Abstract
Sirtuin 6, SIRT6, is critical for both glucose and lipid homeostasis and is involved in maintaining genomic stability under conditions of oxidative DNA damage such as those observed in age-related diseases. There is an intense search for modulators of SIRT6 activity, however, not many specific activators have been reported. Long acyl-chain fatty acids have been shown to increase the weak in vitro deacetylase activity of SIRT6 but this effect is modest at best. Herein we report that electrophilic nitro-fatty acids (nitro-oleic acid and nitro-conjugated linoleic acid) potently activate SIRT6. Binding of the nitro-fatty acid to the hydrophobic crevice of the SIRT6 active site exerted a moderate activation (2-fold at 20 μm), similar to that previously reported for non-nitrated fatty acids. However, covalent Michael adduct formation with Cys-18, a residue present at the N terminus of SIRT6 but absent from other isoforms, induced a conformational change that resulted in a much stronger activation (40-fold at 20 μm). Molecular modeling of the resulting Michael adduct suggested stabilization of the co-substrate and acyl-binding loops as a possible additional mechanism of SIRT6 activation by the nitro-fatty acid. Importantly, treatment of cells with nitro-oleic acid promoted H3K9 deacetylation, whereas oleic acid had no effect. Altogether, our results show that nitrated fatty acids can be considered a valuable tool for specific SIRT6 activation, and that SIRT6 should be considered as a molecular target for in vivo actions of these anti-inflammatory nitro-lipids.
Keywords: activator, sirtuin, SIRT6, fatty acids, nitro-fatty acids, docking, histone deacetylation, cysteine covalent modification, Michael addition, enzyme activation, histone deacetylase (HDAC), enzyme mechanism, enzyme structure, enzyme kinetics, fatty acid, cysteine covalent-modification, enzyme activator, nitro-fatty acid
Sirtuins are a family of NAD+-dependent lysine deacylases critical for maintaining cellular and organ homeostasis (1, 2, 3, 4). This family includes seven sirtuins (SIRT1–7) with different subcellular location, enzymatic activities, substrate proteins, and biological functions, as well as different key roles in metabolism, nutrition, aging, inflammation, and cancer (5, 6, 7, 8). SIRT6 is primarily located in the nucleus along with SIRT1 and SIRT7, whereas SIRT2 is cytosolic, and SIRT3, SIRT4, and SIRT5 are found in the mitochondria (9).
SIRT6 is implicated in the regulation of diverse physiological processes such as maintenance of genomic stability, inflammation, and energy metabolism (10, 11, 12). Overexpression of SIRT6 increased lifespan in male mice, preserved glucose tolerance, and attenuated adipose tissue inflammation (13, 14, 15). More recently, overexpression of SIRT6 showed neuroprotection in an animal model of spinal cord injury (16). Conversely, mice globally deficient in SIRT6 are hypoglycemic, suffer lymphopenia, premature aging, and die within 1 month (17). Tissue-specific SIRT6 ablation in adipocytes, macrophages, or pancreatic beta-cells, sensitizes mice toward high-fat diet-induced obesity and insulin resistance (18, 19, 20). Specific neural SIRT6-deleted mice did not die from hypoglycemia, but displayed postnatal growth retardation and ultimately became obese (21). In these latter mice, hyperacetylation of histone H3 lysine 9 (H3K9) and lysine 56 (H3K56) were observed in various regions of the brain. In fact, H3K9 and H3K56 are the main molecular targets for SIRT6 activity in vivo (22, 23, 24) with recent reports also including H3K18 (25).
In addition to deacetylating histone H3, SIRT6 can also bind and regulate nonhistone proteins such as the transcription factor HIF-1α (26), catalyze intramolecular mono-ADP-ribosylation (27), and mediate deacylation of long-chain fatty acyl groups (e.g. myristoylated tumor necrosis factor-α (28)). As recently reviewed by Klein and Denu (29) the physiological substrates, biological activities, and specific cellular functions of SIRT6 still need further investigation.
All sirtuins share a highly conserved globular catalytic core of 275 amino acids but the N and C termini are highly disordered and differ significantly among family members (30, 31, 32). The active site comprises three structural pockets A, B, and C, with the latter having the most hydrophobic character and the largest number of highly conserved residues. Two domains flank the active site, a large Rossmann-fold where co-substrate NAD+ binds, and a smaller Zn2+-binding motif (Fig. 1) (33). In particular, SIRT6 displays a more open structure in which the two domains are more distant than in other isoforms (34). Accommodation of a long-chain acyl group in the C-pocket favors the domains to close up, thus resulting in better deacylase than deacetylase activity (28, 35). The N- and C-terminal regions in SIRT6 are short, less structured and flexible. The C terminus is required for nuclear localization, whereas the N terminus, rich in positive residues, is important for both chromatin association and for intrinsic catalytic activity because its deletion severely compromised H3K9 and H3K56 deacetylation (27, 36, 37). The mechanism of catalysis includes the binding of cofactor NAD+ and the acylpeptide in the right orientation so that the amide of the acylated lysine can attack the anomeric carbon of the ribose via an SN2-type mechanism, releasing nicotinamide and forming an alkylamidate intermediate (35, 38, 39). A conserved histidine residue in the active site functions as a general base to catalyze the release of the acylated-ADPR and lysine peptide products (40, 41). Thus, in sirtuins, catalysis depends on this histidine residue (His-133 in the human SIRT6 isoform, hSIRT6) and not on Zn2+ as in other histone deacetylases (HDAC class I, II, and IV). In this regard, the zinc atom has a purely structural role and it is coordinated by four cysteine residues in a CXXC motif. In the case of hSIRT6 these are Cys-141, Cys-144, and Cys-166, Cys-177, where the insertion of 10 residues (167 to 176) forming a flexible loop, is unique for SIRT6 (Fig. 1).
Figure 1.

Human SIRT6 structure. Molecular representation of hSIRT6 in complex with a myristoylated H3 peptide at Lys-9 (H3K9Myr) and a co-substrate (PDB code 3ZG6). Structural features such as functional domains, A, B, C pockets and relevant residues are indicated. The location of N and C termini (NT and CT, respectively) is also shown. Continuation of the peptide chain structure toward CT is sketched as a guide for helping visualization.
Given its importance in the maintenance of energy metabolism and DNA stability, the discovery of novel pharmacological agents that boost SIRT6 activity can have great potential for treatment of diabetes, arthritis, cardiovascular, and neurodegenerative diseases (42). As a result, there is an intense search for modulators of SIRT6 activity, however, not many specific activators have been reported. Interestingly, Feldman et al. (43) demonstrated that the low in vitro deacetylase activity of SIRT6 could be activated in the presence of free long-chain fatty acids, particularly oleic and linoleic acid (5-fold increase at 100 μm), and that this activation was not displayed by SIRT1. Similarly, polyphenols were shown to either inhibit (catechins) or weakly activate (anthocyanidins, 2-fold increase at 100 μm) SIRT6 activity but these responses are similarly weak (44, 45). In addition, two synthetic small molecule activators have been identified, a pyrrole[1,2-a]quinoxaline derivative (UBCS039) that achieved a 2-fold increase in activity at 100 μm (46) and MDL-800, a more effective pharmacological activator (22-fold at 100 μm) (47), both binding at a distal region of the fatty acyl substrate. Finally, a targeted screen of fatty acids and lipid-like molecules identified other activators with >15-fold activation at 100 μm such as oleoyl-lysophosphatidic acid (48-fold) and 2-(3-chloro-4-(2,4-dichlorobenzamido)phenyl)-1,3-dioxoisoindoline-5-carboxylic acid (CL-4, 18-fold) (48).
Nitro-fatty acids (NO2-FA) are detected in diverse species, including humans (49, 50, 51) and have been extensively shown to exert cellular protective actions and to protect against metabolic disorders and inflammation (52). NO2-FA are both derived from the diet and can also be formed endogenously by the olefinic nitration of unsaturated fatty acids thus reaching nanomolar concentrations in the plasma of healthy subjects (50, 53). In addition, dietary supplementation with nitrite and CLA (conjugated linoleic acid, also known as rumenic acid) have been shown to increase the levels of NO2-CLA in plasma, urine, and tissues (49, 50). Importantly, the nitration of unsaturated fatty acids results in the formation of an electrophilic nitroalkene moiety that can react via Michael addition with nucleophilic residues, thus resulting in post-translational modification (PTM) of transcription factors and enzymes (52, 54, 55, 56). These post-translational modifications alter protein structure and function, resulting in signaling actions that include Nrf2-regulated gene induction, inhibition of NF-κB-dependent proinflammatory cascades, and modulation of enzyme activities such as xanthine oxidoreductase inhibition and pro-matrix metalloproteinases-7 and -9 activation (57, 58, 59, 60, 61). Considering the presence of a nucleophilic histidine in the active site of sirtuins, as well as of several cysteine residues with known structural roles (zinc-binding motif), we explored the potential of NO2-FA to exert structural and functional changes in these proteins. Herein we report that the nitrated fatty acids nitro-oleic and nitro-conjugated linoleic acid (Fig. 2A) potently activate SIRT6 deacetylase activity via covalent modification of the isoform-specific Cys-18 in the N-terminal domain.
Figure 2.
Effect of fatty acids and nitro-fatty acids on in vitro deacetylase activity of hSIRT6.A, chemical structures of 9- and 10-nitro-oleic acids (NO2-OA), 9- and 12-nitro-conjugated linoleic acids (NO2-CLA). Experiments were performed using the racemic mixtures 9- and 10-NO2-OA, and 9- and 12-NO2-CLA in the presence of 1 μm SIRT6. B, representative runs of deacetylase activity measured using the coupled assay for untreated hSIRT6 (black), hSIRT6 incubated with oleic acid (50 μm, light gray), and hSIRT6 incubated with nitro-oleic acid (20 μm, dark gray). C, fold-change in SIRT6 deacetylase activity after incubation with the indicated concentrations of OA (○), NO2-OA (●), and NO2-CLA (■). Error bars represent S.D. of at least three replicates. *, p < 0.1; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001 ANOVA test.
Results
Nitro-fatty acids stimulate the deacetylase activity of hSIRT6 in vitro
The in vitro deacetylase activity of hSIRT6 is low under basal conditions (kcat = 0.003 s−1, Km = 400 μm) and the activation by oleic acid (OA) was confirmed as previously reported (Fig. 2B). Notably, incubation with a racemic mixture of nitro-oleic acid (NO2-OA) resulted in potent activation of the deacetylase activity, with a fold of activation significantly higher than that achieved by the corresponding non-nitrated fatty acid (Fig. 2C). In this regard, preincubation of SIRT6 with a 20-fold excess of OA increased deacetylase activity (H3K9Ac) by 2-fold, whereas the same treatment with NO2-OA resulted in 40-fold activation. Similar results were obtained with nitro-conjugated linoleic acid (NO2-CLA), the most abundant endogenous NO2-FA in humans (Fig. 2C). No activation was observed for the SIRT6 deacylase (H3K9Myr) activity (kcat = 0.008 s−1, Km = 20 μm). Similarly, no effect was observed by either OA (43) or up to 100 μm NO2-OA on the deacetylase and demyristoylase activities of SIRT1 (data not shown).
Incubation with either OA or NO2-OA did not significantly alter the secondary structure of hSIRT6, but a conformational change was evidenced by near-UV CD. More specifically, a change in the asymmetric environment of the aromatic residues and disulfides was observed, in particular, an ellipticity increase around 260 nm (Fig. 3).
Figure 3.
CD of hSIRT6. Far-UV (A) and near-UV (B) CD spectra of hSIRT6 (3 and 10 μm, respectively) after 30 min incubation at 37 °C in the absence (black) or presence of either 5-fold excess of OA (light gray) or NO2-OA (dark gray). Spectra were normalized for comparison as detailed under “Experimental procedures.”
Reaction of nitro-fatty acids with hSIRT6 cysteine residues
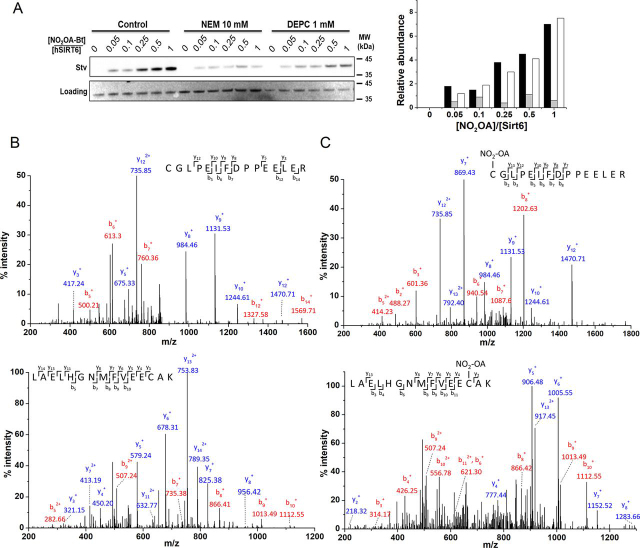
Electrophilic NO2-FA, such as NO2-OA and NO2-CLA, can react with nucleophilic protein residues via reversible Michael addition (54, 56). Because hSIRT6 has a catalytic histidine residue, His-133, the incubation with NO2-FA could result in inactivation. Taking advantage of the change in absorption of NO2-CLA with environment polarity (λmax = 330 nm in water versus λmax = 312 nm in methanol), the binding of this nitro-lipid to hSIRT6 was investigated. As shown in Fig. 4, the blue shift expected in the UV-visible spectrum for NO2-CLA binding to a hydrophobic site in the enzyme was not observed. Instead, a time-dependent decrease in absorption at 330 nm was detected, suggesting a reaction between NO2-CLA and a nucleophilic protein residue. To explore which nucleophilic residues were involved in this reaction, hSIRT6 was treated with NEM or DEPC to block cysteines or histidines, respectively, followed by incubation with biotinylated-NO2-OA (Bt-NO2-OA). Streptavidin-HRP adduct blotting strongly suggested the occurrence of a covalent interaction between NO2-OA and cysteine residues in hSIRT6 (Fig. 5A). ESI ionization MS/MS analysis further confirmed this observation and identified Cys-141 (one of the cysteines coordinating Zn2+) and Cys-18 at the N-terminal domain, as the reactive targets (Fig. 5, B and C). Interestingly, despite the confirmation of adduct formation between NO2-OA and Cys-141, as well as the conformational change evidenced by CD spectra (Fig. 3), no loss of zinc was detected from hSIRT6 upon incubation with either NO2-FA (data not shown).
Figure 4.
Consumption of NO2-CLA by hSIRT6.A, UV-visible spectra of hSIRT6 (solid black line) with a characteristic peak at 280 nm, NO2-CLA alone (dashed line) with a characteristic peak at 330 nm, and of a mixture of 15 μm hSIRT6 with 10 μm NO2-CLA. B, change in absorbance at 330 nm with time after mixing hSIRT6 with NO2-CLA (1.5:1).
Figure 5.
Identification of hSIRT6 residues modified by NO2-OA.A, untreated hSIRT6 (control), and hSIRT6 treated with either NEM or DEPC to block cysteine and histidine residues, respectively, was incubated with the indicated excess of biotinylated-NO2-OA and adduct formation detected using streptavidin-HRP (upper panel). Ponceau S staining was used as a loading control (lower panel) for densitometric analysis: control (black), NEM-treated SIRT6 (gray), and DEPC-treated SIRT6 (white). One of two consistent experiments is shown. B, MS spectra of untreated-hSIRT6 peptides obtained from tryptic digestion (2 h at 37 °C). The upper panel corresponds to the fragmentation of the doubly charged peptide sequence containing Cys-18 (CGLPEIFDPPEELER; monoisotopic m/z: 872.73) and the lower panel corresponds to triply charged sequence containing Cys-141 (LAELHGNMFVEECAK; monoisotopic m/z: 564.34). C, MS/MS spectra of NO2-OA–treated SIRT6 (×5) ions with Δm = 327.24. Upper panel corresponds to the doubly charged peptide sequence containing Cys-18 (CGLPEIFDPPEELER; monoisotopic m/z: 1036.21) and lower panel corresponds to the triply charged sequence containing Cys-141 (LAELHGNMFVEECAK; monoisotopic m/z: 673.56). The spectra are representative of three independent experiments. y- and b-ions detected by MS/MS analysis for each tryptic peptide.
N-terminal Cys-18 is responsible for hSIRT6 nitro-fatty acid activation
Because MS analysis revealed Michael adduct formation with cysteine residues and in particular the N-terminal Cys-18, the effect of NO2-FA was assayed on hSIRT6 mutant C18S. Interestingly, the level of activation achieved on the mutant was similar to the effect of the corresponding non-nitrated fatty acid (2-fold). This result suggested that both nitrated and non-nitrated fatty acids bind to the same pocket in the mutant. However, the significant 40-fold activation observed with the WT protein was lost in the absence of adduct formation with Cys-18 (Fig. 6). Molecular docking of NO2-OA or NO2-CLA into the active site of hSIRT6 in the absence of a H3K9Ac peptide showed that the electrophilic nitroalkene moiety is distant from the catalytic histidine (Fig. 7, A and B), thus providing an explanation for the non-inhibitory activity of NO2-FA on hSIRT6 activity. Furthermore, docking poses of NO2-OA or NO2-CLA into the active site in the presence of H3K9Ac peptide were similar to those observed with OA and CLA, consistent with ∼2-fold activation observed with of the C18S mutant (Fig. 7, C and D). The solvent-exposed conformation of CLA's chromophore group in the resulting poses may agree with observations made in its absorption spectrum (Fig. 4). In all cases, Cys-18 was always found far from the nitroalkene group, thus preventing a possible adduct formation with the docked NO2-FA in the active site of the WT protein. Molecular modeling of a Cys-18 adduct with 9-NO2-OA showed that neither the carboxylic nor the acylic ends of the NO2-FA were able to reach the C-pocket of hSIRT6 (∼20 Å away), on the contrary, they should remain close to the acyl/co-substrate–binding loop (Fig. 8).
Figure 6.
Effect of fatty acids and nitro-fatty acids on in vitro deacetylase activity of WT and C18S hSIRT6. Fold-change in SIRT6 deacetylase activity after incubation of 1 μm WT hSIRT6 (●) or C18S mutant (○) with 20 μm OA, NO2-OA, CLA, or NO2-CLA. Error bars represent S.D. of three replicates. ****, p < 0.0001 ANOVA test.
Figure 7.
In silico docking experiments.A, best poses after docking OA and NO2-OA into the active site of hSIRT6. The carbon backbone is colored according to the docked molecule. Oxygen and nitrogen atoms are red and blue, respectively, whereas unsaturations are shown in gold. The co-substrate is represented with brown sticks. Relevant residues in the protein are pointed out. The molecular surface of the active site is shown in gray, whereas the reminding structural context of the protein is shown as a transparent cartoon. B, same as panel A for CLA and NO2-CLA. C, best poses after docking OA and NO2-OA considering the presence of an acetylated H3 peptide at Lys-9 (H3K9Ac). D, same as panel C for CLA and NO2-CLA.
Figure 8.

Molecular modeling of nitro-fatty acid conjugation to Cys-18.A, molecular representation of one possible conformation of the Michael adduct between Cys-18 and 9-NO2-OA. The nitro-fatty acid is shown in balls and stick, whereas nearby protein residues are drawn in sticks. Residues that may form an exposed hydrophobic patch at the surface of the acyl/co-substrate–binding loop are labeled in magenta. The location and distance to the C-pocket is indicated. B, same as panel A but showing other alternative conformation that the same adduct may adopt.
Binding of hSIRT6 to nitro-fatty acid at the cellular level
To determine whether the reported interaction between NO2-OA and hSIRT6 could take place in a cellular context, HEK293T cells overexpressing FLAG-hSIRT6 were treated with 10 μm Bt-NO2-OA for 1 h. At the end of the incubation period, Bt-NO2-OA-modified proteins were pulled down from whole-cell lysates using streptavidin-conjugated beads and subjected to Western blotting analysis. As show in Fig. 9A, pulldown of Bt-NO2-OA resulted in prominent FLAG-SIRT6 detection, confirming the occurrence of this modification under cellular conditions.
Figure 9.
In situ covalent modification of hSIRT6 by NO2-OA promotes H3K9 deacetylation in cells.A, HEK293T cells expressing FLAG-SIRT6 were treated with 10 μm Bt-NO2-OA for 1 h. After cell lysis, Bt-NO2-OA adducts were affinity-purified using streptavidin-agarose beads followed by FLAG immunodetection (lanes 1–4). FLAG-SIRT6 from the same input lysates used for affinity purification is shown on the right (lanes 5-8). Immunoprecipitation was performed in HEK293T cells overexpressing FLAG-SIRT6 (lanes 3 and 4) and in control HEK293T cells without transfection (lanes 1 and 2). Arrows indicate 55- and 36-kDa molecular mass standards. B, HEK293T cells were treated with 10 μm OA or 10 μm NO2-OA for 1 h in serum-free DMEM and H3K9 acetylation assessed by Western blotting analysis. C, densitometric H3K9Ac quantification versus total amount of H3 (H3K9Ac/H3). Statistically significant differences were observed between control and NO2-OA incubation. Error bars represent S.D. of at least three replicates. **, p < 0.001 as determined by ANOVA.
Nitro-fatty acids induced cellular histone deacetylation
The modulation of the cellular deacetylase activity of hSIRT6 was monitored by assessing changes in H3K9 acetylation levels following 1 h treatment of HEK293T cells overexpressing FLAG-hSIRT6 with OA or NO2-OA (10 μm). In accordance with the potent activation of hSIRT6 deacetylase activity observed in vitro, Western blotting analysis showed a marked decrease in H3K9Ac levels in HEK293T cells incubated with NO2-OA (Fig. 9, B and C). No decrease in H3K9Ac levels was observed upon OA incubation, with H3K9Ac/H3 ratios being identical to control levels. Taken together, these results show that NO2-OA effectively interacts with hSIRT6 in the cellular environment resulting in the activation of its deacetylase activity and the corresponding decrease in H3K9 acetylation levels.
Discussion
The hydrophobic pocket in the active site of hSIRT6 accommodates a long acyl-chain resulting in better deacylase than deacetylase activity in vitro. Previous studies observed that free fatty acids bound to this hydrophobic crevice cause a conformational change that brings closer the two domains of the enzyme, which in turn favors the binding of the substrates. Thus, the poor in vitro deacetylase activity of SIRT6 increases in the presence of free fatty acids such as OA. Considering the enzyme depends on a histidine residue (His-133) for catalysis, we speculated that electrophilic NO2-FA would accommodate in the active site, react with the nucleophilic histidine, and inhibit SIRT6 activity. Surprisingly, incubation with low micromolar concentrations of NO2-OA or NO2-CLA not only resulted in activation of the deacetylase activity but also to a significantly higher degree than that obtained with the corresponding non-nitrated fatty acids (Fig. 2). Molecular docking analyses support the experimental results showing that once accommodated in the active site, the distance and orientation of the reactive nitroalkene moiety do not allow for reaction with the catalytic histidine (Fig. 7). Nevertheless, UV-visible determinations showed that NO2-CLA was consumed upon incubation with hSIRT6 suggesting that a reaction with nucleophilic residues of the enzyme was taking place (Fig. 4). MS analysis confirmed Michael adduct formation with cysteine residues, in particular with Cys-141 (coordinating Zn2+) and Cys-18 at the N terminus (Fig. 5). Interaction of SIRT6 with NO2-OA at the cellular level was confirmed by pulldown with streptavidin after incubating HEK293T cells overexpressing FLAG-SIRT6 with Bt-NO2-OA (Fig. 9). Despite not having a catalytic cysteine, oxidation of sirtuin cysteines has been shown to affect hSIRT6 activity in vitro and in vivo (62). For example, Cys-18 was found to be S-sulfenylated when colon cancer cells were exposed to hydrogen peroxide, resulting in the formation of a covalent complex with HIF1α via disulfide bonding (63). Similarly, work on lipopolysaccharide-activated human monocytes demonstrated SIRT6 sulfenylation on Cys-144 (64), and incubation of hSIRT6 with nitrosating agents resulted in protein S-nitrosation and loss of demyristoylase activity (65).
Molecular docking analysis (Fig. 7) showed that a NO2-FA molecule accommodated in the active site would be unable to react with any of these cysteine residues, indicating that Michael adduct formation likely involves a different reaction site. Importantly, the covalent modification of hSIRT6 by NO2-FA induces a conformational change (Fig. 3) that results in a much larger increase in enzymatic activity than that attained by mere binding of the long acyl chain of the NO2-FA on the active site loop (Fig. 2). In line with this concept, incubation of NO2-FA with the mutant C18S resulted in only a minor activation (“the fatty acid effect”) compared with the much stronger effect achieved with the wt hSIRT6 where the Michael adduct with Cys-18 can be formed (Fig. 6). This result also indicates that, although Cys-141 is susceptible to modification by NO2-FA, this reaction is not responsible for hSIRT6 activation. On the whole, the results suggest that NO2-FA may still activate hSIRT6 by binding to the active site in a similar fashion as non-nitrated fatty acids, the formation of a Michael adduct with Cys-18 induces a much greater activation as a result of the stabilization of the hydrophobic patch constituted by residues Met-3, Tyr-5, Pro-11, Tyr-12, Ala-13, and Lys-15 in the acyl/co-substrate–binding loop (Fig. 8). Both activation mechanisms are expected to be independent and thus could not be mediated by the same NO2-FA molecule. SIRT6 has the shortest N-terminal region among the seven sirtuins and is the only isoform that contains a reactive cysteine in this region (30), suggesting that the potent activation induced by NO2-FA is likely to be specific to SIRT6. In fact, no in vitro activation of SIRT1 was observed by NO2-FA.
Treatment of HEK293T cells with NO2-OA resulted in a significant decrease in the levels of acetylated H3K9, an effect that was not observed when equivalent concentrations of OA were used (Fig. 9, B and C). Importantly, the concentration of NO2-OA required to activate cellular hSIRT6 (10 μm) is comparable with those shown to elicit physiologically relevant signaling events in vitro that then translate successfully to both animal and human studies (52, 66, 67). In this regard, administration of NO2-OA to both genetic and dietary mouse models of obesity results in improved insulin sensitivity and glucose tolerance, as well as in protection against mitochondrial dysfunction (68, 69). These effects have been ascribed to a combination of partial peroxisome proliferator-activated receptor γ agonism, inhibition of pro-inflammatory NF-κB signaling, attenuation of reactive species generation, and prevention of mitochondrial dysfunction in the adipose tissue (68, 69, 70, 71). Notably, these results are also consistent with those obtained upon SIRT6 overexpression in both obese and non-obese mouse models (13, 14, 15), suggesting that posttranslational SIRT6 activation could contribute to the metabolic effects of NO2-OA in vivo.
Considering that SIRT6 is a key enzyme in inflammation and metabolism, effective modulators of its activity are of special interest. Herein we report that nitrated derivatives from known fatty acid activators of hSIRT6 are able to further increase enzyme activity by ∼40-fold. We postulate that a synergistic mechanism may be at play, involving both the binding of a NO2-FA to a pocket in the active site of the enzyme and the formation of a Michael adduct with Cys-18. The complementary effect of Cys-18 nitro-alkylation may shift the N-terminal region of hSIRT6 to its active conformation, similar to other activators, such as MDL-800/801 (45), UBSC039 (44), quercetin, isoquercetin, and cyanidin (48). Taken together, our results indicate that activation of SIRT6-dependent deacetylase activity could significantly contribute to the physiological and pharmacological actions of NO2-FA in vivo.
Experimental procedures
Reagents
Acylpeptide H3K9 (Ac-KQTARKSTGGWW), H3K9Ac (Ac-KQTARK(Ac)STGGWW), and H3K9Myr (Ac-KQTARK(Myr)STGGWW) were custom synthesized at ≥90% purity by United Biosystems, VA, USA. 9- and 10-Nitrooctadec-9-enoic acid (nitro-oleic acid, 9- and 10-NO2-OA), 9- and 12-nitrooctadec-9,11-dienoic acid (nitro-conjugated linoleic acid, 9- and 12-NO2-CLA), and Bt-NO2-OA were synthesized as previously described (58, 72, 73). For both, NO2-OA and NO2-CLA, a 1:1 mixture of the positional isomers in methanol (2 mm) was used as stock and kept at −80 °C. Cell media, isopropyl β-d-1-thiogalactopyranoside (IPTG), β-mercaptoethanol (β-ME), and tris(2-carboxyethyl)phosphine hydrochloride (TCEP) were purchased from Sigma-Aldrich, USA. All other chemicals were of analytical grade.
Recombinant hSIRT6 WT and hSIRT6 C18S expression and purification
Competent BL21-DE3 Escherichia coli cells were transformed with pQE-80L plasmid containing N-terminal His-tagged human SIRT6 sequence (1-355) or His-tagged human SIRT6 C18S and were grown in 2YT medium supplemented with 100 μg/ml of ampicillin to an absorbance of 0.6 at 600 nm. Protein expression was induced overnight by addition of 0.5 mm IPTG at 25 °C. Cells were harvested by centrifugation at 5000 × g and frozen at −80 °C. Frozen cell pellets were thawed on ice and resuspended in lysis buffer: 50 mm sodium phosphate, pH 7.2, containing 250 mm NaCl, 5 mm imidazole, and 1 mm β-ME. Cells were lysed via sonication, centrifuged (18,000 × g), and applied to a nickel-nitrilotriacetic acid column (GE Healthcare), washed with 5 volumes of lysis buffer followed by 5 volumes of 50 mm sodium phosphate, pH 7.5, containing 250 mm NaCl, 10 mm imidazole, and 1 mm β-ME. Elution was carried out using 250 mm imidazole in the buffer. hSIRT6 was further purified by cationic exchange on a HiTrap SP-Sepharose Fast Flow column (GE Healthcare) in 50 mm sodium phosphate, pH 7.2, 50 mm NaCl, and 1 mm β-ME. After washing the column with 150 mm NaCl in buffer, elution was carried out by a linear gradient from 150 to 750 mm NaCl in the same buffer. Fractions containing purified hSIRT6 (containing 1 mol of Zn2+/mol of protein) were concentrated and dialyzed into 50 mm Tris, pH 8.0 (4 °C), 150 mm NaCl, 100 μm TCEP, and 5% (w/v) glycerol, and stored at −80 °C.
Recombinant Pnc1 expression and purification
Competent BL21-DE3 E. coli cells were transformed with pET28a plasmid containing His-tagged Pnc1 nicotinamidase from Saccharomyces cerevisiae (74) and grown in LB medium supplemented with 35 μg/ml of kanamycin to an absorbance of 0.6 at 600 nm. Protein expression was induced by addition of 1 mm IPTG for 4 h at 37 °C. Cells were harvested (5000 × g) and frozen at −80 °C. Frozen cell pellets were thawed on ice and resuspended in lysis buffer: 50 mm Tris, pH 7.2, 250 mm NaCl, 5 mm imidazole, and 1 mm β-ME. Recombinant protein was purified from the clarified cell lysate by nickel affinity chromatography. After loading the column, a 10-volume wash was carried out with 50 mm Tris, pH 7.2, 250 mm NaCl, 30 mm imidazole, and 1 mm β-ME. For the elution, 500 mm imidazole was used in the same buffer. Pnc1 was further purified by gel filtration in 50 mm sodium phosphate, pH 8, 150 mm NaCl, 3 mm β-ME and stored at −80 °C.
Quantification of Zn2+
To determine the stoichiometry of zinc bound to hSIRT6, the metal ion was first released by p-chloromercurybenzoate treatment and complexed with the dye 4-(2-pyridylazo)resorcinol (PAR). A molar extinction coefficient of 66,000 m−1 cm−1 at 500 nm was used for the resulting Zn-PAR complex (75). The reaction mixture contained 2 μm hSIRT6, 20 μm p-chloromercurybenzoate, 40 μm PAR in 50 mm sodium phosphate, pH 7.4, and absorbance at 500 nm was monitored.
Deacetylase activity assay
hSIRT6 deacetylase activity was measured under initial rate conditions using an enzyme-coupled assay as previously reported (76). Assays were performed in 50 mm sodium phosphate, pH 7.4, 150 mm NaCl, at 37 °C, in a final volume of 100 μl. The reaction mixture contained 0.6 mm NAD, 3 mm α-ketogutarate, 0.2 mm NADPH, 3 μm Pnc1, 2 units of glutamic dehydrogenase, 50 μm H3K9Ac, and 0.5 μm hSIRT6 or hSIRT6 C18S. The reaction mixture was preincubated for 20 min at 37 °C until absorbance at 340 nm was stable and deacetylation reaction started by the addition of hSIRT6 or hSIRT6 C18S.
Effect of nitrated and non-nitrated fatty acids on hSIRT6 deacetylase activity
To determine the effect of fatty acids or nitro-fatty acids on deacetylase hSIRT6 activity, the enzyme (WT or mutant, 1 μm) was preincubated, for 30 min at room temperature, with variable excess concentrations of OA, CLA, and their respective nitro-derivatives (NO2-OA, NO2-CLA). Racemic mixtures of the nitro-fatty acids used were: 9- and 10-NO2-OA, and 9- and 12-NO2-CLA (see Fig. 2). Stock solutions of the fatty acids in methanol were diluted in DMSO immediately before the experiment. All fatty acid concentrations used were below the CMC (CMC for OA, 150-200 μm (43), CMC for NO2-OA 40-60 μm).6 Nitro-fatty acid concentrations were determined using ε270 nm = 8220 m−1 cm−1 and ε330nm = 6,490 m−1 cm−1 (in phosphate buffer, pH 7.4) for NO2-OA and NO2-CLA, respectively (77). Excess fatty acid was eliminated by gel filtration before activity measured using the enzyme- coupled assay.
Structural assessment by CD
Protein samples in 10 mm phosphate buffer, pH 7.4, were filtered (0.2 μm) before running the spectra on a Chirascan Q100 spectropolarimeter (Applied Photophysics, UK). A 3 μm protein concentration was used for far-UV CD (0.1-cm light path) and 10 μm for near-UV CD (1-cm light path cuvette). All spectra were normalized using: ellipticity ϴ (mdeg)/(n-1) × (μm protein) × 10 light path (cm), where n is the number of amino acid residues. hSIRT6 was incubated at 37 °C for 30 min in the absence or presence of 5-fold excess of OA or NO2-OA before running the spectra.
In vitro hSIRT6 modification by biotinylated NO2-OA
hSIRT6 was first reduced in batch using a TCEP resin (30 min at 25 °C) followed by incubation with 10 mm NEM or 1 mm DEPC for 30 min at 37 °C to block cysteines and histidines, respectively. Unreacted reagents were removed by gel filtration and protein concentration was determined by UV absorbance (ε280 = 33,460 m−1 cm−1). Variable concentrations of Bt-NO2-OA were incubated with 1.2 μm hSIRT6 for 30 min at 25 °C, followed by SDS-PAGE separation and transfer to a PVDF membrane (1 h, 4 °C). Loaded amounts of hSIRT6 were visualized with Ponceau S staining. Membrane was blocked with PBS, 1% casein, overnight at 4 °C, and incubated with streptavidin-HRP for 1 h at room temperature. After three washes in 0.1% TBS-T buffer, bands were visualized using an ECL Western blotting system (ThermoFisher Scientific). Band intensities were quantified using ImageJ.
ESI-MS/MS analysis of Michael adduct formation between hSIRT6 and NO2-OA
hSIRT6 was reduced with 5 mm DTT (30 min at room temperature) and excess DTT was removed by gel filtration. Reduced hSIRT6 (4 μm) in 100 mm ammonium bicarbonate, pH 8, was incubated with two concentrations of NO2-OA (0.5 and 5×) for 30 min at 25 °C, and unreacted NO2-OA was removed by gel filtration to avoid interference during the analysis. Tryptic digestion was carried out for 2 h at 37 °C and the resulting peptides were analyzed by nano-LC (EASY-nLC 1000, Thermo Scientific) coupled to a linear ion trap mass spectrometer (LTQ Velos, Thermo Scientific). Peptide separation was performed using an in-house packed reverse phase nanocolumn (ReproSil-Pur®, Maisch; inner diameter, 75 mm; length, 28 cm) at a flow rate of 300 nl/min. A 70-min gradient of 0.1% (v/v) formic acid in water, 0.1% (v/v) formic acid in acetonitrile was used (4–55% acetonitrile in 60 min; 55–100% in 10 min). Mass spectrometer was set in a data-dependent acquisition mode (full MS followed by MS/MS of 10 most intense ions, using a 30-s dynamic exclusion list). Peptides were identified by searching a database containing E. coli Uniprot protein sequences (downloaded September 2018) plus hSirt6 sequence. Proteome Discoverer software (version 1.3.0.339, Thermo) with Sequest as search engine were used with the following parameters: precursor mass tolerance: 1.5 Da; fragment mass tolerance 0.8 Da; oxidation of methionine and modification with NO2-OA (+327.24 Da) as dynamic modifications. High-confident peptides (target FDR set to 0.01) were considered. Manual inspection of the MS/MS spectra was performed to corroborate peptide sequence and modification site assignments.
Intracellular SIRT6 modification by NO2-OA
HEK293T cells were grown in DMEM (high-glucose), 10% FBS, 100 units/ml of penicillin, and 100 μg/ml of streptomycin. Cells were transfected with FLAG-SIRT6 plasmid (Addgene plasmid number 13817) with Lipofectamine 2000 (Thermo Fisher Scientific) for 48 h. Cells were then incubated with 10 μm Bt-NO2-OA in serum-free DMEM for 1 h. Cell lysis and immunoprecipitation was performed as described previously (78). Briefly, cells were lysed in NETN buffer (20 mm Tris-HCl, pH 8.0, 100 mm NaCl, 1 mm EDTA, 0.5% Nonidet P-40) supplemented with 5 mm NaF, 50 mm 2-glycerophosphate, and a protease inhibitor mixture (Roche). Homogenates were incubated at 4 °C for 20 min under constant agitation and then centrifuged at 10,000 × g for 10 min at 4 °C. For each immunoprecipitation, 1 mg of protein was used. Samples were incubated with 20 μl of Strep-Tactin Sepharose beads for 1–2 h at 4 °C under constant rotation. Finally, immunoprecipitates were washed 2–3 times with cold NETN before addition of 2× Laemmli buffer followed by SDS-PAGE separation and transfer to a PVDF membrane (1 h, 4 °C). Primary and secondary antibodies utilized were anti-FLAG (Sigma, number F3165) and HRP-conjugated IgG from mouse (Sigma, A9044), respectively. Bands were visualized using an ECL Western blotting system (ThermoFisher Scientific) and images were processed by Photoshop.
Assessment of cellular H3K9 acetylation
HEK293T cells were grown in DMEM (high-glucose), 10% FBS, 100 units/ml of penicillin, and 100 μg/ml of streptomycin. Cells were transfected with FLAG-SIRT6 plasmid (Addgene plasmid number 13817) with Lipofectamine 2000 (Thermo Fisher Scientific) for 48 h. Cells were later incubated with 10 μm NO2-OA (or 10 μm OA) in serum-free DMEM for 1 h. Cell lysis was performed as described previously (79). Briefly, cells were lysed in RIPA buffer (25 mm Tris, pH 8.0, 150 mm NaCl, 1% Nonidet P-40, 0.1% SDS) supplemented with 5 mm NaF, 50 mm 2-glycerophosphate, 1 μm trichostatin A, 5 mm nicotinamide, and a protease inhibitor mixture (Roche). Homogenates were incubated at 4 °C for 20 min under constant agitation and then centrifuged at 10,000 × g for 10 min at 4 °C. Protein lysates were separated on 15% SDS-PAGE followed by transfer to PVDF membrane and incubation with specific primary antibodies (H3K9Ac, H3, FLAG, and tubulin). H3K9Ac and H3 antibodies were from Cell Signaling (9649S and 4620S, respectively). Images were processed using Photoshop and densitometry analysis were done with ImageJ (Rasband, W.S., Bethesda, MD, USA).
Computational modeling
The experimental holo structure of hSIRT6 in complex with a H3K9 myristoyl peptide and ADP-ribose fragment of NAD+ co-substrate (PDB code. 3ZG6 (28)) was used as receptor template. Residues 1 to 12 constituting the acyl-binding loop of hSIRT6 were removed from the structure due to their high flexibility, which is evidenced in apo structures (PDB codes 3PKI (34) and 5X16 (47)) or in presence of inhibitor/activator compounds (46, 80, 81). The co-substrate and water molecules 2006 2018 2019 2035 2036 2038 2040 2042 2053 2080 2099 2103 2104 2110 2111 2123 2124 2132 were kept in the structure to mask unspecific pockets around the protein surface. The coordinates of the receptor were moved prior docking according to the following transformation matrix using VMD 1.9.3 (82): {-0.782000720500946 0.30951544642448425-0.5409945249557495 69.48826599121094} {0.5513688921928406-0.06120478734374046-0.8320134282112122 3.671846389770508} {-0.2906324565410614-0.9489226341247559-0.1227950006723404-6.218472480773926} {0.0 0.0 0.0 1.0}, where braces define row vectors. The ligands corresponded to OA, 9-NO2-OA, 10-NO2-OA, CLA, 9-NO2-CLA, and 12-NO2-CLA (Fig. 2A). Docking calculations were done with AutoDock Vina (83). Each molecule was prepared with AutoDockTools (84) by merging nonpolar hydrogen atoms and computing Gasteiger charges. Histidine residues were assumed to be protonated at Nε. The searching grid was centered at coordinates x = 21.464 Å, y = 3.555 Å, and z = 16.125 Å, and the total size was defined as 50, 70, and 44 Å for each coordinate, respectively. The receptor was considered rigid while ligands were allowed to be full flexible. At each run 20 binding modes were generates using an exhaustiveness of 20 and allowing for energy ranges of 100 kcal/mol. Calculations were replicated 500 times for a total of 10,000 poses per ligand. Poses scoring up to 1 kcal/mol from the best conformation were selected for cluster analysis. Cluster analysis was based on the distance matrix of root mean square deviations, using a cut-off of 4 Å for grouping poses. On each case, the best pose from the top populated cluster was chosen for analysis. Possible conformations for 9-NO2-OA conjugation with Cys-18 were manually build and sculpted using PyMOL 1.8.4.0 (RRID:SCR_000305).
Statistical analysis
Data are presented as mean ± S.D. (experiments repeated at least three times). Statistical comparisons were made using one way-ANOVA followed by Tukey's multiple comparison test (GraphPad Prism 8.02). Difference was considered significant if p < 0.05.
Data availability
All data are contained within the manuscript. Raw data are available upon request (denicola@fcien.edu.uy, escande@pasteur.edu.uy).
Acknowledgments
We are grateful to Unidad de Bioquímica y Proteómica Analíticas, Institut Pasteur de Montevideo for MS analyses, Lucia Turell and Beatriz Alvarez (Universidad de la República) for helpful discussions, and Bruce A. Freeman (University of Pittsburgh) for insightful advice on nitro-fatty acids.
Universidad de la República Uruguay (UdelaR) (CSIC I + D grupo) to Ana Denicola
Agencia Nacional de Investigación e Innovación (ANII) (FCE) to Carlos Escande, and Ana Denicola
HHS | National Institutes of Health (NIH) (K01HL133331) to Darío A. Vitturi
Edited by Ruma Banerjee
Footnotes
Author contributions—M. C., M. B., M. R. M., R. D., D. A. V., C. E., and A. D. formal analysis; M. C., M. B., and M. R. M. investigation; M. C., M. B., M. R. M., L. S., R. D., and D. A. V. methodology; M. R. M., C. E., and A. D. conceptualization; M. R. M. and A. D. writing-original draft; R. D., D. A. V., C. E., and A. D. supervision; D. A. V., C. E., and A. D. funding acquisition; C. E. and A. D. writing-review and editing.
Funding and additional information—This work was supported by grants (to A. D. and C. E.) and fellowships (to M. C., M. B., and L. S.) from Agencia Nacional de Investigación e Innovación (ANII, Uruguay), and National Institutes of Health Grants K01HL133331 and P30DK079307 (to D. A. V.). This work was partially supported by FOCEM (Mercosur), Comisión Sectorial de Investigación Científica (CSIC) Universidad de la República and Programa de Desarrollo de las Ciencias Básicas (PEDECIBA), Uruguay. A. D., C. E., R. D., and M. R. M. are part of the SNI program of ANII.
Conflict of interest—The authors declare that they have no conflict of interest with the contents of this article.
- NO2-FA
- nitro-fatty acids
- CLA
- conjugated linoleic acid
- OA
- oleic acid
- Bt-NO2-OA
- biotinylated-NO2-OA
- IPTG
- isopropyl β-d-1-thiogalactopyranoside
- NEM
- N-ethylmalemide
- HRP
- horseradish peroxidase
- DEPC
- diethyl pyrocarbonate
- TCEP
- tris(2-carboxyethyl)phosphine hydrochloride
- β-ME
- β-mercaptoethanol
- PAR
- 4-(2-pyridylazo)resorcinol
- DMEM
- Dulbecco's modified Eagle's medium
- FBS
- fetal bovine serum
- ANOVA
- analysis of variance
- PDB
- Protein Data Bank
- PVDF
- polyvinylidene difluoride
- CMC
- critical micellar concentration.
M. Carreño and A. Denicola, unpublished results.
Contributor Information
Carlos Escande, Email: escande@pasteur.edu.uy.
Ana Denicola, Email: denicola@fcien.edu.uy.
References
- 1.Chalkiadaki A., Guarente L. Sirtuins mediate mammalian metabolic responses to nutrient availability. Nat. Rev. Endocrinol. 2012;8:287–296. doi: 10.1038/nrendo.2011.225. 22249520. [DOI] [PubMed] [Google Scholar]
- 2.Finkel T., Deng C.X., Mostoslavsky R. Recent progress in the biology and physiology of sirtuins. Nature. 2009;460:587–591. doi: 10.1038/nature08197. 19641587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Imai S., Armstrong C.M., Kaeberlein M., Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. 10693811. [DOI] [PubMed] [Google Scholar]
- 4.Michan S., Sinclair D. Sirtuins in mammals: insights into their biological function. Biochem. J. 2007;404:1–13. doi: 10.1042/BJ20070140. 17447894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Giblin W., Skinner M.E., Lombard D.B. Sirtuins: guardians of mammalian healthspan. Trends Genet. 2014;30:271–286. doi: 10.1016/j.tig.2014.04.007. 24877878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haigis M.C., Sinclair D.A. Mammalian sirtuins: biological insights and disease relevance. Annu. Rev. Pathol. 2010;5:253–295. doi: 10.1146/annurev.pathol.4.110807.092250. 20078221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Houtkooper R.H., Pirinen E., Auwerx J. Sirtuins as regulators of metabolism and healthspan. Nat. Rev. Mol. Cell Biol. 2012;13:225–238. doi: 10.1038/nrm3293. 22395773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rauh D., Fischer F., Gertz M., Lakshminarasimhan M., Bergbrede T., Aladini F., Kambach C., Becker C.F., Zerweck J., Schutkowski M., Steegborn C. An acetylome peptide microarray reveals specificities and deacetylation substrates for all human sirtuin isoforms. Nat. Commun. 2013;4 doi: 10.1038/ncomms3327. 23995836. [DOI] [PubMed] [Google Scholar]
- 9.Michishita E., Park J.Y., Burneskis J.M., Barrett J.C., Horikawa I. Evolutionarily conserved and nonconserved cellular localizations and functions of human SIRT proteins. Mol. Biol. Cell. 2005;16:4623–4635. doi: 10.1091/mbc.e05-01-0033. 16079181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gil R., Barth S., Kanfi Y., Cohen H.Y. SIRT6 exhibits nucleosome-dependent deacetylase activity. Nucleic Acids Res. 2013;41:8537–8545. doi: 10.1093/nar/gkt642. 23892288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kugel S., Mostoslavsky R. Chromatin and beyond: the multitasking roles for SIRT6. Trends Biochem. Sci. 2014;39:72–81. doi: 10.1016/j.tibs.2013.12.002. 24438746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mao Z., Hine C., Tian X., Van Meter M., Au M., Vaidya A., Seluanov A., Gorbunova V. SIRT6 promotes DNA repair under stress by activating PARP1. Science. 2011;332:1443–1446. doi: 10.1126/science.1202723. 21680843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kanfi Y., Naiman S., Amir G., Peshti V., Zinman G., Nahum L., Bar-Joseph Z., Cohen H.Y. The sirtuin SIRT6 regulates lifespan in male mice. Nature. 2012;483:218–221. doi: 10.1038/nature10815. 22367546. [DOI] [PubMed] [Google Scholar]
- 14.Kanfi Y., Peshti V., Gil R., Naiman S., Nahum L., Levin E., Kronfeld-Schor N., Cohen H.Y. SIRT6 protects against pathological damage caused by diet-induced obesity. Aging Cell. 2010;9:162–173. doi: 10.1111/j.1474-9726.2009.00544.x. 20047575. [DOI] [PubMed] [Google Scholar]
- 15.Roichman A., Kanfi Y., Glazz R., Naiman S., Amit U., Landa N., Tinman S., Stein I., Pikarsky E., Leor J., Cohen H.Y. SIRT6 overexpression improves various aspects of mouse healthspan. J. Gerontol. A Biol. Sci. Med. Sci. 2017;72:603–615. doi: 10.1093/gerona/glw152. 27519885. [DOI] [PubMed] [Google Scholar]
- 16.Zhaohui C., Shuihua W. Protective effects of SIRT6 against inflammation, oxidative stress, and cell apoptosis in spinal cord injury. Inflammation. 2020;43:1751–1758. doi: 10.1007/s10753-020-01249-2. [DOI] [PubMed] [Google Scholar]
- 17.Mostoslavsky R., Chua K.F., Lombard D.B., Pang W.W., Fischer M.R., Gellon L., Liu P., Mostoslavsky G., Franco S., Murphy M.M., Mills K.D., Patel P., Hsu J.T., Hong A.L., Ford E. Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell. 2006;124:315–329. doi: 10.1016/j.cell.2005.11.044. 16439206. [DOI] [PubMed] [Google Scholar]
- 18.Kuang J., Zhang Y., Liu Q., Shen J., Pu S., Cheng S., Chen L., Li H., Wu T., Li R., Li Y., Zou M., Zhang Z., Jiang W., Xu G. Fat-specific Sirt6 ablation sensitizes mice to high-fat diet-induced obesity and insulin resistance by inhibiting lipolysis. Diabetes. 2017;66:1159–1171. doi: 10.2337/db16-1225. 28250020. [DOI] [PubMed] [Google Scholar]
- 19.Lee Y., Ka S.O., Cha H.N., Chae Y.N., Kim M.K., Park S.Y., Bae E.J., Park B.H. Myeloid sirtuin 6 deficiency causes insulin resistance in high-fat diet-fed mice by eliciting macrophage polarization toward an M1 phenotype. Diabetes. 2017;66:2659–2668. doi: 10.2337/db16-1446. 28607107. [DOI] [PubMed] [Google Scholar]
- 20.Song M.Y., Wang J., Ka S.O., Bae E.J., Park B.H. Insulin secretion impairment in Sirt6 knockout pancreatic beta cells is mediated by suppression of the FoxO1-Pdx1-Glut2 pathway. Sci. Rep. 2016;6 doi: 10.1038/srep30321. 27457971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schwer B., Schumacher B., Lombard D.B., Xiao C., Kurtev M.V., Gao J., Schneider J.I., Chai H., Bronson R.T., Tsai L.H., Deng C.X., Alt F.W. Neural sirtuin 6 (Sirt6) ablation attenuates somatic growth and causes obesity. Proc. Natl. Acad. Sci. U.S.A. 2010;107:21790–21794. doi: 10.1073/pnas.1016306107. 21098266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kawahara T.L., Michishita E., Adler A.S., Damian M., Berber E., Lin M., McCord R.A., Ongaigui K.C., Boxer L.D., Chang H.Y., Chua K.F. SIRT6 links histone H3 lysine 9 deacetylation to NF-κB-dependent gene expression and organismal life span. Cell. 2009;136:62–74. doi: 10.1016/j.cell.2008.10.052. 19135889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Michishita E., McCord R.A., Berber E., Kioi M., Padilla-Nash H., Damian M., Cheung P., Kusumoto R., Kawahara T.L., Barrett J.C., Chang H.Y., Bohr V.A., Ried T., Gozani O., Chua K.F. SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin. Nature. 2008;452:492–496. doi: 10.1038/nature06736. 18337721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang B., Zwaans B.M., Eckersdorff M., Lombard D.B. The sirtuin SIRT6 deacetylates H3 K56Ac in vivo to promote genomic stability. Cell Cycle. 2009;8:2662–2663. doi: 10.4161/cc.8.16.9329. 19597350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tasselli L., Xi Y., Zheng W., Tennen R.I., Odrowaz Z., Simeoni F., Li W., Chua K.F. SIRT6 deacetylates H3K18ac at pericentric chromatin to prevent mitotic errors and cellular senescence. Nat. Struct. Mol. Biol. 2016;23:434–440. doi: 10.1038/nsmb.3202. 27043296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhong L., D'Urso A., Toiber D., Sebastian C., Henry R.E., Vadysirisack D.D., Guimaraes A., Marinelli B., Wikstrom J.D., Nir T., Clish C.B., Vaitheesvaran B., Iliopoulos O., Kurland I., Dor Y. The histone deacetylase Sirt6 regulates glucose homeostasis via Hif1α. Cell. 2010;140:280–293. doi: 10.1016/j.cell.2009.12.041. 20141841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tennen R.I., Chua K.F. Chromatin regulation and genome maintenance by mammalian SIRT6. Trends Biochem. Sci. 2011;36:39–46. doi: 10.1016/j.tibs.2010.07.009. 20729089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiang H., Khan S., Wang Y., Charron G., He B., Sebastian C., Du J., Kim R., Ge E., Mostoslavsky R., Hang H.C., Hao Q., Lin H. SIRT6 regulates TNF-α secretion through hydrolysis of long-chain fatty acyl lysine. Nature. 2013;496:110–113. doi: 10.1038/nature12038. 23552949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klein M.A., Denu J.M. Biological and catalytic functions of sirtuin 6 as targets for small-molecule modulators. J. Biol. Chem. 2020;295:11021–11041. doi: 10.1074/jbc.REV120.011438. 32518153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Costantini S., Sharma A., Raucci R., Costantini M., Autiero I., Colonna G. Genealogy of an ancient protein family: the Sirtuins, a family of disordered members. BMC Evol. Biol. 2013;13:60. doi: 10.1186/1471-2148-13-60. 23497088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sanders B.D., Jackson B., Marmorstein R. Structural basis for sirtuin function: what we know and what we don't. Biochim. Biophys. Acta. 2010;1804:1604–1616. doi: 10.1016/j.bbapap.2009.09.009. 19766737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao K., Chai X., Clements A., Marmorstein R. Structure and autoregulation of the yeast Hst2 homolog of Sir2. Nat. Struct. Biol. 2003;10:864–871. doi: 10.1038/nsb978. 14502267. [DOI] [PubMed] [Google Scholar]
- 33.Min J., Landry J., Sternglanz R., Xu R.M. Crystal structure of a SIR2 homolog-NAD complex. Cell. 2001;105:269–279. doi: 10.1016/S0092-8674(01)00317-8. 11336676. [DOI] [PubMed] [Google Scholar]
- 34.Pan P.W., Feldman J.L., Devries M.K., Dong A., Edwards A.M., Denu J.M. Structure and biochemical functions of SIRT6. J. Biol. Chem. 2011;286:14575–14587. doi: 10.1074/jbc.M111.218990. 21362626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Feldman J.L., Dittenhafer-Reed K.E., Kudo N., Thelen J.N., Ito A., Yoshida M., Denu J.M. Kinetic and structural basis for acyl-group selectivity and NAD+ dependence in sirtuin-catalyzed deacylation. Biochemistry. 2015;54:3037–3050. doi: 10.1021/acs.biochem.5b00150. 25897714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Frye R.A. Phylogenetic classification of prokaryotic and eukaryotic Sir2-like proteins. Biochem. Biophys. Res. Commun. 2000;273:793–798. doi: 10.1006/bbrc.2000.3000. 10873683. [DOI] [PubMed] [Google Scholar]
- 37.Tennen R.I., Berber E., Chua K.F. Functional dissection of SIRT6: identification of domains that regulate histone deacetylase activity and chromatin localization. Mech. Ageing Dev. 2010;131:185–192. doi: 10.1016/j.mad.2010.01.006. 20117128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Borra M.T., Langer M.R., Slama J.T., Denu J.M. Substrate specificity and kinetic mechanism of the Sir2 family of NAD+-dependent histone/protein deacetylases. Biochemistry. 2004;43:9877–9887. doi: 10.1021/bi049592e. 15274642. [DOI] [PubMed] [Google Scholar]
- 39.Jackson M.D., Denu J.M. Structural identification of 2′- and 3′-O-acetyl-ADP-ribose as novel metabolites derived from the Sir2 family of β -NAD+-dependent histone/protein deacetylases. J. Biol. Chem. 2002;277:18535–18544. doi: 10.1074/jbc.M200671200. 11893743. [DOI] [PubMed] [Google Scholar]
- 40.Jin L., Wei W., Jiang Y., Peng H., Cai J., Mao C., Dai H., Choy W., Bemis J.E., Jirousek M.R., Milne J.C., Westphal C.H., Perni R.B. Crystal structures of human SIRT3 displaying substrate-induced conformational changes. J. Biol. Chem. 2009;284:24394–24405. doi: 10.1074/jbc.M109.014928. 19535340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smith B.C., Denu J.M. Sir2 protein deacetylases: evidence for chemical intermediates and functions of a conserved histidine. Biochemistry. 2006;45:272–282. doi: 10.1021/bi052014t. 16388603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morris B.J. Seven sirtuins for seven deadly diseases of aging. Free Radic. Biol. Med. 2013;56:133–171. doi: 10.1016/j.freeradbiomed.2012.10.525. 23104101. [DOI] [PubMed] [Google Scholar]
- 43.Feldman J.L., Baeza J., Denu J.M. Activation of the protein deacetylase SIRT6 by long-chain fatty acids and widespread deacylation by mammalian sirtuins. J. Biol. Chem. 2013;288:31350–31356. doi: 10.1074/jbc.C113.511261. 24052263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rahnasto-Rilla M., Kokkola T., Jarho E., Lahtela-Kakkonen M., Moaddel R. N-Acylethanolamines bind to SIRT6. ChemBioChem. 2016;17:77–81. doi: 10.1002/cbic.201500482. 26607666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rahnasto-Rilla M., Tyni J., Huovinen M., Jarho E., Kulikowicz T., Ravichandran S., A Bohr V., Ferrucci L., Lahtela-Kakkonen M., Moaddel R. Natural polyphenols as sirtuin 6 modulators. Sci. Rep. 2018;8 doi: 10.1038/s41598-018-22388-5. 29515203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.You W., Rotili D., Li T.M., Kambach C., Meleshin M., Schutkowski M., Chua K.F., Mai A., Steegborn C. Structural basis of sirtuin 6 activation by synthetic small molecules. Angew. Chem. Int. Ed. Engl. 2017;56:1007–1011. doi: 10.1002/anie.201610082. 27990725. [DOI] [PubMed] [Google Scholar]
- 47.Huang Z., Zhao J., Deng W., Chen Y., Shang J., Song K., Zhang L., Wang C., Lu S., Yang X., He B., Min J., Hu H., Tan M., Xu J. Identification of a cellularly active SIRT6 allosteric activator. Nat. Chem. Biol. 2018;14:1118–1126. doi: 10.1038/s41589-018-0150-0. 30374165. [DOI] [PubMed] [Google Scholar]
- 48.Klein M.A., Liu C., Kuznetsov V.I., Feltenberger J.B., Tang W., Denu J.M. Mechanism of activation for the sirtuin 6 protein deacylase. J. Biol. Chem. 2020;295:1385–1399. doi: 10.1074/jbc.RA119.011285. 31822559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Delmastro-Greenwood M., Hughan K.S., Vitturi D.A., Salvatore S.R., Grimes G., Potti G., Shiva S., Schopfer F.J., Gladwin M.T., Freeman B.A., Gelhaus Wendell S. Nitrite and nitrate-dependent generation of anti-inflammatory fatty acid nitroalkenes. Free Radic. Biol. Med. 2015;89:333–341. doi: 10.1016/j.freeradbiomed.2015.07.149. 26385079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bonacci G., Baker P.R., Salvatore S.R., Shores D., Khoo N.K., Koenitzer J.R., Vitturi D.A., Woodcock S.R., Golin-Bisello F., Cole M.P., Watkins S., St Croix C., Batthyany C.I., Freeman B.A., Schopfer F.J. Conjugated linoleic acid is a preferential substrate for fatty acid nitration. J. Biol. Chem. 2012;287:44071–44082. doi: 10.1074/jbc.M112.401356. 23144452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vitturi D.A., Maynard C., Olsufka M., Straub A.C., Krehel N., Kudenchuk P.J., Nichol G., Sayre M., Kim F., Dezfulian C. Nitrite elicits divergent NO-dependent signaling that associates with outcome in out of hospital cardiac arrest. Redox Biol. 2020;32 doi: 10.1016/j.redox.2020.101463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schopfer F.J., Cipollina C., Freeman B.A. Formation and signaling actions of electrophilic lipids. Chem. Rev. 2011;111:5997–6021. doi: 10.1021/cr200131e. 21928855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Salvatore S.R., Vitturi D.A., Baker P.R., Bonacci G., Koenitzer J.R., Woodcock S.R., Freeman B.A., Schopfer F.J. Characterization and quantification of endogenous fatty acid nitroalkene metabolites in human urine. J. Lipid Res. 2013;54:1998–2009. doi: 10.1194/jlr.M037804. 23620137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Baker L.M., Baker P.R., Golin-Bisello F., Schopfer F.J., Fink M., Woodcock S.R., Branchaud B.P., Radi R., Freeman B.A. Nitro-fatty acid reaction with glutathione and cysteine: Kinetic analysis of thiol alkylation by a Michael addition reaction. J. Biol. Chem. 2007;282:31085–31093. doi: 10.1074/jbc.M704085200. 17720974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Buchan G.J., Bonacci G., Fazzari M., Salvatore S.R., Gelhaus Wendell S. Nitro-fatty acid formation and metabolism. Nitric Oxide. 2018;79:38–44. doi: 10.1016/j.niox.2018.07.003. 30006146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Batthyany C., Schopfer F.J., Baker P.R., Duran R., Baker L.M., Huang Y., Cervenansky C., Branchaud B.P., Freeman B.A. Reversible post-translational modification of proteins by nitrated fatty acids in vivo. J. Biol. Chem. 2006;281:20450–20463. doi: 10.1074/jbc.M602814200. 16682416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kansanen E., Bonacci G., Schopfer F.J., Kuosmanen S.M., Tong K.I., Leinonen H., Woodcock S.R., Yamamoto M., Carlberg C., Ylä-Herttuala S., Freeman B.A., Levonen A.L. Electrophilic nitro-fatty acids activate NRF2 by a KEAP1 cysteine 151-independent mechanism. J. Biol. Chem. 2011;286:14019–14027. doi: 10.1074/jbc.M110.190710. 21357422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cui T., Schopfer F.J., Zhang J., Chen K., Ichikawa T., Baker P.R., Batthyany C., Chacko B.K., Feng X., Patel R.P., Agarwal A., Freeman B.A., Chen Y.E. Nitrated fatty acids: Endogenous anti-inflammatory signaling mediators. J. Biol. Chem. 2006;281:35686–35698. doi: 10.1074/jbc.M603357200. 16887803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Villacorta L., Minarrieta L., Salvatore S.R., Khoo N.K., Rom O., Gao Z., Berman R.C., Jobbagy S., Li L., Woodcock S.R., Chen Y.E., Freeman B.A., Ferreira A.M., Schopfer F.J., Vitturi D.A. In situ generation, metabolism and immunomodulatory signaling actions of nitro-conjugated linoleic acid in a murine model of inflammation. Redox Biol. 2018;15:522–531. doi: 10.1016/j.redox.2018.01.005. 29413964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kelley E.E., Batthyany C.I., Hundley N.J., Woodcock S.R., Bonacci G., Del Rio J.M., Schopfer F.J., Lancaster J.R., Jr., Freeman B.A., Tarpey M.M. Nitro-oleic acid, a novel and irreversible inhibitor of xanthine oxidoreductase. J. Biol. Chem. 2008;283:36176–36184. doi: 10.1074/jbc.M802402200. 18974051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bonacci G., Schopfer F.J., Batthyany C.I., Rudolph T.K., Rudolph V., Khoo N.K., Kelley E.E., Freeman B.A. Electrophilic fatty acids regulate matrix metalloproteinase activity and expression. J. Biol. Chem. 2011;286:16074–16081. doi: 10.1074/jbc.M111.225029. 21454668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Santos L., Escande C., Denicola A. Potential modulation of sirtuins by oxidative stress. Oxid. Med. Cell. Longev. 2016;2016 doi: 10.1155/2016/9831825. 26788256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yang J., Gupta V., Carroll K.S., Liebler D.C. Site-specific mapping and quantification of protein S-sulphenylation in cells. Nat. Commun. 2014;5 doi: 10.1038/ncomms5776. 25175731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Long D., Wu H., Tsang A.W., Poole L.B., Yoza B.K., Wang X., Vachharajani V., Furdui C.M., McCall C.E. The oxidative state of cysteine thiol 144 regulates the SIRT6 glucose homeostat. Sci. Rep. 2017;7 doi: 10.1038/s41598-017-11388-6. 28887543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kalous K.S., Wynia-Smith S.L., Summers S.B., Smith B.C. Human sirtuins are differentially sensitive to inhibition by nitrosating agents and other cysteine oxidants. J. Biol. Chem. 2020;295:8524–8536. doi: 10.1074/jbc.RA119.011988. 32371394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Garner R.M., Mould D.R., Chieffo C., Jorkasky D.K. Pharmacokinetic and pharmacodynamic effects of oral CXA-10, a nitro fatty acid, after single and multiple ascending doses in healthy and obese subjects. Clin. Transl. Sci. 2019;12:667–676. doi: 10.1111/cts.12672. 31343124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Villacorta L., Chang L., Salvatore S.R., Ichikawa T., Zhang J., Petrovic-Djergovic D., Jia L., Carlsen H., Schopfer F.J., Freeman B.A., Chen Y.E. Electrophilic nitro-fatty acids inhibit vascular inflammation by disrupting LPS-dependent TLR4 signalling in lipid rafts. Cardiovasc. Res. 2013;98:116–124. doi: 10.1093/cvr/cvt002. 23334216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kelley E.E., Baust J., Bonacci G., Golin-Bisello F., Devlin J.E., St Croix C.M., Watkins S.C., Gor S., Cantu-Medellin N., Weidert E.R., Frisbee J.C., Gladwin M.T., Champion H.C., Freeman B.A., Khoo N.K. Fatty acid nitroalkenes ameliorate glucose intolerance and pulmonary hypertension in high-fat diet-induced obesity. Cardiovasc. Res. 2014;101:352–363. doi: 10.1093/cvr/cvt341. 24385344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Khoo N.K.H., Fazzari M., Chartoumpekis D.V., Li L., Guimaraes D.A., Arteel G.E., Shiva S., Freeman B.A. Electrophilic nitro-oleic acid reverses obesity-induced hepatic steatosis. Redox Biol. 2019;22 doi: 10.1016/j.redox.2019.101132. 30769284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rom O., Khoo N.K.H., Chen Y.E., Villacorta L. Inflammatory signaling and metabolic regulation by nitro-fatty acids. Nitric Oxide. 2018;78:140–145. doi: 10.1016/j.niox.2018.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schopfer F.J., Cole M.P., Groeger A.L., Chen C.S., Khoo N.K., Woodcock S.R., Golin-Bisello F., Motanya U.N., Li Y., Zhang J., Garcia-Barrio M.T., Rudolph T.K., Rudolph V., Bonacci G., Baker P.R. Covalent peroxisome proliferator-activated receptor γ adduction by nitro-fatty acids: selective ligand activity and anti-diabetic signaling actions. J. Biol. Chem. 2010;285:12321–12333. doi: 10.1074/jbc.M109.091512. 20097754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Woodcock S.R., Bonacci G., Gelhaus S.L., Schopfer F.J. Nitrated fatty acids: synthesis and measurement. Free Radic. Biol. Med. 2013;59:14–26. doi: 10.1016/j.freeradbiomed.2012.11.015. 23200809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Woodcock S.R., Salvatore S.R., Bonacci G., Schopfer F.J., Freeman B.A. Biomimetic nitration of conjugated linoleic acid: formation and characterization of naturally occurring conjugated nitrodienes. J. Org. Chem. 2014;79:25–33. doi: 10.1021/jo4021562. 24350701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ghislain M., Talla E., Francois J.M. Identification and functional analysis of the Saccharomyces cerevisiae nicotinamidase gene, PNC1. Yeast. 2002;19:215–224. doi: 10.1002/yea.810. 11816029. [DOI] [PubMed] [Google Scholar]
- 75.Hunt J.B., Neece S.H., Ginsburg A. The use of 4-(2-pyridylazo)resorcinol in studies of zinc release from Escherichia coli aspartate transcarbamoylase. Anal. Biochem. 1985;146:150–157. doi: 10.1016/0003-2697(85)90409-9. 3887984. [DOI] [PubMed] [Google Scholar]
- 76.Smith B.C., Hallows W.C., Denu J.M. A continuous microplate assay for sirtuins and nicotinamide-producing enzymes. Anal. Biochem. 2009;394:101–109. doi: 10.1016/j.ab.2009.07.019. 19615966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Turell L., Vitturi D.A., Coitiño E.L., Lebrato L., Möller M.N., Sagasti C., Salvatore S.R., Woodcock S.R., Alvarez B., Schopfer F.J. The chemical basis of thiol addition to nitro-conjugated linoleic acid, a protective cell-signaling lipid. J. Biol. Chem. 2017;292:1145–1159. doi: 10.1074/jbc.M116.756288. 27923813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Escande C., Chini C.C., Nin V., Dykhouse K.M., Novak C.M., Levine J., van Deursen J., Gores G.J., Chen J., Lou Z., Chini E.N. Deleted in breast cancer-1 regulates SIRT1 activity and contributes to high-fat diet-induced liver steatosis in mice. J. Clin. Invest. 2010;120:545–558. doi: 10.1172/JCI39319. 20071779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Colman L., Caggiani M., Leyva A., Bresque M., Liechocki S., Maya-Monteiro C.M., Mazal D., Batthyany C., Calliari A., Contreras P., Escande C. The protein deleted in breast cancer-1 (DBC1) regulates vascular response and formation of aortic dissection during angiotensin II infusion. Sci. Rep. 2020;10 doi: 10.1038/s41598-020-63841-8. 32317757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.You W., Steegborn C. Structural basis of sirtuin 6 inhibition by the hydroxamate trichostatin A: implications for protein deacylase drug development. J. Med. Chem. 2018;61:10922–10928. doi: 10.1021/acs.jmedchem.8b01455. 30395713. [DOI] [PubMed] [Google Scholar]
- 81.You W., Zheng W., Weiss S., Chua K.F., Steegborn C. Structural basis for the activation and inhibition of Sirtuin 6 by quercetin and its derivatives. Sci. Rep. 2019;9 doi: 10.1038/s41598-019-55654-1. 31844103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. 27–38. 8744570. [DOI] [PubMed] [Google Scholar]
- 83.Trott O., Olson A.J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. 19499576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Morris G.M., Huey R., Lindstrom W., Sanner M.F., Belew R.K., Goodsell D.S., Olson A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. 19399780. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data are contained within the manuscript. Raw data are available upon request (denicola@fcien.edu.uy, escande@pasteur.edu.uy).