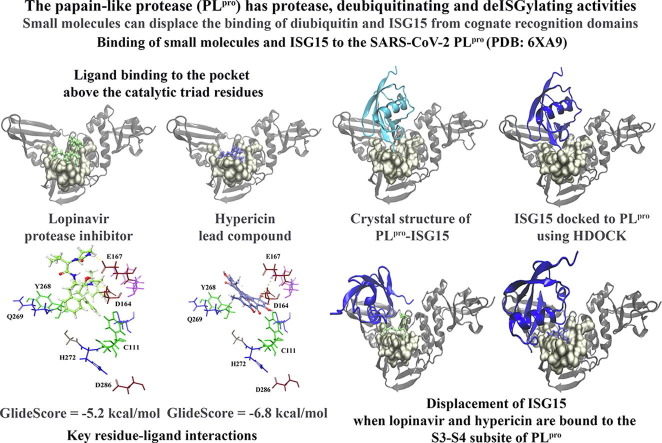
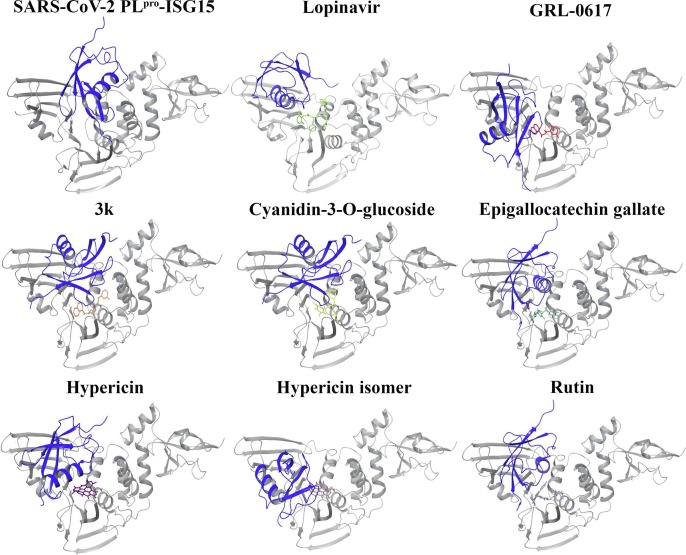
Graphical abstract
Keywords: Coronavirus, COVID-19, SARS-CoV-2, Papain-like protease, deISGylating activity, deubiquitinase activity, Molecular docking, Naphthalene-based inhibitors, Dietary compounds
Abstract
The SARS-CoV-2 papain-like protease (PLpro) is a suitable target for drug development, and its deubiquitinating and deISGylating activities have also been reported. In this study, molecular docking was used to investigate the binding properties of a selection of dietary compounds and naphthalene-based inhibitors to the previously characterised binding site of GRL-0617. The structures of the SARS-CoV-2 and SARS-CoV PLpro in complex with interferon-stimulated gene 15 (ISG15) and lysine 48 (K48)-linked diubiquitin were utilised. To predict whether compounds could potentially interfere with the binding of these cellular modifiers, docking was conducted in the absence and presence of ISG15 and K48-linked diubiquitin.
1. Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the pathogen responsible for causing coronavirus disease (COVID-19) and the resulting pandemic has had a significant impact on all aspects of life [1]. Vaccine trials are already underway and drug repurposing has also formed a crucial part of the response. Computational methods have been increasingly used during the pandemic to screen large libraries of compounds, such as U.S Food and Drug Administration (FDA) approved drugs, against SARS-CoV-2 proteins [2], [3]. In silico methods are utilised in the drug discovery process to explore the mechanisms of action of compounds, identify lead compounds and for structural optimisation [2].
The viral replicase polyproteins, pp1a and pp1ab, are produced via the translation of open reading frames ORF1a and ORF1ab [1], [4]. The replicase proteins are processed further by two cysteine proteases, the main protease (Mpro) and papain-like protease (PLpro), to generate the non-structural proteins [1], [4]. The PLpro domain is a component of non-structural protein 3 (nsp3), the largest multi-domain replicase subunit encoded by the coronavirus genome [5]. PLpro specifically recognises the consensus motif LXGG and is involved in proteolytic cleavage [6]. In addition to the protease activity of the SARS-CoV and SARS-CoV-2 PLpro, this enzyme has been found to possess deubiquitinating and deISGylating activities [6], [7], [8].
Viral proteins and nucleic acids are detected by pattern recognition receptors (PRRs) that are found in various locations including the surface of the cell, endosomal membranes, in the cytosol and extracellularly [9]. This can trigger a cascade of events including the synthesis and release of interferons, the activation of pro-inflammatory responses, the production of signals that can regulate the adaptive immune response, and the induction of apoptosis [9]. Ubiquitin and interferon-stimulated gene 15 (ISG15) are important components of the innate immune system and are involved in the antiviral immune response [10]. These cellular modifiers covalently interact with target proteins and the outcomes can vary [10].
Viruses have consequently developed mechanisms to avoid being detected and destroyed by the host’s immune response, and these evasion strategies continue to be investigated [11]. The C-terminal portion of ubiquitin and ISG15 contains the LXGG motif and this can be recognised by PLpro [6]. The SARS-CoV and SARS-CoV-2 PLpro can therefore remove and remodel ubiquitin or ubiquitin-like proteins from the target substrates [12]. Recent studies have demonstrated that the SARS-CoV-2 PLpro has a preference for ISG15 and it also has the ability to cleave K48-linked ubiquitin [8], [13], [14], [15]. Although ISG15 is able to bind to the SARS-CoV PLpro, this protein has a preference for K48-linked ubiquitin [16]. The S1 and S2 ubiquitin-binding sites (SUb1 and SUb2) in the SARS-CoV and SARS-CoV-2 PLpro are involved in the binding mode of ubiquitin and ISG15 [8], [16].
The deubiquitinase and deISGylating activities of PLpro can be inhibited by compounds, such as GRL-0617 [13], [17]. This can reduce viral replication and increase antiviral signalling [13]. In saying this, a number of studies have assessed the potential of natural compounds to target the SARS-CoV-2 and SARS-CoV cysteine proteases. There is a growing body of literature on the benefits of Traditional Chinese Medicine, as well as phytochemicals with a broad range of biological activities. This includes tea polyphenols, liquorice extract, molecules from Withania somnifera and Alpinia officinarum, anthraquinone derivatives, anthocyanins and curcuminoids to name a few [18], [19], [20]. The antiviral and immunomodulating properties of these plant-based compounds could be useful for the prevention, management and treatment of COVID-19 [21], [22].
In this study, molecular docking was used to predict the interactions that occur between a selection of compounds and the protein residues of the SARS-CoV-2 and SARS-CoV PLpro crystal structures in the presence or absence of ISG15 and diubiquitin. The known binding site of naphthalene-based compounds was of interest and the natural ligands that were examined included (-)-epigallocatechin gallate, hypericin, rutin and cyanidin-3-O-glucoside. The aim was to identify leads that could potentially inhibit the deubiquitinating and deISGylating activities of PLpro, through preventing the binding of ISG15 and polyubiquitin.
2. Materials and methods
2.1. Structures of the proteins and ligands
The SARS-CoV and SARS-CoV-2 PLpro crystal structures in complex with ISG15 and K48-linked diubiquitin were obtained from the RCSB Protein Data Bank (PDB) [23]. This included 6xa9 (SARS-CoV-2 PLpro- ISG15 C-terminal domain propargylamide), 5tl6 (SARS-CoV PLpro – C-terminal domain of human ISG15), 5tl7 (SARS-CoV PLpro – C-terminal domain of mouse ISG15) and 5e6j (SARS-CoV PLpro – K48-linked diubiquitin activity based probe) [7], [8], [24]. The apo PLpro structures were also generated through removing the ISG15 or K48-linked diubiquitin chains. The zinc ion was retained in the 6xa9, 5tl6 and 5tl7 crystal structures, while nickel was retained in 5e6j. GRL-0617 and 3k were the naphthalene-based inhibitors examined in this study and their structures were drawn using Chem3D 19.0 (Perkin Elmer, Massachusetts, USA). Lopinavir, which is a protease inhibitor, and several dietary compounds were also selected for analysis. They included cyanidin-3-O-glucoside, an isomer of hypericin, rutin and (-)-epigallocatechin gallate. The chemical structures of these ligands were obtained from the National Centre for Biotechnology Information PubChem [25]. The structure of hypericin was also downloaded from ChEMBL [26], [27]. We previously identified these compounds as hits in a study that involved the screening of 300 ligands that were sourced from the OliveNet™ database and the pre-existing literature [28], [29].
2.2. Molecular docking using the Schrödinger Suite
The PLpro crystal structures and compounds were imported into Maestro [30]. The crystal structures were prepared using the Protein Preparation Wizard of the Schrödinger Suite (version 2020-2), and the ligands were prepared using the LigPrep tool [31]. The receptor grid for each protein was generated using residues M208, P247, P248, Y264, G266, N267, Y268, Q269, C270, Y273, K157, L162, G163, D164 and E167 in the SARS-CoV-2 structure [32]. These amino acids have been previously found to form a pocket in PLpro [15], [33]. The corresponding residues for the SARS-CoV PLpro were M209, P248, P249, Y265, G267, N268, Y269, Q270, C271, Y274, K158, L163, G164, D165 and E168. The grids were 20 × 20 × 20 Å in size. The default settings were used for each of these steps and the optimized potentials for liquid simulations 3e (OPLS3e) force field was selected [34].
The top-ranking conformer of each compound was used in the molecular docking stage and the ligands were docked using the quantum-polarized ligand docking (QPLD) protocol [35]. The extra precision (XP) mode was selected for the initial docking and redocking stages, and the Jaguar quantum mechanics (QM) level was set to accurate [36], [37]. In the final selection stage of the protocol, the GlideScore option was chosen and this is a measure of the binding affinity in kcal/mol.
2.3. Protein-protein docking and blind docking
The ISG15 and K48-linked diubiquitin chains were isolated from each protein and were docked to the apo PLpro crystal structure using the HDOCK-server [38]. In this study, ab initio protein-protein docking was conducted and the top-ranking model was evaluated further. The K48-linked diubiquitin from the SARS-CoV PLpro was docked to the SARS-CoV-2 PLpro. In order to determine whether the presence of compounds within the target region would affect the binding mode of ISG15 or K48-linked diubiquitin, the ligands that were docked through Schrödinger were retained in this site. ISG15 and K48-linked diubiquitin were subsequently docked to the ligand-bound structures.
Blind docking was performed on the apo SARS-CoV and SARS-CoV-2 PLpro crystal structures, as well as the PLpro-ISG15 and K48-linked diubiquitin complexes. The crystal structures and compounds were prepared as macromolecules and ligands in PyRx, respectively [39]. The receptor grid encompassed the entire protein and an exhaustiveness of 2048 was utilised. Once the configuration file had been generated and the corresponding .pdbqt files were obtained, the jobs were run using AutoDock Vina through the cloud-computing server Galileo (Hypernet Labs) [40].
2.4. Molecular dynamics simulations
Classical MD simulations were performed with SARS-CoV-2 PLpro in complex with ISG15 in the absence or presence of small molecules, as previously described [41]. The starting structure of PLpro and ISG15 in the absence of small molecules was obtained from PDB ID 6xa9 [8]. The docked ligands were used as starting structures, with ligand topologies generated with SwissParam [42]. The structure of the docked ISG15 generated by the HDOCK server in the presence of small molecules bound to the enzyme served as the starting structure for ISG15 [38]. The covalent propargylamide linker was removed. Systems were fully solvated with TIP3P water in a dodecahedral box with a minimum distance of 1.0 nm between protein atoms and the closest box edge. Simulations were performed with a time-step of 2 fs in triplicate for 100 ns.
Free energy calculations were quantified with molecular Mechanics-Poisson Boltzmann Surface Area (MM-PBSA) [43] using the g_mmpbsa tool [44], as previously described [41]. Calculations were performed in triplicate on the final nanosecond of the stabilised trajectories [45].
3. Results and discussion
3.1. Molecular docking to the SARS-CoV-2 PLpro
GRL-0617 is a naphthalene-based compound that was initially identified by Ratia et al. to noncovalently inhibit the SARS-CoV PLpro [17]. GRL-0617 has also been found to inhibit the SARS-CoV-2 PLpro and its mechanisms of action are being investigated further [13], [14]. In the SARS-CoV and SARS-CoV-2 PLpro, the catalytic triad residues are essential for protease activity and a number of compounds function through covalently modifying the catalytic cysteine (C112 in SARS-CoV and C111 in SARS-CoV-2) [17]. In contrast to this, GRL-0617 has been found to bind to a region that is located adjacent to the active site and is positioned in the S3 and S4 subsites of PLpro [15].
When examining the apo SARS-CoV-2 PLpro crystal structure, the GlideScores of GRL-0617 and 3k were found to be −3.8 and −3.5 kcal/mol, respectively (Fig. 1, Fig. 2 ). These naphthalene-based compounds were predominantly surrounded by hydrophobic residues including P247, P248, M208, V165, L162, Y264, Y268, C270, Y112 and Y273. The negatively charged residues E167 and D164, the positively charged residues R166 and K157, the polar residues N267 and T301, and the glycine residues G163 and G271 were also present. GRL-0617 formed no intermolecular bonds with the protein residues, while 3k formed a salt bridge with D164 and a π–π cation with K157 (Fig. 1, Fig. 2).
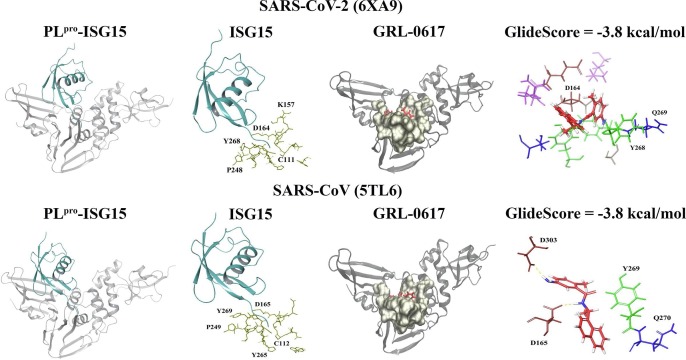
Fig. 1.
Crystal structures of the SARS-CoV-2 (6XA9) and SARS-CoV PLpro (5TL6). The PLpro structures in complex with ISG15 are depicted and the ubiquitin-like protein is coloured cyan. The residues that surround the LXGG motif of ISG15 (5 Å) can be seen and the amino acids are coloured gold. The position of GRL-0617 within the binding site (coloured tan) is shown. The docked ligand can be seen in red and the interactions that were formed with this region are also provided.
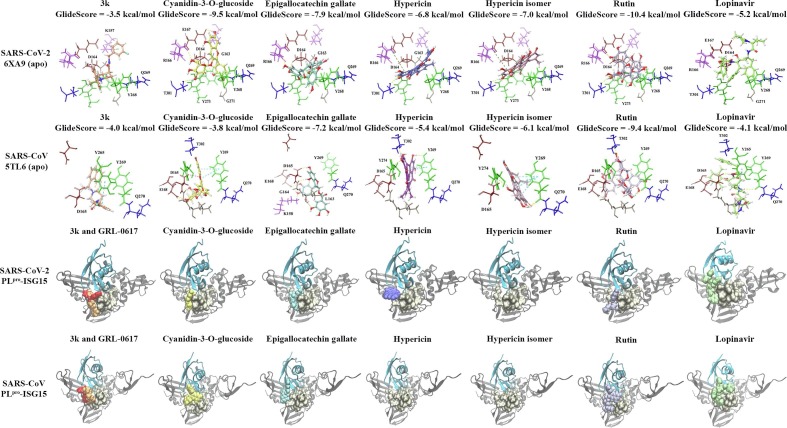
Fig. 2.
Molecular docking results for the apo and PLpro-ISG15 complexes. The protein-ligand interactions for the apo PLpro are provided for 3k (orange), which is a naphthalene-based inhibitor, lopinavir (light green), and the dietary compounds cyanidin-3-O-glucoside (yellow), (-)-epigallocatechin gallate (light blue), hypericin (purple), the hypericin isomer (light pink) and rutin (violet). The polar residues are coloured blue, the hydrophobic residues are coloured green, the positively charged residues are coloured pink, the negatively charged residues are coloured maroon and the glycine residues are coloured brown. The compounds were also docked in the presence of ISG15 and they were found to be positioned adjacent to the binding site of interest. In the SARS-CoV PLpro-ISG15 complex, hypericin was not able to bind and this was also the case for the hypericin isomer in the SARS-CoV-2 and SARS-CoV PLpro (in the presence of ISG15).
Báez-Santos et al. previously identified 3k to be a potent noncovalent inhibitor of the SARS-CoV PLpro and although this compound was found to be less metabolically stable, the authors highlighted how its structure may be optimised further [46]. In a recent study by Bosken et al., the binding properties of 3k and the thiopurine inhibitor 6-mercaptopurine to the SARS-CoV-2 PLpro were evaluated using computational methods [47]. In ligand-free proteins, the blocking loop (BL2) exhibits significant flexibility and most of this motion can be attributed to residues N267, Q269 and Y268 [47]. The molecular dynamics (MD) simulations revealed that upon binding, 3k stabilises the BL2 loop (residues 267-272) in the SARS-CoV-2 PLpro and this has also been found to occur in the ligand-bound structures of the SARS-CoV PLpro [17], [47]. This results in a closed conformation of the BL2 loop and 3k forms hydrogen bonds with D164 and Y268 of the SARS-CoV-2 PLpro [47]. GRL-0617 also induces a conformational change in BL2 that results in its closure and the plasticity of this region has been reported in a number of papers [17], [48]. In the crystal structure of the SARS-CoV-2 PLpro in complex with GRL-0617, this ligand formed hydrogen bonds with D164 and Q269 [48]. In a study conducted by Shin et al. the results from the MD simulations showed that Y268 in the SARS-CoV-2 PLpro and Y269 in the SARS-CoV PLpro were important for the binding mode of GRL-0617 [13].
Lopinavir is a drug that has been approved as a human immunodeficiency virus (HIV) protease inhibitor by the FDA and the role of this compound in the treatment of COVID-19 is being explored [49]. When docked to PLpro, the GlideScore for lopinavir was −5.2 kcal/mol and it formed hydrogen bonds with the amino acids E167 and D164 (Table S1 and Fig. 2). The dietary compounds were binding with a stronger affinity to the binding site and rutin had a GlideScore of −10.4 kcal/mol (Fig. 2). This was followed by cyanidin-3-O-glucoside, (-)-epigallocatechin gallate, the hypericin isomer and hypericin. In comparison to GRL-0617, 3k, and lopinavir, there were a greater number of intermolecular bonds between the functional groups of the natural compounds and the protein residues (Table S1). Rutin, for example, formed inter-atomic contacts with Y268 (π–π interaction), Y273 (H-bond), T301 (H-bond), D164 (H-bond) and R166 (H-bond). Hypericin and its isomer also formed bonds with T301 (H-bonds), Y273 (H-bond for the isomer), R166 (π–π cations) and G163 (H-bond for hypericin). Hydrogen bonds were present for cyanidin-3-O-glucoside and E167, R166, T301, G163, Y273 and G271, while (-)-epigallocatechin gallate formed hydrogen bonds with R166, D164 and G163 (Table S1).
3.2. Molecular docking to the SARS-CoV PLpro
The GlideScores for the SARS-CoV PLpro 5tl6 crystal structure indicated that rutin had the strongest binding affinity, followed by (-)-epigallocatechin gallate, the hypericin isomer, hypericin, and cyanidin-3-O-glucoside (Fig. 2). This was similar to the 5tl7 structure, as rutin had the strongest binding affinity, followed by cyanidin-3-O-glucoside, (-)-epigallocatechin gallate, the hypericin isomer and hypericin (Supplementary Fig. 1). Rutin formed hydrogen bonds with the same protein residues in both crystal structures and they were E168, D165, Y269 and T302. The isomer of hypericin formed intermolecular bonds with Y269 (H-bonds and π–π interactions) and Y274 (H-bond) in the 5tl6 structure and in addition to these amino acids, a hydrogen bond was present with R167 in the 5tl7 SARS-CoV PLpro. Similar to the isomer, hypericin formed bonds with Y269 (H-bonds and π–π interactions) and Y274 (H-bond) in the 5tl6 PLpro structure, as well as T302 (H-bond). For the 5tl7 crystal structure, hypericin formed bonds with Y265 (H-bond), D165 (H-bond), E168 (H-bond) and K158 (π–π cation). In both crystal structures, (-)-epigallocatechin gallate formed hydrogen bonds with K158, E168, D165 and G164. (-)-Epigallocatechin gallate also formed a hydrogen bond with L163 and R167 in the 5tl6 and 5tl7 SARS-CoV PLpro structures, respectively. The protein residues of the 5tl6 structure that were involved in intermolecular bonds (hydrogen bonds) with cyanidin-3-O-glucoside were E168, D165, Y269 and T302. In the 5tl7 structure of the SARS-CoV PLpro, these residues were T302, K158, E168, G164, Y269 and Y265.
With the exception of cyanidin-3-O-glucoside for the 5tl6 crystal structure, the naphthalene-based inhibitors and lopinavir had weaker GlideScores compared to the natural compounds (Fig. 1, Fig. 2, and Supplementary Fig. 1). The 3k inhibitor formed a π–π interaction with Y265 and a salt bridge with D165 in both crystal structures. There was also a π–π interaction present with Y269 in the 5tl7 SARS-CoV PLpro. GRL-0617 formed bonds with D165 and D303 in the 5tl6 SARS-CoV PLpro, whereas bonds were present with Y265, Y269 and Q270 in the 5tl7 crystal structure. In addition to Y269, the residues D165 and Q270 are commonly involved in inter-atomic contacts with naphthalene-based inhibitors in the SARS-CoV PLpro [13], [17], [46]. In the 5tl6 and 5tl7 crystal structures, lopinavir formed hydrogen bonds with D165. The protease inhibitor also formed bonds with Y269 (H-bond) and Y265 (π–π interaction) in the 5tl6 structure, and R167 in the 5tl7 structure.
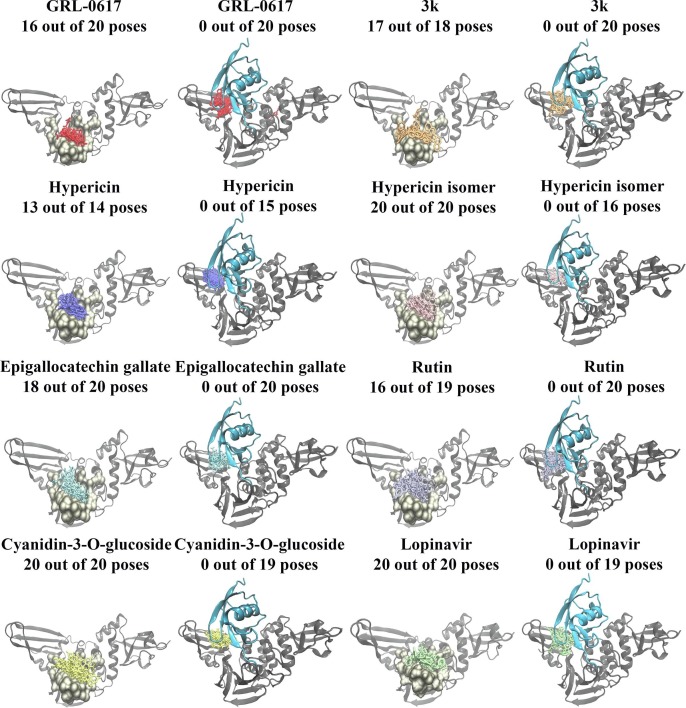
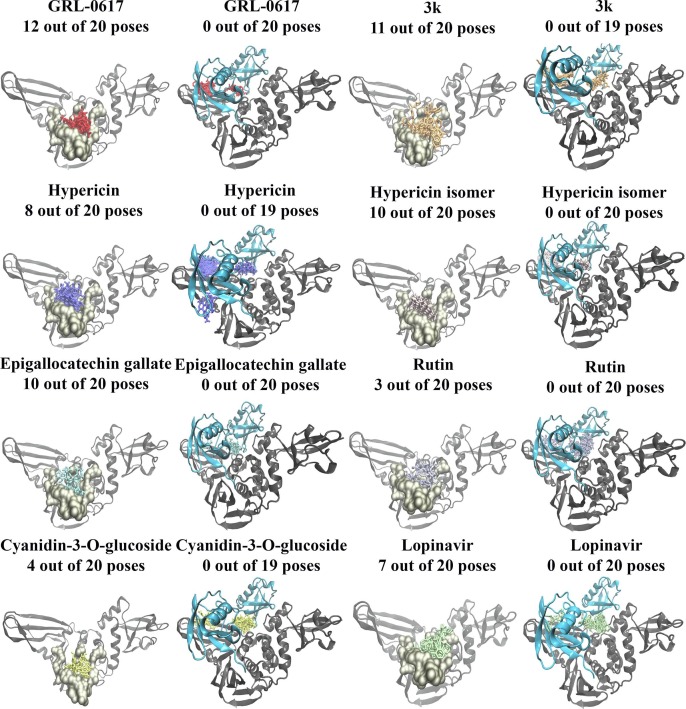
Blind docking was conducted on the SARS-CoV-2 PLpro and the results demonstrated that out of the 20 poses generated for GRL-0617, 16 were in positioned in the binding site of interest (Fig. 3 ). Similarly, there were 17 out of 18 poses for 3k in this site. Interestingly, all of the blind docking poses (20 out of 20 poses) for cyanidin-3-O-glucoside, the hypericin isomer, and lopinavir were found to be in the target binding site. Hypericin had 13 out of 14 poses, rutin had 16 out of 19 poses, and (-)-epigallocatechin gallate had 18 out of 20 poses in this region. Moreover, the blind docking results for the SARS-CoV PLpro revealed that the ligands had fewer poses within the region (Supplementary Figs. 2 and 3).
Fig. 3.
Blind docking results for the SARS-CoV-2 apo and PLpro-ISG15 complexes. Blind docking was conducted in the absence and presence of ISG15 for naphthalene-based inhibitors, lopinavir, and the dietary compounds. The number of poses that were found to be in the target binding region are provided. GRL-0617 is coloured red, 3k is coloured orange, hypericin is coloured purple, the hypericin isomer is coloured light pink, (-)-epigallocatechin gallate is coloured light blue, rutin is coloured violet, cyanidin-3-O-glucoside is coloured yellow, and lopinavir is coloured light green. ISG15 can be seen in cyan, while the main PLpro chain is coloured gray.
The mechanisms of action of phenolic compounds, particularly the flavonoid subclass, and their use as therapeutic agents against coronaviruses are being explored [22]. Several in silico studies have focused on the papain-like protease of coronaviruses and computational methods have been used to identify lead compounds with potential biological activities from plant sources [19], [33]. The antiviral properties of the dietary compounds that were selected for use in this study, as well as their derivatives, have previously been reported and their chemical structures can also be modified to improve their potency, reduce side effects and increase their bioavailability [22]. Additionally, it would be important to gain further insight into the anti-inflammatory and antioxidant properties of rutin, hypericin, cyanidin-3-O-glucoside and (-)-epigallocatechin gallate. Hypericin is classified as an anthraquinone derivative, cyanidin-3-O-glucoside is an anthocyanin cation, rutin is a flavonoid, and (-)-epigallocatechin gallate is a catechin. Based on the results, the compounds hypericin, rutin and cyanidin-3-O-glucoside could be evaluated further. In terms of SARS-CoV-2, these compounds have primarily been tested against the spike protein and Mpro [29], [50].
3.3. Isg15
Daczkowski et al. determined the crystal structure of the SARS-CoV PLpro in complex with the C-terminal domain of human ISG15 and mouse ISG15 propargylamide [24]. The SARS-CoV PLpro had a stronger affinity towards the human ISG15 and when compared to mono-ubiquitin, there was a shift in the global orientation of the ubiquitin-like protein [24]. This rotation was also observed for the C-terminal domain of ISG15 in the SARS-CoV-2 PLpro [8]. Both ISG15 and ubiquitin are positioned on the palm subdomain however, ISG15 interacts with the thumb subdomain of PLpro rather than the fingers subdomain [8]. Most notably, the SUb1 site of PLpro is predominantly involved in ISG15 modifications [8].
When human ISG15 was retained in the SARS-CoV-2 PLpro and the compounds were docked to the protein, it was evident that they were displaced from the binding site. Rather than binding to the SUb1 pocket that the human ISG15 C-terminal residues extend into, the compounds were positioned further to the side (Fig. 2). The ligands were consequently interacting with different residues and there were changes in the GlideScores. The hypericin isomer was unable to dock to PLpro in the presence of ISG15. Likewise, the blind docking results revealed that none of the ligands had poses within the target binding region in the presence of ISG15 and that there were a number of poses in a distinct pocket on the fingers domain of PLpro (Fig. 3). This was also observed for the SARS-CoV PLpro crystal structures in complex with mouse and human ISG15 (Fig. 2, Supplementary Figs. 2 and 3).
Based on the SARS-CoV-2 PLpro crystal structure, the residues that were within 5 Å of L154-AYE157 of ISG15 were W106, N109, N110, C111, Y112, K157, E161, L162, E163, D164, P248, Y264, N267, Y268, Q269, C270, G271, H272, Y273 and T301. When the ISG15 chain was docked to PLpro through the HDOCK server, these same residues were found to surround ISG15 in the top ranked model. The root mean square deviation (RMSD) value between the top ranked model generated from protein-protein docking and the crystallographic structure was 1.1 Å (Fig. 4 ). Furthermore, the RMSD values of the human and mouse ISG15 for the 5tl6 and 5tl7 SARS-CoV PLpro crystal structures were 2.6 Å and 0.4 Å, respectively (Supplementary Figs. 5 and 6).
Fig. 4.
Protein-protein docking results for the SARS-CoV-2 PLpro. The HDOCK server was used to dock ISG15 to the main PLpro chain and the ubiquitin-like protein is coloured dark blue. The displacement of ISG15 can be seen for the ligand-bound structures. GRL-0617 is coloured red, 3k is coloured orange, hypericin is coloured purple, the hypericin isomer is coloured light pink, (-)-epigallocatechin gallate is coloured light blue, rutin is coloured violet, cyanidin-3-O-glucoside is coloured yellow, and lopinavir is coloured light green.
The compounds that were docked to the apo SARS-CoV-2 and SARS-CoV PLpro were also retained in the protein and the ligand-bound structures were used for protein-protein docking with ISG15. The conformation of the docked ubiquitin-like protein differed from that of the crystallised input when each compound was present and the RMSD values were larger (Fig. 4). In terms of the SARS-CoV-2 PLpro-ISG15 complex, the RMSD was 22.5 Å for GRL-0617, 17.5 Å for 3k, 13.3 Å for hypericin, 25.4 Å for the hypericin isomer, 21.6 Å for rutin, 17.4 Å for cyanidin-3-O-glucoside, 21.3 Å for (-)-epigallocatechin gallate, and 20.7 Å for lopinavir. The RMSD values were also greater when the compounds were present in the SARS-CoV PLpro crystal structures. When the mouse ISG15 was docked to the SARS-CoV PLpro, the RMSD values were 22.3 Å for GRL-0617, 21.9 Å for 3k, 14.1 Å for hypericin, 18.9 Å for the isomer of hypericin, 21.3 Å for cyanidin-3-O-glucoside, 18.6 Å for (-)-epigallocatechin gallate, 19.6 Å for rutin, and 42.0 Å for lopinavir (Supplementary Fig. 6). The RMSD values of the SARS-CoV PLpro structure in complex with human ISG15 were 45.9 Å for GRL-0617, 46.1 Å for 3 k, 24.2 Å for rutin, 21.1 Å for cyanidin-3-O-glucoside, 19.2 Å for (-)-epigallocatechin gallate, 19.4 Å for hypericin, 45.2 Å for the hypericin isomer, and 6.4 Å for lopinavir (Supplementary Fig. 5).
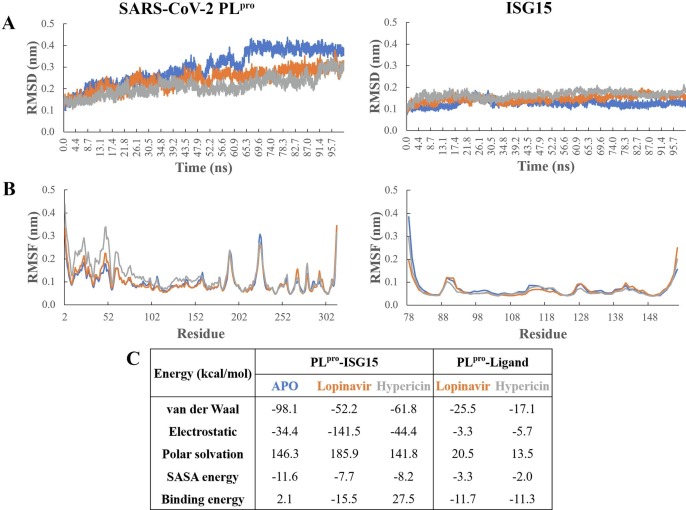
3.4. Stability of PLpro complexed with ISG15 in the presence of lopinavir and hypericin
MD simulations were performed in triplicate for 100 ns to assess the stability of PLpro in complex with ISG15 in the absence or presence of small molecules bound to the naphthalene inhibitor site of the enzyme. Based on root mean square deviation (RMSD) of the protein backbone of PLpro, the enzyme structure reached equilibration after approximately 70 ns (Fig. 5 A). Subsequent analysis was performed on the stabilised trajectory following this timepoint. The average RMSD of PLpro was lower with ligands bound to the naphthalene binding pocket, with values of 0.29 nm for lopinavir-bound and 0.26 nm for hypericin-bound PLpro, compared to 0.38 nm for the PLpro in the apo complex. While the ligand-bound complexes demonstrated slightly lower RMSD values, root mean square fluctuation (RMSF) analysis showed slightly higher values in the first ~ 80 residues of PLpro compared to apo, particularly for hypericin-bound PLpro (Fig. 5B). These residues comprise the N-terminal ubiquitin-like (Ubl) domain (residues 1–62) and a portion of the thumb domain (residues 63–175) in SARS-CoV-2 PLpro, which are located distal to the naphthalene binding pocket and ISG15-binding surface. Residue fluctuations were largely similar at the naphthalene binding pocket between all three systems. The Ub1 domain was observed to be more flexible in general, suggesting that binding of ISG15 stabilises PLpro at its interface residues and surrounding regions. For all three systems, a similar backbone RMSD for the bound ISG15 was observed: 0.12 nm for apo, 0.16 nm for lopinavir-, and 0.17 nm for hypericin-bound (Fig. 5A). Accordingly, backbone RMSF values for ISG15 are low, with the only flexibility in the protein apparent at N- and C-terminal residues (Fig. 5B). This indicates that the structure of ISG15 is similarly stable in all systems with minimal fluctuation.
Fig. 5.
Stability of SARS-CoV-2 PLpro complexed with ISG15 in the presence of lopinavir and hypericin. MD simulations were carried out for 100 ns in triplicate. Systems comprised of PLpro bound to ISG15 in the absence of ligands (blue), or with lopinavir (orange) or hypericin (grey) bound to the naphthalene inhibitor binding site of PLpro. A) Average root mean square deviation (RMSD) for backbone with respect to its initial structure for PLpro (left) and ISG15 (right). B) Average root mean square fluctuation (RMSF) for protein backbone was calculated following stabilisation. C) Average MM-PBSA binding free energy calculations are displayed for the binding of ISG15 and binding of the ligand to PLpro in each system.
MM-PBSA analysis indicates that both lopinavir and hypericin bind strongly to the naphthalene binding site of PLpro with binding energies of −11.7 and −11.3 kcal/mol, respectively (Fig. 5C). While both ligands have a similar affinity to PLpro, there is a marked change in the binding energy of ISG15. Compared to the ligand-free PLpro-ISG15 complex, a 14-fold weaker binding free energy for ISG15 is observed in the presence of hypericin (2.1 kcal/mol for apo, 27.5 kcal/mol for hypericin). Conversely, binding of ISG15 in the presence of lopinavir is stronger, with a binding energy of −15.5 kcal/mol. This suggests that hypericin may be a viable candidate for potential inhibition of de-ISGylating activity in SARS-CoV-2 PLpro, warranting further investigation.
3.5. K48-linked diubiquitin
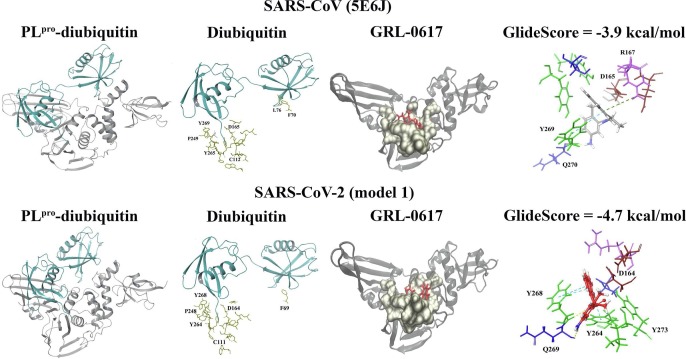
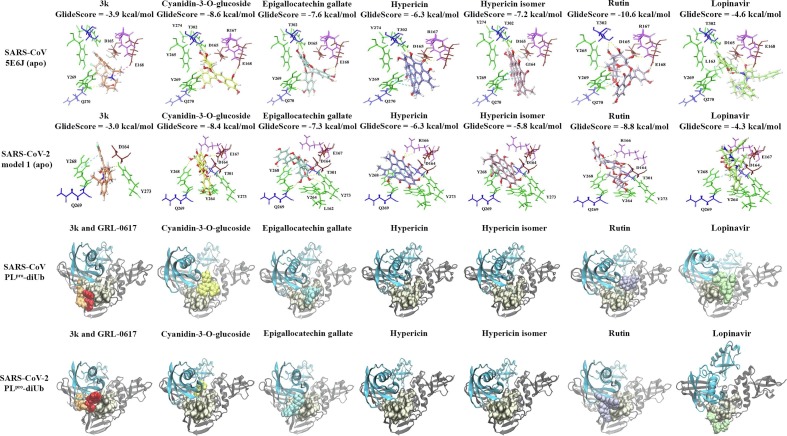
In addition to ISG15, the crystal structure of SARS-CoV in complex with K48-linked diubiquitin was analysed (Fig. 6 ). Protein-protein docking was also used to obtain the structure of SARS-CoV-2 PLpro in complex K48-linked diubiquitin and the top ranked model was investigated further (Fig. 6). When the ligands were docked to the binding site in the apo SARS-CoV and SARS-CoV-2 PLpro, the GlideScores of the dietary compounds were stronger than that of the inhibitors and the residue interactions can be seen in Fig. 7 . In the SARS-CoV and SARS-CoV-2 PLpro, the K48-linked diubiquitin extends into the proximal (SUb1) and distal (SUb2) ubiquitin sites [7], [8]. The compounds were also docked to the PLpro structures with the K48-linked diubiquitin present and were positioned further away from the binding pocket (Fig. 7). This could also be seen in the blind docking results, as the presence of the K48-linked diubiquitin interfered with the ability of compounds to bind to this region (Fig. 8 and Supplementary Fig. 4).
Fig. 6.
Crystal structures of the SARS-CoV (5E6J) and SARS-CoV-2 (model 1) PLpro. The PLpro structures in complex with K48-linked diubiquitin (cyan) are depicted. The main PLpro chain is coloured gray. The HDOCK server was used to dock the SARS-CoV K48-linked diubiquitin to the apo SARS-CoV-2 crystal structure and the top ranked model is shown. Several of the residues (coloured gold) that surround the LXGG motif in the SUb1 site are labelled and the key residues in the SUb2 site can be seen. The docked structure of GRL-0617 (coloured red) and its interactions with this region are provided.
Fig. 7.
Molecular docking results for the apo and PLpro-K48 linked diubiquitin complexes. The protein-ligand interactions for the apo PLpro are provided for 3k (orange), which is a naphthalene-based inhibitor, lopinavir (light green), and the dietary compounds cyanidin-3-O-glucoside (yellow), (-)-epigallocatechin gallate (light blue), hypericin (purple), the hypericin isomer (light pink) and rutin (violet). The polar residues are coloured blue, the hydrophobic residues are coloured green, the positively charged residues are coloured pink, the negatively charged residues are coloured maroon and the glycine residues are coloured brown. The compounds were also docked in the presence of K48-linked diubiquitin. In the SARS-CoV and SARS-CoV-2 PLpro-K48 linked diubiquitin complexes, hypericin was not able to bind and this was also the case for the hypericin isomer.
Fig. 8.
Blind docking results for the SARS-CoV apo and PLpro-K48 linked diubiquitin complexes. Blind docking was conducted in the absence and presence of K48-linked diubiquitin for naphthalene-based inhibitors and the dietary compounds. The number of poses that were found to be in the binding region are provided. GRL-0617 is coloured red, 3k is coloured orange, hypericin is coloured purple, the hypericin isomer is coloured light pink, (-)-epigallocatechin gallate is coloured light blue, rutin is coloured violet, cyanidin-3-O-glucoside is coloured yellow, and lopinavir is coloured light green. K48-linked diubiquitin can be seen in cyan, while the main PLpro chain is coloured gray.
The HDOCK server was used to further evaluate the SARS-CoV PLpro in complex with K48-linked diubiquitin and when comparing the crystallised structure to the top ranked model from protein-protein docking, the RMSD was 0.6 Å (Supplementary Fig. 7). In terms of the SUb2 site, the SARS-CoV PLpro residues F70 and L76 were within 5 Å of the K48-linked diubiquitin residue I44. In the top ranked model of SARS-CoV-2, F69 was found to be within 5 Å of I44. The SUb2 site of PLpro plays an important role in the recognition of K48-polyubiquitin and the N-terminal ubiquitin-like fold in ISG15 [13], [16]. The residues within this region are less conserved when comparing the SARS-CoV-2 and SARS-CoV PLpro, and the SUb2 site contained within the thumb subdomain is dynamic [15], [47]. The interactions that occur between residue I44 of the K48-linked diubiquitin and PLpro have been described in the literature [7], [8], [13]. This includes L76 and F70 in the SARS-CoV PLpro, as well as T75 and F60 in the SARS-CoV-2 PLpro [7], [8], [13].
Like ISG15, the presence of compounds in the crystal structure altered the conformation of the docked K48-linked ubiquitin (Supplementary Fig. 7). These changes were also reflected in the RMSD values as they were 34.4 Å for GRL-0617, 38.9 Å for 3 k, 38.8 Å for rutin, 34.7 Å for hypericin, 38.7 Å for the isomer of hypericin, 38.6 Å for (-)-epigallocatechin gallate, 39.0 Å for cyanidin-3-O-glucoside, and 46.3 Å for lopinavir. This was also the case for the SARS-CoV-2 model and there was a clear displacement of K48-linked diubiquitin when the inhibitors and dietary compounds were bound to PLpro (Supplementary Fig. 8).
4. Conclusions
Together with previous studies, our findings highlight that occupying the S3-S4 subsites on the SARS-CoV-2 PLpro with small molecule ligands can potentially inhibit the protease, deubiquitinating and deISGylating activities of PLpro. Regarding the potential deISGylating inhibitory activities, the lead dietary compounds should be evaluated further using various in vitro and in vivo methods. Hypericin is of particular interest and represents a compound that could undergo further evaluation in antiviral assays.
Author contributions statement
TCK and AH conceptualized the aims and methodology and were involved in supervision. EP performed data analysis, data curation, and was involved in production of the first draft of the manuscript. JL was involved in data analysis and curation and was involved in production of the first draft of the manuscript. All authors contributed to editing and reviewing the manuscript.
Declaration of Competing Interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: Epigenomic Medicine Program (TCK) is supported financially by McCord Research (Iowa, USA), which has a financial interest in dietary compounds described in this work. However, there is no conflict of interest with respect to the inhibition of the SARS-CoV-2 papain-like protease. The remaining co-authors also have no conflicts of interest.’
Acknowledgements
We would like to acknowledge intellectual and financial support by McCord Research (Iowa, USA). JL is supported by an Australian Government Research Training Program Scholarship. We are indebted to Alfonso Perez Escudero and the team at Crowdfight COVID-19 for enabling access to supercomputing facilities, and to Matthew Gasperetti and the team at Hypernet Labs; Galileo, for enabling cloud computing for this project. We thank the National Computing Infrastructure (NCI), and the Pawsey Supercomputing Centre in Australia (funded by the Australian Government). Further, we thank the Spartan High Performance Computing service (University of Melbourne), and the Partnership for Advanced Computing in Europe (PRACE) for awarding the access to Piz Daint, hosted at the Swiss National Supercomputing Centre (CSCS), Switzerland.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.cplett.2021.138468.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Khailany R.A., Safdar M., Ozaslan M. Genomic characterization of a novel SARS-CoV-2. Gene Rep. 2020;19:100682. doi: 10.1016/j.genrep.2020.100682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ojha P.K., Kar S., Krishna J.G., Roy K., Leszczynski J. Therapeutics for COVID-19: from computation to practices—where we are, where we are heading to. Mol. Diversity. 2020 doi: 10.1007/s11030-020-10134-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mukherjee S., Dasgupta S., Adhikary T., Adhikari U., Panja S.S. Structural insight to hydroxychloroquine-3C-like proteinase complexation from SARS-CoV-2: inhibitor modelling study through molecular docking and MD-simulation study. J. Biomol. Struct. Dyn. 2020:1–13. doi: 10.1080/07391102.2020.1804458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Báez-Santos Y.M., St. John S.E., Mesecar A.D. The SARS-coronavirus papain-like protease: structure, function and inhibition by designed antiviral compounds. Antiviral Res. 2015;115:21–38. doi: 10.1016/j.antiviral.2014.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lei J., Kusov Y., Hilgenfeld R. Nsp3 of coronaviruses: structures and functions of a large multi-domain protein. Antiviral Res. 2018;149:58–74. doi: 10.1016/j.antiviral.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barretto N., Jukneliene D., Ratia K., Chen Z., Mesecar A.D., Baker S.C. The papain-like protease of severe acute respiratory syndrome coronavirus has deubiquitinating activity. J. Virol. 2005;79(24):15189–15198. doi: 10.1128/JVI.79.24.15189-15198.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.M. Békés, G.J. van der Heden van Noort, R. Ekkebus, H. Ovaa, T.T. Huang, C.D. Lima, Recognition of Lys48-Linked Di-ubiquitin and Deubiquitinating Activities of the SARS Coronavirus Papain-like Protease, Mol Cell. 62(4) (2016) 572-85. [DOI] [PMC free article] [PubMed]
- 8.Klemm T., Ebert G., Calleja D.J., Allison C.C., Richardson L.W., Bernardini J.P., et al. Mechanism and inhibition of the papain-like protease, PLpro, of SARS-CoV-2. EMBO J. 2020;39(18):e106275. doi: 10.15252/embj.2020106275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Amarante-Mendes G.P., Adjemian S., Branco L.M., Zanetti L.C., Weinlich R., Bortoluci K.R. Pattern recognition receptors and the host cell death molecular machinery. Front. Immunol. 2018;9:2379. doi: 10.3389/fimmu.2018.02379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Perng Y.-C., Lenschow D.J. ISG15 in antiviral immunity and beyond. Nat. Rev. Microbiol. 2018;16(7):423–439. doi: 10.1038/s41579-018-0020-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nelemans T., Kikkert M. Viral innate immune evasion and the pathogenesis of emerging RNA virus infections. Viruses. 2019;11(10):961. doi: 10.3390/v11100961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bailey-Elkin B.A., Knaap R.C.M., Kikkert M., Mark B.L. Structure and function of viral deubiquitinating enzymes. J. Mol. Biol. 2017;429(22):3441–3470. doi: 10.1016/j.jmb.2017.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shin D., Mukherjee R., Grewe D., Bojkova D., Baek K., Bhattacharya A., et al. Papain-like protease regulates SARS-CoV-2 viral spread and innate immunity. Nature. 2020 doi: 10.1038/s41586-020-2601-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Freitas B.T., Durie I.A., Murray J., Longo J.E., Miller H.C., Crich D., et al. Characterization and noncovalent inhibition of the deubiquitinase and deISGylase activity of SARS-CoV-2 papain-like protease. ACS Infect. Dis. 2020;6(8):2099–2109. doi: 10.1021/acsinfecdis.0c00168. [DOI] [PubMed] [Google Scholar]
- 15.Rut W., Lv Z., Zmudzinski M., Patchett S., Nayak D., Snipas S.J., et al. Activity profiling and crystal structures of inhibitor-bound SARS-CoV-2 papain-like protease: a framework for anti–COVID-19 drug design. Sci. Adv. 2020;6(42):eabd4596. doi: 10.1126/sciadv.abd4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ratia K., Kilianski A., Baez-Santos Y.M., Baker S.C., Mesecar A. Structural basis for the ubiquitin-linkage specificity and deISGylating activity of SARS-CoV papain-like protease. PLoS Pathog. 2014;10(5):e1004113. doi: 10.1371/journal.ppat.1004113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ratia K., Pegan S., Takayama J., Sleeman K., Coughlin M., Baliji S., et al. A noncovalent class of papain-like protease/deubiquitinase inhibitors blocks SARS virus replication. Proc. Natl. Acad. Sci. 2008;105(42):16119. doi: 10.1073/pnas.0805240105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tripathi M.K., Singh P., Sharma S., Singh T.P., Ethayathulla A.S., Kaur P. Identification of bioactive molecule from Withania somnifera (Ashwagandha) as SARS-CoV-2 main protease inhibitor. J. Biomol. Struct. Dyn. 2020:1–14. doi: 10.1080/07391102.2020.1790425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.D. Goswami, M. Kumar, S. Ghosh, A. Das, Natural Product Compounds in Alpinia officinarum and Ginger are Potent SARS-CoV-2 Papain-like Protease Inhibitors (2020).
- 20.Khanal P., Patil B.M., Chand J., Naaz Y. Anthraquinone derivatives as an immune booster and their therapeutic option against COVID-19. Nat. Prod. Bioprospect. 2020;10(5):325–335. doi: 10.1007/s13659-020-00260-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xian Y., Zhang J., Bian Z., Zhou H., Zhang Z., Lin Z., et al. Bioactive natural compounds against human coronaviruses: a review and perspective. Acta Pharm. Sin. B. 2020;10(7):1163–1174. doi: 10.1016/j.apsb.2020.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Solnier J., Fladerer J-P. Flavonoids: A complementary approach to conventional therapy of COVID-19? Phytochem. Rev. 2020:1–23. doi: 10.1007/s11101-020-09720-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., et al. The protein data bank. Nucleic Acids Res. 2000;28(1):235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Daczkowski C.M., Dzimianski J.V., Clasman J.R., Goodwin O., Mesecar A.D., Pegan S.D. Structural insights into the interaction of coronavirus papain-like proteases and interferon-stimulated gene product 15 from different species. J. Mol. Biol. 2017;429(11):1661–1683. doi: 10.1016/j.jmb.2017.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim S., Chen J., Cheng T., Gindulyte A., He J., He S., et al. PubChem 2019 update: improved access to chemical data. Nucleic Acids Res. 2019;47(D1):D1102–D1109. doi: 10.1093/nar/gky1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mendez D., Gaulton A., Bento A.P., Chambers J., De Veij M., Félix E., et al. ChEMBL: towards direct deposition of bioassay data. Nucleic Acids Res. 2019;47(D1):D930–D940. doi: 10.1093/nar/gky1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Davies M., Nowotka M., Papadatos G., Dedman N., Gaulton A., Atkinson F., et al. ChEMBL web services: streamlining access to drug discovery data and utilities. Nucleic Acids Res. 2015;43(W1):W612–W620. doi: 10.1093/nar/gkv352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bonvino N.P., Liang J., McCord E.D., Zafiris E., Benetti N., Ray N.B., et al. OliveNet™: a comprehensive library of compounds from Olea europaea. Database (Oxford) 2018;2018:bay016. doi: 10.1093/database/bay016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pitsillou E., Liang J., Karagiannis C., Ververis K., Darmawan K., Ng K., et al. Interaction of small molecules with the SARS-CoV-2 main protease in silico and in vitro validation of potential lead compounds using an enzyme-linked immunosorbent assay. Comput. Biol. Chem. 2020;89:107408. doi: 10.1016/j.compbiolchem.2020.107408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schrödinger, Schrödinger Release 2020-2: Maestro, Schrödinger, LLC, New York, NY, 2020. 2020.
- 31.Madhavi Sastry G., Adzhigirey M., Day T., Annabhimoju R., Sherman W. Protein and ligand preparation: parameters, protocols, and influence on virtual screening enrichments. J. Comput. Aided Mol. Des. 2013;27(3):221–234. doi: 10.1007/s10822-013-9644-8. [DOI] [PubMed] [Google Scholar]
- 32.M. Sacco, C. Ma, J. Wang, Y. Chen, Crystal structure of the native SARS-CoV-2 papain-like protease (PLPro) with inhibitor GRL0617 (2020).
- 33.Alfaro M., Alfaro I., Angel C. Identification of potential inhibitors of SARS-CoV-2 papain-like protease from tropane alkaloids from Schizanthus porrigens: a molecular docking study. Chem. Phys. Lett. 2020;761:138068. doi: 10.1016/j.cplett.2020.138068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harder E., Damm W., Maple J., Wu C., Reboul M., Xiang J.Y., et al. OPLS3: a force field providing broad coverage of drug-like small molecules and proteins. J. Chem. Theory Comput. 2016;12(1):281–296. doi: 10.1021/acs.jctc.5b00864. [DOI] [PubMed] [Google Scholar]
- 35.Cho A.E., Guallar V., Berne B.J., Friesner R. Importance of accurate charges in molecular docking: quantum mechanical/molecular mechanical (QM/MM) approach. J. Comput. Chem. 2005;26(9):915–931. doi: 10.1002/jcc.20222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Friesner R.A., Murphy R.B., Repasky M.P., Frye L.L., Greenwood J.R., Halgren T.A., et al. Extra precision glide: docking and scoring incorporating a model of hydrophobic enclosure for protein−ligand complexes. J. Med. Chem. 2006;49(21):6177–6196. doi: 10.1021/jm051256o. [DOI] [PubMed] [Google Scholar]
- 37.Bochevarov A.D., Harder E., Hughes T.F., Greenwood J.R., Braden D.A., Philipp D.M., et al. Jaguar: a high-performance quantum chemistry software program with strengths in life and materials sciences. Int. J. Quantum Chem. 2013;113(18):2110–2142. [Google Scholar]
- 38.Yan Y., Zhang D., Zhou P., Li B., Huang S.-Y. HDOCK: a web server for protein-protein and protein-DNA/RNA docking based on a hybrid strategy. Nucleic Acids Res. 2017;45(W1):W365–W373. doi: 10.1093/nar/gkx407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dallakyan S., Olson A.J. In: Chemical Biology: Methods and Protocols. Hempel J.E., Williams C.H., Hong C.C., editors. Springer, New York; New York, NY: 2015. Small-Molecule Library Screening by Docking with PyRx; pp. 243–250. [DOI] [PubMed] [Google Scholar]
- 40.Trott O., Olson A.J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010;31(2):455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liang J., Pitsillou E., Karagiannis C., Darmawan K.K., Ng K., Hung A., et al. Interaction of the prototypical α-ketoamide inhibitor with the SARS-CoV-2 main protease active site in silico: molecular dynamic simulations highlight the stability of the ligand-protein complex. Comput. Biol. Chem. 2020;107292 doi: 10.1016/j.compbiolchem.2020.107292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zoete V., Cuendet M.A., Grosdidier A., Michielin O. SwissParam: a fast force field generation tool for small organic molecules. J. Comput. Chem. 2011;32(11):2359–2368. doi: 10.1002/jcc.21816. [DOI] [PubMed] [Google Scholar]
- 43.Baker N.A., Sept D., Joseph S., Holst M.J., McCammon J.A. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. 2001;98(18):10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kumari R., Kumar R., Lynn A. g_mmpbsa—a GROMACS tool for high-throughput MM-PBSA calculations. J. Chem. Inf. Model. 2014;54(7):1951–1962. doi: 10.1021/ci500020m. [DOI] [PubMed] [Google Scholar]
- 45.Hou T., Wang J., Li Y., Wang W. Assessing the performance of the MM/PBSA and MM/GBSA methods. 1. the accuracy of binding free energy calculations based on molecular dynamics simulations. J. Chem. Inf. Model. 2011;51(1):69–82. doi: 10.1021/ci100275a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Báez-Santos Y.M., Barraza S.J., Wilson M.W., Agius M.P., Mielech A.M., Davis N.M., et al. X-ray structural and biological evaluation of a series of potent and highly selective inhibitors of human coronavirus papain-like proteases. J. Med. Chem. 2014;57(6):2393–2412. doi: 10.1021/jm401712t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Y.K. Bosken, T. Cholko, Y.-C. Lou, K.-P. Wu, C.-eA. Chang, Insights Into Dynamics of Inhibitor and Ubiquitin-Like Protein Binding in SARS-CoV-2 Papain-Like Protease 7(174) (2020). [DOI] [PMC free article] [PubMed]
- 48.Gao X., Qin B., Chen P., Zhu K., Hou P., Wojdyla J.A., et al. Crystal structure of SARS-CoV-2 papain-like protease. Acta Pharm. Sin. B. 2020 doi: 10.1016/j.apsb.2020.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Meini S., Pagotto A., Longo B., Vendramin I., Pecori D., Tascini C. Role of lopinavir/ritonavir in the treatment of Covid-19: a review of current evidence, guideline recommendations, and perspectives. J. Clin. Med. 2020;9(7):2050. doi: 10.3390/jcm9072050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Romeo A., Iacovelli F., Falconi M. Targeting the SARS-CoV-2 spike glycoprotein prefusion conformation: virtual screening and molecular dynamics simulations applied to the identification of potential fusion inhibitors. Virus Res. 2020;286:198068. doi: 10.1016/j.virusres.2020.198068. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.