Abstract
Neutrophil elastase is a serine protease released by neutrophils, and its dysregulation has been associated with a variety of debilitating pathologies, most notably cystic fibrosis. This protein is also a prominent component of the so-called neutrophil extracellular traps (NETs), whose formation is a part of the innate immunity response to invading pathogens, but also contributes to a variety of pathologies ranging from autoimmune disorders and inflammation to cancer to thrombotic complications in COVID-19. Retention of neutrophil elastase within NETs is provided by ejected DNA chains, although this protein is also capable of interacting with a range of other endogenous polyanions, such as heparin and heparan sulfate. In this work, we evaluate the feasibility of using native mass spectrometry (MS) as a means of studying interactions of neutrophil elastase with heparin oligomers ranging from structurally homogeneous synthetic pentasaccharide fondaparinux to relatively long (up to twenty saccharide units) and structurally heterogeneous chains produced by partial depolymerization of heparin. The presence of heterogeneous glycan chains on neutrophil elastase and the structural heterogeneity of heparin oligomers render the use of standard MS to study their complexes impractical. However, supplementing MS with limited charge reduction in the gas phase allows meaningful data to be extracted from MS measurements. In contrast to earlier molecular modeling studies where a single heparin-binding site was identified, our work reveals the existence of multiple binding sites, with a single protein molecule being able to accommodate up to three decasaccharides. The measurements also reveal the ability of even relatively short heparin oligomers to bridge two protein molecules, suggesting that characterization of these complexes using native MS can shed light on the structural properties of NETs. Lastly, the use of MS allows the binding preferences of heparin oligomers to neutrophil elastase to be studied with respect to specific structural properties of heparin, such as the level of sulfation (i.e., charge density). All experimental measurements are carried out in parallel with molecular dynamics simulations of the protein/heparin oligomer systems, which are in remarkable agreement with the experimental data and highlight the role of electrostatic interactions as dominant forces governing the formation of these complexes.
Graphical Abstract
Introduction
Human neutrophil elastase (hNE) is a serine protease expressed and stored in the primary granules of neutrophils, and can rapidly degrade connective tissue proteins upon its release.1 While the proteolytic activity of this highly destructive enzyme is tightly controlled by plasma-borne inhibitors, dysregulation of the activity/inhibition balance is associated with a variety of debilitating pathologies,2 most notable of which is cystic fibrosis.3 This protein is also a prominent component of the so-called neutrophil extracellular traps (NETs), elaborate structures produced by neutrophils in response to invading pathogens by releasing chromatin that forms large extracellular DNA networks.4,5 The ability of NETs to entangle and trap invading microorganisms while maintaining high concentrations of antimicrobial and/or degradative agents (such as hNE) is an important part of the innate immunity response. At the same time, the formation of NETs (NETosis) also contributes to the onset and progression of a variety of pathologies ranging from autoimmune disorders and inflammation to cancer.6 Among the studies of NETs related/initiated pathologies, pro-coagulant properties of NETs in general and hNE in particular7 have been coming to light in recent years due to their involvement both in common cardiovascular pathologies,8 and in relatively rare ones (such as the role of hNE activation and NETosis in heparin-induced thrombocytopenia9). Furthermore, the growing realization of the centrality of viral coagulopathy in disease progression among severe COVID-19 patients,10–14 as well as the mounting evidence for the delayed thrombotic complications occurring even among asymptomatic patients,15 is now leading to suggestions that NETosis may also be triggered by the SARS-CoV-2 infection and play a detrimental role in pathophysiology of the disease by contributing to the lung tissue damage16–18 and beyond.19–21 Since hNE binding to extracellular DNA not only increases the local concentration of the protease molecules by confining them to the NETs, but also impairs the action of cognate inhibitors,22 the use of alternative hNE inhibitors has been proposed as a viable therapeutic strategy, both as an intervention23 and prophylaxis.24
Heparin and related glycosaminoglycans (such as heparan sulfate) are cognate polyanions that can also bind to hNE. In fact, the exceptionally high charge density exhibited by heparin makes it a viable competitor to negatively charged DNA vis-á-vis the protease binding (potentially mediated by electrostatic interactions), and in fact heparin has been shown to be capable of degrading NETs.25 Furthermore, there is evidence that heparin has a direct inhibitory effect on hNE,26 while newer studies suggest that heparin-driven hNE activation is also possible.26 Independent of its potential role in regulating the activity of extracellular hNE, heparin has been steadily gaining acceptance as a therapeutic agent in treating/preventing severe COVID-1927,28 due to a range of relevant properties that include (in addition to its traditional role as an anti-coagulant to control thromboembolic complications and other coagulopathies10) anti-inflammatory effects,29 as well as the ability to interfere with the coronavirus entry into the cell.30 These properties, combined with the ability of heparin to modulate the detrimental proteolytic activity of hNE (vide supra), may further advance the use of this century-old medication as a multi-modal therapeutic agent in treating the SARS-CoV-2 infections.31
One distinct feature of heparin that endows it with the ability to interact with a wide range of targets is the inherent structural heterogeneity of this polysaccharide. Unfortunately, this very same feature creates a significant barrier preventing its wider utilization in clinical practice, by making it challenging to (i) identify structural features responsible for the specific physiological effects and (ii) select a subset of structures capable of inducing the desired therapeutic effects while lacking the structural features that may trigger undesirable or indeed outright pathological processes. In fact, the enormous polydispersity of heparin makes even relatively straightforward tasks of monitoring its binding to physiological partners a challenging undertaking.
Native mass spectrometry (MS) became a popular analytical tool capable of monitoring protein/drug interactions for a variety of clinically relevant systems;32,33 however, its applications in the studies of heparin/protein binding had been limited in the past to relatively short heparin oligomers and homogeneous protein targets.34–37 Introduction of limited charge reduction as a means of obtaining meaningful intact mass measurements on highly heterogeneous systems38 allowed the scope of native MS to be expanded to include protein associations with unfractionated heparin.39 Recently, this technique has been applied to study the association of large heparin oligomers with the receptor-binding domain of the SARS-CoV-2 spike protein, and its interference with the docking of the latter to its host cell surface receptor.40 In the present work, we evaluate the feasibility of using native MS combined with limited charge reduction as a means of studying interactions of hNE with a variety of heparin oligomers, ranging from structurally homogeneous synthetic pentasaccharide fondaparinux to relatively long (up to twenty saccharide units) and structurally heterogeneous chains. The measurements allow the stoichiometry of non-covalent complexes to be determined, indicating the existence of multiple binding sites on the protein surface, which can accommodate up to three relatively short heparin oligomers. Native MS also reveals the ability of even relatively short heparin oligomers to bridge two hNE molecules, suggesting that large multi-protein complexes assembled on the polyanionic scaffold can be studied using this technique. Finally, the ability of MS to resolve heparin oligomers based on their level of sulfation (and, therefore, total charge in solution) allows the role of electrostatic interactions as the guiding forces of hNE/heparin complexes assembly to be established.
Materials and Methods
Materials.
Human neutrophil elastase (hNE) was purchased from Athens Research & Technology (Athens, GA) and was reconstituted into 50 mM sodium acetate (pH 5.5) with 150 mM sodium chloride once received. Molecular weights of different glycoforms were measured by reversed-phase LC-MS. The synthetic pentasaccharide (fondaparinux) was acquired from Sigma-Aldrich (St. Louis, MO). Heparin oligomers (dp10 and dp20) produced by partial depolymerization of heparin were purchased from Iduron (Alderley Edge, UK). All chemicals and solvents used in this work were of analytical grade or higher.
LC/MS and native MS/limited charge reduction measurements.
The stock hNE solution was buffer exchanged into 150 mM ammonium acetate solution (pH 6.9) before mixing with heparin molecules and MS analysis. LC/MS measurements were carried out using a reversed-phase C18 column (Agilent, Santa Clara, CA) coupled with a SolariX 7 (Bruker Daltonics, Billerica, MA) Fourier transform ion cyclotron resonance (FT ICR) mass spectrometer. 0.1% formic acid in water and in acetonitrile (v/v) were used as mobile phase A and B respectively with 5-95% B in a 35-min gradient. Native MS measurements were carried out using a Synapt G2-Si (Waters, Milford, MA) hybrid quadrupole/time-of-flight mass spectrometer with a nanospray source. To maintain the stability of hNE-heparin noncovalent complexes, the following parameters were applied: sampling cone, 80V; source offset, 50V; trap CE, 4V; transfer CE, 2V. Precursor ion selection for limited charge reduction measurements was carried out by setting the quadrupole selection parameter, LM resolution, to 4. To trigger limited charge reduction (using 1,3-dicyanobenzene as charge transfer reagent), the trap wave height and velocity were set to 300 m/s and 0.2 V respectively and the discharge current was optimized accordingly.
Heparin depletion measurements.
Heparin (dp10) binding preference was determined by heparin depletion measurements published in a previous paper.37 Briefly, protein/dp10 were mixed at different molar ratios (1:1, 2:1 and 4:1) and the intensity distributions of free dp10 signals at lower m/z range were examined and compared. The depleted dp10 species by protein binding could be determined based on the change of intensity distribution of free dp10 in each protein/dp10 mixture compared to the reference dp10 spectrum.
Molecular modeling.
All computational procedures were performed using applications (Maestro, Desmond, Glide) from the Schrödinger software suite (v. 2019-4, Schrödinger LLC, New York, NY). Docking of synthetic pentasaccharide (extracted from a PF4/pentasaccharide complex (PDB 4R9W)) was performed on a hNE molecule (PDB 2Z7F). The whole protein was used for receptor grid generation (Glide). The OPLS3e force field was used during LigPrep (pre-docking) and post-docking minimization. dp20·hNE, dp10·hNE and hNE·dp10·hNE models were prepared by molecular dynamics simulation considering the complexity of the ligands (longer heparin oligomers). The dp10 models were created by removing 14 saccharide units from the non-reducing end of dp24 (PDB 3IRJ). The 12 ns simulation was set up for each hNE/dpX system (neutralized with Na+) in explicit water and 150 mM NaCl (Desmond). The final frame (Fr-2002) was minimized using the OPLS3e force field to provide models for the hNE/dpX.
Results and Discussion
The polypeptide chain of hNE pre-protein consists of 267 amino acid residues, but the mature protein contains only 218 residues. This truncated form is produced by the removal of both the signal peptide (1-29) from the N-terminus and the C-terminal peptide (248-267).41 Other PTMs include N-glycosylation of three asparagine residues (although one of them remains mostly glycan-free)41 and formation of four disulfide bonds (see Supplementary Material for more detail). Intact mass analysis of hNE carried out under denaturing conditions reveals the presence of seventeen proteoforms with distinct masses ranging from 25,033 to 27,037 Da (Figure 1A), ten of which can be readily detected under near-native conditions as well (Figure 1B). The mass of a neutral, oxidized (i.e., containing four disulfides) polypeptide (30-247) in its glycan-free form is 23,287.6 Da, and the differences between this mass and those measured experimentally (vide supra) allow the glycan compositions for various proteoforms to be established (Table 1).
Figure 1.
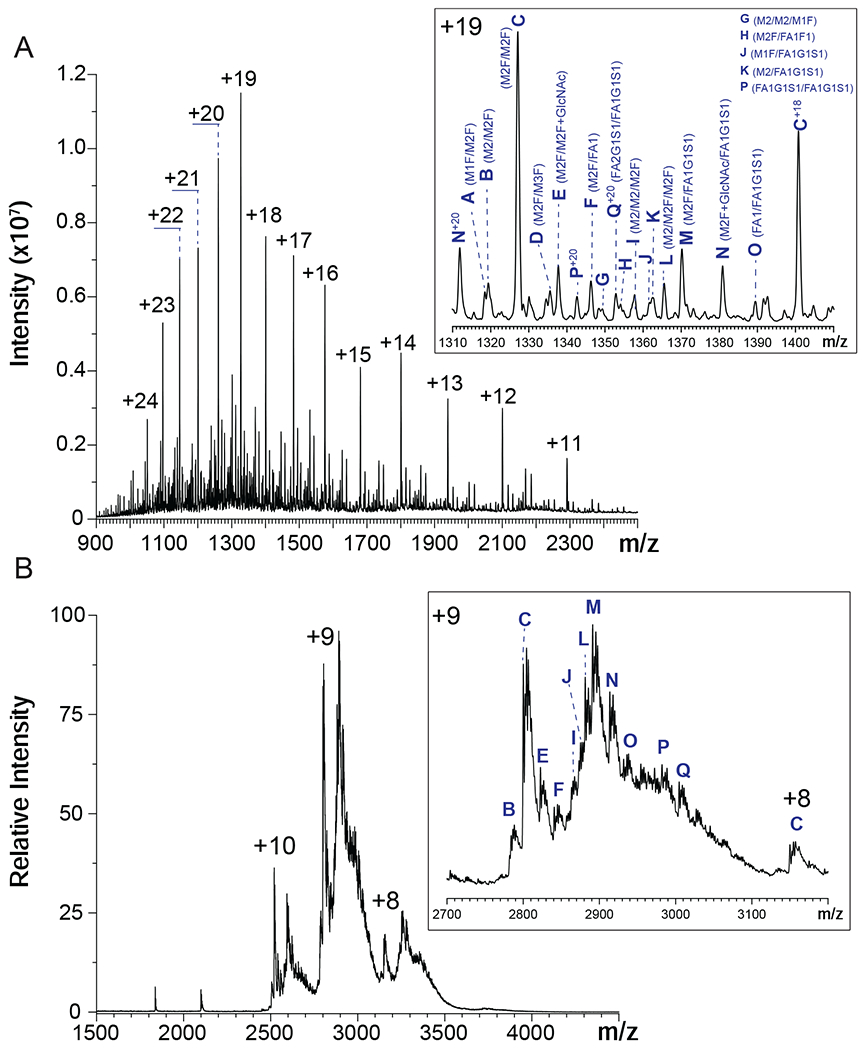
Mass spectra of hNE acquired under denaturing (A) and near-native conditions (B). The insets show zoomed view of the most abundant charge states in each case (+19 in (A) unless labeled with other charge states and +9 in (B)). The hNE glycoforms are labeled according to masses listed in Table 1 and identified referring to the recent study by Loke et al.41
Table 1.
The hNE glycoforms detected by intact-mass MS (the two most abundant glycoforms are in boldface type). The glycan identification is based on the measured masses and the assignments made previously by Loke, et al.41 The underlined glycan compositions correspond to species that were also observed by Loke et al.41 (see also Supplementary Material for the N-glycan structures).
| hNE Glycoform | Mexp. (Da) | Mtheo. (Da) | Δppm | Glycans |
|---|---|---|---|---|
| A | 25033.4 | 25033.9 | 20.0 | M1F/M2F |
| B | 25050.1 | 25050.0 | 4.0 | M2/M2F |
| C | 25196.2 | 25196.2 | 0.0 | M2F/M2F |
| D | 25358.5 | 25358.3 | 5.1 | M2F/M3F |
| E | 25398.8 | 25399.4 | −23.2 | M2F/(M2F+GlcNAc) |
| F | 25561.4 | 25561.5 | −4.7 | M2F/FA1 |
| G | 25619.1 | 25618.6 | 19.5 | M2/M2/M1F |
| H | 25707.5 | 25707.6 | −3.9 | >M2F/FA1F1 |
| I | 25780.5 | 25780.7 | −7.8 | M2/M2/M2F |
| J | 25852.7 | 25852.8 | −1.9 | M1F/FA1G1S1 |
| K | 25868.8 | 25868.8 | 0.0 | M2/FA1G1S1 |
| L | 25926.6 | 25926.9 | −10.4 | M2/M2F/M2F |
| M | 26015.3 | 26014.9 | 15.0 | M2F/FA1G1S1 |
| N | 26218.1 | 26218.1 | −2.3 | (M2F+GlcNAc)/FA1G1S1 |
| O | 26380.2 | 26380.2 | 0.0 | FA1/FA1G1S1 |
| P | 26834.0 | 26833.6 | 13.8 | FA1G1S1/FA1G1S1 |
| Q | 27036.8 | 27036.9 | −3.0 | FA2G1S1/FA1G1S1 |
| # | Not observed | 24887.8 | N/A | M2/M1F |
Note: # represents glycoforms observed by Loke’s et al yet not observed in this work. Nomenclature (adapted from Loke’s et al): monomannosylchitobiose core fucosylated (M1F), bimannosylchitobiose (M2), bimannosylchitobiose core fucosylated (M2F), bimannosylchitobiose core fucosylated with terminal β1,2-linked GlcNAc (M2F+GlcNAc), trimannosylchitobiose core fucosylated (M3F), trimannosylchitobiose core fucosylated with β1,2-linked GlcNAc (FA1), trimannosylchitobiose core fucosylated with β1,2-linked GlcNAc and fucose (FA1F1), trimannosylchitobiose core monoantennary core fucosylated α2,6-monosialylated (FA1G1S1).
The set of glycoforms detected by the intact-mass analysis of hNE in this work significantly overlaps, but is not identical to that reported earlier by Loke et al. for the hNE harvested from blood-derived resting neutrophils collected from healthy donors.41 The two glycoforms that were found to be most abundant in this study, M2F/M2F and M2F/FA1G1S1, were also the most abundant species reported by Loke et al.41 The lower-mass glycoform, M2/M1F observed by Loke et al. was not detected in our studies. On the other hand, a few higher-mass glycoforms detected in this study (e.g., FA2G1S1/FA1G1S1) were not reported previously; overall, out of the eleven different glycoforms detected in intact-mass measurements, three had not been previously reported (see Table 1).
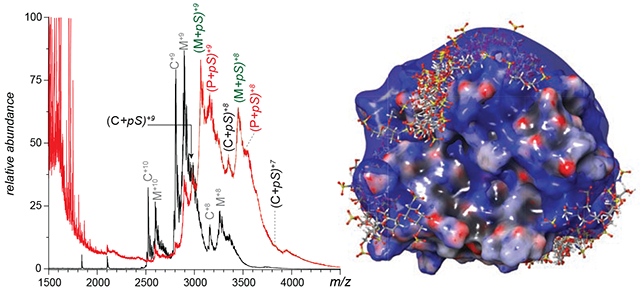
The presence of multiple hNE glycoforms, as well as the incomplete desolvation of the protein ions in native MS give rise to a mass spectrum populated with convoluted, partially resolved peaks (Figure 1B). One intriguing feature of the native mass spectrum is the dramatic change of the relative signal strengths for the two most abundant hNE glycoforms compared to the intensity distributions observed under denaturing solvent conditions, M2F/M2F and M2F/FA1G1S1. The latter generates the most abundant signal under native conditions, which is likely due to the enhanced shielding of less hydrophilic parts of the protein surface by a larger glycan chain (FA1G1S1 vs. M2F). Incubation of hNE with a homogeneous heparin mimetic pentasaccharide (fondaparinux) transforms the native mass spectrum by shifting the ionic peaks to higher m/z region (Figure 2A). The magnitude of this shift appears to be consistent with the addition of a single pentasaccharide (pS) molecule to the protein. Indeed, the most abundant ionic signal in the mass spectrum of the hNE/pS mixture can be readily assigned to ions representing the hNE·pS complex involving the two most abundant glycoforms of the protein (M2F/M2F and M2F/FA1G1S1), as well as one of the lower-abundance glycoforms, FA1G1S1/FA1G1S1 (Figure 2A). However, a closer examination of this mass spectrum reveals features that cannot be assigned to hNE·pS complexes containing any of the glycoforms detected in our intact-mass analysis (Figure 1).
Figure 2.
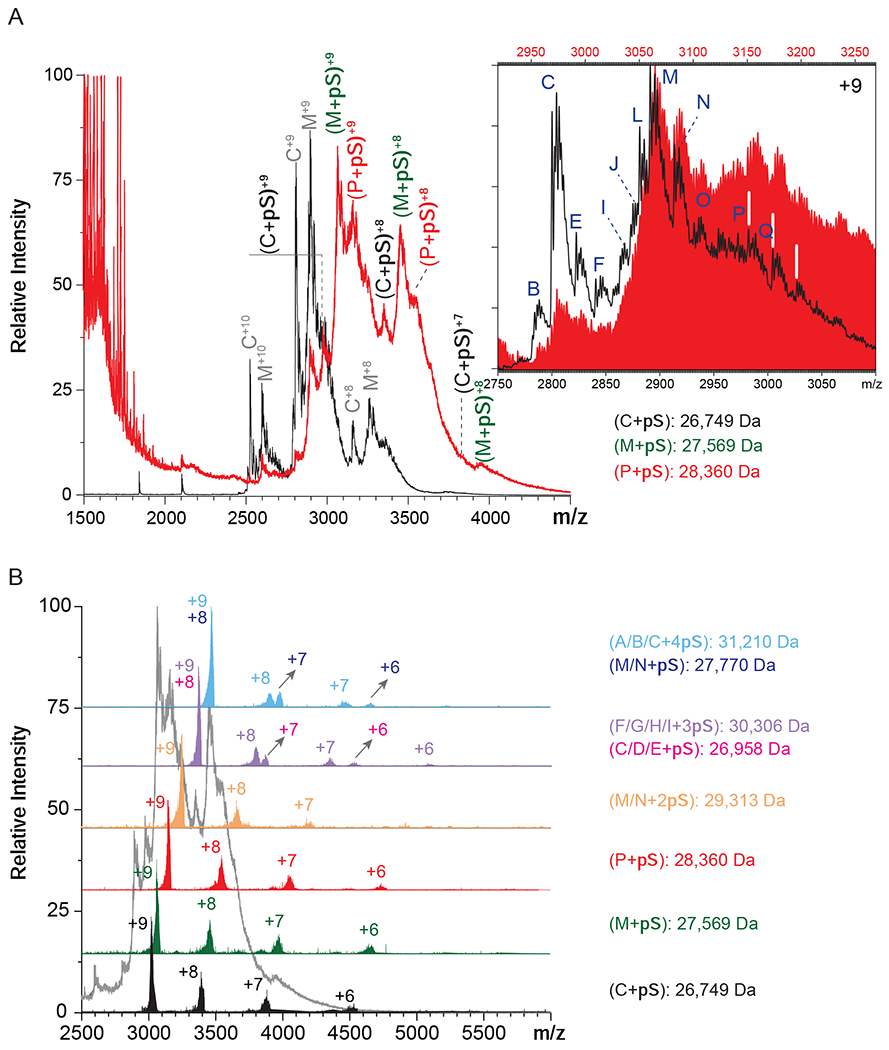
Native MS of a solution containing 25 μM synthetic pentasaccharide fondaparinux (pS) and 5 μM hNE (molar ratio pS:hNE=5:1) (A). The mass spectrum of hNE (black) is used as a reference. The inset shows the overlaid mass spectra of hNE (black) and hNE/pS (red, made by shifting the m/z scale by the value of the mass of pS (1,550 Da)). Charge ladders are generated using limited charge reduction to identify 1:1 and 1:n hNE/pS complexes (B).
In order to identify the ionic species contributing to the convoluted signal in the native MS of the hNE/pS mixture, limited charge reduction measurements were carried out on several ionic populations covering the m/z region 3,000-3,500, which corresponds to the most abundant signal in the mass spectrum. Not surprisingly, limited charge reduction of ionic populations within m/z regions centered around the most abundant and better-defined spectral features (m/z 2,971, 3,066 and 3,152) gave rise to charge ladders corresponding to the following set of masses: 26,749, 27,569 and 28,360 Da which represent complexes formed by association of a single pS molecule with M2F/M2F (25,195 Da), M2F/FA1G1S1 (26,015 Da) and FA1G1S1/FA1G1S1 (26,834 Da) glycoforms of hNE, respectively. This confirmed the initial assignment of the most prominent spectral features in native MS that were made based on shifting the m/z scale by the value of the mass of pS (1550 Da) divided by the presumed charge state, as shown in the inset in Figure 2A. Interestingly, the relative abundance of the hNE·pS complex corresponding to protein isoforms with shorter glycan chains appears to be lower compared to those of the isoforms with larger carbohydrate chains (e.g., compare the relative abundance of the ions representing hNE(M2F/M2F)·pS at m/z 2,800 and hNE(M2F/FA1G1S1)·pS at m/z 2891 in the Figure 2A inset. Furthermore, inspection of the higher m/z region of this mass spectrum reveals the presence of abundant ions corresponding to hNE(FA1G1S1/FA1G1S1)·pS and hNE(FA1G1S1/FA2G1S1)·pS complexes (at m/z 2982 and 3,005, respectively), despite the fact that these glycoforms constitute minor components of the hNE sample. Furthermore, both glycoforms incorporate two sialic acid residues in their glycans, which is expected to make the protein/heparinoid interaction less favorable due to the electrostatic repulsion.
While the prevalence of the 1:1 hNE·pS complexes corresponding to the protein glycoforms with larger/negatively charged carbohydrate chains may seem counterintuitive, consideration of the electrostatic potential distribution on the surface of the protein suggests that it can accommodate more than just a single short polyanionic ligand on its surface. Indeed, the distribution of basic residues on the hNE surface gives rise to a crescent-shaped positive charge basin (Figure 3A), reminiscent of a positive charge “belt” circling another paradigmatic heparin-binding protein, platelet factor 4 (PF4).42 The latter can accommodate multiple pS molecules on its surface,43 suggesting that higher stoichiometry complexes may also be encountered in the case of hNE. Formation of the hNE·pS complexes with 1:n stoichiometry (where n > 1) would give rise to ionic signal in m/z region above 3100, which clearly manifests itself in the mass spectrum shown in the Figure 2A inset as an elevation of the ionic signal intensity level. Not surprisingly, limited charge reduction of ionic populations selected from this region of the mass spectrum reveals the presence of both 1:1 and 1:2 hNE/pS complexes (Figure 2B), the latter incorporating the hNE glycoforms with relatively short glycan chains lacking acidic saccharide units. To identify and assign hNE/pS stoichiometries, different hNE glycoforms were considered for alternative potential assignments. For example, the mass of one observed complex is 29,313 Da, which exceeds 28,570 Da (the mass of the “heaviest” hNE glycoform with one pS molecule), yet is far below 29,683 Da (the “lightest” hNE glycoform with three pS molecules). Therefore, we identified this species as a 1:2 hNE/pS complex confidently. Take the complex of 30,306 Da as another example, the mass falls into the range between 30,136 Da (the “heaviest” hNE glycoform with two pS molecules) to 31,233 Da (the “lightest” hNE glycoform with four pS molecules), leaving this complex as a 1:3 hNE/pS one. Moving the precursor ion selection window further up the m/z scale allows the hNE/pS complexes of even higher stoichiometry to be detected as well (e.g., hNE·pS4 whose presence is revealed by producing the charge ladders for ions confined to the 3450-3480 m/z window, see the blue trace in Figure 2B).
Figure 3.
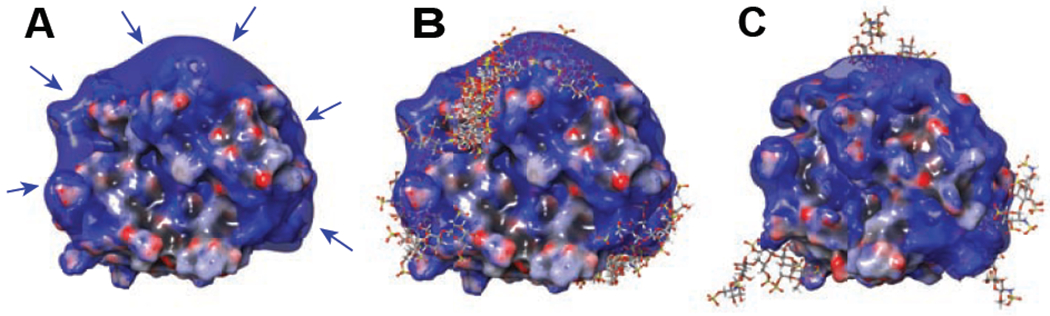
The results of molecular modeling of hNE/pS interaction showing the electrostatic potential surface (negative (red) to positive (blue)) of intact hNE (A), the predicted poses (cumulative structures of pS docking to hNE) from the docking results of 1:1 hNE/pS complexes (B) and the molecular dynamics result of 1:4 hNE/pS complexes (C). Arrows in (A) indicate the potential heparin-binding positive basin.
The existence of the hNE/pS complexes with stoichiometry ranging from 1:1 to 1:4 revealed by native MS and limited charge reduction is consistent with the results of docking and molecular modeling studies. The docking results suggest that the accommodations of pentasaccharide chains are clustered in three areas on the positively charged basin of the protein surface (Figure 3B). At the same time, hNE could accommodate up to 4 pentasaccharide chains with spacing between the adjacent ones sufficient to avoid any steric clashes according to our molecular modeling result (Figure 3C), which corresponds to the interpretation of our measurements. Furthermore, the four pentasaccharide chains are on the three sites suggested in the docking results. While nonspecific interactions during ESI process can occur in the presence of relatively high concentrations of ligand (pS),44 the consistency of MD and ESI-MS proved the reliability of the higher hNE/pS stoichiometries (> 1:1) observed in MS experiments and ruled out the possibilities of nonspecific interactions in this case. On the contrary, since neither ionic interactions nor hydrogen bonds within protein-ligand interactions are compromised in the gas phase, native MS enables the detection of subordinary/lower-affinity protein-ligand interactions.
Substituting the relatively short structurally homogeneous heparin oligomer fondaparinux with a longer fixed-length heparinoid (decasaccharide, dp10) gives rise to convoluted ionic signal in the mass spectra of hNE/oligoheparin mixtures (Figure 4). The shapes of the ionic signal distribution over the m/z scale exhibit strong dependence on the protein/oligoheparin mixing ratio, with discernable spectral features evident only in the mass spectrum of the 4:1 hNE/dp10 mixture while other spectra show near-continuum distribution of the ionic signal (Figure 4A). Once again, the application of limited charge reduction to ionic populations selected within narrow m/z windows allows these spectra to be readily interpreted and the spectral features assigned to specific ions. An example is shown in Figure 4B, where application of limited charge reduction to the 1:1 hNE/dp10 mixture reveals the presence of protein/oligoheparin complexes with binding stoichiometries ranging from 1:1 to 1:3. Although the assignment of the stoichiometries of hNE/oligoheparin was more complicated than hNE/pS complex owing to the heterogeneity of dp10 itself (mass range 2,165-2,965 Da due to various sulfate/acetyl levels), we could still get correct assignment. For example, we observed a species with the mass of 33,249 Da, which exceeds 32,966 Da (the “heavest” hNE glycoform with two highest-sulfated dp10 (10,16,0)) but is still below 33,693 Da (the “lightest” hNE glycoform with four lowest-sulfated dp10 (10,6,0)). Therefore, we confirmed this species to be a 1:3 hNE/dp10 complex. Thus, doubling the length of oligoheparin did not result in elimination of the complexes with 1:2 and even 1:3 stoichiometry, suggesting that the size of the positive charge basin on the protein surface is sufficient to accommodate decameric heparin species. Interestingly, further doubling of the chain length (by substituting dp10 with dp20 species) did result in elimination of protein/oligoheparin complexes with stoichiometries higher than 1:1 (see Supplementary Material for more detail). This leads us to a conclusion that the physical dimensions of this longer polysaccharide chain are such that a single chain can “cover” most of the positive charge basin on the surface of the protein by wrapping around it.
Figure 4.
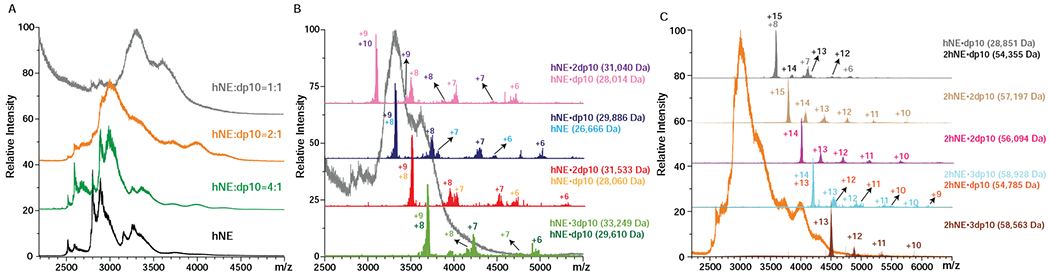
Native MS of 5 μM hNE incubated with dp10 at different molar ratios as shown on the graph (A) and representative limited charge reduction measurements for the molar ratio 1:1 (B) and 2:1 of hNE and dp10 mixtures (C).
Another intriguing feature of the mass spectra of hNE/dp10 mixtures is the presence of complexes containing two protein molecules and a single oligoheparin. These heparin-bridged hNE dimers are detected only in the mixtures that were prepared with molar excess of the protein (an example is shown in Figure 4C). The masses of such complexes range from 54,355 to 54,785 Da since the masses of dp10 molecules spread over the 2,165-2,965 Da range (see Figure 5 and Supplementary Material), it might be possible to estimate the average mass of proteins involved in heparin-bridged dimer formation as 51,390-52,620 Da. However, these estimates implicitly assume that all dp10 chains are equally likely to be recruited as “bridges,” while it is possible that certain structural features (most likely, the extent of sulfation) will result in preferential recruiting of some subpopulations of decasaccharides at the expense of others. In order to check if the binding preferences are indeed determined by the extent of sulfation, a series of heparin depletion measurements37 were carried out. In these measurements the intensity distributions of ionic species representing the free dp10 molecules in solution were determined at different protein/heparin mixing ratios; a protein-free dp10 solution was used as a reference (Figure 5). Analysis of these data provide clear indication that decamers with higher levels of sulfation (> 13 sulfate groups per chain) are preferential binders. Indeed, while the mass spectrum of the reference dp10 sample (the top trace in Figure 5) displays abundant signal for dp10 species carrying 14-15 sulfates, and detectable signal for the decamer carrying 16 sulfate groups, all these species are absent in the mass spectrum of the 1:1 hNE/dp10 mixture. Further shift of the distribution is observed upon increasing the hNE/dp10 molar ratio, with the most abundant ionic signal now corresponding to the decamers with 8-9 sulfate groups. These results echo the conclusions of an earlier study, where highly sulfated heparin oligomers showed a strong binding preference towards fibroblast growth factor, but recruitment of oligomers with lower levels of sulfation also occurred when the protein was present in the mixture in molar excess.37
Figure 5.
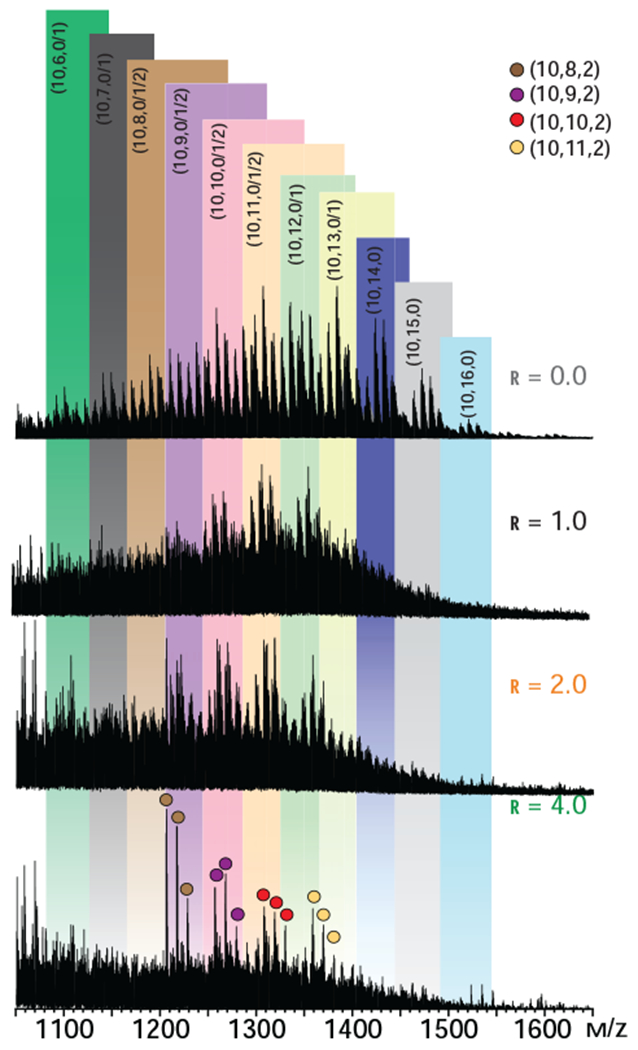
Distributions of unbound dp10 molecules in a 5 μM solution of hNE incubated with dp10 at different molar ratios (as indicated on each graph, r represents the molar ratio of protein to dp10). The bottom trace represents a reference spectrum of dp10 in a protein-free solution. Only ions at +2 charge state are shown for clarity. Ions representing individual heparin oligomers are labeled according to Henriksen’s nomenclature,47 where the three numbers in parentheses represent the oligosaccharide length, the total number of sulfate groups, and the total number of acetyl groups.
The strong preference of hNE for highly sulfated dp10 species narrows down the mass spread of the heparin component of the dp10-bridged hNE dimers to 2,725-2,965 Da (13-16 sulfate groups), allowing a more precise estimate of the protein component mass of such complexes to be made. This comes down to a 51,390-52,060 Da range, which is consistent with the notion of the two protein molecules in the dimer being M2F/FA1G1S1 (the mass of the dimer is 52,030 Da), and excluding the possibility of protein with more extensive glycan chains. It certainly is noteworthy that heparin oligomers as short as decasaccharides can facilitate dimerization of hNE. Although protein bridging by heparin has been reported for a variety of systems, it usually requires longer chains, as is the case for a trapped intermediate formed by factor Xa and antithrombin.45 A recent study of heparin oligomer interactions with platelet factor 4 (PF4) tetramer (a protein having similar dimensions to hNE, and also capable of accommodating multiple polyanionic chains on its surface) failed to reveal any signs of protein condensation on chains as long as eicososaccharides.43 Molecular modeling of the hNE·dp10·hNE species yields a structure where the short heparin oligomer is “sandwiched” between two protein molecule, with the sulfate groups on each side of the dp10 molecule making contacts with the basic side chains of one or the other hNE molecules. This arrangement is strongly dependent on the presence of a large number of sulfate groups within the heparin oligomer, explaining the strong preference of the protein for highly sulfated dp10 under conditions favoring hNE dimerization (vide supra).
In addition to the dp10-bridged hNE dimers, limited charge reduction of ionic populations above m/z 4,000 reveals the presence of 2:2 hNE/dp10 complexes (Figure 4B). The “shared” heparin oligomer in these species is sandwiched between an hNE molecule and an hNE·dp10 complex. The existence of such complexes is not surprising, given the ability of a single hNE molecule to accommodate more than one dp10 species (vide supra). The results of molecular modeling studies of a hNE·dp10·hNE·dp10 complex provide clear indication that such an arrangement is possible, with the “extra” dp10 molecule being localized within the region of the positive charge basin of one of the proteins outside of the dimer interface (Figure 6B). Not surprisingly, no higher stoichiometry complexes have been observed, since the length of the dp10 chain appears to be too short to enable tethering of multiple (> 2) protein molecules.
Figure 6.
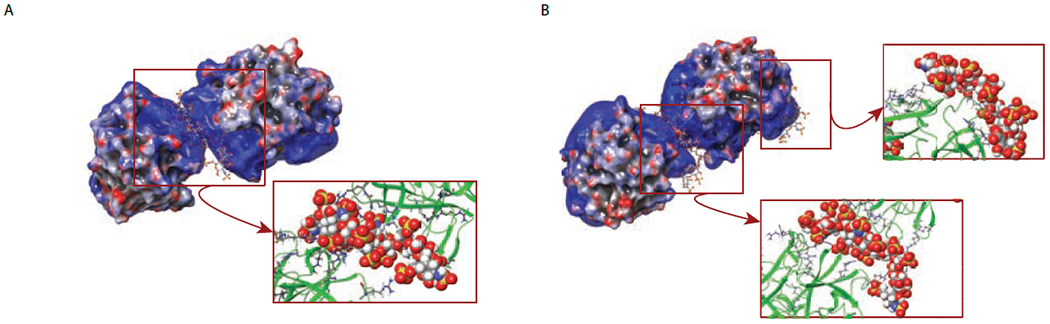
The results of molecular modeling of hNE/dp10 interaction showing the predicted structure of hNE·dp10·hNE complexes (A) and hNE·dp10·hNE·dp10 complexes (B). The hNE/dp10 interaction areas are shown with all arginine residues specified.
The ability of hNE to accommodate multiple oligoheparin molecules observed in this work is important vis-à-vis possible molecular mechanisms underlying the previously reported ability of heparin to degrade NETs.25 Attachment of multiple short heparin oligomers (or a single longer chain) to the protein surface results in a significant reduction or indeed complete elimination of the available binding sites for other endogenous polyanions, such as extra-cellular DNA or polyphosphates. At the same time, the ability of even relatively short heparin chains to induce hNE dimerization observed in our work indicates that this glycosaminoglycan may also participate in NET formation or indeed serve as a scaffold for assembling the NET-like structures. Therefore, the role of heparin in determining the fate of NET appears to be rather nuanced, and further studies are needed in order to understand the details of the heparin influence on NET dynamics. Lastly, it must be mentioned that heparin involvement in NETosis is not limited to the attenuation of the stability of these elaborate extracellular structures. A recent study revealed the ability of heparin to induce NETosis by stimulation of neutrophils, which can apparently uptake this glycosaminoglycan via endocytosis.46 While the molecular mechanisms governing this phenomenon remain unknown, it is feasible that this process involves activation of intracellular hNE. The combined native MS/limited charge reduction approach presented in this work will undoubtedly contribute to better understanding the roles played by heparin in both promoting and arresting NETosis, providing a clearer picture of its in vivo activities and explaining how this polyanionic sugars commonly used as an anti-coagulant can also exert procoagulant effects.
Conclusions
In this work native MS was used to study interaction of hNE with several short heparin oligomers. Since the structural heterogeneity of both the protein and the glycosaminoglycan components of the complexes makes characterization of these systems with native MS a challenging task, limited charge reduction was used to assist the data interpretation. Binding of multiple short oligomers to a single protein molecule is observed, and heparinoids as short as decasaccharides enable protein dimerization. The ability to associate with hNE is not distributed uniformly across all heparin species; instead, highly sulfated chains are clearly favored by the protein. However, heparin oligomers with lower levels of sulfation also associate with hNE (presumably with significantly lower affinity) when the latter is present in solution in molar excess. The results of this initial study have direct impact on the on-going efforts to elucidate the nuanced roles play by heparin in modulating NETosis, one of the ancient immune defense mechanisms that has also been implicated in a range of pathologies (most recently including the progression of severe COVID-19).
Supplementary Material
feasibility of using native mass spectrometry (MS) as a means of studying interactions of neutrophil elastase with heparin oligomers is evaluated
heterogeneity of both the protein and the heparin oligomers render the use of standard MS to study their complexes impractical
supplementing MS with limited charge reduction in the gas phase allows meaningful data to be extracted from MS measurements
in contrast to earlier molecular modeling studies where a single heparin-binding site was identified, our work reveals the existence of multiple binding sites, with a single protein molecule being able to accommodate up to three decasaccharides
the measurements also reveal the ability of even relatively short heparin oligomers to bridge two protein molecules, suggesting that characterization of these complexes using native MS can shed light on the structural properties of the so-called neutrophil extracellular traps (NETs), whose formation is a part of the innate immunity response to invading pathogens, but also contributes to a variety of pathologies ranging from autoimmune disorders and inflammation to cancer to thrombotic complications in COVID-19
Acknowledgements
This work was supported by a grant R01 GM112666 from the National Institutes of Health. All measurements were carried out in the Mass Spectrometry Core facility (UMass-Amherst).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest
The authors declare no conflict of interest.
References
- (1).Travis J Structure, function, and control of neutrophil proteinases. Am. J. Med. 1988, 84, 37–42. [DOI] [PubMed] [Google Scholar]
- (2).Polverino E; Rosales-Mayor E; Dale GE; Dembowsky K; Torres A The Role of Neutrophil Elastase Inhibitors in Lung Diseases. Chest 2017, 152, 249–262. [DOI] [PubMed] [Google Scholar]
- (3).Balfour-Lynn IM The protease-antiprotease battle in the cystic fibrosis lung. J. R. Soc. Med. 1999, 92 (Suppl. 37), 23–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Brinkmann V; Reichard U; Goosmann C; Fauler B; Uhlemann Y; Weiss DS; Weinrauch Y; Zychlinsky A Neutrophil extracellular traps kill bacteria. Science (New York, N.Y.) 2004, 303, 1532–1535. [DOI] [PubMed] [Google Scholar]
- (5).Vorobjeva NV; Chernyak BV NETosis: Molecular Mechanisms, Role in Physiology and Pathology. Biochemistry. Biokhimiia 2020, 85, 1178–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Kaplan MJ; Radic M Neutrophil extracellular traps: double-edged swords of innate immunity. J. Immunol. 2012, 189, 2689–2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Iba T; Miki T; Hashiguchi N; Tabe Y; Nagaoka I Is the neutrophil a ‘prima donna’ in the procoagulant process during sepsis? Crit. Care 2014, 18, 230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Sorvillo N; Cherpokova D; Martinod K; Wagner DD Extracellular DNA NET-Works With Dire Consequences for Health. Circ. Res. 2019, 125, 470–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Perdomo J; Leung HHL; Ahmadi Z; Yan F; Chong JJH; Passam FH; Chong BH Neutrophil activation and NETosis are the major drivers of thrombosis in heparin-induced thrombocytopenia. Nat. Commun. 2019, 10, 1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Costanzo L; Palumbo FP; Ardita G; Antignani PL; Arosio E; Failla G Coagulopathy, thromboembolic complications and the use of heparin in COVID-19 Pneumonia. J. Vasc. Surg. Venous Lymphat. Disord. 2020, 8, 711–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Kipshidze N; Dangas G; White CJ; Kipshidze N; Siddiqui F; Lattimer CR; Carter CA; Fareed J Viral Coagulopathy in Patients With cOviD-19: Treatment and Care. Clin. Appl. Thromb. Hemost. 2020, 26, 1076029620936776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Levi M COVID-19 coagulopathy vs disseminated intravascular coagulation. Blood Adv. 2020, 4, 2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Martín-Rojas RM; Pérez-Rus G; Delgado-Pinos VE; Domingo-González A; Regalado-Artamendi I; Alba-Urdiales N; Demelo-Rodríguez P; Monsalvo S; Rodríguez-Macías G; Ballesteros M; Osorio-Prendes S; Díez-Martin JL; Pascual C COVID-19 coagulopathy: an in-depth analysis of the coagulation system. Eur. J. Haematol. 2020, in press (doi: 10.1111/ejh.13501). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Miesbach W; Makris M COVID-19: Coagulopathy, Risk of Thrombosis, and the Rationale for Anticoagulation. Clin. Appl. Thromb. Hemost. 2020, 26, 1076029620938149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Fan BE; Umapathi T; Chua K; Chia YW; Wong SW; Tan GWL; Chandrasekar S; Lum YH; Vasoo S; Dalan R Delayed catastrophic thrombotic events in young and asymptomatic post COVID-19 patients. J. Thromb. Thrombolysis 2020, in press (doi: 10.1007/s11239-020-02332-z). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Veras FP; Pontelli M; Silva C; Toller-Kawahisa J; de Lima M; Nascimento D; Schneider A; Caetite D; Rosales R; Colon D; Martins R; Castro I; Almeida G; Lopes MI; Benatti M; Bonjorno L; Giannini M; Luppino-Assad R; Almeida S; Vilar F, et al. SARS-CoV-2 triggered neutrophil extracellular traps (NETs) mediate COVID-19 pathology. medRxiv 2020, doi: 10.1101/2020.06.08.20125823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Radermecker C; Detrembleur N; Guiot J; Cavalier E; Henket M; d’Emal C; Vanwinge C; Cataldo D; Oury C; Delvenne P; Marichal T Neutrophil extracellular traps infiltrate the lung airway, interstitial, and vascular compartments in severe COVID-19. J. Exp. Med. 2020, 217, e20201012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Chiang CC; Korinek M; Cheng WJ; Hwang TL Targeting Neutrophils to Treat Acute Respiratory Distress Syndrome in Coronavirus Disease. Front. Pharmacol. 2020, 11, 572009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Makatsariya A; Slukhanchuk E; Bitsadze V; Khizroeva J; Tretyakova M; Tsibizova V; Dobryakov A; Elalamy I; Gris JC COVID-19, neutrophil extracellular traps and vascular complications in obstetric practice. J. Perinat. Med. 2020, 48, 985–994. [DOI] [PubMed] [Google Scholar]
- (20).Ng H; Havervall S; Rosell A; Aguilera K; Parv K; von Meijenfeldt FA; Lisman T; Mackman N; Thalin C; Phillipson M Circulating Markers of Neutrophil Extracellular Traps Are of Prognostic Value in Patients With COVID-19. Arterioscler. Thromb. Vasc. Biol. 2020, in press (doi: 10.1161/ATVBAHA.120.315267), Atvbaha120315267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Arcanjo A; Logullo J; Menezes CCB; de Souza Carvalho Giangiarulo TC; Dos Reis MC; de Castro GMM; da Silva Fontes Y; Todeschini AR; Freire-de-Lima L; Decoté-Ricardo D; Ferreira-Pereira A; Freire-de-Lima CG; Barroso SPC; Takiya C; Conceição-Silva F; Savino W; Morrot A The emerging role of neutrophil extracellular traps in severe acute respiratory syndrome coronavirus 2 (COVID-19). Sci. Rep. 2020, 10, 19630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Barbosa da Cruz D; Helms J; Aquino LR; Stiel L; Cougourdan L; Broussard C; Chafey P; Riès-Kautt M; Meziani F; Toti F; Gaussem P; Anglés-Cano E DNA-bound elastase of neutrophil extracellular traps degrades plasminogen, reduces plasmin formation, and decreases fibrinolysis: proof of concept in septic shock plasma. FASEB J. 2019, 33, 14270–14280. [DOI] [PubMed] [Google Scholar]
- (23).Thierry AR Anti-protease Treatments Targeting Plasmin(ogen) and Neutrophil Elastase May Be Beneficial in Fighting COVID-19. Physiol. Rev. 2020, 100, 1597–1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Mohamed MMA; El-Shimy IA; Hadi MA Neutrophil Elastase Inhibitors: A potential prophylactic treatment option for SARS-CoV-2-induced respiratory complications? Crit. Care 2020, 24, 311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Fuchs TA; Brill A; Duerschmied D; Schatzberg D; Monestier M; Myers DD Jr.; Wrobleski SK; Wakefield TW; Hartwig JH; Wagner DD Extracellular DNA traps promote thrombosis. Proc. Natl. Acad. Sci. U. S. A. 2010, 107, 15880–15885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Spencer JL; Stone PJ; Nugent MA New insights into the inhibition of human neutrophil elastase by heparin. Biochemistry 2006, 45, 9104–9120. [DOI] [PubMed] [Google Scholar]
- (27).Hippensteel JA; LaRiviere WB; Colbert JF; Langouët-Astrié CJ; Schmidt EP Heparin as a Therapy for COVID-19: Current Evidence and Future Possibilities. Am. J. Physiol. Lung. Cell. Mol. Physiol. 2020, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Drago F; Gozzo L; Li L; Stella A; Cosmi B Use of Enoxaparin to Counteract COVID-19 Infection and Reduce Thromboembolic Venous Complications: A Review of the Current Evidence. Front. Pharmacol. 2020, 11, 579886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Paar V; Wernly B; Zhou Z; Motloch LJ; Hoppe UC; Egle A; Lichtenauer M Anticoagulation for COVID-19 treatment: both anti-thrombotic and anti-inflammatory? J. Thromb. Thrombolysis. 2020, in press, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Mycroft-West CJ; Su D; Pagani I; Rudd TR; Elli S; Guimond SE; Miller G; Meneghetti MCZ; Nader HB; Li Y; Nunes QM; Procter P; Mancini N; Clementi M; Forsyth NR; Turnbull JE; Guerrini M; Fernig DG; Vicenzi E; Yates EA, et al. Heparin inhibits cellular invasion by SARS-CoV-2: structural dependence of the interaction of the surface protein (spike) S1 receptor binding domain with heparin. bioRxiv 2020, doi: 10.1101/2020.04.28.066761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Lindahl U; Li JP Heparin - an old drug with multiple potential targets in COVID-19 therapy. J. Thromb. Haemost. 2020, in press (doi: 10.1111/jth.14898). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Tong W; Wang G How can native mass spectrometry contribute to characterization of biomacromolecular higher-order structure and interactions? Methods (San Diego, Calif.) 2018, 144, 3–13. [DOI] [PubMed] [Google Scholar]
- (33).Hannah VV; Atmanene C; Zeyer D; Van Dorsselaer A; Sanglier-Cianferani S Native MS: an ‘ESI’ way to support structure- and fragment-based drug discovery. Fut. Med. Chem. 2010, 2, 35–50. [DOI] [PubMed] [Google Scholar]
- (34).Yu Y; Sweeney MD; Saad OM; Crown SE; Handel TM; Leary JA Chemokine-glycosaminoglycan binding: Specificity for CCR2 ligand binding to highly sulfated oligosaccharides using FT ICR mass spectrometry. J. Biol. Chem. 2005, 280, 32200–32208. [DOI] [PubMed] [Google Scholar]
- (35).Abzalimov RR; Dubin PL; Kaltashov IA Glycosaminoglycans as naturally occurring combinatorial libraries: Developing a mass spectrometry-based strategy for characterization of anti-thrombin interaction with low molecular weight heparin and heparin oligomers. Anal. Chem. 2007, 79, 6055–6063. [DOI] [PubMed] [Google Scholar]
- (36).Harmer NJ; Robinson CJ; Adam LE; Ilag LL; Robinson CV; Gallagher JT; Blundell TL Multimers of the fibroblast growth factor (FGF)-FGF receptor-saccharide complex are formed on long oligomers of heparin. Biochem. J. 2006, 393, 741–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Minsky BB; Dubin PL; Kaltashov IA Electrostatic Forces as Dominant Interactions Between Proteins and Polyanions: an ESI MS Study of Fibroblast Growth Factor Binding to Heparin Oligomers. J. Am. Soc. Mass Spectrom. 2017, 28, 758–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Abzalimov RR; Kaltashov IA Electrospray ionization mass spectrometry of highly heterogeneous protein systems: Protein ion charge state assignment via incomplete charge reduction. Anal. Chem. 2010, 82, 7523–7526. [DOI] [PubMed] [Google Scholar]
- (39).Zhao Y; Abzalimov RR; Kaltashov IA Interactions of Intact Unfractionated Heparin with Its Client Proteins Can Be Probed Directly Using Native Electrospray Ionization Mass Spectrometry. Anal. Chem. 2016, 88, 1711–1718. [DOI] [PubMed] [Google Scholar]
- (40).Yang Y; Du Y; Kaltashov IA The utility of native MS for understanding the mechanism of action of repurposed therapeutics in COVID-19: heparin as a disruptor of the SARS-CoV-2 interaction with its host cell receptor. Anal. Chem. 2020, in press (doi: 10.1021/acs.analchem.0c02449). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Loke I; Østergaard O; Heegaard NHH; Packer NH; Thaysen-Andersen M Paucimannose-Rich N-glycosylation of Spatiotemporally Regulated Human Neutrophil Elastase Modulates Its Immune Functions. Mol. Cell. Proteomics 2017, 16, 1507–1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Zhang X; Chen L; Bancroft DP; Lai CK; Maione TE Crystal structure of recombinant human platelet factor 4. Biochemistry 1994, 33, 8361–8366. [DOI] [PubMed] [Google Scholar]
- (43).Niu C; Yang Y; Huynh A; Nazy I; Kaltashov IA Platelet factor 4 interactions with short heparin oligomers: implications for folding and assembly. Biophys. J. 2020, 118, 1371–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Sun N; Soya N; Kitova E; Klassen J Nonspecific interactions between proteins and charged biomolecules in electrospray ionization mass spectrometry. J. Am. Soc. Mass Spectrom. 2010, 21, 472–481. [DOI] [PubMed] [Google Scholar]
- (45).Minsky BB; Abzalimov RR; Niu C; Zhao Y; Kirsch Z; Dubin PL; Savinov SN; Kaltashov IA Mass Spectrometry Reveals a Multifaceted Role of Glycosaminoglycan Chains in Factor Xa Inactivation by Antithrombin. Biochemistry 2018, 57, 4880–4890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Lelliott PM; Momota M; Shibahara T; Lee MSJ; Smith NI; Ishii KJ; Coban C Heparin induces neutrophil elastase-dependent vital and lytic NET formation. Int. Immunol. 2020, 32, 359–368. [DOI] [PubMed] [Google Scholar]
- (47).Henriksen J; Ringborg LH; Roepstorrf P On-line size-exclusion chromatography/mass spectrometry of low molecular mass heparin. J. Mass Spectrom. 2004, 39, 1305–1312. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


