Abstract
BRCA1/2 genes are the most frequently germline mutated DNA‐repair genes, and the survival of BRCA1/2 carriers has been extensively explored in breast cancer. However, the prevalence of germline mutations in non‐BRCA1/2 DNA‐repair genes and the survival of carriers are largely unknown in a large cohort of unselected breast cancer patients. Germline mutations in 16 DNA‐repair genes were determined using a multigene panel in 7657 BRCA1/2‐negative breast cancer patients who were unselected for family history of cancer or age at diagnosis. Among the 7657 BRCA1/2‐negative breast cancer patients, 257 (3.4%) carried at least 1 pathogenic germline mutation in the 16 DNA‐repair genes. The prevalence of DNA‐repair gene mutations was significantly higher in familial breast cancers (5.2%, P = 0.002) and early‐onset breast cancers (diagnosed at and before the age of 40) (4.5%, P = 0.003) than that of sporadic breast cancers (2.9%) (diagnosed above age of 40), respectively. The DNA‐repair gene mutation carriers were significantly more likely to have a larger tumor (P = 0.04) and axillary lymph node metastasis (P = 0.03). Moreover, DNA‐repair gene mutation was an independent unfavorable factor for recurrence‐free survival (adjusted hazard ratio [HR] = 1.38, 95% CI: 1.00‐1.91, P = 0.05) and disease‐specific survival (adjusted HR=1.63, 95% CI: 1.04‐2.57, P = 0.03) in this cohort. Overall, 3.4% of BRCA1/2‐negative breast cancer patients carried germline mutations in the 16 DNA‐repair genes, and the DNA‐repair gene mutation carriers exhibited an aggressive phenotype and had poor survival compared with noncarriers.
Keywords: breast cancer, cancer susceptibility genes, DNA‐repair genes, germline mutations, survival
In this study, we determined germline mutations in 16 DNA‐repair genes in a large cohort of 7657 BRCA1/2‐negative breast cancer patients. We observed that 3.4% of patients harbored at least 1 pathogenic mutation in DNA‐repair genes in this cohort; moreover, the DNA‐repair gene mutation carriers exhibited an aggressive phenotype and had poor survival compared with noncarriers.

1. INTRODUCTION
DNA repair is a complex process dealing with endogenous and environmental DNA damage; it relies on multiple DNA‐repair pathways, particularly the homologous recombination (HR) pathway. DNA‐repair genes involved in these pathways play a crucial role in the maintenance of genomic stability. Dysfunction of these pathways could, thus, induce genomic instability, which is one of the main forces driving cancer development.1 Most of the known breast cancer susceptibility genes are also key DNA‐repair genes,2 such as BRCA1, BRCA2, TP53, CHEK2, PALB2 and ATM. Germline mutations in DNA‐repair genes may confer a predisposition to breast cancer or other cancers.
Recent studies have indicated that germline mutations in DNA‐repair genes are associated with aggressive clinical behavior and poor prognosis in prostate cancer and pancreatic cancer.3, 4, 5 In breast cancer, the studies conducted by us and other groups have demonstrated that approximately 5.3% of patients carried a BRCA1 or BRCA2 (two key genes involved in the HR pathway) germline mutation in unselected cases and that BRCA1 carriers had worse survival than noncarriers.6, 7, 8 However, to the best of our knowledge, no study has so far investigated the prevalence of germline mutations in non‐BRCA1/2 DNA‐repair genes and the survival of carriers among a large cohort of unselected breast cancer patients.
In this study, we detected germline mutations in important DNA‐repair genes using a multigene panel in a large series of unselected breast cancer patients. Given that BRCA1/2 genes have been extensively explored, they were here excluded from the group of DNA‐repair genes to avoid their effects on prognosis potentially obscuring the effect of other genes.
We investigated the prevalence of germline mutations in 16 DNA‐repair genes in the entire cohort and in subgroups according to a family history of breast cancer, age at diagnosis or molecular subtype; compared the clinical and pathological characteristics of carriers with those of noncarriers; and finally, explored the association between the germline mutations in the 16 DNA‐repair genes and survival in terms of recurrence‐free survival (RFS) and disease‐specific survival (DSS).
2. MATERIALS AND METHODS
2.1. Patients
A total of 8085 consecutive breast cancer patients who were treated at the Breast Center of Peking University Cancer Hospital from October 2003 to May 2015 underwent 62‐gene panel sequencing, as described in our previous report.8 Among them, 428 patients who carried BRCA1/2 germline mutations were excluded. The remaining 7657 BRCA1/2‐negative cases, who were unselected for age at diagnosis and family history of breast cancer, were included in the study (Table S1). Familial breast cancer (FBC) is defined as breast cancer patients who had a family history of breast and/or ovarian cancer; early‐onset breast cancer (EBC) is defined as breast cancer in patients who did not have family history of breast and/or ovarian cancer and were diagnosed at or before the age of 40 years. This study was carried out in accordance with the ethical principles of the Declaration of Helsinki and approved by the Research and Ethics Committee of Peking University Cancer Hospital. Written informed consent was obtained from all participants.
2.2. Panel‐based sequencing assay
The target sequences of genomic DNA extracted from peripheral blood were captured using the SeqCap EZ hybridization and purification kit (Roche). Target sequencing was designed to cover all coding regions and intron–exon boundaries of the 62‐cancer‐gene panel (including the 16 DNA‐repair genes), as described previously.8
2.3. Gene selection and variant classification
Sixteen DNA‐repair genes were selected from the 62‐gene panel based on their roles in the DNA‐repair process and carcinogenesis of breast cancer.8 They play critical roles in DNA damage repair or signaling pathways, based on published literature and Biosystem Databases1, 9 (Table 1).
Table 1.
Sixteen key DNA‐repair genes in DNA damage repair and signaling pathways
| DSR | SSR | Checkpoint | p53 pathway | |||
|---|---|---|---|---|---|---|
| HR | FA | NHEJ | MMR | BER | ||
| RAD51D | FANCC | MER11A | MLH1 | MUTHY | ATM | TP53 |
| PALB2 | RAD51C | NBN | MSH2 | CHEK2 | ||
| BLM | RAD50 | MSH6 | ||||
| PMS2 | ||||||
BER, base excision repair; DSR, double‐strand repair; FA, Fanconi anemia; HR, homologous recombination; MMR, mismatch repair; NHEJ, nonhomologous end joining; SSR, single‐strand repair.
Criteria for classifying variants as described elsewhere were used.8 Variants classified as being pathogenic or likely pathogenic were considered as pathogenic in this study. If a patient carried BRCA1/2 and DNA‐repair gene mutations simultaneously, she was classified as a BRCA1/2 carrier and not included in this analysis.
2.4. Statistical analysis
Categorical variables were compared between mutation carriers and noncarriers using the χ2‐test or Fisher's exact test, where appropriate. Recurrence‐free survival (RFS) was defined as from the time of diagnosis to first recurrence (local or distant), or death from breast cancer (for patients without a record of recurrence), or the date of the last follow up. Disease‐specific survival (DSS) was defined as from the time of breast cancer diagnosis until the date of death from breast cancer or the date of the last follow up. Detailed survival data of each patient were collected from medical records and/or telephone interviews. Death from breast cancer was defined as death from organ failure caused by breast cancer metastasis. Survival was estimated using the Kaplan‐Meier method and differences were tested for statistical significance using the log‐rank test. A Cox proportional hazards model was used to determine whether a factor was associated with survival. Two‐sided P‐values less than 0.05 were considered to be statistically significant. All analyses were performed using SPSS 20.0 software.
3. RESULTS
3.1. Prevalence of germline mutations in DNA‐repair genes
A total of 263 pathogenic germline mutations were identified in 257 (3.4%) of the 7657 BRCA1/2‐negative breast cancer patients (Table 1). Most of the carriers had only 1 mutation, while 6 carriers had 2 mutations simultaneously (Data S1: Table S2). Among these, PALB2 (n = 54), TP53 (n = 36), ATM (n = 31), RAD51D (n = 30), CHEK2 (n = 27), RAD50 (n = 26), ERCC2 (n = 12), BLM (n = 11) and MLH1 (n = 10) were the most frequently mutated genes (Table S3).
The germline mutation rates of the 16 DNA‐repair genes were as follows: 5.2% (34/659) in FBC, 4.5% (45/1241) in EBC and 2.9% (167/5757) in SBC. There were significantly higher prevalence rates of carriers in the FBC group (5.2% vs 2.9%, P = 0.002) and in the EBC group (4.5% vs 2.9%, P = 0.003) compared with that in the SBC group (Table 2).
Table 2.
Prevalence of germline mutations in 16 DNA‐repair genes in this cohort
| Group | Number of patients | Mutation cases | Prevalence (%) | P a |
|---|---|---|---|---|
| All patients | 7657 | 257 | 3.4 | |
| FBC | 659 | 34 | 5.2 | 0.002 |
| EBC | 1241 | 56 | 4.5 | 0.003 |
| SBC | 5757 | 167 | 2.9 |
EBC, early‐onset breast cancer (diagnosed at and before age 40); FBC, familial breast cancer; SBC, sporadic breast cancer.
avs SBC.
We further analyzed the mutation rates in molecular subgroups based on estrogen receptor (ER), progesterone receptor (PR) and human epidermal growth factor receptor (HER2) status. No significant differences in mutation rates were observed among the four molecular subgroups (P = 0.16) (Table S4).
3.2. DNA‐repair gene mutations and clinicopathological characteristics
The average age of breast cancer onset in DNA‐repair gene mutation carriers was significantly younger than that in non‐carriers (mean age ± SD: 48.2 ± 12.0 vs 51.4 ± 11.6, P < 0.001) (Table 3). In addition, the carriers were more likely to have a positive family history of any cancer (38.1% vs 31.4%, P = 0.02), especially breast and/or ovarian cancer family history (13.2% vs 8.4%, P = 0.007), compared with non‐carriers (Table 3).
Table 3.
Comparison of clinicopathological characteristics between DNA‐repair germline mutation carriers and non‐carriers in this cohort
| Characteristics | 16 DNA‐repair gene mutation carriers | Non‐carriers | P | ||
|---|---|---|---|---|---|
| N | % | N | % | ||
| Total | 257 | 7400 | |||
| Age at diagnosis, years | |||||
| Mean ± SD | 48.2 ± 12.0 | 51.4 ± 11.6 | <0.001 | ||
| Median (range) | 7 (22‐83) | 50 (19‐98) | |||
| ≤40 y | 65 | 25.3 | 1276 | 17.2 | 0.001 |
| >40 y | 192 | 74.7 | 6124 | 82.8 | |
| Family history of breast and/or ovarian cancer | |||||
| No | 223 | 86.8 | 6775 | 91.6 | 0.007 |
| Yes | 34 | 13.2 | 625 | 8.4 | |
| Family history of any cancer | |||||
| No | 159 | 61.9 | 5075 | 68.6 | 0.02 |
| Yes | 98 | 38.1 | 2325 | 31.4 | |
| Tumor size | |||||
| ≤2 cm | 95 | 37.4 | 3208 | 43.9 | 0.04 |
| >2 cm | 159 | 62.6 | 4097 | 56.1 | |
| Unknown | 3 | 95 | |||
| Tumor type | |||||
| IDC | 234 | 91.1 | 6597 | 89.1 | 0.35 |
| ILC | 8 | 3.1 | 232 | 3.1 | |
| Medullary | 3 | 1.2 | 36 | 0.5 | |
| Mucinous | 3 | 1.2 | 158 | 2.1 | |
| Others | 9 | 3.5 | 377 | 5.1 | |
| Tumor grade | |||||
| I | 19 | 9.5 | 599 | 10.6 | 0.71 |
| II | 154 | 76.6 | 4188 | 74.0 | |
| III | 28 | 13.9 | 873 | 15.4 | |
| Unknown | 56 | 1740 | |||
| Lymph node status | |||||
| Negative | 168 | 66.7 | 5270 | 73.0 | 0.03 |
| Positive | 84 | 33.3 | 1953 | 27.0 | |
| Unknown | 5 | 177 | |||
| ER status | |||||
| Negative | 67 | 26.8 | 1997 | 28.2 | 0.63 |
| Positive | 183 | 73.2 | 5081 | 71.8 | |
| Unknown | 7 | 322 | |||
| PR status | |||||
| Negative | 78 | 32.4 | 2479 | 35.6 | 0.31 |
| Positive | 163 | 67.6 | 4491 | 64.4 | |
| Unknown | 16 | 430 | |||
| HER2 status | |||||
| Negative | 190 | 78.5 | 5019 | 74.1 | 0.12 |
| Positive | 52 | 21.5 | 1754 | 25.9 | |
| Unknown | 15 | 627 | |||
| Bilateral breast cancer | |||||
| No | 247 | 96.1 | 7216 | 97.5 | 0.16 |
| Yes | 10 | 3.9 | 184 | 2.5 | |
ER, estrogen receptor; HER2, human epidermal growth factor receptor 2; IDC, invasive ductal carcinoma; ILC, invasive lobular carcinoma; PR, progesterone receptor; TNBC, triple negative breast cancer.
In terms of the surgical characteristics of the tumor, the DNA‐repair gene mutation carriers were significantly more likely to have a larger tumor (62.6% vs 56.1%, P = 0.04) and lymph node metastasis (33.3% vs 27.0%, P = 0.03) compared with non‐carriers (Table 3).
3.3. DNA‐repair gene mutations and survival
In this cohort of 7657 BRCA1/2‐negative breast cancer patients, 104 patients with stage IV disease when diagnosed with breast cancer and 71 patients who were lost to follow up were excluded from the survival analysis. Thus, the remaining 7482 (97.7%) primary operable breast cancer patients were analyzed for survival. After the median follow‐up time of 65.2 months (range 1.0‐201.1 months), 887 patients experienced recurrence (local or distant) or died of breast cancer.
Carriers of germline mutations in DNA‐repair genes had significantly worse RFS (unadjusted HR = 1.60, 95% CI: 1.18‐2.15, P = 0.002) and DSS (unadjusted HR = 1.82, 95% CI: 1.19‐2.77, P = 0.005) compared with non‐carriers (Figure 1). Furthermore, DNA‐repair gene germline mutation was a significantly independent unfavorable factor for RFS (adjusted HR = 1.38, 95% CI: 1.00‐1.91, P = 0.05) and DSS (adjusted HR = 1.63, 95% CI: 1.04‐2.57, P = 0.03) after adjustment for age, tumor size, lymph node, tumor grade, ER/PR/HER2 status and treatment (Table 4).
Figure 1.
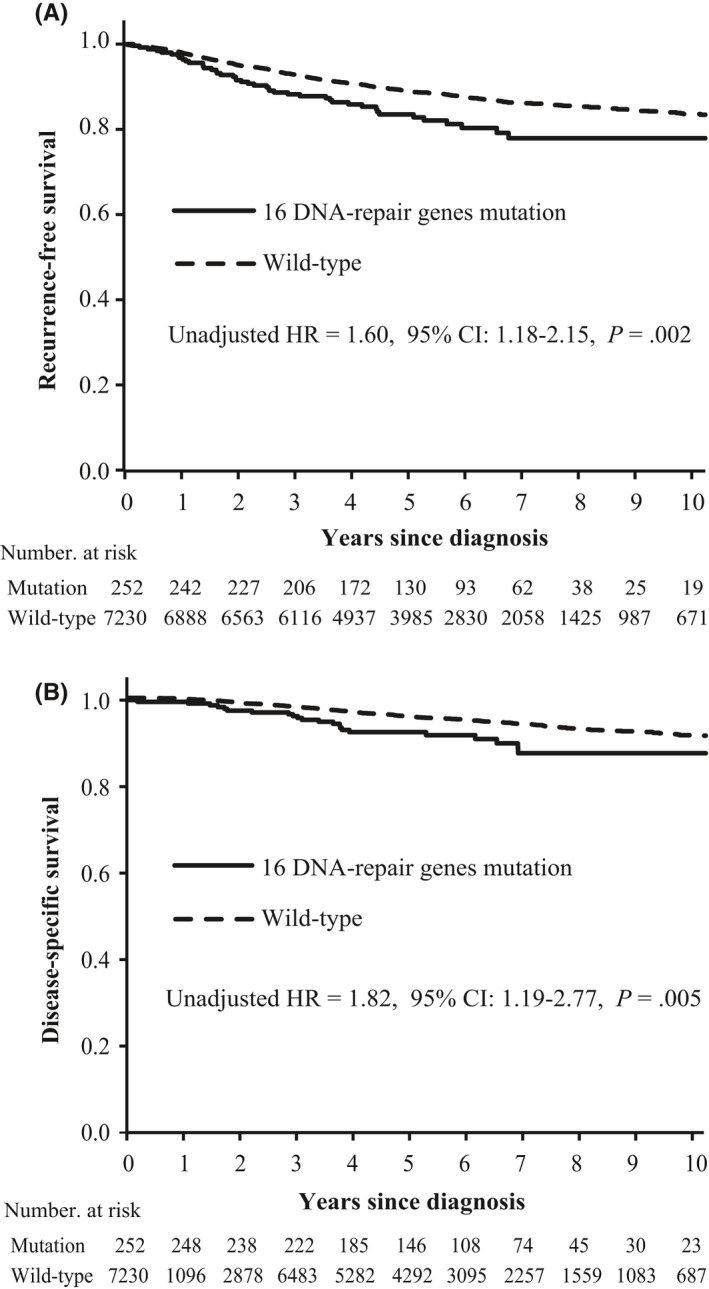
Survival analyses by Kaplan‐Meier according to germline mutation status in 16 DNA‐repair genes. Panel A and B show recurrence‐free survival and disease‐specific survival, respectively
Table 4.
Multivariate analyses of recurrence‐free survival and disease‐specific survival in this cohort
| Variable | RFS | DSS | ||
|---|---|---|---|---|
| HR (95% CI) | P | HR (95% CI) | P | |
| Age | ||||
| ≤40 y | 1.00 | ‐ | 1.00 | ‐ |
| >40 y | 0.80 (0.67‐0.94) | 0.009 | 1.25 (0.93‐1.68) | 0.13 |
| Tumor size | ||||
| ≤2 cm | 1.00 | ‐ | 1.00 | ‐ |
| >2 cm | 1.55 (1.32‐1.82) | <0.001 | 1.74 (1.36‐2.22) | <0.001 |
| Lymph node | ||||
| Negative | 1.00 | ‐ | 1.00 | ‐ |
| Positive | 3.69 (3.18‐4.29) | <0.001 | 5.08 (4.05‐6.37) | <0.001 |
| Grade | ||||
| I | 1.00 | ‐ | 1.00 | ‐ |
| II | 1.37 (1.06‐1.78) | 0.02 | 1.94 (1.23‐3.08) | 0.005 |
| III | 1.49 (1.10‐2.03) | 0.01 | 2.18 (1.30‐3.65) | 0.003 |
| ER status | ||||
| Negative | 1.00 | ‐ | 1.00 | ‐ |
| Positive | 0.72 (0.56‐0.91) | 0.006 | 0.75 (0.45‐1.05) | 0.09 |
| PR status | ||||
| Negative | 1.00 | ‐ | 1.00 | ‐ |
| Positive | 0.73 (0.59‐0.90) | 0.003 | 0.51 (0.38‐0.70) | <0.001 |
| HER2 status | ||||
| Negative | 1.00 | ‐ | 1.00 | ‐ |
| Positive | 1.12 (0.95‐1.31) | 0.18 | 1.13 (0.90‐1.43) | 0.30 |
| Treatment | ||||
| No treatment | 1.00 | ‐ | 1.00 | ‐ |
| C vs no treatment | 1.11 (0.85‐1.47) | 0.44 | 0.83 (0.56‐1.24) | 0.36 |
| E vs no treatment | 1.28 (0.96‐1.70) | 0.09 | 0.97 (0.64‐1.47) | 0.87 |
| C + E vs no treatment | 1.17 (0.84‐1.63) | 0.35 | 0.83 (0.50‐1.37) | 0.46 |
| Mutation | ||||
| Non‐carriers | 1.00 | ‐ | 1.00 | ‐ |
| 16 DNA‐repair genes | 1.38 (1.00‐1.91) | 0.05 | 1.63 (1.04‐2.57) | 0.03 |
C, chemotherapy; CI, confidence interval; DSS, disease‐specific survival; E, endocrine therapy; ER, estrogen receptor; HER2, human epidermal growth factor receptor 2; HR, hazard ratio; PR, progesterone receptor; RFS, recurrence‐free survival.
We further stratified survival analyses for molecular subtypes based on ER, PR and HER2 status. DNA‐repair germline mutations were associated with worse RFS and DSS in the ER+ and/or PR+ subgroup (RFS: P = 0.01; DSS: P = 0.02) and ER−/PR−, HER2+ subgroup (RFS: P = 0.002; DSS: P = 0.04). However, no significant differences in survival were observed between carriers and non‐carriers in the ER−/PR−, HER2− subgroup (Figure S1). Among the 16 DNA‐repair genes, PALB2, ATM, NBN, CHEK2 and TP53 genes are associated with breast cancer risk according to NCCN guideline. No significant differences in survival were observed between patients with these 5 breast cancer susceptibility gene germline mutations and non‐carriers (RFS: unadjusted HR = 1.40, 95% CI: 0.92‐2.11, P = 0.12; DSS: unadjusted HR = 1.47, 95% CI: 0.81‐2.69, P = 0.21) (Figure S2), while germline mutations in 11 other DNA‐repair genes were associated with worse RFS (unadjusted HR = 1.88, 95% CI: 1.23‐2.87, P = 0.003) and DSS (unadjusted HR = 2.31, 95% CI: 1.30‐4.10, P = 0.004) (Figure S2).
4. DISCUSSION
In this study, we determined germline mutations in 16 DNA‐repair genes in a large cohort of 7657 BRCA1/2‐negative breast cancer patients. We observed that 3.4% of patients harbored at least 1 pathogenic mutation in DNA‐repair genes in this cohort; moreover, the DNA‐repair gene mutation carriers exhibited an aggressive phenotype and had poor survival compared with noncarriers.
These 16 DNA‐repair genes play critical roles in core DNA damage repair or signaling pathways, and all of them have been reported to be genes that confer susceptibility to breast or other cancers,10, 11, 12 suggesting the critical role of germline mutations in the 16 DNA‐repair genes in tumorigenesis. Despite mutations in each gene being rare, the overall mutation rate of these 16 DNA‐repair genes was 3.4% in unselected BRCA1/2‐negative cases, which is comparable to the BRCA2 mutation rate (3.5%).8 The carriers of breast cancer susceptibility genes had a trend of clustering in familial and early‐onset patients.13 Similarly, the mutation rates of these DNA‐repair genes in this study were higher in the FBC and EBC groups than in the SBC group. Given the huge number of sporadic breast cancer patients, this low mutation rate in SBC (2.9%) might nonetheless be important for screening. Thus, the screening of DNA‐repair gene mutations could be offered not only to familial and early‐onset breast cancer patients, but also to sporadic breast cancer patients as well.
We found that patients with mutations in these 16 DNA‐repair genes were more likely to have a larger tumor and lymph node metastasis. Moreover, DNA‐repair gene mutation was an independent unfavorable prognosis factor after adjustment for other factors. The associations between germline mutations in a single DNA‐repair gene and survival are reported in previous studies.14, 15, 16, 17 Breast cancer patients carrying PALB2 recurrent mutations tended to present an aggressive tumor phenotype and had worse breast cancer‐specific survival15 and overall survival.14 Similarly, recurrent mutations in CHEK2, a checkpoint factor in the DNA‐repair process, were associated with poor clinical outcome in breast cancer.16, 17 Besides breast cancer, germline mutations in DNA‐repair genes also predicted worse survival in pancreatic cancer patients and prostate cancer patients.4, 5 However, some studies showed that germline and somatic mutations in homologous recombination genes were associated with better survival in ovarian carcinomas.18, 19 This discrepancy in survival may be explained by differences in the cancer type, the selected gene panel, and the applied treatment strategies.20 Notably, survival analysis in this study was based on a large series cohort of breast cancer patients who were unselected for age at diagnosis and family history, and the effect of BRCA1/2 on prognosis has been excluded, so our results objectively reflected the association between germline mutations in DNA‐repair genes and survival in BRCA1/2‐negative breast cancer patients.
A phase II trial revealed that metastatic prostate cancer patients harbored germline mutations in DNA‐repair genes, which may render their tumors sensitive to poly (ADP‐ribose) polymerase (PARP) inhibitors.21 Furthermore, a recent phase III trial showed that PARP inhibitor provided a significant benefit over single‐agent chemotherapy in metastatic breast cancer patients with germline BRCA1/2 mutations.22 Studies in vitro also demonstrated that cell lines or xenograft tumors defective in non‐BRCA1/2 DNA‐repair genes were sensitive to PARP inhibitors.23 These findings suggested that breast cancer patients with germline mutations in DNA‐repair genes may benefit from PARP inhibitor therapy, indicating that carriers of mutations in DNA‐repair genes could be enrolled in future clinical trials for treatment with PARP inhibitors.
There are some limitations in this study. First, the cohort was hospital‐based, so potential selection bias could not be avoided. Second, the panel‐based sequencing cannot identify large structural variants, so the frequency of pathogenic mutations would have been underestimated here.
In summary, we observed that 3.4% of BRCA1/2‐negative breast cancer patients harbored at least 1 pathogenic mutation in 16 DNA‐repair genes. Carriers of mutations in DNA‐repair genes had poor survival and were more likely to have a larger tumor and lymph node metastasis. Given that the multigene assay has become a routine method, our findings may have potential clinical implications and suggest that carriers of DNA‐repair gene mutations could be enrolled in future clinical trials for treatment with PARP inhibitors.
DISCLOSURE
The authors have no conflict of interest.
Supporting information
Fan Z, Hu L, Ouyang T, et al. Germline mutation in DNA‐repair genes is associated with poor survival in BRCA1/2‐negative breast cancer patients. Cancer Sci. 2019;110:3368‐3374. 10.1111/cas.14175
Zhenhua Fan and Li Hu contributed equally to this work.
Contributor Information
Ye Xu, Email: xuye@bjmu.edu.cn.
Yuntao Xie, Email: zlxyt2@bjmu.edu.cn.
REFERENCES
- 1. Jeggo PA, Pearl LH, Carr AM. DNA repair, genome stability and cancer: a historical perspective. Nat Rev Cancer. 2016;16:35‐42. [DOI] [PubMed] [Google Scholar]
- 2. Nielsen FC, van Overeem Hansen T, Sorensen CS. Hereditary breast and ovarian cancer: new genes in confined pathways. Nat Rev Cancer. 2016;16:599‐612. [DOI] [PubMed] [Google Scholar]
- 3. Leongamornlert D, Saunders E, Dadaev T, et al. Frequent germline deleterious mutations in DNA repair genes in familial prostate cancer cases are associated with advanced disease. Br J Cancer. 2014;110:1663‐1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Smith AL, Alirezaie N, Connor A, et al. Candidate DNA repair susceptibility genes identified by exome sequencing in high‐risk pancreatic cancer. Cancer Lett. 2016;370:302‐312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Castro E, Romero‐Laorden N, Del Pozo A, et al. PROREPAIR‐B: a prospective cohort study of the impact of germline DNA repair mutations on the outcomes of patients with metastatic castration‐resistant prostate cancer. J Clin Oncol. 2019;37:490‐503. [DOI] [PubMed] [Google Scholar]
- 6. Robson ME, Chappuis PO, Satagopan J, et al. A combined analysis of outcome following breast cancer: differences in survival based on BRCA1/BRCA2 mutation status and administration of adjuvant treatment. Breast Cancer Res. 2004;6:R8‐R17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Foulkes WD, Chappuis PO, Wong N, et al. Primary node negative breast cancer in BRCA1 mutation carriers has a poor outcome. Ann Oncol. 2000;11:307‐313. [DOI] [PubMed] [Google Scholar]
- 8. Sun J, Meng H, Yao L, et al. Germline mutations in cancer susceptibility genes in a large series of unselected breast cancer patients. Clin Cancer Res. 2017;23:6113‐6119. [DOI] [PubMed] [Google Scholar]
- 9. Curtin NJ. DNA repair dysregulation from cancer driver to therapeutic target. Nat Rev Cancer. 2012;12:801‐817. [DOI] [PubMed] [Google Scholar]
- 10. Graffeo R, Livraghi L, Pagani O, Goldhirsch A, Partridge AH, Garber JE. Time to incorporate germline multigene panel testing into breast and ovarian cancer patient care. Breast Cancer Res Treat. 2016;160:393‐410. [DOI] [PubMed] [Google Scholar]
- 11. Tung N, Battelli C, Allen B, et al. Frequency of mutations in individuals with breast cancer referred for BRCA1 and BRCA2 testing using next‐generation sequencing with a 25‐gene panel. Cancer. 2015;121:25‐33. [DOI] [PubMed] [Google Scholar]
- 12. Thompson ER, Rowley SM, Li N, et al. Panel testing for familial breast cancer: calibrating the tension between research and clinical care. J Clin Oncol. 2016;34:1455‐1459. [DOI] [PubMed] [Google Scholar]
- 13. Tung N, Lin NU, Kidd J, et al. Frequency of germline mutations in 25 cancer susceptibility genes in a sequential series of patients with breast cancer. J Clin Oncol. 2016;34:1460‐1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cybulski C, Kluzniak W, Huzarski T, et al. Clinical outcomes in women with breast cancer and a PALB2 mutation: a prospective cohort analysis. Lancet Oncol. 2015;16:638‐644. [DOI] [PubMed] [Google Scholar]
- 15. Heikkinen T, Karkkainen H, Aaltonen K, et al. The breast cancer susceptibility mutation PALB2 1592delT is associated with an aggressive tumor phenotype. Clin Cancer Res. 2009;15:3214‐3222. [DOI] [PubMed] [Google Scholar]
- 16. Weischer M, Nordestgaard BG, Pharoah P, et al. CHEK2*1100delC heterozygosity in women with breast cancer associated with early death, breast cancer‐specific death, and increased risk of a second breast cancer. J Clin Oncol. 2012;30:4308‐4316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Schmidt MK, Tollenaar RA, de Kemp SR, et al. Breast cancer survival and tumor characteristics in premenopausal women carrying the CHEK2*1100delC germline mutation. J Clin Oncol. 2007;25:64‐69. [DOI] [PubMed] [Google Scholar]
- 18. Pennington KP, Walsh T, Harrell MI, et al. Germline and somatic mutations in homologous recombination genes predict platinum response and survival in ovarian, fallopian tube, and peritoneal carcinomas. Clin Cancer Res. 2014;20:764‐775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Garsed DW, Alsop K, Fereday S, et al. Homologous recombination DNA repair pathway disruption and retinoblastoma protein loss are associated with exceptional survival in high‐grade serous ovarian cancer. Clin Cancer Res. 2018;24:569‐580. [DOI] [PubMed] [Google Scholar]
- 20. Zhong Q, Peng HL, Zhao X, Zhang L, Hwang WT. Effects of BRCA1‐ and BRCA2‐related mutations on ovarian and breast cancer survival: a meta‐analysis. Clin Cancer Res. 2015;21:211‐220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mateo J, Carreira S, Sandhu S, et al. DNA‐repair defects and olaparib in metastatic prostate cancer. N Engl J Med. 2015;373:1697‐1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Robson M, Im SA, Senkus E, et al. Olaparib for metastatic breast cancer in patients with a germline BRCA mutation. N Engl J Med. 2017;377:523‐533. [DOI] [PubMed] [Google Scholar]
- 23. McCabe N, Turner NC, Lord CJ, et al. Deficiency in the repair of DNA damage by homologous recombination and sensitivity to poly(ADP‐ribose) polymerase inhibition. Can Res. 2006;66:8109‐8115. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


