ABSTRACT
Introduction
The COVID-19 pandemic is still escalating and has shaped an extraordinary and pressing need for rapid diagnostics with high sensitivity and specificity. Prompt diagnosis is the key to mitigate this situation. As several diagnostic tools for COVID-19 are already available and others are still under development, mandating a comprehensive review of the efficacy of existing tools and evaluate the potential of others.
Areas Covered
Currently explored platforms for SARS-CoV-2 diagnostics and surveillance centered on qRT-PCR, RT-PCR, CRISPR, microarray, LAMP, lateral flow immunoassays, proteomics-based approaches, and radiological scans are overviewed and summarized in this review along with their advantages and downsides. A narrative literature review was carried out by accessing the freely available online databases to encapsulate the developments in medical diagnostics.
Expert Opinion
An ideal detection method should be sensitive, specific, rapid, cost-effective, and should allow early diagnosis of the infection as near as possible to the point of care that could alter the current situation for the better. Medical diagnostics is a highly dynamic field as no diagnostic method available for SARS-CoV-2 detection offers a perfect solution and requires more attention and continuous R&D to challenge the present-day pandemic situation
KEYWORDS: SARS-CoV-2, nucleocapsid protein, diagnosis, covid-19, naat, serodiagnosis
1. Introduction
In December 2019, cases of enigmatic disease leading to pneumonia in infected individuals of unknown etiology were reported in Wuhan City, Hubei Province of China. The number of cases grew rapidly across the region, ultimately disseminating to 183 countries and 27 territories globally within a short span of around 6 months leading to a full-blown pandemic. There have been 82 million confirmed cases of coronavirus, 1.79 million deaths and around 4,44,437 cases are reported everyday world over, as of 30 December 2020 [1–3]. The newly detected variants of the virus (SARS-CoV-2 VUI-202012/01 UK strain and 501Y.V2 South African strain) are highly transmissible resulting in a spike in the number of COVID-19 cases. Many countries in Europe, North and South America, and Africa continue to report a high incidence of daily new cases and are reinstating lockdown procedures. It was initially speculated that till October 2020, about 10% of the world population may have gotten infected [4–6], which now looks like an overestimate as till Dec 2020 about 1% population got infected and this proved to be saving grace for the mankind.
The causal agent was found out to be a novel coronavirus. This novel coronavirus or SARS-CoV-2 (Severe Acute Respiratory Syndrome Coronavirus 2) is a beta-CoV and through phylogenetic analysis has been placed under the subgenus Sarbecovirus. The genomic sequence of SARS-CoV-2 shares 79% homology to SARS-CoV and 50% to MERS-CoV [7]. It was also found to be closely related to bat-SL-CoVZC45 and bat-SL-CoVZXC21, the bat-derived SARS-like coronaviruses [8].
SARS-CoV and MERS-CoV were transmitted to humans from bats through civet cats and dromedary camels, respectively (Figure 1) [9]. There is still a lot of ambiguity related to the identity of the intermediate host responsible for the transmission of SARS-CoV-2 to humans. Initially, Pangolins were considered as the most probable intermediate host of SARS-CoV-2 due to the high sequence similarity between the two coronaviruses (pangolin coronavirus and novel hCoV). However, the absence of an insertion of four residues motif ‘PRRA’ in the viral genome of pangolin coronaviruses indicates the contrary. From cross-species analysis, animal species such as Minks, ferrets, snakes, turtles, yak, pigs are presumed to be the potential intermediate host between bats and humans [10,11].
Figure 1.

Within 3 months of the appearance of the first case (17 November 2019), on 30 January 2020, COVID-19 was declared as a public health emergency of international concern by the WHO and on 11 March 2020, COVID-19 got categorized as a pandemic. As evidenced by previous epidemics caused by SARS-CoV and MERS-CoV, diagnosis is the most important point in the COVID-19 management not only for the safety of infected individuals but also for the community/population safety as well. Detection methods should be highly sensitive, rapid, cost-effective, and specific. Laboratory diagnosis is essential for effective management and epidemiology of COVID-19 as it serves as a supplementary tool to ascertain not only case identification and treatment but also aids in the contact tracing, finding the animal source and rationalization of control and preventive measures along with the containment of the disease [12,13].
The primary diagnostic tool that is being widely used is Reverse Transcriptase-PCR (RT-PCR), which has good sensitivity and reasonable specificity but also has its own set of shortcomings as for any other diagnostic test. Hindrances like non-availability of kits for all the important genes, limitation of multiplexing, and high cost of instrumentation and reagents are rather common. Another limitation of RT-PCR is the occurrence of false-negative and false-positive test results. A false-negative reporting will contribute to the further spread of the disease within the community whereas false-positive reporting will not only lead to unnecessary treatment but may also undermine the availability of the workforce required for facing this pandemic and cause societal problems.
According to a study published in Emerging Infectious Diseases CDC, the median R0 for COVID-19 is 5.7, which means that one coronavirus infected individual can potentially infect 5–6 people [14]. Hence, investments in R&D for better, more efficient diagnostic techniques that are closer to the point of care and are also cost-effective have become a necessity. As there is a spurt in COVID-19 diagnostics worldwide and the overall landscape include hundreds of different diagnostic kits based on technologies including real-time PCR, CRISPR, microarray, LAMP, lateral flow immunoassays, CT scans, etc., we undertook to systematically review the current strategies on the diagnostics that are employed for detection of SARS-CoV-2 focusing on the pros and cons of each and to delineate the knowledge gaps which require further attention from researchers worldwide.
2. Phases of COVID-19 diagnosis
For COVID-19, early diagnosis is extremely crucial since it will not only increase the chances of the patient’s survival but also guarantee the protection of the unaware contacts. COVID-19 diagnosis is divided into three phases namely pre-analytical, analytical, and post-analytical.
2.1. Pre-analytical phase
It includes the observation of clinical signs and symptoms, specimen collection, and safety measures that should be taken while processing these specimens [9,15]. COVID-19 Symptoms are generally nonspecific and wide-ranging, with no symptoms in some (asymptomatic) to severe pneumonia followed by death. A research on 41 patients who were diagnosed positive for SARS-CoV-2 early during the outbreak (with the date of diagnosis up till 2 January 2020) found myalgia/fatigue (44%), cough (76%), and fever (98%) as the most common symptoms. The atypical symptoms included diarrhea (3%), hemoptysis (5%), headache (8%), and phlegm production (28%). About 50% of the patients developed dyspnea within the first 8 days of infection and about 63% of the patients had Lymphocytopenia. Complications included secondary infections (10%), acute heart injury (12%), and acute respiratory distress syndrome (29%), with 32% of patients required to be treated in the ICUs [16]. Thus, overburdening the healthcare facilities worldwide. Notably, the patients with severe illness might/might not be presented with significant fever demostrating a body temperature of 100.4℉/38°C or higher. The aged patients and those with comorbid conditions like cardiovascular disease, hypertension, and diabetes, have a poor prognosis. Most severe patients die of multiple organ failure, severe respiratory failure, and pneumonia [17]. It has also been reported that infants and young children rarely develop any serious complication of COVID-19 infections and hardly ever suffer loss of life due to it [18].
Another investigation of 1099 confirmed cases (up till 29 January 2020) led by NanShan Zhong, established cough (67.7%) and fever (87.9%) as the most common symptoms. Diarrhea (3.7%) and vomiting (5.0%) were not common and only occurred in a few individuals [9,16]. Nasopharyngeal (NP) swab or Oropharyngeal (OP) swabs are collected as initial respiratory tract specimens. NP swabs are more preferred over OP swabs because sample collection is convenient for the patients and it detects RNA of the virus with more sensitivity in early infection. Sputum sampling or broncho-alveolar lavage (BAL) fluid is collected for the late detection and monitoring of the disease progression of patients with confirmed COVID-19 [15]. Specimens from the lower respiratory tract are useful for diagnosing COVID-19 in patients exhibiting severe or critical symptoms. BAL fluid and sputum give the highest positive rate amongst all other respiratory specimens and are considered to yield the most accurate results [19–21]. However, the use of bronchoscopy as a diagnostic method is not encouraged as the process can generate aerosols that can be a substantial risk for both the patient and the healthcare staff [22]. Sample collection from the upper respiratory tract is preferred because high levels of SARS-CoV-2 RNA can be found right after 3 days of symptom onset. Similar results were obtained for the asymptomatic patients as well [22].
In some patients, high viral load can also be found in the fecal material hence rectal swab can be collected in the late stages. Processing is done either in Class III BSC or Class II BSC with BSL III practices. Guanidinium-based inactivating agent and non-denaturing detergent should be added in lysis buffers for RNA extraction before RT-PCR if the processing is to be done in a Class II BSC [15]. Commonly reported symptoms in COVID-19 patients and the possible clinical samples are depicted in Figure 2.
Figure 2.
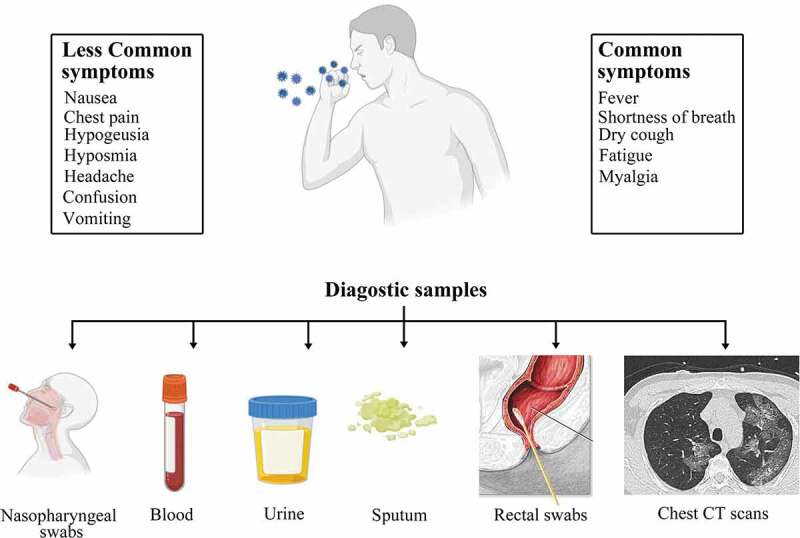
The common symptoms and clinical specimens taken from patients for diagnosis of SARS-CoV-2 [9]
2.2. Analytical phase
It includes the selection of assays to detect SARS-CoV-2 infection. This helps in ruling out the other possible causative agents that are known to cause pneumonia or pneumonia-like symptoms such as SARS, rhino, respiratory syncytial, adeno, parainfluenza, and influenza viruses, etc., and also, from pneumonia caused by mycoplasma, chlamydia, and bacteria. Also, the disease should be distinguishable from other noninfectious diseases including organizing pneumonia, dermatomyositis, and vasculitis. Correct diagnosis aids in observing the proper precautions to avoid the spread of the disease and the effective treatment of the patients [16].
2.2.1. Molecular detection methods
2.2.1.1. PCR based
It involves the amplification of the target gene with the help of primers and DNA polymerase enzyme to 25–35 cycles. In the case of SARS-CoV-2 detection, swabs are tested through qRT-PCR (real-time Reverse transcriptase PCR) [9]. It gives an advantage of simultaneous amplification and analysis of the test sample within a closed system, which prevents the contamination of the amplified products and thereby reduce the probability of false-positive results [15]. Traditionally, the ideal targets of RT-PCR assays are the conserved and/or copiously expressed genes encoding the structural proteins Spike (S), Envelope (E), and Nucleocapsid (N) genes, the non-structural protein RdRp (RNA dependent RNA polymerase) gene, replicase open reading frame (ORF) 1a/b genes, ORF1b-nsp14 genes, hemagglutinin-esterase (HE) and helicase genes. Among all, Hel/RdRp assays have the highest sensitivity and specificity. RdRp is used for confirmation following an analysis of the E gene [13,15,17]. The protocols of several RT-PCR assays have recently been made available online. Nucleocapsid (N) protein gene as a molecular target for real qRT-PCR assay is recommended by the CDC USA. Some regions of the ORF1ab gene are highly conserved in the subgenus Sarbecoviruses and hence are considered to be an appropriate target sequence for RT-PCR [15].
Examples of some of the qRT-PCR kits for diagnostic and non-diagnostic (research) purposes include:
Cobas SARS-CoV-2 Assay
It is an FDA approved automated molecular detection assay for SARS-CoV-2 from Roche [23,24]. It detects both specific SARS-CoV-2 RNA, and highly conserved fragment of the E gene present in all the Sarbecovirus subgenus members. The preferred clinical samples are nasopharyngeal or oropharyngeal swabs. This test is performed on the cobas 6800/8800 systems which has the full process positive, negative, and the internal control to ensure specificity and accuracy. The 6800 and 8800 systems give 384 and 960 results, respectively, in 8 hours whereas both give 96 results in 3–3.5 hours.
RealStar SARS-CoV-2 RT-PCR Kit (Research Use Only)
This research use only RT-PCR technology is meant for the detection of differentiation of the lineage B of the Betacorovirus genus. It consists of positive controls for both the targets of SARS-CoV-2 and B Betacoronavirus [25].
Xpert® Xpress SARS-CoV-2 from Cepheid Inc (USA)
On 21 March 2020, emergency use approval was granted to Xpert® Xpress SARS-CoV-2 from Cepheid Inc (USA) by the FDA. It claims to yield the results within 45 min and encourages hands-off, automated processing of the samples like nasal wash, NP swabs, or aspirate specimens. The result is reported to be positive if more than one target gene is detected. The test delivers point-of-care results with the same level of performance as seen in reference labs [24]. Several other qRT-PCR has been developed as summarized in Table 1.
Table 1.
List of qRT-PCR kits approved by FDA (Food and Drug Administration)
| S. No. | qRT-PCR KIT (Country of approval) MANUFACTURER/CATALOGUE No/REFERENCE | PLATFORM(s) | TARGET gene |
|---|---|---|---|
| 1. | BioCore 2019-nCoV qPCR Kit (USA) by BioCore Co. LTD./BC-01-0099/ [26,27] |
CFX96DX System, Applied Biosystems 7500, SLAN-96P | N and RdRp gene |
| 2. | QIAamp Viral RNA Mini Kit (USA) by Qiagen/52,906/ [28] | QuantStudio6 or QuantStudio7 Real Time PCR System | N gene |
| 3. | Gnomegen COVID-19-RT-Qpcr Detection Kit (USA)by Gnomegen LLC/CV0303/ [29] | Applied Biosystems 7500 Fast Dx Real-Time PCR with SDS version 1.4 software | N gene |
| 4. | Quick SARS-CoV2rRT-PCR Kit(USA) by Zymo Research Corporation/R3011, R3011-1 K, R3011-10 K/ [27,30] | Bio-Rad CFX96 Touch Real-Time PCR Detection System using the Bio-Rad CFX Maestro™ 1.1 Version 4.1.2433.1219 software (or higher). | N gene |
| 5. | LabGun COVID-19 RTPCR Kit (USA) by Lab Genomics Co., Ltd./CV9032B/ [31] | Applied Biosystems™ 7500 fast or BioRad CFX96™ Real-time PCR detection system | E and RdRp gene |
| 6. | RealStar SARS-CoV02 RT-PCR Kits (USA) by Altona Diagnostics GmbH/821,015/ [32] | Mx 3005P™ QPCR System,VERSANT® kPCR Molecular System AD | CoV and SARS-CoV-2 targets |
| 7. | Fosun COVID-19 RTPCR Detection Kit (USA) by Fosun Pharma USA Inc./PCSYHF02-a, PCSYHF03-a/ [27,33] | Applied Biosystems® 7500 RT-PCR software (v1.4, v1.5) | ORF1ab and E gene |
| 8. | GS COVID-19 RT-PCR KIT (USA) by Geno Sensor, LLC/2702-22, 2702–94/ [34] | Applied Biosystems™ 7500 Fast Dx Real-Time PCR system with SDS version 1.4 software. | OFR1ab, E, and N genes. |
| 9. | Xpert Xpress SARS-CoV-2 kit (USA) by Cepheid/302-3562/ [35] | GeneXpert Xpress System | E-gene (Sarbeco specific) and N2-gene (SARS-CoV-2 specific) |
| 10. | 1copy COVID-19 QPCR Kit (Canada)by 1DROP INC. (imported by Luminarie Canada Inc.)(South Korea)/M22MD100M/ [36] | Roche Light Cycler 480 (Product No.05015278001, Software version 1.5) | E, RdRp and N gene |
| 11. | TaqPath Real Time PCR Reagent Kit for SARS-CoV-2 (Japan) by Life Technologies Japan Ltd/A47814/ [37,38] | Applied Biosystems™7500 Fast Dx Real-Time PCR Instrument (used with SDS Software v1.4.1) | ORF1ab, N gene, S gene, |
| 12. | VIASURE SARS-CoV-2 Real Time PCR Detection Kit (Australia) by Cer Test Biotec SL (Spain) (Abacus dx Pty Ltd) /VS-NCO206L/ [39] |
Bio-Rad CFX96™ Real-Time PCR Detection System | ORF1ab and N genes |
| 13. | Allplex™ 2019-nCoV Assay (S Korea) by Seegene Inc/RP10250X/ [27,40] |
CFX96 Real-Time PCR Instrument (Biorad), CFX96 Touch Real Time PCR Detection System (Bio-Rad) | RdRp, N and E Gene |
| 14. | Nucleic Acid reagent test kit for novel coronavirus 2019-nCoV (fluorometric PCR) (China) By Sansure Biotech Inc./S3104E/ [37,41] | ABI 7500 Real-Time PCR System | ORF1ab and N genes |
| 15. | ProTect™ COVID19 RT-qPCR Kit (Singapore) by JN MedsysPte Ltd /10,024/ [42] |
Real time PCR instrument with FAM detection channel | N gene and Human RNase P |
| 16. | BioFire® COVID-19 Test kit (USA) by BioFire Defense, LLC /423,745/ [27,43] |
FilmArray® 2.0 and/or the FilmArray® Torch Instrument Systems | ORF1ab and ORF8 |
| 17. | NxTAG® CoV Extended Panel Assay (USA) by Luminex Molecular Diagnostics, Inc./I054C0463/ [27,44] | Luminex® MAGPIX® instrument including xPONENT | ORF1ab, N and E Gene |
| 18. | NeuMoDx™ SARS-CoV-2 Test Strip (USA) by NeuMoDx Molecular, Inc./300,800/ [27,45] | NeuMoDx™ 288 Molecular System [500,100] orNeuMoDx™ 96 Molecular System [500,200] | Nsp2 and N gene |
| 19. | GeneFinder™ COVID-19 Plus RealAmp Kit (S Korea) by OSANG Healthcare Co., Ltd/IFMR-45/ [27] | Applied Biosystems 7500 & 7500 Fast and Biorad CFX96 Real-Time PCR Instrument |
RdRp, N and E Gene |
| 20. | Cobas SARS-CoV-2 RT-PCR Kit (Canada) by Roche Diagnostics/9,175,431,190/ [27] | cobas 6800/8800 | ORF1 a/b |
| 21. | PhoenixDx® 2019-nCoV (USA) by Procomcure Biotech GmbH (Trax Management Services Inc.)/PCCCSKU1526, PCCSKU15262/ [46] |
Applied Biosystems 7500 fast and BioRadCFX96 Touch Real Time PCR Detection System |
E and RdRp gene |
| 22. | ScienCell™ SARS-CoV-2 Coronavirus Real-time RTPCR (RT-qPCR) Detection Kit (USA) by ScienCell/RX7038/ [27,47] | Light Cycler® 96 Real Time PCR System (Roche) | RdRp and N Gene |
| 23. | Novel Coronavirus (2019nCoV) Real Time Multiplex RT-PCR Kit (Detection of 3 genes)(China) by Shanghai ZJ Bio-Tech Co Ltd (China)/RR-0485-02/ [27] | Applied Biosystems 7500 & 7500 Fast Real Time PCR System | ORF1ab, N and E genes |
| 24. | TaqPath COVID-19 Combo Kit(USA) by Thermo Fisher Scientific Inc/A47814/ [48] | Applied Biosystems 7500 & 7500 Fast and Quant Studio 5 Real Time PCR System | ORF1ab, S and N genes |
| 25. | Quick SARS-CoV-2 rRT-PCR Kit (USA) by Zymo Research Corp/R3011/ [49] | Bio-Rad CFX96 Touch and Applied Biosystems™ Quant Studio 5 Real-Time PCR Detection System, | N gene |
2.2.1.2. LAMP (Loop-mediated isothermal amplification) based method
It is an isothermal nucleic acid amplification technique with high sensitivity and specificity. It is fast and cost-effective due to the non-involvement of high price reagents. The ORF1b gene of SARS-CoV-2 is first amplified and then the product can be detected through gel electrophoresis [9]. The use of the novel RT-LAMP (Reverse Transcription-LAMP) method against the widely recommended standard qRT-PCR has been evaluated through several studies that demonstrated its efficacy. As compared to qRT-PCR, it shows more than 97% sensitivity in targeting the ORF1ab gene [50]. The extreme specificity of LAMP is because this method makes use of multiple primers to identify six different regions on the target DNA simultaneously.
2.2.1.3. Microarray-based methods
It is a fast and high throughput technique wherein the RNA of SARS-CoV-2 is first reverse transcribed to the cDNA which is then labeled with a particular fluorescent probe [9]. The labeled cDNA is loaded and hybridized with an oligonucleotide (60mer oligonucleotide usually) which is fixed on the microarray and then detected through the probes. 60mer oligonucleotide microarray was designed for the detection of SARS-CoV in medical samples [51]. The non-fluorescent, low-value, and low-density oligonucleotide array was designed for the detection of coronaviruses with more sensitivity [52]. Bearing in mind the rapid mutation of SARS Coronaviruses, an advanced microarray was developed [53] that discovers 24 SNPs (Single Nucleotide Polymorphisms) including that of S genes that can be detected with 100% accuracy.
2.2.1.4. NGS (Next-generation sequencing) based methods
This method plays an essential role in the early diagnosis and informs not only about the presence of the virus but also detects if the pathogen underwent mutations or not. For an unfamiliar virus like SARS-CoV-2, genomic sequencing plays an important role in the accurate diagnosis, but it is not as quick as other methods in providing results. NGS-based technologies however, have aided in rapid identification of emerging novel RNA viruses via RNA-Seq. Millions of DNA fragments that are reverse transcribed from RNA can be sequenced by RNA-Seq simultaneously using random primers. The capture-based NGS approach was first employed by Li et al. and it can target most of the CoV species [9,54]. In July 2020, Illumina’s COVIDSeq Test received FDA’s Emergency Use Authorization. This sequencing-based diagnostic tool uses the NovaSeq 6000 System and processes the NP/OP swabs. The workflow can accommodate up to 3072 samples per NovaSeq run and delivers accurate results within 24 hours [55,56].
2.2.1.5. CRISPR (Clustered regularly interspaced short palindromic repeats)
In 2018, Feng Zhang and coworkers developed a technique that uses Cas13, Cas12a, and Csm6 for portable nucleic acid detection. It can detect RNA as well as the DNA of the virus after adding reverse transcriptase to the reaction. Isothermal nucleic acid amplification is performed by the RPA technique to improve the sensitivity of the test. The detection of positive samples is done using a fluorescent reporter and a quencher. Using CRISPR-associated proteins (Cas) and lateral flow chemistry, there is a possibility to develop definitive, highly specific, rapid, and cheap diagnostic kits. It has successfully been used in the development of the Zika virus (ZIKV), human papillomavirus (HPV), and Dengue virus (DENV) molecular diagnostic kits [57] and is explored by various workers across the world for SARS-CoV-2 diagnosis. Some notable examples of CRISPR-based assays for rapid diagnosis of SARS-CoV-2 are SHERLOCK, DETECTR, AIOD-CRISPR, CASdetec, ENHANCE, and FELUDA (Table 2). The Crisper-Cas systems used by different teams are illustrated in Figure 3. Dr. Debojyoti Chakraborty and Dr. Souvik Maiti, at the CSIR-IGIB, New Delhi have developed FELUDA for COVID-19 detection in India. It takes only an hour to give out the results and is very much cost-effective [58].
Table 2.
CRISPR-based assays for rapid detection of SARS-CoV-2
| S. No. | Name of the kit/References | sgRNA target sequences/Cas system involved | Detection technology/Amplification method | Other details |
|---|---|---|---|---|
| 1. | SHERLOCK by Sherlock Biosciences (Cambridge, USA)/ [57] | S and Orf1ab gene/Cas 13a (FDA approved) | Paper strip lateral flow-based detection/RPA | Both SHERLOCK and HUDSON techniques provide more sensitivity. |
| 2. | DETECT by Mammoth Bioscience Inc. (California, USA)/ [57] | N and E gene/Cas 12a | UV or LED based detection/RT-LAMP | It detects SARS-CoV-2 within 30 minutes through lateral flow strip format. |
| 3. | AIOD-CRISPR/ [57] | N gene/Cas 12a | UV or LED based detection/RPA | The ssDNA-FQ reporter is cleaved on binding of Cas12acrRNA to the target and produces fluorescence that is detected. |
| 4. | CASdetec/ [59] | RdRp gene/3ʹDNA7 Cas 12a | Paper based detection/RAA | 7-Nucleotide poly-T reporter is used as it gives better and more specific fluorescence signal. |
| 5. | ENHANCE/ [57] | N gene/Cas 12b | UV or LED based detection/RT-LAMP | Lateral flow assay with FITC-ssDNA-Biotin reporter limits the detection time to 20 minutes. |
| 6. | FELUDA/ [57] | Nsp8 and N gene/Cas 9a ortholog from Francisella novicida fnCAS9 | Paper based detection/RPA | fnCAS9 shows high level of accuracy with significantly reduced off-targeting and faster detection. |
| Abbreviations: (a)SHERLOCK: Specific High-sensitivity Enzymatic Reporter un-LOCKing (b)DETECTR: DNA Endonuclease Targeted CRISPR Trans Reporter (c)AIOD-CRISPR: All-in-One Dual CRISPR (d)CASdetec: CRISPR-Cas12b-mediated DNA detection (e) ENHANCE: Enhanced analysis of nucleic acids with crRNA (CRISPR RNA) extensions (f) FELUDA: FNCAS9 Editor-Linked Uniform Detection Assay | ||||
Figure 3.
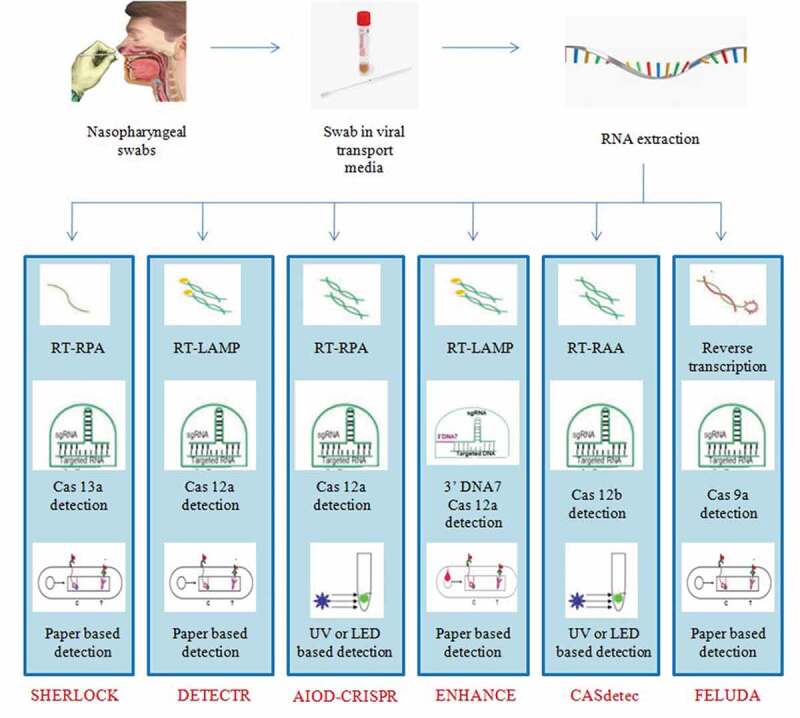
Summary of CRISPR-based assays being developed for the detection of SARS-CoV-2 [57]
qRT-PCR is considered to be the reference standard for the diagnosis of infectious diseases with high accuracy and sensitivity during the acute phase of disease [60]. The presence of viral genome in sufficient amounts at the site of sample collection is the basic requirement for the PCR-based or deep sequencing detection methods. It has been observed that as the disease progresses, the heavy infection of the nasopharyngeal area would possibly become negative (Figure 4). Thus, the nasopharyngeal swab may not be the most appropriate site for sample collection at different stages of the disease. An incorrect sample collection and low viral load during the very early or late stage of the disease, suppressed viral load due to host immunity or in the patients undergoing a preexisting persistent anti-viral treatment (such as anti-HIV drugs) may lead to a false-negative diagnosis. A false-negative diagnosis can have alarming consequences, allowing infected individuals to further spread the disease and hinder the efforts to control the pandemic. Therefore, supplementary diagnostic techniques that can ascertain the presence of infection even when the sample has a low viral titer will be highly valuable and ensure timely diagnosis [61]. But they are costly and time-consuming to perform [62]. Additionally, detection through rRT-PCR method requires professionally trained and skilled personnel to operate sophisticated laboratory equipment, thereby, making it more labor-intensive. The equipment required for RT-PCR is usually located at a central laboratory which is either Biosafety level 2 or above thereby, limiting the accessibility and applicability of these laboratory equipment. The time-consuming nature of PCR-based methods leaves probable cases undiagnosed, which is creating a gap in SARS-CoV-2 containment efforts [50]. Considering the shortcomings of qRT-PCR, immunoassays may prove to be an alternative approach to lessen undiagnosed cases and fulfill the demand for rapidity and economic viability [60].
Figure 4.
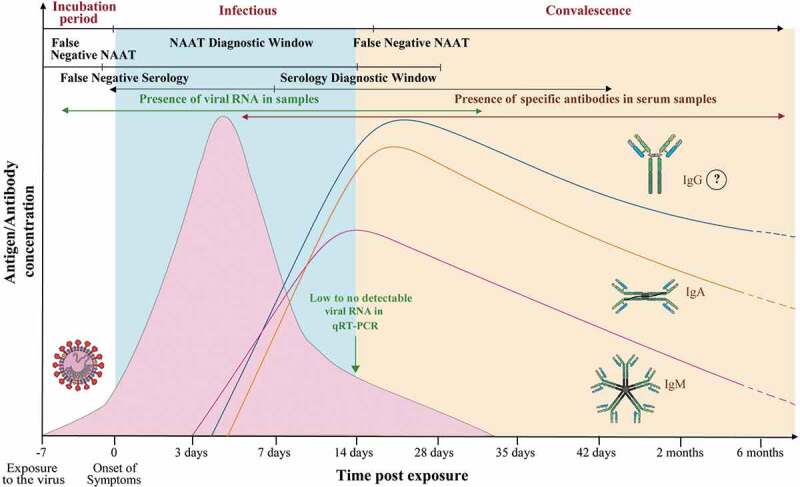
Illustration showing the detection window of SARS-CoV-2 specific viral RNA and antibodies [50]
2.2.2. Immunoassays
Immunoassays are the rapid detection techniques of the pathogen-specific antigens or the antibodies which are produced in response to the infection. These are generally based on rapid lateral flow assays. Progress has been made on high throughput immuno-analyzers for the population-level screening. A lateral flow immunoassay strip (LFIAs) comprises a SARS-CoV-2 antigen and/or anti-human IgG/IgM antibody which has been coupled with colloidal nanoparticles (NPs), usually made up of gold [63]. The advantages of serology-based assays include fast detection, cost-effectiveness, and assessment of herd immunity, while the disadvantages include: poor sensitivity as low viral load cannot be detected [15]and hence are not considered good for early detection.
2.2.2.1. Antigen based
Theoretically, a viral antigen precedes the occurrence of antibodies and hence can be used as a specific marker that can confirm the presence of infection (Figure 4) [12]. In the majority of coronaviruses, including SARS-CoV, the nucleocapsid or N protein is highly immunogenic eliciting the earliest immune response and is expressed abundantly during the early infection [64,65]. In a study conducted by a group of scientists from China, it was found that the tests based on detection of N antigen attained a sensitivity of 94% during the first 5 days after of the illness. When the same test was conducted on another group of patients who had been showing symptoms of SARS-CoV for more than 6 days, the test achieved a sensitivity of 78%. This and the other studies suggest that the N antigen being of highly conserved nature has a high true positive rate and can be optimized for use as an early diagnostic marker for SARS-CoV-2 detection [12]. Spike protein can be another potential antigen for immune-diagnosis of COVID-19, and numerous diagnostic methods have already been developed based on N and/or S proteins.
Fluorescence immuno-chromatographic assay can accurately detect the N protein of SARS-CoV-2 in nasopharyngeal swab samples and urine samples. It is a rapid and simple method and can provide results within 10 minutes. The results are read by the immuno-fluorescent analyzers [12].
However, it is imperative to validate the sensitivity and specificity of the immune assays to prevent false-positive test results. Alpha-coronaviruses (NL63 and 229E), and other beta-coronaviruses (HKU1 and OC42) are quite prevalent and have caused infections in most people. Exposure to these other endemic human coronaviruses can hinder the accuracy of immunoassays meant for SARS-CoV-2 detection and give false-positive outcomes. A 90% similarity between SARS-CoV-2 and SARS-CoV N proteins has been reported. The use of SARS-CoV antigens to diagnose and confirm the presence of COVID-19 infections may be reliable and valid, given that there has not been a single infection of SARS-CoV in humans since 2004. In addition to this, it has been reported that SARS-CoV specific antibodies have been waning and after 6 years, 91% of the patients were shown to be negative [60]. Furthermore, there were only about 9000 survivors of SARS-CoV infections worldwide, making it a remote possibility of reinfection of SARS-CoV patients with SARS-CoV-2.
2.2.2.2. Antibody-based diagnostics
As per Lee et al. 2020[60], it was found that the SARS-CoV-2 viral RNA is detectable in the nasopharyngeal or throat swabs of patients in the first 14 days post-onset of illness. SARS-CoV-2 IgM-specific antibodies are detectable right after the first 3 days post-onset of symptoms, and peaks in the subsequent weeks (during the 2nd and 3rd week). Even after 30 days from initial exposure to the virus, the IgM antibody response was still detectable. IgA and IgG antibodies are distinguishable in the sera samples within 4 days post-onset, and peaks in about 2 weeks. This illustrates the importance of serological assays to retrospectively identify asymptomatic individuals who may still be infected or have had an earlier SARS-CoV-2 exposure that went undiagnosed, for epidemiological lessons [50,60].
In another study, a group of scientists developed a recombinant N protein (rNPs) based indirect ELISA to detect IgM, IgA, and IgG. The results of the western blot analysis revealed no cross-reactivity between SARS-CoV-2 rNP and specific IgG antibodies against HKU1, NL63, 229E, and OC43 strains of human coronaviruses. However, cross-reactivity of anti-N antibodies was observed between SARS-CoV and SARS-CoV-2 [62]. According to Liu and coworkers, out of the 214 serum samples of patients who had been confirmed positive with qRT-PCR, 146 (68.2%) and 150 (70.1%) sera samples tested positive by the rN-based IgM and IgG ELISAs, respectively. In 77.1%, 74.3%, and 82.2% individuals positive results with IgM, IgG, and IgM and/or IgG, respectively, were obtained by rS-based ELISA. This study denoted the significance of rN and rS-based ELISAs in screening for COVID-19 due to its considerable sensitivity [64]. On the contrary, from another set of studies, it was inferred that the test kits based on S protein-specific antibody detection had 91% sensitivity and 100% specificity whereas test kits meant for detecting Anti-N antibodies of SARS-CoV-2 exhibited 100% sensitivity and 100% specificity [66,67].
Hence, more thorough investigations will provide insights into the efficacy of each kind of test kit. Several immunodiagnostic kits for the detection of SARS-CoV-2 have been developed or are under development. Table 3 provides a comprehensive list of FDA approved immunodiagnostic kits.
Table 3.
List of antigen and antibody-based rapid detection kits approved by FDA
| S.No | RAPID DETECTION KIT (Country of approval) |
MANUFACTURER/ REFERENCES |
DETECTION ANTIBODY/ANTIGEN |
|---|---|---|---|
| 1. | Sofia 2 SARS Antigen FIA(USA) | Quidel Corporation/ [68] | N protein |
| 2. | New York SARS-CoV Microsphere Immunoassay for Antibody Detection(USA) | Wadsworth Center, New York State Department of Health/ [69] | IgG, IgM, and IgA antibodies to N protein |
| 3. | LIAISON SARS-CoV-2 S1/S2 IgG(USA) | DiaSorin Inc./ [70] | IgG antibodies to S1 and S2 proteins |
| 4. | Anti-SARS-CoV-2 Rapid Test(USA) | Autobio Diagnostics Co. Ltd./ [71] | IgM and IgG antibodies to the S protein |
| 5. | Cellex qSARS-CoV-2 IgG/IgM Cassette Rapid Test kit(Australia) | Cellex Inc (United States Of America)/ [72] | IgM and IgG antibodies to S and N protein |
| 6. | Wantai SARS-CoV-2 Ab Rapid Test kit(approved for use in Australia) | Beijing Wantai Biological pharmacy Enterprise Co Ltd (China)/ [73] | IgG and IgM antibodies to Spike protein |
| 7. | Wondfo SARS CoV-2 Antibody Test(Singapore) | SkyQuestPte Ltd/ [74] | IgM and IgG antibodies |
| 8. | DPP® COVID-19 IgM/IgG System(Brazil) | CHEMBIO DIAGNOSTICS BRAZIL LTDA/ [75] | IgM and IgG antibodies to N protein |
| 9. | COVID-19 Ag ECO Teste(Brazil) | Eco DiagnosticaLtda/ [37] | SARS-CoV-2 antigens |
| 10. | SGTi-flex COVID-19 IgM/IgG(S Korea) | Sugentech, Inc/ [76] | IgM and IgG antibodies |
| 11. | STANDARD™Q COVID-19 Ag Test (S Korea) |
SD BIOSENSOR/ [77] | SARS-CoV-2 antigens |
| 12. | STANDARD™F COVID19 Ag FIA (S Korea) |
SD BIOSENSOR/ [78] | Monoclonal antiCOVID-19 antibody to N antigen |
| 13. | IgM antibody test kit for novel coronavirus 2019nCoV (colloidal gold method)(China) | Hecin Scientific, Inc./ [37] | IgM antibody |
| 14. | RightSign™ COVID-19 IgG/IgM Rapid Test Cassette(China) | Hangzhou Biotest Biotech Co Ltd (China)/ [79] | IgM and IgG antibodies to the S protein antigen |
| 15. | InnoScreen TM COVID-19 IgG/IgM Rapid Test(Australia) | Innovation Scientific Pvt Ltd (Australia)/ [37] | IgM and IgG antibodies |
| 16. | New Coronavirus (COVID-19) IgG/IgM Rapid Test(India) | Voxtur Bio Ltd, India/ [80] | IgM and IgG antibodies |
| 17. | COVID-19 IgM/IgG Antibody Detection Card Test(India) | VANGUARD Diagnostics, India/ [80] | IgM and IgG antibodies |
| 18. | Makesure COVID-19 Rapid test(India) | HLL Lifecare Limited, India/ [80] | IgM and IgG antibodies |
| 19. | Immuno Quick Rapid Test(India) | Immuno Science India Pvt. Ltd./ [80] | IgM and IgG antibodies |
| 20. | One Step COVID-19 IgM/IgG Antibody(India) | SIDAK Life Care Pvt. Ltd./ [80] | IgM and IgG antibodies |
A serological study on the six members of a family at PUMCH Hospital, Beijing, China revealed that serologically five patients were Corona positive whereas repeated RT-PCR could only report two positives. Observation of ground-glass opacities in the CT scan established three positives. The authors reported that ‘the two patients were diagnosed with COVID-19, two were suspected of COVID-19, and two were considered close contacts’ and advocated the importance of serological testing in the diagnosis of SARS-CoV-2 infections, particularly in contact tracing [81,82,83,84,85,86,87]. While serological assays are rapid, easy to perform, and robust, they also present several challenges. Antibodies take some time to develop post-exposure to the antigen and hence, these immunoassays are unable to detect the presence of the infection during the early stage of the disease. Cross-reactivity could probably be another shortcoming of the immunoassays because it adversely influences the sensitivity and specificity of the test. Table 4 enlists some of the important advantages and disadvantages of immunodiagnostics [60].
Table 4.
Summary of advantages and disadvantages of Immunoassays
| S. No. | Advantages | Disadvantages |
|---|---|---|
| 1. | Ease of perform and interpret: Immunoassays are available in a kit format and do not require training of the operators. Widely accepted and practiced and approved for use in a broad range of applications. Antibodies are more stable as compared to viral RNA, therefore the samples are less prone to deterioration during collection, processing, transportation, storage, and testing as compared to rRT-PCR samples. Moreover, due to the homogeneity of the blood samples, there are less variations observed in contrast to nasopharyngeal specimens [50] | The results can get affected by autoantibodies, human anti reagent antibodies. Cross reactivity between antibodies in multiplexed immunoassays should be prevented to eliminate false reading. Due to the less number of differences among the antigens many antibodies can’t be distinguished easily. These differences in the analytes often have serious diagnostic implications. |
| 2. | It is cost effective as compared to molecular method. Immuno-diagnosis is appropriate for large scale community screening and assessment of herd immunity. | It can’t be used for early diagnosis, as antibodies appear late during infection. |
| 3. | Immunoassays have a high level of sensitivity and can also detect asymptomatic individuals. | Some Rapid tests kits have poor quality and can give false positive and false negative results. |
| 4. | The antibody-antigen based immunoassays are rapid and can give results in 15–20 minutes. | It can give a false sense of immunity because currently there is no evidence to suggest that people who have recovered from COVID-19 are immune to catching it again. |
2.2.3. Radiological study of SARS-COV-2 diagnosis
These are the complementary methods to the molecular methods and make diagnosis more effective without clinical samples. They are also very safe for the community because of the absence of a sample collection step. Radiological images obtained through the chest CT scan, gives the information of infection through the appearance of deviations such as ground glass opacities, bronchial wall thickening, centrilobular nodules, consolidation, vascular enlargement, architectural distortion, crazy paving pattern, reticulation, traction bronchiectasis, subpleural bands, and intrathoracic lymph node enlargement in the image [9]. During the early-stages of infection, Chest CT images of the COVID-19 patients presented with the changes in the lung interstitial tissue and the presence of multiple small plaques [17]. Progression of disease in a patient can be assessed by a chest CT scan as shown in Figure 5. Chest CT is a conventional, noninvasive imaging technology that gives results instantly and has high accuracy. The sensitivity of chest CT scans to detect SARS-CoV-2 is believed to be higher than that qRT-PCR (Table 5)
Figure 5.
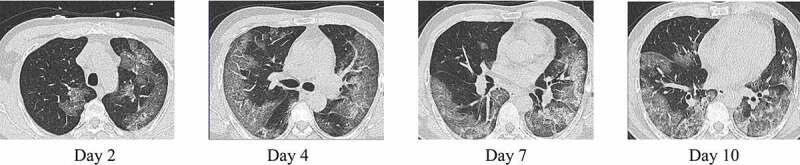
CT-images of a COVID-19 positive young male. The disease progression can be visualized with the help of chest CT scans [102]
Table 5.
Different diagnostic methods and their performance
| Diagnostic Method | Reference | Testing Parameters | Results | Methodological Limitations |
|---|---|---|---|---|
| RT PCR | Fang et al (83) |
Population: 51 patients showing symptoms of acute respiratory distress. Assay: RT-PCR kit by Shanghai ZJ Bio-Tech Co, Ltd Sample: Throat swabs. |
Sensitivity: RT-PCR results revealed that 71% (36/51) of the patients tested positive for SARS-CoV-2 infection in the first round of testing. | The cohort used for this study involved only patients with fevers or acute respiratory symptoms. Used a biased cohort of patients who were suspected to have COVID-19. |
| Ai et al (84) |
Population: 1014 patients showing symptoms of acute respiratory distress. Assay: TaqMan One-Step RT-PCR Kits from Shanghai Huirui Biotechnology Co., Ltd Sample: Throat swab |
Sensitivity: RT-PCR results revealed that 59% (601/1014) of the patients tested positive for SARS-CoV-2 infection. Positive rate: 59% |
||
| RT LAMP | Nawattanapaiboon et al (85) |
Population: 2120 Patients under investigation for COVID-19 Assay: The developed colorimetric RT-LAMP assay could amplify the target gene and enabled visual interpretation in 60 min at 65 °C. Sample: NP and OP swabs |
Sensitivity: 95.74% (With qRT-PCR as reference) Specificity: 99.95% No cross-reactivity with six other common human respiratory viruses (influenza A virus subtypes H1 and H3, influenza B virus, respiratory syncytial virus types A and B, and human metapneumovirus) and five other hCoV (MERS-CoV, HKU-1, OC43, 229E and NL63) was observed. PPV: 97.83%NPV: 99.90% |
The samples of patients under investigation for COVID-19 were used. It hasn’t been specified whether this cohort also included clinical samples from asymptomatic patients. |
| Immunoassays | Porte et al (86) |
Antigen based Population: 127 clinical samples of individuals with respiratory symptoms and/or fever and have an epidemiological risk factor for SARS-CoV-2 infection. Assay: The fluorescence immunochromatographic SARS-CoV-2 antigen test (Bioeasy Biotechnology Co., Shenzhen, China) Sample: NP and OP swabs |
Sensitivity: 93.9% (With RT-PCR as reference) Specificity: 100% PPV: 100%(estimated)NPV: 99.4%(estimated) |
The use of a sample type which is not specifically permitted in the instructions for use. This evaluation was performed during a period of time (late summer in Chile) with a low circulation of other frequent respiratory viruses; therefore the performance of the antigen-based RDT might change in different epidemiological conditions. |
| Whitman et al (87) |
Antibody based Population: The study population included samples from individuals with symptomatic infection and positive RT-PCR results for SARS-CoV-2 infection, pre COVID-19 plasma specimens (collected prior to July 2018). A total of 288 samples were analysed. Assay: Ten Immunochromatographic Lateral Flow Assays (LFAs) and two ELISAs that detect the presence of IgM and IgG Ab (specific to the RBD protein of the virus) in the clinical samples. Sample: Serum and Plasma Samples |
Specificity: >95% for four assays (Bioperfectus, Premier, Wondfo, in-house ELISA) IgM detection was more variable than IgG for nearly all assays. No consistent cross-reactivity was observed. |
This study focused on comparisons of percent positivity by time interval, rather than reporting the “sensitivity” of each assay. | |
| CRISPR | Patchsung, Maturada et al. (88) |
Population: 154 clinical samples collected at Siriraj Hospital, Thailand. Assay: 1. SHERLOCK with lateral-flow readout. 2. SHERLOCK with fluorescence readout. Sample: NP and throat swabs |
Positive RT-qPCR samples: 81 Negative RT-qPCR samples: 73 1. SHERLOCK with lateral-flow readout (With RT-qPCR as reference) Sensitivity: 97.14% Specificity:100.00% PPA: 100.00% NPA: 97.33% ln(DOR): 8.30 2. SHERLOCK with fluorescence readout (With RT-qPCR as reference) Sensitivity:100.00% Specificity:100.00% PPA: 100.00% NPA: 100.00% ln(DOR): 9.94 |
The SHERLOCK protocol involves the RNase inhibitors and negative control to ensure the inactivation of nucleases and absence of contamination. But an in-strip confirmation either by fluorescence readout or lateral flow readout could be a cause of contamination. |
| CT Scan | Fang et al (83) |
Population: 51 patients showing symptoms of acute respiratory distress. | Sensitivity: Chest CT scans revealed that 98% (50/51) of patients had abnormalities that were consistent with viral pneumonia. | Since no asymptomatic COVID-19 positive patients were included,these studies may have exaggerated the sensitivity of CT. |
| Ai et al (84) |
Population: 1014 patients showing symptoms of acute respiratory distress. Systems: uCT 780, United Imaging, China; Optima 660, GE, America; Somatom Definition AS+, Siemens Healthineers, Germany |
Sensitivity: The sensitivity of chest CT was 88% (888/1014) (With RT-PCR as reference) Specificity: The reported specificity was 25% PPV: 65% NPV: 83% |
Abbreviations:
PPV: Positive Predictive Values
NPV: Negative Predictive Values
NPA: Negative Percent Agreement
ln(DOR): Natural logarithm of Diagnostic Odds Ratio
PPA: Positive Percent Agreement
Low-cost techniques such as Chest X-rays and ultrasounds have also been considered as a means of diagnosis but due to their low sensitivity and specificity, they are not recommended. Nevertheless, they do help in monitoring disease progression [22].
The diagnostic performance of the different diagnostic methods has been encapsulated in Table 5.
2.2.4. SARS-CoV-2 diagnosis using nonspecific parameters
Many parameters increase or decrease in response to COVID-19 infection. At an early stage of the disease patients were presented with lymphopenia, leukopenia, elevated C-reactive protein (CRP), aspartate aminotransferase, and RBC sedimentation rate. Levels of Procalcitonin in the serum of most patients were normal. Additionally, the severe cases had greater levels of lactate dehydrogenase, alanine aminotransferase, D-dimer, and ferritin along with considerably increased levels of TNF-α, IL-6, IL-2 R, and IL-10 [17]. Elevated levels of troponin have also been reported in patients who subsequently passed away due to fulminant myocarditis [17].
2.2.5. Virus neutralization assays (VNA)
Virus neutralization assay (VNA) is a method classically used to investigate the neutralizing antibody response to a virus and demonstrate the inhibition of viral replication. Extreme sensitivity and specificity of VNA are determined by the fact that it only detects antibodies that can inhibit the virus. This is crucial because related groups of viruses may share common antigens, but only some of these antigens are recognized and targeted by neutralizing antibodies. Virus serotyping based on its neutralization can be used for serotyping a virus [50].
Pseudovirus-based neutralization assays (PBNA) has an edge over the conventional VNA because of their versatility and safety. In contrast with the VNA, PBNA is a specific, sensitive, robust, and reproducible method that is more objective and less labor-intensive. Convalescent COVID-19 patient exhibited higher neutralization activity against the pseudovirus, highlighting its potential as a possible therapeutics [88].
GenScript has developed a diagnostic test kit called cPass™ SARS-CoV-2 Neutralization Antibody Detection kit or SARS-CoV-2 surrogate Virus Neutralization Test (sVNT) Kit. It is claimed to be an automatable, faster, and more scalable alternative to the traditional neutralizing antibody tests. Unlike the conventional test kits that yield accurate results in days, the cPass kits can measure functional neutralizing antibodies (nAbs) within an hour and do not involve live biological specimens or stringent BSL3 facility for diagnosis. It detects SARS-CoV-2 neutralizing antibodies that block the contact between the ACE2 receptor and the receptor-binding domain of the S protein [89]. The potency of such assays has been ascertained through several studies assessing and validating the confirmatory testing of MERS-CoV and was able to detect the negatives that were reported positive by other serological tests including ELISA. It was reported that the samples which tested positive for IgG ELISA produced negative results with PBNA [50]. Another set of studies revealed that the incorporation of VNA with the serological testing of MERS-CoV improved the accuracy of the result and was able to detect even the subclinical infections [50]. The highly specific and sensitive nature of this assay makes it suitable for use as a reference or a confirmatory test.
Although VNA is highly sensitive and offers several advantages, it is quite labor-intensive as it requires skilled professionals to conduct the assay, and is time-consuming as well. Therefore, VNA is primarily used for research purposes [50].
2.2.6. Targeted proteomics: an alternative to antibody-based assays
Targeted proteomics is another modality of diagnosis that has gained significant popularity in the scientific community over the past few years. Mass spectrometry (MS) (using the triple quadrupole (QqQ) mass spectrometer) based targeted proteomics offers high sensitivity, quantitative accuracy, rapidity, and reproducibility over other traditional methods as the analysis only focuses on the subsets of proteins that are important and needed to the line of inquiry. Some processing techniques such as ‘selected reaction monitoring’ (SRM) also known as ‘multiple reaction monitoring (MRM)’ focus the mass spectrometer to detect pre-selected proteins. Conceptually SRM approach is quite similar to immunoassays but what makes it powerful is that it takes both antibody-based detection and discovery-based MS into consideration and bridges the gap between the two. In SRM, the protein of interest is selected and the triple-quadrupole mass spectrometer is programmed to detect only signature protein or peptide known as a proteotypic peptide. Mass spectrometer then fragments and analyzes only the selected protein from the samples [90] (Figure 6). It offers the advantage of high sequence-based selectivity of MS/MS (tandem mass spectrometry in which more than one mass analyzer is combined) and ‘rapid duty cycle’ that is compatible with contemporary and high-efficiency liquid chromatography (LC) technique called ‘ultra-high performance liquid chromatography (UHPLC)’. Moreover, the sensitivity and simplicity with which the QqQ (triple quadrupole mass spectrometer) devices renders the results, makes it exceptional to any other type of mass analyzer. However, the data can only be acquired for a fraction of peptides and the workflow of detection involved is quite complex [91].
Figure 6.
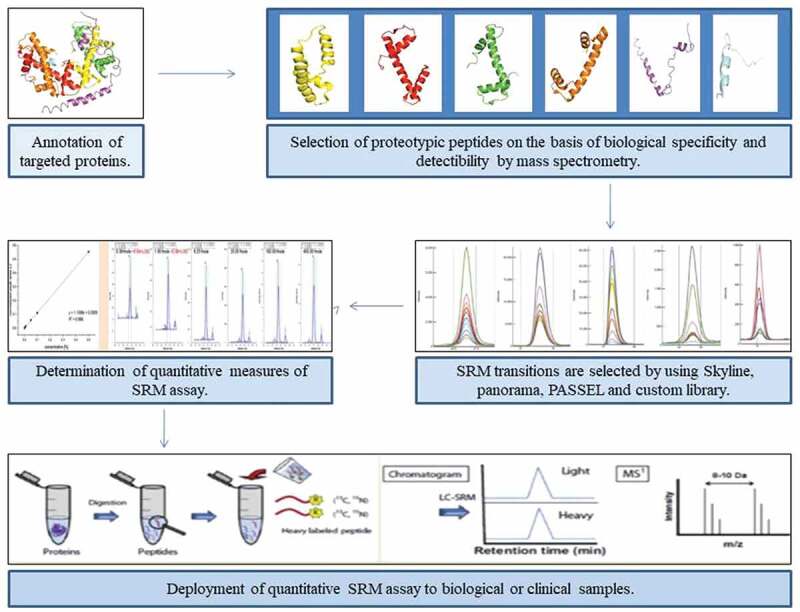
Summary of the targeted proteomics quantitative SRM assays under development for COVID-19 [91]
Use of targeted LC-MS (Liquid Chromatography-Mass Spectrometry) along with quantitative immunoassays such as densitometric western blots and ELISA can enhance the efficiency of the process by making it more selective, allowing a wider dynamic range and enabling it to be more amenable in multiplexing. Integration of immune enriching target proteins or peptides followed by targeted LC-MS can also be an alternative for diagnosis [92]. LC-MS technology offers advantages of high selectivity, sensitivity, sample throughput, sample volumes, cost-per-sample, reproducibility, multiplexing, and extended compound range but the equipment cost, complexity, and sample complexity limit its uses [93]. LC-MS technology can be the next best option for immunoassays but there is still a lot of work to be done [93].
PEPperCHIP® SARS-CoV-2 Proteome Microarray (PEPperPRINT) is a newly developed modality for diagnosis of SARS-CoV-2 involving qualitative and quantitative proteomic screening of the samples. It can serologically screen about 5000 individual peptides. In this method, the chip captures the proteins (antibodies, enzymes, or ligands) and immobilizes them to use them as a probe. Protein analyte which is to be tested is added to the matrix and modified by using different markers such as luminescence, radioisotopes, fluorescence markers, etc. The interaction between micro-matrix proteins and analyte produces an analytical signal and helps in the detection [94].
2.3. Postanalytical phase
This phase includes the interpretation of the assay results. Ct (Cycle threshold) refers to the number of cycles required for the amplification of viral RNA to reach the level for detection. It is a semi-quantitative indicator that detects the concentration of viral genetic material in a patient sample with the simple rule of inverse proportionality. Serial Ct values are valuable in the interpretations for clinical management of patients in the hospital settings. Ct values are only applied for clinical interpretations and may not be reported by all RT-PCR platforms. It is also not directly comparable between assays. Clinical history is a must to interpret a single positive Ct value in case of staging infectious course, prognosis, infectivity, or as an indicator [95]. If CT values are >40 then the test is negative. If we have two target proteins and the CT value of only one protein is <40 then the result is indeterminate and requires confirmation through retesting [15].
Every immunoassay comes tagged with a label molecule that is responsible for producing a quantifiable signal indicating the binding of the antibody with the analyte of interest. The results of qualitative immunoassays can be interpreted through visual cues like the emergence of color and lines after the reaction. The presence of lines/bands in the test and control region/zone of a Rapid test kit indicate a positive result whereas the absence of a line from the test region indicates a negative result. The results of quantitative immunoassays can be obtained by using an appropriate plate reader which measures the intensity of the signal and provides values corresponding to concentrations of the analyte present in the sample. It is worth noting that each type of immunoassay has its standard values and unique mode of result interpretation [96].
In the Fluorescence immuno-chromatographic assay immunofluorescence analyzer directly gives the result by comparing the detection value with the reference cutoff or threshold value which is already set within the internal parameter of the kit’s ID chip [9]. The possible platforms for SARS-CoV-2 diagnosis either already in use or in developmental stages are illustrated in Figure 7.
Figure 7.
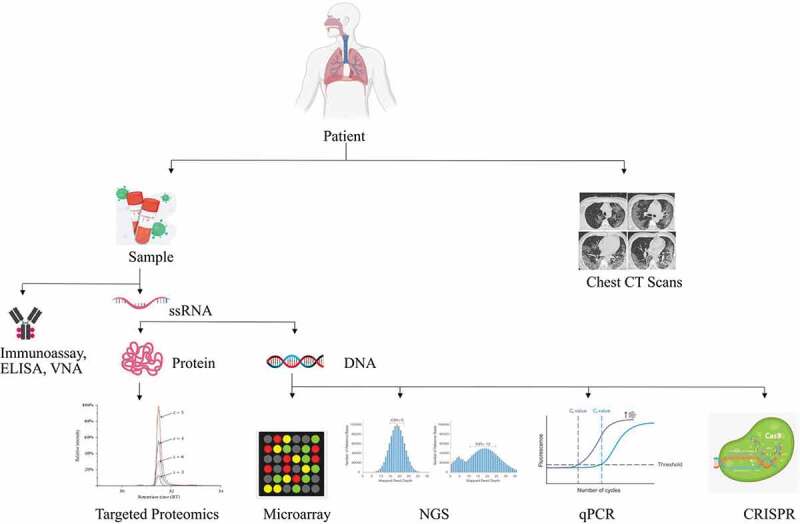
An overview of the diagnostic methods for SARS-CoV-2
3. Methodology
We carried out a comprehensive narrative literature review to encapsulate the developments in the medical diagnosis of SARS-CoV-2 by accessing the freely available online databases. The research papers and articles were mainly retrieved from PubMed using keywords like ‘SARS-CoV-2 Diagnosis’, COVID-19, ‘Coronavirus Diagnosis’ and ‘SARS-CoV-2’. The publications that have been included as references in this article, date from 2005 to 2020. The data about the presently deployed and approved diagnostic kits were obtained from the WHO, FDA, and ICMR websites. The literature search was limited to publications in English and excluded abstracts from commentaries. Research papers focusing on the diagnosis of SARS-CoV-2 were only included. Literature search, abstract screening, and study selection were performed by the aforementioned authors. Any discrepancies were resolved through consensus. The websites that were searched are as follows: https://pubmed.ncbi.nlm.nih.gov/, https://www.who.int/, https://www.cdc.gov/, https://www.fda.gov/home, https://www.icmr.gov.in/.
4. Expert opinion
Pandemics being large-scale infection outbreaks, gravely distress the world at large and have far-reaching consequences by causing significant social, economic, and political disarray. The universal disparity that already exists and is prevalent around the world also extends to this catastrophe. The Low-and-Middle income countries (LMICs) are much more vulnerable to the deleterious repercussions and face disproportionately higher morbidity and mortality during pandemics. The lack of infrastructure and resources curbs their domestic capacity to manufacture diagnostic test kits making them heavily dependent on imports. Accessibility and affordability are some of the crucial variables that should be considered while devising any diagnostic method and easy acquisition of effective diagnostic test kits to all the affected countries is imperative [96–100].
Several detection methods are available for the diagnosis of SARS-CoV-2 but neither is ideal in all situations. Molecular methods aren’t swift and cost-effective with a high likelihood of false-negative results which can jeopardize the health of the patient and the community [9]. A sensitivity of only 30 ~ 60% is achieved by the RT-PCR of SARS-CoV-2 RNA, determined by the stage of the disease, condition of the patient, the protocol followed while collecting clinical specimens and the type and number of specimens procured [17]. Immunodiagnostic methods based on IgM/IgG antibody detecting N and S antigens can partially overcome the shortcomings of RT-PCR in some cases and are rapid and cost-effective but the sensitivity is poor [15]. Whereas, detection of anti-N or anti-S antibodies is suitable for diagnosis in the later stage during the infection, contact tracing, and retrospective community screening [15]. Immunodiagnostics aids the diagnosis of COVID-19 and increases the positive rate of detection when combined with qRT-PCR, including the detection of subclinical cases [62].
Radiological methods are complementary to other methods and are not suitable for the screening of the population as differentiation of similar symptoms caused by the different etiological agents cannot be made. Hence further research into COVID-19 diagnostics is binding. Research is underway to determine the efficacy of other methods such as CRISPR, multiplex isothermal amplification followed by microarray detection, and immunological kits with great sensitivity. More data is required on pseudovirus-based neutralizing assays (PBNA), as far as neutralizing antibody detection is concerned. LC-MS technology for targeted proteomics can be an alternative to immunoassays but improvements in technology adoption, assay development, and validation are still required. Nanosensors and aptamer technology are also in development which will serve as an alternative to thermal screening guns and will help in the screening of the people at public places such as airports, malls, restaurants, and the territories which are more prone to crowding [101].
Although significant developments have been made in all the countries for diagnosis of SARS-CoV-2 in a very short time which matches the blinding speed at which the virus has disseminated, the limitations of available diagnostics represent the need for preparedness and long-term investments in SARS-CoV-2 diagnostics. Several vaccines are under phase 3 clinical trials with the Pfizer mRNA based vaccine BNT162 already completing the trials and reporting encouraging results, the possibility of vaccine availability soon is high. But manufacturing of required doses and distribution across the world is not anticipated any sooner. An accurate diagnosis will remain the forerunner in the management of the pandemic. Moreover, the phase 4 efficacy of these vaccines will rely heavily on appropriate detection of the protective immune response and reactogenicity elicited by the vaccine further increasing the diagnostic burden. So far, a combination of NAAT and immuno-diagnostics along with radiological diagnostics remains the preferred options and we hope to see a change with the development of better technologies toward a single multipurpose diagnostic test. To be sufficiently prepared for any future pandemics, healthcare institutions and systems should be essentially reinforced.
Funding Statement
This paper was not funded by any funding agency.
Declaration of interest
The authors have no relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties.
Reviewer disclosures
Peer reviewers on this manuscript have no relevant financial or other relationships to disclose.
References
Papers of special note have been highlighted as either of interest (•) or of considerable interest (••) to readers.
- 1.World Health Organization . Coronavirus Disease (COVID-19) Situation Reports [Internet]. 2020. [cited 2020 Dec21]. Available from: https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports
- 2.COVID-19 pandemic [Internet] . [cited 2020. Dec 30]. Available from: https://en.wikipedia.org/wiki/COVID-19_pandemic
- 3.Coronavirus Update (Live) on COVID-19 Virus Pandemic - Worldometer [Internet]. [cited 2020. Dec 30]. Available from: https://www.worldometers.info/coronavirus/
- 4.Weekly Update : Global Coronavirus Impact and Implications [Internet]. [cited 2020. October 21]. Available from: https://www.counterpointresearch.com/coronavirus-weekly-update/
- 5.Weekly epidemiological update 22 December 2020. [Internet]. [cited 2020 Dec 31]. Available from: https://www.who.int/publications/m/item/weekly-epidemiological-update—22-december-2020
- 6.Weekly epidemiological update 29 December 2020. [Internet]. [cited 2020 Dec 31]. Available from: https://www.who.int/publications/m/item/weekly-epidemiological-update—29-december-2020
- 7.Ioannidis JPA Global perspective of COVID-19 epidemiology for a full-cycle pandemic [Internet]. Vol. 50, Eur J Clin Invest. Blackwell Publishing Ltd; 2020. [cited 2021 Jan 2]. 12; Doi: 10.1111/eci.13423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zheng J, SARS-coV-2: an emerging coronavirus that causes a global threat. Int J Biol Sci. [Internet]. 2020. [cited 2020 Sep 12];16(10):1678–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]; • This paper gives the information on the origin, signs and symptoms, and the chemotherapeutic options of the SARS-CoV-2 infection.
- 9.Lu R, Zhao X, Li J, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet [Internet]. 2020. February 22 [cited 2020 Sep 12];395(10224):565–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ozma MA, Maroufi P, Khodadadi E et al. Clinical manifestation, diagnosis, prevention and control of SARS-CoV-2 (COVID-19) during the outbreak period. Le infezioni in medicina. 2020. [cited 2020 Sep 12]. Available from: https://pubmed.ncbi.nlm.nih.gov/32275257/ 28 2 153–165 [PubMed] [Google Scholar]; •• An important review article with essential details on the diagnosis of SARS-CoV-2.
- 11.Li X, Zai J, Zhao Q, et al. Evolutionary history, potential intermediate animal host, and cross-species analyses of SARS-CoV-2. J Med Virol. [Internet]. 2020. Jun 1 [cited 2020 Dec 29];92(6):602–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao J, Cui W, Tian B, The Potential Intermediate Hosts for SARS-CoV-2. Front Microbiol. [Internet]. 2020. Sep 30 [cited 2020 Dec 29];11:2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Diao B, Wen K, Chen J, et al. Diagnosis of Acute Respiratory Syndrome Coronavirus 2 Infection by Detection of Nucleocapsid Protein. medRxiv [Internet]. 2020. March 13 [cited 2020 Sep 12];2020.03.07.20032524. Available from: Doi: 10.1101/2020.03.07.20032524 [DOI]
- 14.Chan JF-W, Yip CC-Y, To KK-W, et al. Improved Molecular Diagnosis of COVID-19 by the Novel, Highly Sensitive and Specific COVID-19-RdRp/Hel Real-Time Reverse Transcription-PCR Assay Validated In Vitro and with Clinical Specimens. Vol. 58, J Clin Microbiol. American Society for Microbiology; 2020. [cited 2020 Sep 12]. 5 Doi: 10.1128/JCM.00310-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sanche S, Lin YT, Xu C, et al. High Contagiousness and Rapid Spread of Severe Acute Respiratory Syndrome Coronavirus 2 - Vol 26, Number 7—July 2020. - Emerging Infectious Diseases journal - CDC. [cited 2020 Oct 14]. 7 1470–1477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tang Y-W, Schmitz JE, Persing DH, et al. Laboratory diagnosis of COVID-19: current issues and challenges [Internet]. Vol. 58, J Clin Microbiol. American Society for Microbiology; 2020. [cited 2020 Sep 12].(6) Doi: 10.1128/JCM.00512-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu D, Wu T, Liu Q, et al. The SARS-CoV-2 outbreak: what we know [Internet]. Inter J Infect Dis Elsevier B.V.; 2020. [cited 2020 Sep 12]. p. 44–48. Available from. . 94:. https://pubmed.ncbi.nlm.nih.gov/32171952/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang H, Li X, Li T, et al. The genetic sequence, origin, and diagnosis of SARS-CoV-2 [Internet]. Eur J Clin Microbiol Infect Dis. Springer; 2020. [cited 2020 Sep 12]. 39(9); p. 1629–1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Root-Bernstein R. Why Infants Rarely Die of COVID-19 and Morbidity and Mortality Rates Vary by Location: pneumococcal and Hib Vaccinations as Possible Means to Mitigate Future Pandemics. 2020. April 15 [cited 2020 Sep 17]; Available from: www.preprints.org
- 20.Yang Y, Yang M, Shen C, et al. Evaluating the accuracy of different respiratory specimens in the laboratory diagnosis and monitoring the viral shedding of 2019-nCoV infections. medRxiv [Internet]. 2020. February 17 [cited 2020 Dec 30];2020.02.11.20021493. Available from: Doi: 10.1101/2020.02.11.20021493 [DOI]
- 21.Ravi N, Cortade DL, Ng E, et al. Diagnostics for SARS-CoV-2 detection: A comprehensive review of the FDA-EUA COVID-19 testing landscape. Biosens Bioelectron [Internet]. 2020. October 1 [cited 2020 Dec 30];165:112454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang W, Xu Y, Gao R, et al. Detection of SARS-CoV-2 in Different Types of Clinical Specimens. JAMA. American Medical Association; 2020. [cited 2020 Dec 30]. 323(18); p. 1843–1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pascarella G, Strumia A, Piliego C, et al. COVID-19 diagnosis and management: a comprehensive review [Internet]. J Intern Med. Blackwell Publishing Ltd; 2020. [cited 2020 Sep 13]. p. 192–206. 288(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roche Roche’s cobas SARS-CoV-2 Test to detect novel coronavirus receives FDA Emergency Use Authorization and is available in markets accepting the CE mark [Internet]. [cited 2020. Aug 25]. Available from: https://www.roche.com/media/releases/med-cor-2020-03-13.htm
- 25.Tu Y-F, Chien C-S, Yarmishyn AA, et al. A review of sars-cov-2 and the ongoing clinical trials [Internet]. Int J Mol Sci. MDPI AG; 2020. [cited 2020 Sep 12]. 21(7)2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.RealStar® SARS-CoV-2 RT-PCR Kit RUO - Altona-Diagnostics EN [Internet]. [cited 2020. Aug 25]. Available from: https://www.altona-diagnostics.com/en/products/reagents-140/reagents/realstar-real-time-pcr-reagents/realstar-sars-cov-2-rt-pcr-kit-ruo.html
- 27.BioCore 2019-nCoV Real Time PCR Kit [Internet]. [cited 2020. October 14]. Available from: http://www.bio-core.com/biocore/kr/common/Brochure_(ENG).pdf
- 28.The Global Fund Report on the list of SARS-CoV-2 Diagnostic test kits [Internet]. 2020. [cited 2020 Aug26]. Available from: https://www.theglobalfund.org/media/9629/covid19_diagnosticproducts_list_en.pdf?u=637261896210000000
- 29.QIAamp Viral RNA Mini Kit - QIAGEN Online Shop [Internet]. [cited 2020. Aug 27]. Available from: https://www.qiagen.com/us/products/diagnostics-and-clinical-research/sample-processing/qiaamp-viral-rna-mini-kit/?clear=true#orderinginformation
- 30.Food and Drug Administration . COVID-19 RT-qPCR Detection Kit Instructions for Use For Emergency Use Authorization Only. 2020.
- 31.Quick SARS-CoV-2 rRT-PCR Kit | ZYMO RESEARCH [Internet]. [cited 2020. Aug 27]. Available from: https://www.zymoresearch.com/products/quick-sars-cov-2-rrt-pcr-kit
- 32.Food and Drug Administration . Instructions for LabGunTM COVID-19 RT-PCR Kit [Internet]. 2020. September [cited 2020 Aug28]. Available from: www.labgenomics.co.kr
- 33.RealStar® SARS-CoV-2 RT-PCR Kit - Altona-Diagnostics EN [Internet]. [cited 2020. Aug 28]. Available from: https://altona-diagnostics.com/en/products/reagents-140/reagents/realstar-real-time-pcr-reagents/realstar-sars-cov-2-rt-pcr-kit.html
- 34.Food and Drug Administration . Instruction for Use Fosun COVID-19 RT-PCR Detection Kit Rx Only For Emergency Use Authorization (EUA) only [Internet]. 2020. [cited 2020 Aug28]. Available from: www.fosunpharmausa.com/covid19/pcr/
- 35.Food and Drug Administration . GenoSensor GS TM COVID-19 Real-Time PCR Kit For Emergency Use Authorization Only Instructions for Use (IFU) Issue 1.0. 2020.
- 36.Food and Drug Administration . FACT SHEET FOR HEALTHCARE PROVIDERS [Internet]. 2020. [cited 2020 Aug28]. Available from: https://www.accessdata.fda.gov/scripts/medwatch/index.cfm?action=reporting.home
- 37.Food and Drug Administration . copyTM COVID-19 qPCR Multi Kit (Cat no. M22MD100M) Instructions for Use For in vitro diagnostic use For Emergency Use Authorization Only Prescription Use Only. 2020. May.
- 38.World Health Organization . List of COVID-19 diagnostic kits with the information of manufacturer and country of approval [Internet]. 2020. Mar [cited 2020 Aug29]. Available from: https://www.fda.gov/emergency-preparedness-and-response/mcm-legal-regulatory-and-policy-
- 39.Food and Drug Administration . TaqPath TM COVID-19 Combo Kit and TaqPath TM COVID-19 Combo Kit Advanced* INSTRUCTIONS FOR USE Multiplex real-time RT-PCR test intended for the qualitative detection of nucleic acid from SARS-CoV-2. 2020. March.
- 40.VIASURE SARS-CoV-2 Real Time PCR Detection Kit [Internet]. 2020. [cited 2020 Aug30]. Available from: https://www.abacusdx.com/media/CT_CerTest_VIASURE_2020.pdf
- 41.Food and Drug Administration .AllplexTM 2019-nCoV Assay [Internet]. 2020. [cited 2020 Aug30]. Available from: http://www.seegene.com/upload/product/IFU_FDA_COVID19_Seegene.pdf
- 42.Food and Drug Administration . Novel Coronavirus (2019-nCoV) Nucleic Acid Diagnostic Kit (PCR-Fluorescence Probing) For Emergency Use Only Instructions for Use (24 Tests/kit and 48 Tests/kit) For in vitro Diagnostic (IVD) Use For Prescription Use only For Emergency Use Authorization only [Internet]. 2020. [cited 2020 Aug30]. Available from: http://eng.sansure.com.cn/
- 43.ProTectTM COVID-19 RT-qPCR Kit [Internet]. [cited 2020. Aug 30]. Available from: http://www.camtech.org/assets/ProTect_COVID19_Kit_BrochureN.pdf
- 44.BioFire-COVID-19-Test-Info-Sheet-FLM2-PRT-0263 [Internet]. [cited 2020. Augt 30]. Available from: https://www.biofiredefense.com/wp-content/uploads/2020/04/BioFire-COVID-19-Test-Info-Sheet-FLM2-PRT-0263.pdf
- 45.Food and Drug Administration . NxTAG ® CoV Extended Panel Assay Package Insert [Internet]. 2020. March [cited 2020 Aug30]. Available from: www.luminexcorp.com
- 46.Food and Drug Administration . NeuMoDxTM SARS-CoV-2 Assay Instructions For Use. 2020. April.
- 47.Food and Drug Administration . INSTRUCTIONS FOR USE For Use under Emergency Use Authorization For In Vitro Diagnostic Use RX Only PhoenixDx® SARS-CoV-2 Multiplex for invitro diagnostic use qualitative RT-PCR-based detection of SARS-CoV-2 50 Tests PCCSKU15262 v 2.0. 2020.
- 48.SARS-Cov-2 Coronavirus Real-time RT-PCR (RT-qPCR) Detection Kit (CVPD) [Internet]. [cited 2020 Aug31]. Available from: https://www.sciencellonline.com/PS/7038.pdf
- 49.TaqPathTM COVID-19 Combo Kit [Internet]. [cited 2020 Aug31]. Available from: https://www.thermofisher.com/order/catalog/product/A47814#/A47814
- 50.Food and Drug Administration. Quick SARS-CoV-2 rRT-PCR Kit Instructions for Use. 2020. [Google Scholar]
- 51.Younes N, Al-Sadeq DW, Jighefee HAL, et al. Challenges in laboratory diagnosis of the novel coronavirus SARS-CoV-2 [Internet]. Viruses. MDPI AG; 2020. [cited 2020 Sep 12]. 12(6)582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shi R, Ma W, Wu Q, et al. Design and application of 60mer oligonucleotide microarray in SARS coronavirus detection. Chinese Sci Bull. [Internet]. 2003. Jun [cited 2020 Sep 13];48(12):1165–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.De Souza Luna LK, Heiser V, Regamey N, et al. Generic detection of coronaviruses and differentiation at the prototype strain level by reverse transcription-PCR and nonfluorescent low-density microarray. J Clin Microbiol. [Internet]. 2007. [cited 2020 Sep 13];45(3):1049–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Guo X, Geng P, Wang Q, et al. Development of a Single Nucleotide Polymorphism DNA Microarray for the Detection and Genotyping of the SARS Coronavirus. J Microbiol Biotechnol [Internet]. 2014. October 28 [cited 2020 Sep 13];24(10):1445–1454. Available from: [DOI] [PubMed] [Google Scholar]
- 55.Li B, Si H-R, Zhu Y, et al. Discovery of Bat Coronaviruses through Surveillance and Probe Capture-Based Next-Generation Sequencing. mSphere. 2020;5(1):e00807-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Illumina Receives First FDA Emergency Use Authorization for a Sequencing-Based COVID-19 Diagnostic Test [Internet]. [cited 2020. Aug 31]. Available from: https://www.illumina.com/company/news-center/press-releases/2020/8cd141fb-68d0-4144-8922-45693ac3f453.html
- 57.Sheridan C. COVID-19 spurs wave of innovative diagnostics. Nat Biotechnol. 2020;38(7):769–772. [DOI] [PubMed] [Google Scholar]
- 58.VS J, Kancharla N, Bhadra B, et al. CRISPR-based assays for rapid detection of SARS-CoV-2. Preprints [Internet]. 2020;(June):1–10. Available from: www.preprints.org [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sharma S. Low-cost ‘Feluda’ test to detect virus in an hour likely in 4 weeks - india news - Hindustan Times [Internet]. hindustantimes. 2020. [cited 2020 Sep 01]. Available from: https://www.hindustantimes.com/india-news/low-cost-feluda-test-to-detect-virus-in-an-hour-likely-in-4-weeks/story-q1Q8ATDRNnwEGuRbhWVI2O.html
- 60.Guo L, Sun X, Wang X, et al. SARS-CoV-2 detection with CRISPR diagnostics [Internet]. Cell Discov. Springer Nature; 2020. [cited 2020 Sep 13]. 6(1); p. 1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lee CY-P, Lin RTP, Renia L, et al. Serological Approaches for COVID-19: epidemiologic Perspective on Surveillance and Control [Internet]. Vol. 11, Front Immunol. Frontiers Media S.A.; 2020. [cited 2020 Sep 12]. Doi: 10.3389/fimmu.2020.00879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pan Y, Li X, Yang G, et al. Serological immunochromatographic approach in diagnosis with SARS-CoV-2 infected COVID-19 patients. J Infect [Internet]. 2020. July 1 [cited 2020 Sep 12];81(1):e28–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Guo L, Ren L, Yang S, et al. Profiling Early Humoral Response to Diagnose Novel Coronavirus Disease (COVID-19). Clin Infect Dis [Internet]. 2020. July 28 [cited 2020 Sep 12];71(15):778–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wen T, Huang C, Shi F-J, et al. Development of a lateral flow immunoassay strip for rapid detection of IgG antibody against SARS-CoV-2 virus. Analyst [Internet]. 2020. August 7 [cited 2020 Sep 13];145(15):5345–5352. [DOI] [PubMed] [Google Scholar]
- 65.Liu W, Liu L, Kou G, et al. Evaluation of nucleocapsid and spike protein-based enzyme-linked immunosorbent assays for detecting antibodies against SARS-CoV-2. J Clin Microbiol [Internet]. 2020. June 1 [cited 2020 Sep 12];58(6). Doi: 10.1128/JCM.00461-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gupta V, Tabiin TM, Sun K, et al. SARS coronavirus nucleocapsid immunodominant T-cell epitope cluster is common to both exogenous recombinant and endogenous DNA-encoded immunogens. Virology [Internet]. 2006. March 30 [cited 2020 Sep 13];347(1):127–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Haljasmägi L, Remm A, Rumm AP, et al. LIPS method for the detection of SARS-CoV-2 antibodies to spike and nucleocapsid proteins. Eur J Immunol [Internet]. 2020. August 6 [cited 2020 Sep 13];50(8):1234–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.PD B, FX R, Morishima C, et al. Detection of nucleocapsid antibody to SARS-CoV-2 is More Sensitive than Antibody to Spike Protein in COVID-19 Patients. medRxiv Prepr Serv Heal Sci [Internet]. 2020. April 24 [cited 2020 Sep 13];2020.04.20.20071423. Available from: http://www.ncbi.nlm.nih.gov/pubmed/32511445
- 69.Sofia SARS Antigen FIA [Internet]. 2020. [cited 2020 Sep02]. Available from: https://www.quidel.com/sites/default/files/product/documents/CL1438902EN00.pdf
- 70.Food and Drug Administration . ACCELERATED EMERGENCY USE AUTHORIZATION (EUA) SUMMARY NEW YORK SARS-COV MICROSPHERE IMMUNOASSAY FOR ANTIBODY DETECTION (WADSWORTH CENTER AT THE NEW YORK STATE DEPARTMENT OF HEALTH) for in vitro diagnostic use Rx only for use under emergency use authorization (EUA) only. 2020.
- 71.LIAISON ® SARS-CoV-2 S1/S2 IgG The fully automated serology test for the detection of SARS-CoV-2 IgG Antibodies [Internet]. [cited 2020. October 15]. Available from: https://www.diasorin.com/sites/default/files/allegati/liaisonr_sars-cov-2_s1s2_igg_brochure.pdf.pdf
- 72.Food and Drug Administration . Anti-SARS-CoV-2 Rapid Test. 2020.
- 73.Food and Drug Administration . Cellex qSARS-CoV-2 IgG/IgM Rapid Test. 2020.
- 74.Food and Drug Administration . Wantai SARS-CoV-2 Diagnostics WANTAI SARS-CoV-2 Ab Rapid Test Rapid Test for Detection of Total Antibodies to SARS-CoV-2 FOR SERUM/PLASMA/VENIPUNCTURE WHOLE BLOOD SPECIMEN INSTRUCTIONS FOR USE. 2020. [cited 2020 Sep03]; Available from: 10.1101/2020.04.09.20056325
- 75.SARS-CoV-2 antibody test (lateral flow method) [Internet]. [cited 2020. Sep 03]. Available from: https://www.bilcare.com/SARS-CoV-2 Antibody Test (Lateral Flow Method).pdf
- 76.Food and Drug Administration . DPP® COVID-19 IgM/IgG System [Internet]. 2020. [cited 2020 Sep04]. Available from: https://www.fda.gov/media/136963/download
- 77.SGTi-flex COVID-19 IgM/IgG Kit - Sugentech, Inc. [Internet]. [cited 2020. Sep 04]. Available from: https://sugentech.com/products/products-view.php?ct=7&target=32%27
- 78.STANDARD Q COVID-19 Ag Test Kit [Internet]. [cited 2020. Sep 04]. Available from: http://sdbiosensor.com/xe/product/7672
- 79.STANDARD F COVID-19 Ag FIA Kit [Internet]. [cited 2020. Sep 04]. Available from: http://sdbiosensor.com/xe/product/7677
- 80.Food and Drug Administration . RightSign TM COVID-19 IgG/IgM rapid test cassette package insert [Internet]. 2020. [cited 2020 Sep05]. Available from: https://www.fda.gov/medical-devices/emergency-situations-medical-devices/emergency-use-
- 81.Indian Council of Medical Research , New Delhi [Internet]. [cited 2020. October 15]. Available from: https://www.icmr.gov.in/
- 82.Xu Y, Xiao M, Liu X, et al. Significance of serology testing to assist timely diagnosis of SARS-CoV-2 infections: implication from a family cluster. Emerg Microbes Infect [Internet]. 2020. January 1 [cited 2020 Sep 10];9(1):924–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Fang Y, Zhang H, Xie J, et al. Sensitivity of Chest CT for COVID-19: comparison to RT-PCR. Radiology. Radiological Society of North America Inc.; 2020. [cited 2020 Dec 31]. 296(2); p. E115–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ai T, Yang Z, Hou H, et al. Correlation of chest CT and RT-PCR testing for coronavirus disease 2019 (COVID-19) in China: A report of 1014 cases. Radiology [Internet]. 2020. August 1 [cited 2020 Dec 31];296(2):E32–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Nawattanapaiboon K, Pasomsub E, Prombun P, et al. Colorimetric reverse transcription loop-mediated isothermal amplification (RT-LAMP) as a visual diagnostic platform for the detection of the emerging coronavirus SARS-CoV-2. Analyst [Internet]. 2020. [cited 2020 Dec 29]; Available from: https://pubs.rsc.org/en/content/articlehtml/2021/an/d0an01775b [DOI] [PubMed]
- 86.Porte L, Legarraga P, Vollrath V, et al. Evaluation of a novel antigen-based rapid detection test for the diagnosis of SARS-CoV-2 in respiratory samples. International Journal of Infectious Diseases. 2020. October 1;99:328–333. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Whitman JD, Hiatt J, Mowery CT, et al. Test performance evaluation of SARS-CoV-2 serological assays. medRxiv [Preprint]. 2020. May 17. Doi: 10.1101/2020.04.25.20074856. Update in: Nat Biotechnol. 2020 August 27;: PMID: 32511497; PMCID: PMC7273265. [DOI]
- 88.Patchsung M, Jantarug K, Pattama A et al. Clinical validation of a Cas13-based assay for the detection of SARS-CoV-2 RNA. Nat Biomed Eng [Internet]. 2020. Dec 1 [cited 2020 December30];4(12). Available from: https://pubmed.ncbi.nlm.nih.gov/32848209/ [DOI] [PubMed] [Google Scholar]
- 89.Nie J, Li Q, Wu J, et al. Establishment and validation of a pseudovirus neutralization assay for SARS-CoV-2. Emerging Microbes & Infections. 2020;9(1):680–686. [cited 2020 Sep 12]; Available from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.COVID-19 Detection | SARS-CoV-2 Neutralization Antibody Detection Kit (RUO) [Internet]. [cited 2020. October 14]. Available from: https://www.genscript.com/covid-19-detection-svnt.html
- 91.Marx V, Targeted proteomics | nature methods [Internet]. Nat Methods. 2013.[cited 2020 Sep 13]. Available from. https://www.nature.com/articles/nmeth.2285 [Google Scholar]
- 92.Vidova V, Spacil Z. A review on mass spectrometry-based quantitative proteomics: targeted and data independent acquisition [Internet]. Anal Chim Acta Elsevier B.V.; 2017. [cited 2020 Sep 13]. p. 7–23. Available from. . 964:. https://pubmed.ncbi.nlm.nih.gov/28351641/. [DOI] [PubMed] [Google Scholar]
- 93.Manes NP, Nita-Lazar A. Application of targeted mass spectrometry in bottom-up proteomics for systems biology research. J Proteomics [Internet]. 2018. October 30 [cited 2020 Sep 13];189:75–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Cross TG, Hornshaw MP. Can LC and LC-MS ever replace immunoassays? J Appl Bioanal [Internet]. 2016. October 13 [cited 2020 Sep 13];2(4):108–116. [Google Scholar]
- 95.Kubina R, Dziedzic A. Molecular and Serological Tests for COVID-19 a Comparative Review of SARS-CoV-2 Coronavirus Laboratory and Point-of-Care Diagnostics. Diagnostics (Basel). 2020. Jun 26;10(6):434. https://10.3390/diagnostics10060434. PMID: 32604919; PMCID: PMC7345211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Understanding cycle threshold (Ct) in SARS-CoV-2 RT-PCR A guide for health protection teams Understanding cycle threshold (Ct) in SARS-CoV-2 RT-PCR 2.
- 97.Darwish IA . Immunoassay Methods and their Applications in Pharmaceutical Analysis: Basic Methodology and Recent Advances. Int J Biomed Sci. 2006. Sep;2(3):217–35. PMID: 23674985; PMCID: PM C3614608 [PMC free article] [PubMed] [Google Scholar]
- 98.SARS-CoV-2 Antigen detecting rapid diagnostic test implementation projects [Internet] . [cited 2020. November 15]. Available from: https://www.who.int/news-room/articles-detail/sars-cov-2-antigen-detecting-rapid-diagnostic-test-implementation-projects
- 99.Developing countries face diagnostic challenges as the COVID-19 pandemic surges [Internet] . [cited 2020. November 18]. Available from: https://cen.acs.org/analytical-chemistry/diagnostics/Developing-countries-face-diagnostic-challenges/98/i27
- 100.Pandemics: Risks, Impacts, and Mitigation - Disease control priorities: improving health and reducing poverty - NCBI Bookshelf [Internet]. [cited 2020. November 18]. Available from: https://www.ncbi.nlm.nih.gov/books/NBK525302/
- 101.Coronavirus (SARS-CoV-2) : Test kits to detect the causative agent of COVID-19 [Internet]. [cited 2020. September 01]. Available from: https://www.rapidmicrobiology.com/test-method/testing-for-the-wuhan-coronavirus-a-k-a-covid-19-sars-cov-2-and-2019-ncov
- 102.Radiology Assistant - COVID-19 Imaging findings. [Internet]. [cited2020Oct14]. Available from: https://radiologyassistant.nl/chest/covid-19/covid19-imaging-findings


