Abstract
Background
Age-related changes in adaptive and innate immune cells have been associated with a decline in effective immunity and chronic, low-grade inflammation. Epigenetic, transcriptional, and functional changes in monocytes occur with aging, though most studies to date have focused on differences between young adults and the elderly in populations with European ancestry; few data exist regarding changes that occur in circulating monocytes during the first few decades of life or in African populations. We analyzed DNA methylation profiles, cytokine production, and inflammatory gene expression profiles in monocytes from young adults and children from western Kenya.
Results
We identified several hypo- and hyper-methylated CpG sites in monocytes from Kenyan young adults vs. children that replicated findings in the current literature of differential DNA methylation in monocytes from elderly persons vs. young adults across diverse populations. Differentially methylated CpG sites were also noted in gene regions important to inflammation and innate immune responses. Monocytes from Kenyan young adults vs. children displayed increased production of IL-8, IL-10, and IL-12p70 in response to TLR4 and TLR2/1 stimulation as well as distinct inflammatory gene expression profiles.
Conclusions
These findings complement previous reports of age-related methylation changes in isolated monocytes and provide novel insights into the role of age-associated changes in innate immune functions.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12979-021-00223-2.
Keywords: Monocyte, DNA methylation, Epigenetic, Ageing, Aging, Innate immune
Background
Development of the human immune system is a highly dynamic process influenced by genetic and environmental factors, including early-life exposures which can impact long-term risk for immune-mediated diseases [1–3]. Susceptibility to infection varies according to age, with newborns and the elderly generally at the highest risk [4, 5]. The effects of an aging immune system are well characterized in the elderly (aged 65 years or older), which include weakened responses to pathogens and vaccines (immunosenescence) and chronic, low-grade inflammation (inflamm-aging) [6–8].
Monocytes are innate immune cells important to phagocytosis, inflammatory cytokine production, antigen presentation, and tissue repair, and play a central role in dysregulated innate immune responses that characterize inflamm-aging [9]. Age-related changes in monocytes from older vs. young adults include several altered gene expression programs, including those linked to metabolic homeostasis, antigen presentation, authophagy, and protein synthesis [10–13]. Cytokine production in response to pattern recognition receptor (PRR) engagement is dysregulated in monocytes from older vs. young adults. Studies have shown increased TNF and decreased CCL20, IFN-γ, and IL-1β production in response to LPS treatment in elderly monocytes [11, 14, 15]. Additional studies have shown decreased type I IFN production after stimulation with 5’pppRNA and influenza [11, 16]; decreased IL-1β and IFN-γ with CL097 stimulation [11]; and decreased IL-6 and TNF with Pam3Cys stimulation [17]. Other age-associated changes in elderly monocytes include decreased phagocytosis and altered cellular metabolism, including reduced mitochondrial capacity and increased glucose consumption [10, 14].
The development, differentiation, and functions of immune cells are regulated by dynamic epigenetic modifications, including histone modifications and DNA methylation of cytosines in CpG dinucleotides [18]. DNA methylation patterns are influenced by genetic sequence and a host of environmental factors, including nutrition and infectious exposures [19]. Age-related DNA methylation patterns have been well described in multiple tissue types in adults, and methylation patterns at age-associated CpG sites have been analyzed to produce highly accurate and precise estimates of chronological age in older age groups [20, 21]. DNA methylation changes associated with aging include genomic regions with increased methylation with older age (hyper-methylation) as well as regions with decreased methylation with older age (hypo-methylation) [13, 22–25]. Previous studies of both children and adults have shown that CpG sites that are hypo-methylated with age are located in CpG island shores and outside of CpG islands [24, 26, 27]. Several studies in adult populations have shown that CpG sites that are hyper-methylated with age are enriched in CpG islands [13, 26–28], although a study in children did not find enrichment of age-associated hyper-methylated sites within CpG islands [24]. There are limited data available regarding age-associated DNA methylation changes in pediatric populations, although existing evidence suggests that age-related methylation changes in peripheral blood occur more rapidly in childhood than adulthood, and that changes in many age-associated CpG sites follow a logarithmic rather than linear lifelong trend [24].
Most studies of age-related DNA methylation changes have used whole blood or other mixed cell samples, and few have studied methylation changes in purified monocyte samples [10, 12, 13, 29–31]. Studies of age-associated methylation patterns in purified T cells and monocytes from the same individuals showed distinct methylation changes in T cells vs. monocytes and emphasize the importance of studying isolated cell populations [12, 13]. Additionally, it is important to take into account the influence that genetic factors and gene-environment interactions have on the aging epigenome, yet the majority of studies on age-related methylation markers have been performed in populations with European ancestry, and few have been performed in African populations [32, 33]. Here, we aimed to analyze DNA methylation profiles and innate immune phenotypes and functions of purified monocytes from healthy individuals living in western Kenya, comparing children aged 1–9 years to adults aged 19–35 years. Participants for this study were enrolled in a larger observational cohort study of naturally acquired immunity to malaria [34, 35]. Malaria transmission in this area is perennially high, and malaria-related morbidity and mortality is highest among children under 5 years [36, 37]. By young adulthood, individuals living in this region develop clinical immunity in which they are protected against severe malarial disease but occasionally have episodes of uncomplicated malaria as well as subclinical parasitemia. Findings from this study reveal age-related differences in monocyte DNA methylation, innate immune responses, and inflammatory gene expression patterns that provide insights into differential age-related risks for both infectious and non-infectious diseases in populations living in malaria-endemic areas. Monocytes from healthy malaria-naïve adults from the United States were examined as historic controls to highlight the important considerations of genetic and ecologic diversity in studies of the developing and aging immune system.
Results
Age-related differences in monocyte DNA methylation in Kenyan adults vs. children
We analyzed genome-wide DNA methylation profiles in negatively selected monocytes from 8 healthy children (aged 3.6–9.3 years) and 8 healthy young adults (aged 26–30 years) from western Kenya (Additional file 1: Table S1). Methylation profiles were determined using the Infinium MethylationEPIC bead array, which interrogates methylation at > 850,000 sites across the genome at single-nucleotide resolution. In the analysis of differentially methylated positions (DMPs) and differentially methylated regions (DMRs), we filtered out probe sites located at known single nucleotide polymorphisms (SNPs) and those on the X and Y chromosomes. Eighteen percent of children in the overall cohort had asymptomatic Pf parasitemia (a common finding in this malaria endemic region), so we included sex and asymptomatic Pf parasitemia status as covariates to adjust for potential confounding effects.
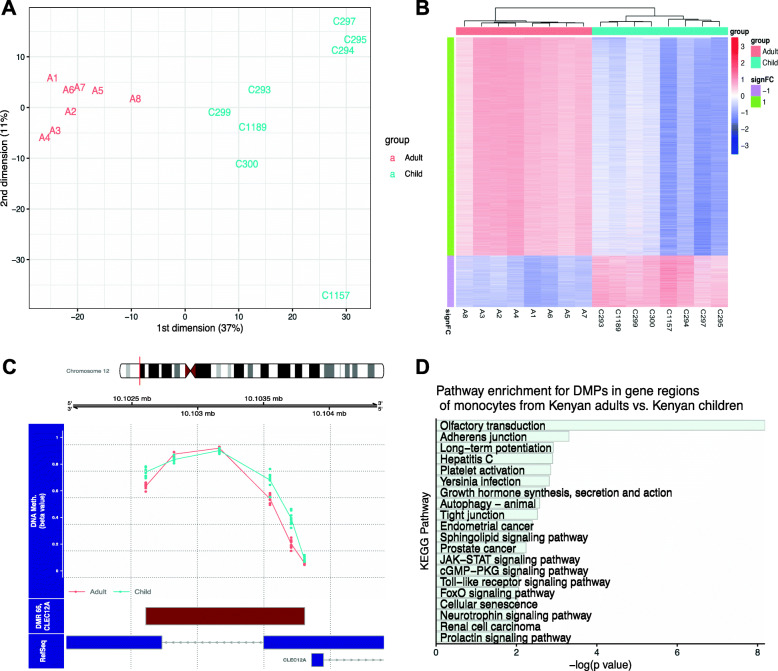
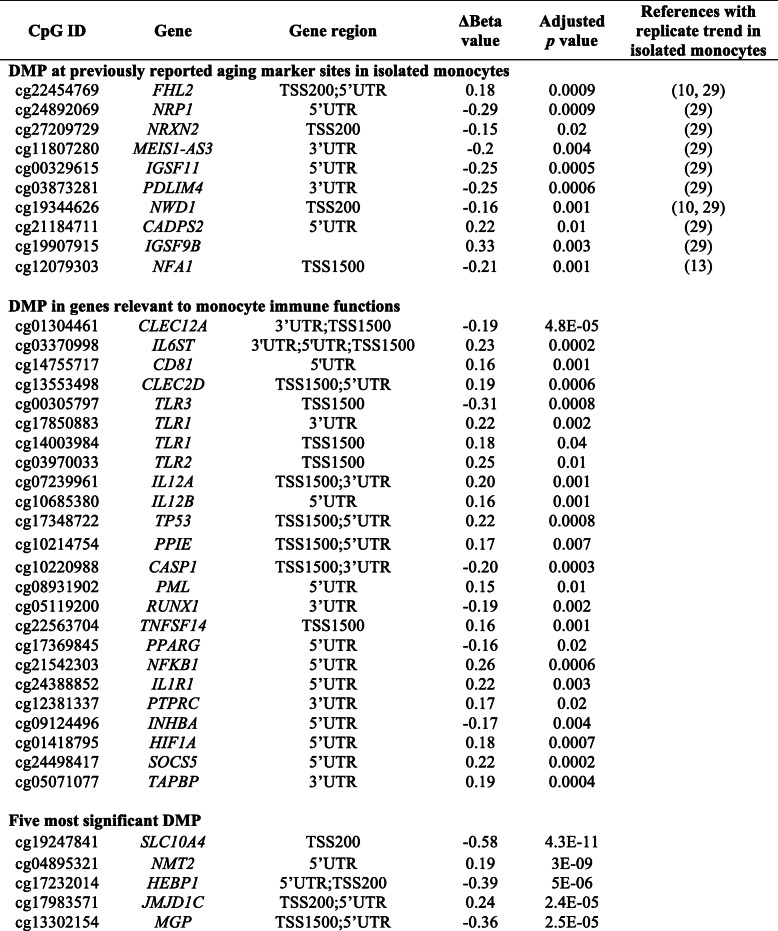
Principal components analysis (PCA) showed that principal component 1 (PC1) accounted for 37% of variance in the data and was significantly associated with age group (p = 0.0008) (Fig. 1a). Using a threshold for absolute beta value difference of 0.15, we identified 14,259 differentially methylated CpG sites, with 11,535 sites hyper-methylated in adults vs. children and 2724 hypo-methylated in adults vs. children (P < 0.01, FDR-adjusted P < 0.05) (Fig. 1b; Additional file 2). Table 1 highlights DMPs in our study that replicate findings in the literature of age-related DMPs in isolated monocytes, DMPs that are relevant to monocyte immune functions, and the top five most significant DMPs.
Fig. 1.
Age-related differences in monocyte DNA methylation in Kenyan adults and children. Monocytes were negatively selected from fresh venous blood samples from healthy Kenyan children (n = 8) and adults (n = 8). a Principal components analysis (PCA) plot of DNA methylation results for Kenyan adult (red) and child (blue) monocytes. b Heatmap of DNA methylation results showing differentially methylated positions between Kenyan adult and child monocytes, includes all CpG probe sites meeting differential methylation criteria (absolute beta value difference > 0.15, p < 0.01, and FDR-adjusted p < 0.05). c Plot of differentially methylated region in the CLEC12A gene locus, showing beta values for Kenyan adult (red) and child (blue) monocytes at 6 CpG probe sites within the region. d KEGG pathway enrichment analysis of genes associated with differentially methylated positions in monocytes from Kenyan adults vs. children, showing the most significantly enriched pathways
Table 1.
Differentially methylated positions in monocytes from Kenyan adults vs. children
We found DMPs at CpG sites previously reported to have age-associated differential methylation in isolated monocytes [10, 13, 29] as well as whole blood [20, 21, 33] (Table 1; Additional file 2). For example, we found hyper-methylation in adults vs. children at the site cg22454769 in the FHL2 locus, and similar findings of hyper-methylation with older age have been reported in whole blood samples [20, 21] as well as in studies of isolated monocytes by Tserel et al. [29] and Saare et al. [10]. Several DMPs in our study showed similar patterns in adults vs. children as those found by Tserel et al. in isolated monocytes from old vs. young adults [29], including hypo-methylation at sites cg24892069 in the NRP1 locus and cg27209729 in the NRXN2 locus. We also found hypo-methylation at the site cg18334392 in the SNORD123 locus (Δbeta = − 0.2, adj. p = 0.03), and both DMPs in the NRXN2 and SNORD123 loci were previously identified in an epigenome wide association study (EWAS) meta-analysis (using samples from multiple tissue types) of an African hunter-gatherer group and also hypo-methylated with older age [33]. Well-known CpG sites in the ELOVL2 promoter are associated with hyper-methylation with increasing age [20, 38], and this correlation has been consistent across tissue types [39] and populations [33]. In our study, three CpG sites in ELOVL2 followed this trend with hyper-methylation in adults vs. children but did not meet our 0.15 beta-difference threshold (cg16867657, Δbeta = 0.12, adj. p = 0.06; cg07901130, Δbeta = 0.14, adj. p = 0.01; cg08683008, Δbeta = 0.06, adj. p = 0.01). We found that CpG probe sites that were hypo-methylated in adults vs. children were more likely to be located in CpG shores (22.2% of hypo-methylated sites were in CpG shores, compared to assay coverage of 17.8%, p < 0.0001), while CpG probe sites that were hyper-methylated in adults vs. children were not enriched in CpG islands.
Several DMPs were contained in gene regions important to monocyte inflammatory and immune responses (Table 1; Additional file 2). These include CpG sites in genes for PRRs (CLEC12A, CLEC2D, TLR2, TLR1, TLR3), inflammasome and cytokine signaling (CASP1, IL6ST, IL12A, IL12B, IL1R1), and downstream signaling molecules and transcription factors (TP53, PPARG, NFKB1, PTPRC, HIF1A). In addition, several DMRs (regions containing multiple DMPs) were in gene regions relevant to immune function, including CLEC12A (6 CpG sites, combined p = 1.3E-07) (Fig. 1c), IL6ST (3 CpGs, p = 1.8E-08), TLR1 (9 CpGs, p = 0.01), TLR3 (8 CpGs, p = 0.0008), and IL12A (15 CpGs, p = 0.02) (Additional file 3).
We performed enrichment analysis of DMPs in gene regions of monocytes from Kenyan adults vs. children for Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways, which was notable for enrichment in several important signaling pathways (including Sphingolipid, JAK-STAT, cGMP-PKG, Toll-like receptor, and FoxO signaling pathways), “Cellular senescence,” and several malignancy-related pathways (Fig. 1d; Additional file 4).
Monocyte subset proportions are similar in Kenyan adults and children
Circulating monocytes are a heterogeneous population that are classified into three subsets according to CD14 and CD16 expression (classical CD14++CD16−, intermediate CD14++CD16+, and nonclassical CD14+CD16++). The subsets have characteristic phenotypic, functional, transcriptomic, and epigenetic profiles [40, 41], including subset-specific miRNA expression [42] and DNA methylation profiles linked to distinct immunological processes [43]. Several studies have shown an increase in circulating CD16+ subsets in elderly vs. young adult monocytes [14, 15, 17], though others have shown no difference in proportions of subsets between elderly and young adults [11]. To determine if there were baseline differences in proportions of monocytes subsets in Kenyan young adults vs. children, we performed flow cytometry to calculate percentages of classical, intermediate, and nonclassical subsets in PBMC samples from 17 healthy children (aged 1.2–9.6 years) and 14 healthy adults (aged 18–35 years). We found no differences between the two groups in proportions of the three subsets (Additional file 1: Fig. S1).
Increased TLR4 and TLR2/1 responsiveness in monocytes from Kenyan adults vs. children
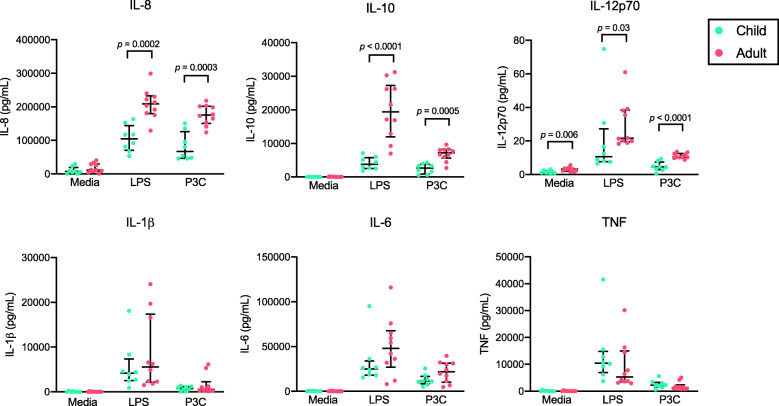
We analyzed monocyte cytokine production using negatively isolated monocytes from fresh venous blood samples obtained from 8 healthy children (aged 3.9–9.8 years) and 10 healthy adults (aged 26–30 years). Cells were cultured 18 h, and constitutive cytokine production (media alone) was compared to stimulation with a TLR4 agonist (LPS 10 ng/ml) and a TLR2/1 agonist (Pam3CSK4 [P3C] 100 ng/ml). Monocytes from adults showed increased production of IL-8, IL-10, and IL-12p70 in response to both TLR4 and TLR2/1 stimulation compared to monocytes from children (Fig. 2). Constitutive production of IL-12p70 was slightly higher in adult monocytes (median 3.2 pg/ml) vs. child monocytes (median 1.1 pg/ml) (p = 0.006). Production of IL-1β, IL-6, and TNF in response to TLR4 and TLR2/1 agonists was robust and equal in monocytes from Kenyan adults and children (Fig. 2).
Fig. 2.
Monocyte responsiveness to TLR4 and TLR2/1 stimulation in Kenyan adults and children. Monocytes were negatively selected from fresh venous blood samples from healthy Kenyan children (n = 8) and adults (n = 10). Cells were cultured for 18 h with media alone, 10 ng/ml LPS, or 100 ng/ml Pam3CSK4 (P3C), and cytokine concentrations were measured in culture supernatants. Mann Whitney U test was used to compare the groups. Data are shown as medians with interquartile ranges
Differential inflammatory gene expression in monocytes from Kenyan adults vs. children
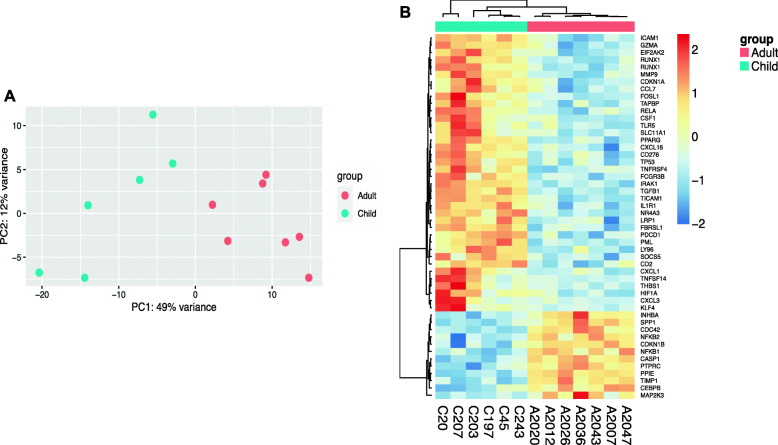
We examined inflammatory gene expression profiles of negatively isolated monocytes from 6 healthy Kenyan children (aged 3.3–4.9 years) and 7 healthy Kenyan adults (aged 28–33 years) using a targeted digital RNA sequencing panel, which measured expression of 508 genes involved in inflammation and immunity. PCA analysis showed separation of the two groups along the first principal component, which explained 49% of variance in the data (Fig. 3a). Of the 508 genes included in the panel, 178 were differentially expressed between adult and child monocytes (73 genes had increased expression in adult vs. child monocytes, 105 genes had decreased expression in adult vs. child monocytes, adj. p < 0.05) (Additional file 5). The top 50 differentially expressed genes are shown in Fig. 3b and include several of the genes in which differential methylation was observed, such as CASP1, IL1R1, TP53, PPARG, NFKB1, PTPRC, and HIF1A. Of the 508 inflammation-associated genes analyzed, 135 (26.6%) had differential methylation observed in promoter or gene body regions in Kenyan adults vs. children. The three most significantly enriched KEGG pathways for differential gene expression included “Cytokine-cytokine receptor interaction” (45 genes in list, FDR p = 1.1E-45), “Toll-like receptor signaling pathway” (27 genes in list, FDR p = 2.1E-32), and “IL-17 signaling pathway” (25 genes in list, FDR p = 1.5E-30). Of note, monocyte samples for DNA methylation and gene expression analyses were obtained from different individuals, so direct correlation between differential methylation and mRNA levels could not be performed.
Fig. 3.
Monocyte inflammatory gene expression profiles in Kenyan adults and children. Targeted digital RNA sequencing was performed on monocytes isolated from cryopreserved PBMCs via negative selection. A customized panel targeted 508 genes important in inflammation and immunity (QIAseq Human Inflammation and Immunity Transcriptome Panel, Qiagen). a Principal components analysis (PCA) plot of targeted digital RNA sequencing results for Kenyan adult (red) and child (blue) monocytes. b Heatmap of the top 50 most significantly differentially expressed genes between Kenyan adult and child monocytes
Differences in DNA methylation, immune phenotypes, and gene expression in monocytes from Kenyan adults vs. US adults
To examine the potential influence of differences in geographic location and ancestry on age-related differences in monocytes, we also compared monocyte DNA methylation, subset proportions, TLR responses, and inflammatory gene expression profiles of healthy Kenyan adults (median age 28.5 years, range 18–35) to healthy adults from the United States (median age 29 years, range 24–69) (Additional file 1: Table S1).
A PCA plot of monocyte DNA methylation profiles of Kenyan children, Kenyan adults, and US adults showed separation of the Kenyan child group (n = 8) from both groups of adults (n = 8 Kenyan adults and n = 8 US adults) along PC1 (Fig. 4a). There were a far greater number of DMPs between Kenyan children and US adults (20,919 DMPs) than between Kenyan adults and US adults (288 DMPs) (Additional files 6 and 7). Differential methylation at known aging marker sites was consistent when comparing Kenyan children to both Kenyan adults and US adults. For example, methylation at cg22454769 in FHL2 was hyper-methylated in both Kenyan and US adult groups compared to Kenyan children (average beta values 0.49, 0.49, and 0.32, respectively, adj. p = 9.3E-05), as was methylation at cg16867657 in ELOVL2 (average beta values 0.44, 0.51, and 0.32, respectively, adj. p = 0.02). KEGG pathway enrichment analysis of DMPs in gene regions of monocytes from Kenyan adults vs. US adults was notable for enrichment in several pathways related to immune function and metabolism (such as “RIG-I-like receptor signaling pathway,” “Type II diabetes mellitus,” and “Mannose type O-glycan biosynthesis”) (Fig. 4b; Additional file 8). These data suggest that age may be a larger driver of differential methylation in monocytes than geographic location or ancestry, though the differences in methylation profiles between Kenyan adults vs. US adults may be explained in part by differences in genetic background as well as cumulative environmental exposures, such as malaria.
Fig. 4.
Epigenetic, phenotypic, functional, and transcriptional differences in monocytes from Kenyan adults vs. US adults. a Principal components analysis (PCA) plot of DNA methylation results for Kenyan adult (red; n = 8), Kenyan child (green; n = 8), and US adult (blue; n = 8) monocytes. b KEGG pathway enrichment analysis of genes associated with differentially methylated positions in monocytes from Kenyan adults vs. US adults, showing the most significantly enriched pathways. c Proportions of all circulating monocytes that are classical, intermediate, and nonclassical for PBMC samples from Kenyan adults (n = 14) and US adults (n = 4). Mann Whitney U test was used to compare the groups. d Monocyte cytokine production in response to 18 h of TLR4 (LPS 10 ng/ml) and TLR2/1 (P3C 100 ng/ml) stimulation in Kenyan adults (n = 10) and US adults (n = 14). Monocytes were negatively selected from fresh venous blood samples. Mann Whitney U test was used to compare the groups. Data are shown as medians with interquartile ranges. e PCA plot of targeted digital RNA sequencing results for Kenyan adult (red; n = 7) and US adult (blue; n = 4) monocytes. f Heatmap of the top 35 most significantly differentially expressed genes between Kenyan adult (blue) and US adult (purple) monocytes
We compared proportions of circulating classical, intermediate, and nonclassical monocyte subsets in 14 Kenyan adults and 4 US adults, and found that Kenyan adults had overall greater variability, higher proportions of the CD14+CD16++ nonclassical subset, and lower proportions of the CD++CD16− classical subset (Fig. 4c). We then compared monocyte cytokine production in response to TLR2/1 and TLR4 stimulation in 10 Kenyan adults and 10 US adults. Monocytes from healthy Kenyan adults showed consistently higher TLR2/1 responses compared to monocytes from healthy US adults, with increased production of IL-8, IL-10, IL-12p70, IL-1β, IL-6, and TNF after stimulation with P3C (Fig. 4d). In addition, monocytes from Kenyan adults vs. US adults produced higher levels of IL-12p70 in response to TLR4 stimulation with LPS (Fig. 4d).
Using the targeted digital RNA sequencing panel of 508 genes involved in inflammation and immunity, we compared gene expression profiles of monocytes from 7 healthy Kenyan adults to 4 healthy US adults. PCA analysis showed separation of the two groups along the first principal component, which explained 46% of variance in the data (Fig. 4e). Of the 508 genes included in the panel, 121 were differentially expressed between Kenyan adult vs. US adult monocytes (80 genes had increased expression in Kenyan adult vs. US adult monocytes, 41 genes had decreased expression in Kenyan adult vs. US adult monocytes, adj. p < 0.05) (Additional file 9). The top 35 differentially expressed genes are shown in the heatmap in Fig. 4f, including upregulation of several important cytokines, chemokines, and transcription factors. Of the 508 inflammation-associated genes analyzed, five (0.98%) had differential methylation observed in promoter or gene body regions in Kenyan adults vs. US adults. The three most significantly enriched KEGG pathways for differential gene expression included “Cytokine-cytokine receptor interaction” (32 genes in list, FDR p = 4.8E-31), “Hematopoietic cell lineage” (19 genes in list, FDR p = 3.4E-23), and “IL-17 signaling pathway” (17 genes in list, FDR p = 2.7E-20). Of note, the 17th most significantly enriched KEGG pathway was “Malaria” (FDR p = 4.6E-13) and included differential expression of the following genes in that pathway: CXCL8, IL6, CCL2, IL1A, IL1B, TNF, IFNG, VCAM1, ICAM1, CD40, and LRP1. Taken together these data on monocyte subset proportions, TLR responses, and gene expression profiles suggest a hyper-inflammatory state in Kenyan adult monocytes that is distinct from both Kenyan child monocytes as well as US adult monocytes, likely secondary to complex gene-environment interactions and their influence on the aging epigenome.
We examined the overlap in DMPs and differentially expressed genes between Kenyan adults vs. Kenyan children and Kenyan adults vs. US adults. We found 12 CpG sites that were differentially methylated in monocytes from Kenyan adults compared to both Kenyan children and US adults. There were 62 genes in the panel of 508 inflammatory genes that were differentially expressed in Kenyan adults compared to the two other groups. Of these 62 genes, 12 were higher in Kenyan adults and 8 were lower in Kenyan adults compared to both Kenyan children and US adults. The remaining 42 genes were expressed at an intermediate level in Kenyan adults compared to Kenyan children and US adults. One of the 12 overlapping DMPs (cg23061725, located in the 5’UTR of CASP8; average beta values 0.57 in Kenyan adults, 0.34 in Kenyan children, and 0.36 in US adults) was noted alongside altered monocyte CASP8 gene expression among the three groups (average normalized expression values 286 in Kenyan adults, 181 in Kenyan children, and 495 in US adults). CASP8 encodes Caspase-8, a cysteine-aspartate protease involved in several cellular processes, including programmed cell death, autophagy, and inflammasome activation [44].
Discussion
In this study, we investigated genome-wide DNA methylation patterns of purified monocytes obtained from children and young adults living in a malaria-endemic area of Kenya. Our findings replicate several of the strongest markers for age-related changes in DNA methylation and identify differentially methylated CpG sites at gene loci relevant to innate immune responses. We also found that while monocyte subset proportions were similar between young adults and children, monocyte cytokine production in response to TLR agonists was increased in adults compared to children, and adult monocytes had a distinct upregulated inflammatory gene expression profile.
Several of the age-related changes in DNA methylation in our study occur at CpG sites that are part of the “epigenetic clock” and correlate with chronological aging across tissue and cell types, and have the potential to be used to predict disease states when this correlation is altered [20, 21]. In older adults, patterns for age-associated methylation changes at many CpG sites appear to follow a linear trend [21], however a meta-analysis of age-related DNA methylation changes in pediatric and adult cohorts revealed that changes in methylation occur at a three- to four-fold higher rate in children than in adults and that lifelong trends for most age-associated CpG sites are best modelled by a logarithmic relationship between beta value and age [24]. A similar finding was reported in a study that included Central African children, in which CpG sites that were hypo-methylated with age were better modeled by a function of logarithmic age [33]. Our analyses were limited to comparisons between children and young adults. Important areas for future study include further defining the relationship between DNA methylation and age across the lifespan.
The relationship between DNA methylation and gene expression is complex [45, 46]. Limitations of our study include small sample size, measurement of monocyte gene expression using a targeted panel rather than the entire transcriptome, and that the samples used for gene expression and DNA methylation measurements were not from the same individual. Reynolds et al. integrated DNA methylation and transcriptome profiles of CD14+ monocytes (sample size > 1000) and found that 4.7% of age-associated differentially methylated sites correlated with cis-gene expression changes [13]. With a smaller sample size, Saare et al. examined the effect of age on purified monocyte DNA methylation, gene expression, and metabolic functions and found a weak correlation between DNA methylation and gene expression [10]. The functional consequences of age-associated differential DNA methylation and transcription have yet to be elucidated. Saare et al. found age-related differences in cellular metabolic fitness [10], which could plausibly underlie age-related impairment in monocyte functions such as phagocytosis, antigen presentation, and cytokine production [11, 47, 48]. Further research is also needed to determine the transcriptional and epigenetic changes that regulate monocyte differentiation and how differences among the monocyte subsets relate to both age and immunity.
Studies of aging monocytes have shown generally weakened PRR responses in elderly vs. young adults [11, 14–17]. In this study, monocyte cytokine production in response to TLR stimulation was increased in Kenyan young adults compared to children. These differences among age groups may reflect continual epigenetic changes that occur in response to a lifetime of environmental exposures, including infections such as malaria. The relationship between altered DNA methylation and functional changes in monocytes was recently explored in the context of bacterial sepsis [49]. Monocytes from patients with sepsis had altered DNA methylation profiles compared to controls, with several hyper- and hypo-methylated CpG sites in monocytes from patients with sepsis correlating with increased constitutive production of IL-10 and IL-6. In vitro models of LPS-induced monocyte tolerance revealed changes in DNA methylation that reflected changes observed in monocytes from septic patients [49]. In another study of the role of DNA methylation in the regulation of innate immune responses to bacterial infection, Pacis et al. found that monocyte-derived dendritic cell exposure to Mycobacterium tuberculosis led to transcriptional activation and downstream active demethylation at thousands of CpG sites [50, 51]. The potential regulatory role that DNA methylation may play in innate immune memory and responses to subsequent stimuli has yet to be determined [52–55].
Epigenetic, transcriptional, and functional changes that occur in innate immune cells during the first few decades of life are likely to have a substantial impact on immunity to pathogens that disproportionately affect young children, most notably malaria. The importance of intrinsic age-related differences in immune responses to malaria infection was highlighted in studies of a migrant population of malaria-naïve children and adults who moved from Java, where malaria was rare, to a malaria-holoendemic area of Indonesian Papua [56–58]. Compared to children, newly arrived adults were much more likely to suffer from severe malaria and require evacuation to a hospital for clinical support [56, 57], yet adults appeared to develop clinical immunity more quickly than did children [58]. Further study of age-related differences in monocyte responses to malaria could provide new insights into disease pathogenesis and the acquisition of clinical immunity.
Conclusions
In summary, we identified DNA methylation patterns in isolated monocytes from Kenyan young adults that include known markers of aging as well as changes in gene loci important to innate immune and inflammatory responses. Monocytes from this cohort of young adults and children also show differences in TLR responses and inflammatory gene expression profiles. These results will inform future studies of the functional significance of altered monocyte DNA methylation in the aging process and immune system development.
Methods
Study site and study participants
Participants in this study were enrolled in an observational cohort study of naturally acquired immunity to malaria conducted at the Chulaimbo Sub-County Hospital in Kisumu County, Kenya. Enrollment and sample collection were conducted between June 2013 and April 2016. The study population were of Luo ethnicity. Malaria transmission in this area is perennially high with peaks coinciding with seasonal rains [36], and Plasmodium falciparum (Pf) is the primary malaria parasite species [59]. All healthy children and adults included in this study were afebrile and asymptomatic at the time of sample collection. Subclinical, asymptomatic Pf parasitemia was determined by either positive blood smear or positive Pf PCR. A blood smear slide was deemed negative when no parasites were seen after counting microscopic fields containing at least 200 leukocytes. Submicroscopic blood-stage infections were detected by a conventional nested PCR assay targeting 18S Pf–specific small subunit ribosomal RNA genes performed as previously described [60].
Healthy malaria-naïve US adult controls were enrolled in this study as an additional comparison group. The characteristics (age, sex, and Pf parasitemia status) of all study participants included in all assays for this study are summarized in Additional file 1: Table S1. This includes Kenyan children (n = 33), median age 5.1 years (range 1.2–9.8 years), 66.7% male, and 6 (18.2%) with subclinical asymptomatic Pf parasitemia; Kenyan adults (n = 30), median age 28.5 (range 18–35 years), 36.7% male, and none with asymptomatic Pf parasitemia; and US adults (n = 17), median age 28 years (range 24–69 years), 52.9% male, and none with asymptomatic Pf parasitemia.
DNA methylation profiles
Monocytes were isolated from whole blood via negative selection with the RosetteSep Human Monocyte Enrichment Cocktail (Stemcell Technologies, 15,068). Infinium MethylationEPIC BeadChip arrays (Illumina, Inc.) were used to perform epigenome-wide DNA methylation analysis of isolated monocytes. This platform interrogates methylation at > 850,000 sites across the genome at single-nucleotide resolution. Genomic DNA from monocyte samples was bisulfite-converted (Zymo EZ DNA Methylation kit, D5001) and then hybridized to the array according to manufacturer’s instructions. Illumina methylation array data may show batch effects secondary to technical artifacts from different runs. Our data were collected over two different runs with four separate slides, which included 8 healthy children from western Kenya and the same 8 Kenyan children during a preceding episode of acute uncomplicated malaria (run 1, samples evenly dispersed over slides A and B); and 8 healthy adults from western Kenya and 8 malaria-naïve US adults (run 2, samples evenly dispersed over slides C and D). All quality control and normalization procedures were performed to include all four groups, and principal components analyses did not show significant association with slide for run 1 or run 2. This does not, however, exclude the potential for batch effects that may be present or that may affect only a portion of the genome.
Quality control and normalization
Data quality were assessed using the standard Bioconductor/R minfi pipeline (version 1.28.14) [61]. We filtered out probe sites with intensities below background (detection P values > 0.01; 4849 probe sites removed), probes at known SNPs, and probes on X and Y chromosomes. We normalized signal intensity using the functional normalization procedure [62], which accounts for between-array technical variation and background variation. After normalization and filtering, 809,761 probe sites remained for differential methylation analysis.
Differential methylation analysis
Normalized probe intensity values were used to calculate beta and M-values, where the beta value is the ratio of methylated probe intensity to the overall intensity and the M-value is log2 of the ratio of intensities of methylated vs. unmethylated probes [log2(β/(1-β))]. Differentially methylated positions (DMPs) were determined using a linear model from the limma package (v. 3.38.3) [63], with age group as the primary predictor variable, adjusting for sex and asymptomatic Pf infection status. Probe sites were considered differentially methylated if the absolute beta value difference was > 0.15. Statistical significance was set at p < 0.01 and false discovery rate (FDR)-adjusted p < 0.05. Differentially methylated regions (DMRs) were determined using the DMRcate package (v. 1.18.0) [64], which extracts and annotates DMRs using a kernel-smoothed estimate. Gaussian kernel bandwidth for smoothed-function estimation was set at lambda = 1000 nucleotides, with the scaling factor for bandwidth C = 2. Statistical significance was reported by the Stouffer combined p value, which is the transformation of the group of FDR-adjusted p values for individual CpG sites as DMR constituents [65]. Enrichment of probe sites in CpG islands, shores, or shelves was analyzed by comparing the proportions of age-associated hyper- or hypo-methylated probe sites in these regions to the MethylationEPIC assay coverage of probe sites in these regions.
Pathway enrichment analysis
KEGG pathway enrichment analysis was performed using the missMethyl package (v. 1.16.0) [66], which takes into account the bias in methylation arrays for gene set testing due to differing numbers of probes per gene. Input for enrichment analyses were the top differentially methylated probe sites with FDR-adjusted p values < 0.05 and absolute beta value difference > 0.15 (14,259 probe sites), tested against a background of all probe sites tested (809,761 probe sites).
Data availability
The DNA methylation data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession number GSE157611 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE157611).
Monocyte subset proportions by flow cytometry
Proportions of monocyte subsets were determined for 17 healthy children and 14 healthy adults from western Kenya and for 4 healthy malaria-naïve US adults, as previously described [35]. Briefly, peripheral blood mononuclear cells (PBMC) were separated by standard Ficoll-Hypaque density gradient centrifugation and cryopreserved. PBMC were gently thawed and resuspended at a concentration of 106 cells/ml in RPMI-1640 (GIBCO). LIVE/DEAD Fixable Violet stain was used to assess cell viability (ThermoFisher Scientific). PBMC were directly stained for 20 min at 4 °C with Alexa Fluor 700–labeled anti-CD14 (clone 61D3; eBioscience) and APC-Cy 7–labeled anti-CD16 (clone 3G8; BioLegend). The stained cells were washed twice, fixed, and subjected to flow analysis on LSRII flow cytometer (BD Biosciences). FlowJo v8 software (Tree Star) was used for data analysis. The gating strategy put forth by the Nomenclature Committee of the International Union of Immunological Societies was used to determine proportions of monocyte subsets (classical CD14++CD16−, intermediate CD14++CD16+, and nonclassical CD14+CD16++) [67].
Monocyte cytokine production
Monocyte cytokine production was analyzed using freshly isolated monocytes from 8 healthy Kenyan children, 10 healthy Kenyan adults, and 10 healthy US adults, as previously described [35]. Monocytes were isolated from whole blood via negative selection with the RosetteSep Human Monocyte Enrichment Cocktail (Stemcell Technologies, 15,068). Cells were suspended in culture medium (RPMI-1640 supplemented with 2 mM L-glutamine, 10% FBS, 10 mM HEPES, 1 mM sodium pyruvate, 4.5 g/l glucose, 1.5 g/l sodium bicarbonate, and 0.05 mM 2-ME) and placed in 96-well polypropylene plates at 5 × 104 cells per well (concentration 5 × 105 cells/ml). Cells were stimulated with 10 ng/ml LPS (Sigma-Aldrich) and 100 ng/ml Pam3CSK4 (P3C) (Invivogen) and compared with a media-alone control; each condition was performed in duplicate. Cells were cultured for 18 h at 37 °C, in 5% CO2, on an orbital shaker. Supernatants were harvested and stored at − 80 °C. A multiplex magnetic bead–based immunoassay was used to measure concentrations of IL-1β, IL-6, IL-8, IL-10, IL-12p70, and TNF (EMD Millipore) in the culture supernatants immediately after initial thawing.
Targeted digital RNA sequencing
Monocyte inflammatory gene expression profiles were analyzed in samples from 6 Kenyan children, 7 Kenyan adults, and 4 US adults, as previously described [35]. Monocytes were isolated from cryopreserved PBMC via negative selection (Pan Monocyte Isolation Kit, Miltenyi Biotec, 130–096-537). Cells were lysed in RNAprotect Cell Reagent (Qiagen), and total RNA was prepared using the RNeasy kit (Qiagen) as per the manufacturer’s instructions. Total RNA integrity was assessed by an Agilent 2100 Bioanalyzer, and the RNA integrity number was calculated (≥8.5 for all samples). Targeted digital RNA sequencing was performed using a customized QIAseq Targeted RNA Panel (Human Inflammation and Immunity Transcriptome Panel, 508 genes) by the manufacturer (Qiagen).
Differential gene expression analysis
Raw QIAseq output in read counts per gene per sample were used as input for differential gene expression analysis using DESeq2 (v. 1.22.2) [68]. Significance for differential gene expression was set at FDR-adjusted p < 0.05. KEGG pathway enrichment analysis of differentially expressed genes was performed using [69].
Statistics
Monocyte DNA methylation and gene expression statistical analyses were performed as described above using Bioconductor/R (v. 3.5.3). For the remainder of the assays, Mann-Whitney U test was used to compare continuous variables between groups. Kruskal-Wallis test was used to compare categorical variables (sex, Pf infection status) among healthy Kenyan children, Kenyan adults, and US adults. Differences were considered significant at p < 0.05. Graphs were constructed and statistical analyses were performed using Prism software (v. 8.2.1; GraphPad).
Study approval
Informed consent was obtained from all participants or their guardians in the appropriate local language (English for US adults, Luo for Kenyan participants). Ethical approval was obtained from the Institutional Review Board of University Hospitals Cleveland Medical Center, Cleveland, Ohio, USA and from the Scientific and Ethical Review Unit (SERU) of the Kenya Medical Research Institute Ethical Review Committee.
Supplementary Information
Additional file 1 Table S1. Study participant characteristics. Figure S1. Monocyte subset proportions in Kenyan adults and children.
Additional file 2. DMPs_Kenyan_child_vs_Kenyan_adult.csv.
Additional file 3. DMRs_Kenyan_child_vs_Kenyan_adult.csv.
Additional file 4. KEGG_meth_Kenyan_child_vs_Kenyan_adult.csv.
Additional file 5. DESeq2_Kenyan_child_vs_Kenyan_adult.csv.
Additional file 6. DMPs_Kenyan_child_vs_US_adult.csv.
Additional file 7. DMPs_Kenyan_adult_vs_US_adult.csv.
Additional file 8. KEGG_meth_Kenyan_adult_vs_US_adult.csv.
Additional file 9. DESeq2_Kenyan_adult_vs_US_adult.csv.
Acknowledgements
This manuscript was submitted with permission from the director of KEMRI. We thank all the parents and guardians for giving their consent and all study participants for their cooperation. We thank all members of the study and field teams at Chulaimbo hospital and the Kisian research station.
Abbreviations
- PRR
Pattern recognition receptor
- DMP
Differentially methylated position
- DMR
Differentially methylated region
- SNP
Single nucleotide polymorphism
- PCA
Principal components analysis
- EWAS
Epigenome wide association study
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- PBMC
Peripheral blood mononuclear cells
- P3C
Pam3CSK4
Authors’ contributions
KRD designed, performed, and analyzed all experiments and drafted the manuscript. PE designed and performed all experiments. EK and SO developed and coordinated the cohort study and collected participant samples. SM designed and coordinated the cohort study. JWK designed and coordinated the project and cohort study, evaluated the data, provided funding for all experiments, and prepared the manuscript. AED designed and coordinated the project and cohort study, evaluated the data, and prepared the manuscript. All authors read and approved the final manuscript.
Funding
Funding for this work was provided by the NIH/National Institute of Allergy and Infectious Diseases (R01-AI095192 to JWK and K23-AI132644 to KRD) and by the Clinical and Translational Science Collaborative of Cleveland, 4UL1TR000439 from the National Center for Advancing Translational Sciences (NCATS) component of the National Institutes of Health and NIH roadmap for Medical Research. The contents of the publication are solely the responsibility of the authors and do not necessarily represent the official views of the NIH.
Availability of data and materials
The DNA methylation data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession number GSE157611 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE157611). Supporting DNA methylation and gene expression data are included as supplementary material. All other supporting data are available from the corresponding author on reasonable request.
Declarations
Ethics approval and consent to participate
Informed consent was obtained from all participants or their guardians in the appropriate local language (English for US adults, Luo for Kenyan participants). Ethical approval was obtained from the Institutional Review Board of University Hospitals Cleveland Medical Center, Cleveland, Ohio, USA and from the Scientific and Ethical Review Unit (SERU) of the Kenya Medical Research Institute Ethical Review Committee.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Olin A, Henckel E, Chen Y, Lakshmikanth T, Pou C, Mikes J, et al. Stereotypic immune system development in newborn children. Cell. 2018;174(5):1277–1292. doi: 10.1016/j.cell.2018.06.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arrieta MC, Stiemsma LT, Dimitriu PA, Thorson L, Russell S, Yurist-Doutsch S, et al. Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci Transl Med. 2015;7(307):307ra152. doi: 10.1126/scitranslmed.aab2271. [DOI] [PubMed] [Google Scholar]
- 3.Vatanen T, Kostic AD, d'Hennezel E, Siljander H, Franzosa EA, Yassour M, et al. Variation in microbiome LPS immunogenicity contributes to autoimmunity in humans. Cell. 2016;165(6):1551. doi: 10.1016/j.cell.2016.05.056. [DOI] [PubMed] [Google Scholar]
- 4.Kollmann TR, Kampmann B, Mazmanian SK, Marchant A, Levy O. Protecting the newborn and young infant from infectious diseases: lessons from immune ontogeny. Immunity. 2017;46(3):350–363. doi: 10.1016/j.immuni.2017.03.009. [DOI] [PubMed] [Google Scholar]
- 5.Crossley KB, Peterson PK. Infections in the elderly. Clin Infect Dis. 1996;22(2):209–215. doi: 10.1093/clinids/22.2.209. [DOI] [PubMed] [Google Scholar]
- 6.Giefing-Kroll C, Berger P, Lepperdinger G, Grubeck-Loebenstein B. How sex and age affect immune responses, susceptibility to infections, and response to vaccination. Aging Cell. 2015;14(3):309–321. doi: 10.1111/acel.12326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sansoni P, Vescovini R, Fagnoni F, Biasini C, Zanni F, Zanlari L, et al. The immune system in extreme longevity. Exp Gerontol. 2008;43(2):61–65. doi: 10.1016/j.exger.2007.06.008. [DOI] [PubMed] [Google Scholar]
- 8.Baylis D, Bartlett DB, Patel HP, Roberts HC. Understanding how we age: insights into inflammaging. Longev Healthspan. 2013;2(1):8. doi: 10.1186/2046-2395-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Albright JM, Dunn RC, Shults JA, Boe DM, Afshar M, Kovacs EJ. Advanced age alters monocyte and macrophage responses. Antioxid Redox Signal. 2016;25(15):805–815. doi: 10.1089/ars.2016.6691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saare M, Tserel L, Haljasmagi L, Taalberg E, Peet N, Eimre M, et al. Monocytes present age-related changes in phospholipid concentration and decreased energy metabolism. Aging Cell. 2020;19(4):e13127. doi: 10.1111/acel.13127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Metcalf TU, Wilkinson PA, Cameron MJ, Ghneim K, Chiang C, Wertheimer AM, et al. Human monocyte subsets are transcriptionally and functionally altered in aging in response to pattern recognition receptor agonists. J Immunol. 2017;199(4):1405–1417. doi: 10.4049/jimmunol.1700148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reynolds LM, Ding J, Taylor JR, Lohman K, Soranzo N, de la Fuente A, et al. Transcriptomic profiles of aging in purified human immune cells. BMC Genomics. 2015;16:333. doi: 10.1186/s12864-015-1522-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reynolds LM, Taylor JR, Ding J, Lohman K, Johnson C, Siscovick D, et al. Age-related variations in the methylome associated with gene expression in human monocytes and T cells. Nat Commun. 2014;5:5366. doi: 10.1038/ncomms6366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hearps AC, Martin GE, Angelovich TA, Cheng WJ, Maisa A, Landay AL, et al. Aging is associated with chronic innate immune activation and dysregulation of monocyte phenotype and function. Aging Cell. 2012;11(5):867–875. doi: 10.1111/j.1474-9726.2012.00851.x. [DOI] [PubMed] [Google Scholar]
- 15.Sadeghi HM, Schnelle JF, Thoma JK, Nishanian P, Fahey JL. Phenotypic and functional characteristics of circulating monocytes of elderly persons. Exp Gerontol. 1999;34(8):959–970. doi: 10.1016/S0531-5565(99)00065-0. [DOI] [PubMed] [Google Scholar]
- 16.Pillai PS, Molony RD, Martinod K, Dong H, Pang IK, Tal MC, et al. Mx1 reveals innate pathways to antiviral resistance and lethal influenza disease. Science. 2016;352(6284):463–466. doi: 10.1126/science.aaf3926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nyugen J, Agrawal S, Gollapudi S, Gupta S. Impaired functions of peripheral blood monocyte subpopulations in aged humans. J Clin Immunol. 2010;30(6):806–813. doi: 10.1007/s10875-010-9448-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao M, Wang Z, Yung S, Lu Q. Epigenetic dynamics in immunity and autoimmunity. Int J Biochem Cell Biol. 2015;67:65–74. doi: 10.1016/j.biocel.2015.05.022. [DOI] [PubMed] [Google Scholar]
- 19.van Dongen J, Nivard MG, Willemsen G, Hottenga JJ, Helmer Q, Dolan CV, et al. Genetic and environmental influences interact with age and sex in shaping the human methylome. Nat Commun. 2016;7:11115. doi: 10.1038/ncomms11115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garagnani P, Bacalini MG, Pirazzini C, Gori D, Giuliani C, Mari D, et al. Methylation of ELOVL2 gene as a new epigenetic marker of age. Aging Cell. 2012;11(6):1132–1134. doi: 10.1111/acel.12005. [DOI] [PubMed] [Google Scholar]
- 21.Horvath S. DNA methylation age of human tissues and cell types. Genome Biol. 2013;14(10):R115. doi: 10.1186/gb-2013-14-10-r115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rakyan VK, Down TA, Maslau S, Andrew T, Yang TP, Beyan H, et al. Human aging-associated DNA hypermethylation occurs preferentially at bivalent chromatin domains. Genome Res. 2010;20(4):434–439. doi: 10.1101/gr.103101.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hernandez DG, Nalls MA, Gibbs JR, Arepalli S, van der Brug M, Chong S, et al. Distinct DNA methylation changes highly correlated with chronological age in the human brain. Hum Mol Genet. 2011;20(6):1164–1172. doi: 10.1093/hmg/ddq561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alisch RS, Barwick BG, Chopra P, Myrick LK, Satten GA, Conneely KN, et al. Age-associated DNA methylation in pediatric populations. Genome Res. 2012;22(4):623–632. doi: 10.1101/gr.125187.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hannum G, Guinney J, Zhao L, Zhang L, Hughes G, Sadda S, et al. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol Cell. 2013;49(2):359–367. doi: 10.1016/j.molcel.2012.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Day K, Waite LL, Thalacker-Mercer A, West A, Bamman MM, Brooks JD, et al. Differential DNA methylation with age displays both common and dynamic features across human tissues that are influenced by CpG landscape. Genome Biol. 2013;14(9):R102. doi: 10.1186/gb-2013-14-9-r102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Christensen BC, Houseman EA, Marsit CJ, Zheng S, Wrensch MR, Wiemels JL, et al. Aging and environmental exposures alter tissue-specific DNA methylation dependent upon CpG island context. PLoS Genet. 2009;5(8):e1000602. doi: 10.1371/journal.pgen.1000602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johansson A, Enroth S, Gyllensten U. Continuous aging of the human DNA Methylome throughout the human lifespan. PLoS One. 2013;8(6):e67378. doi: 10.1371/journal.pone.0067378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tserel L, Limbach M, Saare M, Kisand K, Metspalu A, Milani L, et al. CpG sites associated with NRP1, NRXN2 and miR-29b-2 are hypomethylated in monocytes during ageing. Immun Ageing. 2014;11(1):1. doi: 10.1186/1742-4933-11-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Austin MK, Chen E, Ross KM, McEwen LM, Maclsaac JL, Kobor MS, et al. Early-life socioeconomic disadvantage, not current, predicts accelerated epigenetic aging of monocytes. Psychoneuroendocrinology. 2018;97:131–134. doi: 10.1016/j.psyneuen.2018.07.007. [DOI] [PubMed] [Google Scholar]
- 31.Salpea P, Russanova VR, Hirai TH, Sourlingas TG, Sekeri-Pataryas KE, Romero R, et al. Postnatal development- and age-related changes in DNA-methylation patterns in the human genome. Nucleic Acids Res. 2012;40(14):6477–6494. doi: 10.1093/nar/gks312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Husquin LT, Rotival M, Fagny M, Quach H, Zidane N, McEwen LM, et al. Exploring the genetic basis of human population differences in DNA methylation and their causal impact on immune gene regulation. Genome Biol. 2018;19(1):222. doi: 10.1186/s13059-018-1601-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gopalan S, Carja O, Fagny M, Patin E, Myrick JW, McEwen LM, et al. Trends in DNA methylation with age replicate across diverse human populations. Genetics. 2017;206(3):1659–1674. doi: 10.1534/genetics.116.195594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weber GE, White MT, Babakhanyan A, Sumba PO, Vulule J, Ely D, et al. Sero-catalytic and antibody acquisition models to estimate differing malaria transmission intensities in Western Kenya. Sci Rep. 2017;7(1):16821. doi: 10.1038/s41598-017-17084-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dobbs KR, Embury P, Vulule J, Odada PS, Rosa BA, Mitreva M, et al. Monocyte dysregulation and systemic inflammation during pediatric falciparum malaria. JCI Insight. 2017;2:18. doi: 10.1172/jci.insight.95352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou G, Afrane YA, Vardo-Zalik AM, Atieli H, Zhong D, Wamae P, et al. Changing patterns of malaria epidemiology between 2002 and 2010 in Western Kenya: the fall and rise of malaria. PLoS One. 2011;6(5):e20318. doi: 10.1371/journal.pone.0020318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reyburn H, Mbatia R, Drakeley C, Bruce J, Carneiro I, Olomi R, et al. Association of transmission intensity and age with clinical manifestations and case fatality of severe plasmodium falciparum malaria. JAMA. 2005;293(12):1461–1470. doi: 10.1001/jama.293.12.1461. [DOI] [PubMed] [Google Scholar]
- 38.Bacalini MG, Deelen J, Pirazzini C, De Cecco M, Giuliani C, Lanzarini C, et al. Systemic age-associated DNA Hypermethylation of ELOVL2 gene: in vivo and in vitro evidences of a cell replication process. J Gerontol A Biol Sci Med Sci. 2017;72(8):1015–1023. doi: 10.1093/gerona/glw185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Slieker RC, Relton CL, Gaunt TR, Slagboom PE, Heijmans BT. Age-related DNA methylation changes are tissue-specific with ELOVL2 promoter methylation as exception. Epigenetics Chromatin. 2018;11(1):25. doi: 10.1186/s13072-018-0191-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cormican S, Griffin MD. Human monocyte subset distinctions and function: insights from gene expression analysis. Front Immunol. 2020;11:1070. doi: 10.3389/fimmu.2020.01070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schmidl C, Renner K, Peter K, Eder R, Lassmann T, Balwierz PJ, et al. Transcription and enhancer profiling in human monocyte subsets. Blood. 2014;123(17):e90–e99. doi: 10.1182/blood-2013-02-484188. [DOI] [PubMed] [Google Scholar]
- 42.Duroux-Richard I, Robin M, Peillex C, Apparailly F. MicroRNAs: fine tuners of monocyte heterogeneity. Front Immunol. 2019;10:2145. doi: 10.3389/fimmu.2019.02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zawada AM, Schneider JS, Michel AI, Rogacev KS, Hummel B, Krezdorn N, et al. DNA methylation profiling reveals differences in the 3 human monocyte subsets and identifies uremia to induce DNA methylation changes during differentiation. Epigenetics. 2016;11(4):259–272. doi: 10.1080/15592294.2016.1158363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mandal R, Barron JC, Kostova I, Becker S, Strebhardt K. Caspase-8: the double-edged sword. Biochim Biophys Acta Rev Cancer. 1873;2020(2):188357. doi: 10.1016/j.bbcan.2020.188357. [DOI] [PubMed] [Google Scholar]
- 45.Wagner JR, Busche S, Ge B, Kwan T, Pastinen T, Blanchette M. The relationship between DNA methylation, genetic and expression inter-individual variation in untransformed human fibroblasts. Genome Biol. 2014;15(2):R37. doi: 10.1186/gb-2014-15-2-r37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hannon E, Gorrie-Stone TJ, Smart MC, Burrage J, Hughes A, Bao Y, et al. Leveraging DNA-methylation quantitative-trait loci to characterize the relationship between Methylomic variation, gene expression, and complex traits. Am J Hum Genet. 2018;103(5):654–665. doi: 10.1016/j.ajhg.2018.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pence BD, Yarbro JR. Aging impairs mitochondrial respiratory capacity in classical monocytes. Exp Gerontol. 2018;108:112–117. doi: 10.1016/j.exger.2018.04.008. [DOI] [PubMed] [Google Scholar]
- 48.Shen-Orr SS, Furman D, Kidd BA, Hadad F, Lovelace P, Huang YW, et al. Defective signaling in the JAK-STAT pathway tracks with chronic inflammation and cardiovascular risk in aging humans. Cell Syst. 2016;3(4):374–384. doi: 10.1016/j.cels.2016.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lorente-Sorolla C, Garcia-Gomez A, Catala-Moll F, Toledano V, Ciudad L, Avendano-Ortiz J, et al. Inflammatory cytokines and organ dysfunction associate with the aberrant DNA methylome of monocytes in sepsis. Genome Med. 2019;11(1):66. doi: 10.1186/s13073-019-0674-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pacis A, Tailleux L, Morin AM, Lambourne J, MacIsaac JL, Yotova V, et al. Bacterial infection remodels the DNA methylation landscape of human dendritic cells. Genome Res. 2015;25(12):1801–1811. doi: 10.1101/gr.192005.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pacis A, Mailhot-Leonard F, Tailleux L, Randolph HE, Yotova V, Dumaine A, et al. Gene activation precedes DNA demethylation in response to infection in human dendritic cells. Proc Natl Acad Sci U S A. 2019;116(14):6938–6943. doi: 10.1073/pnas.1814700116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ostuni R, Piccolo V, Barozzi I, Polletti S, Termanini A, Bonifacio S, et al. Latent enhancers activated by stimulation in differentiated cells. Cell. 2013;152(1–2):157–171. doi: 10.1016/j.cell.2012.12.018. [DOI] [PubMed] [Google Scholar]
- 53.Saeed S, Quintin J, Kerstens HH, Rao NA, Aghajanirefah A, Matarese F, et al. Epigenetic programming of monocyte-to-macrophage differentiation and trained innate immunity. Science. 2014;345(6204):1251086. doi: 10.1126/science.1251086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kaufmann E, Sanz J, Dunn JL, Khan N, Mendonca LE, Pacis A, et al. BCG educates hematopoietic stem cells to generate protective innate immunity against tuberculosis. Cell. 2018;172(1–2):176–190. doi: 10.1016/j.cell.2017.12.031. [DOI] [PubMed] [Google Scholar]
- 55.Quintin J, Cheng SC, van der Meer JW, Netea MG. Innate immune memory: towards a better understanding of host defense mechanisms. Curr Opin Immunol. 2014;29:1–7. doi: 10.1016/j.coi.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 56.Baird JK, Basri H, Weina P, MaGuire JD, Barcus MJ, Picarema H, et al. Adult Javanese migrants to Indonesian Papua at high risk of severe disease caused by malaria. Epidemiol Infect. 2003;131(1):791–797. doi: 10.1017/S0950268803008422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Krisin, Basri H, Fryauff DJ, Barcus MJ, Bangs MJ, Ayomi E, et al. Malaria in a cohort of Javanese migrants to Indonesian Papua. Ann Trop Med Parasitol. 2003;97(6):543–556. doi: 10.1179/000349803225001454. [DOI] [PubMed] [Google Scholar]
- 58.Baird JK, Krisin, Barcus MJ, Elyazar IR, Bangs MJ, Maguire JD, et al. Onset of clinical immunity to plasmodium falciparum among Javanese migrants to Indonesian Papua. Ann Trop Med Parasitol. 2003;97(6):557–564. doi: 10.1179/000349803225001472. [DOI] [PubMed] [Google Scholar]
- 59.Munyekenye OG, Githeko AK, Zhou G, Mushinzimana E, Minakawa N, Yan G. Plasmodium falciparum spatial analysis, western Kenya highlands. Emerg Infect Dis. 2005;11(10):1571–1577. doi: 10.3201/eid1110.050106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Singh B, Bobogare A, Cox-Singh J, Snounou G, Abdullah MS, Rahman HA. A genus- and species-specific nested polymerase chain reaction malaria detection assay for epidemiologic studies. Am J Trop Med Hyg. 1999;60(4):687–692. doi: 10.4269/ajtmh.1999.60.687. [DOI] [PubMed] [Google Scholar]
- 61.Fortin JP, Triche TJ, Jr, Hansen KD. Preprocessing, normalization and integration of the Illumina HumanMethylationEPIC array with minfi. Bioinformatics. 2017;33(4):558–560. doi: 10.1093/bioinformatics/btw691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fortin JP, Labbe A, Lemire M, Zanke BW, Hudson TJ, Fertig EJ, et al. Functional normalization of 450k methylation array data improves replication in large cancer studies. Genome Biol. 2014;15(12):503. doi: 10.1186/s13059-014-0503-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Peters TJ, Buckley MJ, Statham AL, Pidsley R, Samaras K, R VL, et al. De novo identification of differentially methylated regions in the human genome Epigenetics Chromatin. Epigenetics Chromatin. 2015;8:6. doi: 10.1186/1756-8935-8-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kim SC, Lee SJ, Lee WJ, Yum YN, Kim JH, Sohn S, et al. Stouffer's test in a large scale simultaneous hypothesis testing. PLoS One. 2013;8(5):e63290. doi: 10.1371/journal.pone.0063290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Phipson B, Maksimovic J, Oshlack A. missMethyl: an R package for analyzing data from Illumina's HumanMethylation450 platform. Bioinformatics. 2016;32(2):286–288. doi: 10.1093/bioinformatics/btv560. [DOI] [PubMed] [Google Scholar]
- 67.Ziegler-Heitbrock L, Ancuta P, Crowe S, Dalod M, Grau V, Hart DN, et al. Nomenclature of monocytes and dendritic cells in blood. Blood. 2010;116(16):e74–e80. doi: 10.1182/blood-2010-02-258558. [DOI] [PubMed] [Google Scholar]
- 68.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ge SX, Jung D, Yao R. ShinyGO: a graphical gene-set enrichment tool for animals and plants. Bioinformatics. 2020;36(8):2628–2629. doi: 10.1093/bioinformatics/btz931. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1 Table S1. Study participant characteristics. Figure S1. Monocyte subset proportions in Kenyan adults and children.
Additional file 2. DMPs_Kenyan_child_vs_Kenyan_adult.csv.
Additional file 3. DMRs_Kenyan_child_vs_Kenyan_adult.csv.
Additional file 4. KEGG_meth_Kenyan_child_vs_Kenyan_adult.csv.
Additional file 5. DESeq2_Kenyan_child_vs_Kenyan_adult.csv.
Additional file 6. DMPs_Kenyan_child_vs_US_adult.csv.
Additional file 7. DMPs_Kenyan_adult_vs_US_adult.csv.
Additional file 8. KEGG_meth_Kenyan_adult_vs_US_adult.csv.
Additional file 9. DESeq2_Kenyan_adult_vs_US_adult.csv.
Data Availability Statement
The DNA methylation data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession number GSE157611 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE157611).
The DNA methylation data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession number GSE157611 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE157611). Supporting DNA methylation and gene expression data are included as supplementary material. All other supporting data are available from the corresponding author on reasonable request.