear cells. J Int Med Res. 2017;45:948–963. doi: 10.1177/0300060516652762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matsumoto Y., Ikezumi Y., Kondo T. Urinary monocyte chemotactic protein 1 as a predictive marker of steroid responsiveness in children with idiopathic nephrotic syndrome. Fujita Med J. 2018;4:17–22. [Google Scholar]
- 20.Cuzzoni E., Franca R., De Iudicibus S. MIF plasma level as a possible tool to predict steroid responsiveness in children with idiopathic nephrotic syndrome. Eur J Clin Pharmacol. 2019;75:1675–1683. doi: 10.1007/s00228-019-02749-3. [DOI] [PubMed] [Google Scholar]
- 21.Turner M.D., Nedjai B., Hurst T. Cytokines and chemokines: At the crossroads of cell signalling and inflammatory disease. Biochim Biophys Acta. 2014;1843:2563–2582. doi: 10.1016/j.bbamcr.2014.05.014. [DOI] [PubMed] [Google Scholar]
- 22.Klimontov V.V., Korbut A.I., Orlov N.B. Multiplex bead array assay of a panel of circulating cytokines and growth factors in patients with albuminuric and non-albuminuric diabetic kidney disease. J Clin Med. 2020;9:3006. doi: 10.3390/jcm9093006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Croghan C.W., Egeghy P.P. Southeastern SAS User Group; St. Petersburg, FL: 2003. Methods of Dealing With Values Below the Limit of Detection Using SAS. [Google Scholar]
- 24.Kim J.S., Bellew C.A., Silverstein D.M. High incidence of initial and late steroid resistance in childhood nephrotic syndrome. Kidney Int. 2005;68:1275–1281. doi: 10.1111/j.1523-1755.2005.00524.x. [DOI] [PubMed] [Google Scholar]
- 25.Garin E.H., Laflam P., Chandler L. Anti-interleukin 8 antibody abolishes effects of lipoid nephrosis cytokine. Pediatr Nephrol. 1998;12:381–385. doi: 10.1007/s004670050470. [DOI] [PubMed] [Google Scholar]
- 26.Garin E.H., Blanchard D.K., Matsushima K. IL-8 production by peripheral blood mononuclear cells in nephrotic patients. Kidney Int. 1994;45:1311–1317. doi: 10.1038/ki.1994.171. [DOI] [PubMed] [Google Scholar]
- 27.Al-Eisa A.A., Al Rushood M., Al-Attiyah R.J. Urinary excretion of IL-1beta, IL-6 and IL-8 cytokines during relapse and remission of idiopathic nephrotic syndrome. J Inflamm Res. 2017;10:1–5. doi: 10.2147/JIR.S124947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jafar T., Agrawal S., Mahdi A.A. Cytokine gene polymorphism in idiopathic nephrotic syndrome children. Indian J Clin Biochem. 2011;26:296–302. doi: 10.1007/s12291-011-0126-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fry T.J., Mackall C.L. Interleukin-7: from bench to clinic. Blood. 2002;99:3892–3904. doi: 10.1182/blood.v99.11.3892. [DOI] [PubMed] [Google Scholar]
- 30.Kanai T., Shiraishi H., Yamagata T. Elevated serum interleukin-7 level in idiopathic steroid-sensitive nephrotic syndrome. Pediatr Int. 2011;53:906–909. doi: 10.1111/j.1442-200X.2011.03380.x. [DOI] [PubMed] [Google Scholar]
- 31.Xiong T., Attar M., Gnirck A.C. Interleukin-9 protects from early podocyte injury and progressive glomerulosclerosis in Adriamycin-induced nephropathy. Kidney Int. 2020;98:615–629. doi: 10.1016/j.kint.2020.04.036. [DOI] [PubMed] [Google Scholar]
- 32.Lin Q., Menon M.C., He J.C. IL-9: a novel pro-podocyte survival cytokine in FSGS. Kidney Int. 2020;98:541–543. doi: 10.1016/j.kint.2020.05.045. [DOI] [PubMed] [Google Scholar]
- 33.Deshmane S.L., Kremlev S., Amini S. Monocyte chemoattractant protein-1 (MCP-1): an overview. J Interferon Cytokine Res. 2009;29:313–326. doi: 10.1089/jir.2008.0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Besbas N., Kalyoncu M., Cil O. MCP1 2518 A/G polymorphism affects progression of childhood focal segmental glomerulosclerosis. Ren Fail. 2015;37:1435–1439. doi: 10.3109/0886022X.2015.1074474. [DOI] [PubMed] [Google Scholar]
- 35.Varner J.D., Matory A., Gbadegesin R.A. Genetic basis of health disparity in childhood nephrotic syndrome. Am J Kidney Dis. 2018;72:S22–S25. doi: 10.1053/j.ajkd.2018.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Decker M.L., Gotta V., Wellmann S. Cytokine profiling in healthy children shows association of age with cytokine concentrations. Sci Rep. 2017;7:17842. doi: 10.1038/s41598-017-17865-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Decker M.L., Grobusch M.P., Ritz N. Influence of age and other factors on cytokine expression profiles in healthy children—a systematic review. Front Pediatr. 2017;5:255. doi: 10.3389/fped.2017.00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.