Abstract
The COVID19 pandemic has caused more than a million of deaths worldwide, primarily due to complications from COVID19-associated acute respiratory distress syndrome (ARDS). Controversy surrounds the circulating cytokine/chemokine profile of COVID19-associated ARDS, with some groups suggesting that it is similar to patients without COVID19 ARDS and others observing substantial differences. Moreover, although a hyperinflammatory phenotype associates with higher mortality in non-COVID19 ARDS, there is little information on the inflammatory landscape’s association with mortality in patients with COVID19 ARDS. Even though the circulating leukocytes’ transcriptomic signature has been associated with distinct phenotypes and outcomes in critical illness including ARDS, it is unclear whether the mortality-associated inflammatory mediators from patients with COVID19 are transcriptionally regulated in the leukocyte compartment. Here, we conducted a prospective cohort study of 41 mechanically ventilated patients with COVID19 infection using highly calibrated methods to define the levels of plasma cytokines/chemokines and their gene expressions in circulating leukocytes. Plasma IL1RA and IL8 were found positively associated with mortality, whereas RANTES and EGF negatively associated with that outcome. However, the leukocyte gene expression of these proteins had no statistically significant correlation with mortality. These data suggest a unique inflammatory signature associated with severe COVID19.
Keywords: chemokines, COVID19, cytokines, IL1RA, mortality
INTRODUCTION
The COVID19 pandemic has so far caused over a million deaths worldwide, primarily due to complications from acute respiratory distress syndrome (ARDS) (1). Controversy surrounds the circulating cytokine/chemokine profile of COVID19-associated ARDS, with some groups suggesting that it is similar to patients without COVID19 ARDS (2) and others observing substantial differences (3). Patients without COVID ARDS who develop a hyperinflammatory signature have a higher mortality than those demonstrating a hypoinflammatory phenotype; furthermore, these subgroups have different responses to specific interventions such as positive end expiratory pressure (PEEP), administration of simvastatin, and fluid management (4–8). However, the clinical effects of the inflammatory profile on severe COVID19 infection remains controversial (9, 10). Moreover, most descriptions of the inflammatory signatures in patients with COVID19 have been reported using standard clinical laboratory tests (11–17) or multiplex systems (3, 15, 18) that lack adequate methodological calibration of individual cytokines (10, 19). A more precise characterization of circulating cytokine/chemokine expression profile in COVID19-associated ARDS could lead to the development of personalized therapies directed against specific inflammatory signatures (20, 21). Also, because distinct transcriptional signatures in circulating leukocytes have been associated with mortality in critical illness including ARDS (22, 23), it is rational to postulate that COVID19 mortality-associated cytokines are transcriptionally regulated in leukocytes or other sources. Thus, cytokine gene expression levels in circulating leukocytes could provide important insights into the organ-specific regulation of the inflammatory response in patients with COVID19.
In this study, we prospectively interrogated the plasma cytokine/chemokine levels and their leukocyte gene expression in patients with severe COVID19 infection using validated methods. We hypothesized that specific signatures would be associated with mortality in this setting. Portions of this study have been recently reported in a preprint form (24).
METHODS
We conducted a prospective cohort study involving 41 patients with diagnosis of COVID19 who were admitted to the Albany Medical Center Intensive Care Unit (ICU) and required mechanical ventilation due to ARDS. These patients were enrolled between March 20 and April 17, 2020. Ethical approval was obtained from Albany Medical College Committee on Research Involving Human Subjects (IRB No. 5670-20). Patients were considered for enrollment if they were older than 18 years and were admitted to the ICU for invasive mechanical ventilation due to COVID19-associated respiratory failure. Exclusion criteria included likely imminent death or inability to provide consent. Informed consent was obtained from the patient or a legally authorized representative. Acute illness severity was assessed using the Sequential Organ Failure Assessment (SOFA) score, Simplified Acute Physiology Score (SAPS II), and the Acute Physiology and Chronic Health Evaluation (APACHE II) score (25). Chronic disease burden information was aggregated with the Charlson Comorbidity index (26). COVID19 diagnosis was confirmed with regular polymerase chain reaction (PCR) method. Ventilatory system static compliance was obtained at the time of enrollment with patients supported with volume-control ventilatory mode and under full sedation.
Cytokines Measurements
At the time of enrollment, blood samples were obtained and immediately centrifuged for plasma fractionation. Samples were frozen in multiple individual aliquots for later cytokine/chemokine determinations using a Human XL Cytokine Magnetic Luminex-Discovery Fixed Panel (R&D Systems #LKTM014) performed simultaneously in duplicates within a week of enrollment completion. Only one freeze-thaw cycle occurred. Cytokines that were found associated with mortality with the multiplex system were later individually measured with enzyme-linked immunosorbent assay (ELISA) using a plate reader (Cytation 5 Imager, BioTek, Winooski, VT). Samples used for ELISA were obtained from the original individual aliquots, so all the procedures involved only one thawing step. Reagents manufacturers and catalogue numbers can be found in Supplemental Table S1 (all Supplemental material is available at https://doi.org/10.6084/m9.figshare.13369097.v1).
Leukocyte mRNA Expression Determination
In addition to plasma samples, whole blood samples were simultaneously collected from all participants, and leukocytes were isolated using LeukoLOCK filtering system (Cat. No. AM1923; Thermo Fisher). RNA was then extracted from peripheral leukocytes with TRIzol reagent (Cat. No. 15596018; Thermo Fisher), and total RNA was isolated following manufacturer’s instructions. Four hundred nanograms of total RNA was used to prepare cDNA using Primescript RT Master Mix at 42°C (Cat. No. RR036A; Takara) following manufacturer’s instructions. The cDNA was diluted 10-fold in nuclease-free water, and then, 2 µL of cDNA was used per PCR reaction. qPCR was performed in a StepOnePlus (Applied Biosystems) instrument using SYBR green-based iTaq supermix (Cat. No. 1725125; Bio-Rad) and 2 pmol primers (Eurofins or Thermo Fisher). Fold induction was calculated using the ΔΔCt method using GAPDH as the reference gene. Each sample was assayed in duplicate, and a negative control was included in each experiment. All methods were carried out in accordance with relevant guidelines and regulations, and primers sequences can be found in Supplemental Table S2.
Statistical Analysis
Because of the long hospitalization stays from COVID19 infection (27), especially in patients receiving mechanical ventilation, the analyzed outcome variable was mortality at day 45 postenrollment. Median [interquartile ranges (IQRs)] and frequency (%) were used to report ICU patient baseline characteristics for continuous and categorical variables, respectively. Associations between mortality and cytokine/chemokine levels obtained with the multiplex Luminex system were analyzed with ANOVA, with P value <0.05 considered significant. As multiplex systems require a second inferential step to adjust for false discovery rate (FDR), and because that step can lead to false negative and positive results (19), any cytokine/chemokine that were found statistically significantly associated with mortality before adjustment for FDR were individually measured with ELISA. For that test, comparison between cytokines/chemokines soluble levels in surviving versus nonsurviving patients was accomplished with Student’s t test, and a P value of <0.05 was considered significant. Association of leukocyte gene products magnitudes in leukocytes with patients’ mortality was done using with Student’s t test, and a P value <0.05 was considered significant.
RESULTS
A total of 41 patients were enrolled. The average age of the cohort was 60 years old, and there was a significant male sex predominance (68%). A total of 27% were Caucasians, 12% African Americans, 31% Hispanics, and 30% other ethnicities. Consistent with previous reports, we found that African Americans had significantly higher mortality than Caucasians (28). Average APACHE II, SAPS II, and SOFA scores were 24, 60, and 9.5, respectively. Other important baseline characteristics are presented in Table 1.
Table 1.
Patients’ baseline characteristics in the whole cohort
| Variables | Entire Cohort | Survived | Died | P Value |
|---|---|---|---|---|
| Sex, N (%) | 41 | 26 | 15 | |
| Male | 28 (68.3%) | 19 (73.1%) | 9 (60.0%) | 0.38 |
| Female | 13 (31.7%) | 7 (26.9%) | 6 (40.0%) | 0.38 |
| Age (IQR) | ||||
| 59.9 (50–72) | 57.3 (50–66) | 64.3 (62–73) | 0.07 | |
| Ethnicity, N (%) | ||||
| White | 11 (26.8%) | 6 (23.1%) | 5 (33.3%) | 0.48 |
| African American | 5 (12.2%) | 1 (3.8%) | 4 (26.7%) | 0.03* |
| Asian | 2 (5.9%) | 1 (3.8%) | 1 (6.7%) | 0.69 |
| Hispanic | 13 (31.7%) | 1 (3.8%) | 4 (26.7%) | 0.03* |
| Other | 10 (24.4%) | 9 (34.9%) | 1 (6.7%) | 0.04* |
| Comorbidities, N (%) | ||||
| Congestive heart failure | 1 (2.4%) | 1 (3.8%) | 0 (0%) | 0.44 |
| Pulmonary disease | 6 (14.6%) | 4 (15.4%) | 2 (13.3%) | 0.85 |
| Diabetes mellitus | 16 (39.1%) | 10 (38.5%) | 6 (40.0%) | 0.92 |
| Renal disease | 4 (9.8%) | 1 (3.8%) | 3 (20.0%) | 0.09 |
| Cancer | 2 (4.9%) | 1 (3.8%) | 1 (6.7%) | 0.69 |
| HIV/AIDS | 1 (2.4%) | 1 (3.8%) | 0 (0%) | 0.44 |
| Charlson score index | 3.05 (1.0–4.0) | 2.69 (1.0–3.0) | 3.67 (2.0–5.5) | 0.10 |
| Severity scores (IQR) | ||||
| APACHEII | 24.17 (18.50–28.50) | 22.04 (18.25–24.75) | 27.87 (23.00–33.50) | <0.01** |
| SAPS II | 59.85 (50.50–68.50) | 57.11 (51.00–59.00) | 64.60 (54.00–74.00) | 0.02* |
| SOFA | 9.54 (7.00–12.00) | 8.81 (7.00–10.75) | 10.80 (7.50–12.50) | 0.04* |
| Biomarkers (IQR) | ||||
| Ferritin (ng/mL) | 895.33 (376.50–1259.00) | 810.53 (374.50–1271.75) | 1052.79 (681.25–1111.25) | 0.13 |
| CRP (mg/L) | 179.31 (73.10–256.50) | 184.73 (75.28–258.45) | 168.47 (79.50–242.60) | 0.33 |
| D-dimer (mg/L FEU) | 21.93 (2.12–25.65) | 19.53 (1.87–20.84) | 26.12 (4.89–37.10) | 0.25 |
| Procalcitonin (ng/mL) | 6.35 (0.47–4.34) | 5.83 (0.28–2.78) | 7.34 (0.91–11.53) | 0.38 |
| Lactate (mmol/L) | 1.25 (0.91–1.51) | 1.30 (0.89–1.52) | 1.17 (0.95–1.29) | 0.24 |
| Fibrinogen (mg/DL) | 556.53 (411.50–685.75) | 555.04 (411.50–685.75) | 559.07 (443.00–657.75) | 0.48 |
| Respiratory variables (IQR) | ||||
| P/F ratio (mmHg) | 159.15 (98-202.5) | 173.46 (128.25–214.25) | 134.33 (84.50–155.00) | 0.06 |
| Static compliance (mmHg) | 41.05 (28.38–45.1) | 43.11 (29.25–44.98) | 37.46 (28.50–44.84) | 0.20 |
| Treatment—N (%) | ||||
| ECMO | 2 (4.9%) | 1 (3.8%) | 1 (6.7%) | 0.69 |
| Antibiotics | 41 (100%) | 26 (100%) | 15 (100%) | NA |
| Hydroxychloroquine | 38 (92.7%) | 24 (92.3%) | 14 (93.3%) | 0.90 |
| Antiviral | 1 (2.4%) | 1 (3.8%) | 0 (0%) | 0.44 |
| IL6–Antagoinist | 2 (4.9%) | 1 (3.8%) | 1 (6.7%) | 0.69 |
| Convalescent plasma | 16 (39.1%) | 8 (30.8%) | 8 (53.3%) | 0.15 |
| Steroids | 28 (68.3%) | 19 (73.1%) | 9 (60.0%) | 0.38 |
Comparison of clinical and laboratory variables between surviving vs. nonsurviving patients is indicated. Data are presented as median with interquartile range (IQR), unless otherwise indicated. APACHE II, Acute Physiology and Chronic Health Evaluation II; CRP, C-reactive protein; ECMO, extracorporeal membrane oxygenation; SAPS II, Simplified Acute Physiology Score; SOFA, Sequential Organ Failure Assessment. Significant P values are indicated in bold and italic font. P values for categorical variables are from χ2 tests of association or Fisher’s exact tests when the expected value in any cell is five or less. P values for continuous variables are from Student’s t test or for nonnormal distributed variables by Mann–Whitney rank test.
Association of Cytokine/Chemokine Circulating Levels with Mortality
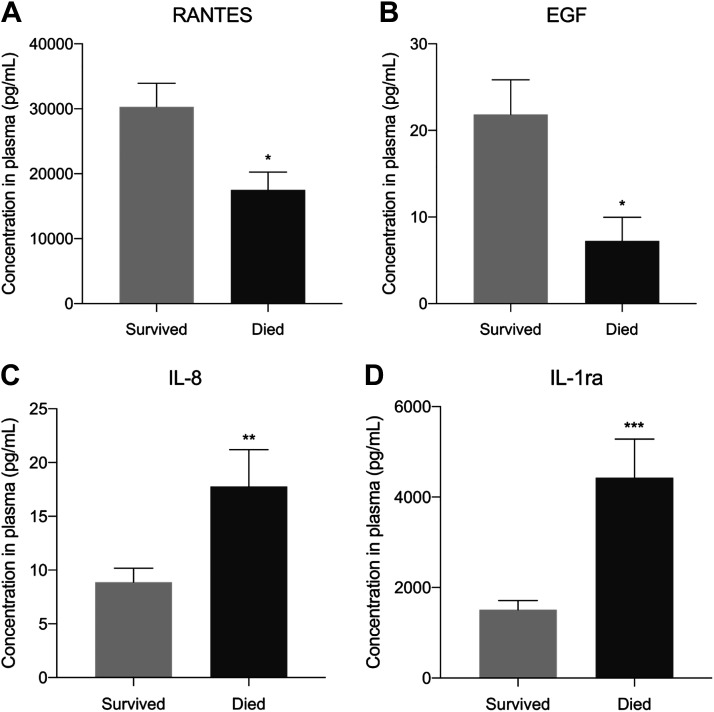
A total of 45 cytokines/chemokines were measured, and seven were found associated with mortality with the multiplex system, with RANTES, EGF, IL1a, and IL5 associated with lower mortality, and IL1RA, IL8, and CCL19 associated with greater mortality (Fig. 1). IL6, which is associated with worse outcomes in non-COVID19 ARDS, was found unassociated with mortality in this cohort. Out of the seven cytokines/chemokines that were associated with mortality, individual ELISA determinations confirmed the positive associations of IL1RA and IL8 and negative associations of RANTES and EGF with mortality (Fig. 2). IL5 and CCL19 levels were unassociated with mortality, and IL1a was not detected by the assay (Supplemental Fig. S1).
Figure 1.
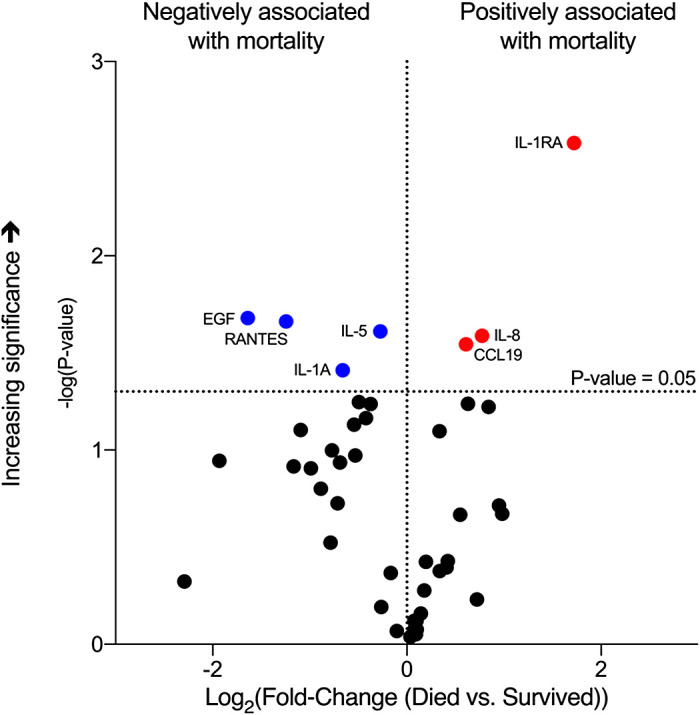
Cytokine determinations at the time of enrollment and association with mortality based on multiplex system determination. Volcano plot showing cytokines/chemokines significantly associated with mortality: blue dots, negatively associated; red dots, positively associated. Black dots identify entities not statistically significantly associated with mortality. Threshold of significance was established at a P value of 0.05 before adjustment for false discovery rate. IL-1ra, IL1 receptor antagonist.
Figure 2.
A–D: cytokines associated with mortality based on ELISA tests performed individually. Bar graphs showing the cytokines/chemokines that were found associated with mortality by ELISA testing. *P < 0.05; **P < 0.01; ***P < 0.001. IL-1ra, IL1 receptor antagonist.
Association of Cytokine/Chemokine Gene Expression Levels in Circulating Leukocytes with Their Respective Protein Product and with Mortality
Cytokine/chemokine gene expression levels in circulating leukocytes were not significantly associated with mortality. EGF and RANTES gene expression were modestly but significantly correlated with their respective protein products’ circulating levels, whereas mRNA expression and circulating levels of the other factors were not (Table 2).
Table 2.
Correlation between cytokine/chemokine gene expression in circulating leukocytes with soluble protein and with mortality
| With Soluble Protein |
With Mortality | ||
|---|---|---|---|
| Cytokine | R2 | P Value | P Value |
| RANTES | 0.2318 | <0.01** | 0.28 |
| EGF | 0.3277 | <0.001*** | 0.25 |
| IL-1ra | 0.0729 | 0.10 | 0.86 |
| IL-8 | 0.0007 | 0.87 | 0.54 |
IL-1ra, IL1 receptor antagonist. Significant P values are indicated in bold and italic font.
DISCUSSION
We describe a cytokine/chemokine landscape associated with COVID19 mortality, which is different from the one observed in non-COVID19-associated ARDS: Higher IL1RA and IL8 and reduced levels of RANTES and EGF were associated with mortality at 45 days.
The soluble cytokines and chemokines profile associated with COVID19 ARDS and its relationship to mortality has been recently debated and remains incompletely understood (9, 10). Multiple large ARDS trials have found robust elevations of interleukin (IL)-6 (4, 7, 8), and both IL6 and IL8 associate with worse outcomes in that setting (29). We identified IL8, but not IL6, to be associated with higher mortality in our cohort, suggesting an overlapping but distinct inflammatory signature of COVID19-driven respiratory failure. Importantly, recent evidence demonstrates that IL6 receptor blockade is not associated with better outcomes in patients with COVID19 (30, 31). Consistent with a very recent report (18), we found IL1 receptor antagonist (IL1RA) to be associated with higher mortality in COVID19. Significantly, although we found that IL1RA levels positively associate with mortality, multiple previous reports indicate that it confers protection against ARDS in patients without COVID19. IL1RA is a natural inhibitor of the proinflammatory IL1β, which is a highly active cytokine found in the lungs of patients with non-COVID19 ARDS (29, 32, 33). IL1β has been found to increase alveolar permeability both in animals and in humans (34, 35), and experimental models of IL1β-induced lung injury are mitigated by IL1RA administration (36). Reduced levels of IL1RA in bronchioalveolar lavage samples are associated with higher mortality of ARDS (37), and IL1RA administration has been proposed as a measure to attenuate lung injury (38, 39). Moreover, a single nucleotide polymorphism (SNP) leading to higher plasma IL1RA levels is associated with reduced ARDS susceptibility (40). These data suggest that IL1RA elevation represents a feature of COVID19-induced respiratory failure that differs from non-COVID19 ARDS, and the potential pharmacologic modulation of IL1RA has been recently suggested for patients with COVID19 (41, 42).
Our finding that RANTES (CCL5) is negatively associated with mortality in our cohort is consistent with a recent report indicating that it may protect patients with COVID19 from developing severe disease (3). RANTES is produced by CD8+ T cells after antigen stimulation and competes with the HIV virus for the CCR5 receptor binding (43); RANTES could operate in a similar fashion in COVID19 infections. Indeed, a recent study demonstrates that a clonal expansion of CD8+ T cells in broncho-alveolar fluid is associated with milder forms of COVID-19 infection (44).
We also found a negative association between epidermal growth factor (EGF) and COVID19 mortality. It has been previously suggested that acute lung injury elicits growth factor responses that initiate repair mechanisms to restore lung integrity (45). EGF regulates bronchial and alveolar epithelial repair after lung injury (46) and attenuates alveolar epithelial junctional permeability increasing lung fluid clearance (47). Moreover, EGF gene polymorphisms associate with ARDS risk (48). Although it is possible that EGF plays a beneficial role in the COVID19-induced ARDS pathophysiology, its role should be directly investigated in future studies.
There is a growing interest in the value of leukocyte gene expression profiles to define ARDS and sepsis phenotypes and in establishing their association with mortality (23, 49). Indeed, recent data indicate that corticosteroids could be beneficial in certain patient subgroups demonstrating specific leukocyte transcriptomic signatures (20, 50). To gain insight into the regulation of mortality-associated inflammatory mediators in COVID19, we determined the expression levels of the different mortality-associated cytokine and chemokine genes in circulating leukocytes and found that although some of the interrogated gene expression levels were modestly correlated with the magnitude of circulating soluble protein, none of them were associated with mortality. Although future research could elucidate the source of the mortality-associated inflammatory mediators in COVID19, we speculate that the cytokine profile in these patients could be contributed by cells other than circulating leukocytes such as the bronchus-associated lymphoid tissue (BALT) (51), alveolar epithelial or endothelial cells (52, 53), or by subpopulations of circulating immune cells such as NK cells and others (54), whose gene expression levels may not be captured with the global leukocyte mRNA analysis. It is also possible that the turnover of these proteins is regulated at the posttranscriptional level and thus their expression may not be reflected by the magnitude of mRNA expression.
A main strength of this work is that we describe a prospective cohort of patients with COVID19 ARDS and analyzed the association between circulating cytokines and chemokines with mortality. In addition, we determined plasma cytokine levels with ELISA, as opposed to relying on less calibrated clinical laboratory or multiplex testing systems. Indeed, the lack of individual ELISA assays in most of the current COVID19 literature is considered an important factor that complicates comparing these studies with the non-COVID-associated ARDS trials (10). Our study has some limitations. First, this is a single-center study that may not be universally reflective of patients with COVID19 ARDS. Second, we did not have access to an external validation cohort. However, the associations between IL1RA and worse outcomes and RANTES with better outcomes in COVID-ARDS have been replicated by two recently published studies (3, 18). Third, although we enrolled the patients at the time of hospital admission, we had no control over the period elapsed between the disease development and the blood sampling, which may have affected the inflammatory landscape. Fourth, we had no control of different drugs administered to the patients, which could have impacted their inflammatory profile, which was caused by the overwhelming pandemic environment and by the fact that at the time the patients were enrolled there were no formal recommendation by medical societies guidelines as of whether corticosteroids and other drugs were required for patients with COVID19 in respiratory failure. However, even though the administration of different drugs including corticosteroids could interact with the expression levels with circulating cytokines and chemokines, we found that corticosteroids were similarly distributed between survivors and nonsurvivors who indeed showed differential expression of soluble cytokines/chemokines offers (Supplemental Table S3). Future larger studies could multivariably adjust for medications administration and determine potential interaction not identified in the present work. Fifth, although IL1a was found in the multiplex assay to negatively correlate with mortality (Table 3) but could not be detected in the used ELISA assay, we cannot rule out the possibility that the lack of detection could be caused by a difference in sensitivity between the two assays, and thus our results do not invalidate IL1a’s potential role in COVID19 outcomes, which could be investigated in future studies.
Table 3.
Luminex multiplex panel data
| Ligand Mean Value (SD) | Survived | Died | P Value |
|---|---|---|---|
| n = 26 | n = 15 | ||
| CD40 Ligand, pg/mL | 761.10 (882.98) | 391.10 (335.67) | 0.15 |
| EGF, pg/mL | 26.16 (26.70) | 8.60 (7.50) | 0.02* |
| Eotaxin, pg/mL | 84.71 (38.72) | 79.10 (48.14) | 0.70 |
| FGF basic, pg/mL | 7.07 (12.09) | 10.75 (12.18) | 0.37 |
| Flt-3 ligand, pg/mL | 53.64 (52.35) | 57.75 (20.79) | 0.78 |
| Fractalkine, pg/mL | 573.20 (229.58) | 719.80 (287.87) | 0.10 |
| G-CSF, pg/mL | 18.36 (12.79) | 12.78 (722.96) | 0.18 |
| GM-CSF, pg/mL | 118.30 (102.83) | 149.80 (82.01) | 0.34 |
| Granzyme B, pg/mL | 14.90 (9.58) | 14.56 (10.81) | 0.92 |
| GROa, pg/mL | 60.22 (24.30) | 87.42 (108.18) | 0.25 |
| GROb, pg/mL | 336.00 (196.92) | 267.20 (154.30) | 0.28 |
| IFN-a, pg/mL | 4.78 (4.21) | 5.58 (9.92) | 0.73 |
| IFN-B, pg/mL | 0.96 (0.54) | 0.56 (1.24) | 0.26 |
| IFN-y, pg/mL | 4.90 (5.68) | 4.13 (4.68) | 0.67 |
| IL-1a, pg/mL | 8.42 (4.79) | 5.08 (2.84) | 0.03* |
| IL-1B, pg/mL | 1.66 (0.79) | 1.47 (0.40) | 0.43 |
| IL-1ra, pg/mL | 1616.00 (1255.74) | 5801.00 (6030.25) | 0.001** |
| IL-2, pg/mL | 1.72 (2.03) | 0.89 (1.27) | 0.19 |
| IL-3, pg/mL | 10.38 (11.08) | 11.06 (10.87) | 0.86 |
| IL-4, pg/mL | 0.87 (0.98) | 0.41 (0.29) | 0.10 |
| IL-5, pg/mL | 4.71 (1.19) | 3.86 (0.79) | 0.03* |
| IL-6, pg/mL | 697.80 (2297.27) | 1231.00 (2807.93) | 0.53 |
| IL-7, pg/mL | 3.49 (2.21) | 2.70 (1.62) | 0.26 |
| IL-8, pg/mL | 7.30 (4.69) | 13.87 (9.61) | 0.01* |
| IL-10, pg/mL | 232.70 (161.94) | 278.80 (162.29) | 0.41 |
| IL-12 p70, pg/mL | 2.20 (2.30) | 1.72 (1.94) | 0.53 |
| IL-13, pg/mL | 42.69 (18.22) | 31.22 (14.33) | 0.06 |
| IL-15, pg/mL | 3.70 (1.84) | 4.33 (1.85) | 0.32 |
| IL-17A, pg/mL | 2.03 (1.92) | 1.80 (2.14) | 0.75 |
| IL-17E, pg/mL | 27.76 (44.65) | 7.56 (10.01) | 0.11 |
| IL-33, pg/mL | 9.18 (4.84) | 6.63 (2.35) | 0.08 |
| IP-10, pg/mL | 790.60 (900.84) | 1168.00 (992.29) | 0.24 |
| MCP-1, pg/mL | 281.40 (301.84) | 357.50 (218.36) | 0.42 |
| MIP-1a, pg/mL | 9.28 (4.61) | 10.82 (6.84) | 0.42 |
| MIP-1B, pg/mL | 158.20 (56.29) | 129.80 (48.19) | 0.13 |
| MIP-3a, pg/mL | 54.48 (74.26) | 65.01 (191.42) | 0.64 |
| MIP-3B, pg/mL | 152.90 (91.82) | 226.00 (122.35) | 0.04* |
| PDGF-AA, pg/mL | 1729.00 (1389.01) | 959.80 (861.68) | 0.07 |
| PDGF-AB/BB, pg/mL | 610.30 (635.42) | 273.10 (216.61) | 0.07 |
| PD-L1/B7-H1, pg/mL | 841.30 (3516.13) | 171.80 (123.95) | 0.49 |
| RANTES, pg/mL | 27005.00 (23226.68) | 12112.00 (10793.81) | 0.03* |
| TGF-a, pg/mL | 7.31 (5.79) | 4.10 (3.51) | 0.07 |
| TNF-a, pg/mL | 13.76 (11.97) | 21.15 (23.51) | 0.12 |
| TRAIL, pg/mL | 10.32 (7.71) | 13.54 (14.58) | 0.38 |
| VEGF, pg/mL | 202.40 (150.49) | 308.30 (201.76) | 0.08 |
IL-1ra, IL1 receptor antagonist. Significant P values are indicated in bold and italic font.
Perspectives and Significance
Higher IL1RA and IL8 levels in plasma are positively associated with mortality in a cohort of critically ill patients with COVID19. Elevated RANTES and EGF protein are negatively associated with mortality in these patients. We found no association between gene expression of these cytokines in circulating leukocytes with mortality. Although our data indicate that a unique inflammatory signature is associated with severe COVID19 mortality, future research could determine the value of these mediators’ measurements as mortality-predictive biomarkers and define if their pharmacologic modulation could improve severe COVID19 prognosis.
GRANTS
Part of the results reported herein have been funded by National Heart, Lung, and Blood Institute (NHLBI) of the National Institutes of Health (NIH) under the award number K01-HL130704 (A. Jaitovich), and by the Collins Family Foundation Endowment (A. Jaitovich); NIH/NHLBI 5R01HL049426 (H. A. Singer); NIH/National Institute of General Medical Sciences Grant 1R01GM124133 (A. P. Adam); P41 GM108538 (K. A. Overmyer, E. Shishkova, and J. J. Coon).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
J.B., E.S., J.J.C., H.A.S., M.A.J., A.J., A.P.A., A.C., H.C.C., N.M., R.B.R., P.J.F., and K.A.O. conceived and designed research; J.B., E.S., J.J.C., M.A.J., A.J., A.P.A., A.C., H.C.C., L.A.D., N.M., R.B.R., P.J.F., and K.A.O. performed experiments; J.B. and A.J. analyzed data; J.B. and A.J. interpreted results of experiments; J.B. and A.J. prepared figures; J.B., M.A.J., and A.J. drafted manuscript; J.B., E.S., J.J.C., H.A.S., M.A.J., A.J., A.P.A., A.C., H.C.C., L.A.D., N.M., R.B.R., P.J.F., and K.A.O. edited and revised manuscript; J.B., E.S., J.J.C., H.A.S., M.A.J., A.J., A.P.A., A.C., H.C.C., L.A.D., N.M., R.B.R., P.J.F., and K.A.O. approved final version of manuscript.
REFERENCES
- 1.Guan WJ, Ni ZY, Hu Y, Liang WH, Ou CQ, He JX, et al . Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med 382: 1708–1720, 2020. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wilson JG, Simpson LJ, Ferreira A-M, Rustagi A, Roque J, Asuni A, Rananath T, Grant PM, Subramanian A, Rosenberg-Hasson Y, Maecker HT, Holmes SP, Levitt JE, Blish CA, Roers AJ. Cytokine profile in plasma of severe COVID-19 does not differ from ARDS and sepsis. JCI Insight 5: e140289, 2020. doi: 10.1172/jci.insight.140289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhao Y, Qin L, Zhang P, Li K, Liang L, Sun J, Xu B, Dai Y, Li X, Zhang C, Peng Y, Feng Y, Li A, Hu Z, Xiang H, Ogg G, Ho L-P, McMichael A, Jin R, Knight JC, Dong T, Zhang Y. Longitudinal COVID-19 profiling associates IL-1ra and IL-10 with disease severity and RANTES with mild disease. JCI Insight 5: e139834, 2020. doi: 10.1172/jci.insight.139834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Calfee CS, Delucchi K, Parsons PE, Thompson BT, Ware LB, Matthay MA. Subphenotypes in acute respiratory distress syndrome: latent class analysis of data from two randomised controlled trials. Lancet Respir Med 2: 611–620, 2014. doi: 10.1016/S2213-2600(14)70097-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Calfee CS, Delucchi KL, Sinha P, Matthay MA, Hackett J, Shankar-Hari M, McDowell C, Laffey JG, O’Kane CM, McAuley DF; Irish Critical Care Trials Group. Acute respiratory distress syndrome subphenotypes and differential response to simvastatin: secondary analysis of a randomised controlled trial. Lancet Respir Med 6: 691–698, 2018. doi: 10.1016/S2213-2600(18)30177-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Delucchi K, Famous KR, Ware LB, Parsons PE, Thompson BT, Calfee CS; ARDS Network. Stability of ARDS subphenotypes over time in two randomised controlled trials. Thorax 73: 439–445, 2018. doi: 10.1136/thoraxjnl-2017-211090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Famous KR, Delucchi K, Ware LB, Kangelaris KN, Liu KD, Thompson BT, Calfee CS; ARDS Network. Acute respiratory distress syndrome subphenotypes respond differently to randomized fluid management strategy. Am J Respir Crit Care Med 195: 331–338, 2017. doi: 10.1164/rccm.201603-0645OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sinha P, Delucchi KL, Thompson BT, McAuley DF, Matthay MA, Calfee CS; NHLBI ARDS Network. Latent class analysis of ARDS subphenotypes: a secondary analysis of the statins for acutely injured lungs from sepsis (SAILS) study. Intensive Care Med 44: 1859–1869, 2018. doi: 10.1007/s00134-018-5378-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hedrick TL, Murray BP, Hagan RS, Mock JR. COVID-19: clean up on IL-6. Am J Respir Cell Mol Biol 63: 541–543, 2020. doi: 10.1165/rcmb.2020-0277LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sinha P, Matthay MA, Calfee CS. Is a "cytokine storm" relevant to COVID-19? JAMA Intern Med 180: 1152, 2020. doi: 10.1001/jamainternmed.2020.3313. [DOI] [PubMed] [Google Scholar]
- 11.Cummings MJ, Baldwin MR, Abrams D, Jacobson SD, Meyer BJ, Balough EM, Aaron JG, Claassen J, Rabbani LE, Hastie J, Hochman BR, Salazar-Schicchi J, Yip NH, Brodie D, O’Donnell MR. Epidemiology, clinical course, and outcomes of critically ill adults with COVID-19 in New York City: a prospective cohort study. Lancet 395: 1763–1770, 2020. doi: 10.1016/S0140-6736(20)31189-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fraser DD, Cepinskas G, Slessarev M, Martin C, Daley M, Miller MR, O’Gorman DB, Gill SE, Patterson EK, Dos Santos CC. Inflammation profiling of critically ill coronavirus disease 2019 patients. Crit Care Explor 2: e0144, 2020. doi: 10.1097/CCE.0000000000000144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mo P, Xing Y, Xiao Y, Deng L, Zhao Q, Wang H, Xion Y, Chen Z, Gao S, Lian K, Luo M, Chen T, Song S, Ma Z, Chen X, Zheng R, Cao Q, Wang F, Zhang Y. Clinical characteristics of refractory COVID-19 pneumonia in Wuhan, China. Clin Infect Dis ciaa270, 2020. doi: 10.1093/cid/ciaa270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qin C, Zhou L, Hu Z, Zhang S, Yang S, Tao Y, Xie C, Ma K, Shang K, Wang W, Tian D-S. Dysregulation of immune response in patients with coronavirus 2019 (COVID-19) in Wuhan, China. Clin Infect Dis 71: 762–768, 2020. doi: 10.1093/cid/ciaa248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Y, Lu X, Li Y, Chen H, Chen T, Su N, Huang F, Zhou J, Zhan B, Yan F, Wang J. Clinical course and outcomes of 344 intensive care patients with COVID-19. Am J Respir Crit Care Med 201: 1430–1434, 2020. doi: 10.1164/rccm.202003-0736LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu C, Chen X, Cai Y, Xia J, Zhou X, Xu S, Huang H, Zhang L, Zhou X, Du C, Zhang Y, Song J, Wang S, Chao Y, Yang Z, Xu J, Zhou X, Chen D, Xiong W, Xu L, Zhou F, Jiang J, Bai C, Zheng J, Song Y. Risk factors associated with acute respiratory distress syndrome and death in patients with coronavirus disease 2019 pneumonia in Wuhan, China. JAMA Intern Med 180: 934–943, 2020. doi: 10.1001/jamainternmed.2020.0994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou F, Yu T, Du R, Fan G, Liu Y, Liu Z, Xiang J, Wang Y, Song B, Gu X, Guan L, Wei Y, Li H, Wu X, Xu J, Tu S, Zhang Y, Chen H, Cao B. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet 395: 1054–1062, 2020. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lucas C, Wong P, Klein J, Castro TBR, Silva J, Sundaram M, , et al. Longitudinal analyses reveal immunological misfiring in severe COVID-19. Nature 584: 463–469, 2020. doi: 10.1038/s41586-020-2588-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Benjamini Y. Simultaneous and selective inference: current successes and future challenges. Biom J 52: 708–721, 2010. doi: 10.1002/bimj.200900299. [DOI] [PubMed] [Google Scholar]
- 20.Antcliffe DB, Burnham KL, Al-Beidh F, Santhakumaran S, Brett SJ, Hinds CJ, Ashby D, Knight JC, Gordon AC. Transcriptomic signatures in sepsis and a differential response to steroids. From the VANISH randomized trial. Am J Respir Crit Care Med 199: 980–986, 2019. doi: 10.1164/rccm.201807-1419OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Collins FS, Varmus H. A new initiative on precision medicine. N Engl J Med 372: 793–795, 2015. doi: 10.1056/NEJMp1500523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bos LD, Schouten LR, van Vught LA, Wiewel MA, Ong DSY, Cremer O, Artigas A, Martin-Loeches I, Hoogendijk AJ, van der Poll T, Horn J, Juffermans N, Calfee CS, Schultz MJ; MARS consortium. Identification and validation of distinct biological phenotypes in patients with acute respiratory distress syndrome by cluster analysis. Thorax 72: 876–883, 2017. doi: 10.1136/thoraxjnl-2016-209719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Davenport EE, Burnham KL, Radhakrishnan J, Humburg P, Hutton P, Mills TC, Rautamen A, Gordon AC, Garrard C, Hill AV, Hinds CJ, Kinght JC. Genomic landscape of the individual host response and outcomes in sepsis: a prospective cohort study. Lancet Respir Med 4: 259–271, 2016. doi: 10.1016/S2213-2600(16)00046-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Balnis J, Adam AP, Chopra A, Chieng HC, Drake LA, Martino N, Ramos RB, Feustel PJ, Overmyer KA, Shishkova E, Coon JJ, Singer HA, Judson MA, Jaitovich A. Higher plasma levels of chemokine CCL19 are associated with poor SARS-CoV-2 acute respiratory distress syndrome (ARDS) outcomes (Preprint). medRxiv, 2020. doi: 10.1101/2020.05.21.20051300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zou X, Li S, Fang M, Hu M, Bian Y, Ling J, Yu S, Jing L, Li D, Huang J. Acute physiology and chronic health evaluation ii score as a predictor of hospital mortality in patients of coronavirus disease 2019. Crit Care Med 48: e657–e665, 2020. doi: 10.1097/CCM.0000000000004411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Charlson ME, Pompei P, Ales KL, MacKenzie CR. A new method of classifying prognostic comorbidity in longitudinal studies: development and validation. J Chronic Dis 40: 373–383, 1987. doi: 10.1016/0021-9681(87)90171-8. [DOI] [PubMed] [Google Scholar]
- 27.Li X, Ma X. Acute respiratory failure in COVID-19: is it "typical" ARDS? Crit Care 24: 198, 2020. doi: 10.1186/s13054-020-02911-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yancy CW. COVID-19 and African Americans. JAMA 323: 1891, 2020. doi: 10.1001/jama.2020.6548. [DOI] [PubMed] [Google Scholar]
- 29.Shankar-Hari M, Fan E, Ferguson ND. Acute respiratory distress syndrome (ARDS) phenotyping. Intensive Care Med 45: 516–519, 2019. doi: 10.1007/s00134-018-5480-6. [DOI] [PubMed] [Google Scholar]
- 30.Hermine O, Mariette X, Tharaux P-L, Resche-Rigon M, Porcher R, Ravaud P; CORIMUNO-19 Collaborative Group. Effect of tocilizumab vs usual care in adults hospitalized with COVID-19 and moderate or severe pneumonia: a randomized clinical trial. JAMA Intern Med 181: 32–40, 2020. doi: 10.1001/jamainternmed.2020.6820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stone JH, Frigault MJ, Serling-Boyd NJ, Fernandes AD, Harvey L, Foulkes AS, etal. Efficacy of tocilizumab in patients hospitalized with COVID-19. N Engl J Med 383: 2333–2344, 2020. doi: 10.1056/NEJMoa2028836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pugin J, Ricou B, Steinberg KP, Suter PM, Martin TR. Proinflammatory activity in bronchoalveolar lavage fluids from patients with ARDS, a prominent role for interleukin-1. Am J Respir Crit Care Med 153: 1850–1856, 1996. doi: 10.1164/ajrccm.153.6.8665045. [DOI] [PubMed] [Google Scholar]
- 33.Pugin J, Verghese G, Widmer MC, Matthay MA. The alveolar space is the site of intense inflammatory and profibrotic reactions in the early phase of acute respiratory distress syndrome. Crit Care Med 27: 304–312, 1999. doi: 10.1097/00003246-199902000-00036. [DOI] [PubMed] [Google Scholar]
- 34.Lee YM, Hybertson BM, Cho HG, Terada LS, Cho O, Repine AJ, Repine JE. Platelet-activating factor contributes to acute lung leak in rats given interleukin-1 intratracheally. Am J Physiol Lung Cell Mol Physiol 279: L75–L80, 2000. doi: 10.1152/ajplung.2000.279.1.L75. [DOI] [PubMed] [Google Scholar]
- 35.Roux J, Kawakatsu H, Gartland B, Pespeni M, Sheppard D, Matthay MA, Canessa CM, Pittet J-F. Interleukin-1beta decreases expression of the epithelial sodium channel alpha-subunit in alveolar epithelial cells via a p38 MAPK-dependent signaling pathway. J Biol Chem 280: 18579–18589, 2005. doi: 10.1074/jbc.M410561200. [DOI] [PubMed] [Google Scholar]
- 36.Herold S, Tabar TS, Janssen H, Hoegner K, Cabanski M, Lewe-Schlosser P, Albrecht J, Driever F, Vadasz I, Seeger W, Steinmueller M, Lohmeyer J. Exudate macrophages attenuate lung injury by the release of IL-1 receptor antagonist in gram-negative pneumonia. Am J Respir Crit Care Med 183: 1380–1390, 2011. doi: 10.1164/rccm.201009-1431OC. [DOI] [PubMed] [Google Scholar]
- 37.Donnelly SC, Strieter RM, Reid PT, Kunkel SL, Burdick MD, Armstrong I, Mackenzie A, Haslett C. The association between mortality rates and decreased concentrations of interleukin-10 and interleukin-1 receptor antagonist in the lung fluids of patients with the adult respiratory distress syndrome. Ann Intern Med 125: 191–196, 1996. doi: 10.7326/0003-4819-125-3-199608010-00005. [DOI] [PubMed] [Google Scholar]
- 38.Chada M, Nogel S, Schmidt AM, Ruckel A, Bosselmann S, Walther J, Papadopoulos T, von der Hardt K, Dotsch J, Rascher W, Kandler MA. Anakinra (IL-1r antagonist) lowers pulmonary artery pressure in a neonatal surfactant depleted piglet model. Pediatr Pulmonol 43: 851–857, 2008. doi: 10.1002/ppul.20851. [DOI] [PubMed] [Google Scholar]
- 39.Ortiz LA, Dutreil M, Fattman C, Pandey AC, Torres G, Go K, Phinney DG. Interleukin 1 receptor antagonist mediates the antiinflammatory and antifibrotic effect of mesenchymal stem cells during lung injury. Proc Natl Acad Sci USA 104: 11002–11007, 2007. doi: 10.1073/pnas.0704421104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meyer NJ, Feng R, Li M, Zhao Y, Sheu CC, Tejera P, Gallop R, Bellamy S, Rushefski M, Lanken PN, Aplenc R, O'Keefe GE, Wurfel MM, Christiani DC, Christie JD. IL1rn coding variant is associated with lower risk of acute respiratory distress syndrome and increased plasma IL-1 receptor antagonist. Am J Respir Crit Care Med 187: 950–959, 2013. doi: 10.1164/rccm.201208-1501OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cauchois R, Koubi M, Delarbre D, Manet C, Carvelli J, Blasco VB, Jean R, Fouche L, Bornet C, Pauly V, Mazodier K, Pestre V, Jarrot P-A, Dinarello CA, Kaplanski G. Early IL-1 receptor blockade in severe inflammatory respiratory failure complicating COVID-19. Proc Natl Acad Sci USA 117: 18951–18953, 2020. doi: 10.1073/pnas.2009017117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cavalli G, De Luca G, Campochiaro C, Della-Torre E, Ripa M, Canetti D, Oltolini C, Castiglioni B, Din CT, Boffini N, Tomelleri A, Farina N, Ruggeri A, Rovere-Querini P, Lucca GD, Martinenghi S, Scotti R, Tresoldi M, Ciceri F, Landoni G, Zangrillo A, Scarpellini P, Dagna L. Interleukin-1 blockade with high-dose anakinra in patients with COVID-19, acute respiratory distress syndrome, and hyperinflammation: a retrospective cohort study. Lancet Rheumatol 2: e325–e331, 2020. doi: 10.1016/S2665-9913(20)30127-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cocchi F, DeVico AL, Garzino-Demo A, Arya SK, Gallo RC, Lusso P. Identification of RANTES, MIP-1 alpha, and MIP-1 beta as the major HIV-suppressive factors produced by CD8+ T cells. Science 270: 1811–1815, 1995. doi: 10.1126/science.270.5243.1811. [DOI] [PubMed] [Google Scholar]
- 44.Liao M, Liu Y, Yuan J, Wen Y, Xu G, Zhao J, Cheng L, Li J, Wang X, Wang F, Liu L, Amit I, Zhang S, Zhang Z. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat Med 26: 842–844, 2020. doi: 10.1038/s41591-020-0901-9. [DOI] [PubMed] [Google Scholar]
- 45.Desai TJ, Cardoso WV. Growth factors in lung development and disease: friends or foe? Respir Res 3: 2, 2002. doi: 10.1186/rr169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ryan RM, Mineo-Kuhn MM, Kramer CM, Finkelstein JN. Growth factors alter neonatal type II alveolar epithelial cell proliferation. Am J Physiol 266: L17–L22, 1994. doi: 10.1152/ajplung.1994.266.1.L17. [DOI] [PubMed] [Google Scholar]
- 47.Sznajder JI, Ridge KM, Yeates DB, Ilekis J, Olivera W. Epidermal growth factor increases lung liquid clearance in rat lungs. J Appl Physiol (1985) 85: 1004–1010, 1998. doi: 10.1152/jappl.1998.85.3.1004. [DOI] [PubMed] [Google Scholar]
- 48.Sheu CC, Zhai R, Su L, Tejera P, Gong MN, Thompson BT, , et al. Sex-specific association of epidermal growth factor gene polymorphisms with acute respiratory distress syndrome. Eur Respir J 33: 543–550, 2009. doi: 10.1183/09031936.00091308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bos LDJ, Scicluna BP, Ong DSY, Cremer O, van der Poll T, Schultz MJ. Understanding heterogeneity in biologic phenotypes of acute respiratory distress syndrome by leukocyte expression profiles. Am J Respir Crit Care Med 200: 42–50, 2019. doi: 10.1164/rccm.201809-1808OC. [DOI] [PubMed] [Google Scholar]
- 50.Wong HR, Cvijanovich NZ, Anas N, Allen GL, Thomas NJ, Bigham MT, Weiss SL, Fitzgerald J, Checchia PA, Meyer K, Shanley TP, Quasney M, Hall M, Gedeit R, Freishtat RJ, Nowak J, Shekhar RS, Gertz S, Dawson E, Howard K, Harmon K, Beckman E, Frank E, Lindsell CJ. Developing a clinically feasible personalized medicine approach to pediatric septic shock. Am J Respir Crit Care Med 191: 309–315, 2015. doi: 10.1164/rccm.201410-1864OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rangel-Moreno J, Moyron-Quiroz JE, Hartson L, Kusser K, Randall TD. Pulmonary expression of CXC chemokine ligand 13, CC chemokine ligand 19, and CC chemokine ligand 21 is essential for local immunity to influenza. Proc Natl Acad Sci USA 104: 10577–10582, 2007. doi: 10.1073/pnas.0700591104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Damiani S, Fiorentino M, De Palma A, Foschini MP, Lazzarotto T, Gabrielli L, Viale PL, Attard L, Riefolo M, D’Errico A. Pathological post-mortem findings in lungs infected with SARS-CoV-2. J Pathol 253: 31. –. 2020. doi: 10.1002/path.5549. [DOI] [PubMed] [Google Scholar]
- 53.Dorward DA, Russell CD, Um IH, Elshani M, Armstrong SD, Penrice-Randal R, Millar T, Lerpiniere CE, Tagliavini G, Hartley CS, Randle NP, Gachanja NN, Potey PM, Dong X, Anderson AM, Campbell VL, Duguid AJ, Al Qsous W, BouHaidar R, Baillie JK, Dhaliwal K, Wallace WA, Bellamy CO, Prost S, Smith C, Hiscox JA, Harrison DJ, Lucas CD. Tissue-specific immunopathology in fatal COVID-19. Am J Respir Crit Care Med 203: 192–201, 2021. doi: 10.1164/rccm.202008-3265OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Spits H, Artis D, Colonna M, Diefenbach A, Di Santo JP, Eberl G, Koyasu S, Locksley RM, McKenzie AN, Mebius RE, Powrie F, Vivier E. Innate lymphoid cells–a proposal for uniform nomenclature. Nat Rev Immunol 13: 145–149, 2013. doi: 10.1038/nri3365. [DOI] [PubMed] [Google Scholar]