Abstract
Dipeptidyl peptidase 4 (DPP4) and α-glucosidase inhibitors have been developed as anti-diabetic agents for the treatment of diabetes mellitus. In the present study, the anti-diabetic effects of the lupinalbin A compound isolated from Apios americana was investigated by measuring its inhibitory activity against DPP4 and α-glucosidase. To detect the inhibitory effect of lupinalbin A, DPP4 and α-glucosidase assays were performed in vitro. Molecular docking analysis was performed using AutoDock 4.2. The IC50 values of lupinalbin A against DPP4 and α-glucosidase were 45.2 and 53.4 µM, respectively. Analysis of the enzyme kinetics revealed that lupinalbin A interacted with the active site of DPP4 in a competitive manner, with an inhibition constant (Ki) value of 35.1±2.0 µM, whereas the lupinalbin A interaction with α-glucosidase was non-competitive, with a Ki value of 45.0 µM. Molecular docking analysis revealed a binding pose between the DPP4 enzyme and lupinalbin A. Taken together, these data suggest lupinalbin A is more effective against DPP4 than α-glucosidase, with regard to its anti-diabetic effects.
Keywords: Apios americana, lupinalbin A, dipeptidyl peptidase 4, α-glucosidase, molecular docking
Introduction
Diabetes mellitus is a metabolic disease characterized by chronic hyperglycemia due to dysfunctional insulin sensitivity. The incidence of diabetes mellitus is predicted to double worldwide by 2030 compared to 2000(1). Type II diabetes mellitus (T2DM) accounts ~90% of diabetic patients and is well-established to be associated with low levels of physical activity, increased stress and obesity (2,3). Inhibition of the activity of various enzymes, such as α-glucosidase, protein tyrosine phosphatase 1B and dipeptidyl peptidase 4 (DPP4), is involved in regulating blood glucose, and analysis of molecular docking has been used as a potential method for improving treatment of T2DM (4,5). In general, α-glucosidase inhibitors delay the digestion and absorption of carbohydrates in the small intestine, alleviating postprandial hyperglycemia.
The family of DPP proteins comprises structurally homologous enzymes, including DPP4, DPP8, DPP9 and fibroblast activation protein (FAP) (6). Among these, DPP4, also known as CD26, was first discovered in 1966 by Hopsu-Havu and Glenner (7). The DPP4 enzyme is expressed in the epithelial cells of the liver, lung, kidney and spleen (8). In addition to its role in T-cell immune responses (9), DPP4 has been previously shown to be involved in incretin hormone metabolism and has been used as a target for the treatment of diabetes mellitus (10,11). Effective inhibition of DPP4 is key for the development of natural anti-diabetic agents.
Apios americana (A. americana) Medik is a perennial vine of the Leguminosae family that is frequently consumed by Native Americans (12). Apios tubers contain isoflavonoids, such as genistein, barpisoflavone A, 2'-hydroxygenistein, 5-methylgenistein and 2'-hydroxy genistein-7-O-gentibioside (13,14); these have been shown to exhibit antioxidant, soluble epoxide hydrolase inhibitory and tyrosinase inhibitory activities, which are beneficial for the treatment of hypertension and diabetes (14-18). In our previous study, it was reported that lupinalbin A isolated from A. americana exhibits anti-inflammatory effects in LPS-induced RAW264.7 cells (19). Lupinalbin A has also been shown to possess an inhibitory effect against α-glucosidase (20). Nonetheless, the mechanism underlying the inhibition of α-glucosidase by lupinalbin A and the molecular interactions of lupinalbin A with DPP4 are not fully understood.
The aim of the present study was to propose determine the suitability of lupinalbin A (isolated from A. americana) as a potentially novel compound for management of diabetes mellitus, and its effects on inhibition of DPP4 and α-glucosidase, by measuring the inhibitory activities and enzyme kinetics, as well as performing molecular docking simulation studies.
Materials and methods
General experimental procedures
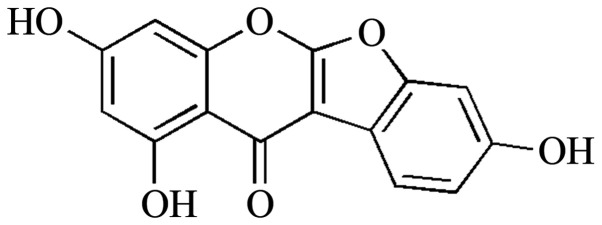
Nuclear magnetic resonance experiments were performed using an ECA500 (JEOL, Ltd.). Mass spectra were measured using an Agilent LC-MS 6100 (Agilent Technologies, Inc.). Thin-layer chromatography analysis was performed using Kieselgel 60 F254 plates (Merck KGaA). The compound was visualized by dipping the plates into 10% (v/v) H2SO4 (Sigma-Aldrich; Merck KGaA) and then heating at 300˚C for 30 sec using a 230-400 mesh silica gel (Merck KGaA), sephadex LH-20 (GE Healthcare) and ODS-A silica gel (YMC, Co.) resins were used for open column chromatography. Lupinalbin A was isolated from A. americana tubers, as described previously (Fig. 1) (19).
Figure 1.
Structure of lupinalbin A isolated from Apios Americana.
DPP4 inhibitory assay and kinetic analysis
DPP4 inhibitory activity was measured using a DPP(IV) inhibitor screening assay kit (Cayman Chemical Company) according to the manufacturer's protocol, with minor modifications. Briefly, the enzyme solution (120 µl) was dissolved in 480 µl DPP assay buffer [20 mM Tris-HCI (pH 8.0), 100 mM NaCl and 1 mM EDTA] and was used as the enzyme solution. The substrate, 5 mM H-Gly-Pro conjugated with aminomethylcoumarin (AMC), was prepared in the same buffer. The assay procedure was performed according to the manufacturer's protocols, and is briefly described as follows: Diluted assay buffer (30 µl) and diluted enzyme solution (10 µl) were added to the 96-well plates containing 10 µl solvent (blank) or solvent-dissolved test compounds (0-75 µM). The reaction was initiated by adding 50 µl diluted substrate solution, the reaction was measured using fluorometric determination (excitation wavelength, 350 nm; emission wavelength, 450 nm) using a plate reader (Tecan Group, Ltd.) every 30 sec for 20 min at 37˚C. Sitagliptin, which was included in the kit, was used as a positive control. Various concentrations (0-75 µM) of the enzyme inhibitor (tested compounds) and substrate were used in the reactions, the initial rate of reaction (v) based on free AMC, and the release rate were calculated using Lineweaver-Burk [1/v vs. 1/(substrate)] or Dixon plots [1/v vs. (inhibitor)].
α-glucosidase inhibitory assay
The α-glucosidase inhibitory assay was performed as described previously (21), with minor modifications. A total of 130 µl enzyme (0.16 U/ml) in phosphate buffer [0.1 mM phosphate (pH 6.8)], 20 μl methanol (control) or ligand (0.062-1 mM) in methanol and 50 µl substrate (2.5 mM p-nitrophenyl-α-D-glucopyranoside) were mixed in a 96-well plate. After starting each reaction at 37˚C, p-nitrophenol was quantified using a UV-vis spectrophotometer at 405 nm. Acarbose (20 µl) provided as ready to use in the kit was used as a positive control.
Molecular docking
The molecular simulations for the interaction between the inhibitors and enzyme were generated using AutoDock software version 4.2 (The Scripps Research Institute). The 3D structure of the ligand was built and minimized using MM2 in Chem 3D Pro (version 14.0). The flexible bonds of the ligand were assigned with AutoDockTools. The 3D structure of DPP4 (PDB ID: 5T4E) was obtained from Research Collaboratory for Structural Bioinformatics Protein Data Bank after substrates were removed from the original enzyme. The grid for docking was formed using X, Y and Z axes, all of which were 180 units in length. Default settings were used for docking, except for the Lamarckian genetic algorithms (run 50) and maximum number of evaluations (long). The results of the molecular simulation were prepared using LigPlot and Chimera software (22).
Statistical analysis
All inhibitory assays were performed in triplicate. Data are presented as mean ± standard deviation, and were analyzed using SigmaPlot (Systat Software Inc.) or GraphPad Prism version 6 (GraphPad Software Inc.). Differences were compared using a one-way ANOVA followed by a post-hoc Dunnett's multiple comparison test. P<0.05 was considered to indicate a statistically significant difference.
Results
Inhibition of DPP4 and α-glucosidase activities by lupinalbin A
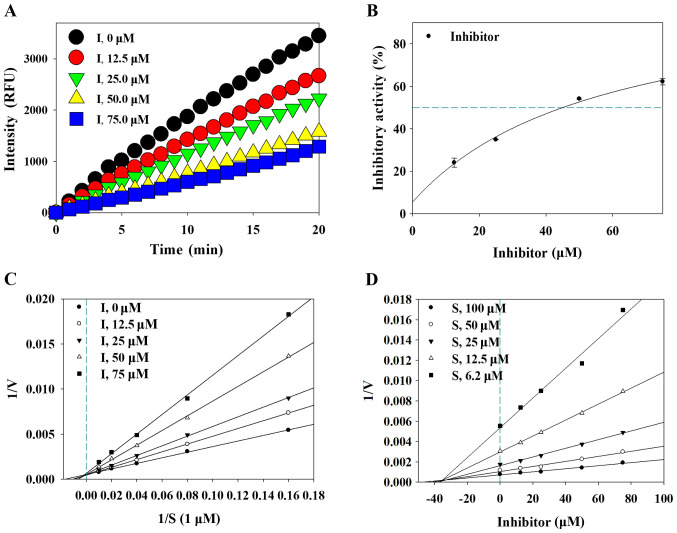
To investigate the inhibitory effect of lupinalbin A, the inhibitory activity of lupinalbin A against DPP4 and α-glucosidase were measured. Results showed that lupinalbin A inhibited DPP4 activity in a time- and dose-dependent manner (Fig. 2A and B). The IC50 values of lupinalbin A against DPP4 and sitagliptin (positive control) were 45.2±0.8 µM and 70.7±4.3 nM, respectively (Table I). Furthermore, the IC50 values of lupinalbin A against α-glucosidase and acarbose (positive control) were 53.4±1.2 and 240.5±2.1 µM, respectively.
Figure 2.
Enzyme kinetic analysis of DPP4 on the lupinalbin A. (A) Progression curves of DPP4 inhibition by lupinalbin A. (B) Inhibitory activity of the lupinalbin A on DPP4. (C) Lineweaver-Burk plot and (D) Dixon plot showing the inhibition of lupinalbin A. DPP4, dipeptidyl peptidase 4; RFU, relative fluorescence units; V, velocity; S, substrate.
Table I.
Inhibitory activities of lupinalbin A on DPP4 and α-glucosidase.
| DPP4 | α-glucosidase | |||
|---|---|---|---|---|
| Compound | IC50, µMa | Binding mode (Ki, µM)a | IC50, µMa | Binding mode (Ki, µM)a |
| Lupinalbin A | 45.2±0.8 | Competitive, (35.1±2.0) | 53.4±1.2 | Non-competitive (45.0±4.1) |
| Sitagliptinb | 70.7±4.3 nM | - | ||
| Acarbosec | - | 240.5±2.1 | ||
aData are presented as the mean ± standard deviation.
bPositive control for DPP4.
cPositive control for α-glucosidase. DPP4, dipeptidyl peptidase 4; Ki, inhibition constant.
Evaluation of enzyme kinetics
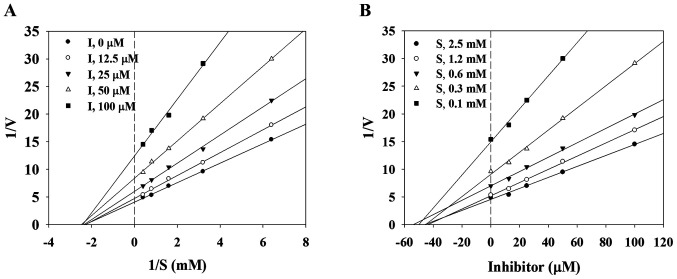
To determine whether lupinalbin A inhibits DPP4 and α-glucosidase by interacting with the active site of these enzymes, the enzyme kinetics of DPP4 and α-glucosidase were examined. An association between lupinalbin A and the enzyme substrate was confirmed using the Lineweaver-Burk plot, and the inhibition constant (Ki) was determined using a Dixon plot. The slope and X-axis of lupinalbin A changed in a dose-dependent manner, whereas the Y-axis was intersected at only one point (Fig. 2C). This suggests that lupinalbin A competitively suppressed the activity of DPP4. Analysis of α-glucosidase kinetics using the Lineweaver-Burk plot showed that the slope and Y-axis of lupinalbin A changed in a dose-dependent manner, whereas the line only intersected the X-axis at one point, indicating non-competitive inhibition (Fig. 3A). According to the Dixon plot, the Ki values of lupinalbin A for DPP4 and α-glucosidase were 35.1 and 45.0 µM, respectively, indicating that lupinalbin A was a competitive inhibitor of DPP4 and a non-competitive inhibitor of α-glucosidase (Table I; Figs. 2D and 3B).
Figure 3.
Inhibitory activity of lupinalbin A on α-glucosidase. (A) Lineweaver-Burk plot and (B) Dixon plot of the inhibition of lupinalbin A. V, velocity; S, substrate.
Molecular docking simulation of lupinalbin A with DPP4
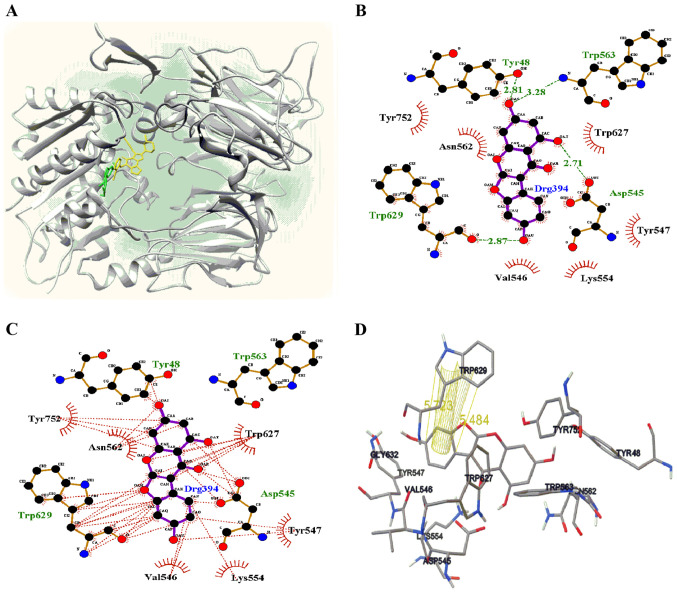
Molecular docking simulations are used to facilitate the development of enzyme inhibitors (23,24), determine the binding position of a ligand in an enzyme, and identify key amino acids that participate in ligand-receptor binding (25,26). Molecular docking was performed in the present study to set up a grid containing the activity site based on the results of the enzyme kinetic study. The binding pose of a ligand with a receptor, as determined using AutoDock, was suggested at the lowest energy values. Thus, lupinalbin A (1) occupied the right site next to the active site and a small portion of the active site (Fig. 4A), with a binding energy of -7.32 kcal/mol. Lupinalbin A formed four hydrogen bonds with Tyr48 (2.81 Å), Asp545 (2.71 Å), Trp563 (3.28 Å) and Trp629 (2.87 Å) residues surrounding the active site of DPP4 (Fig. 4B). A total of 9 amino acids of the receptor participated in hydrophobic interactions to maintain a stable binding pose with the ligand (Fig. 4C). Moreover, the B-ring of lupinalbin A interacted two π-π stacking (5.48 and 5.72 Å) with the indole ring of tryptophan (Fig. 4D).
Figure 4.
Predicted molecular docking model of lupinalbin A with DPP4. (A) X-ray structure of linagliptin (yellow) and docking poses of lupinalbin A (green) with DPP4. (B) Green text (Tyr48, 2.81 Å; Asp545, 2.71 Å; Trp563, 3.28 Å; Trp629, 2.87 Å) represent the amino acids involved in forming hydrogen bonds. (C) Schematic representations of hydrophobic interactions and (D) π-π interactions between lupinalbin A and the receptor. DPP4, dipeptidyl peptidase 4.
Discussion
In the present study, the potential anti-diabetic effects of lupinalbin A, which is isolated from A. americana, were determined using enzyme assays. The IC50 value of isoflavonoids isolated from the leaves of Smilax china L. against DPP4 is greater than 100 µM (4); thus, the DPP4 inhibitory activity of lupinalbin A (IC50, 45.2 µM) is stronger than these isoflavonoids. Moreover, Bai et al (20) reported that lignans and flavonoids isolated from mung beans exhibited inhibitory activity against α-glucosidase, and lupinalbin A, a type of isoflavonoid was shown to be more potent in their study compared with the present study, with an IC50 value of 10.73±2.01 µM. Taken together, lupinalbin A isolated from A. americana inhibited the activity of DPP4 and α-glucosidase.
Through enzyme kinetics analysis, the ability of lupinalbin A to inhibit the activity of different enzymes was determined. Fan et al (27) showed that phenolic compounds, including resveratrol and flavone, competitively inhibited DPP4 activity, which is consistent with the results of the present study. The binding force of flavone is stronger than that of lupinalbin A; the Ki value of flavone is 18.6 µM (23). Generally, the lower the Ki value, the stronger the binding of the enzyme and inhibitor; thus, an inhibitor with a lower Ki value is more effective than an inhibitor with a higher Ki value. On the basis of the Ki values of lupinalbin A for DPP4 and α-glucosidase, lupinalbin A binds to DPP4 more strongly to α-glucosidase. Thus, the DPP4 inhibitory activity of lupinalbin A is stronger than its α-glucosidase inhibitory activity, which is consistent with the results of the enzyme inhibition assay.
In conclusion Lupinalbin A, a compound isolated from A. americana, exhibited anti-diabetic activity, and more potently inhibited DPP4 compared with α-glucosidase. As a result, it is suggested that lupinalbin A may be one of the active components underlying the anti-diabetic effects of A. americana.
Acknowledgements
Not applicable.
Funding Statement
Funding: This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIP) (grant no. 2017M2A2A6A05018541).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
CHJ designed the experiments. HYK performed the experiments. HYK, JHK and CHJ analyzed the data. JHK isolated lupinalbin A. HYK, JHK and CHJ wrote the manuscript. HGJ assisted in designing the experiment and revising the manuscript. All authors have read and approved the final manuscript. HYK, JHK and CHJ confirm the authenticity of all the raw data.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Wild S, Roglic G, Green A, Sicree R, King H. Global prevalence of diabetes: estimates for the year 2000 and projections for 2030. Diabetes Care. 2004;27:1047–1053. doi: 10.2337/diacare.27.5.1047. [DOI] [PubMed] [Google Scholar]
- 2.Kim SM, Lee JS, Lee J, Na JK, Han JH, Yoon DK, Baik SH, Choi DS, Choi KM. Prevalence of diabetes and impaired fasting glucose in Korea: Korean National Health and Nutrition Survey 2001. Diabetes Care. 2006;29:226–231. doi: 10.2337/diacare.29.02.06.dc05-0481. [DOI] [PubMed] [Google Scholar]
- 3.DeFronzo RA. Pathogenesis of type 2 diabetes mellitus. Med Clin North Am. 2004;88:787–835. doi: 10.1016/j.mcna.2004.04.013. ix. [DOI] [PubMed] [Google Scholar]
- 4.Zhao BT, Le DD, Nguyen PH, Ali MY, Choi JS, Min BS, Shin HM, Rhee HI, Woo MH. PTP1B, α-glucosidase, and DPP-IV inhibitory effects for chromene derivatives from the leaves of Smilax china L. Chem Biol Interact. 2016;253:27–37. doi: 10.1016/j.cbi.2016.04.012. [DOI] [PubMed] [Google Scholar]
- 5.Li ZP, Song YH, Uddin Z, Wang Y, Park KH. Inhibition of protein tyrosine phosphatase 1B (PTP1B) and α-glucosidase by xanthones from Cratoxylum cochinchinense, and their kinetic characterization. Bioorg Med Chem. 2018;26:737–746. doi: 10.1016/j.bmc.2017.12.043. [DOI] [PubMed] [Google Scholar]
- 6.Yazbeck R, Howarth GS, Abbott CA. Dipeptidyl peptidase inhibitors, an emerging drug class for inflammatory disease? Trends Pharmacol Sci. 2009;30:600–607. doi: 10.1016/j.tips.2009.08.003. [DOI] [PubMed] [Google Scholar]
- 7.Hopsu-Havu VK, Glenner GG. A new dipeptide naphthylamidase hydrolyzing glycyl-prolyl-beta-naphthylamide. Histochemie. 1966;7:197–201. doi: 10.1007/BF00577838. [DOI] [PubMed] [Google Scholar]
- 8.Gorrell MD, Gysbers V, McCaughan GW. CD26: A multifunctional integral membrane and secreted protein of activated lymphocytes. Scand J Immunol. 2001;54:249–264. doi: 10.1046/j.1365-3083.2001.00984.x. [DOI] [PubMed] [Google Scholar]
- 9.Morimoto C, Schlossman SF. The structure and function of CD26 in the T-cell immune response. Immunol Rev. 1998;161:55–70. doi: 10.1111/j.1600-065x.1998.tb01571.x. [DOI] [PubMed] [Google Scholar]
- 10.Drucker DJ. Dipeptidyl peptidase-4 inhibition and the treatment of type 2 diabetes: Preclinical biology and mechanisms of action. Diabetes Care. 2007;30:1335–1343. doi: 10.2337/dc07-0228. [DOI] [PubMed] [Google Scholar]
- 11.Mulvihill EE, Drucker DJ. Pharmacology, physiology, and mechanisms of action of dipeptidyl peptidase-4 inhibitors. Endocr Rev. 2014;35:992–1019. doi: 10.1210/er.2014-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wilson PW, Pichardo FJ, Liuzzo JA, Blackmon WJ, Reynolds BD. Amino acids in the american groundnut (Apios americana) J Food Sci. 1987;52:224–225. [Google Scholar]
- 13.Ichige M, Fukuda E, Miida S, Hattan J, Misawa N, Saito S, Fujimaki T, Imoto M, Shindo K. Novel isoflavone glucosides in groundnut (Apios americana Medik) and their antiandrogenic activities. J Agric Food Chem. 2013;61:2183–2187. doi: 10.1021/jf305233t. [DOI] [PubMed] [Google Scholar]
- 14.Kaneta H, Koda M, Saito S, Imoto M, Kawada M, Yamazaki Y, Momose I, Shindo K. Biological activities of unique isoflavones prepared from Apios americana Medik. Biosci Biotechnol Biochem. 2016;80:774–778. doi: 10.1080/09168451.2015.1127132. [DOI] [PubMed] [Google Scholar]
- 15.Kim JH, Kim HY, Kang SY, Kim YH, Jin CH. Soluble epoxide hydrolase inhibitory activity of components isolated from Apios americana Medik. Molecules. 2017;22(1432) doi: 10.3390/molecules22091432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim JH, Kim HY, Kang SY, Kim JB, Kim YH, Jin CH. Chemical constituents from Apios americana and their inhibitory activity on tyrosinase. Molecules. 2018;23(232) doi: 10.3390/molecules23010232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iwai K, Matsue H. Ingestion of Apios americana Medikus tuber suppresses blood pressure and improves plasma lipids in spontaneously hypertensive rats. Nutr Res. 2007;27:218–224. [Google Scholar]
- 18.Gilbert ER, Liu D. Anti-diabetic functions of soy isoflavone genistein: Mechanisms underlying its effects on pancreatic β-cell function. Food Funct. 2013;4:200–212. doi: 10.1039/c2fo30199g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim HY, Kim JH, So Y, Kang SY, Jeong HG, Jin CH. Anti-inflammatory effect of lupinalbin A isolated from Apios americana on lipopolysaccharide-treated RAW264.7 cells. Molecules. 2018;23(583) doi: 10.3390/molecules23030583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bai Y, Xu Y, Chang J, Wang X, Zhao Y, Yu Z. Bioactives from stems and leaves of mung beans (Vigna radiata L.) J Funct Foods. 2016;25:314–322. [Google Scholar]
- 21.Yang D, Xie H, Jiang Y, Wei X. Phenolics from strawberry cv. Falandi and their antioxidant and α-glucosidase inhibitory activities. Food Chem. 2016;194:857–863. doi: 10.1016/j.foodchem.2015.08.091. [DOI] [PubMed] [Google Scholar]
- 22.Kim JH, Ryu YB, Lee WS, Kim YH. Neuraminidase inhibitory activities of quaternary isoquinoline alkaloids from Corydalis turtschaninovii rhizome. Bioorg Med Chem. 2014;22:6047–6052. doi: 10.1016/j.bmc.2014.09.004. [DOI] [PubMed] [Google Scholar]
- 23.Meng XY, Zhang HX, Mezei M, Cui M. Molecular docking: A powerful approach for structure-based drug discovery. Curr Comput Aided Drug Des. 2011;7:146–157. doi: 10.2174/157340911795677602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.de Ruyck J, Brysbaert G, Blossey R, Lensink MF. Molecular docking as a popular tool in drug design, an in silico travel. Adv Appl Bioinform Chem. 2016;9:1–11. doi: 10.2147/AABC.S105289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heitz MP, Rupp JW. Determining mushroom tyrosinase inhibition by imidazolium ionic liquids: A spectroscopic and molecular docking study. Int J Biol Macromol. 2018;107 (Pt B):1971–1981. doi: 10.1016/j.ijbiomac.2017.10.066. [DOI] [PubMed] [Google Scholar]
- 26.Seong SH, Ali MY, Kim HR, Jung HA, Choi JS. BACE1 inhibitory activity and molecular docking analysis of meroterpenoids from Sargassum serratifolium. Bioorg Med Chem. 2017;25:3964–3970. doi: 10.1016/j.bmc.2017.05.033. [DOI] [PubMed] [Google Scholar]
- 27.Fan J, Johnson MH, Lila MA, Yousef G, de Mejia EG. Berry and citrus phenolic compounds inhibit dipeptidyl peptidase IV: Implications in diabetes management. Evid Based Complement Alternat Med. 2013;2013(479505) doi: 10.1155/2013/479505. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.