Abstract
Stabilization of the metastable envelope glycoprotein (Env) of HIV-1 is hypothesized to improve induction of broadly neutralizing antibodies. We improved the expression yield and stability of the HIV-1 envelope glycoprotein BG505SOSIP.664 gp140 by means of a previously described automated sequence and structure-guided computational thermostabilization approach, PROSS. This combines sequence conservation information with computational assessment of mutant stabilization, thus taking advantage of the extensive natural sequence variation present in HIV-1 Env. PROSS is used to design three gp140 variants with 17–45 mutations relative to the parental construct. One of the designs is experimentally observed to have a fourfold improvement in yield and a 4 °C increment in thermostability. In addition, the designed immunogens have similar antigenicity profiles to the native flexible linker version of wild type, BG505SOSIP.664 gp140 (NFL Wt) to major epitopes targeted by broadly neutralizing antibodies. PROSS eliminates the laborious process of screening many variants for stability and functionality, providing a proof of principle of the method for stabilization and improvement of yield without compromising antigenicity for next generation complex, highly glycosylated vaccine candidates.
Keywords: Rosetta, Protein stability, Protein design, Vaccine, Immunogen, Virus, Automated
Graphical abstract
Highlights
-
•
One-step stabilization of HIV-1 Env gp140.
-
•
One-step yield improvement of HIV-1 Env gp140.
-
•
Native-like oligomeric conformation of designed vaccine candidates.
-
•
Unaltered antigenicity of designed vaccine candidates.
1. Introduction
The functional envelope (Env) glycoprotein of HIV-1 is a trimer, with each monomer comprising two subunits, gp120 and gp41. Env is the principal target of the humoral immune response, making it an important vaccine candidate (Burton et al., 2012, Kwong et al., 2011). Despite extensive efforts spanning nearly three and half decades, however, an HIV-1 vaccine is still not available. The RV144 clinical trial conducted a decade ago reported ~31% efficacy with an ALVAC-AIDSVAX B/E gp120 prime-boost regime (Rerks-Ngarm et al., 2009), renewing hope in a universal HIV-1 vaccine candidate. Until recently, however, the lack of a near-native like structure of Env stalled the progress of rational vaccine design. The structure elucidation of the Env ectodomain derivative, BG505SOSIP.664 gp140 (PDB: 4TVP), reinvigorated interest in rational, structure-based protein engineering and a number of complexes of the Env ectodomain with broadly neutralizing antibodies have facilitated immunogen design efforts (Aldon et al., 2018, Bianchi et al., 2018, de Taeye et al., 2018, de Taeye et al., 2015, Garces et al., 2015, Georgiev et al., 2015, Gristick et al., 2016, Guenaga et al., 2015, Havenar-Daughton et al., 2016, Joyce et al., 2017, Klasse et al., 2018, Kulp et al., 2017, Liu et al., 2017, Pancera et al., 2014, Rutten et al., 2018, Sanders et al., 2015, Sharma et al., 2015, Torrents de la Pena et al., 2017, Xu et al., 2018, Yang et al., 2018, Zhang et al., 2018).The Env ectodomain truncated at residue 664 in gp41 incorporates two cysteine mutations in gp120 (A501C) and gp41 (T605C) termed SOS to covalently link the two subunits, and an additional mutation (Ile559Pro), termed IP, in gp41 to stabilize the ectodomain in its prefusion state. This design, derived from the clade BG505 isolate, is termed BG505SOSIP.664 (or SOSIP) (Pancera et al., 2014). A single-chain derivative of the Env ectodomain lacking SOS and with the furin cleavage site (R6) replaced with a ten amino acid flexible linker (G4S)2 that shows similar properties as SOSIP is termed BG505 NFL (Native Flexibly Linked or NFL) (Sharma et al., 2015). Stabilization of difficult to neutralize (tier-2) viral Env ectodomain gp140 derivatives from diverse clades (A, B, C, G and CRF) including BG505SOSIP.664 has been attempted by elaborate structure-guided rational methods (Stewart-Jones et al., 2016, Boliar et al., 2015, Joyce et al., 2017, Rutten et al., 2018). These involved incorporation of numerous disulphides (intra, inter subunit and inter protomer) (Aldon et al., 2018, de Taeye et al., 2015, de Taeye et al., 2018, Joyce et al., 2017, Sanders et al., 2013, Sanders et al., 2015, Torrents de la Pena et al., 2017, Yang et al., 2018), screening of pre-fusion stabilizing mutations (Kesavardhana and Varadarajan, 2014, Aldon et al., 2018), better re-packing of cavities with large hydrophobic residues (de Taeye et al., 2018, de Taeye et al., 2015), reducing/eliminating the binding to CD4 receptor (Kulp et al., 2017, Zhang et al., 2018), and screening of high yielding variants by sequence and structure guided transfer of stabilizing residues from BG505SOSIP.664 (Leaman and Zwick, 2013, Steichen et al., 2016, Sullivan et al., 2017, Guenaga et al., 2015, Rutten et al., 2018). In addition, alternative immunogen design approaches have been applied to address the issue of the large global HIV-1 Env diversity, comprising diverse clades and circlating recombinant forms, so as to induce better cell mediated Potential T cell Epitopic (PTE) responses by the design of mosaic Env gp140 (Mos M), and to improve humoral responses by the design of consensus group M Env gp140 (Con-S) (Barouch et al., 2010, Eugene et al., 2013, Hulot et al., 2015, Liao et al., 2013). The reported MosM gp140 and ConS/ConM gp140 Envs were not stabilized versions and hence some versions had low yields (Aldon et al., 2018, Sliepen et al., 2019, Barouch et al., 2010, Nkolola et al., 2014, Eugene et al., 2013, Hulot et al., 2015, Liao et al., 2013). Recently, stabilization of membrane anchored and soluble uncleaved prefusion optimized (UFO) versions of consensus HIV-1 Env trimer was attempted, for example in ConSOSL UFO.716 (Aldon et al., 2018) though introduction of consensus mutations into a WT background, did not result in improvement of either stability or yield (Sliepen et al., 2019). Stabilization of consensus group M HIV-1 Env ectodomain (ConM) in the SOSIP.664 background was performed by the transfer of additional previously identified stabilizing mutations from BG505SOSIP.664 stabilized versions into ConM SOSIP.664 (Sliepen et al., 2019). Another interesting design strategy to focus immune responses is by either grafting epitopes onto heterologous protein scaffolds or stabilizing designed fragments of the protein of interests (Correia et al., 2010, Azoitei et al., 2011, Bhattacharyya et al., 2013, Jardine et al., 2013, Rathore et al., 2018).
Immunization studies carried out in rabbits and macaques with BG505SOSIP.664 as well as derivatives of SOSIP or NFL trimers from diverse clades have so far largely resulted in autologous tier-2 neutralizing antibodies with very weak and sporadic occurrences of heterologous tier-2 neutralizing antibodies (Feng et al., 2016, Havenar-Daughton et al., 2016, Kulp et al., 2017, Sanders et al., 2015, Schiffner et al., 2018, Torrents de la Pena et al., 2017, Zhang et al., 2018, Sliepen et al., 2019). This autologous neutralizing antibody response is possibly due to the inherent metastability and dynamic flexibility of the Env protein (Guttman et al., 2014, Munro et al., 2014) as well as particular features, such as a glycan hole present in the Env of the specific BG505 isolate that has been used for most immunogen designs (Stewart-Jones et al., 2016, Klasse et al., 2018). Thus, an optimized version of the Env with reduced flexibility, higher thermo-tolerance and yield, that displays the major neutralizing antibody epitopes while masking non-neutralizing epitopes will likely be a superior vaccine candidate. BG505SOSIP.664 Env immunogens and similar derivatives from major clades have been reported to adopt native-like trimer conformations in both cleaved (requiring furin cleavage between gp120 and gp41) and cleavage-independent Native Flexibly Linked (NFL) platforms (Guenaga et al., 2017, Guenaga et al., 2015, Sharma et al., 2015). Chemical crosslinking strategies designed to rigidify Env did not result in an improvement in heterologous response towards HIV-1 (Feng et al., 2016, Schiffner et al., 2018). In the present study, we have utilized the PROSS (Protein Repair One Stop Shop) protein stability-design algorithm to improve the thermal stability and production yield of the BG505SOSIP.664 Env ectodomain gp140 (Campeotto et al., 2017, Goldenzweig et al., 2016). This method was previously used to improve protein-production yields and stabilize a number of other proteins by introducing dozens of stabilizing mutations and eliminates the use of extensive mutational screening for stability, yield and binding to major neutralizing antibodies. Importantly, however, our study represents the first example of using such an automated stability design method to such a complex glycoprotein such as gp140.
2. Material and methods
2.1. Sequence curation, atomistic Rosetta design
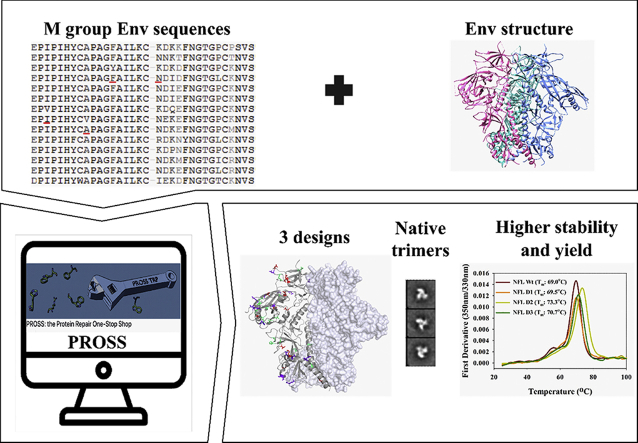
Envelope (Env) sequences from the HIV-1 M group were obtained from the 2014 version of the HIV LANL database and renumbered according to the HxB2 reference numbering scheme. First, a sequence alignment of 4174 non-redundant sequences, was retrieved from the HIVLANL database (Fig. 1A) from which we generated a Position specific Scoring Matrix (PSSM). Only sequences that had the identical loop length to BG505 were considered for generation of PSSM scores for the amino acid substitutions in the loops. PROSS proceeds in two steps: initially, PROSS filters out all single-point mutations for which the PSSM scores are <0 or which Rosetta atomistic modelling calculations predict to be destabilizing to predict the tolerated sequence space for the target protein. In the second step, the Rosetta sequence optimization calculations are conducted within the above tolerated sequence space. In this step, Rosetta combinatorial sequence design steps were interspersed with backbone and sidechain relaxation steps while maintaining C3 symmetry throughout all simulations (Goldenzweig et al., 2016, DiMaio et al., 2011).
Fig. 1.
A. Schematic of the PROSS computational stabilization algorithm. The approach involves alignment of sequences, filtering for residues with PSSM≥0, Rosetta screening of stabilizing residues (R.e.u: Rosetta energy units), combinatorial optimization of the Rosetta screened stabilizing residues leading to the prediction of three designs with most to least permissive energies in R.e.u). B. Structural mapping of resulting mutations on to BG505SOSIP.664 (PDBID: 4TVP) structure with Red, Violet and Green highlighted mutations exclusively pertaining to Design 1, 2 and 3 respectively.
2.2. Cloning of BG505SOSIP.664 designs and BG505SOSIP.664 NFL designs
The mutations resulting from the sequence and structure guided all atom Rosetta computational thermostabilization algorithm were introduced in both BG505SOSIP.664 gp140 and BG505SOSIP.664 NFL backgrounds. Sequences were codon optimized for human cell expression, and the resulting nucleotide sequences were synthesized and subcloned between the BglII and KpnI sites in the V1jns vector (Varadarajan et al., 2005) by GeneArt (Germany).
2.3. Purification of proteins
2.3.1. HEK293F protein purification
Transfections were performed according to the manufacturer's guidelines. Briefly, one day prior to transfection, cells were passaged at a density 1 × 106 cells/ml. On the day of transfection, cells were resuspended at 2.5 × 106 cells/ml in prewarmed fresh Freestyle 293F media. Desired plasmids (3 μg/ml of 293F cells) were transiently transfected into HEK293F cells using PEI (9 μg/ml of 293F cells). Twenty four hours post transfection, the medium volume was doubled with prewarmed Freestyle 293F media (Gibco). After an additional 72hr, supernatants were collected and proteins were affinity purified by using Lentil-Lectin Sepharose 4B (GE Healthcare). Supernatant was bound to a column equilibrated with PBS (pH7.4) supplemented with 1 mM EDTA or HBS (5 mM HEPES, 150 mM NaCl) (pH7.5) supplemented with 1 mM EDTA for negative stain EM sample preparation. A four column volume wash of PBS (pH7.4), supplemented with 500 mM NaCl and 1 mM EDTA or HBS (5 mM HEPES, 150 mM NaCl) (pH7.5) supplemented with 500 mM NaCl and 1 mM EDTA were given. Bound protein was eluted with 1M α-d-methylmannopyranoside in PBS (pH 7.4), supplemented with 1 mM EDTA or HBS (5 mM HEPES, 150 mM NaCl) (pH7.5) supplemented with 1 mM EDTA for negative stain EM sample preparation. The eluted fractions were pooled, and concentrated using a 100 kDa centricon Molecular Weight Cut-off (MWCO) (Amicon Ultra 15, Millipore). Concentrated protein was subjected to SEC purification on a Superdex 200 column (GE Healthcare) in PBS/HBS (pH 7.4). Trimer fractions were collected, pooled and concentrated using VIVASPIN 500 30 kDa MWCO microfuge concentrators (Sartorius, UK). Protein concentration was determined by absorbance (A280) using the theoretical molar extinction coefficient using the ProtParam tool (ExPASy).
2.3.2. Expi293F protein purification
Transfections were performed according to the manufacturer's guidelines. Briefly, one day prior to transfection cells, were passaged at a density of 2 × 106 cells/ml. On the day of transfection, cells were diluted to 2.5 × 106 cells/ml. Desired plasmids (1 μg/ml of Expi293F cells) were complexed with ExpiFectamine293 (2.6 μl/ml of Expi293F cells) and transiently transfected into Expi293F cells. Post 16hr, Enhancer 1 and Enhancer 2 were added according to the manufacturer's protocol. Seven days post transfection, culture supernatant was collected, filtered and proteins were purified as described above in HEK293F protein purification.
2.4. SDS-PAGE, BN-PAGE and Western blot analysis
SDS-PAGE and BN-PAGE (blue native-PAGE) were performed to determine the oligomeric states of proteins. All purified proteins were analysed under non-denaturing conditions using BN-PAGE. BN-PAGE, protein samples were mixed with sample buffer (250 mM MOPS, 250 mM Tris–HCL, 0.1% Coomassie brilliant blue G250 and 40% glycerol). Samples were loaded onto a 4–12% gradient bis-tris NuPAGE gel (Invitrogen). Electrophoresis was done at 4 °C for 3 h at 150 V. The cathode buffer comprised of 50 mM MOPS, 50 mM Tris (pH 7.7) and 0.002% Coomassie brilliant blue G250. The anode buffer contained 50 mM MOPS and 50 mM Tris (pH 7.7). SDS-PAGE was performed using an 8% polyacrylamide gel. Protein samples were denatured by boiling with sample buffer containing SDS. Samples were then loaded onto an 8% gel with and without DTT. For western blotting, following SDS-PAGE, proteins were electrophoretically transferred onto an Immobilon-P membrane (Millipore). After transfer, the membrane was blocked with 5% non-fat milk. The membrane was washed with PBST (PBS with 0.05% Tween) and incubated with anti-JRCSF gp120 polyclonal sera generated in rabbits, at 1:1000 dilution. The membranes were washed again with PBST and then incubated with anti-rabbit IgG conjugated to HRP (horseradish peroxidase) (Sigma). After washing with PBST, an enhanced chemiluminescence (ECL) method was used to develop the blot using HRP substrate and luminol in a 1:1 ratio (Biorad).
2.5. SEC-MALS (size exclusion chromatography – multi angle light scattering)
Briefly, a Superdex-200 10/300 GL analytical gel filtration column (GE healthcare) equilibrated in PBS (pH 7.4) buffer was utilized for characterizing BG505SOSIP, BG505SOSIP D1, D2, D3 and NFL Wt, D1, D2, and D3 using a SHIMADZU liquid chromatography FPLC system connected in-line with UV (SHIMADZU), refractive index (WATERS corp.) and MALS detectors (mini DAWN TREOS, Wyatt Technology corp.) for molecular weight characterization. UV, MALS and RI data were collected and analysed using ASTRA™ software (Wyatt Technology).
2.6. Sample preparation and EM imaging of negative stained samples
For negative stain electron microscopy, trimers were purified by lentil lectin affinity chromatography, following which trimer fractions were collected by size exclusion chromatography using a Superdex 200 (GE Healthcare) column pre-equilibrated with buffer containing 5 mM HEPES, 150 mM NaCl (pH 7.5).
The SEC purified samples were prepared for negative staining in the following conditions – (i) NFL Wt, (ii) designed trimer NFL D1, (iii) designed trimer NFL D2, (iv) designed trimer NFL D3. All samples were prepared for Transmission Electron Microscopy imaging by conventional negative staining method. Briefly, 3.5 μl of sample (for all the constructs) was applied to glow discharged carbon coated copper grid for 30 s followed by blotted out excess buffer and sample. Negative staining was performed using 2% uranyl acetate for 25 s. All negative stained samples were visualized and imaged at room temperature using Tecnai T12 electron microscope equipped with a LaB6 filament operated at 120 kV. All images were recorded using a side-mounted Olympus VELITA (2Kx2K) CCD camera at 2.54 Å/pixel on the specimen level.
2.7. 2D classification using single particle analysis
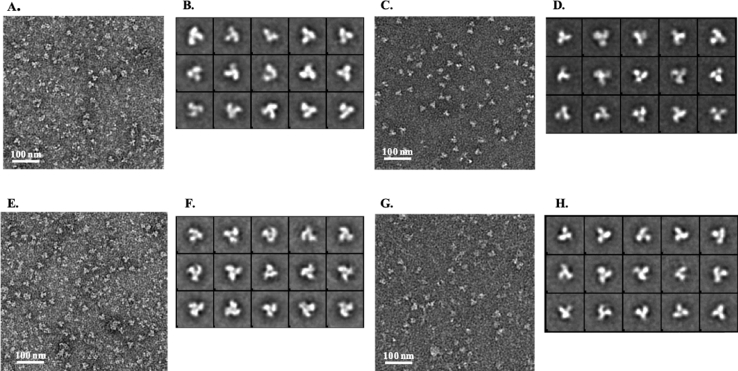
Reference free 2D alignment and classification of different projections of particles were done with EMAN 2.1 (Bell et al., 2016) and SIMPLE 2.1 (Reboul et al., 2018) software. Particles were picked manually for all four different samples and extracted using e2boxer.py in EMAN2.1 software. Total number of particles were extracted for NFL Wt, NFL D1, NFL D2, NFL D3 are 2715, 2638, 3181 and 2868 respectively. The reference-free 2D class averages were calculated using simple_prime2D in SIMPLE 2.1 software and 2D particle projections were classified into 50 classes (Representative 2D class averages are shown in Fig. 3).
Fig. 3.
Negative Stain Electron Micrographs and 2D Class averages of the designs. A, C, E and G. Raw micrograph of NFL Wt, NFL D1, NFL D2, NFL D3 respectively. B, D, F and H. Selected Class averages of NFL Wt, NFL D1, NFL D2, NFL D3 respectively.
2.8. nanoDSF studies
Thermal unfolding experiments of BG505SOSIP, BG505SOSIP D1, D2, D3 and NFL Wt, D1, D2, and D3 were carried out by nanoDSF (Prometheus NT.48) (Chattopadhyay & Varadarajan, 2019).The assays were carried out in triplicate with 1 μM of protein in the temperature range of 20–100 °C at 60% LED power and initial discovery scan counts (350 nm) ranging between 5000 and 10,000.
2.9. Fab preparation and purification
Briefly, to generate antigen binding fragment monomer (Fab), the IgG was cleaved at 37 °C overnight with 1:100 w/w of 2x crystalline Papain (Sigma) in the presence of 1 mM EDTA (Sigma), 10 mM Cysteine (Sigma) in 20 mM phosphate buffer (pH 6.3). The digestion reaction was quenched by adding 75 mM of freshly prepared iodoacetamide. The undigested IgG and IgG Fc fragments were affinity removed by incubation with ProetinG agarose resin by incubation at 4 °C kept at constant shaking. Beads were washed with 1X PBS to remove excess Fab. The resultant Fab was buffer exchanged with 1X PBS (pH 7.4).
2.10.1. SPR - Proteon XPR36 protein interaction array
The binding studies were carried out using the ProteOn XPR36 Protein Interaction Assay V.3.1 from Bio-Rad. The GLM sensor chip was activated by reaction with EDC (1-Ethyl-3-[3-dimethylaminopropyl] carbodiimide hydrochloride) and sulfo-NHS (N-hydroxysulfosuccinimide) (Sigma). Following this, 10 μg/ml of various bnAbs (VRC01, 2G12, PGDM1400, PGT145, PG9, PGT151,PGT128) and non-nAbs (17b, b6) were coupled in the presence of 10 mM sodium acetate buffer pH 4.5 at 30 μl/min for 100 s in various channels, leaving one channel as blank that acts as a reference. The Response Units for coupling were monitored till ~1500-2000RU was immobilized. Finally, the excess sulfo-NHS esters were quenched using 1M ethanolamine. NFL Wt, D1, D2 and D3 were passed at a flow rate of 30 μl/min for 300 s over the chip surface, followed by a dissociation step of 600 s. A lane without any immobilization but blocked with ethanolamine was also used to monitor non-specific binding. After each kinetic assay, the chip was regenerated in 4M MgCl2. Various concentrations of the NFL Wt, D1, D2 and D3 (250 nM, 125 nM, 62.2 nM, 31.1 nM, 15.6 nM) in 1X PBST were used for binding studies. The kinetic parameters were obtained by fitting the data to the simple 1:1 Langmuir interaction model using Proteon Manager.
2.10.2. SPR-binding of immobilized immunogens to fab analytes
The binding studies were carried out using the ProteOn XPR36 Protein Interaction Assay V.3.1 from Bio-Rad. The GLM sensor chip was activated by reaction with EDC (1-Ethyl-3-[3-dimethylaminopropyl] carbodiimide hydrochloride) and sulfo-NHS (N-hydroxysulfosuccinimide) (Sigma). Following this, 10 μg/ml of anti-His monoclonal antibody was coupled in the presence of 10 mM sodium acetate buffer pH 4.0 at 30 μl/min for 100 s in various channels, leaving one reference channel blank. The Response Units (RU) for coupling were monitored till ~3500-4000RU was immobilized. Finally, the excess sulfo-NHS esters were quenched using 1M ethanolamine. C-termial Octa-His tagged designs: NFL Wt, D1, D2 and D3 were captured onto immobilized anti-His monoclonal antibody at ~800–900 RU at a flow rate of 30 μl/min. Purified VRC01 Fab, and PGT145 Fab were passed as analytes at a flow rate of 30 μl/min for 300 s over the chip surface, followed by a dissociation step of 600 s. A lane without any immobilization but blocked with ethanolamine was also used to monitor non-specific binding. After each kinetic assay, the chip was regenerated in 4M MgCl2 and reimmobilized with NFL Wt, designs. Various concentrations of the VRC01 Fab (145 nM, 72.5 nM, 36 nM, 18 nM, 9 nM) and PGT145 Fab (320 nM, 160 nM, 80 nM, 40 nM, 20 nM) in 1X PBST were used for binding studies. The kinetic parameters were obtained by fitting the data to the simple 1:1 Langmuir interaction model using Proteon Manager.
3. Results
3.1. PROSS design of thermostabilizing variants of HIV-1 Env glycoprotein
The rich, diverse and extensive sequence information available for HIV-1 Env from multiple subtypes makes it an attractive target for sequence-based stabilization. Additionally, the recent crystal structure of HIV-1 envelope of clade A BG505, BG505SOSIP.664 (PDB: 4TVP) provides atomic and structural constraints for PROSS stability design.
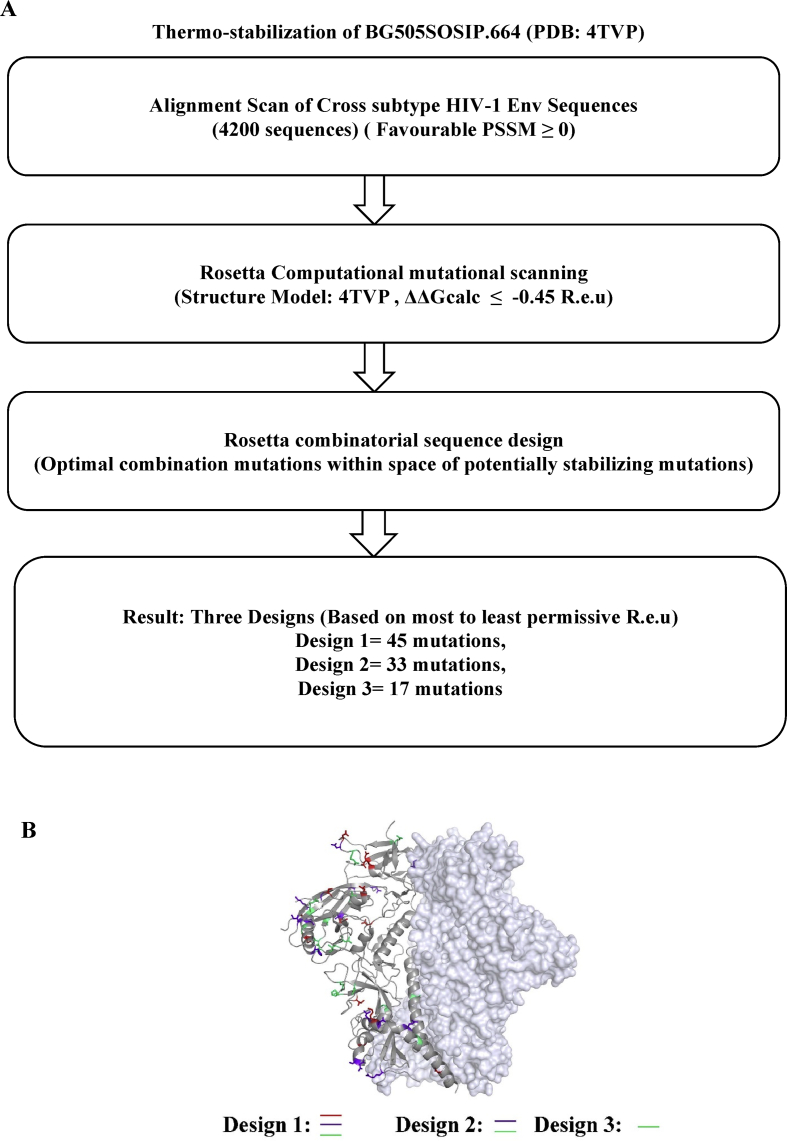
Based on the PROSS calculations, three potentially stabilizing designs were shortlisted for further experimental testing. These designs have the following number of mutations: 45, 33 and 17 in Design 1 (D1), Design 2 (D2) and Design 3 (D3) respectively (Fig. 1A and B). These PROSS designs containing the maximum (D1: 45) to minimum number (D3: 17) of mutations correspond to the highest to lowest Rosetta stabilizing energy. The PROSS designs utilized diverse amino acid substitutions ranging from the first to sixth most frequently occurring residues with the most frequently occurring amino acid accounting for ~60% of the substitutions (Supplementary Table 2).
We visually inspected the designs to verify that they did not include any mutations and potential N-linked glycosylation sites (PNGS) in the epitope of VRC01 (CD4 binding site bnAb). The PGT121 class supersite glycan (N332) was also retained as in BG505SOSIP.664. The mutations were observed to be distributed across the entire Env ectodomain, covering regions of the gp120 subunit (C1, V1/V2 loop, V4 loop) and gp41 subunits (N-heptad repeat (NHR) and C-heptad repeat (CHR)) (Fig. 1B, Table 1). The mutations in the loop between NHR and CHR were eliminated as there was missing density in the solved crystal structure (PDB: 4TVP). Also, two residues that incorporated additional glycosylation sites at 334 and 362 were eliminated after visualizing the mutations.
Table 1.
Mutations incorporated in the designs. Numbering is with respect to the HxB2 reference. The corresponding residue in BG505SOSIP.664 (Wt), the gp120 subunit residues range 31–507 and gp41 subunit residues range 512–664, secondary structural element, and residue accessibility information are indicated. The residues with % accessibility ≥10 are considered exposed and % accessibility <10 are considered buried. Mutations to the consensus (most frequently occurring) residue are underlined in bold.
| HxB2 Number | BG505SOSIP.664 Wt Residue |
Design 1 | Design 2 | Design 3 | Secondary Structural Region of Env | Accessibility |
|---|---|---|---|---|---|---|
| 84 | I | V | – | – | β0 | Exposed |
| 92 | E | N | N | N | β1 | Exposed |
| 130 | Q | N | N | N | A | Exposed |
| 140 | D | N | – | – | V1-hypervariable | Exposed |
| 141 | D | N | N | – | V1-hypervariable | Exposed |
| 151 | R | N | N | N | V1-hypervariable | Exposed |
| 154 | L | M | – | – | B V1 loop | Buried |
| 161 | M | I | V | – | B V2 loop | Buried |
| 178 | R | K | – | – | C V2 loop | Exposed |
| 211 | E | D | – | – | Exposed | |
| 229 | K | N | N | N | β5 | Exposed |
| 240 | P | K | K | K | β6 | Exposed |
| 241 | S | N | N | N | β7 | Exposed |
| 270 | V | I | I | I | Buried | |
| 277 | I | F | F | F | loop D | Exposed |
| 285 | L | I | I | I | β11 | Buried |
| 290 | T | E | K | K | Exposed | |
| 293 | Q | E | E | E | β12 | Exposed |
| 335 | K | R | R | – | α2 | Exposed |
| 336 | A | T | – | – | α2 | Exposed |
| 337 | T | Q | K | – | α2 | Exposed |
| 343 | G | K | K | K | α2 | Exposed |
| 344 | K | R | – | – | α2 | Exposed |
| 352 | H | Y | Y | – | α2 | Exposed |
| 358 | I | T | T | – | β14 | Exposed |
| 360 | R | I | I | I | β14 | Exposed |
| 369 | L | P | – | – | α3 | Exposed |
| 373 | T | M | M | M | α3 | Exposed |
| 388 | S | T | – | – | α4 | Exposed |
| 389 | G | – | Q | – | α4 | Exposed |
| 440 | Q | A | E | – | Exposed | |
| 442 | V | K | K | – | Exposed | |
| 446 | V | T | T | – | β22 | Exposed |
| 447 | S | A | – | – | β22 | Exposed |
| 500 | R | K | K | – | Exposed | |
| 531 | G | A | – | – | α6 | Exposed |
| 543 | N | Q | Q | Q | α6 HR1 | Exposed |
| 583 | V | M | M | M | α7 HR1 | Buried |
| 618 | N | S | S | – | α8 | Exposed |
| 620 | S | D | D | – | α8 HR2 | Exposed |
| 629 | L | M | M | – | α9 HR2 | Exposed |
| 632 | D | E | E | – | α9 HR2 | Exposed |
| 633 | K | R | – | – | α9 HR2 | Exposed |
| 640 | Q | N | N | – | α9 HR2 | Exposed |
| 644 | G | N | N | N | α9 HR2 | Exposed |
| 658 | Q | R | – | – | α9 HR2 | Exposed |
| Total No. of mutations | 45 | 33 | 17 |
These designs were tested against two backgrounds, namely the cleavage-competent version of gp140, which requires furin to cleave at the “RRRRRR” (R6) cleavage site between gp120 and gp41, and the native flexible linker (NFL) version, in which R6 is replaced by a (G4S)2 linker (Sharma et al., 2015). In addition, the SOS disulfide (A501C, T605C) mutations were retained in the NFL designs. Retention of SOS does not affect the conformation of the soluble or the membrane-bound-full-length Env even in the presence of the (G4S)2 linker (Aldon et al., 2018). The nomenclature of the designs is as follows: Wt, D1, D2, D3 for the designs in SOSIP.664 background and NFL Wt, NFL D1, NFL D2, NFL D3 for designs in native flexible linker (NFL) background. Most preliminary characterizations were performed in both cleavage-competent and NFL backgrounds. The NFL based platform was chosen for further biophysical and antigenic characterization of the designs as it avoids an additional furin-cleavage process between gp120 and gp41.
3.2. PROSS designs improved yield, increased stability and did not alter trimer formation
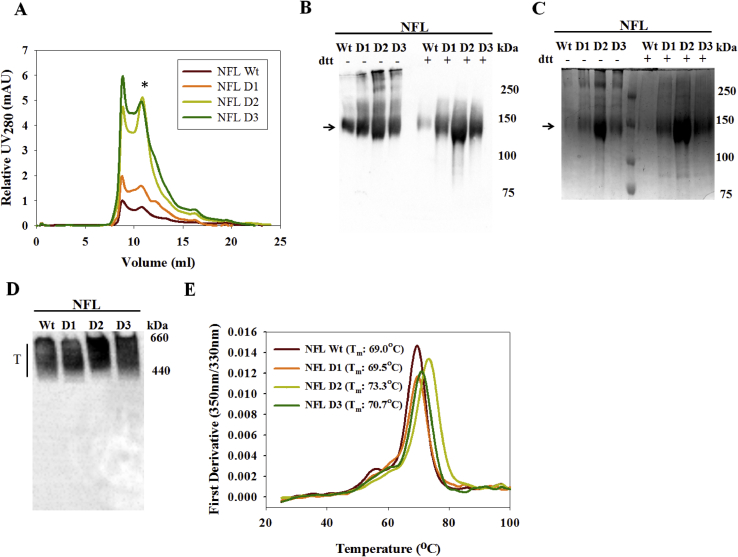
Trimer fractions of the NFL designs were purified from transiently transfected 293F or Expi293F suspension cell culture supernatants by a two-step process involving lentil lectin affinity purification followed by SEC purification. The trimer fraction was higher for NFL D2 and D3 as compared to NFL D1 and NFL Wt (Fig. 2A). The purity of the SEC purified trimers was confirmed by reducing and non-reducing SDS-PAGE and Western blot analysis (Fig. 2B and C). NFL D2 had the highest yield with nearly four-fold improvement compared to NFL Wt, as evident from SDS-PAGE and Western blot analysis (Fig. 2). NFL D1 and NFL D3 also exhibited up to two fold improvement in their yield. The total yields of the trimers purified from 293F cells were estimated to be ~0.5 mg/L, ~0.8 mg/L, ~2.0 mg/L and ~1.2 mg/L for the NFL Wt, NFL D1, NFL D2 and NFL D3 respectively. The average yields of SEC purified trimers from Expi293F for NFL Wt, NFL D1, NFL D2 and NFL D3 are 23.8 ± 1.9 mg/L, 25.2 ± 2.0 mg/L, 42.0 ± 3.3 mg/L and 26.7 ± 2.1 mg/L respectively.
Fig. 2.
Biophysical characterization of NFL designs. A: Relative UV280 chromatograms of lentil lectin purified designs subjected to SEC purification on an S200 column. ∗ Trimer faction peak ~10.8 ml. B, C: Reducing (+) and non-reducing (−) SDS-PAGE western and SDS-PAGE Coomassie brilliant blue stained gels respectively. Arrow indicating the size of the monomeric band resulting from denaturation (~140 kDa). D: Blue Native PAGE western of the SEC purified trimer. E: nanoDSF thermal melt of the SEC purified trimers.
The oligomeric profile of the SEC purified trimeric immunogens was confirmed by BN-PAGE Western blot analysis (Fig. 2D). The molecular weights of these immunogens correspond to that of a trimer (MW: 480–620 kDa), consistent with previously reported results (Sharma et al., 2015). In addition, Multi Angle Light Scattering (SEC-MALS) of lentil lectin affinity purified immunogens confirmed the molecular weight of the putative trimer peak. Marginal variability in the molecular weight of each of the designs compared to NFL Wt can be due to the difference in their glycosylation profiles (Supplementary Figure 1B). The average molecular weight of trimer immunogens was estimated to be ~563 kDa (Supplementary Figure. 1B).
The thermal stability of the purified trimers was estimated by nanoDSF which confirmed an increment in Tm of NFL D2 with respect to NFL Wt of 4 °C (Fig. 2E). The other two designs were observed to have comparable stability as NFL Wt.
2D reference free class averages from negative staining transmission electron microscopy reveal the formation of tri-lobed propeller shaped trimers for all three designs, consistent with native-like trimer assembly (Fig. 3).
We conclude that the designed trimers fold properly into native like envelope trimers and exhibit improved yields. NFL D1 and NFL D3 did not show considerable improvement in stability but resulted in twofold improved yield. NFL Design 2 showed fourfold improvement in yield and a 4 °C increment in thermostability.
3.3. Antigenicity of the PROSS predicted designs
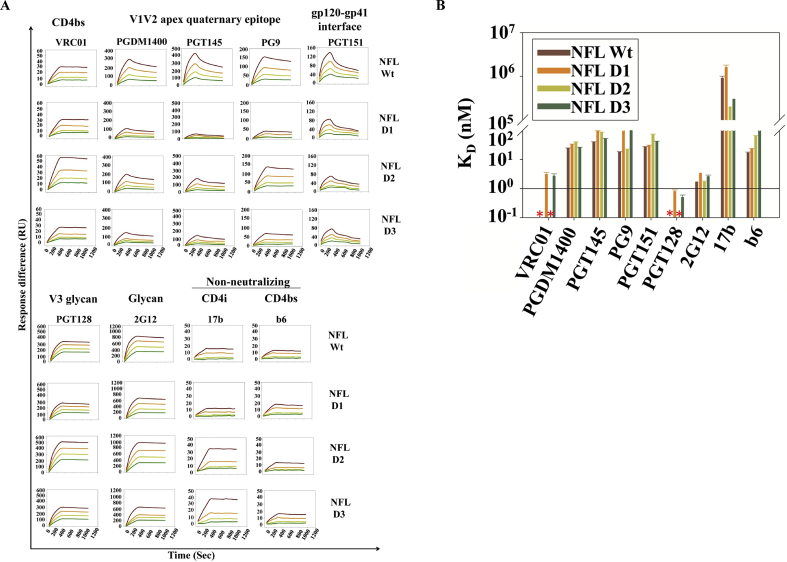
The antigenicity of the PROSS predicted envelope designs was tested against a panel of broadly neutralizing antibodies (bnAbs) and non-neutralizing antibodies (non-nAbs) covering most of the characterized epitopes on gp140. These antibodies include VRC01 (CD4 binding site (CD4bs)), PGDM1400 (quaternary CD4bs), PGT145 and PG9 (V1V2 quaternary apex), PGT128 (V3 apex N332 glycan dependent), PGT151 (gp120-gp41 interface), and 2G12 (envelope glycan). The non-nAbs include 17b (CD4i) and b6 (CD4 binding site (CD4bs)). The PROSS designed trimers were purified from transiently transfected Expi293F cells using the procedure mentioned in the previous section. The designs bind VRC01 to similar Response Units (RU) with NFL D2 and NFL Wt showing no dissociation (Fig. 4A). Quaternary epitope antibody (PGDM1400, PGT145 and PG9) binding, which is considered the gold standard for native trimer formation, was observed for all the designs with NFL D2 showing the maximum binding compared to the other designs but ~1.5–2 fold lower binding affinity compared to NFL Wt (Fig. 4A and B, Supplementary Table 1). The NFL designs were observed to bind PGT128 and 2G12 with negligible dissociation (Fig. 4A). NFL D2 design showed the highest binding to both the above antibodies. The gp120-gp41 interface recognising bNAb PGT151 binding was observed for all the designs but NFL D2 showed twofold reduction in binding compared to NFL Wt (Fig. 4B, Supplementary Table 1). Additionally, visual inspection of the gp140-PGT151 complex (PDB ID: 5FUU, 6MAR) suggested that mutations in the epitopic residues 84, 543, 640 (Table 1) led to reduction in PGT151 binding to the designs. The designs showed binding approximately comparable to Wt to both the non-neutralizing antibodies 17b and b6. Encouragingly, D2 showed the lowest binding to b6 and the highest binding to VRC01 of all the four proteins tested.
Fig. 4.
Proteon XPR36 binding studies with panel of bnAbs and nNAb. A) The binding studies performed with well characterized broadly neutralizing antibodies (bNAbs) against CD4bs:VRC01, V1V2 apex: PGDM1400, PGT145 and PG9, intersubunit gp120:gp41 interface PGT151, V3 glycan apex: PGT128, Glycan dependent: 2G12 and non-neutralizing antibodies against CD4i: 17b and CD4bs: b6. Each of the antibodies were immobilized at ~1500–2000 RU for performing binding studies. The trimeric analyte concentrations of each design used are 250 nM, 125 nM, 62.2 nM and 31.1 nM (Curves from top to bottom respectively). B) KD (nM) of NFL Wt and designs for each of bNAbs and non-NAbs, ∗ ND: No dissociation, hence KD could not be determined.
The apparent differences in binding of a given antibody to different immunogens might be due to either avidity effects or changes in immunogen molecular weight because of glycosylation. To rule out these possibilities, in two cases (VRC01 and PGT145) binding was also measured by immobilizing equivalent RUs of NFL Wt and designed trimers onto an anti-His coupled GLM chip and measuring the kinetics of binding to corresponding Fab analytes. VRC01 and PGT145 Fab were chosen as they target two different epitopes in the native trimer. While VRC01 targets the conserved non-contiguous CD4 binding site, PGT145 targets the quaternary epitope at the apex of the native trimer. We observe that the overall trends in binding and kinetic data remained similar with the monovalent Fab, relative to when the bivalent antibody was used as ligand (compare VRC01 and PGT45 binding data in Fig. 4A and Supplementary Figure 2).
We conclude that antigenicity was well retained with respect to the major bNAbs, with design D2 performing the best in terms of antigenicity.
4. Discussion
We have attempted to stabilize BG505SOSIP.664 gp140 Env by a novel method PROSS, which utilizes sequence conservation analysis and atomistic Rosetta computational design to stabilize proteins (Goldenzweig et al., 2016). PROSS predicted three designs that were experimentally observed to form native-like trimers with substantially improved yield and increased or unchanged thermostability, comparing favourably to previous studies to thermostabilize Env in which yield was compromised (Torrents de la Pena et al., 2017). The PROSS designs retained binding to a wide range of bnAbs, confirming that the major neutralizing epitopes were maintained despite the large number of mutations. The NFL version of the Env glycoprotein is shown to form native-like Env trimers (Sharma et al., 2015). We note that the present designs incorporated an increased number of asparagines relative to NFL Wt (D1:9, D2:8, D3:6). This is most likely due to the overrepresentation of asparagines in the sequence dataset, arising from the fact that HIV-1 Env uses N-linked glycosylation sites to evade the immune response. The glycan hole present in Clade A BG505 was absent in the designs because of incorporation of the N241 PNGS. This hole results in the elicitation of autologous neutralizing antibodies in rabbits and macaques (Stewart-Jones et al., 2016, Klasse et al., 2018). Therefore, we speculate that the PROSS designed variants may eliminate such isolate specific protein-interaction surfaces, resulting in a broader protective antibody response. The present designs retain binding to trimer-specific bnAbs (PDGM1400, PGT145, PG9) targeting the Cap/apex region as well as to the gp120-gp41 interface trimer specific bnAb PGT151. The NFL based platform was previously shown to have reduced binding affinity to PGT151 compared to BG505SOSIP.664 due to the presence of a linker that sterically compromises its binding (Georgiev et al., 2015, Sharma et al., 2015). Minimal binding to 17b and b6 non-nAbs was observed for NFL D1 and NFL D3 compared to NFL D2. NFL D1 did not have an increment in thermostability but exhibited twofold improvement in yield and showed the least binding to 17b. Unlike previous studies the present thermostabilization methodology did not involve extensive screening of variants (Julien et al., 2015, Guenaga et al., 2015, Joyce et al., 2017, Kulp et al., 2017, Leaman and Zwick, 2013, Rutten et al., 2018, Sullivan et al., 2017, Torrents de la Pena et al., 2017, Zhang et al., 2018). In addition, this method does not involve the incorporation of additional non-native disulphide bonds that might result in reduction in overall yield (Joyce et al., 2017, Torrents de la Pena et al., 2017, Zhang et al., 2018) –and have the potential to lead to misfolded conformations with non-native disulphide pairing. There have been past attempts to improve the breadth and protective efficacy of antibodies elicited by HIV-1 immunogens using mosaic and consensus sequence approaches. However, the inherently flexible consensus group M Env gp140 (Con M/Con-S gp140) has a low trimer yield (Sliepen et al., 2019). Con-S Env gp140 induced higher binding antibody titers in guinea pigs and rhesus macaques compared to the transmited-founder (T/F) clade B, C and consensus Env of clades A, B and C (Meyerhoff et al., 2017, Hulot et al., 2015, Liao et al., 2013). Additionally, the resultant antisera from guinea pigs and rhesus macaques was weakly protective against tier-2 virus isolates (Eugene et al., 2013, Liao et al., 2013). Stabilization of the ConM gp140 by the transfer of a dozen stabilizing mutations known from previous stabilization efforts of BG505SOSIP.664 led to ~5 fold improvement of ConM gp140 yield (Sliepen et al., 2019). As discussed above mosaic Env immunogens are an alternative approach to induce improved breadth of protection. Some previously designed mosaic MosM Env trimers have low stability and expression, particularly in MosM 3.2 and Mos M 3.3 backgounds (Nkolola et al., 2014). It is not possible to comment on the yields of the reported Mosaic Envs (Mos M) and previous ConS gp140s due to absence of literature (Barouch et al., 2010, Eugene et al., 2013, Liao et al., 2013, Nkolola et al., 2014, Hulot et al., 2015, Meyerhoff et al., 2017). Our approach, using the consensus group M sequence and atomistic Rosetta design has led to significant yield improvement and a small amount of thermostabilization, without the laborious process of first designing and validating individual putative stabilizing mutations. In contrast to the traditional consensus design approach that utilizes the consensus sequence alone, PROSS designs utilized mutations ranging from first to sixth most frequent residues that would not be selected by traditional approaches. PROSS designs incorporated consensus mutations at only ~60% of the mutated sites, consistent with earlier observations that only ~50–60% of the consensus residues are stabilizing (Magliery, 2015, Steipe et al., 1994, Sternke et al., 2019). The present designs will next be characterized for immunogenicity in small animals.
HIV-1 Env glycoprotein is a very challenging target for thermostabilization. It is a flexible, highly glycosylated protein with multiple disulfides. To date, a general, automated method for stabilizing Env glycoprotein, the major target of the humoral immune response in HIV-1 was lacking. With structures of major clades of HIV-1 Env ectodomains being solved, the PROSS stabilization approach can be utilized to produce optimized versions of other major clades of HIV-1 Env glycoprotein immunogens based on the NFL platform with improved yields (Bartesaghi et al., 2013, Julien et al., 2015, Pancera et al., 2014, Stewart-Jones et al., 2016). These optimized immunogens can be further tested for improved breadth of the humoral immune response based on multivalent cocktail/serial immunization regimes and also be used as B cell probes for isolating novel bnAbs. In addition, these mutations can be additionally combined with other stabilizing mutations (Aldon et al., 2018, de Taeye et al., 2018, de Taeye et al., 2015, Guenaga et al., 2015, Joyce et al., 2017, Kesavardhana and Varadarajan, 2014, Kulp et al., 2017, Leaman and Zwick, 2013, Rutten et al., 2018, Sanders et al., 2013, Sanders et al., 2015, Steichen et al., 2016, Sullivan et al., 2017, Torrents de la Pena et al., 2017, Yang et al., 2018, Zhang et al., 2018). Recently, PROSS was used to stabilize the malarial antigen protein PfRH5 (Plasmodium falciparum reticulocyte binding protein homolog 5) (Campeotto et al., 2017). The present work provides the first proof of principle that the method can also be used to optimize one of the most challenging viral glycoprotein targets to date.
5. Conclusion
The highly complex and metastable envelope glycoprotein of HIV-1 was optimized by a one-step sequence and structure guided protein stability design algorithm PROSS. The optimized envelope glycoproteins formed native like trimer with substantially improved yields and improved or similar antigenicity to broadly neutralizing antibodies as the wild type protein. Our work highlights the utility of this one step automated protein design approach to enhance protein expression, stability and yield for one of the most challenging and complex viral glycoprotein targets to date.
CRediT authorship contribution statement
Sameer Kumar Malladi: Investigation, Methodology, Validation, Writing - original draft, Writing - review & editing. David Schreiber: Investigation, Methodology, Validation. Ishika Pramanick: Investigation, Methodology. Malavika Abhineshababu Sridevi: Investigation. Adi Goldenzweig: Investigation, Methodology, Validation. Somnath Dutta: Investigation, Supervision, Writing - review & editing. Sarel Jacob Fleishman: Conceptualization, Funding acquisition, Supervision, Writing - review & editing. Raghavan Varadarajan: Conceptualization, Funding acquisition, Supervision, Writing - review & editing.
Declaration of Competing Interest
None declared
Acknowledgements
We thank the Neutralizing Antibody Consortium of IAVI and the NIH AIDS Research and Reference Program for various HIV-1 directed monoclonal antibodies. This work was funded by the UGC-Israel Science Funding (UGC-ISF) (grant number: F.No. 6-3/2016(IC) UC/SE), and a grant from National Institutes of Health (grant number R01AI118366-01, DT.15/7/2015) to RV. Research in the Fleishman lab was additionally supported by a generous donation from Sam Switzer and family. SD and RV thank DBT-IISc partnership programme for negative staining imaging T12 Tecnai microscope facility. We also acknowledge funding for infrastructural support from the following programs of the Government of India: DST-FIST, UGC Center for Advanced Study, MHRD-FAST, the DBT-IISc Partnership Program, and of a JC Bose Fellowship from DST to RV. S.K.M acknowledges the support of MHRD-IISc doctoral fellowship. We are thankful to Dr. Sivaramaiah Nallapeta (Head, Business Operations, NanoTemper Technologies) and Dr. Saji Menon (Application Specialist, NanoTemper Technologies) for use of their nanoDSF facility. We thank Dr. Likhesh Sharma (Project Managar at Cytiva (GE Healthcare)) for helping with troubleshooting the SPR kinetic data analysis. We also thank all the members of the R.V. and S.J.F labs for their valuable suggestions.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.crstbi.2020.04.001.
Contributor Information
Sarel Jacob Fleishman, Email: sarel.fleishman@weizmann.ac.il.
Raghavan Varadarajan, Email: varadar@iisc.ac.in.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Aldon Y., McKay P.F., Allen J., Ozorowski G., Felfodine Levai R., Tolazzi M., Rogers P., He L., de Val N., Fabian K., Scarlatti G., Zhu J., Ward A.B., Crispin M., Shattock R.J. Rational design of DNA-expressed stabilized native-like HIV-1 envelope trimers. Cell Rep. 2018;24:3324–3338 e5. doi: 10.1016/j.celrep.2018.08.051. http://www.ncbi.nlm.nih.gov/pubmed/30232012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azoitei M.L., Correia B.E., Ban Y.-E.A., Carrico C., Kalyuzhniy O., Chen L., Schroeter A., Huang P.-S., McLellan J.S., Kwong P.D., Baker D., Strong R.K., Schief W.R. Computation-guided backbone grafting of a discontinuous motif onto a protein scaffold. Science. 2011;334:373–376. doi: 10.1126/science.1209368. http://www.ncbi.nlm.nih.gov/pubmed/22021856 [DOI] [PubMed] [Google Scholar]
- Barouch D.H., O'Brien K.L., Simmons N.L., King S.L., Abbink P., Maxfield L.F., Sun Y.-H., La Porte A., Riggs A.M., Lynch D.M., Clark S.L., Backus K., Perry J.R., Seaman M.S., Carville A., Mansfield K.G., Szinger J.J., Fischer W., Muldoon M., Korber B. Mosaic HIV-1 vaccines expand the breadth and depth of cellular immune responses in rhesus monkeys. Nat. Med. 2010;16:319–323. doi: 10.1038/nm.2089. http://www.nature.com/articles/nm.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartesaghi A., Merk A., Borgnia M.J., Milne J.L.S., Subramaniam S. Prefusion structure of trimeric HIV-1 envelope glycoprotein determined by cryo-electron microscopy. Nat. Struct. Mol. Biol. 2013;20:1352–1357. doi: 10.1038/nsmb.2711. http://www.ncbi.nlm.nih.gov/pubmed/24154805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell J.M., Chen M., Baldwin P.R., Ludtke S.J. 2016. High resolution single particle refinement in EMAN2.1. Methods 100; pp. 25–34.http://www.ncbi.nlm.nih.gov/pubmed/26931650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya S., Singh P., Rathore U., Purwar M., Wagner D., Arendt H., DeStefano J., LaBranche C.C., Montefiori D.C., Phogat S., Varadarajan R. Design of an Escherichia coli expressed HIV-1 gp120 fragment immunogen that binds to b12 and induces broad and potent neutralizing antibodies. J. Biol. Chem. 2013;288:9815–9825. doi: 10.1074/jbc.M112.425959. http://www.ncbi.nlm.nih.gov/pubmed/23430741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi M., Turner H.L., Nogal B., Cottrell C.A., Oyen D., Pauthner M., Bastidas R., Nedellec R., McCoy L.E., Wilson I.A., Burton D.R., Ward A.B., Hangartner L. Electron-microscopy-based epitope mapping defines specificities of polyclonal antibodies elicited during HIV-1 BG505 envelope trimer immunization. Immunity. 2018;49:288–300 e8. doi: 10.1016/j.immuni.2018.07.009. http://www.ncbi.nlm.nih.gov/pubmed/30097292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boliar S., Das S., Bansal M., Shukla B.N., Patil S., Shrivastava T., Samal S., Goswami S., King C.R., Bhattacharya J., Chakrabarti B.K. An efficiently cleaved HIV-1 clade C Env selectively binds to neutralizing antibodies. PloS One. 2015;10 doi: 10.1371/journal.pone.0122443. http://www.ncbi.nlm.nih.gov/pubmed/25822521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton D.R., Ahmed R., Barouch D.H., Butera S.T., Crotty S., Godzik A., Kaufmann D.E., McElrath M.J., Nussenzweig M.C., Pulendran B., Scanlan C.N., Schief W.R., Silvestri G., Streeck H., Walker B.D., Walker L.M., Ward A.B., Wilson I.A., Wyatt R. A blueprint for HIV vaccine discovery. Cell Host Microbe. 2012;12:396–407. doi: 10.1016/j.chom.2012.09.008. http://www.ncbi.nlm.nih.gov/pubmed/23084910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campeotto I., Goldenzweig A., Davey J., Barfod L., Marshall J.M., Silk S.E., Wright K.E., Draper S.J., Higgins M.K., Fleishman S.J. One-step design of a stable variant of the malaria invasion protein RH5 for use as a vaccine immunogen. Proc. Natl. Acad. Sci. U. S. A. 2017;114:998–1002. doi: 10.1073/pnas.1616903114. http://www.ncbi.nlm.nih.gov/pubmed/28096331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyay G., Varadarajan R. Facile measurement of protein stability and folding kinetics using a nano differential scanning fluorimeter. Protein Sci. 2019;28:1127–1134. doi: 10.1002/pro.3622. http://www.ncbi.nlm.nih.gov/pubmed/30993730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correia B.E., Ban Y.-E.A., Holmes M.A., Xu H., Ellingson K., Kraft Z., Carrico C., Boni E., Sather D.N., Zenobia C., Burke K.Y., Bradley-Hewitt T., Bruhn-Johannsen J.F., Kalyuzhniy O., Baker D., Strong R.K., Stamatatos L., Schief W.R. Computational design of epitope-scaffolds allows induction of antibodies specific for a poorly immunogenic HIV vaccine epitope. Structure. 2010;18:1116–1126. doi: 10.1016/j.str.2010.06.010. http://www.ncbi.nlm.nih.gov/pubmed/20826338 [DOI] [PubMed] [Google Scholar]
- de Taeye S.W., de la Pena A.T., Vecchione A., Scutigliani E., Sliepen K., Burger J.A., van der Woude P., Schorcht A., Schermer E.E., van Gils M.J., LaBranche C.C., Montefiori D.C., Wilson I.A., Moore J.P., Ward A.B., Sanders R.W. Stabilization of the gp120 V3 loop through hydrophobic interactions reduces the immunodominant V3-directed non-neutralizing response to HIV-1 envelope trimers. J. Biol. Chem. 2018;293:1688–1701. doi: 10.1074/jbc.RA117.000709. http://www.ncbi.nlm.nih.gov/pubmed/29222332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Taeye S.W., Ozorowski G., Torrents de la Pena A., Guttman M., Julien J.P., van den Kerkhof T.L., Burger J.A., Pritchard L.K., Pugach P., Yasmeen A., Crampton J., Hu J., Bontjer I., Torres J.L., Arendt H., DeStefano J., Koff W.C., Schuitemaker H., Eggink D., Berkhout B. Immunogenicity of stabilized HIV-1 envelope trimers with reduced exposure of non-neutralizing epitopes. Cell. 2015;163:1702–1715. doi: 10.1016/j.cell.2015.11.056. http://www.ncbi.nlm.nih.gov/pubmed/26687358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiMaio F., Leaver-Fay A., Bradley P., Baker D., André I. Modeling symmetric macromolecular structures in Rosetta 3. PloS One. 2011;6 doi: 10.1371/journal.pone.0020450. http://www.ncbi.nlm.nih.gov/pubmed/21731614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eugene H.S., Pierce-Paul B.R., Cragio J.K., Ross T.M. Rhesus macaques vaccinated with consensus envelopes elicit partially protective immune responses against SHIV SF162p4 challenge. Virol. J. 2013;10:102. doi: 10.1186/1743-422X-10-102. http://www.ncbi.nlm.nih.gov/pubmed/23548077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y., Tran K., Bale S., Kumar S., Guenaga J., Wilson R., de Val N., Arendt H., DeStefano J., Ward A.B., Wyatt R.T. Thermostability of well-ordered HIV spikes correlates with the elicitation of autologous tier 2 neutralizing antibodies. PLoS Pathog. 2016;12 doi: 10.1371/journal.ppat.1005767. http://www.ncbi.nlm.nih.gov/pubmed/27487086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garces F., Lee J.H., de Val N., de la Pena A.T., Kong L., Puchades C., Hua Y., Stanfield R.L., Burton D.R., Moore J.P., Sanders R.W., Ward A.B., Wilson I.A. Affinity maturation of a potent family of HIV antibodies is primarily focused on accommodating or avoiding glycans. Immunity. 2015;43:1053–1063. doi: 10.1016/j.immuni.2015.11.007. http://www.ncbi.nlm.nih.gov/pubmed/26682982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgiev I.S., Joyce M.G., Yang Y., Sastry M., Zhang B., Baxa U., Chen R.E., Druz A., Lees C.R., Narpala S., Schon A., Van Galen J., Chuang G.Y., Gorman J., Harned A., Pancera M., Stewart-Jones G.B., Cheng C., Freire E., McDermott A.B. Single-chain soluble BG505.SOSIP gp140 trimers as structural and antigenic mimics of mature closed HIV-1 Env. J. Virol. 2015;89:5318–5329. doi: 10.1128/JVI.03451-14. http://www.ncbi.nlm.nih.gov/pubmed/25740988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldenzweig A., Goldsmith M., Hill S.E., Gertman O., Laurino P., Ashani Y., Dym O., Unger T., Albeck S., Prilusky J., Lieberman R.L., Aharoni A., Silman I., Sussman J.L., Tawfik D.S., Fleishman S.J. Automated structure- and sequence-based design of proteins for high bacterial expression and stability. Mol. Cell. 2016;63:337–346. doi: 10.1016/j.molcel.2016.06.012. http://www.ncbi.nlm.nih.gov/pubmed/27425410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gristick H.B., von Boehmer L., West A.P., Jr., Schamber M., Gazumyan A., Golijanin J., Seaman M.S., Fatkenheuer G., Klein F., Nussenzweig M.C., Bjorkman P.J. Natively glycosylated HIV-1 Env structure reveals new mode for antibody recognition of the CD4-binding site. Nat. Struct. Mol. Biol. 2016;23:906–915. doi: 10.1038/nsmb.3291. http://www.ncbi.nlm.nih.gov/pubmed/27617431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenaga J., Dubrovskaya V., de Val N., Sharma S.K., Carrette B., Ward A.B., Wyatt R.T. Structure-guided redesign increases the propensity of HIV Env to generate highly stable soluble trimers. J. Virol. 2015;90:2806–2817. doi: 10.1128/JVI.02652-15. http://www.ncbi.nlm.nih.gov/pubmed/26719252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenaga J., Garces F., de Val N., Stanfield R.L., Dubrovskaya V., Higgins B., Carrette B., Ward A.B., Wilson I.A., Wyatt R.T. Glycine substitution at helix-to-coil transitions facilitates the structural determination of a stabilized subtype C HIV envelope glycoprotein. Immunity. 2017;46:792–803 e3. doi: 10.1016/j.immuni.2017.04.014. http://www.ncbi.nlm.nih.gov/pubmed/28514686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M., Garcia N.K., Cupo A., Matsui T., Julien J.P., Sanders R.W., Wilson I.A., Moore J.P., Lee K.K. CD4-induced activation in a soluble HIV-1 Env trimer. Structure. 2014;22:974–984. doi: 10.1016/j.str.2014.05.001. http://www.ncbi.nlm.nih.gov/pubmed/24931470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havenar-Daughton C., Carnathan D.G., Torrents de la Pena A., Pauthner M., Briney B., Reiss S.M., Wood J.S., Kaushik K., van Gils M.J., Rosales S.L., van der Woude P., Locci M., Le K.M., de Taeye S.W., Sok D., Mohammed A.U.R., Huang J., Gumber S., Garcia A., Kasturi S.P. Direct probing of germinal center responses reveals immunological features and bottlenecks for neutralizing antibody responses to HIV Env trimer. Cell Rep. 2016;17:2195–2209. doi: 10.1016/j.celrep.2016.10.085. http://www.ncbi.nlm.nih.gov/pubmed/27880897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulot S.L., Korber B., Giorgi E.E., Vandergrift N., Saunders K.O., Balachandran H., Mach L.V., Lifton M.A., Pantaleo G., Tartaglia J., Phogat S., Jacobs B., Kibler K., Perdiguero B., Gomez C.E., Esteban M., Rosati M., Felber B.K., Pavlakis G.N., Parks R. Comparison of immunogenicity in rhesus macaques of transmitted-founder, HIV-1 group M consensus, and trivalent mosaic envelope vaccines formulated as a DNA prime, NYVAC, and envelope protein boost. J. Virol. 2015;89:6462–6480. doi: 10.1128/JVI.00383-15. http://www.ncbi.nlm.nih.gov/pubmed/25855741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jardine J., Julien J.-P., Menis S., Ota T., Kalyuzhniy O., McGuire A., Sok D., Huang P.-S., MacPherson S., Jones M., Nieusma T., Mathison J., Baker D., Ward A.B., Burton D.R., Stamatatos L., Nemazee D., Wilson I.A., Schief W.R. Rational HIV immunogen design to target specific germline B cell receptors. Science. 2013;340:711–716. doi: 10.1126/science.1234150. http://www.ncbi.nlm.nih.gov/pubmed/23539181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce M.G., Georgiev I.S., Yang Y., Druz A., Geng H., Chuang G.Y., Kwon Y.D., Pancera M., Rawi R., Sastry M., Stewart-Jones G.B.E., Zheng A., Zhou T., Choe M., Van Galen J.G., Chen R.E., Lees C.R., Narpala S., Chambers M., Tsybovsky Y. Soluble prefusion closed DS-SOSIP.664-Env trimers of diverse HIV-1 strains. Cell Rep. 2017;21:2992–3002. doi: 10.1016/j.celrep.2017.11.016. http://www.ncbi.nlm.nih.gov/pubmed/29212041 [DOI] [PubMed] [Google Scholar]
- Julien J.P., Lee J.H., Ozorowski G., Hua Y., Torrents de la Pena A., de Taeye S.W., Nieusma T., Cupo A., Yasmeen A., Golabek M., Pugach P., Klasse P.J., Moore J.P., Sanders R.W., Ward A.B., Wilson I.A. Design and structure of two HIV-1 clade C SOSIP.664 trimers that increase the arsenal of native-like Env immunogens. Proc. Natl. Acad. Sci. U. S. A. 2015;112:11947–11952. doi: 10.1073/pnas.1507793112. http://www.ncbi.nlm.nih.gov/pubmed/26372963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kesavardhana S., Varadarajan R. Stabilizing the native trimer of HIV-1 Env by destabilizing the heterodimeric interface of the gp41 postfusion six-helix bundle. J. Virol. 2014;88:9590–9604. doi: 10.1128/JVI.00494-14. http://www.ncbi.nlm.nih.gov/pubmed/24920800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klasse P.J., Ketas T.J., Cottrell C.A., Ozorowski G., Debnath G., Camara D., Francomano E., Pugach P., Ringe R.P., LaBranche C.C., van Gils M.J., Bricault C.A., Barouch D.H., Crotty S., Silvestri G., Kasturi S., Pulendran B., Wilson I.A., Montefiori D.C., Sanders R.W. Epitopes for neutralizing antibodies induced by HIV-1 envelope glycoprotein BG505 SOSIP trimers in rabbits and macaques. PLoS Pathog. 2018;14 doi: 10.1371/journal.ppat.1006913. http://www.ncbi.nlm.nih.gov/pubmed/29474444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulp D.W., Steichen J.M., Pauthner M., Hu X., Schiffner T., Liguori A., Cottrell C.A., Havenar-Daughton C., Ozorowski G., Georgeson E., Kalyuzhniy O., Willis J.R., Kubitz M., Adachi Y., Reiss S.M., Shin M., de Val N., Ward A.B., Crotty S., Burton D.R. Structure-based design of native-like HIV-1 envelope trimers to silence non-neutralizing epitopes and eliminate CD4 binding. Nat. Commun. 2017;8:1655. doi: 10.1038/s41467-017-01549-6. http://www.ncbi.nlm.nih.gov/pubmed/29162799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong P.D., Mascola J.R., Nabel G.J. Rational design of vaccines to elicit broadly neutralizing antibodies to HIV-1. Cold Spring Harb. Perspect. Med. 2011;1 doi: 10.1101/cshperspect.a007278. http://www.ncbi.nlm.nih.gov/pubmed/22229123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leaman D.P., Zwick M.B. Increased functional stability and homogeneity of viral envelope spikes through directed evolution. PLoS Pathog. 2013;9 doi: 10.1371/journal.ppat.1003184. http://www.ncbi.nlm.nih.gov/pubmed/23468626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao H.X., Tsao C.Y., Alam S.M., Muldoon M., Vandergrift N., Ma B.J., Lu X., Sutherland L.L., Scearce R.M., Bowman C., Parks R., Chen H., Blinn J.H., Lapedes A., Watson S., Xia S.M., Foulger A., Hahn B.H., Shaw G.M., Swanstrom R. Antigenicity and immunogenicity of transmitted/founder, consensus, and chronic envelope glycoproteins of human immunodeficiency virus type 1. J. Virol. 2013;87:4185–4201. doi: 10.1128/JVI.02297-12. http://www.ncbi.nlm.nih.gov/pubmed/23365441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q., Acharya P., Dolan M.A., Zhang P., Guzzo C., Lu J., Kwon A., Gururani D., Miao H., Bylund T., Chuang G.Y., Druz A., Zhou T., Rice W.J., Wigge C., Carragher B., Potter C.S., Kwong P.D., Lusso P. Quaternary contact in the initial interaction of CD4 with the HIV-1 envelope trimer. Nat. Struct. Mol. Biol. 2017;24:370–378. doi: 10.1038/nsmb.3382. http://www.ncbi.nlm.nih.gov/pubmed/28218750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magliery T.J. Protein stability: computation, sequence statistics, and new experimental methods. Curr. Opin. Struct. Biol. 2015;33:161–168. doi: 10.1016/j.sbi.2015.09.002. http://www.ncbi.nlm.nih.gov/pubmed/26497286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyerhoff R.R., Scearce R.M., Ogburn D.F., Lockwood B., Pickeral J., Kuraoka M., Anasti K., Eudailey J., Eaton A., Cooper M., Wiehe K., Montefiori D.C., Tomaras G., Ferrari G., Alam S.M., Liao H.-X., Korber B., Gao F., Haynes B.F. HIV-1 consensus envelope-induced broadly binding antibodies. AIDS Res. Hum. Retrovir. 2017;33:859–868. doi: 10.1089/aid.2016.0294. http://www.ncbi.nlm.nih.gov/pubmed/28314374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro J.B., Gorman J., Ma X., Zhou Z., Arthos J., Burton D.R., Koff W.C., Courter J.R., Smith A.B., 3rd, Kwong P.D., Blanchard S.C., Mothes W. Conformational dynamics of single HIV-1 envelope trimers on the surface of native virions. Science. 2014;80:759–763. doi: 10.1126/science.1254426. http://www.ncbi.nlm.nih.gov/pubmed/25298114 346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nkolola J.P., Bricault C.A., Cheung A., Shields J., Perry J., Kovacs J.M., Giorgi E., van Winsen M., Apetri A., Brinkman-van der Linden E.C.M., Chen B., Korber B., Seaman M.S., Barouch D.H. Characterization and immunogenicity of a novel mosaic M HIV-1 gp140 trimer. J. Virol. 2014;88:9538–9552. doi: 10.1128/JVI.01739-14. http://www.ncbi.nlm.nih.gov/pubmed/24965452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pancera M., Zhou T., Druz A., Georgiev I.S., Soto C., Gorman J., Huang J., Acharya P., Chuang G.Y., Ofek G., Stewart-Jones G.B., Stuckey J., Bailer R.T., Joyce M.G., Louder M.K., Tumba N., Yang Y., Zhang B., Cohen M.S., Haynes B.F. Structure and immune recognition of trimeric pre-fusion HIV-1 Env. Nature. 2014;514:455–461. doi: 10.1038/nature13808. http://www.ncbi.nlm.nih.gov/pubmed/25296255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathore U., Purwar M., Vignesh V.S., Das R., Kumar A.A., Bhattacharyya S., Arendt H., DeStefano J., Wilson A., Parks C., La Branche C.C., Montefiori D.C., Varadarajan R. Bacterially expressed HIV-1 gp120 outer-domain fragment immunogens with improved stability and affinity for CD4-binding site neutralizing antibodies. J. Biol. Chem. 2018;293:15002–15020. doi: 10.1074/jbc.RA118.005006. http://www.ncbi.nlm.nih.gov/pubmed/30093409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reboul C.F., Eager M., Elmlund D., Elmlund H. Single-particle cryo-EM-Improved ab initio 3D reconstruction with SIMPLE/PRIME. Protein Sci. 2018;27:51–61. doi: 10.1002/pro.3266. http://www.ncbi.nlm.nih.gov/pubmed/28795512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rerks-Ngarm S., Pitisuttithum P., Nitayaphan S., Kaewkungwal J., Chiu J., Paris R., Premsri N., Namwat C., de Souza M., Adams E., Benenson M., Gurunathan S., Tartaglia J., McNeil J.G., Francis D.P., Stablein D., Birx D.L., Chunsuttiwat S., Khamboonruang C., Thongcharoen P. Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N. Engl. J. Med. 2009;361:2209–2220. doi: 10.1056/NEJMoa0908492. http://www.ncbi.nlm.nih.gov/pubmed/19843557 [DOI] [PubMed] [Google Scholar]
- Rutten L., Lai Y.-T., Blokland S., Truan D., Bisschop I.J.M., Strokappe N.M., Koornneef A., van Manen D., Chuang G.-Y., Farney S.K., Schuitemaker H., Kwong P.D., Langedijk J.P.M. A universal approach to optimize the folding and stability of prefusion-closed HIV-1 envelope trimers. Cell Rep. 2018;23:584–595. doi: 10.1016/j.celrep.2018.03.061. http://www.ncbi.nlm.nih.gov/pubmed/29642014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders R.W., Derking R., Cupo A., Julien J.P., Yasmeen A., de Val N., Kim H.J., Blattner C., de la Pena A.T., Korzun J., Golabek M., de Los Reyes K., Ketas T.J., van Gils M.J., King C.R., Wilson I.A., Ward A.B., Klasse P.J., Moore J.P. A next-generation cleaved, soluble HIV-1 Env trimer, BG505 SOSIP.664 gp140, expresses multiple epitopes for broadly neutralizing but not non-neutralizing antibodies. PLoS Pathog. 2013;9 doi: 10.1371/journal.ppat.1003618. http://www.ncbi.nlm.nih.gov/pubmed/24068931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders R.W., van Gils M.J., Derking R., Sok D., Ketas T.J., Burger J.A., Ozorowski G., Cupo A., Simonich C., Goo L., Arendt H., Kim H.J., Lee J.H., Pugach P., Williams M., Debnath G., Moldt B., van Breemen M.J., Isik G., Medina-Ramirez M. HIV-1 VACCINES. HIV-1 neutralizing antibodies induced by native-like envelope trimers. Science. 2015;80:349. doi: 10.1126/science.aac4223. http://www.ncbi.nlm.nih.gov/pubmed/26089353 aac4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiffner T., Pallesen J., Russell R.A., Dodd J., de Val N., LaBranche C.C., Montefiori D., Tomaras G.D., Shen X., Harris S.L., Moghaddam A.E., Kalyuzhniy O., Sanders R.W., McCoy L.E., Moore J.P., Ward A.B., Sattentau Q.J. Structural and immunologic correlates of chemically stabilized HIV-1 envelope glycoproteins. PLoS Pathog. 2018;14 doi: 10.1371/journal.ppat.1006986. http://www.ncbi.nlm.nih.gov/pubmed/29746590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S.K., de Val N., Bale S., Guenaga J., Tran K., Feng Y., Dubrovskaya V., Ward A.B., Wyatt R.T. Cleavage-independent HIV-1 Env trimers engineered as soluble native spike mimetics for vaccine design. Cell Rep. 2015;11:539–550. doi: 10.1016/j.celrep.2015.03.047. http://www.ncbi.nlm.nih.gov/pubmed/25892233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sliepen K., Han B.W., Bontjer I., Mooij P., Garces F., Behrens A.-J., Rantalainen K., Kumar S., Sarkar A., Brouwer P.J.M., Hua Y., Tolazzi M., Schermer E., Torres J.L., Ozorowski G., van der Woude P., de la Peña A.T., van Breemen M.J., Camacho-Sánchez J.M., Burger J.A. Structure and immunogenicity of a stabilized HIV-1 envelope trimer based on a group-M consensus sequence. Nat. Commun. 2019;10:2355. doi: 10.1038/s41467-019-10262-5. http://www.ncbi.nlm.nih.gov/pubmed/31142746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steichen J.M., Kulp D.W., Tokatlian T., Escolano A., Dosenovic P., Stanfield R.L., McCoy L.E., Ozorowski G., Hu X., Kalyuzhniy O., Briney B., Schiffner T., Garces F., Freund N.T., Gitlin A.D., Menis S., Georgeson E., Kubitz M., Adachi Y., Jones M. HIV vaccine design to target germline precursors of glycan-dependent broadly neutralizing antibodies. Immunity. 2016;45:483–496. doi: 10.1016/j.immuni.2016.08.016. http://www.ncbi.nlm.nih.gov/pubmed/27617678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steipe B., Schiller B., Plückthun A., Steinbacher S. Sequence statistics reliably predict stabilizing mutations in a protein domain. J. Mol. Biol. 1994;240:188–192. doi: 10.1006/jmbi.1994.1434. http://www.ncbi.nlm.nih.gov/pubmed/8028003 [DOI] [PubMed] [Google Scholar]
- Sternke M., Tripp K.W., Barrick D. Consensus sequence design as a general strategy to create hyperstable, biologically active proteins. Proc. Natl. Acad. Sci. USA. 2019;116:11275–11284. doi: 10.1073/pnas.1816707116. http://www.ncbi.nlm.nih.gov/pubmed/31110018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart-Jones G.B., Soto C., Lemmin T., Chuang G.Y., Druz A., Kong R., Thomas P.V., Wagh K., Zhou T., Behrens A.J., Bylund T., Choi C.W., Davison J.R., Georgiev I.S., Joyce M.G., Kwon Y.D., Pancera M., Taft J., Yang Y., Zhang B. Trimeric HIV-1-Env structures define glycan shields from clades A, B, and G. Cell. 2016;165:813–826. doi: 10.1016/j.cell.2016.04.010. http://www.ncbi.nlm.nih.gov/pubmed/27114034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan J.T., Sulli C., Nilo A., Yasmeen A., Ozorowski G., Sanders R.W., Ward A.B., Klasse P.J., Moore J.P., Doranz B.J. High-throughput protein engineering improves the antigenicity and stability of soluble HIV-1 envelope glycoprotein SOSIP trimers. J. Virol. 2017;91 doi: 10.1128/JVI.00862-17. http://www.ncbi.nlm.nih.gov/pubmed/28878072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torrents de la Pena A., Julien J.P., de Taeye S.W., Garces F., Guttman M., Ozorowski G., Pritchard L.K., Behrens A.J., Go E.P., Burger J.A., Schermer E.E., Sliepen K., Ketas T.J., Pugach P., Yasmeen A., Cottrell C.A., Torres J.L., Vavourakis C.D., van Gils M.J., LaBranche C. Improving the immunogenicity of native-like HIV-1 envelope trimers by hyperstabilization. Cell Rep. 2017;20:1805–1817. doi: 10.1016/j.celrep.2017.07.077. http://www.ncbi.nlm.nih.gov/pubmed/28834745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varadarajan R., Sharma D., Chakraborty K., Patel M., Citron M., Sinha P., Yadav R., Rashid U., Kennedy S., Eckert D., Geleziunas R., Bramhill D., Schleif W., Liang X., Shiver J. Characterization of gp120 and its single-chain derivatives, gp120-CD4D12 and gp120-M9: implications for targeting the CD4i epitope in human immunodeficiency virus vaccine design. J. Virol. 2005;79:1713–1723. doi: 10.1128/JVI.79.3.1713-1723.2005. http://www.ncbi.nlm.nih.gov/pubmed/15650196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K., Acharya P., Kong R., Cheng C., Chuang G.Y., Liu K., Louder M.K., O'Dell S., Rawi R., Sastry M., Shen C.H., Zhang B., Zhou T., Asokan M., Bailer R.T., Chambers M., Chen X., Choi C.W., Dandey V.P., Doria-Rose N.A. Epitope-based vaccine design yields fusion peptide-directed antibodies that neutralize diverse strains of HIV-1. Nat. Med. 2018;24:857–867. doi: 10.1038/s41591-018-0042-6. http://www.ncbi.nlm.nih.gov/pubmed/29867235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Sharma S.K., Cottrell C., Guenaga J., Tran K., Wilson R., Behrens A.J., Crispin M., de Val N., Wyatt R.T. Structure-guided redesign improves NFL HIV Env trimer integrity and identifies an inter-protomer disulfide permitting post-expression cleavage. Front. Immunol. 2018;9:1631. doi: 10.3389/fimmu.2018.01631. http://www.ncbi.nlm.nih.gov/pubmed/30065725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P., Gorman J., Geng H., Liu Q., Lin Y., Tsybovsky Y., Go E.P., Dey B., Andine T., Kwon A., Patel M., Gururani D., Uddin F., Guzzo C., Cimbro R., Miao H., McKee K., Chuang G.Y., Martin L., Sironi F. Interdomain stabilization impairs CD4 binding and improves immunogenicity of the HIV-1 envelope trimer. Cell Host Microbe. 2018;23:832–844 e6. doi: 10.1016/j.chom.2018.05.002. http://www.ncbi.nlm.nih.gov/pubmed/29902444 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.