Abstract
Background:
Breast cancer (BC) is a major public health concern, and its prognosis is very poor once metastasis occurs. The tumor microenvironment and chemical pollution have been suggested recently to contribute, independently, to the development of metastatic cells. The BC microenvironment consists, in part, of adipocytes and preadipocytes in which persistent organic pollutants (POPs) can be stored.
Objectives:
We aimed to test the hypothesis that these two factors (2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD), an extensively studied, toxic POP and the microenvironment) may interact to increase tumor aggressiveness.
Methods:
We used a co-culture model using BC MCF-7 cells or MDA-MB-231 cells together with hMADS preadipocytes to investigate the contribution of the microenvironment and 2,3,7,8-tetrachlorodibenzo-p-dioxin TCDD on BC cells. Global differences were characterized using a high-throughput proteomic assay. Subsequently we measured the BC stem cell–like activity, analyzed the cell morphology, and used a zebrafish larvae model to study the metastatic potential of the BC cells.
Results:
We found that coexposure to TCDD and preadipocytes modified BC cell properties; moreover, it induced the expression of ALDH1A3, a cancer stem cell marker, and the appearance of giant cancer cells with cell-in-cell structures (CICs), which are associated with malignant metastatic progression, that we demonstrated in vivo.
Discussion:
The results of our study using BC cell lines co-cultured with preadipocytes and a POP and an in vivo zebrafish model of metastasis suggest that the interactions between BC cells and their microenvironment could affect their invasive or metastatic potential. https://doi.org/10.1289/EHP7102
Introduction
Breast cancer (BC) has a high incidence and mortality among women in addition to critical socioeconomic impacts (Bray et al. 2018). Over the past 50 y, BC has become a major health problem for women worldwide with, in 2018, more than new cases diagnosed (12% of all new cancer cases and 25% of all cancers in women) and more than 600,000 deaths (Bray et al. 2018). If the cancer is located only in the breast, the 5-y survival rate of people with BC is 99%, but this rate decreases to 85% if it has spread to lymph nodes and, more dramatically decreases to only 26% if distant metastases are present at diagnosis (Cronin et al. 2018). The metastatic process, or the dissemination of tumor cells throughout the body, is responsible for about 90% of cancer patient deaths (Chaffer and Weinberg 2011) and represents a major challenge of tumor oncology.
The progression of BC is a complex process that involves hormones as well as genetic and epigenetic alterations and the tumor microenvironment with the peritumoral stromal fraction, which is composed of adipose tissue, cancer-associated fibroblasts, endothelial cells, and immune cells such as macrophages and leukocytes (Lim et al. 2018). During tumor progression, cancer cells will modify their microenvironment extensively, which, in turn, will favor the promotion and dissemination of the tumor (Allen and Jones 2011; Polanska and Orimo 2013). Adipose tissue, which consists mainly of mature adipocytes and progenitors (preadipocytes and adipose-derived stem cells), is one of the most abundant components surrounding BC cells (Kothari et al. 2020). Adipose tissue exerts a major endocrine and secretory role and thus represents an essential actor in the inflammatory, angiogenic, and remodeling responses of the extracellular matrix that influence tumor behavior (Kothari et al. 2020). Several studies have shown that stromal cells, including preadipocytes and adipocytes that are influenced by factors produced by tumor cells in a constant and dynamic intercellular communication, may promote migration and invasion of BC cells participating in local inflammation via the production of interleukins and chemokines (Ham and Moon 2013).
Environmental pollutants, in particular persistent organic pollutants (POPs), have been suspected to contribute to the development of breast cancer, although in many cases the mechanisms of carcinogenicity are elusive (Cohn et al. 2019; Warner et al. 2011; Rodgers et al. 2018). Studies of the effects of POPs are challenging because of the long-term effects of these pollutants and the delay between exposure initiation and health outcome (La Merrill et al. 2013). Recently, clinical evidence has suggested that increased POP concentrations are associated with increased risk of progression of BC and metastasis (Koual et al. 2019). Despite current knowledge of the critical role of the tumor microenvironment, little is known about possible interactions between pollutants and the microenvironment. In particular, it is unclear whether the impact of pollutants on cancer progression results from direct effects on cancer cells or from tumor microenvironment-mediated effects. This is particularly relevant for POPs because they are known to interact with adipocytes. Indeed, these cells store POPs such as dioxins, furans, polychlorinated biphenyl flame retardants, and organochlorine pesticides for long periods of time (Jackson et al. 2017), as do preadipocytes which, in addition, have higher secretory capacities (Kim et al. 2012). In addition to their toxic effects, POPs remain a major public health concern due to their persistence in the environment and in organisms. Due to their lipophilicity, POPs have been found to accumulate in tissues such as liver, brain, and adipose, as was observed with 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) in mice (Joffin et al. 2018), polychlorinated biphenyls (PCBs) and Polybrominated diphenyl ethers (PBDEs) in human postmortem brain samples (Mitchell et al. 2012) and Polychlorinated dibenzodioxins (PCDD), Polychlorinated dibenzofurans (PCDF), and Organochlorine pesticides (OCPs) in liver and brain tissues of herring gulls (Falkowska et al. 2016). In a previous study, we showed in human obese subjects who underwent bariatric surgery that adipose tissue released POPs in blood slowly and chronically (Kim et al. 2011). This constant internal release and recontamination may also be responsible for the chronic toxicity associated with POPs exposure, as was shown using a xenograft mouse model where fat pads of TCDD-exposed mice were grafted onto naïve mice (Joffin et al. 2018). Although it was reported that TCDD impairs the epithelial-to-mesenchymal transition (EMT) during mouse development (a cellular developmental phenomenon that is observed during gastrulation in triploblastic organisms) (Abbott et al. 1994), we recently found that, from a mechanistic standpoint, POPs promoted the EMT in human epithelial cancer cell lines (Bui et al. 2009). EMT is involved in pathological processes such as fibrosis (Pierre et al. 2014) or the formation of metastatic cells (Diepenbruck and Christofori 2016; Li and Kang 2016). POP TCDD (Seveso dioxin) is the most toxic congener of the dioxin family (Fernandes and Falandysz 2021). It is also one of the most potent activators of the aryl hydrocarbon receptor (AhR) (Barouki et al. 2012; Fernandes and Falandysz 2021), a transcription factor that regulates xenobiotic metabolism and the EMT. The role of AHR in tumorigenesis and its involvement in cancer aggressiveness remains uncertain (Wang et al. 2011). Several studies suggest that it may be involved in cancer progression (Bui et al. 2016; Bui et al. 2009).
Because of a) the suspected role of POPs in BC metastasis, b) the interaction of POPs with preadipocytes and adipocytes, and c) the role of these cells in the tumor microenvironment, we hypothesized that the co-occurrence of preadipocytes and a persistent organic pollutant (TCDD), which mimics a contaminated microenvironment in the proximity of breast cancer cells, favors the acquisition of proinvasive properties by tumor cells as compared with TCDD or preadipocytes alone. Here, we aimed to study the evolution of the properties of mammary cancer epithelial cells following a) co-culture with preadipocytes (to study their paracrine role), b) exposure to TCDD, and c) coexposure (). To study the first and third conditions (a, c), we developed an original in vitro co-culture model that consists of human mammary tumor cells (MCF-7) and human preadipocytes (hMADS, human multipotent adipose-derived stem cells). We also used the human mammary tumor MDA-MB-231 cells (which do not have the same properties as MCF-7 cells with respect to estrogen and progesterone receptors) in co-culture and coexposure conditions to analyze the influence of the BC cell initial phenotype on the most relevant outcomes observed with MCF-7 cells. Then we used a well-characterized in vivo zebrafish xenograft model to explore the metastatic capacities of the BC.
Methods
Cell Culture
Human BC cell line MCF-7 (ATCC® HTB-22) cells were cultured at 37°C in a humidified atmosphere in 5% in Dulbecco’s modified essential medium (DMEM) high glucose, complemented with nonessential amino acids, human recombinant insulin, 10% fetal bovine serum (FBS), penicillin, streptomycin (Invitrogen), and fungizone. Human breast cancer cell line MDA-MB-231 (ATCC® HTB-26) cells were cultured at 37% in DMEM complemented with nonessential amino acids, 10% FBS, penicillin, streptomycin (Invitrogen). The hMADS cell line (human multipotent adipose-derived stem cells) has been described previously (Rodriguez et al. 2005) and was provided by Christian Dani (Institut de Biologie Valrose/Université Côte d’Azur, UMR CNRS/INSERM, Faculté de Médecine, Nice, France). These cells were isolated from human adipose tissue, and they are able to maintain their properties after several passages (Rodriguez et al. 2005). Proliferating hMADS cells were seeded into proliferating medium consisting of DMEM low glucose, with 10% fetal calf serum, hFGF2, HEPES buffer, penicillin, and streptomycin and incubated at 37°C in 5% . hFGF2 was removed for all the experiments.
The Co-Culture Model
MCF-7 cells or MDA-MB-231 cells and hMADS preadipocytes were co-cultured in Transwell culture plates in proliferating medium without FGF (fibroblast growth factor). Briefly, 400,000 BC cells (MCF-7 or MDA-MB 231 cells) were seeded into the wells of a 6-well companion (lower part, see Figure 1A), and 400,000 hMADS preadipocyte cells were seeded onto polyester membrane inserts ( pore size, upper part; see Figure 1A) in 6-well culture dishes. The two cell types shared the same culture medium, which diffuses through the inserts. MCF-7 and MDA-MB-231 cells also were grown alone as controls. After 24 h of incubation at 37°C, the medium was replaced, and the cells were treated with TCDD or vehicle for 48 h. The TCDD stock solution at in nonane was first diluted at in the culture medium before treating the cells. Vehicle control cells were treated with 0.016% nonane and were called “control.” The MCF-7 or MDA-MB-231 cells grown in the presence of the hMADS cells were called “co-culture,” and the MCF-7 or MDA-MB-231 grown in the presence of the hMADS cells treated with TCDD were called “coexposure” (). We also collected the conditioned media of the lower and upper chambers of the co-culture system after 2 d of treatment and immediately froze at for short storage or for long storage. This conditioned media was used to stimulate BC cells for the spheroid formation, alamarBlue cell viability, and the zebrafish larva metastasis assays. The number of different experiments with the co-culture model for all assays was at least three.
Figure 1.
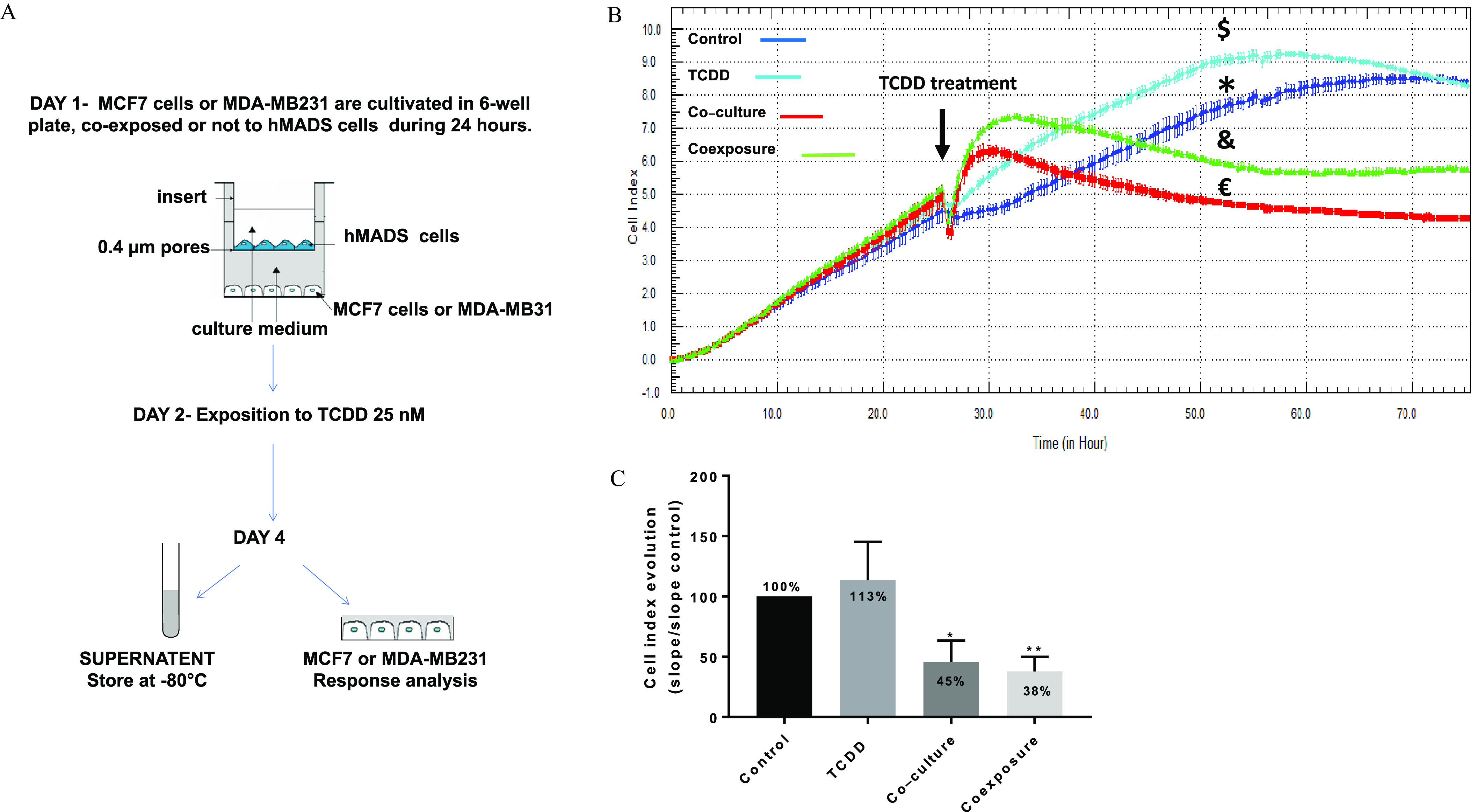
Co-culture model and real time MCF7 cells analysis. (A) Presentation of the 2D co-culture system and the protocol. (B) xCELLigence dynamic monitoring of MCF-7 cells, A representative graph from xCELLigence system: cell index (CI) profiles are the (duplicate) of each condition: Control (*, vehicle MCF-7 cells, alone), TCDD (, MCF-7 cells treated with TCDD), co-culture (, MCF-7 co-cultured with hMADS) and coexposure (&, co-culture with TCDD). (C) The evolution of the CI for each condition was determined by analyzing the slope of the line in the interval (26–48 h). Each graph represents the mean slope (in bold) compared with the for six measurements. The numerical information and -values are provided in Table S1. (Kruskal–Wallis’s H test (nonparametric comparison of independent series) followed by a 1-factor ANOVA test (parametric comparison of k independent series), **; *). Note: ANOVA, analysis of variance; SEM, standard error of the mean; TCDD, 2,3,7,8-tetrachlorodibenzo-p-dioxin.
Chemicals—Antibodies
TCDD (#ED-901, CAS: 1746-01-6) was purchased from LGC Standards, and nonane was purchased from Sigma-Aldrich.
Rabbit polyclonal anti-beta Catenin (dilution 1/1,000) (Ab16051) and rabbit monoclonal anti-Paxillin (dilution 1/200) (Ab32084) were from Abcam. AlexaFluor®488 and AlexaFluor®568 Goat anti-rabbit secondary dilution (1/200) were from Invitrogen.
xCELLigence® assay/monitoring of cell growth using real-time cell analysis (RTCA).
ACEA’s xCELLigence® Real-Time Cell Analysis (RTCA) instruments use specific 16-well plates (E-plates) with gold microelectrodes embedded in the bottom of wells. The impedance value of each well was automatically monitored by the xCELLigence® system and expressed as a cell index (CI) value. The magnitude of this impedance is dependent on the number of cells, the size of the cells, and the cell–substrate attachment quality, initial attachment, and spreading.
MCF-7 cells were seeded in duplicate for each condition into the E-plates at 40,000 cells/well in a final volume of , and immediately an E-plate insert containing hMADS cells (40 000 cells/well) was added. One day after seeding, the medium was removed, and the cells were treated with TCDD or vehicle. The CI value of each well was automatically monitored by the xCELLigence® system (ACEA Biosciences, Inc). The CI evolution for the different conditions was determined by analyzing the slope of the line between 26 and 48 h. Six replicates were performed.
Sample preparation for mass spectrometry analysis.
After 48 h of treatment with TCDD, MCF7 cells were lysed in RIPA Buffer. The FASP (filter-aided sample preparation) procedure for protein digestion was applied using of each lysate as described previously (Lipecka et al. 2016) using 30 kDa MWCO centrifugal filter units (Microcon, Millipore, Cat. No. MRCF0R030). Proteins were reduced with dithiothreitol (DTT) for 30 min at 60°C, then applied to the filters, mixed with of urea, Tris-HCl pH 8.8 (UA buffer), and centrifuged for 15 min at . The filters were washed two times with of UA buffer to remove dithiothreitol (DTT) and detergents. Alkylation was performed by incubation for 20 min in the dark with iodoacetamide. Filters were then washed two times with of UA buffer ( for 15 min), followed by two washes with of ABC buffer ( for 10 min) to remove urea. All centrifugation steps were performed at 25°C. For the final step, digestion with trypsin (in a 1:30 ratio) was used (incubation at 37°C overnight).
nanoLC-MS/MS protein identification and quantification.
Each sample was dried with a vacuum and resuspended in of 0.1% trifluoroacetic acid, 10% acetonitrile for liquid chromatography–tandem mass spectrometry (LC-MS/MS). For each run, was injected into a nanoRSLC-Q Exactive PLUS (RSLC Ultimate 3000; Thermo Scientific). Separation of the peptides were obtained with a reversed-phase LC column (Pepmap C18, Thermo Scientific). The solvents were a) 0.1% formic acid in water, and b) 0.08% formic acid, 80% acetonitrile. Elution of the peptides was performed with the following gradient: 5% to 40% B (120 min), 40% to 80% B (10 min). At 131 min, the gradient was returned to 5% B to re-equilibrate the column for 30 min (before performing the next injection). Two blank conditions were run between each triplicate to prevent sample carryover. The analysis of the eluted peptides was performed by data-dependent MS/MS (the top-10 acquisition method was applied). The resolution was set to 70,000 [mass spectrometric (MS) scans] and 17,500 (data-dependent MS/MS scans). The MS AGC target was set to 3.106 counts (whereas MS/MS AGC target was set to 1.105 counts). We used an MS scan range from 400 to . Records of both MS and MS/MS scans were performed in the profile mode. Dynamic exclusion was set to 30 s. duration. For each sample, three replicates were analyzed by the nanoLC-MS/MS.
Data processing following nanoLC-MS/MS acquisition.
The MaxQuant software (version 1.5.8.30; Max-Planck-Institute of Biochemistry) was used to process the MS files; the files were searched with an Andromeda search engine against the Uniprot Human database. To search fragment ions and parent mass, we set a mass deviation of 20 and , respectively. The minimum length of a peptide was set to seven residues and a strict specificity related to the trypsin cleavage was imposed, allowing up to two missed sites of cleavage. Carbamidomethylation (Cysteine) was set as a fixed modification; on the contrary, oxidation (Methionine) and acetylation of the N-terminal extremity, were set as variable modifications. The false discovery rates (FDRs) (at the protein and peptide level) were set to 1%. Scores were calculated in MaxQuant (Cox and Mann 2008). The reverse and common contaminants hits were removed from MaxQuant output. The quantification of the proteins was performed according to the MaxQuant label-free algorithm [we used label-free quantification (LFQ) intensities] (Cox et al. 2014). Bioinformatic and statistical analyses, including profile plots, heat maps, and clustering, were performed with Perseus software (freely available version 1.5.5.3; http://www.perseus-framework.org) (Tyanova et al. 2016). For statistical comparisons, we defined four groups, each containing biological quadruplicates. Each sample was run as well in technical triplicates. The data were a) filtered to keep only the proteins with at least four valid values out of four in at least one group, b) imputed to fill the missing data points (a Gaussian distribution of random numbers was created with a standard deviation (SD) of 33% relative to the SD of the measured values and 2.5 SD downshift of the mean to simulate the distribution of low signal values). We performed t-tests using , , for all pertinent pairs and represented the results with volcano plots.
The data have been deposited in the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD014328 (Perez-Riverol et al. 2019).
ALDEFLUOR assay.
We used the ALDEFLUOR kit (Stem Cell Technologies) to isolate the cells with low and high ALDH activities (Pearce et al. 2005). After 48h of treatment, MCF-7 cells were harvested, washed, and incubated in the ALDEFLUOR assay buffer containing BODIPY-aminoacetaldehyde (BAAA), the substrate of the ALDH, at 37°C for 30 min. cells are the ones able to catalyze BAAA to BODIPY-aminoacetate (BAA), its fluorescent product. A specific inhibitor of ALDH, diethylaminobenzaldehyde (DEAB), was also used, providing a negative control. After incubation, the cells were resuspended in fresh assay buffer. and cells were sorted by a BD Canto II flow cytometer. Percentage of ALDH positive cell was set according to the gate of DEAB control cells.
Spheroid formation.
MCF-7 cells (input of 3,000 cells) were transferred to conditioned medium. Cell suspensions () were transferred to each well of a 6-well plate coated with 1% agarose to allow spontaneous spheroid formation within 7 d. After 7 d, the number of tumorspheres per microscopic field for five fields was determined using Nikon TMS-F microscope with camera DS-Fi2, [10 times] magnification. The area of the tumorspheres were measured with Image J software (freeware, NIH Image, http://rsb.info.nih.gov/ij/). Technical triplicates were used for statistical analyses (, t-test). After 7 d, cells were trypsinized and replated in growing medium (second passage).
Zebrafish larva metastasis assays.
The experiments were performed (according to the IACUC-approved protocols) at the zebrafish facility at the Boston University School of Medicine. fish (Danio rerio strain) breeders were crossed, and overexpressing-GFP embryos (in the vasculature) were obtained. RFP-labeled MDA-MB-231 cells (ATCC® HTB-26MET) and MCF-7 cells were preseeded at 400,000 cells into a 6-well plate and treated with the different conditioned medium for 48 h. The four conditioned media used were the medium derived from cultures of MCF-7 cells alone (control), MCF-7 cells exposed to TCDD (TCDD), MCF-7 cells co-cultured with hMADS cells (co-culture), and co-cultured MCF-7 cells exposed to TCDD (coexposed). Each experiment was repeated in triplicate (MCF-7) or quadruplicate (MDA-MB-231).
For CM-Dil labeling, MCF-7 cells were trypsinized and centrifuged to obtain a pellet and resuspended in CM-DIL dilution (Invitrogen), incubated in the dye at 37° at 5% for 10 min and then rinsed once with PBS. For micro-injections and transplantation, the RFP-labeled MDA-MB-231 cells and CM-Dil-labeled MCF-7 were resuspended at in DMEM with 10% FBS. The 1-d postfertilization (1 dpf) larvae were dechorionated using the enzyme Pronase (Roche Diagnostics); they were allowed to recover in the dark, overnight (temperature: 28°C), in sterilized egg water. The 2-dpf zebrafish larvae were subsequently anesthetized with tricaine 5%; just before the injections, they were individually mounted in a low-melting 1.5% agarose gel (Fisher) for side view immobilization using a thin eyelash. Borosilicate glass capillaries ( outside ) (World Precision Instruments) were pulled using 500 V (, ) in a capillary machine (Sutter Instrument). The volume of microinjections of MCF-7 or MDA-MB-231 cells was ( or ). The transplantation was made directly into the perivitelline space of the larvae using a microinjection station (World Precision Instruments) and a needle holder (Nicoli and Presta 2007). The xenografted larvae were rescued in sterile egg water and subsequently transferred to a standard 6-well plate (incubation in the dark at 33°C for 24 h). Forty-eight hours after injection, the larvae were anesthetized a second time with tricaine 5% and individually placed on a microscope slide for imaging. Quantification of migrating cells was manually performed using an inverted fluorescence microscope (Leica DMIL LED). The total number of fish injected with CM-Dil-labeled MCF-7 was control (), TCDD (), co-culture (), and coexposure (). The number of fish injected with RFP-labeled MDA-MB-231 cells was control (), TCDD (), co-culture (), and coexposure ().
Immunocytochemistry and confocal microscopy.
MCF-7 cells were seeded onto coverslips, cultured with or without preadipocyte hMADS cells and treated with TCDD or vehicle. After 2 d or 7 d of co-culture, the cells were fixed (20 min, 4% paraformaldehyde) and subsequently permeabilized with 0.2% Triton X-100 (3 min, room temperature). The cells were then washed with PBS and incubated in a “blocking” solution ( PBS-glycine bovine serum albumin) for 1 h. The incubation with the primary antibody (PBS-1% bovine serum albumin) was performed for 1 h at room temperature. The cells were washed with PBS containing 0.1% Tween-20 (PBS-T) and then incubated for 1 h at room temperature with a secondary antibody (conjugated with a fluorescent dye). For nuclei and actin staining, TO-PRO-3 (Invitrogen) and FITC-conjugated phalloidin were included during the incubation with the secondary antibody. Sealing of the coverslips was performed with Dako Faramount Aqueous Mounting Medium Ready-to-Use (Invitrogen), and images were recorded using a confocal microscope (Zeiss LSM 510, Carl Zeiss Meditec France SAS) using a 40× Plan-Neofluar 1.3 NA oil objective or 20× Plan-Neofluar (for the nuclei and cell measurement) and LSM Image Browser (Zeiss). Quantitation of MCF7 cell number and giant cells (very large cells with multiple nuclei) per field were done on three fields per conditions. Graphs represent of three experiments.
Apoptosis assay—Annexin V-FITC/propidium iodide (PI) dual staining assay.
The Annexin V-FITC/PI dual staining assay was carried out using FITC Annexin V Apoptosis Detection Kit (Biolegend). After 48 h of co-culture and treatment, MCF-7 cells were collected by trypsinization. The collected cells were washed twice with ice-cold PBS and then incubated for 5 min at room temperature in the dark in binding buffer containing Annexin V-FITC and PI. Cells were analyzed for apoptosis after 15 min of incubation with a BD Canto II flow cytometer. The data derive from at least five different experiments. Values are expressed as ; * and ** .
AlamarBlue cell viability assay.
Cells were plated at cells per well in a 96-well plate, grown for 24 h and treated with the different conditioned media. Plates were incubated at 37°C for 48 h. According to the manufacturer’s protocol, alamarBlue was added to each well. Cells were incubated for 2 h at 37°C, and the absorbance at was recorded using as a reference wavelength on a PowerWaveX spectrophotometer using KC4 software (BioTek). Data were expressed as the (SEM). Data are representative of at least 3 different experiments and are expressed as the .
activity.
A staining kit was used to assess enzymatic activity (Senescence Cell Staining Kit; Sigma-Aldrich). Briefly, 10,000 MCF7 cells were plated in 12-well plates, subsequently treated with the conditioned mediums, incubated overnight at 37°C, washed with PBS (pH 6.0), fixed, and stained overnight at 37°C with 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside (X-Gal). Only senescent cells stain at pH 6.0. The blue-stained cells and the total number of cells were observed under a Nikon TMS-F microscope with camera DS-Fi2, 10× magnification (1 field per well). The pictures are representative of at least three different experiments.
Preparation of RNA and reverse transcription quantitative PCR (qRT-PCR).
RNA was prepared using the RNeasy mini kit from Qiagen. Reverse transcription was performed using the high-capacity cDNA reverse transcription Kit (Applied Biosystems) as described. Then quantitative PCR was performed using 20 ng of cDNA per reaction on CFX 384 thermocycler (Bio-Rad). Duplicated reactions of each sample were performed using Takyon SYBR® 2X qPCR Mastermix Blue (Eurogentec). The qRT-PCR program was: a) polymerase activation (3 min at 95°C), b) amplification (40 cycles: 10 s at 95°C followed by 60 s at 60°C), and c) dissociation curve (from 60°C to 95°C).
The human primers were: RPL13A forward AAGGTCGTGCGTCTGAAG and reverse GAGTCCGTGGGTCTTGAG, ATG5 forward GTTGCCGGTTGTATTCGCTG and reverse ACCACACATCTCGAAGCACA, ATG7 forward CACCAGATCCGGGGATTTCT and reverse TGCAGCAATGTAAGACCAGTC, Bax forward CCGAGAGGTCTTTTTCCG and reverse GCCTTGAGCACCAGTTTG, and E-cadherin forward GGACAGGGAGGATTTTGGC and reverse GTGAAGGGAGATGTATTGGG. The relative amounts of mRNA were estimated compared with the control condition using the delta-delta Ct method with RPL13A as the reference. Data are representative of at least three different experiments and are expressed as the .
Statistics.
Each experiment was performed at least in triplicate. The results of three or more independent experiments are expressed as the . Statistical analysis was performed with GraphPad Prism software using Kruskal–Wallis’s H test (nonparametric comparison of k independent series) followed by a 1-factor analysis of variance (parametric comparison of k independent series). A value of was considered statistically significant; *, **, and ***.
Results
MCF-7 Adhesion Properties under Control, TCDD, Co-Culture, or Coexposure Conditions Using the xCELLigence® System
We designed an easy-to-implement co-culture model (adipose cells and human tumor cells). It was used to produce conditioned media and to study the communication between both types of cells. Figure 1A describes, schematically, this model, which was used to investigate the influence of TCDD on the phenotype of MCF-7 tumor cells. Briefly, MCF-7 or MDA-MB 231 cells were seeded onto the bottom of 6-well culture dishes and hMADS preadipocyte cells were seeded onto polyester membrane inserts. Both cell types were cultured in the same medium, which can diffuse through the inserts. Twenty-four hours later, the cells were treated with TCDD or vehicle for 48 h. The tumor cells grown in the presence of the hMADS cells was called “co-culture” (no pollutant); the MCF-7 or MDA-MB-231 grown in the presence of the hMADS cells treated with TCDD was called “coexposure” (therefore, ). Moreover, conditioned media derived from cultures of MCF-7 cells alone (control), MCF-7 cells exposed to TCDD (TCDD), MCF-7 cells co-cultured with hMADS cells (co-culture), and co-cultured MCF-7 cells exposed to TCDD (coexposed) were collected (and used for subsequent spheroid formation and zebrafish larva metastasis assays; see below).
We first monitored MCF-7 adhesion properties under control, TCDD, co-culture, or coexposure conditions using the xCELLigence® system, which measures impedance values and converts into a CI (Figures 1B and 1C). We found that cells treated with TCDD alone did not have a significantly higher CI than those in the control condition. MCF-7 cells co-cultured with hMADS cells (co-culture) exhibited a lower CI value. Cells in the coexposure condition also exhibited a lower CI, similar to the co-culture condition.
To identify the most relevant biomarkers for each culture condition, we then used an untargeted approach (proteomics as proteins represent the active functional entities of the cells).
Proteomic Analysis, ALDH Activity, and Tumorsphere Formation of MCF-7 Cells under Control, TCDD, Co-Culture, or Coexposure Conditions
Considering the preceding results, we performed a high-throughput proteomic analysis of the extracts of cells grown under the different conditions. We quantified 2,238 proteins (Excel Table S1). The differences among the different conditions in comparison with the control condition were very low except for ALDH1A3 [aldehyde dehydrogenase 1A3, a cancer stem cell (CSC) marker], CYP1A1 and CYP1B1; as expected, CYP1A1 and CYP1B1 were induced in the presence of TCDD. The co-culture condition did not have a significant impact. However, high levels of ALDH1A3 protein were found only in the coexposure condition (Figure 2A, third panel). We then measured ALDH activity (which is not only representative of ALDH1A3) in MCF-7 cells by FACS analysis, which was found to be significantly higher in the presence of TCDD and in the coexposure condition in comparison with the control (Figure 2B). Overall, these results demonstrated that the protein levels of ALDH1A3 were higher specifically in the coexposure condition, suggesting a potential impact of both factors (TCDD, hMADS) on the acquisition of CSC properties by MCF-7.
Figure 2.
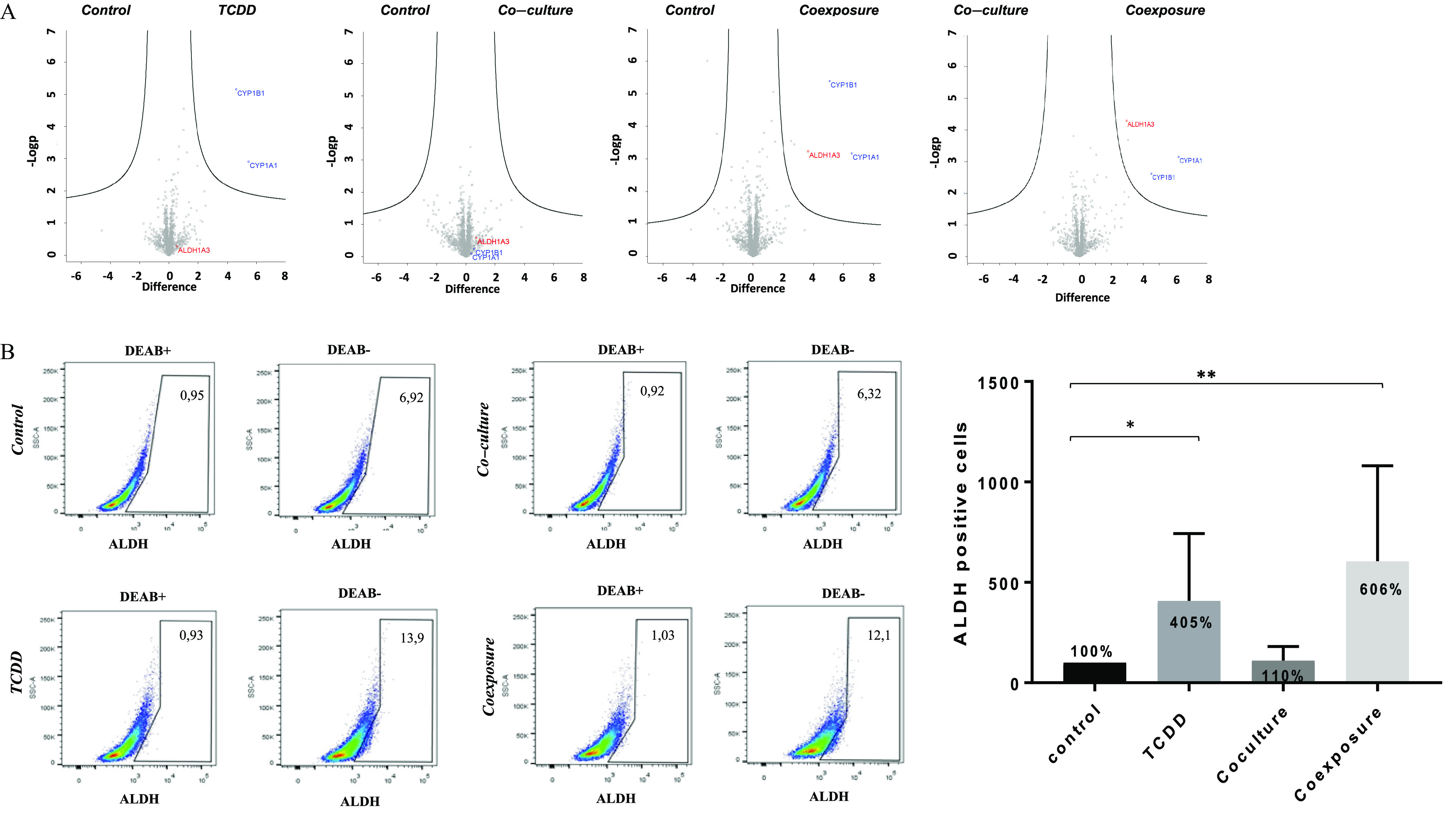
Proteomics analysis and ALDH enzymatic analysis of MCF7 cells growth in co-culture with hMADS and exposed to TCDD for 48 h. (A) High-throughput proteomic analysis of MCF7 cells [Control (vehicle MCF-7 cells, alone), TCDD (MCF-7 cells treated with TCDD), co-culture (MCF-7 co-cultured with hMADS), and coexposure (co-culture with TCDD)]. The plots show the mean of biological quadruplicates and technical triplicates for each sample. CYP1A1, CYP1B1, and ALDH1A3 were induced when they appeared in the upper right part of the representation. (B) ALDH (aldehyde dehydrogenase) enzymatic activity was detected in MCF7 cells using the ALDEFLUOR assay (FACS analysis). DEAB was used to inhibit the reaction of ALDH with the ALDEFLUOR reagent, providing a negative control. Percentage of ALDH positive cell was set according to the gate of DEAB control cells. Graph represents means of the percentage of ALDH positive cells (in bold) compared with the of six measurements. The numerical information and p-values are provided in Table S2. (Kruskal–Wallis’s H test (nonparametric comparison of k independent series) followed by a 1-factor ANOVA test (parametric comparison of k independent series, **; *). Note: ANOVA, analysis of variance; SEM, standard error of the mean; TCDD, 2,3,7,8-tetrachlorodibenzo-p-dioxin.
We then explored the ability of MCF-7 cells to form tumorspheres. We showed that cells exposed to media from both co-culture and coexposure conditions formed spheres with larger areas (Figures 3A and B).
Figure 3.
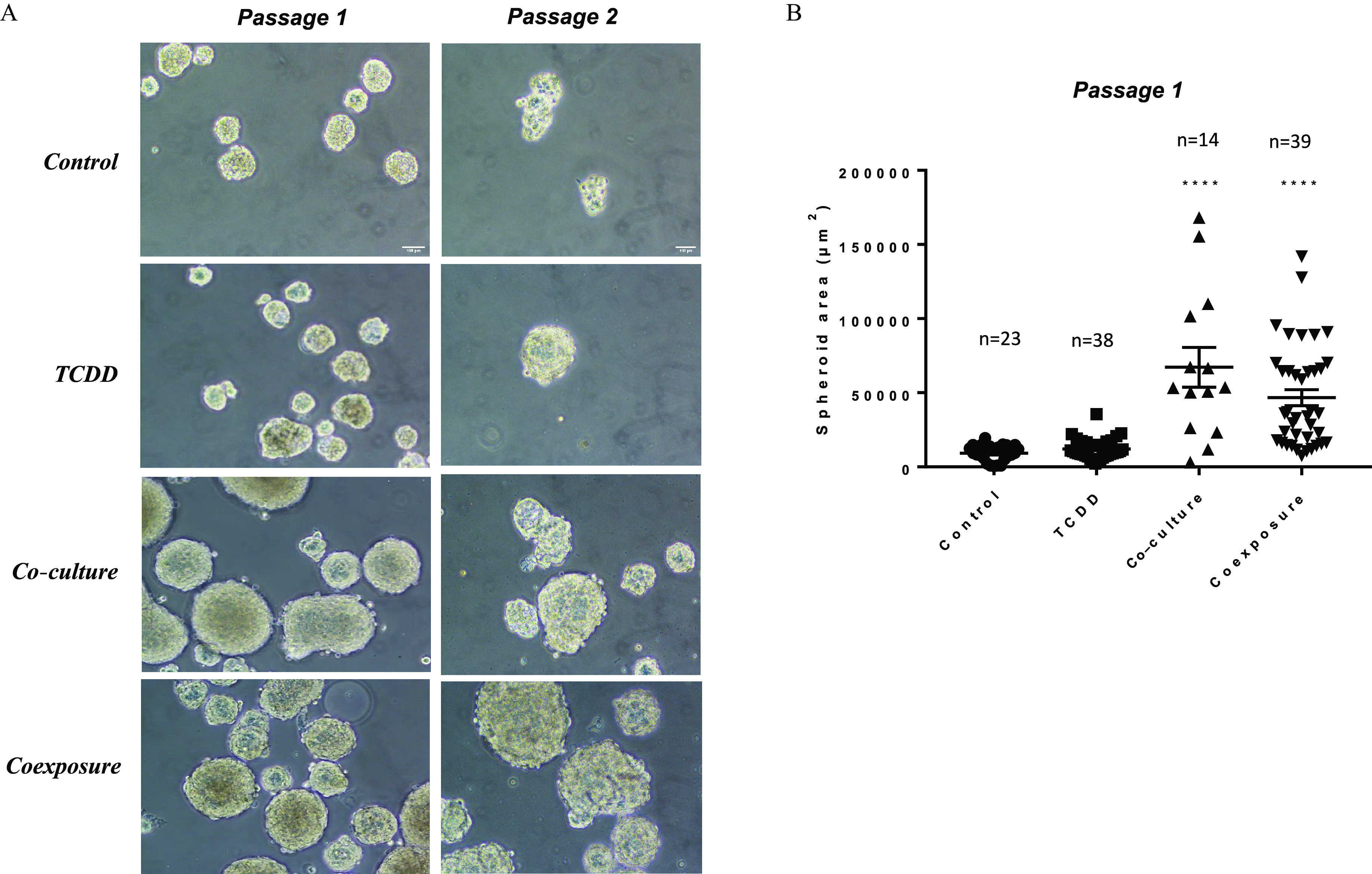
Sphere formation assay. (A) Representative images of sphere formation taken on day 7 on two consecutive generations. Only breast stem/progenitor cells can self-renew and grow into a spheroid structure. Control (vehicle MCF-7 cells, alone), TCDD (MCF-7 cells treated with TCDD), co-culture (MCF-7 co-cultured with hMADS) and coexposure (co-culture with TCDD). Scale bar . (B) Spheroid area (in ) in passage 1 using media from the various conditions. Graph represents of n (see figure) measurements. The numerical information and p-values are provided in Table S3. Untreated MCF-7 cells alone represent the control condition (Kruskal–Wallis’s H test (nonparametric comparison of k independent series) followed by a 1-factor ANOVA test (parametric comparison of k independent series, ****). Note: ANOVA, analysis of variance; SEM, standard error of the mean; TCDD, 2,3,7,8-tetrachlorodibenzo-p-dioxin.
It was therefore necessary to perform a more extensive analysis of the morphological differences that occur in each condition by focusing on the cytoskeleton and the focal adhesion sites, which are indicative of aggressive properties.
Evaluation of Properties Associated with Cell-in-Cell Structures in MCF-7 and MDA-MB-231 Cells under Control, TCDD, Co-Culture, or Coexposure Conditions
We next performed immunofluorescence staining on both MCF-7 and MDA-MB-231 cells in our cell co-culture model. Antibodies to paxillin and actin were used to examine the focal adhesion sites and cytoskeleton, respectively. In the presence of TCDD, the cells dissociated. The localization of paxillin became punctuated with the formation of focal adhesions and stress fibers in comparison with the control condition (Figure 4A). A lower mRNA expression of E-cadherin (Figure S1) was also observed. Markedly altered cellular morphologies were also observed under both co-culture and coexposure conditions. When co-cultured with hMADS, MCF-7 cells were spread out and had extended lamellipods (Figure 4A). Moreover, the coexposure condition () induced the formation of giant cells characterized by a polynucleation (very large cells with multiple nuclei) (Figure 4B). The polynuclear giant cells were defined by several irregular nuclei with a cell size at least two to five times larger than regular diploid cancer cells (Zhang et al. 2014). All these features were observed with MCF-7 cells (Figure 4A) but also with another type of human BC cell line, MDA-MB-231, cultivated under the same conditions (Figure 4C).
Figure 4.
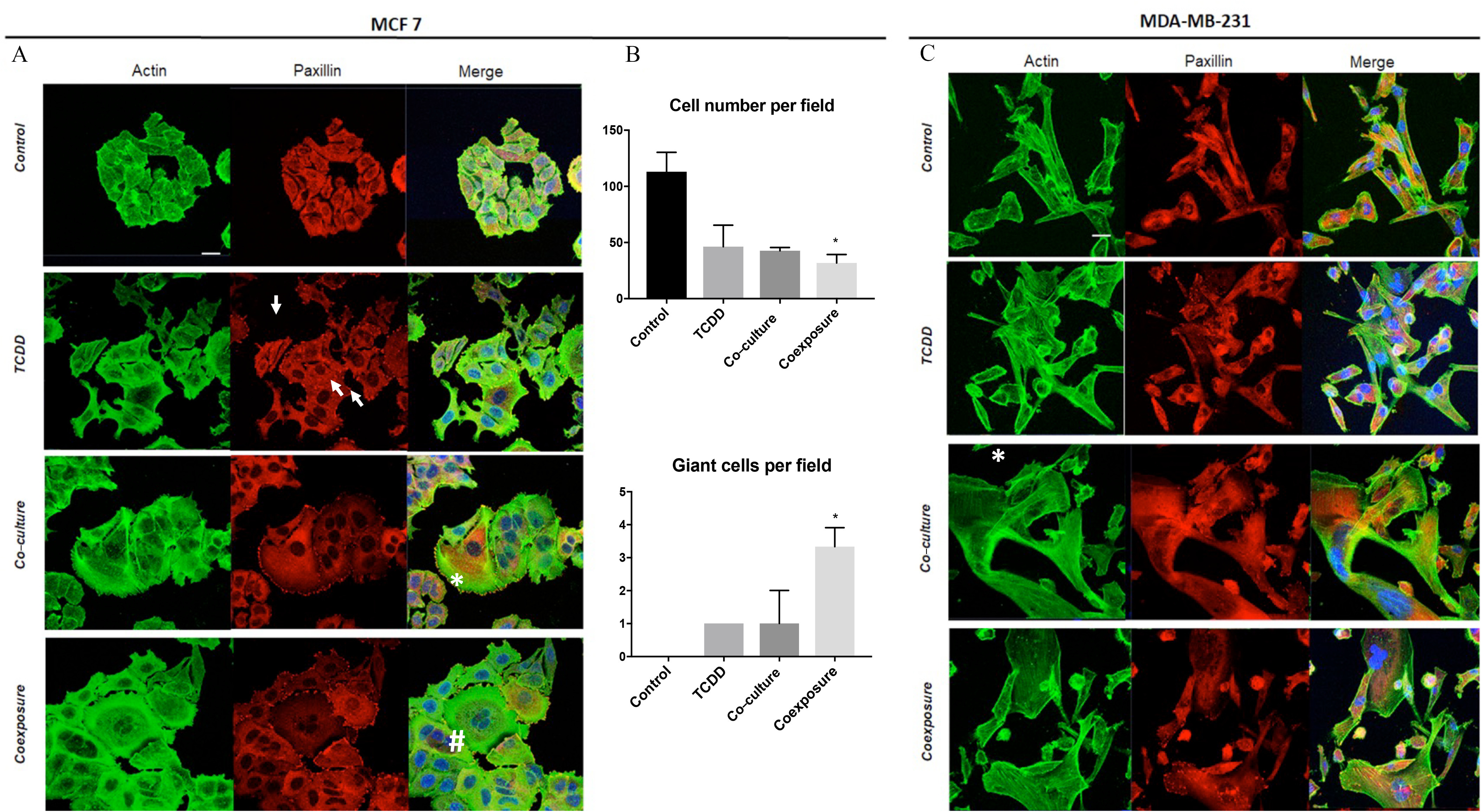
Morphological differences of MCF7 cells and MDA-MB-231 cells following exposure to TCDD, co-culture, or coexposure. Cells were grown with hMADS and/or treated with TCDD [Control (vehicle MCF-7 cells, alone), TCDD (MCF-7 cells treated with TCDD), co-culture (MCF-7 co-cultured with hMADS), and coexposure (co-culture with TCDD)]. After 48 h treatment, cells were fixed and stained for paxillin, actin, and nucleus (blue). (A) Staining of MCF7 cells. Scale bar . (B) Quantitation of MCF7 cell number and giant cells (very large cells with multiple nuclei) per field () field per conditions. Graph represents of three experiments. The numerical information and p-values are provided in Table S4. Kruskal–Wallis’s H test (nonparametric comparison of k independent series) followed by a 1-factor ANOVA test (parametric comparison of k independent series, *). (C) Staining of MDA-MB-231. Scale bar . Symbols were used to point out the focal adhesions (arrow), lamellipods (*) and giant cells (#). Note: ANOVA, analysis of variance; SEM, standard error of the mean; TCDD, 2,3,7,8-tetrachlorodibenzo-p-dioxin.
The formation of these cells led us to investigate cellular processes such as cell death, senescence, proliferation, or autophagy and we did not observe difference among the 4 different conditions (Figure S2). Thus, we explored the detailed morphology of these polynucleated cells co-exposed for 48h to the hMADS cells and to TCDD (coexposure condition) assessing, for example, the localization of beta-catenin (Figure 5A, fourth panel). In the coexposure condition, beta-catenin was found to surround cells inside other cells, which were sometimes polyploid (Figure 5B). In comparison, this was not observed in the TCDD condition or in the co-culture condition; these features are characteristic of ‘Cell-In-Cell’ structures (CICs, see discussion) which were generally associated with malignant metastatic progression. We then investigated the metastatic potential of our cells using a validated, in vivo tool, the zebrafish xenograft model.
Figure 5.
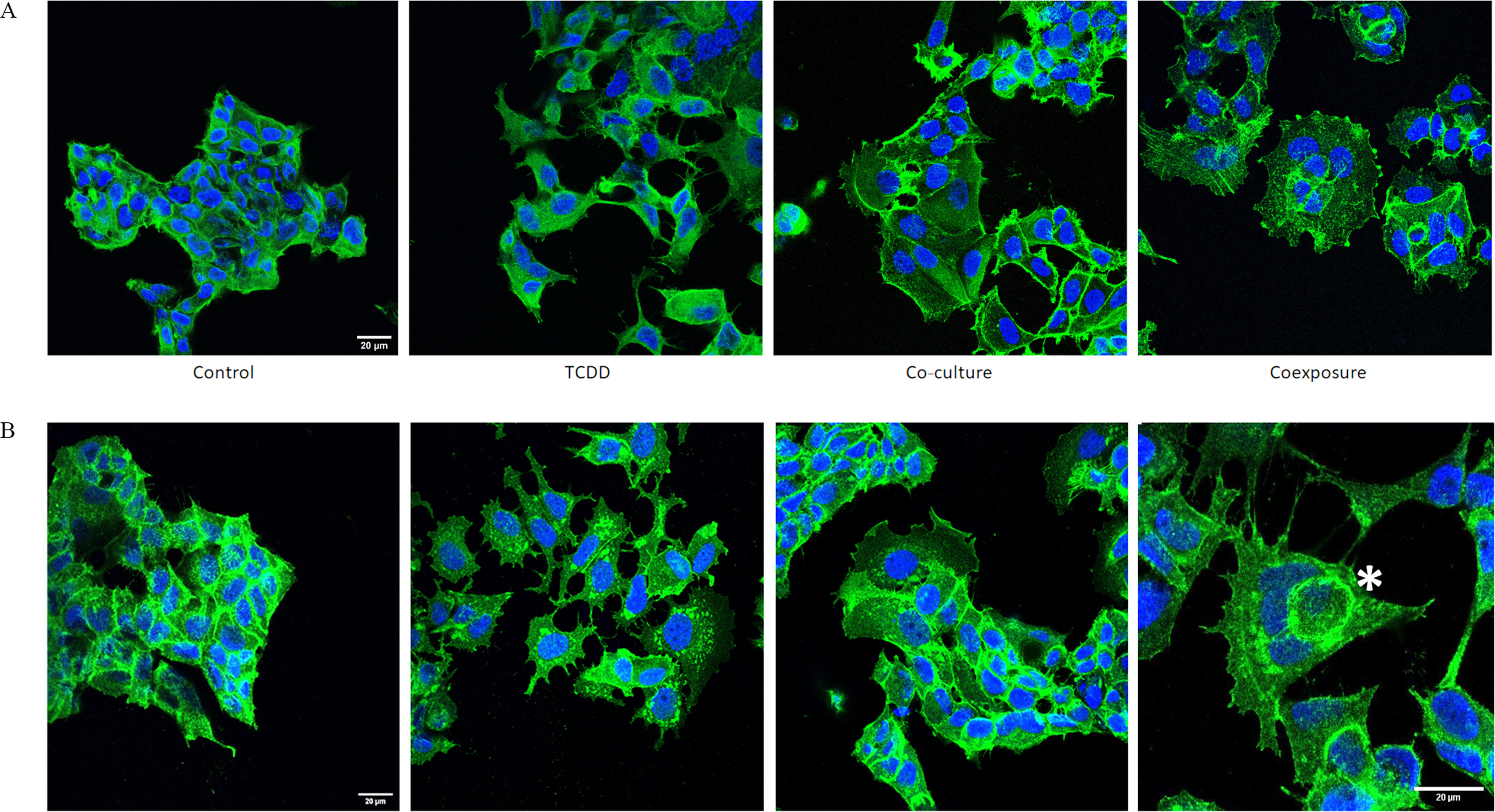
Protein (A-actin, B-beta-catenin) localization & nucleus staining in MCF7 cells and MDA-MB-231 cells. MCF7 cells were grown with hMADS and/or treated with TCDD for 48h [Control (vehicle MCF-7 cells, alone), TCDD (MCF-7 cells treated with TCDD), co-culture (MCF-7 co-cultured with hMADS), and coexposure (co-culture with TCDD)]. (A) MCF7 cells were stained for actin (green) and nucleus (blue). Scale bar . (B) MCF7 cells were stained for beta-catenin (green) and nucleus (blue). One representative cell with cell-in-cell structures was marked with an asterisk (*). Scale bar . Note: TCDD, 2,3,7,8-tetrachlorodibenzo-p-dioxin.
Evaluation of Metastatic Spreading in an in Vivo Zebrafish Model of MCF-7 Cells under Control, TCDD, Co-Culture, or Coexposure Conditions
We next explored the ability of both MCF-7 and MDA-MB-231 cells to metastasize in vivo. We used a well-characterized zebrafish xenograft model of human cancer cell metastasis. Since the larvae are optically transparent, the implementation of fluorescent cancer cells (which are xenografts) and the follow-up of their growth or intra-/extravasation (early metastatic events) is facilitated. This model now is considered as a powerful tool for the study such events (Cornet et al. 2019; Cabezas-Sáinz et al. 2020). The different conditioned media were used, individually, to treat MCF-7 cells labeled with CM-Dil and RFP-MDA-MB-231 cells for 48 h and then injected into the perivitelline space of two-day-old zebrafish larvae (Figure 6). Fish were imaged 24h later. The number of fish with metastases in the tail (vascular plexus) or in the cephalic pole (“head”) and the number of metastases per fish were counted for each type of medium used. Formation of metastases using MCF-7 cells was observed in 53% of fish using media from the control condition (), 43% from the TCDD condition (), 66% from the co-culture condition (), and 76% from the coexposure condition (). The number of metastases per fish was significantly higher in the coexposure condition as compared to the control (). Metastases in the cephalic pole were observed only from the co-culture and coexposure conditions with, respectively, 8% and 21% of fish affected. Similar results were found with MDA-MB-231 cells. Metastasis formation was observed in 50% of fish using media from the control condition (), 51% from the TCDD condition (), 58% from the co-culture condition (), and 79% from the coexposure condition (). The number of metastases per fish was significantly higher from the coexposure condition (), and metastases in the cephalic pole were observed only in the coexposure condition (40% of fish with “head” metastases).
Figure 6.
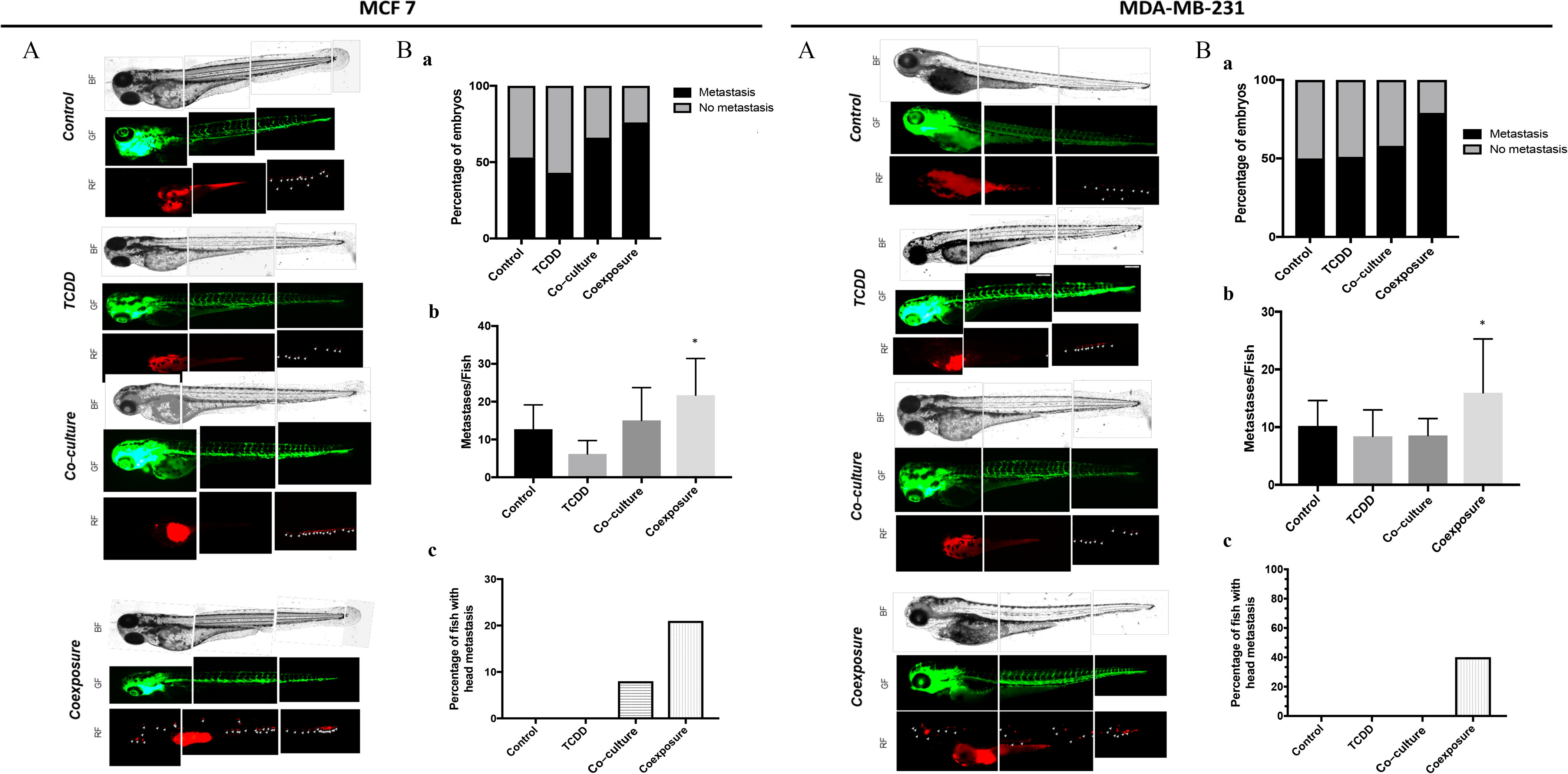
Measurement of metastatic spread of MCF7 and MDA-MB231 cells in zebrafish larvae in vivo models Human RFP-MCF7 (left) or RFP-MDA-MB-231 (right) cells cultured with conditioned media from different conditions, were injected into the perivitelline space of 2-day-old zebrafish larvae [Control (vehicle MCF-7 cells, alone), TCDD (MCF-7 cells treated with TCDD), co-culture (MCF-7 co-cultured with hMADS), and coexposure (co-culture with TCDD)]. Fish were imaged 24 h later at magnification by fluorescence microscopy. (A) Representative images from 21–46 fish/group. (B) Quantitation of (a) the number of fish with one or more metastases and (b) the average number of . Graph represents of three measurements. The numerical information and p-values are provided in Table S8. (Kruskal–Wallis’s H test (nonparametric comparison of k independent series) followed by a 1-factor ANOVA test (parametric comparison of k independent series, * ). (C) The percentage of fish with metastasis in the head region. Each experiment was repeated in triplicate (MCF-7) or quadruplicate (MDA-MB-231) with 7–12 fish per condition in each experiment. Note: ANOVA, analysis of variance; SEM, standard error of the mean; TCDD, 2,3,7,8-tetrachlorodibenzo-p-dioxin.
Discussion
In this study, we used a co-culture model using MCF-7 cells or MDA-MB-231 cells and hMADS preadipocytes (to investigate the contribution of the microenvironment) and 2,3,7,8-tétrachlorodibenzo-p-dioxin (TCDD, to investigate the role of a POP) on BC cells. We showed that each condition impacted differentially the cellular properties of MCF-7 (adhesion, migration, or proliferation). We also showed that human BC cells acquired pro-metastatic features, in vitro and in vivo, when cultured with both preadipocytes and exposed to TCDD (a persistent organic pollutant), what we named the ‘co-exposure’ condition. Using zebrafish larvae, we demonstrated a significantly higher number of brain metastases with both types of human breast cell lines (MDA-MB-231 and MCF-7 cell lines) using media specifically produced under the coexposure condition. We showed that this coexposure lead, in vitro, to the appearance of the giant cells and the acquisition of cellular features such as polyploidy and ‘cell-in-cell’ structures (CICs). We also observed the generation of irregular nuclei, identified as cells with nuclei two to five times larger than those of regular diploid cancer cells as described in Zhang et al. (2014).
CICs were reported in various cancer types and they mostly were associated with a poor diagnosis for the patients (Ruan et al. 2019). We used beta-catenin staining to characterize the coronation of engulfed cells, a process which was associated with entosis or cannibalism (Durgan and Florey 2018). Entosis was first described in 2007, as a nonapoptotic cell death program characterized by the invasion of one cell into another one. The invasive cell is alive, transiently, before being degraded or released (Overholtzer et al. 2007). Cell cannibalism is, to the contrary, the ability of a cell to engulf another living cell. Cell cannibalism was described as a process enabling aggressive tumor cells to be continuously fed with cellular neighbors (Fais and Overholtzer 2018). CICs were generally associated with malignant metastatic progression and their presence has been demonstrated in vivo in human samples (Fais and Overholtzer 2018; Ruan et al. 2019). Nuclear morphometric parameters can help to identify aggressive properties (Kronqvist et al. 1998) and provide significant prognostic data for survival and risk of tumor progression (Abdalla et al. 2009). The localization of beta-catenin, which is mainly cytoplasmic in these giant cells, was also a useful predictor of metastasis in colorectal (Maruyama et al. 2000) and breast (López-Knowles et al. 2010) cancers. Further investigations will be necessary to characterize the molecular processes which lead to the generation of CICs and to determine what we observed exactly, entosis or cell cannibalism.
The cells also acquired properties reminiscent of cancer stem cells (CSCs). Markers of CSCs include aldehyde dehydrogenase 1A3 (ALDH1A3), CD44 and CD133. Cells with these markers are also more resistant to chemotherapy (Mirzayans et al. 2018). CSCs were identified for the first time in blood mononuclear cells in human acute myeloid leukemia (Bonnet and Dick 1997). They possess characteristics that promote tumor metastasis (Peitzsch et al. 2017; Chang 2016). In the current work, we found that cells in the coexposure condition had higher levels of ALDH1A3 specifically. In BC, the ALDH activity of CSCs has been shown to be due to the isoform ALDH1A3, which is a marker for cells with an increased propensity to metastasize especially in BC (Peitzsch et al. 2017; Marcato et al. 2011; 2015). ALDH1A3 expression can be used as a prognostic factor (Motomura et al. 2020). ALDH1A3 is not only a cancer cell biomarker but also a causal factor in the occurrence of CSCs (Vassalli 2019). Downregulation of ALDH1A3 in breast cancer cell lines did not impact cell proliferation but specifically affected cell migration and the potency of the cells to form metastases (Croker et al. 2017). It was also suspected to impact cell survival (Kashii-Magaribuchi et al. 2016). However, the role, in all these processes, of retinoic acid, an ALDH1A3 enzyme metabolite, is still unknown (Marcato et al. 2015; Ginestier et al. 2009).
Our work supports the hypothesis that the concomitant action of preadipocytes and TCDD leads to the emergence of a new population of tumor cells with properties similar to CSCs. We focused on the contribution of the preadipocyte cells, hMADS: previous work in our laboratory has shown that this cell line was a valuable human model for the evaluation of pollutant toxicity (Kim et al. 2012). Further, it was particularly useful for investigations of the regulation of the inflammatory pathway as exemplified by studies of its response to dioxin (Kim et al. 2012). Very few in vitro studies of the role of toxicology in the tumorigenic process have been performed using co-culture models. These models have the disadvantage of requiring the characterization of one common medium upstream of the functional experiments in order to conserve the properties of each cell type. In our hands, the hMADS cells could not be maintained in the MCF-7 medium. However, we were able to maintain the tumor cells in the preadipocyte medium. The properties of the MCF-7 cells were slightly different but they conserved important features of certain processes of interest. One such feature was the decreased lower of E-cadherin in cells exposed to TCDD (Figure S2), a characteristic of the EMT. Moreover, the relocalization of paxillin, the formation of focal adhesions and stress fibers, observed previously (Bui et al. 2009; Diry et al. 2006) were also typical of the EMT, a phenomenon linked to cancer metastasis (Zhang and Weinberg 2018).
In spite of some disadvantages, we believe that it is important to develop co-culture models because they are easier to generate than organoids, another complementary tool that allows the study of xenobiotic effects (including drugs) through an integrated approach implicating many cell types. Organoids, however, present the disadvantage of not being able to characterize which cell-to-cell communications are the most significant for the explanation of the biological effects of a toxicant. It is important that we used two BC cell lines (MCF-7 and MDA-MB-231 cells) in our co-culture model; MCF-7 cells express estrogen and progesterone receptors, a wild-type p53, whereas MDA-MB-231 cells lack those features. This difference might be important to consider because several articles have reported demonstrations of the anti-estrogenicity of TCDD (induction of cytochromes that transform estrogens into catechols, degradation of estrogen receptors by a ubiquitin complex regulated by the AhR involving CUL4B…) (Ohtake et al. 2007; Coumoul et al. 2001). Therefore, we suspected, initially, that the absence of the estrogen receptors in MDA-MB-231 might have an impact on the effects displayed by TCDD in MCF-7 cells. Despite their differences, several important outcomes were conserved within each model, including the potential to metastasize in vivo (zebrafish experiments). Our models allowed the study of these contributions while focusing on tumor cells and one key mesenchymal cell type in its microenvironment, the preadipocyte. Preadipocytes are the precursors of adipocytes, a major component of the adipose tissue (Kothari et al. 2020). This is the reason for using the hMADS cell line, a preadipocyte line that can be differentiated. We recently showed, in a clinical study, that TCDD was associated with a significant risk for the development of BC metastasis specifically in women who were overweight, which suggests that adipose tissue may exert a significant effect (Koual et al. 2019). This finding reinforces the hypothesis that adipocytes and inflammation may be significant contributors to the development of aggressive tumors.
TCDD, which is lipophilic and resistant to xenobiotic metabolism, as well as other persistent organic pollutants, is stored in adipose tissue. We used TCDD, the most potent dioxin, on C57BL/6J mice to mimic long-term bioaccumulating molecules that also stimulate the AhR signaling pathway (Duval et al. 2017). In a previous study, we showed that TCDD led to an EMT in human hepatocarcinoma cells and to liver fibrosis in an animal model (Pierre et al. 2014). Liver fibrosis is suspected to involve EMT processes, which also are observed during cancer metastasis. However, it is important to note that the role of the AhR in tumor progression remains controversial, possibly due to the plasticity of this receptor, which is able to bind a variety of ligands and, potentially, to act in a ligand-dependent manner (Gouédard et al. 2004; Guyot et al. 2013). Although several studies, including ours, have showed that TCDD favors cancer cell migration, invasion, and metastasis (Bui et al. 2009; Biswas et al. 2008; Seifert et al. 2009), others suggest that TCDD (and also 3,3′-diindolylmethane) could inhibit irregular colony formation in Matrigel and block metastasis in vivo while accelerating cell migration (Narasimhan et al. 2018). In our previous studies, we showed that manipulation of the AhR level, using inhibitors or SiRNA knockdown gene appeared to reduce human cell invasion, migration, and metastasis in several BC cell lines (Bui et al. 2009; Bui et al. 2016). This controversial issue concerning the effects of the receptor might be due to several factors (cell culture condition, tumor microenvironment, different actions of the various AhR ligands); therefore, as the action of the AhR, like many nuclear receptors, is context-dependent (e.g., breast, prostate), further investigation will be needed to decipher the complex role of the AhR in cancer progression and metastasis.
Because daily life involves exposure to multiple POPs, it is important to delineate the effects of mixtures of POPs in our model to determine to what extent they stimulate tumor progression. Also, mixtures of nonpersistent organic pollutants, such as phthalates and bisphenols or heavy metals, should be evaluated, even though these xenobiotics are nonpersistent. Our study provides evidence, to the best of our knowledge, that TCDD can favor the occurrence of metastases of breast cancer cells in zebrafish by in part targeting the tumor microenvironment and by stimulating previously unsuspected molecular pathways. This finding reinforces the idea that more complex models such as co-culture models or organoids should be used to study toxicological mechanisms. They could, potentially, be used as well for chemical assessment of drugs or pollutants. This knowledge should be relevant, generally, in terms of public health for the management of tumors, for which the generation of aggressive forms remains poorly studied and assessed.
Data Availability
The mass spectrometry proteomics data have been deposited in the ProteomeXchange Consortium via the Proteomics Identifications Database (PRIDE) (Perez-Riverol et al., 2019) partner repository with the data set identifier PXD014328.
Supplementary Material
Acknowledgments
This work was supported by the Association pour la recherche sur le cancer (ARC) fondation, Fondation de France/Unicancer, French Program on Endocrine Disruption (PNRPE), the French National Academy of Medicine, and the French National Group of Gynecology and Obstetrics (CNGOF) through doctoral and international mobility grants. The authors thank L. Aggerbeck for his critical reading of our manuscript. The authors acknowledge the cytometry core facility and S. Dupuy of BioMedTech Facilities, Université de Paris, for assistance with the generation of cytometry data. The authors also acknowledge the molecular biology core facility of BioMedTech Facilities, Université de Paris, for assistance with the generation of molecular biology data».
Author contributions: M.K. designed and performed experiments, raised funds, and wrote the article. C.T. designed and performed experiments and wrote parts of the “Materials and Methods” section. I.C.C. performed experiments and wrote parts of the “Materials and Methods” section. D.S. designed experiments. R.B. designed experiments and wrote the article. X.C. designed experiments and wrote the article.
References
- Abbott BD, Perdew GH, Birnbaum LS. 1994. Ah receptor in embryonic mouse palate and effects of TCDD on receptor expression. Toxicol Appl Pharmacol 126(1):16–25, PMID: 8184424, 10.1006/taap.1994.1085. [DOI] [PubMed] [Google Scholar]
- Abdalla F, Jamela B, Rabia M, Hussein H, Abdelbaset B, Yrjö C. 2009. Correlation of nuclear morphometry of breast cancer in histological sections with clinicopathological features and prognosis. Anticancer Res 29(5):1771–1776. PMID: 19443402. [PubMed] [Google Scholar]
- Allen M, Jones JL. 2011. Jekyll and Hyde: the role of the microenvironment on the progression of cancer. J Pathol 223(2):162–176, PMID: 21125673, 10.1002/path.2803. [DOI] [PubMed] [Google Scholar]
- Barouki R, Aggerbeck M, Aggerbeck L, Coumoul X. 2012. The aryl hydrocarbon receptor system. Drug Metabol Drug Interact 27(1):3–8, PMID: 22718620, 10.1515/dmdi-2011-0035. [DOI] [PubMed] [Google Scholar]
- Biswas G, Srinivasan S, Anandatheerthavarada HK, Avadhani NG. 2008. Dioxin-mediated tumor progression through activation of mitochondria-to-nucleus stress signaling. Proc Natl Acad Sci U S A 105(1):186–191, PMID: 18172213, 10.1073/pnas.0706183104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnet D, Dick JE. 1997. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med 3(7):730–737, PMID: 9212098, 10.1038/nm0797-730. [DOI] [PubMed] [Google Scholar]
- Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. 2018. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 68(6):394–424, PMID: 30207593, 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- Bui L-C, Tomkiewicz C, Chevallier A, Pierre S, Bats A-S, Mota S, et al. 2009. Nedd9/Hef1/Cas-L mediates the effects of environmental pollutants on cell migration and plasticity. Oncogene 28(41):3642–3651, PMID: 19648964, 10.1038/onc.2009.224. [DOI] [PubMed] [Google Scholar]
- Bui L-C, Tomkiewicz C, Pierre S, Chevallier A, Barouki R, Coumoul X. 2016. Regulation of aquaporin 3 expression by the AhR pathway is critical to cell migration. Toxicol Sci 149(1):158–166, PMID: 26454884, 10.1093/toxsci/kfv221. [DOI] [PubMed] [Google Scholar]
- Cabezas-Sáinz P, Pensado-López A, Sáinz B, Sánchez L. 2020. Modeling cancer using zebrafish xenografts: drawbacks for mimicking the human microenvironment. Cells 9(9):1978, PMID: 32867288, 10.3390/cells9091978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaffer CL, Weinberg RA. 2011. A perspective on cancer cell metastasis. Science 331(6024):1559–1564, PMID: 21436443, 10.1126/science.1203543. [DOI] [PubMed] [Google Scholar]
- Chang JC. 2016. Cancer stem cells: role in tumor growth, recurrence, metastasis, and treatment resistance. Medicine (Baltimore) 95 (1 suppl 1):S20–S25, PMID: 27611935, 10.1097/MD.0000000000004766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohn BA, Cirillo PM, La Merrill MA. 2019. Correlation of body mass index with serum DDTs predicts lower risk of breast cancer before the age of 50: prospective evidence in the Child Health and Development Studies. J Expo Sci Environ Epidemiol 29(3):302–309, PMID: 30224754, 10.1038/s41370-018-0072-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornet C, Dyballa S, Terriente J, Di Giacomo V. 2019. ZeOncoTest: refining and automating the zebrafish xenograft model for drug discovery in cancer. Pharmaceuticals 13(1):1, PMID: 31878274, 10.3390/ph13010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coumoul X, Diry M, Robillot C, Barouki R. 2001. Differential regulation of cytochrome P450 1A1 and 1B1 by a combination of dioxin and pesticides in the breast tumor cell line MCF-7. Cancer Res 61(10):3942–3948, PMID: 11358810. [PubMed] [Google Scholar]
- Cox J, Hein MY, Luber CA, Paron I, Nagaraj N, Mann M. 2014. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol Cell Proteomics 13(9):2513–2526, PMID: 24942700, 10.1074/mcp.M113.031591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J, Mann M. 2008. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol 26(12):1367–1372, PMID: 19029910, 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- Croker A, Rodriguez-Torres M, Xia Y, Pardhan S, Leong H, Lewis J, et al. 2017. Differential functional roles of ALDH1A1 and ALDH1A3 in mediating metastatic behavior and therapy resistance of human breast cancer cells. Int J Mol Sci 18(10):2039, PMID: 28937653, 10.3390/ijms18102039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronin KA, Lake AJ, Scott S, Sherman RL, Noone A-M, Howlader N, et al. 2018. Annual Report to the Nation on the Status of Cancer, part I: national cancer statistics. Cancer 124(13):2785–2800, PMID: 29786848, 10.1002/cncr.31551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diepenbruck M, Christofori G. 2016. Epithelial-mesenchymal transition (EMT) and metastasis: yes, no, maybe?” Curr Opin Cell Biol 43:7–13, PMID: 27371787, 10.1016/j.ceb.2016.06.002. [DOI] [PubMed] [Google Scholar]
- Diry M, Tomkiewicz C, Koehle C, Coumoul X, Bock KW, Barouki R, et al. 2006. Activation of the dioxin/aryl hydrocarbon receptor (AhR) modulates cell plasticity through a JNK-Dependent mechanism. Oncogene 25(40):5570–5574, PMID: 16619036, 10.1038/sj.onc.1209553. [DOI] [PubMed] [Google Scholar]
- Durgan J, Florey O. 2018. Cancer cell cannibalism: multiple triggers emerge for entosis. Biochim Biophys Acta Mol Cell Res 1865(6):831–841, PMID: 29548938, 10.1016/j.bbamcr.2018.03.004. [DOI] [PubMed] [Google Scholar]
- Duval C, Teixeira-Clerc F, Leblanc AF, Touch S, Emond C, Guerre-Millo M, et al. 2017. Chronic exposure to low doses of dioxin promotes liver fibrosis development in the C57BL/6J Diet-induced obesity mouse model. Environ Health Perspect 125(3):428–436, PMID: 27713108, 10.1289/EHP316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fais S, Overholtzer M. 2018. Cell-in-cell phenomena in cancer. Nat Rev Cancer 18(12):758–766, PMID: 30420767, 10.1038/s41568-018-0073-9. [DOI] [PubMed] [Google Scholar]
- Falkowska L, Reindl AR, Grajewska A, Lewandowska AU. 2016. Organochlorine contaminants in the muscle, liver and brain of seabirds (Larus) from the coastal area of the Southern Baltic. Ecotoxicol Environ Saf 133 (November):63–72, PMID: 27414257, 10.1016/j.ecoenv.2016.06.042. [DOI] [PubMed] [Google Scholar]
- Fernandes AR, Falandysz J. 2021. Polybrominated dibenzo-p-dioxins and furans (PBDD/Fs): contamination in food, humans and dietary exposure. Sci Total Environ 761:143191. PMID: 33160676, 10.1016/j.scitotenv.2020.143191. [DOI] [PubMed] [Google Scholar]
- Ginestier C, Wicinski J, Cervera N, Monville F, Finetti P, Bertucci F, et al. 2009. Retinoid signaling regulates breast cancer stem cell differentiation. Cell Cycle 8(20):3297–3302, PMID: 19806016, 10.4161/cc.8.20.9761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouédard C, Barouki R, Morel Y. 2004. Induction of the paraoxonase-1 gene expression by resveratrol. Arterioscler Thromb Vasc Biol 24(12):2378–2383, PMID: 15458977, 10.1161/01.ATV.0000146530.24736.ce. [DOI] [PubMed] [Google Scholar]
- Guyot E, Chevallier A, Barouki R, Coumoul X. 2013. The AhR twist: ligand-dependent AhR signaling and pharmaco-toxicological implications. Drug Discov Today 18(9–10):479–486, PMID: 23220635, 10.1016/j.drudis.2012.11.014. [DOI] [PubMed] [Google Scholar]
- Ham M, Moon A. 2013. Inflammatory and microenvironmental factors involved in breast cancer progression. Arch Pharm Res 36(12):1419–1431, PMID: 24222504, 10.1007/s12272-013-0271-7. [DOI] [PubMed] [Google Scholar]
- Jackson E, Robin S, Nika L, Lisa C. 2017. Adipose tissue as a site of toxin accumulation. Compr Physiol 7(4):1085–1135, 10.1002/cphy.c160038, PMID: 28915320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joffin N, Noirez P, Antignac J-P, Kim M-J, Marchand P, Falabregue M, et al. 2018. Release and toxicity of adipose tissue-stored TCDD: direct evidence from a xenografted fat model. Environ Int 121(Pt 2):1113–1120, PMID: 30390924, 10.1016/j.envint.2018.10.027. [DOI] [PubMed] [Google Scholar]
- Kashii-Magaribuchi K, Takeuchi R, Haisa Y, Sakamoto A, Itoh A, Izawa Y, et al. 2016. Induced expression of cancer stem cell markers ALDH1A3 and Sox-2 in hierarchical reconstitution of apoptosis-resistant human breast cancer cells. Acta Histochem Cytochem 49(5):149–158, PMID: 27917009, 10.1267/ahc.16031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MJ, Pelloux V, Guyot E, Tordjman J, Bui L-C, Chevallier A, et al. 2012. Inflammatory pathway genes belong to major targets of persistent organic pollutants in adipose cells. Environ Health Perspect 120(4):508–514, PMID: 22262711, 10.1289/ehp.1104282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M-J, Marchand P, Henegar C, Antignac J-P, Alili R, Poitou C, et al. 2011. Fate and complex pathogenic effects of dioxins and polychlorinated biphenyls in obese subjects before and after drastic weight loss. Environ Health Perspect 119(3):377–383, PMID: 21156398, 10.1289/ehp.1002848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kothari C, Diorio C, Durocher F. 2020. The importance of breast adipose tissue in breast cancer. Int J Mol Sci 21(16):5760, PMID: 32796696, 10.3390/ijms21165760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koual M, Cano-Sancho G, Bats A-S, Tomkiewicz C, Kaddouch-Amar Y, Douay-Hauser N, et al. 2019. Associations between persistent organic pollutants and risk of breast cancer metastasis. Environ Int 132:105028, PMID: 31382183, 10.1016/j.envint.2019.105028. [DOI] [PubMed] [Google Scholar]
- Kronqvist P, Kuopio T, Collan Y. 1998. Morphometric grading of invasive ductal breast cancer. I. Thresholds for nuclear grade. Br J Cancer 78(6):800–805, PMID: 9743304, 10.1038/bjc.1998.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Merrill M, Emond C, Kim MJ, Antignac J-P, Le Bizec B, Clément K, et al. 2013. Toxicological function of adipose tissue: focus on persistent organic pollutants. Environ Health Perspect 121(2):162–169, PMID: 23221922, 10.1289/ehp.1205485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Kang Y. 2016. Probing the fifty shades of EMT in metastasis. Trends Cancer 2(2):65–67, PMID: 27042694, 10.1016/j.trecan.2016.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim B, Woodward WA, Wang X, Reuben JM, Ueno NT. 2018. Inflammatory breast cancer biology: the tumour microenvironment is key. Nat Rev Cancer 18(8):485–499, PMID: 29703913, 10.1038/s41568-018-0010-y. [DOI] [PubMed] [Google Scholar]
- Lipecka J, Chhuon C, Bourderioux M, Bessard M-A, van Endert P, Edelman A, et al. 2016. Sensitivity of mass spectrometry analysis depends on the shape of the filtration unit used for filter aided sample preparation (FASP). Proteomics 16(13):1852–1857, PMID: 27219663, 10.1002/pmic.201600103. [DOI] [PubMed] [Google Scholar]
- López-Knowles E, Zardawi SJ, McNeil CM, Millar EK, Crea P, Musgrove EA, Sutherland RL, et al. 2010. Cytoplasmic localization of beta-catenin is a marker of poor outcome in breast cancer patients. SA Cancer Epidemiol Biomarkers Prev. 119(1):301–309, PMID: 20056651, 10.1158/1055-9965.EPI-09-0741. [DOI] [PubMed] [Google Scholar]
- Marcato P, Dean CA, Pan Da, Araslanova R, Gillis M, Joshi M, Helyer L, et al. 2011. Aldehyde dehydrogenase activity of breast cancer stem cells is primarily due to isoform ALDH1A3 and its expression is predictive of metastasis. Stem Cells (Cells 29(1):32–45, PMID: 21280157, 10.1002/stem.563. [DOI] [PubMed] [Google Scholar]
- Marcato P, Dean CA, Liu R-Z, Coyle KM, Bydoun M, Wallace M, et al. 2015. Aldehyde dehydrogenase 1A3 influences breast cancer progression via differential retinoic acid signaling. Mol Oncol 9(1):17–31, PMID: 25106087, 10.1016/j.molonc.2014.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama K, Ochiai A, Akimoto S, Nakamura S, Baba S, Moriya Y, et al. 2000. Cytoplasmic beta-catenin accumulation as a predictor of hematogenous metastasis in human colorectal cancer. Oncology 59(4):302–309, PMID: 11096342, 10.1159/000012187. [DOI] [PubMed] [Google Scholar]
- Mirzayans R, Andreas B, Murray D. 2018. Roles of polyploid/multinucleated giant cancer cells in metastasis and disease relapse following anticancer treatment. Cancers (Basel) 10(4):118, PMID: 29662021, 10.3390/cancers10040118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell MM, Woods R, Chi L-H, Schmidt RJ, Pessah IN, Kostyniak PJ, et al. 2012. Levels of select PCB and PBDE congeners in human postmortem brain reveal possible environmental involvement in 15q11-Q13 duplication autism spectrum disorder. Environ Mol Mutagen 53(8):589–598, PMID: 22930557, 10.1002/em.21722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motomura H, Nozaki Y, Onaga C, Ozaki A, Tamori S, Shiina T-A, et al. 2020. High expression of c-Met, PKCλ and ALDH1A3 predicts a poor prognosis in late-stage breast cancer. Anticancer Res 40(1):35–52, PMID: 31892551, 10.21873/anticanres.13924. [DOI] [PubMed] [Google Scholar]
- Narasimhan S, Stanford Zulick E, Novikov O, Parks AJ, Schlezinger JJ, Wang Z, et al. 2018. Towards resolving the pro- and anti-tumor effects of the aryl hydrocarbon receptor. Int J Mol Sci 19(5):1388, PMID: 29735912, 10.3390/ijms19051388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicoli S, Presta M. 2007. The zebrafish/tumor xenograft angiogenesis assay. Nat Protoc 2(11):2918–2923, PMID: 18007628, 10.1038/nprot.2007.412. [DOI] [PubMed] [Google Scholar]
- Ohtake F, Baba A, Takada I, Okada M, Iwasaki K, Miki H, et al. 2007. Dioxin receptor is a ligand-dependent E3 ubiquitin ligase. Nature 446(7135):562–566, PMID: 17392787, 10.1038/nature05683. [DOI] [PubMed] [Google Scholar]
- Overholtzer M, Mailleux AA, Mouneimne G, Normand G, Schnitt SJ, King RW, et al. 2007. A nonapoptotic cell death process, entosis, that occurs by cell-in-cell invasion. Cell 131(5):966–979, PMID: 18045538, 10.1016/j.cell.2007.10.040. [DOI] [PubMed] [Google Scholar]
- Pearce DJ, Taussig D, Simpson C, Allen K, Rohatiner AZ, Lister TA, et al. 2005. Characterization of cells with a high aldehyde dehydrogenase activity from cord blood and acute myeloid leukemia samples. Stem Cells 23(6):752–760, PMID: 15917471, 10.1634/stemcells.2004-0292. [DOI] [PubMed] [Google Scholar]
- Peitzsch C, Tyutyunnykova A, Pantel K, Dubrovska A. 2017. Cancer stem cells: the root of tumor recurrence and metastases. Semin Cancer Biol 44:10–24, PMID: 28257956, 10.1016/j.semcancer.2017.02.011. [DOI] [PubMed] [Google Scholar]
- Perez-Riverol Y, Csordas A, Bai J, Bernal-Llinares M, Hewapathirana S, Kundu DJ, et al. 2019. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res 47(D1):D442–50, PMID: 30395289, 10.1093/nar/gky1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierre S, Chevallier A, Teixeira-Clerc F, Ambolet-Camoit A, Bui L-C, Bats A-S, et al. 2014. Aryl hydrocarbon receptor-dependent induction of liver fibrosis by dioxin. Toxicol Sci 137(1):114–124, PMID: 24154488, 10.1093/toxsci/kft236. [DOI] [PubMed] [Google Scholar]
- Polanska UM, Orimo A. 2013. Carcinoma-associated fibroblasts: non-neoplastic rumour-promoting mesenchymal cells. J Cell Physiol 228(8):1651–1657, PMID: 23460038, 10.1002/jcp.24347. [DOI] [PubMed] [Google Scholar]
- Rodgers KM, Udesky JO, Rudel RA, Brody JG. 2018. Environmental chemicals and breast cancer: an updated review of epidemiological literature informed by biological mechanisms. Environ Res 160:152–182, PMID: 28987728, 10.1016/j.envres.2017.08.045. [DOI] [PubMed] [Google Scholar]
- Rodriguez A-M, Elabd C, Amri E-Z, Ailhaud G, Dani C. 2005. The human adipose tissue is a source of multipotent stem cells. Biochimie 87(1):125–128, PMID: 15733747, 10.1016/j.biochi.2004.11.007. [DOI] [PubMed] [Google Scholar]
- Ruan B, Niu Z, Jiang X, Li Z, Tai Y, Huang H, et al. 2019. High frequency of cell-in-cell formation in heterogeneous human breast cancer tissue in a patient with poor prognosis: a case report and literature review. Front Oncol 9:1444, PMID: 31921689, 10.3389/fonc.2019.01444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert A, Rau S, Küllertz G, Fischer B, Santos AN. 2009. TCDD induces cell migration via NFATc1/ATX-signaling in MCF-7 cells. Toxicol Lett 184(1):26–32, PMID: 19028555, 10.1016/j.toxlet.2008.10.026. [DOI] [PubMed] [Google Scholar]
- Tyanova S, Temu T, Sinitcyn P, Carlson A, Hein MY, Geiger T, et al. 2016. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat Methods 13(9):731–740, PMID: 27348712, 10.1038/nmeth.3901. [DOI] [PubMed] [Google Scholar]
- Vassalli G. 2019. Aldehyde dehydrogenases: not just markers, but functional regulators of stem cells. Stem Cells Int 2019:3904645, PMID: 30733805, 10.1155/2019/3904645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Wyrick KL, Meadows GG, Wills TB, Vorderstrasse BA. 2011. Activation of the aryl hydrocarbon receptor by TCDD inhibits mammary tumor metastasis in a syngeneic mouse model of breast cancer. Toxicol Sci 124(2):291–298, PMID: 21948867, 10.1093/toxsci/kfr247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner M, Mocarelli P, Samuels S, Needham L, Brambilla P, Eskenazi B. 2011. Dioxin exposure and cancer risk in the Seveso Women’s Health Study. Environ Health Perspect 119(12):1700–1705, PMID: 21810551, 10.1289/ehp.1103720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Mercado-Uribe I, Xing Z, Sun B, Kuang J, Liu J. 2014. Generation of cancer stem-like cells through the formation of polyploid giant cancer cells. Oncogene 33(1):116–128, PMID: 23524583, 10.1038/onc.2013.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Weinberg RA. 2018. Epithelial-to-mesenchymal transition in cancer: complexity and opportunities. Front Med 12(4):361–373, PMID: 30043221, 10.1007/s11684-018-0656-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited in the ProteomeXchange Consortium via the Proteomics Identifications Database (PRIDE) (Perez-Riverol et al., 2019) partner repository with the data set identifier PXD014328.


