ABSTRACT
Introduction: The current SARS-CoV-2 pandemic urgently demands for both prevention and treatment strategies. RNA-dependent RNA-polymerase (RdRp), which has no counterpart in human cells, is an excellent target for drug development. Given the time-consuming process of drug development, repurposing drugs approved for other indications or at least successfully tested in terms of safety and tolerability, is an attractive strategy to rapidly provide an effective medication for severe COVID-19 cases.
Areas covered: The currently available data and upcoming studies on RdRp which can be repurposed to halt SARS-CoV-2 replication, are reviewed.
Expert opinion: Drug repurposing and design of novel compounds are proceeding in parallel to provide a quick response and new specific drugs, respectively. Notably, the proofreading SARS-CoV-2 exonuclease activity could limit the potential for drugs designed as immediate chain terminators and favor the development of compounds acting through delayed termination. While vaccination is awaited to curb the SARS-CoV-2 epidemic, even partially effective drugs from repurposing strategies can be of help to treat severe cases of disease. Considering the high conservation of RdRp among coronaviruses, an improved knowledge of its activity in vitro can provide useful information for drug development or drug repurposing to combat SARS-CoV-2 as well as future pandemics.
KEYWORDS: COVID-19, drug repurposing, nucleoside inhibitors, rna polymerase, sars-CoV-2
1. Introduction
Coronaviruses are positive sense, single-stranded, enveloped RNA viruses with a propensity to cross species barriers and causing disease in humans and animals [1]. In the past two decades, two zoonotic pathogenic coronaviruses belonging to the β-coronavirus genus, caused epidemics of severe respiratory infection among humans, including severe acute respiratory syndrome coronavirus (SARS-CoV) in 2003 and Middle East respiratory syndrome coronavirus (MERS-CoV) in 2012. Coronavirus disease 2019 (COVID-19) is an acute respiratory infection disease caused by a novel coronavirus (2019-nCoV, later renamed SARS-CoV-2) which was first identified in December 2019 in Wuhan, Hubei, China. The incidence of COVID-19 has grown dramatically in China and the virus has rapidly spread to more than 200 countries since late February 2020. On 11 March 2020 the World Health Organization declared the COVID-19 outbreak a global pandemic and as of 12 January 2021, over 88 million cases and 1.9 million deaths have been reported globally, with global death-to-cases ratio 2.2%, but these numbers are still on the rise in most countries. (https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports).
SARS-CoV-2 virions are spherical, with a diameter of 70–100 nm conforming to the typical coronaviruses diameter [2]. A prominent feature of the virion is the club-shaped spike projections protruding from the surface of the virion and resembling the appearance of a solar corona. Coronaviruses have helically symmetric nucleocapsids, which is uncommon among positive-sense RNA viruses, but far more common for negative-sense RNA viruses [3]. The genome of SARS-CoV-2 is a single‐stranded positive‐sense RNA of about 29.9 kb in length with the structure typical of known CoVs, as shown by (i) a highly conserved genomic organization with a large replicase gene, (ii) expression of many non-structural genes by ribosomal frameshifting, (iii) several enzymatic activities encoded within the large replicase-transcriptase polyprotein, (iv) expression of downstream genes by synthesis of 3ʹ nested subgenomic mRNAs. The genome is flanked by two untranslated regions (UTRs), similar to those of other β-coronaviruses, with nucleotide identities of > 83.6% [4].
The clinical spectrum of COVID-19 ranges from asymptomatic infection to fatal disease. The disease is usually mild in children, but severe infection in immunocompromised and elderly patients, particularly in the presence of significant comorbidity, may be associated with an increased fatality rate in this most vulnerable population. The most important risk factors for progression to severe disease include age, hypertension, diabetes, immunodeficiency and chronic cardiovascular and pulmonary diseases and cancer. Patients with severe COVID-19 may develop acute respiratory distress syndrome (ARDS) and hypoxia with oxygen saturation levels under 90%, multiorgan dysfunction syndrome, and other extrapulmonary manifestations [2]. As exhaustively described in recent reviews [5,6], severe COVID-19 symptoms are mostly the consequence of immune dysregulation and uncontrolled inflammatory response. Indeed, the rapid activation of the cell-mediated response leads to an increased pro-inflammatory status with a massive release of cytokines, particularly IL-6, IL-2, IL-10, and TNF-α, causing acute lung injury and contributing to the COVID-19 pathology. The accumulating evidence of dysregulated pro-inflammatory responses during SARS-CoV-2 infections has led to the use of immune modulators to mitigate COVID-19 immunopathology, including corticosteroids and the IL-6 receptor antagonist tolicizumab [7]. However, these agents can delay viral clearance and enhance the risk of secondary infections [2,8]. Overall, treatment of severe or critical COVID-19 remains supportive including high flow nasal oxygen, continuous positive airway pressure or mechanical ventilation, as well as continuous renal replacement therapy and extracorporeal membrane oxygenation [5].
Considering the devastating economic and social impact of the SARS-CoV-2 pandemic, there is an unprecedently urgent need for effective therapeutics to reduce the clinical consequences of SARS-CoV-2 infection. Drug repurposing has been used in response to emerging infectious diseases to rapidly identify potential therapeutics. The repurposing of drugs approved for treatment of other pathogens is a feasible and attractive strategy to deliver at least a partially effective agent(s) and mitigate SARS-CoV-2 pathology and spread. Indeed, recycling FDA-approved compounds dramatically shortens the development time and cost allowing immediate drug testing in clinical trials [9].
Viral enzymes are favorite targets for drug repurposing because specific domains are conserved within the coronaviridae family and may show homology with other positive-sense RNA viruses [10]. The non-structural protein (nsp) 12 is the central component of the SARS-CoV-2 replication/transcription machinery responsible for full virus genome replication and multiple subgenomic mRNAs synthesis, with nsp7 and nsp8 acting as cofactors to increase processivity [11]. Nsp8 is capable of de-novo initiating the replication process and has been proposed to operate as a primase, similarly to nsp7 [12]. Nsp12 needs to associate with nsp7 and nsp8 to activate its capability to replicate long RNA templates. The structure of the SARS-CoV-2 full-length nsp12 (residues 1–932) complexed with nsp7 (residues 1–83) and nsp8 (residues 1–198) cofactors has recently been solved by high-resolution cryo-electron microscopy [13,14]. The replication/transcription complex [12] is similar to those formed by SARS-CoV, including two monomers (nsp12 and nsp8) and one heterodimer (nsp7 and nsp8) showing three distinct domains: a ‘right hand’ RNA-dependent RNA polymerase (RdRp) domain (residues 367–920), a nidovirus-unique N-terminal extension domain (residues 4–28 and 69–249) harboring the nucleotidyltransferase activity (NiRAN) and an interface domain (residues 250–365) (Figure 1). SARS-CoV nsp12, nsp7 and nsp8 show high homology with SARS‐CoV‐2 counterparts sharing 96.35%, 98.8% and 97.5% similarity, respectively [15].
Figure 1.
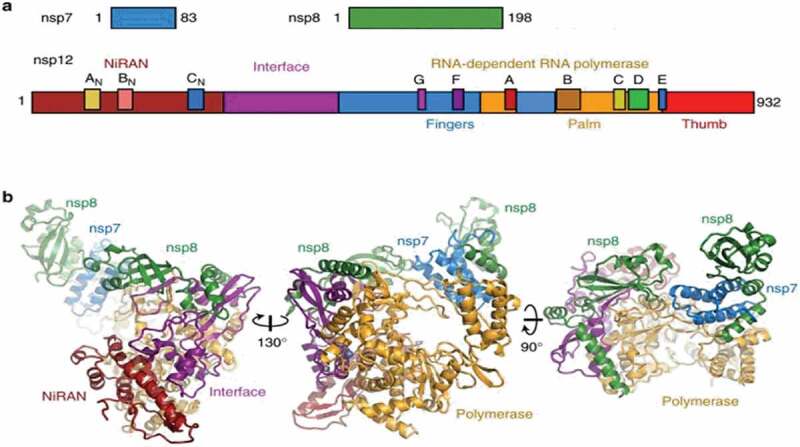
Color-coded scheme and structure of the SARS-CoV nsp12 RdRp bound to nsp7 and nsp8 co-factors. (a) Diagram of the SARS-CoV nsp7, nsp8, and nsp12 proteins indicating domains and conserved motifs. (b) SARS-CoV nsp12 contains a large N-terminal extension composed of the NiRAN domain (dark red) and an interface domain (purple) adjacent to the polymerase domain (orange). nsp12 binds to a heterodimer of nsp7 (blue) and nsp8 (green) as well as to a second subunit of nsp8. Adapted (http://creativecommons.org/licenses/by/4.0/) from Kirchdoerfer et al. [12]. Color figure.
The RdRp domain displays the canonical arrangement of the viral polymerases family [16] and consists of three subdomains: the finger subdomain (residues 366–581 and 621–679), the palm subdomain (residues 582–620 and 680–815), and the thumb subdomain (residues 816–920). RdRp contains all conserved motifs (from A to F) of RNA viruses RdRp [17] and the polymerase active site (Ser-Asp-Asp within motif C) is conserved among nidoviruses [18]. Nsp12 also carries the motif G [19], which is a signature sequence of RdRp that initiates RNA synthesis in a primer-dependent manner [20]. The active site of SARS-COV-2 RdRp, encompassing motifs A to G in the palm domain, is highly conserved not only among coronaviruses but among different RNA positive-stranded viruses [13,21]. Indeed, motif A carries the classic divalent-cation–binding residue D618, which is conserved in most viral polymerases including Hepatitis C virus (HCV) NS5B (residue D220) and poliovirus (PV) polymerase (residue D233). Motif C, which binds to the RNA 3ʹ end, contains the catalytic residues (from 759 to 761) required for RNA synthesis and conserved in most viral RdRps (from 317 to 319 in HCV and from 327 to 329 in PV). The configuration of the template-primer entry paths, the NTP entry channel, and the nascent strand exit path are similar to those described for SARS-CoV and for other RNA polymerases, such as HCV and PV polymerase [13]. Other accessory proteins involved in the replication complex machinery are the helicase (nsp13), carrying an N-terminal domain conserved among all nidoviruses which can unwind DNA or RNA in an NTP-dependent manner [22,23] and the exoribonuclease (nsp14 also called ExoN) responsible for increased fidelity of virus replication [24]. The proofreading activity of the coronavirus replication complex could indeed reduce the activity of nucleoside analogs through discrimination or excision of the candidate antiviral agent. It has been already observed that ExoN is responsible for the intrinsic resistance of coronavirus species to ribavirin and several other nucleoside analogs [25,26]. Thus, this feature must be considered in drug design or repurposing.
The conservation of RdRp among evolutionary distant RNA viruses and the absence of host homologs clearly make it an ideal target for drug repurposing [9,27]. Indeed, 130 clinical trials involving 65,263 patients are ongoing (last updated at January 2021; https://covdb.stanford.edu/clinical-trials/) to evaluate RdRp inhibitors alone or in combinations; 10 of them have completed phase III. Concomitantly, several studies evaluating the activity of RdRp in cell culture using different cell systems and readout methods have been published (https://covdb.stanford.edu/search/?target=Polymerase&virus=SARS-CoV-2#invitro-cells).
The most promising, broad-spectrum class of viral RdRp inhibitors are nucleoside and nucleotide analogs (NAs). Upon delivery into the host cell, nucleoside/nucleotide prodrugs are metabolized into an active 5ʹ-triphosphate form (5ʹ-TP) which competes with endogenous nucleotides for the incorporation into the nascent viral RNA operated by the RdRp. NAs terminate the RNA synthesis acting as obligate chain terminators, due to the lack of the 3ʹ-OH required for RNA chain elongation. Lack of the 3ʹ-OH group is not always essential for the chain termination activity of NAs. Indeed, in the case of sofosbuvir, a key antiviral drug approved for the treatment of Hepatitis C [28], the chain termination is due to the steric clashes of the 2ʹ-methyl group. In addition, the inhibitory activity of sofosbuvir appeared to be enhanced by resistance to the excision activity exerted by the SARS-CoV-2 exonuclease [29]. With remdesivir, another broad range antiviral initially developed to combat Ebola virus and eventually approved under emergency use for the treatment of SARS-CoV-2, the presence of the 3ʹ-OH group allows the formation of the phosphodiester bond to the next incoming nucleotide and the elongation of the growing RNA chain. However, viral RNA synthesis is blocked following the addition of another three NTPs as a result of the steric hindrance caused by the 1ʹ-CN group with the S861 residue [30]. Other NAs (i.e. favipiravir, ribavirin) are incorporated in the nascent viral RNA at high rate, but they are not recognized as endogenous nucleotides in the subsequent round of replication, increasing the mutation rate and leading to the generation of non-viable genomes, a process referred to as ‘lethal mutagenesis’ [31].
In light of the massive in vitro testing of molecules aiming at the identification of SARS-CoV-2 antivirals, the intent of this review is to provide an overview of the most promising SARS-CoV-2 RdRp inhibitors under clinical investigation that have been identified according to the drug repurposing strategy (Figure 2 and Table 1).
Figure 2.
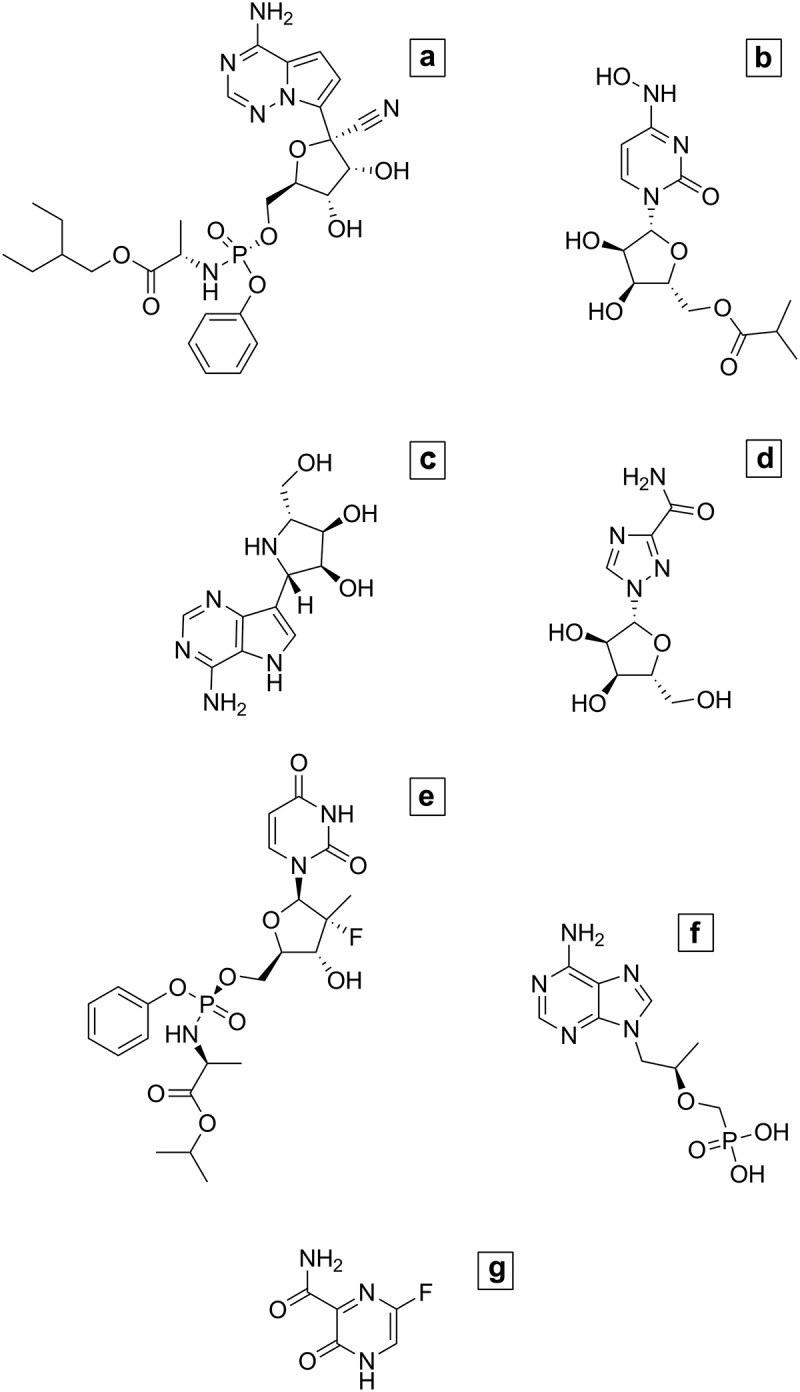
Chemical structure of potential SARS-CoV-2 RdRp inhibitors under clinical investigation. (A) Remdesivir; (B) Molnupiravir; (C) Galidesivir; (D) Ribavirin; (E) Sofosbuvir; (F) Tenofovir; (G) Favipiravir
Table 1.
Overview of the potential SARS-CoV-2 RdRp inhibitors under clinical investigation that have been identified according to the drug repurposing strategy. NT: not tested; NA: Not approved; RSV: Respiratory Syncytial Virus; HCV: Hepatitis C virus; HBV: Hepatitis B; TDF: Tenofovir disoproxil fumarate; TAF: Tenofovir alafenamide. References are indicated in square brackets. Additional data based on non-peer-reviewed manuscripts can be found at the Coronavirus Antiviral Research Database website [https://covdb.stanford.edu/ [32]
| SARS-COV-2 | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Drug (alternative names) | Original patent application number | FDA approval | In vitro activity (range IC50, µM) | In vivo activity | Number of clinical trials | ||||
| Non-human cells | Human cells | Host | Lung titer | Swab titer | Completed | Ongoing | |||
| Remdesivir (GS-5734; Veklury) | WO 2016161176 | SARS-COV-2 | 0.3–27[33–46] | 0.003–1.3[35,39,40,47,48] | Mice [35] and rhesus macaque[49] | Decreased | Decreased | 5 | 27 |
| Molnupiravir (EIDD-2801) | WO 2017156380 | NA | 0.3 [38,50] | 0.02–0.09 [50] | Mice [50] | Decreased | Decreased | 1 | 2 |
| Favipiravir (Avigan; Favipira; T-705) | WO 200010569 | Influenza* SARS-CoV-2**; | 62 [33] | not active [39] | NT | NT | NT | 3 | 25 |
| Galidesivir (BCX4430) | WO 1999019338 | NA | >100 [34] | NT | NT | NT | NT | 1 | 2 |
| Ribavirin (Virazole) | US 3976545 | RSV; HCV; Lassa fever virus; Crimean Congo and other hemorrhagic fever viruses, New World arenaviruses | >20 [33,34,38] | Not active [39] | NT | NT | NT | 1 | 5 |
| Sofosbuvir (Sovaldi) | WO 2008121634 | HCV | >20 [38] | Not active [39] | NT | NT | NT | 1 | 8 |
| Tenofovir (TDF/Viread, TAF/Vemlidy) | US 4808716 | HIV; HBV | NT | NT | Ferret [51] | NT | Decreased | 0 | 3 |
* Approved against influenza in China, Japan and Russia; **approved against SARS-CoV-2 in China and Russia.
2. SARS-CoV-2 polymerase inhibitors
2.1. Remdesivir
Remdesivir (RDV, formerly GS-5734, brand name Veklury) is a prodrug form of the monophosphate adenosine analog GS-441524 initially developed to fight Ebola Virus infection due to its potent antiviral activity in vitro [52]. Despite treatment with RDV was associated with a 100% survival in non-human primates, the use of RDV in humans was discontinued due to a higher incidence of deaths as compared to treatments with monoclonal antibodies in a clinical trial conducted in the Democratic Republic of Congo, seriously affected by Ebola virus outbreaks in recent years [53].
Further, cell-based screening revealed that RDV had a considerable broad-spectrum antiviral activity against RNA viruses belonging to the Filoviridae, Paramyxoviridae (except for mumps virus), Coronaviridae, Picornaviridae and Pneumoviridae families, while little or no activity was detected against members of the Flaviviridae, Bunyaviridae, Arenaviridae and Rhabdoviridae families [54–58]. However, two recent studies showed that the triphosphate form of RDV can efficiently inhibit several flaviviral polymerases in a biochemical assay, suggesting that the optimization of the chemical structure of RDV might lead to a more potent inhibition of flavivirus replication also in cell-based assays and in vivo[56,59].
Biochemical studies indicated that RDV is incorporated into the nascent RNA with similar or increased efficacy as compared to the natural nucleoside adenosine in the presence of viral RdRp but not of human DNA-dependent RNA polymerases or DNA-dependent DNA polymerases [15,60–62]. Focusing on coronaviruses, the proposed mechanism of action is a delayed chain termination where the RNA synthesis is terminated three nucleotides after the addition of RDV, thus reducing the removal of RDV through the proof-reading activity of the viral exonuclease nsp14 [61,63,64]. Indeed, experiments with the β-coronavirus murine hepatitis virus model demonstrated that the lack of exonuclease activity caused a 6-fold increase in the susceptibility to RDV [64]. In the same model of infection, resistance selection experiments allowed to identify two emerging mutations (F476L and V553L) in the fingers domain of RdRp resulting in decreased susceptibility to RDV and reduced replication capacity. Homologous mutations in SARS-CoV (F480L + V557L) led to resistance to RDV and are hypothesized to cause a conformational change favoring the incorporation of adenosine instead of RDV [65].
Due to early studies demonstrating its antiviral activity against coronaviruses circulating among humans and animal hosts [54] RDV was soon considered as a promising treatment option for SARS-CoV-2 infection. Indeed, in vitro testing showed that RDV was able to inhibit SARS-CoV-2 infection in VeroE6 cells in the low micromolar range, while a more potent inhibition was observed in human lung cell lines and primary human epithelial airway cells [33–36, 66]. Moreover, RDV inhibited SARS-CoV, MERS-CoV and bat CoVs in the nanomolar range in human epithelial cell culture models of infection with minimal cytotoxicity [54,67]. Prophylactic treatment of mice infected with SARS-CoV was effective in reducing viral titers in the lung as well as lung injury, while therapeutic treatment was able to reduce viral load but not the progression of pulmonary disease [54]. By contrast, both prophylactic and therapeutic treatments with RDV in mice and rhesus macaques infected with MERS-CoV resulted in a mitigation of pathological lung injuries [67,68]. Similar results were achieved in non-human primates infected with Marburg virus, where treatment with RDV after the onset of the symptoms was associated with higher survival and ameliorated clinical parameters [69]. In vivo testing on mouse models infected with chimeric SARS-CoV-2 virus [35] and on rhesus macaques infected with SARS-CoV-2 [49] demonstrated that early treatment with RDV was associated with clinical benefits, including reduced lung damage and lower viral titers.
Synergistic activity of RDV and molecules targeting both nsp12 and other viral or cellular proteins has also been observed, suggesting that combination therapy may inhibit viral replication at lower effective drug concentrations [34,66].
Based on in vitro activity and subsequent in vivo evidences in humans, RDV received the Emergency Use Authorization for the treatment of hospitalized patients affected by COVID-19 irrespective of the severity of the disease. Indeed, preliminary evidences indicated that intravenous administration of RDV is associated with a reduced time to recovery or clinical improvement, although with statistical significance only in patients not requiring intensive care unit hospitalization [70–76]. To date, more than fifty clinical trials have been registered to comprehensively evaluate the efficacy of RDV, either alone or in combination with other antiviral or immunomodulatory drugs (source clinicaltrials.gov).
2.2. Molnupiravir
Molnupiravir (formerly EIDD-2801) is the isopropylester prodrug of the ribonucleoside analog β-D-N4-hydroxycytidine (EIDD-1931, or N-hydroxycytidine). EIDD-1931 was found to inhibit the replication of several viruses (including Chikungunya virus, Venezuelan Equine Encephalitis virus (VEEV), Respiratory Syncytial virus (RSV), HCV, Norovirus, Influenza A and B viruses, EBOV and human coronaviruses) with minimal cytotoxicity and high genetic barrier to resistance [77–84]. Incorporation of EIDD-1931 in place of cytosine or uracil during RNA synthesis was found to cause lethal mutagenesis throughout the whole genome of several viruses, inducing G to A and C to U transitions in a dose-dependent manner [77,78,80]. Development of resistance to EIDD-1931 was observed only after several passages of VEEV in the presence of increasing concentrations of the drug, resulting in the selection of three mutations (P187S, A189V, I190T) located in the index finger domain of the viral nsP4 RdRp that caused a low-level resistance [80].
The prodrug molnupiravir was developed to overcome the issue of poor bioavailability observed in animal models and demonstrated both in vitro and in vivo efficacy against influenza and multiple coronaviruses [50,85]. For these reasons, the safety, tolerability and pharmacokinetics of molnupiravir have been investigated in healthy volunteers in a phase 1 trial (NCT04392219), while two-phase IIa clinical trials are recruiting hospitalized patients with diagnosis of COVID-19 to evaluate the time of viral clearance after the administration of molnupiravir (NCT04405739) or the safety and efficacy compared to placebo and according to the time of the start of therapy with respect to the onset of symptoms (NCT04405570).
2.3. Favipiravir
Favipiravir (FPV; T-705, Avigan, Favipira; 6-fluoro-3-hydroxy-2-pyrazine carboxamide) is a guanine analog, as synthesized by modifying its pyrazine analog. It was approved in Japan, China and Russia for the treatment of influenza and it is effective against all subtypes of influenza viruses, both sensitive and resistant to the marketed neuraminidase inhibitors. FPV is not reported to have significant adverse effects; however, it may increase the risk for teratogenicity and embryotoxicity [86]. FPV is converted throughout phosphoribosylation and phosphorylation in FPV-ribofuranosyl-5′-triphosphate (F-RTP) by human cells and recognized as a purine nucleotide by RdRp blocking viral RNA synthesis [87]. The docking analysis revealed that F-RTP forms five hydrogen bond and seven hydrophobic interactions with the crucial amino acids of SARS‐CoV‐2 RdRp, including Arg553 acting on rNTP binding and Asp760-Asp761 positioned near the catalytic center of functional motif C. Considering the conservation of catalytic domain of viral RdRp, FPV has a potential of broad range activity on different viruses. Previous studies showed in vitro and in vivo antiviral activities of FPV against different RNA viruses including influenza A, B, and C, EBOV and Lassa viruses [86]. FPV showed protective effect against a wide range of RNA virus infections in animal models [88–91] but a reduced or absent selectivity against SARS‐CoV‐2 in monkey cell lines (SI = 1 [1–4.5]) [34,37,38,92] and in human cell lines engineered to overexpress ACE-2 [39]. Considering that monkey cell lines (VERO and VEROE6) are not amenable to assess the antiviral activity of RdRp inhibitors [35,93], the lack of activity in the A549 human cell line expressing the human ACE2 receptor is the first data excluding a protective role of FPV against SARS-CoV-2 [39]; further in vitro experiments using primary or human cell lines with different metabolic profiles are necessary to confirm this observation. Indeed, several clinical trials are evaluating the efficacy of FPV alone or in combination with other drugs. The first study conducted in China (ChiCTR2000029600) was an open-label, nonrandomized trial involving 80 hospitalized patients without severe COVID-19 [94]. A significant reduction in the median time to viral clearance was observed in the group treated with FPV with respect to the control arm, treated with lopinavir/ritonavir (P < 0.001). By day 14, the FPV group had a higher rate of improved chest computed tomography scans in comparison with the lopinavir/ritonavir control group (p = 0.004). In both arms, aerosolized IFN-α was administered until viral clearance, partly confounding the assessment of FPV efficacy. Despite unfavorable pharmacokinetic (PK) profile [95] and the lack of conclusive efficacy data, FPV was approved for marketing in the treatment of COVID-19 patients in China in March 2020 and in the Russian Federation in May 2020.
2.4. Galidesivir
Galidesivir (BCX4430) is an adenosine analog that has a substitution of carbon for nitrogen at position 7 on the base and a substitution of nitrogen for oxygen at position 1 on the ribose ring [96]. When the RdRp incorporates galidesivir triphosphate, the structural change alters its electrostatic interaction, resulting in premature termination of the elongating RNA strand. In vitro and in animal models, galidesivir can inhibit viral RNA polymerases of a wide array of RNA viruses including flaviviruses [97], filoviruses [96,98] and coronaviruses [96]. In docking simulation, Galidesivir inhibits SARS‐CoV‐2 by tightly binding to RdRp with six hydrogen bonds and four hydrophobic interactions (binding energy of −7.0 kcal/mol) [92]; it establishes strong connections with 10 different RdRp amino acid residues (Thr455, Arg553, Lys621, Arg624, Asp452, Ala554, Asp623, Asn691, Ser759, Asp760) [99–101]. Galidesivir has a rapid pharmacokinetics (below 5 minutes half‐life) which is extended to six hours in vivo for galidesivir triphosphate. Galidesivir was safe and generally well tolerated in a Phase 1 double-blind clinical trial (NCT02319772) evaluating the safety, tolerability and pharmacokinetics of intramuscular administration versus placebo in healthy subjects [98]. Confirmation of these data for the intravenous route of administration is pending upon completion of a recent trial (NCT03800173). Galidesivir displayed poor activity in monkey cell-line models [34], similarly to the others NI, and activity as a SARS-CoV-2 inhibitor has not yet been tested in animal models. However, in April 2020, a Phase 1b randomized, double-blind, placebo-controlled study has been started to evaluate the pharmacokinetics, safety, and antiviral effects of galidesivir administered intravenously vs. placebo in 132 hospitalized adult subjects with either Yellow Fever disease or severe COVID-19.
2.5. Ribavirin
Ribavirin (RBV, 1‐β‐D‐ribofuranosyl‐1,2,4‐triazole‐3‐carboxamide) is a guanosine analog with broad-spectrum antiviral activity, firstly synthesized in 1970 [102] and approved by FDA for the treatment of chronic HCVinfection, RSV infection and certain viral hemorrhagic fevers, including Lassa fever, hemorrhagic fever with renal syndrome caused by Hantavirus infection, New World arenaviruses infections, and Crimean-Congo hemorrhagic fever [103–105]. RBV inhibits a broad spectrum of RNA viruses including RSV, Influenza virus, several coronaviruses, flaviviruses and herpesviruses in animal and human cell lines [106–111].
As other nucleoside inhibitors, RBV is converted by host kinases into RBV triphosphate which pairs to the cytidine triphosphate or uridine triphosphate in the RNA template with equal efficiency blocking viral RNA elongation. RBV also increases the rate of viral mutations leading to accumulation of defective virions. Indeed, alternative, not mutually exclusive mechanisms of action have been proposed: (i) the RBV monophosphate metabolite inhibits the host inosine monophosphate dehydrogenase enzyme halting de novo synthesis of guanine nucleotides; (ii) the triphosphate form prevents the cap methylation step of the 5′ end of viral mRNA inhibiting the viral RNA guanylyltransferase and the mRNA 2′-O-methyltransferase; (iii) RBV exerts an immunomodulatory action enhancing the induction of interferon-related genes [112]. RBV is usually administered in combination with interferon or other antivirals; it is not prescribed in pregnant women and in the elderly population because of fetal toxicity and reduction of calcium and magnesium blood levels in the two populations, respectively [113].
Against MERS-CoV, RBV reduced viral replication in cell culture [106] and in combination with IFN-α2b in infected rhesus macaques improving the clinical outcome and the host response [114]. Similar results were not obtained against SARS-CoV [108,115]. RBV has potential activity against SARS-CoV-2, as shown by tightly binding to RdRp active site in molecular docking studies [116,117]. In cell-line models, RBV was not active against SARS-CoV-2 in VERO [33,34] cells and in the A549 human cell line expressing the ACE-2 receptor [39]. As previously explained, VERO cells are not adequate for the screening of polymerase inhibitors, as shown by lack of activity of RBV and other NIs, due to the inability to convert NIs into the tri-phosphate active form [118].
Five clinical trials including RBV alone (NCT04356677, Phase I), RBV in combination with IFN-β (NCT04494399, Phase II) or with different antivirals (NCT04497649 and NCT04460443, Phase II; NCT04392427, Phase III) are ongoing. The only open-label, randomized phase 2 trial completed (NCT04276688), recruited 127 adults with severe COVID-19 which were randomly assigned to a 14-day triple combination of lopinavir/ritonavir, RBV, and interferon beta-1b (combination group) or to 14 days of lopinavir/ritonavir (control group). The combination group had a significantly shorter median time to reach negative nasopharyngeal swab (7 days [IQR 5–11]) than the control group (12 days [IQR 8–15]; hazard ratio 4 · 37 [95% CI 1 · 86–10 · 24], p = 0.0010) [119]. Except for the phase I trial, the efficacy of RBV against SARS-CoV-2 in vivo has been, or is being, evaluated only in combination with other antivirals, thus to date its use is thought only in association with other active molecules, similar to clinical use in the treatment of HCV infection.
2.6. Sofosbuvir
Sofosbuvir (SOF, β-d-2′-deoxy-2′-α-fluoro-2′-β-C-methyluridine) is a uridine analog licensed for the treatment of HCV infection, targeting the NS5B HCV polymerase [120]. SOF is efficiently taken up by hepatocytes where it is converted by phosphorylation in the active form, the nucleoside triphosphate. SOF triphosphate competes with uridine to be incorporated by the HCV RNA polymerase into the elongating RNA strand, resulting in chain termination. The combination of SOF with velpatasvir, an HCV NS5A inhibitor, is effective against different HCV genotypes [121]. Previous work has shown that SOF has in vitro and/or in vivo antiviral activity against other flaviviruses [122–125], alphaviruses [126] and Hepatitis A virus [127].
In silico, SOF can tightly bind to SARS‐CoV‐2 RdRp with a − 7.5 kcal/mol binding energy, forming seven H‐bonds with four RdRp residues (W508 (3), K512 (2), A653, and W691) and two hydrophobic contacts (Y510 and D651) [117]. The binding between SARS-CoV-2 RdRp and SOF has been confirmed in different in silico and biochemical studies [128–131]. In addition, Chien et al. [130], demonstrated that SARS-CoV-2 RdRp is also permanently blocked by SOF triphosphate, leading to irreversible blocking of polymerase mediated RNA extension. Despite modeling and docking studies suggesting a potential role of SOF in the inhibition of SARS-CoV-2 nsp12 activity, SOF activity was not confirmed in a human cell-line model [39].
However, 9 randomized clinical trials are investigating the role of SOF in combination with RBV (NCT04460443, Phase II/III), daclatasvir (another HCV NS5A inhibitor) (NCT04460443, NCT04497649 and NCT04468087, Phase II/III; NCT04561063 and NCT04532931, Phase II; NCT04535869, Phase III), daclatasvir plus hydroxychloroquine (NCT04443725, Phase II/III) and ledipasvir (also an HCV NS5A inhibitor) (NCT04530422, Phase III; NCT04498936, Phase IV). The only clinical trial with published results (https://www.irct.ir/trial/46463) conducted in Tehran, was not included in the list of clinical trials involving SOF available at www.clinicaltrials.gov. This open-label, multicentre, and randomized clinical trial (IRCT20200128046294N2) was conducted on 66 in adults with moderate or severe COVID-19. Patients were randomized into a treatment arm receiving SOF and daclatasvir plus SOC, or a control arm receiving SOC alone. The primary endpoint was clinical recovery within 14 days of treatment. Clinical recovery, defined as normalization of fever, respiratory rate, and oxygen saturation (≥94%) by day 14 occurred in 29 (88%) of the SOF/DAC group compared with 22/33 (67%) of the SOC group. The treatment arm had a significantly shorter median duration of hospitalization [6 days (IQR 4–8)] than the control group [8 days (IQR 5–13)]; P = 0.029 [132].
Considering the excellent safety profile and oral availability of SOF, the information deriving from cell models and in vivo studies will be useful to clarify the potential of this drug against SARS-CoV-2.
2.7. Tenofovir
Tenofovir disoproxil fumarate (TDF) and tenofovir alafenamide (TAF) are both prodrug formulations of an acyclic nucleotide adenosine analog widely used in combination with other agents in the therapy of the Human Immunodeficiency virus (HIV) infection and as single agent in Hepatitis B virus (HBV) infection. TAF has been recently developed to replace TDF due to its improved pharmacokinetics properties and lower renal and bone toxicity [133]. Tenofovir acts as an immediate chain terminator when incorporated during the synthesis of viral DNA, synergizing with HIV-1 nucleoside and non-nucleoside reverse transcriptase inhibitors that prevent the pyrophosphorolysis of tenofovir and the consequent excision from the nascent DNA chain [134].
Molecular docking analysis carried out on a model of SARS-CoV-2 RdRp showed that tenofovir has a binding energy comparable to those of the native nucleotides, suggesting a possible inhibitory activity [99]. While data on the antiviral activity in cell-based systems are still awaited, the combination of tenofovir and emtricitabine, which is included in most of the currently recommended HIV-1 treatments and used as pre-exposure prophylaxis to prevent HIV-1 infection, demonstrated to reduce the viral titer and clinical symptoms as compared to a control group in an animal model of SARS-CoV-2 infection [51].
Considering the promising data from animal testing, the favorable pharmacokinetic properties and safety, the efficacy of combination of tenofovir and emtricitabine in the prevention of SARS-CoV-2 infection in healthcare personnel is currently under clinical investigation in three studies (NCT04519125, NCT04405271, NCT04334928).
3. Conclusions
The pressing need of clinically effective drugs against SARS-CoV-2 infection has led to an extensive in silico and in vitro screening of drug candidates among those already licensed for other viral diseases or compounds that have been previously tested in phase I/II clinical studies. Similar to other pathogenic viruses, SARS-CoV-2 RdRp represents an attractive drug target for antiviral therapy since viral polymerases are key enzymes for viral replication. Indeed, most of the recommended treatments of viral infections include at least one polymerase inhibitor, although the majority of these drugs target viral DNA-dependent DNA-polymerases and HIV and HBV RNA-dependent DNA-polymerases [135]. Before the SARS-CoV-2 pandemic, SOF (for HCV, worldwide) and FPV (for influenza, only in Japan) have been the only two approved drugs targeting RdRp. Due to the structural homology with other viral polymerases, particularly those of other members of the Coronaviridae family such as SARS-CoV-1, MERS-CoV and other human coronaviruses, in silico and biochemical studies soon identified possible SARS-CoV-2 RdRp inhibitors [116,117,130,136]. Among the molecules showing promising antiviral activity at nontoxic concentration in cell-based assays, RDV is the only one drug that recently received the authorization for the emergency use in hospitalized patients with COVID-19 based on preliminary data indicating reduced progression of the disease and faster time to recovery. FPV also obtained approval as an anti-SARS-CoV-2 medication in China and Russia, despite lack of conclusive clinical data, highlighting the desperate need for COVID-19 treatment. Other polymerase inhibitors showing encouraging in vitro activity against SARS-CoV-2, including the guanosine nucleotide analog AT-527 recently demonstrating inhibitory activity against HCV RdRp [137], are currently under clinical investigations and comprehensive data regarding in vivo efficacy are eagerly awaited in the next few months. In most of the cases, these drugs are being tested in combination with other compounds targeting other viral proteins (e.g., main and serine proteases, envelope spike protein) or with immunomodulators aiming to counteract the systemic inflammatory response generated during SARS-CoV-2 infection in an attempt to halt virus replication and curb the immunopathogenic mechanisms underlying the disease.
4. Expert opinion
The dramatic spread of SARS-CoV-2 is seriously affecting all human activities and health systems worldwide. While forecasting the course of the epidemic may be subject to gross error, there is no doubt cascading effects will impact global economy and politics in the next several months and possibly beyond. Immediate response to decrease the virus spread must rely on different forms and rules of social distancing. However, vaccination and antiviral treatment are the only means to halt the pandemic and cure severe cases. An unprecedented effort joining private and public resources is being spent along this line. Drug repurposing has been soon considered as the fastest strategy to find candidate drugs among those already licensed to treat other diseases or at least with acceptable safety and tolerability according to phase I/II clinical trials. It must be emphasized that even a partially effective treatment could prove of great values because reduced viral replication could substantially decrease the consequent immunopathogenic response and avoid a number of severe or critical diseases.
Based on genetic and structural similarities, the search for SARS-CoV-2 antivirals lays its foundation on previous work carried out on other coronaviruses affecting humans and animal species, such as SARS-CoV-1, MERS-CoV and bat coronaviruses. Among viral proteins, RdRp is being considered as the most strategic drug target for two main reasons: (i) the availability of clinically approved nucleoside analogs developed for other viruses; (ii) the rapid development of biochemical and cell-based assays that have accelerated the in vitro screening of candidate SARS-CoV-2 inhibitors. Indeed, the first drug that has received the emergency use authorization for COVID-19 treatment is the adenosine analog RDV, while other RdRp inhibitors showing promising in vitro activity, such as molnupiravir, are currently under clinical investigation. Data from a large clinical trial indicated that clinical benefits derived from the use of remdesivir were higher in patients treated as soon as possible after the onset of symptoms [70]. This finding supports that halting viral replication with an early antiviral intervention may be beneficial by reducing viral spread to the lower respiratory tract and/or decreasing the viral burden and the consequent triggering of immunopathology. Availability of more potent anti-SARS-CoV-2 antivirals could thus contribute substantially to improve the clinical course of the disease in patients at high risk of developing severe COVID-19, provided treatment can be started early.
Despite possible short-term reward from drug repurposing, further development of RdRp inhibitors is clearly necessary to identify compounds with improved potency and pharmacokinetics as well as reduced side effects. The experiments conducted to elucidate the mechanism of action of RDV suggest that delayed chain termination is an essential prerequisite for the inhibition of SARS-CoV-2 RNA synthesis by nucleoside analogs. Indeed, among the viral proteins involved in the coronavirus replication machinery, the nsp14 exonuclease exerts a key proof-reading activity, excising misincorporated nucleotides at the 3ʹ end of the nascent RNA chain and resuming RNA synthesis. Consequently, immediate chain terminators such as those lacking the 3ʹ-OH group might be easily removed by exonuclease when incorporated into the nascent RNA chain, while the addition of natural bases after the incorporation of a nucleoside analog can protect from nsp14 mediated excision. This suggests that future RdRp inhibitors should be preferably endowed with the ability to cause delayed, rather than immediate, chain termination coupled with a mechanism to block translocation or impair RNA polymerization later. In addition, it must be considered that all competitive RdRp inhibitors must be triphosphorylated to be incorporated into nascent RNA and phosphorylation may differ among different cell lines and particularly be suboptimal or yield misleading results when using non-human cell lines.
Although drug repurposing and combination therapies can help to reduce mortality in patients with moderate to severe COVID-19, the goal of future intervention is the containment of SARS-CoV-2 spread at an early stage. Several candidate prophylactic vaccines are under clinical evaluation and few of them have been recently approved for emergency use by one or more national/international regulatory authorities. In particular, the SARS-CoV-2 spike protein RNA-based BNT162b2 vaccine (brand name Comirnaty) developed by BioNTech and Pfizer has been licensed in 48 countries based on the demonstration of 95% efficacy in preventing COVID-19 after a two-dose schedule [138]. More recently, a similar SARS-CoV-2 spike protein RNA-based vaccine, manufactured by Moderna, has been licensed by FDA and EMA after achieving the same efficacy as the BNT162b2 vaccine in the field trial [139]. Although only short-term data are available, both vaccines have not shown serious side effects. However, mass vaccination will require considerable time and the duration of immunity in vaccinated people is still under investigation. In addition, surveillance of circulating viral strains has recently revealed the emergence of variants with aminoacidic substitutions in the spike coding region with unknown impact on the immunity in vaccine recipients (https://www.cdc.gov/coronavirus/2019-ncov/more/science-and-research/scientific-brief-emerging-variants.html). For these reasons, alternative therapeutic approaches based on combinations of drugs targeting multiple viral targets (e.g., spike protein, RdRp, helicase, protease) and/or reducing cytokine-mediated pathology must be developed. The clinical usefulness of candidate COVID-19 therapies should be evaluated on appropriate animal models to assess pharmacokinetic properties, safety, and efficacy. Several animal species are permissive for the replication of SARS-CoV-2; however, the clinical hallmarks of the COVID-19 are variable among species. For example, mice, hamsters, ferrets, and minks support viral replication and develop a variable spectrum of the clinical symptoms and pathology as those occurring in humans, while none of the non-human primate models closest to humans appear to recapitulate all the clinical manifestations of COVID-19 [140,141]. Moreover, future treatments should be conceived to be administered through inhalation to facilitate the penetration in the upper and lower respiratory tract and possibly increase ease of use across different patient populations. Finally, once SARS-CoV-2 antiviral drug(s) have substantially entered clinical use, plausible development of resistance to these drugs should also be monitored. Although treatment of an acute infection is administered for few days, thus limiting the possibility of emergent drug resistance, the huge number of patients eventually undergoing treatment can boost significantly this probability or drug-resistant viral variants may appear due to genetic drift, as shown with influenza neuraminidase inhibitors [142]. Ideally, testing for the genetic barrier to resistance should be an integral part of the drug development process with any candidate drug targeting a viral function. Integrating the medicinal chemistry, virology and immunology domains remains the best pathway to discover novel drugs and contribute to an effective response to SARS-CoV-2 as well as to future pandemics.
Funding Statement
This work was partially supported by funds from Ministero dell'Istruzione, dell'Università e della Ricerca, project PRIN 2017, ORIGINALE CHEMIAE in Antiviral Strategy - Origin and Modernization of Multi-Component Chemistry as a Source of Innovative Broad-Spectrum Antiviral Strategy (cod. 2017BMK8JR).
Article highlights
• RNA‐dependent RNA polymerase (RdRp) plays an essential role in SARS‐CoV‐2 viral replication.
• RdRp motifs are highly conserved among coronaviruses and RNA positive-stranded viruses, offering an attractive target for broad-spectrum antivirals
• This review focuses on most promising repurposed drugs inhibiting the SARS-COV-2 RdRp
• For each drug, in vitro and in vivo data against SARS-COV-2 are summarized
• The review provides an expert opinion of the clinical use of nucleoside analogs in COVID-19 treatment
This box summarizes key points contained in the article.
Declaration of interest
M Zazzi has received grants and personal fees from Gilead Sciences, Janssen-Cilag, Merck Sharp and Dohme, ViiV Healthcare, outside the current work. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties.
Reviewer disclosures
Peer reviewers on this manuscript have no relevant financial or other relationships to disclose.
References
Papers of special note have been highlighted as either of interest (•) or of considerable interest (••) to readers.
- 1.Chan JFW, To KKW, Tse H, et al. Interspecies transmission and emergence of novel viruses: lessons from bats and birds. Trends Microbiol. 2013;21(10):544–555. Epub 2013 Jun 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Neerukonda SN, Katneni U.. A review on SARS-CoV-2 virology, pathophysiology, animal models, and anti-viral interventions. Pathogens. 2020;9(6):426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fehr AR, Perlman S. Coronaviruses: an overview of their replication and pathogenesis. Methods Mol Biol. 2015;1282:1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen Y, Liu Q, Guo D. Emerging coronaviruses: genome structure, replication, and pathogenesis. J Med Virol. 2020;92(4):418–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kumar R, Harilal S, Al-Sehemi AG, et al. The chronicle of COVID-19: possible strategies to curb the pandemic. Curr Med Chem. 2020;27. DOI: 10.2174/0929867327666200702151018. [DOI] [PubMed] [Google Scholar]
- 6.Khalaf K, Papp N, Chou JT, et al. SARS-CoV-2: pathogenesis, and Advancements in Diagnostics and Treatment. Front Immunol. 2020;11:570927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.V’kovski P, Kratzel A, Steiner S, et al. Coronavirus biology and replication: implications for SARS-CoV-2. Nat Rev Microbiol. 2020;18:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McCreary EK, Pogue JM. Coronavirus disease 2019 treatment: a review of early and emerging options. Open Forum Infect Dis. 2020;7(4):ofaa105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pushpakom S, Iorio F, Eyers PA, et al. Drug repurposing: progress, challenges and recommendations. Nat Rev Drug Discov. 2018;18(1): 41–58. . [DOI] [PubMed] [Google Scholar]; •• Drug repurposing strategies.
- 10.Amin SA, Jha T. Fight against novel coronavirus: a perspective of medicinal chemists. Eur J Med Chem. 2020;201:112559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tan YW, Fung TS, Shen H, et al. Coronavirus infectious bronchitis virus non-structural proteins 8 and 12 form stable complex independent of the non-translated regions of viral RNA and other viral proteins. Virology. 2018;513:75–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kirchdoerfer RN, Ward AB. Structure of the SARS-CoV nsp12 polymerase bound to nsp7 and nsp8 co-factors. Nat Commun. 2019;10(1):2342. . [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Cryo–electron microscopy structure of COVID-19 nsp12 bound to nsp7 and nsp8 cofactors
- 13.Gao Y, Yan L, Huang Y, et al. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science. 2020;368(6492): 779–782. . [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Cryo–electron microscopy structure of COVID-19 virus full-length nsp12.
- 14.Chand GB, Banerjee A, Azad GK. Identification of novel mutations in RNA-dependent RNA polymerases of SARS-CoV-2 and their implications on its protein structure. PeerJ. 2020;8:e9492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Q, Wu J, Wang H, et al. Structural basis for RNA replication by the SARS-CoV-2 polymerase. Cell. 2020;182(2):417–428.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mcdonald SM. RNA synthetic mechanisms employed by diverse families of RNA viruses. Wiley Interdiscip Rev RNA. 2013;4(4):351–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Poch O, Sauvaget I, Delarue M, et al. Identification of four conserved motifs among the RNA-dependent polymerase encoding elements. Embo J. 1989;8(12):3867–3874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smertina E, Urakova N, Strive T, et al. Calicivirus RNA-dependent RNA polymerases: evolution, structure, protein dynamics, and function. Front Microbiol. 2019;10:1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gorbalenya AE, Pringle FM, Zeddam JL, et al. The palm subdomain-based active site is internally permuted in viral RNA-dependent RNA polymerases of an ancient lineage. J Mol Biol. 2002;324(1):47–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Te Velthuis AJW, Arnold JJ, Cameron CE, et al. The RNA polymerase activity of SARS-coronavirus nsp12 is primer dependent. Nucleic Acids Res. 2009;38(1):203–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hillen HS, Kokic G, Farnung L, et al. Structure of replicating SARS-CoV-2 polymerase. Nature. 2020;584(7819):154–156. [DOI] [PubMed] [Google Scholar]
- 22.Lee NR, Kwon HM, Park K, et al. Cooperative translocation enhances the unwinding of duplex DNA by SARS coronavirus helicase nsP13. Nucleic Acids Res. 2010;38(21):7626–7636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sevajol M, Subissi L, Decroly E, et al. Insights into RNA synthesis, capping, and proofreading mechanisms of SARS-coronavirus. Virus Res. 2014;194:90–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Minskaia E, Hertzig T, Gorbalenya AE, et al. Discovery of an RNA virus 3′→5′ exoribonuclease that is critically involved in coronavirus RNA synthesis. Proc Natl Acad Sci USA. 2006;103(13):5108–5113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ferron F, Subissi L, De Morais ATS, et al. Structural and molecular basis of mismatch correction and ribavirin excision from coronavirus RNA. Proc Natl Acad Sci USA. 2017;115:E162–E171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith EC, Blanc H, Vignuzzi M, et al. Coronaviruses lacking exoribonuclease activity are susceptible to lethal mutagenesis: evidence for proofreading and potential therapeutics. PLoS Pathog. 2013;9(8):e1003565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.de Farias ST, Dos Santos AP, Rêgo TG, et al. Origin and evolution of RNA-dependent RNA polymerase. Front Genet. 2017;8:125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lam AM, Murakami E, Espiritu C, et al. PSI-7851, a pronucleotide of beta-D-2ʹ-deoxy-2ʹ-fluoro-2ʹ-C-methyluridine monophosphate, is a potent and pan-genotype inhibitor of hepatitis C virus replication. Antimicrob Agents Chemother. 2010;54(8):3187–3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jockusch S, Tao C, Li X, et al. Sofosbuvir terminated RNA is more resistant to SARS-CoV-2 proofreader than RNA terminated by Remdesivir. Sci Rep. 2020;10(1):16577. Published 2020 Oct 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Picarazzi F, Vicenti I, Saladini F, et al. Targeting the RdRp of emerging RNA viruses: the structure-based drug design challenge. Molecules. 2020;25(23):5695. Published 2020 Dec 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Crotty S, Maag D, Arnold JJ, et al. The broad-spectrum antiviral ribonucleoside ribavirin is an RNA virus mutagen. Nat Med. 2000;6(12):1375–1379. [DOI] [PubMed] [Google Scholar]
- 32.Tzou PL, Tao K, Nouhin J, et al. Coronavirus antiviral research database (CoV-RDB): an online database designed to facilitate comparisons between candidate anti-coronavirus compounds. Viruses. 2020;12(9):1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang M, Cao R, Zhang L, et al. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30(3):269–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Choy KT, Wong AYL, Kaewpreedee P, et al. Remdesivir, lopinavir, emetine, and homoharringtonine inhibit SARS-CoV-2 replication in vitro. Antiviral Res. 2020;178:104786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pruijssers AJ, George AS, Schäfer A, et al. Remdesivir inhibits SARS-CoV-2 in human lung cells and chimeric SARS-CoV expressing the SARS-CoV-2 RNA polymerase in mice. Cell Rep. 2020;32(3):107940. . [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Anti-SARS-COV-2 activity of remdesivir in vitro and in animal models
- 36.Pizzorno A, Padey B, Julien T, et al. Characterization and treatment of SARS-CoV-2 in nasal and bronchial human airway epithelia. Cell Rep Med. 2020;1(4):100059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jeon S, Ko M, Lee J, et al. Identification of antiviral drug candidates against SARS-CoV-2 from FDA-approved drugs. Antimicrob Agents Chemother. 2020;64(7):e00819–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zandi K, Amblard F, Musall K, et al. Repurposing nucleoside analogs for human coronaviruses. Antimicrob Agents Chemother. 2020;65(1):e01652–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xie X, Muruato AE, Zhang X, et al. A nanoluciferase SARS-CoV-2 for rapid neutralization testing and screening of anti-infective drugs for COVID-19.. Nat Commun. 2020;11(1):5214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Riva L, Yuan S, Yin X, et al. Discovery of SARS-CoV-2 antiviral drugs through large-scale compound repurposing. Nature. 2020;586(7827):113–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bouhaddou M, Memon D, Meyer B, et al. The global phosphorylation landscape of SARS-CoV-2 infection. Cell. 2020;182(3):685–712.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cho J, Lee YJ, Kim JH, et al. Antiviral activity of digoxin and ouabain against SARS-CoV-2 infection and its implication for COVID-19. Sci Rep. 2020;10(1):16200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fu L, Ye F, Feng Y, et al. Both Boceprevir and GC376 efficaciously inhibit SARS-CoV-2 by targeting its main protease. Nat Commun. 2020;11(1):4417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hahn F, Wangen C, Häge S, et al. IMU-838, a developmental DHODH inhibitor in phase II for autoimmune disease, shows anti-SARS-CoV-2 and broad-spectrum antiviral efficacy in vitro. Viruses. 2020;12(12):1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hattori SI, Higshi-Kuwata N, Raghavaiah J, et al. GRL-0920, an indole chloropyridinyl ester, completely blocks SARS-CoV-2 infection. mBio. 2020;11(4):e01833–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Touret F, Gilles M, Barral K, et al. In vitro screening of a FDA approved chemical library reveals potential inhibitors of SARS-CoV-2 replication. Sci Rep. 2020;10(1):13093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ko M, Jeon S, Ryu WS, et al. Comparative analysis of antiviral efficacy of FDA-approved drugs against SARS-CoV-2 in human lung cells. J Med Virol. 2020. DOI: 10.1002/jmv.26397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.De Meyer S, Bojkova D, Cinatl J, et al. Lack of antiviral activity of darunavir against SARS-CoV-2. Int J Infect Dis. 2020;97:7–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Williamson BN, Feldmann F, Schwarz B, et al. Clinical benefit of remdesivir in rhesus macaques infected with SARS-CoV-2. Nature. 2020;585(7824):273–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sheahan TP, Sims AC, Zhou S, et al. An orally bioavailable broad-spectrum antiviral inhibits SARS-CoV-2 in human airway epithelial cell cultures and multiple coronaviruses in mice. Sci Transl Med. 2020;12(541):eabb5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Park SJ, Yu KM, Kim Y Il, et al. Antiviral efficacies of FDA-approved drugs against SARS-COV-2 infection in ferrets. MBio. 2020;11(3):e01114–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Warren TK, Jordan R, Lo MK, et al. Therapeutic efficacy of the small molecule GS-5734 against Ebola virus in rhesus monkeys. Nature. 2016;531(7594):381–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mulangu S, Dodd LE, Davey RT, et al. A randomized, controlled trial of ebola virus disease therapeutics. N Engl J Med. 2019;381(24):2293–2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sheahan TP, Sims AC, Graham RL, et al. Broad-spectrum antiviral GS-5734 inhibits both epidemic and zoonotic coronaviruses. Sci Transl Med. 2017;9(396):eaal3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lo MK, Jordan R, Arvey A, et al. GS-5734 and its parent nucleoside analog inhibit Filo-, Pneumo-, and Paramyxoviruses. Sci Rep. 2017;7:43395 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Siegel D, Hui HC, Doerffler E, et al. Discovery and synthesis of a phosphoramidate prodrug of a pyrrolo[2,1-f][triazin-4-amino] adenine C-nucleoside (GS-5734) for the treatment of ebola and emerging viruses. J Med Chem. 2017;60(5):1648–1661. [DOI] [PubMed] [Google Scholar]
- 57.Brown AJ, Won JJ, Graham RL, et al. Broad spectrum antiviral remdesivir inhibits human endemic and zoonotic deltacoronaviruses with a highly divergent RNA dependent RNA polymerase. Antiviral Res. 2019;169:104541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ye W, Yao M, Dong Y, et al. Remdesivir (GS-5734) impedes enterovirus replication through viral RNA synthesis inhibition. Front Microbiol. 2020;11(1):105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Konkolova E, Dejmek M, Hřebabecký H, et al. Remdesivir triphosphate can efficiently inhibit the RNA-dependent RNA polymerase from various flaviviruses. Antiviral Res. 2020;182:104899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tchesnokov EP, Feng JY, Porter DP, et al. Mechanism of inhibition of ebola virus RNA-dependent RNA polymerase by remdesivir. Viruses. 2019;11(4):326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gordon CJ, Tchesnokov EP, Feng JY, et al. The antiviral compound remdesivir potently inhibits RNAdependent RNA polymerase from Middle East respiratory syndrome coronavirus. J Biol Chem. 2020;295(15):4773–4779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yin W, Mao C, Luan X, et al. Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir. Science. 2020;368(6498):1499–1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gordon CJ, Tchesnokov EP, Woolner E, et al. Remdesivir is a direct-acting antiviral that inhibits RNA-dependent RNA polymerase from severe acute respiratory syndrome coronavirus 2 with high potency. J Biol Chem. 2020;295(20):6785–6797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Agostini ML, Andres EL, Sims AC, et al. Coronavirus susceptibility to the antiviral remdesivir (GS-5734) is mediated by the viral polymerase and the proofreading exoribonuclease. MBio. 2018;9(2):e00221–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shannon A, Le NTT, Selisko B, et al. Remdesivir and SARS-CoV-2: structural requirements at both nsp12 RdRp and nsp14 Exonuclease active-sites. Antiviral Res. 2020;178:104793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pizzorno A, Padey B, Dubois J, et al. In vitro evaluation of antiviral activity of single and combined repurposable drugs against SARS-CoV-2. Antiviral Res. 2020;181:104878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sheahan TP, Sims AC, Leist SR, et al. Comparative therapeutic efficacy of remdesivir and combination lopinavir, ritonavir, and interferon beta against MERS-CoV. Nat Commun. 2020;11(1):222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.de Wit E, Feldmann F, Cronin J, et al. Prophylactic and therapeutic remdesivir (GS-5734) treatment in the rhesus macaque model of MERS-CoV infection. Proc Natl Acad Sci USA. 2020;117(12):6771–6776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Porter DP, Weidner JM, Gomba L, et al. Remdesivir (GS-5734) Is efficacious in cynomolgus macaques infected with marburg virus. J Infect Dis. 2020;222(11):1894–1901. [DOI] [PubMed] [Google Scholar]
- 70.Beigel JH, Tomashek KM, Dodd LE, et al. Remdesivir for the treatment of Covid-19 — final report. N Engl J Med. 2020;383:1813–1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Goldman JD, Lye DCB, Hui DS, et al. Remdesivir for 5 or 10 days in patients with severe Covid-19. N Engl J Med. 2020;383:1827–1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Spinner CD, Gottlieb RL, Criner GJ, et al. Effect of remdesivir vs standard care on clinical status at 11 days in patients with moderate COVID-19: a randomized clinical trial. J Am Med Assoc. 2020;324(11):1048–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Grein J, Ohmagari N, Shin D, et al. Compassionate use of remdesivir for patients with severe Covid-19. N Engl J Med. 2020;382(24):2327–2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang Y, Zhang D, Du G, et al. Remdesivir in adults with severe COVID-19: a randomised, double-blind, placebo-controlled, multicentre trial. Lancet. 2020;395(10236):1569–1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Antinori S, Cossu MV, Ridolfo AL, et al. Compassionate remdesivir treatment of severe Covid-19 pneumonia in intensive care unit (ICU) and Non-ICU patients: clinical outcome and differences in post-treatment hospitalisation status. Pharmacol Res. 2020;158:104899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Olender SA, Perez KK, Go AS, et al. Remdesivir for severe coronavirus disease 2019 (COVID-19) versus a cohort receiving standard of care. Clin Infect Dis. 2020. DOI: 10.1093/cid/ciaa1041.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Agostini ML, Pruijssers AJ, Chappell JD, et al. Small-molecule antiviral β-D- N 4 -hydroxycytidine inhibits a proofreading-intact coronavirus with a high genetic barrier to resistance. J Virol. 2019;93(24):e01348–1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yoon JJ, Toots M, Lee S, et al. Orally efficacious broad-spectrum ribonucleoside analog inhibitor of influenza and respiratory syncytial viruses. Antimicrob Agents Chemother. 2018;62(8):e00766–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Painter GR, Bowen RA, Bluemling GR, et al. The prophylactic and therapeutic activity of a broadly active ribonucleoside analog in a murine model of intranasal venezuelan equine encephalitis virus infection. Antiviral Res. 2019;171:104597. [DOI] [PubMed] [Google Scholar]
- 80.Urakova N, Kuznetsova V, Crossman DK, et al. β-D- N 4 -hydroxycytidine is a potent anti-alphavirus compound that induces a high level of mutations in the viral genome. J Virol. 2018;92(3):e01965–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ehteshami M, Tao S, Zandi K, et al. Characterization of β-D-N4-hydroxycytidine as a novel inhibitor of Chikungunya virus. Antimicrob Agents Chemother. 2017;61(4):e02395–1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Reynard O, Nguyen XN, Alazard-Dany N, et al. Identification of a new ribonucleoside inhibitor of ebola virus replication. Viruses. 2015;7(12):6233–6240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Costantini VP, Whitaker T, Barclay L, et al. Antiviral activity of nucleoside analogues against norovirus. Antivir Ther. 2012;17(6):981–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Stuyver LJ, Whitaker T, McBrayer TR, et al. Ribonucleoside analogue that blocks replication of bovine viral diarrhea and hepatitis C viruses in culture. Antimicrob Agents Chemother. 2003;47(1):244–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Toots M, Yoon JJ, Cox RM, et al. Characterization of orally efficacious influenza drug with high resistance barrier in ferrets and human airway epithelia. Sci Transl Med. 2019;23(515). DOI: 10.1126/scitranslmed.aax5866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Furuta Y, Komeno T, Nakamura T. Favipiravir (T-705), a broad spectrum inhibitor of viral RNA polymerase. Proc Jpn Acad Ser. 2017;93(7):449–463. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kiso M, Takahashi K, Sakai-Tagawa Y, et al. T-705 (favipiravir) activity against lethal H5N1 influenza A viruses. Proc Natl Acad Sci USA. 2010;107(2):882–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dawes BE, Kalveram B, Ikegami T, et al. Favipiravir (T-705) protects against Nipah virus infection in the hamster model. Sci Rep. 2018;8(1):7604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Oestereich L, Lüdtke A, Wurr S, et al. Successful treatment of advanced Ebola virus infection with T-705 (favipiravir) in a small animal model. Antiviral Res. 2014;105:17–21. [DOI] [PubMed] [Google Scholar]
- 90.Hawman DW, Haddock E, Meade-White K, et al. Efficacy of favipiravir (T-705) against Crimean-Congo hemorrhagic fever virus infection in cynomolgus macaques. Antiviral Res. 2020;181:104858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Fang QQ, Huang WJ, Li XY, et al. Effectiveness of favipiravir (T-705) against wild-type and oseltamivir-resistant influenza B virus in mice. Virology. 2020;545:1–9. [DOI] [PubMed] [Google Scholar]
- 92.Wang Y, Anirudhan V, Du R, et al. RNA-dependent RNA polymerase of SARS-CoV-2 as a therapeutic target. J Med Virol. 2020. Epub ahead of print. DOI: 10.1002/jmv.26264.. [DOI] [PubMed] [Google Scholar]; •• Possible therapeutic interventions for the treatment of SARSCoV-2.
- 93.Mumtaz N, Jimmerson LC, Bushman LR, et al. Cell-line dependent antiviral activity of sofosbuvir against Zika virus. Antiviral Res. 2017;146:161–163. [DOI] [PubMed] [Google Scholar]
- 94.Cai Q, Yang M, Liu D, et al. Experimental treatment with favipiravir for COVID-19: an open-label control study. Engineering (Beijing). 2020;6(10):1192–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gowen BB, Sefing EJ, Westover JB, et al. Alterations in favipiravir (T-705) pharmacokinetics and biodistribution in a hamster model of viral hemorrhagic fever. Antiviral Res. 2015;121:132–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Warren TK, Wells J, Panchal RG, et al. Protection against filovirus diseases by a novel broad-spectrum nucleoside analogue BCX4430. Nature. 2014;508(7496):402–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Julander JG, Siddharthan V, Evans J, et al. Efficacy of the broad-spectrum antiviral compound BCX4430 against Zika virus in cell culture and in a mouse model. Antiviral Res. 2017;137:14–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Taylor R, Kotian P, Warren T, et al. BCX4430 - A broad-spectrum antiviral adenosine nucleoside analog under development for the treatment of Ebola virus disease. J Infect Public Health. 2016;9(3):220–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Elfiky AA, Ribavirin R, Sofosbuvir G. Tenofovir against SARS-CoV-2 RNA dependent RNA polymerase (RdRp): a molecular docking study. Life Sci. 2020;253:117592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Aftab SO, Ghouri MZ, Masood MU, et al. Analysis of SARS-CoV-2 RNA-dependent RNA polymerase as a potential therapeutic drug target using a computational approach. J Transl Med. 2020;18(1):275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhang WF, Stephen P, Stephen P, et al. Novel coronavirus polymerase and nucleotidyl-transferase structures: potential to target new outbreaks. J Phys Chem Lett. 2020;11(11):4430–4435. [DOI] [PubMed] [Google Scholar]
- 102.Witkowski JT, Robins RK, Sidwell RW, et al. Design, synthesis, and broad spectrum antiviral activity of l-(3-D-Ribofuranosyl-l,2,4-triazole-3-carboxamidef and related nucleosides. J Med Chem. 1972;15(11):1150–1154. [DOI] [PubMed] [Google Scholar]
- 103.Lau JYN, Tam RC, Liang TJ, et al. Mechanism of action of ribavirin in the combination treatment of chronic HCV infection. Hepatology. 2002;35(5):1002–1009. [DOI] [PubMed] [Google Scholar]
- 104.Fernandez H, Banks G, Smith R. Ribavirin: a clinical overview. Eur J Epidemiol. 1986;2(1):1–14. [DOI] [PubMed] [Google Scholar]
- 105.Khan JA, Rehman S, Fisher-Hoch SP, et al. Crimean congo-haemorrhagic fever treated with oral ribavirin. Lancet. 1995;346(8973):472–475. [DOI] [PubMed] [Google Scholar]
- 106.Falzarano D, De Wit E, Martellaro C, et al. Inhibition of novel β coronavirus replication by a combination of interferon-α2b and ribavirin. Sci Rep. 2020;3:1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Swaminathan S, Schlaberg R, Lewis J, et al. Fatal zika virus infection with secondary nonsexual transmission. N Engl J Med. 2016;375(19):1907–1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Day CW, Baric R, Cai SX, et al. A new mouse-adapted strain of SARS-CoV as a lethal model for evaluating antiviral agents in vitro and in vivo. Virology. 2009;395(2):210–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kim HY, Kong S, Oh S, et al. Benzothiazepinecarboxamides: novel hepatitis C virus inhibitors that interfere with viral entry and the generation of infectious virions. Antiviral Res. 2016;129:39–46. [DOI] [PubMed] [Google Scholar]
- 110.Morgenstern B, Michaelis M, Baer PC, et al. Ribavirin and interferon-β synergistically inhibit SARS-associated coronavirus replication in animal and human cell lines. Biochem Biophys Res Commun. 2005;326(4):905–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wilson SZ, Knight V, Wyde PR, et al. Amantadine and ribavirin aerosol treatment of influenza A and B infection in mice. Antimicrob Agents Chemother. 1980;17(4):642–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Te HS, Randall G, Jensen DM. Mechanism of action of ribavirin in the treatment of chronic hepatitis C. Gastroenterol Hepatol. 2007;3(3):218–225. [PMC free article] [PubMed] [Google Scholar]
- 113.Brochot E, Castelain S, Duverlie G, et al. Ribavirin monitoring in chronic hepatitis C therapy: anaemia versus efficacy. Antivir Ther. 2010;15(5):687–695. [DOI] [PubMed] [Google Scholar]
- 114.Falzarano D, De Wit E, Rasmussen AL, et al. Treatment with interferon-α2b and ribavirin improves outcome in MERS-CoV-infected rhesus macaques. Nat Med. 2013;19(10):1313–1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.de Wilde AH, Falzarano D, Zevenhoven-Dobbe JC, et al. Alisporivir inhibits MERS- and SARS-coronavirus replication in cell culture, but not SARS-coronavirus infection in a mouse model. Virus Res. 2017;228:7–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Elfiky AA. SARS-CoV-2 RNA dependent RNA polymerase (RdRp) targeting: an in silico perspective. J Biomol Struct Dyn. 2020;1–9. DOI: 10.1080/07391102.2020.1761882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Elfiky AA. Anti-HCV, nucleotide inhibitors, repurposing against COVID-19. Life Sci. 2020;248:117477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Shah NR, Sunderland A, Grdzelishvili VZ. Cell type mediated resistance of vesicular stomatitis virus and sendai virus to ribavirin. PLoS One. 2010;5(6):e11265 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Hung IFN, Lung KC, Tso EYK, et al. Triple combination of interferon beta-1b, lopinavir–ritonavir, and ribavirin in the treatment of patients admitted to hospital with COVID-19: an open-label, randomised, phase 2 trial. Lancet. 2020;395(10238):1695–1704. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Götte M, Feld JJ. Direct-acting antiviral agents for hepatitis C: structural and mechanistic insights. Nat Rev Gastroenterol Hepatol. 2016;13(6):338–351. [DOI] [PubMed] [Google Scholar]
- 121.Greig SL. Sofosbuvir/Velpatasvir: a review in chronic hepatitis C. Drugs. 2016;76(16):1567–1578. [DOI] [PubMed] [Google Scholar]
- 122.Xu HT, Colby-Germinario SP, Hassounah SA, et al. Evaluation of Sofosbuvir (β-D-2′-deoxy-2′-α-fluoro-2′-β-C-methyluridine) as an inhibitor of Dengue virus replication. Sci Rep. 2017;7(1):6345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Mesci P, Macia A, Moore SM, et al. Blocking Zika virus vertical transmission. Sci Rep. 2018;8(1):1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Dragoni F, Boccuto A, Picarazzi F, et al. Evaluation of sofosbuvir activity and resistance profile against West Nile virus in vitro. Antiviral Res. 2020;175:104708. [DOI] [PubMed] [Google Scholar]
- 125.Mendes ÉA, Pilger DRB, Santos Nastri ACS, et al. Sofosbuvir inhibits yellow fever virus in vitro and in patients with acute liver failure. Ann Hepatol. 2019;18(6):816–824. [DOI] [PubMed] [Google Scholar]
- 126.Ferreira AC, Reis PA, de Freitas CS, et al. Beyond members of the Flaviviridae family, sofosbuvir also inhibits chikungunya virus replication. Antimicrob Agents Chemother. 2019;63(2):e01389–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Jiang W, Muhammad F, Ma P, et al. Sofosbuvir inhibits hepatitis A virus replication in vitro assessed by a cell-based fluorescent reporter system. Antiviral Res. 2018;154:51–57. [DOI] [PubMed] [Google Scholar]
- 128.Ahmad M, Dwivedy A, Mariadasse R, et al. Prediction of small molecule inhibitors targeting the severe acute respiratory syndrome coronavirus-2 RNA-dependent RNA polymerase. ACS Omega. 2020;5(29):18356–18366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Perišić O. Recognition of potential covid-19 drug treatments through the study of existing protein–drug and protein–protein structures: an analysis of kinetically active residues. Biomolecules. 2020;10(9):1–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Chien M, Anderson TK, Jockusch S, et al. Nucleotide analogues as inhibitors of SARS-CoV-2 polymerase, a key drug target for COVID-19. J Proteome Res. 2020;19(11):4690–4697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Elfiky AA, Azzam EB, Shafaa MW. The anti-HCV, Sofosbuvir, versus the anti-EBOV Remdesivir against SARS-CoV-2 RNA dependent RNA polymerase in silico. Mol Divers. 2021;1–11. DOI: 10.1007/s11030-020-10178-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Sadeghi A, Ali Asgari A, Norouzi A, et al. Sofosbuvir and daclatasvir compared with standard of care in the treatment of patients admitted to hospital with moderate or severe coronavirus infection (COVID-19): a randomized controlled trial. J Antimicrob Chemother. 2020;75(11):3379–3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.De Clercq E. Tenofovir alafenamide (TAF) as the successor of tenofovir disoproxil fumarate (TDF). Biochem Pharmacol. 2016;119:1–7. [DOI] [PubMed] [Google Scholar]
- 134.Feng JY, Ly JK, Myrick F, et al. The triple combination of tenofovir, emtricitabine and efavirenz shows synergistic anti-HIV-1 activity in vitro: a mechanism of action study. Retrovirology. 2009;6:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.De Clercq E, Li G. Approved antiviral drugs over the past 50 years. Clin Microbiol Rev. 2016;29(3):695–747. . [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Comprehensive overview on antivirals
- 136.Jockusch S, Tao C, Li X, et al. A library of nucleotide analogues terminate RNA synthesis catalyzed by polymerases of coronaviruses that cause SARS and COVID-19. Antiviral Res. 2020;180:104857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Good SS, Moussa A, Zhou XJ, et al. Preclinical evaluation of AT-527, a novel guanosine nucleotide prodrug with potent, pan-genotypic activity against hepatitis C virus. PLoS One. 2020;15(1):e0227104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Polack FP, Thomas SJ, Kitchin N, et al. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N Engl J Med. 2020;383(27):2603–2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Baden LR, El Sahly HM, Essink B, et al. Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N Engl J Med. 2020;NEJMoa2035389. DOI: 10.1056/NEJMoa2035389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Younes S, Younes N, Shurrab F, et al. Severe acute respiratory syndrome coronavirus-2 natural animal reservoirs and experimental models: systematic review. Rev Med Virol. 2020;e2196. DOI: 10.1002/rmv.2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Lu S, Zhao Y, Yu W, et al. Comparison of nonhuman primates identified the suitable model for COVID-19. Signal Transduct Target Ther. 2020;5(1):157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Lee N, Hurt AC. Neuraminidase inhibitor resistance in influenza: a clinical perspective. Curr Opin Infect Dis. 2018;31(6):520–526. [DOI] [PubMed] [Google Scholar]


