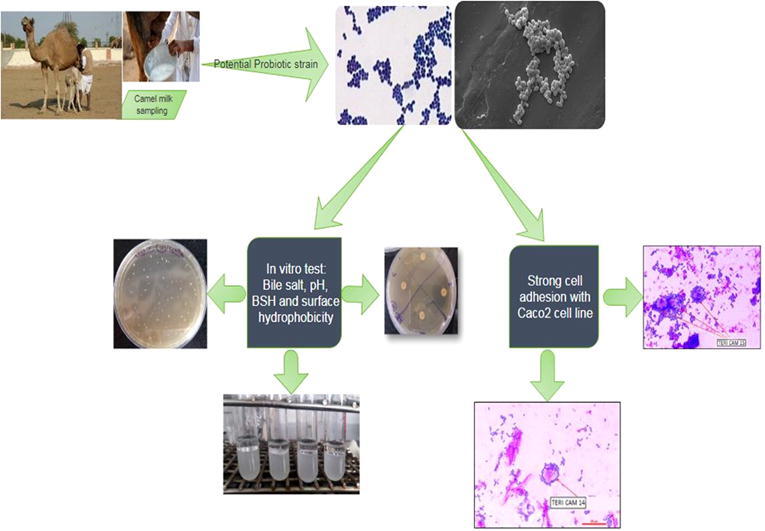
Graphical abstract
Keywords: Camel milk, Lactic acid bacteria, Probiotics, In vitro studies, Caco-2 cell line
Abstract
In the present study, a total of 80 presumed lactic acid bacteria (LAB) were isolated from camel milk. Selected LAB were identified as Lactococcus lactis (cam 12), Enterococcus lactis (cam 14) and Lactobacillus plantarum (cam 15) and their potential were tested by tolerance & de-conjugation of bile salts, antimicrobial activity, surface hydrophobicity and adhesion potential) along with this of probiotics were evaluated for curd formation and assessed for sensory properties and syneresis. Selected LABs showed antimicrobial activity against wide range of pathogenic bacteria (Staphylococcus aureus, Pseudomonas aeruginosa, Bacillus cereus and Escherchiaia. coli). LAB (cam 12, cam 14 and cam15) were highly sceptible to chloramphenicol, vancomycin, and tetracyclin. In vitro adhesion studies with Caco-2 cells demonstrated strong adhesion activity with hydrophobicity (99%) was observed. Acute oral toxicity of E. lactis and L. plantarum showed non-toxic, non-virulent and safe for industrial application. The study provides potential LAB which may act as a substitute of functional food, synthetic feed and industrial curd formulation with in the shortest span (240 min at 28–32 °C).
1. Introduction
The Camelus dromedarius, is a large, even-toed ungulate with one hump on its back and also known as the Arabian camel. Camel milk is nutritious, rich in vitamins, proteins, and minerals. In comparison to bovine milk, camel milk contains a greater amount of natural antimicrobial compounds (El-agamy et al., 1996). There are a total of approximately 23.9 million camels in the world. Out of this, India has 0.45 million which is almost 1.9% of the total world camel population (BAHS, 2012). Among this estimated world population, 17 million are believed to be one-humped dromedary camels (Camelus dromedaries) and 2 million are two-humped (Camelus bactrianus). Different Indian camel breeds, like Mewari, Bikaneri, Kachchi and Jaisalmeri possess the milk production potential of about 4.190 ± 0.12, 3.22 ± 0.15, 3.94 ± 0.13 and 2.17 ± 0.16 litres/day, respectively with a lactation period of 14–16 months (Singh et al., 2017a, Singh et al., 2017b). Camel milk has been widely recognized for its extraordinary medicinal properties. It is known to have therapeutic potential against many diseases including cancer. Camel milk is a nutritive substitute with enhanced functional food values in it (Alebie et al., 2017). It also has medicinal attributes which play an essential role in improving the immune system. Its major protein content consists of lactoferrin, peptidoglycan, antibodies, immunoglobulins and enzymes (lysozyme and lactoperoxidase) which have a valuable effect against major disorders. Daily consumption of camel milk might improve the defense mechanism of our immune system. As compared to other ruminant’s milk camel milk is superior with all these vital components present in it with add on low values of bad fat (i.e. cholesterol) and sugars. It also consists of a high amount of vitamin C and insulin which also have a beneficial impact on the health of human beings.
Etymologically, probiotics are pro (for), and bios (life). According to FAO probiotics are defined as “live microorganisms which when administered in adequate amounts confer a health benefit on the host” (FAO/WHO, 2002). Despite camel milk’s physiochemical composition, it also has beneficial microbiota which is mostly represented by LABs (lactic acid bacteria). Camel milk is in demand because of its wholesome nature as food. Many attempts have been made earlier to identify LAB from camel milk and its products (Fguiri et al., 2016, Mahmoudi et al., 2016). The present study was conducted to: (i) identify and characterize potential LAB from camel milk and their probiotic ability. (ii) Formulation of curd with potential probiotic dairy starters.
2. Materials and methods
2.1. Sample collection
In the present study four camel breeds namely Mewari, Bikaneri, Kachchi and Jaisalmeri were considered. Milk samples (four from each breed with variable lactation period i.e. 1, 4 & 12 months) were collected from National Research Centre on Camel (NRCC) Bikaner, Rajasthan by traditional milking method in sterile containers under suitable conditions.
2.2. Physicochemical analysis of milk
Camel milk samples were analyzed for fat, SNF (solids not fat), protein, lactose, density, temperature, pH and freezing point by using lactoscan (Milkotronic ltd. Bulgaria) (AOAC, 1995).
2.3. Enrichment, isolation and screening of LAB
The samples were enriched by adding 1% volume to 50 mL MRS (de Man, Rogosa and Sharpe) broth. The enriched samples were then incubated at 37 °C for one week in the orbital rotary shaker (Brunswick). Lactic acid bacteria were isolated from milk sample and spread over solidified MRS medium (de Man et al., 1960) for enumeration. The plates were incubated at 37 °C for 24–48 h. The colonies with distinct morphology were picked and purified by further sub-culturing. All the experiments presented in the study have been performed in triplicates. The data labels are thus, the average of triplicate values ± standard deviation (less than 5% of average). The results obtained were statically significant (p < 0.05).
2.4. Characterization and identification of bacterial strains
A two-stage screening was performed to select bacterial isolates with probiotic activity; primary and secondary screening. The primary screening involved morphological and physiological assessment (Gram staining and catalase test) followed by exposure to abiotic stress conditions (salinity and temperature tolerance). The morphological characterization was performed through Gram staining kit (Hi-Media, India). The cultures were examined under a bright field microscope (Olympus, Japan). The strains were then assessed for the presence of catalase enzyme using 3% H2O2. To study the morphology of selected bacterial strains showing probiotic nature, 24 h cultures were used and observed under a scanning electron microscope (SEM). In order to preserve the surface morphology CPD (critical point drying) method was used (Prasanna and Charalampopolous, 2018). Biological samples were then examined under 10KV in scanning electron microscope (Zeiss EVO MA 10). Biochemical tests of selected bacterial strains were performed with HiBacillusTM kit (KB013 Himedia) and results were interpreted as per the instructions given. Further genomic DNA was isolated by employing DNA Kit (QIAamp DNA kit, Qiagen). Presence of genomic DNA was confirmed by 0.8% agarose gel electrophoresis. 16S rRNA gene amplification was performed with PCR (Eppendorf, Germany) by employing the universal specific forward and reverse oligonucleotide PCR primers (Sigma-Aldrich); 27F (5′- AGAGTTTGATCMTGGCTCAG-3′) and 1492R (5′-TACGGYTACCTTGTTACGACTT-3′). PCR reaction mixture (25 µL) consisted of 0.5 µL of forward and reverse primer each (20 pmol µl−1), 0.5 µL dNTP, 2 µL of template, 2.5 µL 10X buffer, 2.5 µL MgCl2, 0.2 µL Taq polymerase and 16.3 µL molecular grade water. The PCR products were analyzed by electrophoresis on 1% agarose gel. PCR amplified 16S rRNA gene product was purified using a gel extraction kit (Real Genomics, RBC, India). Further the purified PCR amplified DNA was processed for 16S rRNA gene sequence analysis (Macrogen Korea). 16S rRNA gene sequence of selected bacterial strains was compared with reference sequences available in the NCBI database using the Basic Local Alignment Search Tool (BLAST) algorithm. Closely related sequences were retrieved and aligned using the Clustal W program. The phylogenetic tree was constructed from evolutionary distance matrix process using MEGA 7.0 software (Batta et al., 2013).
2.5. Screening of selected bacterial strains for probiotic attributes
To explore the probiotic attributes of selected nine (based on primary screening) strains; each strain was subjected to secondary screening. Three species of LAB were used (Lactobacillus plantarum, Lactococcus lactis and Enterococcus lactis) and their probiotic properties were studied in detail. As per the FAO/WHO guidelines; two a bacterial strain needs to pass all the tests involved in this stage to prove its value as a potential probiotic candidate (FAO/WHO, 2002). This screening involves the performance of various in vitro assays which are as described below in the following sections
2.5.1. Acid tolerance
An acid tolerance was studied for the selected bacterial strain (s). The MRS broth with pepsin enzyme (3 mg/mL) was used as a medium. The pH of broth was adjusted at different pH values (2.0, 3.0 and 4.0) with 1.0 N HCl and a control set (pH 7.0) was used along with the same (Pereira and Gibson, 2002). Further, the broth was inoculated with overnight grown cultures of putative strains and incubated at 37 °C for 24 h. Sampling was performed at an interval of 6 h. The optical density was measured at 620 nm and viable counts were also considered.
2.5.2. Bile salt tolerance
Strains were investigated to test for their ability to grow in presence of bile salt at different concentrations. This was performed using three selected salts (sodium deoxycholate, sodium taurocholate and cholic acid, HiMedia) at different concentrations of 0.1%, 0.2% and 0.3% (w/v) (Kuda et al., 2016). These different bile salt concentrations were prepared in 100 mL flasks, containing 20 mL sterile MRS broth. Control was maintained using MRS broth. The flasks were then inoculated with overnight grown LAB culture and incubated at 37 °C for 15 h. Aliquots were taken at a time interval of 6 h and OD620 of samples was measured to check the viability of cells using UV–visible spectrophotometer (Shimadzu, Japan).
2.5.3. Bile salt hydrolase activity
To investigate the BSH activity of strains, plate assay method was performed (Gallego et al., 2013). To test the activity, overnight grown cultures were plated on MRS agar plates using spread plate method on MRS agar plates containing different bile salts (as mentioned in bile salt tolerance). The plates were incubated at 37 °C for 72 h. Presence of precipitated halos around the colonies confirmed BSH activity.
2.5.4. Cell surface hydrophobicity
The fundamental criterion for theadhesion process is the ability of organisms to adhere to hydrocarbon which was determined by modified protocol of (Vinderola and Reinheimer, 2003). The LAB culture grown in MRS broth was harvested by centrifugation at 6000 g for 10 min washed twice in 0.05 M K2HPO4 and was suspended in the same buffer to obtain an OD of approximately 1.0. Around 3 mL of this bacterial suspension was allowed to come in contact with 0.6 mL of three different hydrocarbons (n-hexadecane, toluene and xylene) by whirling on vortex for 2 min. The phases were allowed to separate by decantation at 37 °C for 1 h and the aqueous phase was decanted in a clean test tube/flask. The OD560 nm was measured for the removed aqueous phase. The decrease in the value of absorbance of aqueous phase was considered to be equivalent to the cell surface hydrophobicity (H%), which was calculated with the given formula.
where A0 and A are the absorbance before and after extraction with hydrocarbons
2.6. Safety evaluation of LAB (lactic acid bacteria)
2.6.1. Antibiotic susceptibility
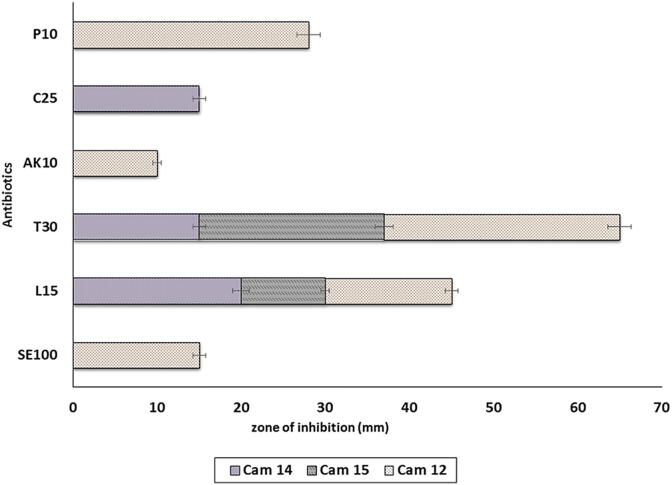
Antibiotic susceptibility of bacterial strain (s) was determined by agar disc diffusion method as published by (Bauer et al., 1966). MRS agar plates were prepared, in which 100 µL of freshly grown culture was mixed with 10 mL media. Antibiotic discs were placed on the solidified agar surface. And the plates were incubated at 37 °C for 48 h. Resistance against following antibiotics, namely penicillin G (10 µg), streptomycin (100 µg), lincomycin (15 µg), amikacin (10 µg), tetracycline (30 µg), chloramphenicol (25 µg) was tested. The zone of inhibition was measured in millimetres (mm).
2.6.2. Antimicrobial activity test
Antimicrobial activity of bacterial strain (s) was checked using agar well diffusion method (Balouiri et al., 2016). Strains were screened for production of antimicrobial against Staphylococcus aureus (ATCC- 6538), E. coli (ATCC- 11775), Bacillus cereus (ATCC-BAA-512) and Pseudomonas aeruginosa (ATCC- 19429). Mueller Hinton agar plates were prepared and seeded with indicator bacteria. Wells (5 mm) were made in the agar plates and filled with 100 µL of tested strain culture. Plates were incubated at 37 °C for 24–48 h. A clear zone of inhibition was measured in mm.
2.6.3. Haemolytic activity test
To test the activity, the overnight grown LAB strain were streaked on blood agar plate and further incubated at 37 °C for 48 h. The plates were observed for the formation of any β-haemolysis (clean) or α-haemolysis (greenish) and γ-haemolysis (no such haemolytic zones) around the colonies (Wang et al., 2016).
2.6.4. Biogenic amines
Production of biogenic amines was evaluated by modified protocol (Bover-cid and Holzapfel, 1999). Strains were streaked on MRS plates substituted with amino acids (tyrosine, lysine, arginine, histidine and ornithine) procured from Sigma-Aldrich. Plates were incubated at 37 °C for 72 h. Production of biogenic amines was confirmed by the colour change of the indicator.
2.6.5. In-vitro cell adhesion assay
Cell adhesion is the leading criterion to examine the potential of a probiotic. The adhesion ability of a probiotic bacterium to adhere to the epithelial cells of the intestinal tract is a prerequisite for establishing colonization. In the present study, intercellular adhesion was determined by using CaCo2 cell line and adherences junction where the cell interact with the intracellular junction of interact were monitored and adhesion score was determined by using Giemsa stain and observed 20 different microscopic fields (Han et al., 2017). Bacterial adhesion potential towards the CaCo2 was examined for L. plantarum and E. lactis. The cell line was maintained under recommended condition (at 37 °C in a humidified atmosphere of 5% CO2 and 95% air). In vitro adhesion studies were conducted at the National Toxicology Centre (NTC) Pune.
2.7. Fermentation of milk by isolated LAB strains
Milk was procured from the local market. Milk samples were boiled (80–90 °C) and cooled to (40 °C). Milk was inoculated with 1% of the strains (strains used in this study: L. lactis, L. plantarum and E. lactis.) maintaining the cell count of 1 × 106 and the milk was poured into sterilized evaporating glass dishes. The volume of milk used for the fermentation was 50 mL. Fermentations were performed at 28–32 °C for 4 h. Formulated curd samples were analyzed for the biochemical properties like pH, acidity (AOAC, 1995) and syneresis with a modified protocol (Hickisch et al., 2016). Viable count of microorganism was also evaluated after fermentation by plating serial dilutions of sample on MRS plates incubated for 24–48 h at 37 °C. Viable cell count was expressed in log of mean colony forming units (CFU).
2.8. Taste evaluation
Formulated curd samples were analyzed for colour/appearance, texture, odour, taste and overall preference. The experiment was conducted during March 2018 with the help of laboratory members (age group belongs from early 40 s to late 50 s with an equal number of male and female candidates). Group of ten members had trained and performed taste analysis (Training was given to members internally with in laboratory itself based on guidelines described by Sensory Evaluation Practices, 2nd edition by Stone and Sidel, 1993). Samples were provided to members in a cup 5–10 mL per cup for taste analysis.
As per the guidelines taste analysis of formulated curd was observed and rated as described (Stone and Sidel, 1993). The taste scores included; Like extremely = 9, Like very much = 8, Like moderately = 7, Like slightly = 6, Neither like nor dislike = 5, Dislike slightly = 4, Dislike moderately 3, Dislike very much = 2, Dislike extremely = 1.
All the experiments presented in the study have been performed in triplicates. The data labels are thus, the average of triplicate values ± standard deviation (less than 5% of average). The results obtained were statically significant (p < 0.05).
Ethical committee of National Research Centre on Camel, Bikaner approved the protocol and all members gave written consent before participation in the study.
3. Results and discussion
3.1. Characterization of camel milk sample
In the present study, all Indian breeds of camel (Mewari, Kachchi, Bikaneri and Jaishalmeri) with a variable lactation period were taken into consideration for screening the potential of LABs. In India, camel breeds are named after the region, in which they have originated. Camel milk of all Indian breeds with a lactation period of 1–12 months was examined for physicochemical characterization where fat, solid not fat (SNF), protein, ash content and pH were studied (Table 1). Camel milk is rich in vitamin C due to which its pH is low and this eventually helps in absorption in the GI tract. It is also rich in minerals like Ca, Fe and Zn (Table 2). As the role of Zn is crucial in the development and maintenance of a functioning immune system and its deficiency may cause complications in the overall functioning of the system (Gizachew et al., 2014). Total fat (2.82%) and SNF (6.61%) values were more in Bikaneri as compared to other camel breeds. The milk fat consists mainly of long-chain polyunsaturated fatty acids (PUFA) (Wernery, 2007). Protein is the core constituent of camel milk and has a major nutritional value. The percentage of protein varies from 2.01 to 2.95%. Presence of β lacto globulin was not found instead of which β casein was present (Table 2). Milk proteins are casein complexes and whey protein fractions and are a heterogeneous group of compounds. Whey proteins constitute about 20–25% of the total protein and α - lactalbumin is the main component of camel milk.
Table 1.
Showing milk composition (percent) value of different breeds of camel in India.
| Camel breed | Longitude and latitude | pH |
Milk composition (percent) |
|||||
|---|---|---|---|---|---|---|---|---|
| Water | Fat | SNF | Protein | Lactose | Ash | |||
| Bikaneri | 28.0229° N, 73.3119° E | 6.35 ± 0.002 | 83.97 ± 0.02 | 2.82 ± 0.001 | 6.61 ± 0.002 | 2.16 ± 0.001 | 3.68 ± 0.001 | 0.76 ± 0.0001 |
| Jaislmeri | 26.9157° N, 70.9083° E | 6.35 ± 0.002 | 85.13 ± 0.01 | 2.50 ± 0.001 | 6.18 ± 0.003 | 2.01 ± 0.001 | 3.47 ± 0.001 | 0.71 ± 0.0002 |
| Kachchhi | 23.7337° N, 69.8597° E | 6.39 ± 0.001 | 84.13 ± 0.03 | 2.68 ± 0.001 | 6.60 ± 0.002 | 2.14 ± 0.001 | 3.69 ± 0.001 | 0.76 ± 0.0001 |
| Mewari | 29.9917° N, 78.5931° E | 6.38 ± 0.002 | 84.51 ± 0.02 | 2.72 ± 0.001 | 6.38 ± 0.003 | 2.10 ± 0.001 | 3.56 ± 0.001 | 0.73 ± 0.0003 |
Values are given as mean ± standard deviation (SD) from triplicate experiments.
Table 2.
Fatty acid and mineral profile of camel milk (average value) composition.
| Fatty acid | Butyric acid C3H7COOH | Caproic acid C6H12O2 | Caprylic acid C8H16O2 | Capric acid C10H20O2 | Lauric acid C12H24O2 | Myristic acid C14H28O2 | Myristoleic acid C14H26O2 | Palmitic acid C16H32O2 | Palmitoleic acid C16H30O2 | Stearic acid C18H36O2 | Oleic acid C18H34O2 | Linoleic acid C18H32O2 | Arachidic acid C20H40O2 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Value % by weight | 0.31–0.75 | 0.2–0.6 | 0.2–0.3 | 0.2–0.4 | 1–1.8 | 15.9–25.2 | 1.7–4.5 | 25–29.5 | 6.1–19.1 | 1.9–11.7 | 6.8–24.9 | 0.9–0.2 | 0.6–3.4 |
| Mineral profile | Na | K | Ca | P | Mg | Fe | Zn | Cu | α-casein - | β-casein - | κ-casein - | – | – |
| Values | 29.70 ± 0.53 mEq/L | 50.74 ± 0.51 mEq/L | 94.06 ± 0.75 mg% | 41.68 ± 0.55 mg% | 11.82 ± 0.22 mg% | 1.00 ± 0.12 mg/dl | 2.00 ± 0.02 mg/dl | 0.44 ± 0.04 mg/dl | 21% | 65% | 3.47% | – | – |
Values are given as average in percentages (%).
Camel milk is the best natural adjunct for mother milk as it lacks the β lacto globulins which is a protein normally found in other ruminant milk and sometime may cause an allergic reaction. The major functional constituent of camel milk is the presence of immunoglobulins widely, and they are meant to be helpful in the reduction of allergic reactions by improving the immune system (Shabo et al., 2005).
3.2. Isolation and characterization of LAB (lactic acid bacteria)
Isolation of LAB from camel milk was performed on MRS Agar. A total of eighty presumed LABs showed gram positive and catalase negative were considered for further testing. All eighty LAB isolates were studied for their morphological, physiological characteristics. Based on the temperature (37–45 °C) and salt tolerance (6–8% NaCl), pH (2–5) tolerance, only nine LAB isolates were considered for detailed probiotic characterization out of which only three LABs (cam 12 L. lactis, cam 14 E. lactis & cam 15 L. plantarum showed bile salt tolerance. These three LAB were further studied for identification, biochemical characterization and probiotic properties (Table S1).
Maximum sugar utilization was observed with cam 14 and was able to utilize all the tested sugars which include lactose, glucose, sucrose, mannitol, galactose, arabinose & trehalose. All the tested sugar utilization was seen in all the selected LAB (cam12, cam14 & cam15) however arabinose utilization was observed only in cam 14. The sugar utilization was performed by using HiBacillus kit (Himedia). Further nitrate reduction test showed positive in cam 12 L. lactis. In the Voges – Proskauer (VP) test, cam 14 showed positive, VP test determines acetoin production from glucose which is a precursor for the synthesis of 2, 3 Butane-diol. Other researchers have also studied and reported similar results for Enterococcus species (Morandi et al., 2013).
The micrographs of strains revealed coccus (329.0–787.5 nm), rods (584.2–943.1 nm) and occurred in pairs or chains. Morphologically strains were gram-positive and non-motile, under SEM imaging, no flagella or polar fibrils were observed (Fig. 1). Thus E. lactis (cam 14), L. lactis (cam 12) and L. plantarum (cam 15) share the similarity and morphological resemblance to members of the group LABs with potential budding probiotic species as well (Nuryshev et al., 2016).
Fig. 1.
SEM images of selected strains.
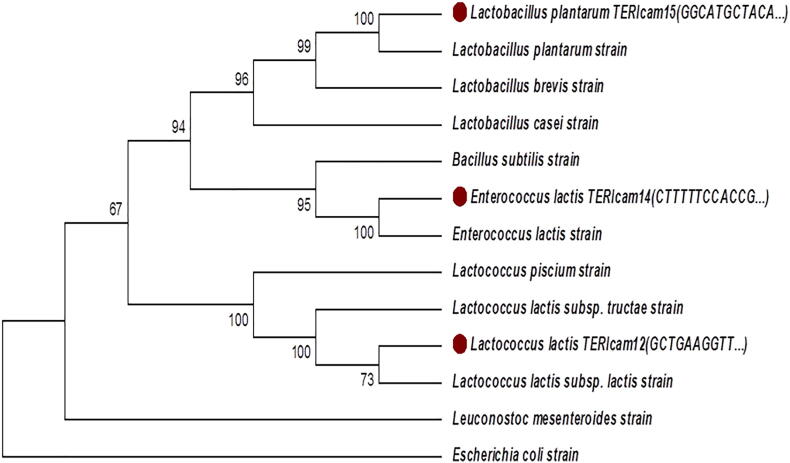
Based on the probiotic dynamics of LAB isolated from different Indian breeds of camel’s identification was performed. Further selected potential candidates (cam14, cam 12 and cam 15) were studied for genetic characterization by 16S rRNA gene. The strains were identified as E. lactis, L. lactis and L. plantarum and the accession numbers were MF143551, MF143552, and MF143553. Leading existence of L. lactis and E. species in camel milk have been reported in the past also (Khay et al., 2011, Hamed and Elattar, 2013). As isolated from camel milk the majority of strains were identified as Enterococcus, Lactococcus and Lactobacillus species, the dominance of E. lactis and L. lactis in milk has also been highlighted by authors in cow’s milk (Zamfir et al., 2006) and also in goats milk (Badis et al., 2004). It was also observed that the presence of Enterococcus in milk was more and it is directly linked to the milking practices and surroundings of the animal sheds or farms. Earlier the same observations were also made that there is a direct contact between milking parlor and the presence of hay in the bedding which seems to promote the inoculation of milk with Enterococcus faecalis or Enterococcus faecium (Tormo et al., 2015).
Phylogenetic analysis with MEGA (version 7.0 packages) suggests that cam 14, cam 12 L. lactis and cam 15; L. plantarum might represent strains that form a distinct lineage from the known type LAB (Fig. 2).
Fig. 2.
Phylogenetic analysis of selected strains.
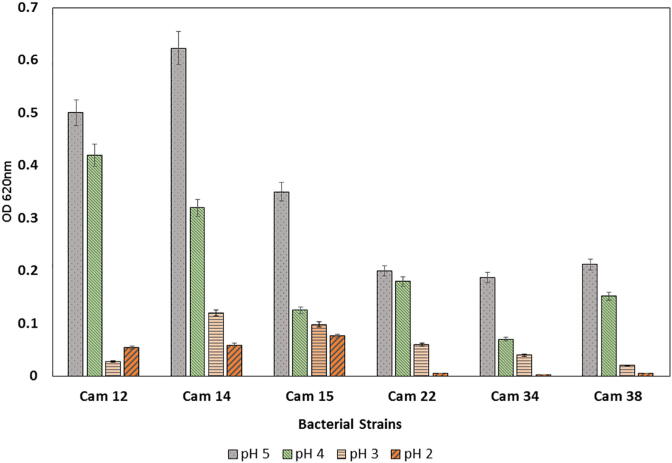
3.3. Acid tolerance
One of the foremost benchmarks for probiotic is, to be resistant to the acidic environment. Virtuous probiotics withstood low pH (ranging up to pH –2.0) and also showed tolerance to high acid levels, which exists in human, stomachs (Das et al., 2016). Selected potential LAB were screened for their ability to tolerate acidic conditions in MRS broth in the presence of pepsin enzyme with pH adjusted at 5.0, 4.0, 3.0 and 2.0. Survival at pH 3.0 was favourable for all selected LAB. All selected LAB showed survival at pH –3.0 whereas E. lactis (cam 14) and L. plantarum (cam 15) continued its growth up to pH 2.0 (Fig. 3 & Table S2). Earlier also observations were made for three strains of E. faecium survived at pH 3.0 after 3 h (Strompfova et al., 2004). To the best of our understanding, in our study the strain E. lactis (cam 14) from camel milk showed the maximum pH tolerance even at pH 2.0 among other Enterococcus sp. Among Enterococci, E. faecium is the most commonly used species in commercial probiotics because E. faecium is used as efficient probiotic and it has potential to defend animals from disease caused by E. coli, Salmonellae or Clostridia. Even the well-studied probiotic L. rhamnosus GG is only tolerant to acid conditions at pH 3.0 whereas at a lower pH it lost its viability in gastric juices (Goldin et al., 1992).
Fig. 3.
Acid tolerance of selected strains in different pH.
In our study, the pattern of acid tolerance is strain- dependent. E. lactis showed the tolerance even at pH 2.0 whereas L. lactis and L. plantarum showed tolerance at pH 3.0. Similar results were observed by researchers that acid tolerance in Lactobacilli and Bifidobacteria are also specific to strains even at a pH range of 1.5–3.0 (Lankaputhra and Shah, 1995).
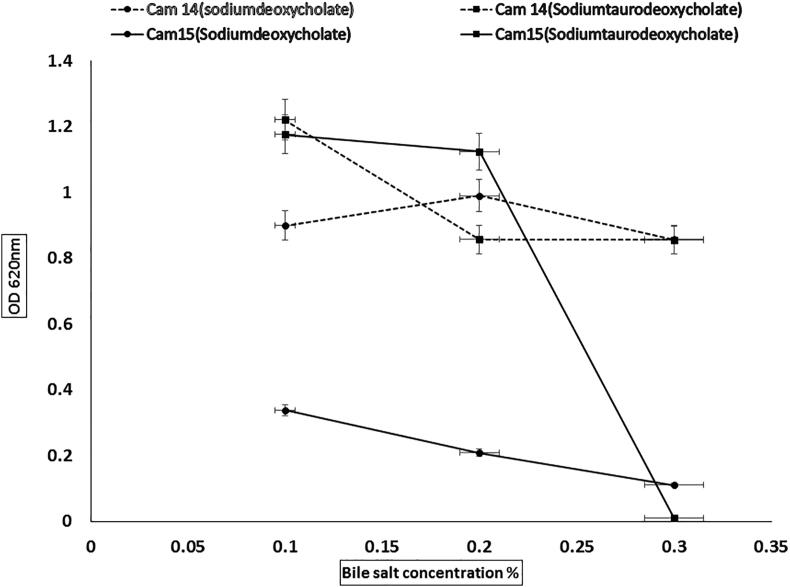
3.4. Bile salt tolerance and bile salt hydrolysis
Resistance to bile salts is one of the most significant qualities of probiotics as they dissolve membrane lipids leading to cell leakage and death (Choi and Chang, 2015). In the present study, bile salt tolerance and hydrolysis was studied with sodium deoxycholate, sodium taurocholate and sodium cholate. E. lactis and L. plantarum (cam 14 and cam 15) showed growth and survival against bile salts (sodium deoxycholate and sodium taurocholate) in the range of 0.1, 0.2 and 0.3% (Fig. 4) whereas none of the LAB isolated from camel milk showed tolerance towards sodium cholate bile salt. Acid and bile salt tolerance of all LAB varied significantly due to specific nature (Pitino et al., 2012). It has also been reported that resistant mechanism towards low pH or bile concentration is strain and species dependent (Aarti et al., 2017). Similar results were reported for Enterococcus species strains from dogs. It could also tolerate the bile salt media conditions and survive in intestinal conditions (Strompfova et al., 2004, Lee et al., 2011). Our study and results also suggest the acceptable survival of L. plantarum and E. lactis in bile salt environment even in a high percentage (0.3%) of bile concentrations. Previous studies on lactobacilli also suggested a high tolerance of probiotic L. fermentum, L. plantarum and L. paracasei strains to bile salts (Zoumpopoulou et al., 2008). Whereas Bile salt hydrolase action is an important feature for probiotic candidates, as with this integral property of LAB they might endure the toxic environment present in the intestine due to conjugated bile salts.
Fig. 4.
Bile salt tolerance data of selected strains.
Bile salt hydrolysis test was performed with selected LAB and it was observed that LAB from camel milk were able to grow and conjugate the bile salts. Potential hydrolysis activity was seen in Enterococcus lactis (cam 14) with sodium deoxycholate, sodium taurodeoxycholate and sodium cholate whereas Lactobacillus plantarum (cam 15) showed hydrolysis activity with sodium deoxycholate, sodium taurodeoxycholate followed by Lactococcus lactis (cam 12) showed BSH activity with sodium taurodeoxycholate (Table 3).
Table 3.
Bile salt hydrolysis activity.
| Strains | Sodium deoxycholate (log10CFU/ml) | Sodium tauro deoxy cholate (log10CFU/ml) | Cholic acid log10CFU/ml |
|---|---|---|---|
| Lactococcus lactis (cam 12) | No precipitated colonies were observed | 7.0 ± 0.001 | No precipitated colonies were observed |
| Enterococcus lactis (cam 14) | 8.25 ± 0.003 | 7.85 ± 0.002 | 7.89 ± 0.002 |
| Lactobacillus plantarum (cam 15) | 7.21 ± 0.002 | 7.90 ± 0.001 | No precipitated colonies were observed |
Values are given as mean ± standard deviation (SD) from triplicate experiments
Scientists reported and demonstrated the presence of taurodeoxycholate sodium salt hydrolase activity in all the strains of L. acidophilus and L. johnsonii isolated from probiotic yoghurts (Schillinger et al., 2005). The positive indications were showed by the salt precipitated around the colonies by the LABs which is a good indication for probiotics as it can reduce the cholesterol accumulation in humans. Similar studies were also reported on Lp9 which showed positive BSH activity against sodium glycocholate and sodium taurodeoxycholate (Kaushik et al., 2009).
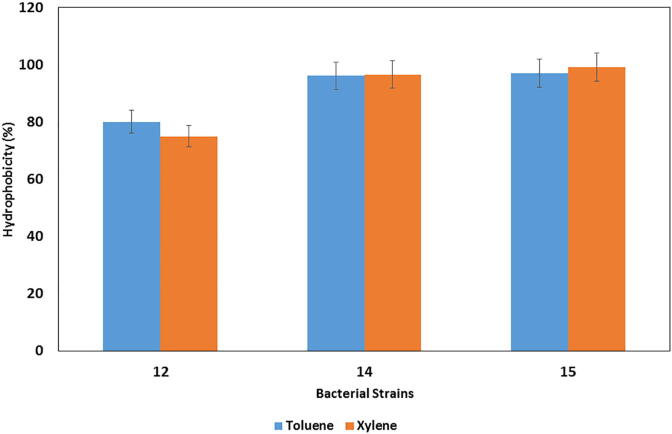
3.5. Cell surface hydrophobicity
In the present study, in vitro cell surface hydrophobicity was studied and the maximum cell hydrophobicity found for L. plantarum (cam 15) with xylene (99%) followed by toluene (97%). Similarly, E. lactis showed cell hydrophobicity 96% with toluene and xylene (Fig. 5). In our study L. lactis showed the less surface hydrophobicity with xylene and toluene and none of the strains showed the hydrophobicity against n-hexadecane. Strains simply showed the variation in hydrophobicity (H%) patterns with different hydrocarbons which proves that hydrophobicity is related to the cell surface proteins which are strain specific. Similar observations have been reported for L. animalis TSU 4 and L. gasseri TSU 3 which showed strong hydrophobicity against xylene and toluene whereas the later one showed hydrophobicity only for xylene (Sahoo et al., 2015). Earlier researchers suggested that the difference in the level of expression of surface proteins by bacteria may result in a huge variation of cell surface hydrophobicity among LAB (Schillinger et al., 2005).Various reports also showed a correlation between hydrophobicity and adhesion ability (Ehrmann et al., 2002).
Fig. 5.
Percentage of surface hydrophobicity (H%).
3.6. Antibiotic susceptibility
The genus Lactobacillus is the largest group among the lactic acid bacteria (LAB) and likely the most commonly used as a probiotic in a variety of foods, mainly fermented dairy products. In the present study potential LAB from camel milk were further taken up for antibiotic susceptibility (penicillin, amikacin, lincomycin, streptomycin, tetracycline and chloramphenicol) using disc diffusion method on MRS agar plate under aerobic conditions, and results are reported in Fig. 6. Within the groups of antimicrobial agents that inhibit the cell wall and nucleic acid synthesis, all LAB (100%) were found to be resistant to chloramphenicol and lincomycin. Only 30% LAB intermediate to streptomycin chloramphenicol, and amikacin.
Fig. 6.
Antibiotic evaluation of potential strains.
With regard to genetic mechanisms for antibiotic susceptibility in Lactobacillus is limited, although plasmid-encoded antibiotic genes have been reported in both L. reuteri and L. plantarum (Solieri et al., 2014). Lactobacilli are usually sensitive to the cell wall-targeting penicillin. Similarly, Lactobacilli are usually susceptible to antibiotics that inhibit protein synthesis, such as chloramphenicol, lincomycin and tetracycline. Different cat genes, encoding a chloramphenicol acetyltransferase, have previously been found on a plasmid in a L. plantarum strain isolated from pork (Egervarn et al., 2009). Tetracyclin also encoded for protein protecting on the ribosome and distributed in lactobacillus. Based on the data it is suggested that antibiotic susceptibility and resistance of LAB is also varying with different species (Solieri et al., 2014).
3.7. Antimicrobial activity test
While selecting probiotics it is of great importance strains must inhibit pathogenic bacterial growth in the gastro-intestinal tract. All the strains used in this study indicated a significant inhibitory effect on the growth of pathogenic microorganisms. Strains cam 14 E. lactis and cam 15 L. plantarum showed a stronger effect on S.aureus, E. coli, B. cereus whereas cam 12 Lactococcus lactis had a normal inhibitory effect on E. coli. Zone of inhibition of strains was shown (Table S3). One of the significant features for probiotic bacteria is to have the ability to inhibit the growth of pathogenic bacteria. In our study results depicted that selected LAB strains can produce antimicrobial substances (organic acids and bacteriocins) and they have the potential to be used as a food preservative. Earlier also it was observed that the source of isolation of LAB played a substantial role in inhibiting against a broad range of pathogens, which is in support of our outcomes (Annuk et al., 2003).
3.8. Haemolytic and biogenic amines activity
In the present study all the strains showed no haemolysis (γ-haemolysis) of blood cells. Results were shown (Table S4). Similar results were reported in which L. plantarum and Pediococcus strains showed no haemolytic activity (Oh and Jung, 2015). As per guidelines no haemolysis of blood cells proves the safety of probiotics (FAO/WHO, 2002).
Biogenic amines were produced during the decarboxylation of amino acids. Earlier also reported the presence of high levels of biogenic amines in foods had a severe impact on humans after consumption (Bover-Cid and Holzapfel, 1999, Karovičová and Kohajdová, 2005). Therefore, the absence of biogenic amines is a criterion for food safety. In our study, biogenic amines were not produced by cam 15 Lactobacillus plantarum, cam 12 Lactococcus lactis and cam 14 E. lactis. Since there was no colour change was found proving that no biogenic amines were produced (Data not shown). Thus, cam 15 L. plantarum, cam 12 L. lactis and cam 14 E. lactis validates to be safe as starter cultures.
3.9. In-vitro cell adhesion assay
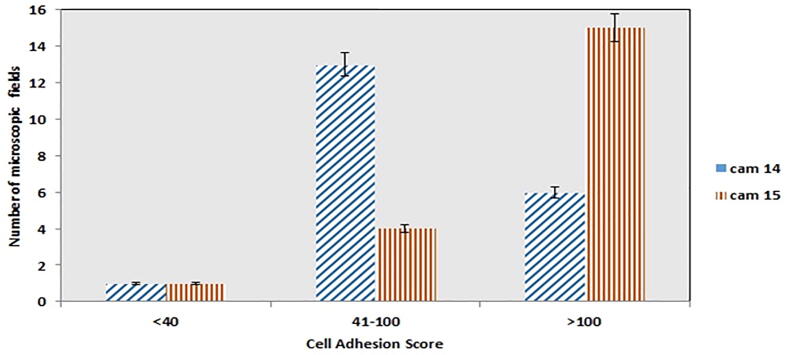
Probiotics potential is invariably dependent on their endurance in gastrointestinal tracts. Thus, adhesion ability considered as a standard for selecting a potential probiotic (Duary et al., 2011). In the present study, selected potential LAB L. plantrum (cam 15) and Enterococcus lactis (cam 14) were tested for adhesion ability with Caco-2 cell lines. L. plantrum (cam 15) showed strong adhesion with the Caco-2 cell line and adhesion scores (>100 Bacteria/ 15 microscopic fields) are shown in Fig. 7 and Table S5. Whereas E. lactis (cam 14) also showed an adhesion score of 100 Bacteria/ 6 microscopic fields. (Fig. 8A, B). On comparative evaluation L. reuteri showed 100% adhesion score. These results agree with the findings of another study, which found that high auto-aggregation ability is related to strong adhesion ability (Wang et al., 2018).
Fig. 7.
Adhesion score of strains (no. of bacterial cells adhere to Caco-2 cell line).
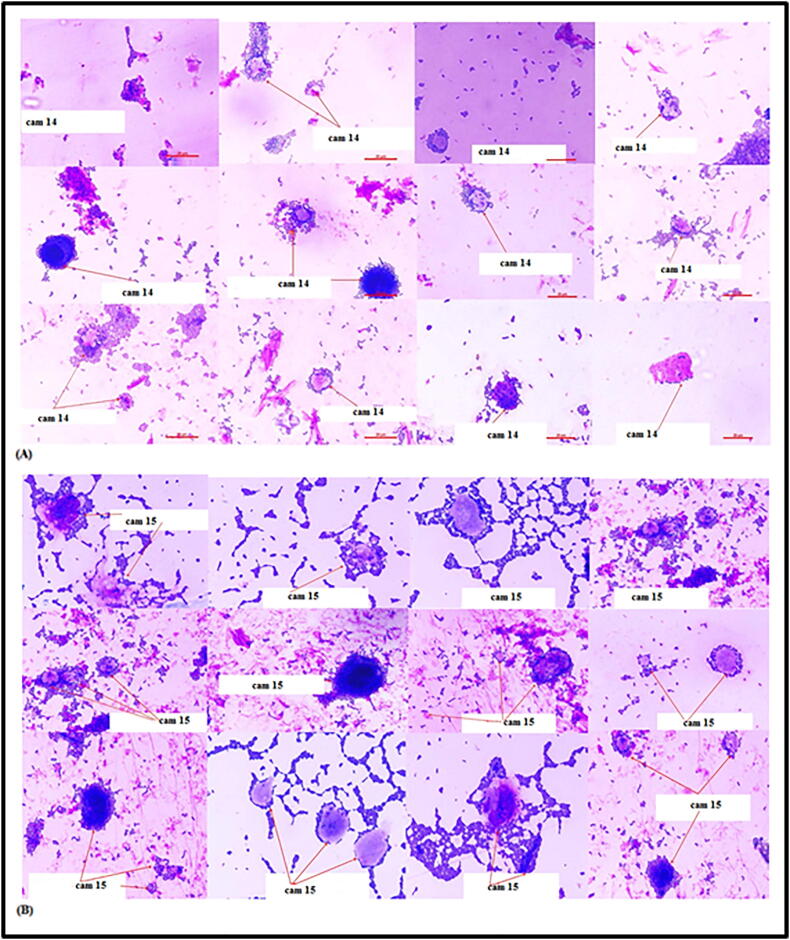
Fig. 8.
(A) TERI cam 14 Enterococcus lactis cells adhered to CaCo2 cells (B) TERI cam15 Lactobacillus plantarum cells adhered to CaCo2 cells.
The mechanism of adhesion involves the interaction between lipids, peptidoglycan and surface proteins present on the bacterial cell wall. Bacterial cell wall linked proteinaceous constituents facilitating bacterial adhesion to intestinal epithelial cells has been demonstrated for many Lactobacillus species (Singh et al., 2017a, Singh et al., 2017b). The results of this study demonstrate that probiotic strains Lactobacillus plantrum (cam 15) Enterococcus lactis (cam 14) tested were strongly adhesive with intestinal epithelial cells which might help compete with enteropathogens. However, it is significant that the mechanism of adhesion and adhesion ability was highly specific to probiotic strains.
Recently many reports suggested that hydrophobicity of a cell is linked to adhesion property of bacterial cells which favours the colonization towards epithelial cells (Caggia et al., 2015). Thus colonization of probiotics in epithelial cells is strain specific and particularly depends on the secretion of extracellular proteins by bacteria.
3.10. Post-fermentation properties of formulated curd
Titratable acidity and pH values of fermented milk (curd) samples were determined at 4 h (Table S6). The pH value of all the three probiotic curd samples reduced. The decrease of pH is the highest in curd inoculated with E. lactis and the lowest decrease was found with L. lactis. Titratable acidity values varied from 0.90 to 0.95% as compared to probiotic curd formulations lower levels of acidity were observed in control samples. The syneresis values varied from 20 to 30% in inoculated curd samples as compared to the control sample the value increases to 50%. As syneresis is an undesirable characteristic in curd formation since it leads to separation of the liquid phase from the gel (Muganga et al., 2015). From the above observation it can be assumed that syneresis and texture of formulated curd were influenced by probiotic strains used and the fermentation time. The role of microorganism is of significance in dairy and fermented products as the starter culture play a part in giving the originality to the final product in terms of texture, flavour and overall preference (Cocolin et al., 2018). Viable cell count of L. plantrum (cam 15) E. lactis (cam 14) and Lactococcus lactis (cam 12) are presented in Table S6.
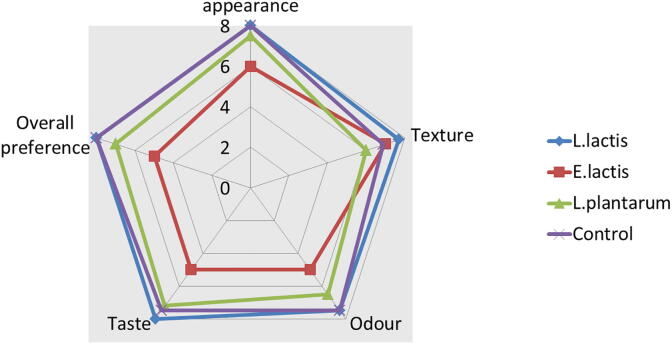
3.11. Taste evaluation of formulated curd
The scores were recorded for colour/appearance, texture, taste, odour and overall preference are summarized (Fig. 9). All formulated probiotic curd was assessed with comparable values by the trained members compared to control/plain curd sample. In this study, formulated curd with L. lactis and L. plantarum is overall preferred over the formulated curd with probiotic starter Enterococcus lactis. It is due to the specific probiotic strains were used as starters because they produce various flavouring compounds and acids during the fermentation process which is strain specific. From the study the formulated curd with E. lactis as starter lacked the authentic curd flavor. Similarly, researcher reported that curd formulated with starter E.faecium was buff coloured, lacked the curd flavour and had a fatty mouth feel (Ramakrishnan et al., 2014). Low preference score obtained for E. lactis formulated curd is may be due to the different palate test and lack of consumption of curd with probiotic starters. Also, a pathogenicity test for cam14 & cam15 were performed. Acute oral toxicity results suggested that the presence of LAB were not seen in any of the organs therefore cultures are a non-toxic, non-virulent and safe substitute for probiotic formulations.
Fig. 9.
Taste attributes presented as a radar chart of Taste evaluation data of formulated probiotic curd (L. lactis, L. plantarum and E. lactis and control sample).
4. Conclusion
The milk of Indian camel breeds like Mewari, Bikaneri, Kachchi and Jaisalmeri was screened for the efficient LAB to check their probiotic potential. According to morphological, physiological and biochemical properties and confirmation through the 16S rRNA gene sequences, potential LAB were identified as L. lactis, E. lactis and L.plantarum. Results showed that Enterococcus lactis was highly resistant to low pH and high bile salt whereas Lactobacillus plantarum adhered strongly to Caco2 human epithelial cell line. Strains were highly susceptible to chloramphenicol, vancomycin, and tetracyclin. Among all LAB from camel milk, L. plantarum, L. lactis and E. lactis were found to be an efficient probiotic. Even the strains were used to formulate the curd and taste analysis was done. And it shows the overall acceptability of curd samples formulated by using cam15 L. plantarum and L. lactis. Therefore, L. plantarum and L. lactis (which refers to cam15 L. plantarum and cam12 L. lactis respectively) as a bioactive dynamic probiotic and might be a viable substitute for synthetic feed for infants. Hence, appropriate clinical trials will be studied to validate the potency of developed probiotics.
Author contributions
AS performed the entire lab- scale experiments, and also wrote the manuscrip as. ML had designed the research framework. RS helped in taste analysis. BL and ML critically reviewed the manuscript. All the authors have read and approve the manuscript.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We are thankful to TERI for providing the infrastructural facilities to accomplish the present study. We are grateful to Department of Biotechnology (DBT grant No. BT/PR10482/PFN/20/866/2013), government of India for financial support in collaboration with project partner Dr. Raghvendar Singh, National Research Centre on Camel. We would also like to acknowledge Director, NRCC for their support and guidance related to taste analysis. We would also like to acknowledge Mr. Sanjiv Kumar, TERI for sample collection from NRCC. We would also like to thank the technical staff of our laboratory for all sorts of help during the study.
Footnotes
Peer review under responsibility of King Saud University.
References
- Aarti C., Khusro A., Varghese R., Arasu M.V., Agastian P., Al-Dhabi N., Ilavenil S., Choi K. In vitro studies on probiotic and antioxidant properties of Lactobacillus brevis strain LAP2 isolated from Hentak, a fermented fish product of north-east India. LWT - Food Sci. Technol. 2017;86:438–446. doi: 10.1016/j.lwt.2017.07.055. [DOI] [Google Scholar]
- Alebie G., Yohannes S., Worku A. Therapeutic applications of camel’s milk and urine against cancer: current development efforts and future perspectives. J. Cancer Sci. Ther. 2017;9:468–478. doi: 10.4172/1948-5956.1000461. [DOI] [Google Scholar]
- Annuk H., Shchepetova J., Kullisaar T., Songisepp E., Zilmer M., Mikelsaar M. Characterization of intestinal lactobacilli as putative probiotic candidates. J. Appl. Microbiol. 2003;94:403–412. doi: 10.1046/j.1365-2672.2003.01847.x. [DOI] [PubMed] [Google Scholar]
- AOAC, 1995. Official methods of analysis of the association of official analytical chemists. Arlintong, VA.
- Badis, A., Guetarni, D., Boudjema, B.M., Henni, D.E., K.I.H.A.L., M., 2004. Identification and technological properties of lactic acid bacteria isolated from raw goat milk of four Algerian races. Food Microbiol. 21, 579 – 588. doi: 10.1016/j.fm.2003.11.006
- BAHS, 2012. Basic Animal Husbandry Statistics, http//www.dahd.nic.in.
- Balouiri M., Sadiki M., Ibnsouda S.K. Methods for in vitro evaluating anti-microbial activity: A review. J. Pharmaceutical Anal. 2016;6:71–79. doi: 10.1016/j.pha.2015.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batta N., Subudhi S., Lal B., Devi A. Isolation of a lead tolerant novel bacteria species, Achromobactersp. TL-3: Assessment of bioflocculant activity. Indian J. Exp. Biol. 2013;51:1004–1011. doi: hdl.handle.net/123456789/23472. [PubMed] [Google Scholar]
- Bauer A.W., Kirby W.M., Sherris J.C., Turck M. Antibiotic susceptibility testing by a standardized single disk method. Am. J. Clin. Pathol. 1966;45:493–496. doi: 10.1093/ajcp/45.4_ts.493. [DOI] [PubMed] [Google Scholar]
- Bover-Cid S., Holzapfel W.H. Improved screening procedure for biogenic amine production by lactic acid bactéria. Int. J. Food Microbiol. 1999;53:33–41. doi: 10.1016/s0168-1605(99)00152-x. [DOI] [PubMed] [Google Scholar]
- Caggia C., De Angelis M., Pitino I., Pino A., Randazzo C.L. Probiotic features of Lactobacillus strains isolated from Ragusano and Pecorino Siciliano cheeses. Food Microbiol. 2015;50:109–117. doi: 10.1016/j.fm.2015.03.010. [DOI] [PubMed] [Google Scholar]
- Choi E.A., Chang H.C. Cholesterol-lowering effects of a putative probiotic strain L. plantarum isolated from kimchi. LWT - Food Sci. Technol. 2015;62:210–217. doi: 10.1016/j.lwt.2015.01.019. [DOI] [Google Scholar]
- Das P., Khowala S., Biswas S. In Vitro probiotic characterization of Lactobacillus casei isolated from marine samples. LWT – Food Sci. Technol. 2016;73:383–390. doi: 10.1016/j.lwt.2016.06.029. [DOI] [Google Scholar]
- De Man J.D., Rogosa M., Sharpe M.E. A medium for the cultivation of lactobacilli. J. Appl. Bacteriol. 1960;23:130–135. doi: 10.1111/j.1365-2672.1960.tb00188.x. [DOI] [Google Scholar]
- Ehrmann M.A., Kurzak P., Bauer J., Vogel R.F. Characterization of lactobacilli towards their use as probiotic adjuncts in poultry. J. Appl. Microbiol. 2002;92:966–975. doi: 10.1046/j.1365-2672.2002.01608.x. [DOI] [PubMed] [Google Scholar]
- Duary R.K., Rajput Y.S., Batish V.K., Grover S. Assessing the adhesion of putative indigenous probiotic lactobacilli to human colonic epithelial cells. Indian J. Med. Res. 2011;134:664–671. doi: 10.4103/0971-5916.90992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egervarn M., Roos S., Lindmark H. Identification and characterization of antibiotic resistance genes in Lactobacillus reuteri and Lactobacillus plantarum. J. Appl. Microbiol. 2009;107:1658–1668. doi: 10.1111/j.1365-2672.2009.04352.x. [DOI] [PubMed] [Google Scholar]
- El-Agamy E.I., Ruppanner R., Ismail A., Champagne C.P., Assaf R. Purification and characterization of lactoferrin, lactoperoxidase, lysozyme and immunoglobulins from camel’s milk. Int. Dairy J. 1996;6:129–145. doi: 10.1016/0958-6946(94)00055-7. [DOI] [Google Scholar]
- FAO/WHO Working Group Report, 2002. Guidelines for the evaluation of probiotics in food.
- Fguiri I., Ziadi M., Ayeb N., Arroum S., Assadi M., Khorchani T. Isolation and characterization of lactic acid bacteria strains from raw camel milk for potential use in the production of fermented Tunisian dairy products. Int. J. Dairy Technol. 2016;69:103–113. doi: 10.1111/1471-0307.12226. [DOI] [Google Scholar]
- Gallego J.B., Lopez F.N.A., Rantsiou K., Diaz R.J., Fernandez A.G., Cocolin L. Screening of lactic acid bacteria isolated from fermented table olives with probiotic potential. Food Res. Int. 2013;50:135–142. doi: 10.1016/j.foodres.2012.10.004. [DOI] [Google Scholar]
- Gizachew A., Teha J., Birhanu T. Review on medicinal and nutritional values of camel milk. Nat. Sci. 2014;12:35–40. [Google Scholar]
- Goldin B.R., Gorbach S.L., Saxelin M., Barakat S., Gualtieri L., Salminen S. Survival of Lactobacillus species (Strain GG) in human gastrointestinal tract. Dig. Dis. Sci. 1992;37:121–128. doi: 10.1007/BF01308354. [DOI] [PubMed] [Google Scholar]
- Hamed E., Elattar A. Identification and some probiotic potential of Lactic Acid Bacteria isolated strains from Egyptian camel’s milk. Life Sci. J. 2013;10:1952–1957. [Google Scholar]
- Han Q., Kong B., Chen Q., Sun F., Zhang H. In vitro comparison of probiotic properties of lactic acid bacteria isolated from harbin dry sausages and selected probiotics. J. Funct. Foods. 2017;32:391–400. doi: 10.1016/j.jff.2017.03.020. [DOI] [Google Scholar]
- Hickisch A., Beer R., Vogel R.F., Toelstede S. Influence of lupin milk heat treatment and exopolysaccharide-producing lactic acid bacteria on the physical characteristics of lupin-based yogurt alternatives. Food Res. Int. 2016;84:180–188. doi: 10.1016/j.foodres.2016.03.037. [DOI] [Google Scholar]
- Karovičová J., Kohajdová Z. Biogenic amines in foods. Chem. Pap. 2005;59:70–79. [Google Scholar]
- Kaushik J.K., Kumar A., Duary R.K., Mohanty A.K., Grover S., Batish V.K. Functional and probiotic attributes of an indigenous isolated strain of Lactobacillus plantarum. PLoS ONE. 2009;4 doi: 10.1371/journal.pone.0008099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khay E., Idaomar M., Castro L.P., Bernárdez P.F., Senhaji N.S., Abrini J. Antimicrobial activities of the bacteriocin-like substances produced by lactic acid bacteria isolated strains from Moroccan dromedary milk. Afr. J. Biotechnol. 2011;10:10447–10455. doi: 10.5897/AJB11.1328. [DOI] [Google Scholar]
- Kuda T., Masuko Y., Kawahara M., Kondo S., Nemoto M., Nakata T., Kataoka M., Takahashi H., Kimura B. Bile acid – lowering properties of Lactobacillus plantarum Sanriku – SU3 isolated from Japanese surfperch fish. Food Biosci. 2016;14:41–46. doi: 10.1016/j.fbio.2016.02.004. [DOI] [Google Scholar]
- Lankaputhra W.E.V., Shah N.P. Survival of Lactobacillus acidophilus and Bifido bacterium spp in the presence of acid and bile salts. Cult Dairy Prod. J. 1995;30:2–7. [Google Scholar]
- Lee H., Yoon H., Ji Y., Kim H., Park H., Lee J., Shin H., Holzapfel W. Functional properties of Lactobcillus strains isolated from kimchi. Int. J. Food Microbiol. 2011;145:155–161. doi: 10.1016/j.ijfoodmicro.2010.12.003. [DOI] [PubMed] [Google Scholar]
- Mahmoudi I., Moussa O.B., Khaldi T.E.M., Kebouchi M., Soligot C., Le Roux Y., Hassouna M. Functional in vitro screening of Lactobacillus strains isolated strains from Tunisian camel raw milk toward their selection as probiotic. Small Rumin Res. 2016;137:91–98. doi: 10.1016/j.smallrumres.2016.03.016. [DOI] [Google Scholar]
- Morandi S., Silvetti T., Brasca M. Biotechnological and safety characterization of Enterococcus lactis, a recently described species of dairy origin. Antonie Van Leeuwenhoek. 2013;103:239–249. doi: 10.1007/s10482-012-9806-z. [DOI] [PubMed] [Google Scholar]
- Muganga L., Liu X., Tian F., Zhao J., Zhang H., Chen W. Screening for lactic acid bacteria based on antihyperglycaemic and probiotic potential and application in symbiotic set yoghurt. J. Funct. Foods. 2015;16:125–136. doi: 10.1016/j.jff.2015.04.030. [DOI] [Google Scholar]
- Nuryshev M.Z., Stoyanova L.G., Netrusov A.I. New probiotic culture of Lactococcus lactis ssp. Lactis: effective opportunities and prospects. J. Microbial. Biochem. Technol. 2016;8:290–295. doi: 10.4172/1948-5948.1000299. [DOI] [Google Scholar]
- Oh Y.J., Jung D.S. Evaluation of probiotic properties of Lactobacillus and Pediococcus strains isolated from Omegisool, a traditionally fermented millet alcoholic beverage in Korea. LWT - Food Sci. Technol. 2015;63:437–444. doi: 10.1016/j.lwt.2015.03.005. [DOI] [Google Scholar]
- Pereira D.I., Gibson G.R. Effects of consumption of probiotics and prebiotics on serum lipid levels in humans. Crit. Rev. Biochem. Mol. Biol. 2002;37:259–281. doi: 10.1080/10409230290771519. [DOI] [PubMed] [Google Scholar]
- Pitino I., Randazzo C.L., Cross K.L., Parker M.L., Bisignano C., Wickham M.S.J., Mandalari G., Caggia C. Survival of Lactobacillus rhamnosusstrains inoculated in cheese matrix during simulated human digestion. Food Microbiol. 2012;31:57–63. doi: 10.1016/j.fm.2012.02.013. [DOI] [PubMed] [Google Scholar]
- Prasanna P.H.P., Charalampopolous D. Encapsulation of Bifidobacterium longum in alginate-dairy matrices and survival in simulated gastrointestinal conditions, refrigeration, cow milk and goat milk. Food Biosci. 2018;21:72–79. doi: 10.1016/j.fbio.2017.12.002. [DOI] [Google Scholar]
- Ramakrishnan V., Goveas L.C., Prakash M., Halami P.M., Narayan B. Optimization of conditions for probiotic curd formulation by Enterococcus faecium MTCC 5695 with probiotic properties using response surface methodology. J. Food Sci. Technol. 2014;51:3050–3060. doi: 10.1007/s13197-012-0821-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahoo T.K., Jena P.K., Nagar N., Patel A.K., Seshadri S. In vitro evaluation of probiotic properties of lactic acid bacteria from the gut of Labeo rohita and Catla catla. Probiotics Antimicro Prot. 2015;7:126–136. doi: 10.1007/s12602-015-9184-8. [DOI] [PubMed] [Google Scholar]
- Schillinger U., Guigas C., Holzapfel W.H. In vitro adherence and other properties of lactobacilli used in probiotic yoghurt-like products. Int. Dairy J. 2005;15:1289–1297. doi: 10.1016/j.idairyj.2004.12.008. [DOI] [Google Scholar]
- Shabo Y., Barzel R., Margoulis M., Yagil R. Camel milk for food allergies in children. Isr. Med. Assoc. J. 2005;7:796–798. [PubMed] [Google Scholar]
- Singh R., Mal G., Kumar D., Patil N.V., Pathak K.M.L. Camel milk: An important natural adjuvant. Agric. Res. 2017;6:327–340. doi: 10.1007/s40003-017-0284-4. [DOI] [Google Scholar]
- Singh T.P., Kaur G., Kapila S., Malik R.K. Antagonistic activity of lactobacillus reuteri strains on the adhesion characteristics of selected pathogens. Front. Microbiol. 2017;8:486. doi: 10.3389/fmicb.2017.00486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solieri L., Bianchi A., Mottolese G., Lemmetti F., Giudici P. Tailoring the probiotic potential of non-starter Lactobacillus strains from ripened Parmigiano Reggiano cheese by in vitro screening and principal component analysis. Food Microbiol. 2014;38:240–249. doi: 10.1016/j.fm.2013.10.003. [DOI] [PubMed] [Google Scholar]
- Stone, H., Sidel, J.L. Sensory evaluation practices. 1993; New York: Academic Press, Inc.
- Strompfova V., Laukova A., Ouwehand A.C. Selection of enterococci for potential canine probiotic additives. Vet. Microbiol. 2004;100:107–114. doi: 10.1016/j.vetmic.2004.02.002. [DOI] [PubMed] [Google Scholar]
- Tormo H., Lekhal A.H.D., Roques C. Phenotypic and genotypic characterisation of lactic acid bacteria isolated from raw goat milk and effect of farming practices on the dominant species of lactic acid bacteria. Int. J. Food Microbiol. 2015;210:9–15. doi: 10.1016/j.ijfoodmicro.2015.02.002. [DOI] [PubMed] [Google Scholar]
- Vinderola C., Reinheimer J. Lactic acid starter and probiotic bacteria: a comparative “in vitro” study of probiotic characteristics and biological barrier resistance. Food Res. Int. 2003;36:895–904. doi: 10.1016/S0963-9969(03)00098-X. [DOI] [Google Scholar]
- Wang Y., Zhou J., Xia X., Zhao Y., Shao W. Probiotic potential of Lactobacillus paracasei FM-LP-4 isolated from Xinjiang camel milk yoghurt. Int. Dairy J. 2016;62:28–34. doi: 10.1016/j.idairyj.2016.07.001. [DOI] [Google Scholar]
- Wang G., Zhang M., Zhao J., Xia Y., Phoency F.H., Ai L. A surface protein from Lactobacillus plantarum increases the adhesion of Lactobacillus Strains to human epithelial cells. Front. Microbiol. 2018;9:2858. doi: 10.3389/fmicb.2018.02858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernery, U., 2007. Camel milk - new observations. In T.K. Gahlot. Proceedings of the International Camel Conference, CVAS, Bikaner. pp. 200 – 204
- Zamfir M., Vancanneyt M., Makras L., Vaningelgem F., Lefebvre K., Pot B., Swings J., De Vuyst L. Biodiversity of lactic acid bacteria in Romanian dairy products. Sys. Appl. Microbiol. 2006;29:487–495. doi: 10.1016/j.syapm.2005.10.002. [DOI] [PubMed] [Google Scholar]
- Zoumpopoulou G., Foligne B., Christodoulou K., Grangette C., Pot B., Tsakalidou E. Lactobacillus fermentum ACA-DC 179 displays probiotic potential in vitro and protects against trinitrobenzene sulfonic acid (TNBS)-induced colitis and Salmonella infection in murine models. Int. J. Food Microbiol. 2008;121:18–26. doi: 10.1016/j.ijfoodmicro.2007.10.013. [DOI] [PubMed] [Google Scholar]