Abstract
The recent structural elucidation of ex vivo Drosophila Orb2 fibrils revealed a novel amyloid formed by interdigitated Gln and His residue side chains belonging to the prion-like domain. However, atomic-level details on the conformational transitions associated with memory consolidation remain unknown. Here, we have characterized the nascent conformation and dynamics of the prion-like domain (PLD) of Orb2A using a nonconventional liquid-state NMR spectroscopy strategy based on 13C detection to afford an essentially complete set of 13Cα, 13Cβ, 1Hα, and backbone 13CO and 15N assignments. At pH 4, where His residues are protonated, the PLD is disordered and flexible, except for a partially populated α-helix spanning residues 55–60, and binds RNA oligos, but not divalent cations. At pH 7, in contrast, His residues are predominantly neutral, and the Q/H segments adopt minor populations of helical structure, show decreased mobility and start to self-associate. At pH 7, the His residues do not bind RNA or Ca2+, but do bind Zn2+, which promotes further association. These findings represent a remarkable case of structural plasticity, based on which an updated model for Orb2A functional amyloidogenesis is suggested.
Keywords: nuclear magnetic resonance, NMR, memory consolidation, functional amyloid, prion-like domain, amyloidogenic proteins, amyloid, intrinsically disordered protein, Drosophila, metal ion-protein interaction, zinc, polyglutamine, pH regulation
The comprehension of how the nervous system encodes memory has been an important goal since the beginning of modern neuroscience. At the turn of the twentieth century, memory was proposed to be encoded as alterations in the neurite network (1) as a persistent chemical change (2). This hypothesis has gained support over the decades (3, 4, 5, 6), and in 2003, Si et al. (7) published a seminal paper providing the first evidence for the molecular basis of memory consolidation. In that study, Si and co-workers proposed that functional aggregation of the protein CPEB into ordered structures is key to memory consolidation in Aplysia. Numerous studies have corroborated the key functional role of CPEB aggregation as well as that of its homologs in memory consolidation in Drosophila melanogaster and mammals (8).
The Drosophila CPEB homolog, named Orb2, has been investigated in the most detail. Two Orb2 isoforms, called Orb2A and Orb2B, are relevant for long-term memory in Drosophila (9, 10). Although they differ at the N terminus, both Orb2A and Orb2B contain a prion-like domain (PLD) rich in Gln and His residues, followed by a stretch of Gly and Ser residues (Fig. S1A). Although the Orb2A isoform is rare, it is essential for triggering the aggregation of the more abundant Orb2B and to maintain the memory trace (10, 11). The first nine residues of Orb2A are unique to this isoform (Fig. S1A). Rich in hydrophobic side chains, these nine residues have been reported to interact with membrane mimetics (12), to be crucial for aggregation (11), and to adopt a β-strand conformation within an Orb2 amyloid structure formed in vitro as characterized by solid-state NMR and EPR (13).
Both Orb2A and Orb2B contain a 31-residue-long Q/H-rich stretch, followed by a short amphiphilic segment (residues N55LSAL59) and a second modest Q/H-stretch (H60HHHQQQQQ68) (Fig. S1A). Because of its similarity to amyloid-forming polyQ stretches in huntingtin (14) and the androgen receptor (15), the main Q/H-rich stretch (residues 23–53) was suspected to be key for Orb2 amyloid formation. Indeed, while both Q/H-rich stretches appear rather disordered in Orb2 amyloids formed in vitro (13), Orb2 aggregation and memory consolidation can be blocked in Drosophila by a peptide which inhibits polyglutamine aggregation (16). Very recently, the elucidation of the structure of physiological Orb2 amyloid isolated directly from Drosophila fly brains has shown that the main Q/H-rich stretch does in fact form the physiologically relevant amyloid structure (17). This amyloid features 3-fold symmetry and side chain to main chain H-bonds (Fig. S1B). The participation of numerous His residues in the amyloid core provides a singular mechanism for amyloid destabilization through acidification. Intriguingly, the first nine residues of Orb2A, although relevant for triggering aggregation (11), do not contribute to the amyloid core, which is composed of the more abundant Orb2B isoform (17) (Fig. S1).
Following the second Q/H-rich stretch, there is a 200-residue, presumptively disordered region with an elevated content of Gly, Ser, Pro, and Asn (Figs. S1A and S2). The conserved C-terminal half of Orb2 is composed of two RRM domains, which bind Orb2-specific mRNA targets, and a ZZ-type zinc finger domain (Fig. S1A). ZZ-type zinc fingers are a special subclass that binds two Zn2+ ions and generally mediate protein/protein interactions, not nucleic acid binding (18). In Orb2, the conserved C-terminal domains are crucial for binding mRNAs containing a uridine-rich 3′ UTR, whose consensus sequence is 5′UUUUAU3′ (19), and potentially different translation complexes (17). This binding suppresses the translation of mRNAs coding for factors promoting synapse formation, synapse growth, and proteases (19). Upon neuronal stimulation, Orb2 forms amyloid aggregates leading to the activation of these mRNAs (20). Because of its physiological importance, Orb2 aggregation is finely controlled. Studies have reported key roles of intron retention (21), phosphorylation and protein stability (22), and chaperones (23) in tightly regulating Orb2 functional amyloid formation. Besides the mentioned control of Orb2 aggregation via acidification (17), Siemer and co-workers proposed that the spacing of His residues within the long Q/H-rich stretch would position them on the same face of a hypothetical α-helix (24). This may contribute to the binding of Ni2+, Cu2+, and Zn2+ which might impact aggregation (24). Interestingly, the longer Q/H-rich region of Orb2 may be sequestered into pathological amyloids formed by expanded polyQ segments of huntingtin, providing a novel explanation for memory loss in dementia (16, 25).
Whereas all these results have provided molecular insight into Orb2's role in memory consolidation, much is still unclear regarding the atomic-level details of the amyloid formation process. In particular, it is unknown whether the nascent protein contains elements of partial structure which predispose the formation of the functional amyloid, or alternatively, act as a safety mechanism to prevent premature or excess amyloid formation. The objective of this study is to characterize the atomic-level conformation and dynamics of Orb2A PLD and the first residues of the G/S-rich region using solution NMR spectroscopy. Because pH is proposed to impact amyloid formation and stability (17), we have characterized the conformation of Orb2A both at pH 4, where His residues are positively charged, which impedes aggregation, as well as pH 7, where neutral His are compatible with amyloid formation. In addition, we have probed the interaction of RNA ligands, Ca2+ and Zn2+ with the Q/H-rich regions of Orb2A. The structural transitions observed may represent nascent structures in the process of aggregation into functional amyloids.
Results
The Orb2A PLD is disordered at pH 4.0, except for a partly populated α-helix spanning residues 55–60
Initial 1D 1H and 2D 1H-15N HSQC spectra recorded in PBS at pH 7.0 revealed broad lines, which are not optimal for spectral analysis and assignment. Therefore, following the low pH, low ionic strength strategy of Song and coworkers (26), which we have recently applied to assign the snow flea antifreeze protein (27) and a RepA winged helix domain (28), spectra of Orb2A PLD were recorded at pH 4.0 in 1 mm acetic acid buffer at 25°C. Under these conditions, the signals were sharper and gave excellent quality 2D and 3D spectra, despite significant peak overlap. The 1Hα, 1HN, 13Cα, 13Cβ, and backbone 15N and 13CO chemical shifts at pH 4.0, 25°C, obtained through an unconventional strategy of proton-less 3D NMR spectra (see “Experimental procedures”), have been deposited in the BMRB data bank under accession number 50274. The assignments are complete except for the 1Hα of proline residues and the 13CO and 13Cβ of terminal Ser-88.
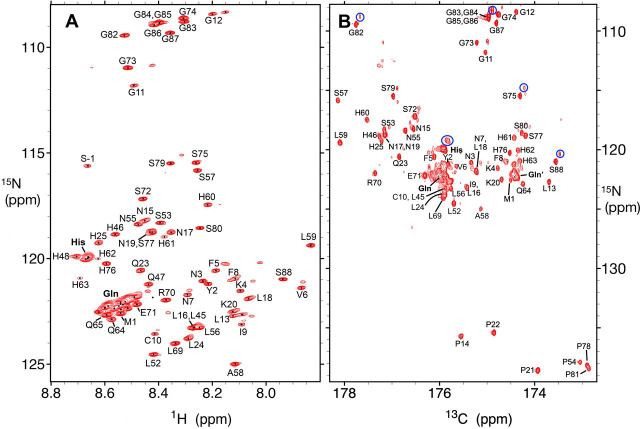
The assigned 2D 1H-15N HSQC and 2D 13C-15N CON spectra registered at pH 4.0, 25°C, are shown in Fig. 1, A and B, respectively. The resonances are generally sharp and well defined except for clusters of overlapped Gln and His residues belonging to the amyloidogenic tract (Fig. S1B). The low 1HN signal dispersion is a hallmark of disordered proteins and an assessment of the 13Cα, 13Cβ, 13CO, and 15N chemical shifts using the TALOS+ program confirmed the overall dearth of structure in the Orb2A PLD under these conditions. With regard to the glutamine side chain amide groups, their 1H2-15N signals are also clumped into overlapped peaks (Fig. S3). Notably, they do not show the striking pattern of disperse signals observed for the glutamine side chain resonances of the androgen receptor, which participate in side chain to backbone hydrogen bonds (29). Orb2A PLD contains a single Cys residue. Its 13Cβ chemical shift indicates that it is reduced at pH 4.0 in 1 mm acetic acid buffer in the absence of reducing agent. Regarding the proline residues, all Xaa-Pro peptide bonds are mainly in the trans conformation, which is expected for a disordered chain (30), as judged by their observed 13Cβ and 13CO chemical shifts.
Figure 1.
Assigned 2D 1H-15N HSQC and 2D 13C-15N CON spectra of Orb2A PLD recorded at pH 4.0, 25°C.A, 2D 1H-15N HSQC spectrum of Orb2A PLD recorded at pH 4.0, 25 °C. Individual assigned signals are labeled. S-1 corresponds to the Ser residue resulting from cloning and tag cleavage that precedes M1. The bold His and Gln labels refer to overlapped His and Gln peaks; these arise from residues in the amyloidogenic segment. The multiplication factor between contour levels is 1.4 here and in panel B as well as Fig. 4A. B, 2D 13C-15N CON spectrum of Orb2A PLD recorded at pH 4.0, 25°C. The labels shown correspond to the 15N signals; the 13CO correlations arise from the preceding residue. Note that the y axis scale is longer in panel B than A to include Pro residues. The bold His, Gln, and Gln′ labels are for overlapped His and Gln residues; the Gln′ cluster comes from i His 13CO/i + 1 Gln 15N correlations. For intense or overlapped peaks, a satellite peak is often seen slightly to the right and above the main peak. These satellite peaks arise from deuteration and some examples are indicated with blue circles.
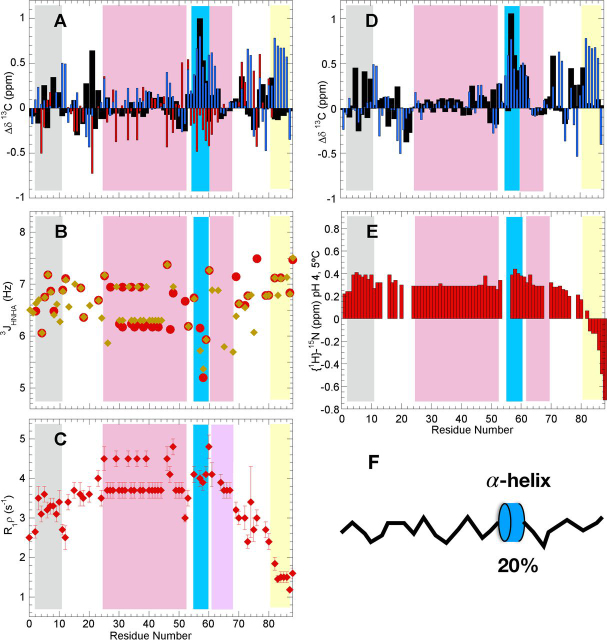
The 13Cα, 13Cβ, and 13CO Δδ values, which reveal partially populated elements of secondary structure, are plotted in Fig. 2A. The segment composed of residues 55–60, which follows the amyloidogenic Q/H-rich stretch, adopts a minor population of α-helix. Based on the conformational chemical shift (Δδ) values expected for 100% α-helix of 3.1, −0.4 and 2.2 ppm for 13Cα, 13Cβ, and 13CO, respectively (31, 32), the population of α-helix for these residues is ∼20%. Otherwise, the N-terminal hydrophobic stretch, the Q/H-rich segments and the C-terminal G-rich element do not contain detectable populations of secondary structure under these conditions. Whereas the Δδ 13CO values of terminal G82GGGGG87 are anomalously high at pH 4 (and also pH 7, see below), which suggests α-helix formation, they are not corroborated by high Δδ 13Cα values, so this may rather reflect a shortcoming in the prediction of secondary structure from 13CO δ values for polyG repeats. As an orthogonal measure of secondary structure, the 3JHNHA coupling constants were measured and analyzed using the Karplus equation to obtain backbone ϕ angles. Some 3JHNHA coupling constants appear to be somewhat lower for residues 55–60 (Fig. 2B), which may reflect the minor population of α-helix. Next, the µs/ms dynamics were assessed by R1ρ relaxation measurements. The obtained values are consistent with high mobility, except for residues 55–60, whose slightly elevated R1ρ rates reflect a modestly increased rigidity (Fig. 2C). Therefore, the NMR data indicate formation of a small population of α-helical structure in residues 55–60 of the Orb2A PLD.
Figure 2.
Orb2A PLD is largely disordered at pH 4, save a short α-helix in residues 55–60. In all panels, the different zones of the Orb2 PLD are shaded as follows: N-terminal hydrophobic stretch, gray; Q/H-rich, cyan; residues 55–60, blue; H·Q-rich, cyan, G-rich, yellow. In the Q/H or H·Q-rich regions, most of the values shown come from averaging of overlapped His or Gln peaks. A, conformational chemical shifts (Δδ) of Orb2 PLD at pH 4, 25°C. 13Cα, black bars; 13CO, blue bars; 13Cβ, red bars. Experimental uncertainties are ≤ ± 0.04, 0.02, and 0.08 ppm for 13Cα, 13CO, and 13Cβ, respectively. B, 3JHNHA coupling constants at pH 4, 25°C. Values of 5 Hz or less are indicative of α-helix; higher values, coil or extended conformations. Values shown in gold were measured by peak height using the Sparky program. Values in red were obtained using the peak integral function of Topspin 4.0. C, transverse relaxation rates in the rotating frame (R1ρ) at pH 4.0, 25°C. D, conformational chemical shifts (Δδ) of Orb2 PLD at pH 4, 5°C. 13Cα, black bars; 13CO, blue bars. The experimental uncertainties are generally ≤ ± 0.05 ppm for both 13Cα and 13CO. E, {1H}-15N NOE at pH 4.0, 5.0°C. Values approaching 0.85 indicate stiffness on fast ps/ns and are typical of well-folded rigid proteins; values less than 0 indicate a high flexibility. Uncertainties are about ± 0.01. F, schematic diagram of Orb2 PLD at pH 4 featuring a flexible and disordered conformational ensemble with a short, modestly populated α-helix spanning residues 55–60.
The population of marginally stable elements of secondary structure in polypeptides is generally enhanced by cooling. To check if this is the case for Orb2A PLD, additional spectra were recorded at 15°C and 5°C, pH 4.0. Nevertheless, the analysis of the conformational chemical shifts did not reveal any additional elements of secondary structure, beyond the 55-60 helix already detected at 25 °C (Fig. 2D). Next, the fast ps/ns dynamics were characterized by {1H}-15N NOE measurements at 5°C. Low values indicative of flexibility are seen throughout the domain and are only slightly higher for the hydrophobic segment and α-helix spanning residues 55–60 (Fig. 2E). The G/S-rich C-terminal residues are especially mobile. Finally, the presence of helical conformations by residues Asn-55–His-60 is further supported by the observation of HNi-HNi+1 NOES between Ala-58–Leu-59, Leu-59–His-60, and His-60–His-61 and Hβi-HNi+1 NOEs between Asn-55–Leu-56, Leu-56–Ser-57, and Ala-58–Leu-59, in a 3D 1H-1H-15N NOESY·HSQC spectrum (Fig. S4). Overall, we can conclude that at pH 4.0, where the His residues of Orb2A's PLD are chiefly positively charged, the protein is disordered and flexible, except for the minor population of α-helix formed by residues 55–60 (Fig. 2F).
The amyloidogenic Q/H-rich segment adopts partly populated helical conformations and rigidifies at neutral pH
A visible precipitate formed when the pH of the sample was increased from 4.0 to 7.0. Nevertheless, the concentration of protein remaining in solution was sufficient to record good quality 2D 1H-15N and 1H-13C HSQC as well as 3D 1H-detected HNCO, HNCA, and CBCAcoNH spectra. The 2D 1H-15N HSQC spectrum (Fig. 3A) is significantly broadened. The average 1HN peak width is 27 ± 4 Hz at pH 7.0 compared with 17 ± 2 Hz at pH 4.0 (Fig. 3B). This is an indication of conformational interconversion, oligomerization, or both (33). The assignments at pH 7.0 are complete for 13Cα, 13Cβ, 1HN, and backbone 13CO and 15N nuclei save the 13Cβ of Ser-77, the 13CO of Ser-79, the 13Cβ and 13CO of Ser-88, as well as the 13Cβ and 13CO of Leu-15 and other residues preceding proline residues. Moreover, all assignments are missing for His-63, His-64, and His-78 as these residues' 1HN signals may be adversely affected by exchange broadening. The assigned chemical shift values, like those measured at pH 4.0, have been deposited in the BMRB under accession number 50274.
Figure 3.
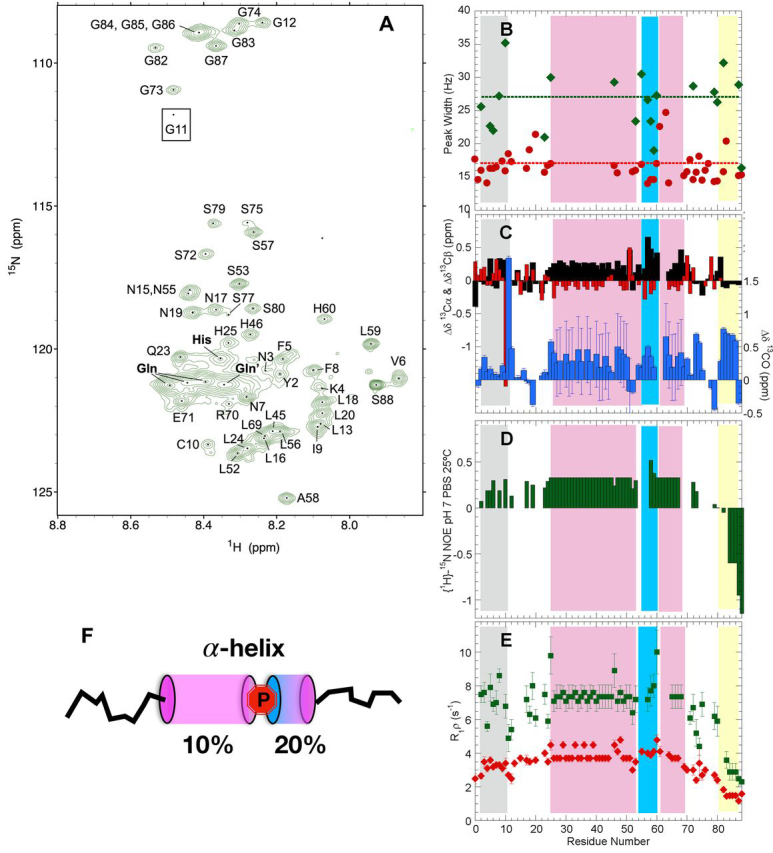
The Q/H-stretch of Orb2A partially adopts α-helical conformations and rigidifies at pH 7.B–E, the different zones of the Orb2 PLD are shaded as follows: N-terminal hydrophobic stretch, gray; Q/H-rich, light magenta; residues 55–60, blue; H·Q-rich, light magenta; G-rich, yellow. In the Q/H or H·Q-rich regions most of the values shown come from averaging of overlapped His or Gln peaks. A, 2D 1H-15N HSQC spectrum of Orb2A PLD registered at pH 7.0, 25°C. The bold His, Gln, and Gln′ labels refer to overlapped His and Gln peaks; these arise from residues in the amyloidogenic segment. The Gln′ peak arises from Gln, which follows His along the sequence. The signal of Gly-11 (boxed) is beneath the lowest selected contour level. To afford a fair appreciation of the wider peak widths observed here relative to the spectrum at pH 4, the same axes, aspect ratio, and multiplication factor between contour levels (1.4) are used here as in Fig. 2A. B, 1H peak width of Orb2 PLD 1H-15N HSQC peaks at 25°C and pH 7 (green diamonds) and pH 4 (red circles). The dotted lines represent the average peak widths at pH 7 (green) and pH 4 (red). Overlapped peaks are excluded. The uncertainties are approximately ± 1.5 Hz. C, conformational chemical shifts for 13Cα (black bars), 13Cβ (red bars), and 13CO (blue bars) at pH 7.0, 25°C. For clarity, the y axis scale is shifted for the 13CO Δδ values. The 13CO of Gly-11 and the 13Cβ of Cys are outliers. The 13CO Δδ values of the Q-rich region have large uncertainties because of the broad nature of the overlapped peak of Q residues. D, fast ps/ns dynamics assessed by the heteronuclear {1H}-15N NOE. Uncertainties are about ± 0.015. E, transverse relaxation rates in the rotating frame (R1ρ) for Orb2 PLD at pH 7.0, 25°C (green squares). For comparison, the R1ρ value obtained at pH 4.0, 25°C and already shown in Fig. 3C are also shown here by red diamonds. F, at pH 7, the Q/H segments (magenta cylinders), and the 55–60 segment (blue cylinder) adopts a small but detectable population of α-helical conformers separated by proline 54 (red).
Remarkably, the assessment of the 13Cα, 13Cβ, and 13CO Δδ values shows that at neutral pH, the main Q/H-rich stretch partly adopts α-helical conformations, whose population is ∼10% (Fig. 3C). Residues Ser-53–Pro-54 separate this helix from that formed by residues 55–60. The shorter Q/H segment also appears to adopt a minor population of α-helix. The nine N-terminal residues do not show a clear tendency to adopt a preferred secondary structure at pH 7. Cys-10 remains reduced and Xaa-Pro peptide bonds continue to be mainly trans at pH 7.
To corroborate the pH-dependent conformational changes, a series of 2D 1H-15N HSQC spectra were recorded on a fresh 15N-labeled sample of Orb2A PLD, at 25°C and pH values ranging from 3.5 to 7.3 (Fig. S5A). Interestingly enough, the 15N conformational chemical shifts for residues in the Q/H-rich segments become more negative at pH values where His residues are neutral (Fig. S5B), which suggests an increase in α-helical conformers (34). This observation is consistent with the increase in α-helical structure detected by the Δδ13Cα, Δδ13Cβ, and Δδ13CO values (Fig. 3C). By contrast, the Δδ15N values of Gly residues near the C terminus show small changes. Those of Ala-58 and Leu-59 suggest a decreased helicity at pH 7 (Fig. S5), which may be because of the loss of a favorable charge/helix macrodipole interaction (35) when the His residues following Leu-59 lose their positive charge.
Relaxation measurements at pH 7 yielded {1H}-15N NOE and R1ρ values that are elevated for most residues (Fig. 3, D and E) relative to values obtained at pH 4.0 (Fig. 2, C and E). These measurements reflect stiffening on ps/ns and µs/ms time scales and are consistent with the formation of preferred conformers (Fig. 3F) and associative processes. However, the G/S-rich segment located C-terminal to the PLD continues to be highly dynamic at pH 7, both on faster ps/ns time scales as well as slower µs/ms time scales.
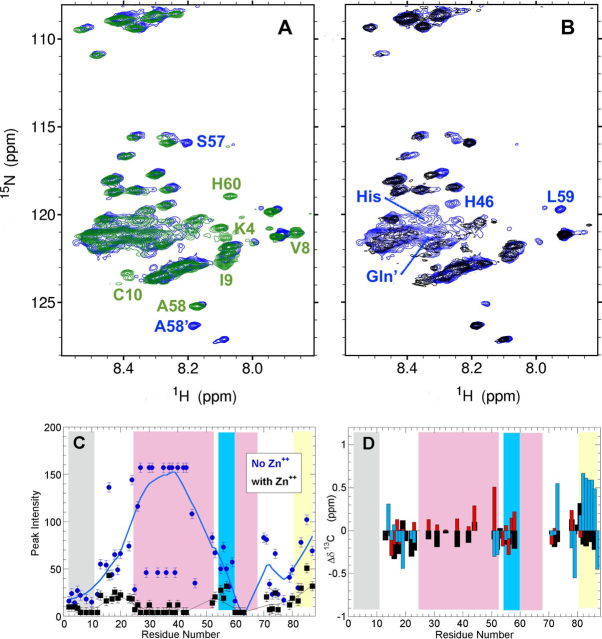
After a 2-month incubation at 4°C, Orb2A PLD's 1H-15N NMR spectra at 25°C in PBS revealed that despite some changes, most peaks retained their positions; this suggests that slow conformational changes or aggregative processes are minimal (Fig. 4A). Zinc chloride was then added to a final concentration of 2 mm. In the presence of this divalent cation, the peak intensities of His and Gln 1H-15N resonances drop practically to the signal/noise limit (Fig. 4, B and C). Strong decreases are also seen for the 1H-13Cβ and 1H-13C Cα signals of His in 2D 1H-13C HSQC spectra (Fig. S6). This strongly suggests that Zn2+ binds to His residues and promotes association processes of nearby Gln residues which are manifested as a loss of signal intensity. The signal intensity of other residues also decreased following Zn2+ addition, but sufficed to record good quality 3D HNCO, HNCA and CBCAcoNH spectra. The analysis of these spectra revealed that the conformational ensemble of the molecules remaining visible to NMR is poor in helical structures and slightly enriched in extended conformations (Fig. 4D). Thus, Zn2+ binds the His residues and drives the association of helical conformations of Orb2A PLD into large assemblies invisible to liquid state NMR.
Figure 4.
Zn2+ binding to His residues at pH 7 promotes further oligomerization.A, 1H-15N HSQC before (green) and after (blue) the 2-month pause. Some distinct residues are labeled. B, 1H-15N before (blue) and after (black) the addition of ZnCl2. His and Gln signals (labeled) weaken sharply after addition of Zn2+. C, the largest drop in peak intensity upon Zn addition occurs for residues of the Q/H-rich segments (rose band). D, conformational chemical shifts for 13Cα (black), 13Cβ (red), and 13CO (blue) strongly suggest that the molecules remaining in solution lack helical zones and are disordered.
Orb2A PLD binding to Zn2+ and RNA oligos is pH dependent
To confirm that Zn2+ binds Orb2A PLD His residues, we performed H/D exchange to remove all 1HN signals, leaving the His, Phe, and Tyr aromatic ring 1H as the only signals in the downfield region. Because His 1Hδ2 and 1Hε1 lie close to the side chain nitrogen atom that putatively ligates Zn2+, their NMR signals should be sensitive probes of this binding. In addition to Zn2+, the entry of Ca2+ is important for nerve impulses. No changes in the His 1Hδ2 and 1Hε1 resonances were observed under acidic conditions (pH 4.0, 25°C), following the addition of a 10-fold excess of Ca2+ and Zn2+ (Fig. S7A). Therefore, we conclude that these cations do not bind to the His residues of Orb2A PLD when they are in the imidazolium form. The charged state of His residues may affect their capacity to bind Ca2+ and Zn2+. Upon titration with Ca2+ to a 10-fold excess relative to a fresh Orb2A PLD sample at neutral pH, 25°C, no visible alterations in the His 1Hδ2 and 1Hε1 signals were seen (Fig. S7B). In contrast, significant changes in these resonances were observed upon adding a 10-fold excess of Zn2+ at pH 7.0, 25°C.
Whereas the RRM and ZnF domains of Orb2 have been reported to bind to specific RNA sequences in the 3′ UTR (19) because of their charged nature at low pH, the Q/H-rich segments might also be able to bind RNA. To test this idea, 2D 1H-15N HSQC spectra of Orb2A PLD were recorded and show significant changes at pH 4 (where His residues are mostly cationic) in the presence of 5′UUUUUAU3′, the consensus ligand, as well as 5′CCCCCGC3′, the opposite sequence studied as a negative control (Fig. S8A). In particular, many peaks corresponding to residues near the N terminus and in the residues 55–60 α-helix disappear while those arising from residues in the Q/H-rich stretches shift and weaken. These changes are consistent with observation of visible aggregates upon mixing Orb2 PLD with the RNA oligos. However, residues beyond Arg-70 show much smaller shifts and remain relatively strong. This suggests that the Gly-rich C-terminal region does not participate in RNA binding or aggregation. The RNA-induced alterations in Orb2A PLD's HSQC spectra are much less striking at pH 7, where His residues are chiefly neutral (Fig. S8B). These observations lead us to suggest that the Orb2A PLD can bind RNA nonspecifically at pH 4 and that the binding is mostly mediated by electrostatic interactions.
Discussion
The key findings of this study are that the amyloidogenic Q/H-rich region of Orb2's PLD is disordered and flexible at low pH but adopts a minor population of α-helix at pH 7.0, where the His residues are mostly in the neutral state. The hypothesis that Orb2 aggregation triggers enduring synapse-specific alterations in translation which are key for memory consolidation received strong support by the elucidation of its amyloid structure from adult Drosophila heads (17). Gln and His side chains are interlocked in this amyloid (Fig. S1B). The relative instability of Orb2 amyloid at acidic conditions, as shown in Fig. S15 of Ref. 17, can be attributed to His side chains becoming positively charged and producing electrostatic repulsion at low pH. In vivo, this may facilitate the disintegration of Orb2 amyloid fibrils in the lysosome. Considering the results reported here, is it possible that Orb2 PLD histidines could be initially maintained in a charged state in vivo and then neutralized to help trigger amyloid formation?
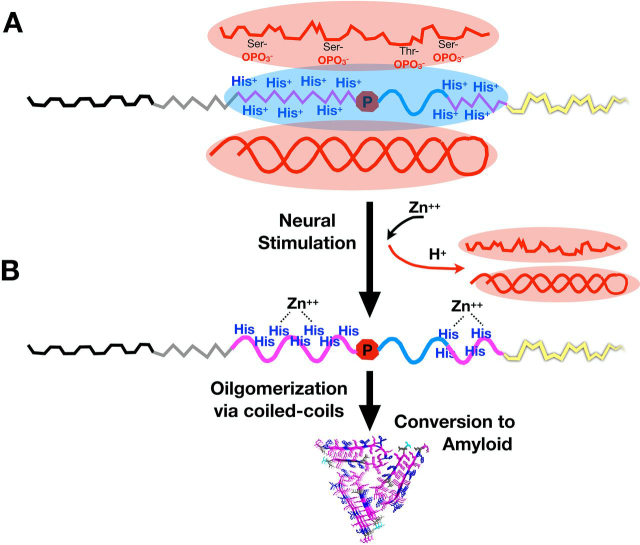
In the Drosophila synapse, Orb2 monomers are initially bound to target mRNA molecules via their RRM domains. The proximity of polyanionic mRNA, phosphorylated Tob (22), and/or possibly other anions of the neuronal granule (36) could tend to stabilize the charged imidazolium form of His side chains. This possibility is supported by our observations that the PLD of Orb2 can bind to RNA under acidic conditions (Fig. S8). Upon neural stimulation, the influx of Ca2+ and Zn2+ cations could displace the imidazolium H+ and bind neutral His. This scenario is supported by ITC measurements demonstrating that the Q/H-rich region of Orb2 binds transition metal cations like Zn2+, but not Ca2+ (24), which we corroborate here by NMR (Fig. 4 and Figs. S6 and S7). Those researchers also proposed that if this segment were to adopt an α-helical conformation (just as we have shown here), then the His residues, which are mostly spaced i, i + 3 and i, i + 4, would be mainly positioned on the same side of the α-helix (24). The binding of cations such as Zn2+ to such an array of His residues could increase the formation of α-helix, although this point is difficult to test because of the Orb2A PLD aggregation that accompanies cation binding, even at the low concentrations used for CD spectroscopy (24). The resulting helical conformations may drive association processes such as the formation of coiled coils, as seen for other proteins which undergo liquid-liquid phase separation via intermolecular helical associations (37). Q-rich coiled coils and higher oligomeric species have been proposed as intermediates in amyloidogenesis of Aplysia and mammalian CPEBs (38, 39, 40, 41), which are probably heterogeneous as strongly suggested by kinetic modeling (42). A hypothetical working model that incorporates these ideas is shown in Fig. 5.
Figure 5.
His neutralization unleashes α-helix formation, oligomerization and amyloidogenesis.A, the first N-terminal hydrophobic segment (black) may interact with membranes which promote partial α-helix formation as reported by Soria et al. (12). The proximity of phosphorylated Tob protein (top, red), RNA (bottom, red), and/or possibly other polyanions, creates a negatively charged milieu which favors the cationic form of the 12 His residues (His +) of the Q/H-rich segments (magenta). This keeps the Q/H-rich segments disordered. Pro-54 (red hexagon) would also act to prevent premature amyloidogenesis. The 55–60 residue segment (blue) adopts a partial α-helix whereas the G/S-rich segment (yellow) remains disordered and flexible. B, following neural stimulation, the entry of Zn2+ and other cations could displace H+ and bind to the neutral form of His residues (blue) as proposed by Bajakian et al. (24); releasing Tob and RNA (red). The neutral form of His allows partial α-helix formation of the Q/H-rich segments, unlocking pathways to amyloid formation through coiled-coil intermediates.
The role of the initial hydrophobic segment (Fig. 1A) in Orb2A on amyloidogenesis has been debated. It was found to be important for Orb2-mediated memory, as even the conservative substitution of Phe-5 for Tyr affected activity (11). Moreover, this segment, and not the amyloidogenic Q/H-rich stretch, was found to adopt an α-helix that later evolved into amyloid in vitro as monitored by circular dichorism spectroscopy and solid state NMR spectroscopy (13). However, this segment was not observed in the physiological amyloid, which is composed chiefly of Orb2B, formed in Drosophila heads (17). Instead, the amyloid is composed of the longer Q/H-rich stretch (Fig. S1). Here, this Orb2A N-terminal hydrophobic segment was found to be devoid of detectable populations of secondary structure, at both pH 4.0 and pH 7.0. By contrast, this segment was reported to form an amphiphilic α-helix in the presence of membrane mimetics (12). Therefore, it is possible that under in vivo conditions, this segment may form an α-helix that facilitates functional amyloid formation.
Although Aplysia and mammalian CPEB homologs share a similar domain organization with Drosophila Orb2, they lack the multiple His residues interspersed in their Q-rich segments (40, 43, 44), which are found in Orb2. This suggests that the pH-regulated nature of Orb2 amyloid formation and dissociation may be substituted by other mechanisms. Proline 54, which interrupts the helices formed by the Q/H-rich segments and residues 55–60, may play an important role in preventing premature amyloid formation. Similar proline residues that delimit α-helices have been recently described in human CPEB3 (41).
Overall, our observations reveal remarkable structural plasticity in the region of Orb2A which is key for amyloid formation. The characterization of nascent Orb2A conformers presented here, together with the recent structural elucidation of Orb2 native amyloid fibrils (17), provides the cadre for understanding Orb2 amyloidogenesis. Further investigation is required to fully comprehend this relevant process.
Experimental procedures
DNA coding for the first 88 residues of Orb2A is M1YNKFV-NFIC10-GGLPNLNLNK20-PPQLHQQQHQ30-QQHQQHQQH-Q40-QQQQLHQHQQ50-QLSPNLSALH60-HHHQQQQQLR70-ESGGSHSPSS80-PGGGGGGS88, which comprises the PLD domain (underlined) and initial residues of the Gly/Ser-rich region (shown in italics above), was subcloned into a pET28(a) vector containing a TXA fusion protein, a His6 tag for IMAC purification and a TEV protease tag. After TEV cleavage, Gly and Ser residues remained attached to the N terminus of the Orb2A protein construct. Following expression in minimal media enriched with 15NH4Cl or 15NH4Cl and 13C-glucose as the sole sources of nitrogen and carbon, the resulting 15N- or 15N,13C-labeled protein was purified by Ni2+ affinity chromatography using a GE Biosciences ÄKTA FPLC system, using the buffer 20 mm Tris HCl, 500 mm NaCl, pH 8.0, adding imidazole (500 mm) for the elution. Next, the protein was cleaved with TEV protease, using 20 mm Tris HCl, 150 mm NaCl, 1 mm DTT, 0.5 mm EDTA, pH 7.0, and re-purified to remove the cut TXA and His tag. Because of the high number of His residues in Orb2A, the protein remained attached to the column and co-eluted with TXA. To remove the fusion protein, we performed anion exchange chromatography using 20 mm Tris HCl, 8 m urea, 1 mm DTT, pH 7.0, where the elution was achieved applying a linear gradient until 1 m NaCl was present in the buffer. The presence of 8 M urea prevents premature aggregation during purification and storage and is a common practice for amyloidogenic proteins, including the Orb2A (12, 13). The resulting protein is electrophoretically pure and was stored in concentrated urea at −80°C until use. Its purity and identity were subsequently corroborated by NMR spectroscopy.
Just prior to NMR spectroscopy, the urea was removed and the protein was transferred to 1.0 mm deuterated acetic acid buffer containing 15% D2O (99.96% atom D, Euroisotop), pH 4.0, using a PD-10 (GE Biosciences) desalting column. This concentration of D2O (15%) is somewhat higher than the 10% typically used; it was necessary to avoid losing the lock during the acquisition of long spectra. Following the renovation of the NMR consoles in April 2020, we recorded spectra with 10% D2O and obtained similar Orb2A spectra. By refractive index, NMR samples were found to contain less than 10 mm residual urea. This concentration of urea has a negligible effect on protein conformational stability (45). The concentration of Orb2A PLD samples was measured by UV absorbance using an extinction coefficient of 1490 cm−1·M−1 at 280 nm based on its content of one Tyr residue (46).
Following NMR spectral acquisition at pH 4.0, the sample was transferred to pH 7.0 by adding a 10× buffer stock of PBS prepared in D2O. Upon adding the PBS stock, the initial pH was 6.2; it was increased to 7.01 by adding small aliquots of 0.3 M KOH. The final buffer content of the pH 7 sample was 0.9 mm deuterated acetate, 25 mm Na2HPO4, 25 mm NaH2PO4, 100 mm KCl, and 1 mm TCEP as the reducing agent. The final sample volumes were 0.201 ml (pH 4) and 0.226 ml (pH 7). A final series of 2D and 3D spectra was recorded following the addition of ZnCl2 (Fluka) to a final concentration of 2 mm. All spectra were recorded in a water-matched, 5 mm diameter Shigemi NMR tube.
NMR spectral assignments
All NMR spectra were recorded on a Bruker Avance NMR spectrometer operating at 800 MHz (1H), except a 3D 1H-1H-15N NOESY·HSQC spectrum and those examining the effects of pH, and the binding of RNA, Ca2+, and Zn2+, which were recorded with a Bruker 800 MHz Neo Avance NMR spectrometer. The spectrometer is equipped with a triple resonance (1H, 13C, 15N) cryoprobe and Z-gradients. The program Topspin (Bruker Biospin) versions 2.1 and 4.0.8 were used to record, process, and analyze the spectra. Because the Orb2A's N-terminal region is disordered and its sequence is of low complexity and contains many repeated consecutive residues, it presents special challenges for NMR spectral assignment. Therefore, instead of the standard approach based by 1HN excitation and 13Cα and 13Cβ connectivities (47), we have applied an unconventional proton-less approach based on a pair of 3D 13C-detected spectra which provide consecutive 13CO and 15N backbone connectivities as these nuclei retain more dispersion in IDPs (48). Even with this approach, some cases of sequential residues with identical 13CO and 15N chemical shift values were observed. Therefore, to overcome these ambiguities, an additional 3D 1H-detected spectrum that yields consecutive 1HN-1HN connectivities was recorded (49, 50). The analysis of these data led to the essentially complete spectral assignment at pH 4.0 as described in “Results.” After measuring relaxation measurements at pH 4.0, the sample pH was increased to pH 7.0. As described “Results,” the sample tends to aggregate at neutral pH. Because of its lower solution concentration, the assignment was carried out by conventional 1H-detected 2D 1H-15N HSQC and 3D CBCAcoNH, HNCO and HNCA spectra, and comparison with the pH 4 spectra. A complete list of the NMR experiments and their parameters is shown in Table S1. 1H peak widths in 1H-15N HSQC spectra were measured using a Gaussian function in the program NMRFAM Sparky 1.4. 3JHNHA coupling constants were calculated based on the relative signal intensities of the 1HN/1Hα crosspeak: 1HN/1HN diagonal peak using either peak integration in Topspin 4.0 or peak heights in NMRFAM Sparky 1.4 as described previously (51).
The expected chemical shift values for the 13Cα, 13Cβ, 1Hα, 1HN, 15N, and 13CO nuclei in statistical coil ensembles were calculated using the parameters tabulated by Kjaergaard and co-workers (52, 53) and implemented on the RRID:SCR_019189 server at the Bax laboratory. These values were subtracted from the experimentally measured chemical shift values (δ) to calculate conformational chemical shifts (Δδ).
3D 1H-1H-15N NOESY·HSQC spectrum.
To obtain additional evidence for secondary structure formation, a 3D 1H-1H-15N NOESY·HSQC spectrum was recorded at 25°C, pH 4.10, with a 100 ms mixing time. The sample contained 1 mm deuterated acetic acid and 1.17 mm15N-labeled Orb2A PLD. Because of severe signal overlap, we forwent analysis of the Q/H-rich stretches and focused on residues 55–60.
pH titration followed by 2D 1H-15N HSQC spectroscopy.
Further testing of the assignments and helical structure was performed by recording a series of five 2D 1H-15N HSQC spectra at 25°C over a range of pH values spanning 3.5 to 7.3. The original sample contained 270 μm15N-Orb2A PLD and 1.0 mm deuterated acetic acid as a minimal buffer and was titrated with microliter additions of 100 mm sodium carbonate.
NMR relaxation measurements.
The dynamics on the ps/ns time scales were probed by measuring the heteronuclear {1H}-15N NOE of backbone amide groups as the ratio of spectra recorded with and without saturation in an interleaved mode. An 11-s recycling delay was employed. Uncertainties in peak integrals were estimated from the standard deviation of intensities from spectral regions lacking signal and containing only noise. R1ρ relaxation rates, which sense dynamic processes on slower µs/ms time scales, were measured by recording two sets of 10 1H-15N correlation spectra with relaxation delays at 8, 300, 36, 76, 900, 100, 500, 156, 200, and 700 ms. The R1ρ relaxation rates were calculated by least-squares fitting of an exponential decay function to peak integral data, which were obtained using Topspin 4.0.8.
Ca2+ and Zn2+ binding experiments followed by 1D 1H NMR.
Two aliquots of 15N-labeled Orb2A PLD were lyophilized overnight and then redissolved in 100% D2O and 0.20 mm DSS containing 12.5 mm KH2PO4 + 12.5 mm K2HPO4, pH* 7.0 or 3.6 mm deuterated sodium acetate (d3, 99% atom D, Cambridge Isotope Laboratory) + 21.4 mm deuterated acetic acid (d4, >99.5% atom D, Aldrich), pH* 4.0. “pH*” is the pH meter reading without correction for the deuterium isotope effect. The Orb2A PLD concentration was 0.20 mm. H/D exchange was monitored by 1D 1H and 2D 1H-15N HSQC spectra and found to be essentially complete within minutes at pH* 7.0 and within an hour at pH* 4.0. Considering the parameters of Englander and co-workers (54), implemented on the webpage Sphere HX (RRID:SCR_019188), this quick exchange is consistent with a disordered protein containing no or unstable elements of secondary structure. Following exchange, only the aromatic 1H of His, Tyr, and Phe are present in the downfield region of the 1D 1H NMR spectrum of Orb2A PLD. Changes in these signals were monitored at 25°C following the addition of Ca2+ and Zn2+ from stock solutions of 100 mm CaCl2 (Pancrea) and 100 mm ZnCl2 (Fluka) prepared in D2O.
Binding to RNA oligos.
RNA oligos with sequences 5′UUU-UUAU3′, the consensus ligand, as well as 5′CCCCCGC3′, the opposite sequence, were purchased from Integrated DNA Technologies. Following the cleaning of pipettes and surfaces with RNase Zap® (Sigma), they were dissolved as concentrated stock solutions in 50 μl of mQ H2O, which had been previously boiled and cooled. RNA stock solutions' concentrations were calculated by UV absorbance at 260 nm and the stocks were incubated on ice until use. Samples containing 200 μm15N-labeled Orb2A PLD in either 25 mm K2HPO4/KH2PO4 buffer, pH 7.0, or 25 mm deuterated sodium acetate/deuterated acetic acid buffer, pH 4.0, with 0.20 mm DSS and 10% D2O/90% mQ H2O were prepared and their 1H-15N HSQC spectra were recorded at 25°C. Then, an RNA oligo was added to the Orb2A PLD sample to give a final molar ratio of 1:1 and another 1H-15N HSQC spectrum was recorded. Experiments were repeated for each pH and type of RNA oligo. Upon completion, the pH of each sample was measured and found to be 3.98 for the Orb2 + UUUUUAU sample, 3.96 for the Orb2 + CCCCCGC sample, and 6.87 for both samples prepared in potassium phosphate buffer.
Bioinformatic analysis.
Disorder predictions for the Orb2A sequence were performed with PONDR-XSL2 and PONDR-XL1-XT (55).
Data availability
NMR chemical shifts of the Orb2A PLD recorded at pH 4 and at pH 7 have been deposited in the BMRB data bank under file number 50274. The raw NMR data can be requested from D. V. Laurents at dlaurents@iqfr.csic.es. All other data presented and discussed are contained within the article and its supporting material.
Acknowledgments
We are grateful to Daniel Ramírez de Mingo, Rubén Hervás, and Mariano Carrión-Vázquez for critical comments on the manuscript and to José Manuel Pérez Cañadillas for the pET28-TXA-His cDNA vector. NMR experiments were performed in the “Manuel Rico” NMR Laboratory (LMR) of the Spanish National Research Council (CSIC), a node of the Spanish Large-Scale National Facility (ICTS R-LRB).
Fundación BBVA (BBVA Foundation) (BBM_TRA_0203) to Javier Oroz
FCT-Portugal (PD/BD/148028/2019) to Sara S. Félix, and Eurico J. Cabrita
PCT-Portugal (UIDB/04378/2020) to Sara S. Félix, and Eurico J. Cabrita
Ministerio de Economía y Competitividad (MEC) (SAF2016-76678-C2-2-R) to Douglas V. Laurents
Edited by Paul E. Fraser
Footnotes
This article contains supporting information.
Author contributions—J. O. and D. V. L. conceptualization; J. O., S. S. F., and D. V. L. data curation; J. O., S. S. F., and D. V. L. formal analysis; J. O., E. J. C., and D. V. L. supervision; J. O., E. J. C., and D. V. L. funding acquisition; J. O., S. S. F., E. J. C., and D. V. L. validation; J. O., S. S. F., and D. V. L. investigation; J. O. and S. S. F. methodology; J. O., S. S. F., and D. V. L. writing-original draft; J. O., E. J. C., and D. V. L. project administration; J. O., S. S. F., E. J. C., and D. V. L. writing-review and editing; E. J. C. visualization.
Funding and additional information—This work was supported by Leonardo Grant BBM_TRA_0203 from the BBVA Foundation (to J. O.) and by FCT-Portugal PhD studentship PD/BD/148028/2019 (to S. S. F.). This study was also supported by Spanish Ministry of Economy and Competitivity project SAF2016-76678-C2-2-R (AEI/FEDER, UE to D. V. L.). This work was also partially supported by the Applied Molecular Biosciences Unit—UCIBIO, which is financed by national funds from the Portuguese FCT UIDB/04378/2020. J. O. is a Ramón y Cajal fellow from the Spanish AEI-Ministry of Science and Innovation.
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
- CPEB
- cytoplasmic polyadenylation element binding
- IDR
- intrinsically disordered region
- PLD
- prion-like domain
- TEV
- tobacco etch virus.
Supplementary Material
References
- 1.Cajal S.R. The Croonian Lecture: La fine structure des centres nerveux. Proc. R. Soc. London Ser. B. 1894;55:444–467. [Google Scholar]
- 2.Semon R. In: Die Mneme als erhaltendes Prinzip im Wechsel des organischen Geschehens. Engelmann W., editor. 1904. Leipzig. [Google Scholar]
- 3.Hebb D.O. John Wiley & Sons; New York: 1949. The Organisation of Behaviour. [Google Scholar]
- 4.Frey U., Morris R.G. Synaptic tagging and long-term potentiation. Nature. 1997;385:533–536. doi: 10.1038/385533a0. 9020359. [DOI] [PubMed] [Google Scholar]
- 5.Kandel E.R. W. W. Norton & Co.; New York: 2006. In Search of Memory: The Emergence of a New Science of Mind. [Google Scholar]
- 6.Josselyn S.A., Tonegawa S. Memory engrams: Recalling the past and imagining the future. Science. 2020;367 doi: 10.1126/science.aaw4325. 31896692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Si K., Lindquist S., Kandel E.R. A neuronal isoform of the Aplysia CPEB has prion-like properties. Cell. 2003;115:879–891. doi: 10.1016/S0092-8674(03)01020-1. 14697205. [DOI] [PubMed] [Google Scholar]
- 8.Si K., Kandel E.R. The role of functional prion-like proteins in the persistence of memory. Cold Spring Harb. Perspect. Biol. 2016;8 doi: 10.1101/cshperspect.a021774. 27037416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Keleman K., Krüttner S., Alenius M., Dickson B.J. Function of the Drosophila CPEB protein Orb2 in long-term courtship memory. Nat. Neurosci. 2007;10:1587–1593. doi: 10.1038/nn1996. 17965711. [DOI] [PubMed] [Google Scholar]
- 10.Krüttner S., Stepien B., Noordermeer J.N., Mommaas M.A., Mechtler K., Dickson B.J., Keleman K. Drosophila CPEB Orb2A mediates memory independent of Its RNA-binding domain. Neuron. 2012;76:383–395. doi: 10.1016/j.neuron.2012.08.028. 23083740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Majumdar A., Cesario W.C., White-Grindley E., Jiang H., Ren R., Khan M.R., Li L., Choi E.M.K., Kannan K., Guo F., Unruh J., Slaughter B., Si K. Critical role of amyloid-like oligomers of Drosophila Orb2 in the persistence of memory. Cell. 2012;148:515–529. doi: 10.1016/j.cell.2012.01.004. 22284910. [DOI] [PubMed] [Google Scholar]
- 12.Soria M.A., Cervantes S.A., Bajakian T.H., Siemer A.B. The functional amyloid Orb2A binds to lipid membranes. Biophys. J. 2017;113:37–47. doi: 10.1016/j.bpj.2017.05.039. 28700922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cervantes S.A., Bajakian T.H., Soria M.A., Falk A.S., Service R.J., Langen R., Siemer A.B. Identification and structural characterization of the N-terminal amyloid core of Orb2 isoform A. Sci. Rep. 2016;6 doi: 10.1038/srep38265. 27922050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Scherzinger E., Lurz R., Turmaine M., Mangiarini L., Hollenbach B., Hasenbank R., Bates G.P., Davies S.W., Lehrach H., Wanker E.E. Huntingtin-encoded polyglutamine expansions form amyloid-like protein aggregates in vitro and in vivo. Cell. 1997;90:549–558. doi: 10.1016/S0092-8674(00)80514-0. 9267034. [DOI] [PubMed] [Google Scholar]
- 15.Eftekharzadeh B., Piai A., Chiesa G., Mungianu D., García J., Pierattelli R., Felli I.C., Salvatella X. Sequence context influences the structure and aggregation behavior of a PolyQ tract. Biophys. J. 2016;110:2361–2366. doi: 10.1016/j.bpj.2016.04.022. 27276254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hervás R., Li L., Majumdar A., Fernández-Ramírez M., Unruh J.R., Slaughter B.D., Galera-Prat A., Santana E., Suzuki M., Nagai Y., Bruix M., Casas-Tintó S., Menéndez M., Laurents D.V., Si K. Molecular basis of Orb2 amyloidogenesis and blockade of memory consolidation. PLoS Biol. 2016;14 doi: 10.1371/journal.pbio.1002361. 26812143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hervás R., Rau M.J., Park Y., Zhang W., Murzin A.G., Fitzpatrick J., Scheres S., Si K. Cryo-EM structure of a neuronal functional amyloid implicated in memory persistence in Drosophila. Science. 2020;367:1230–1234. doi: 10.1126/science.aba3526. 32165583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Legge G.B., Martinez-Yamout M.A., Hambly D.M., Trinh T., Lee B.M., Dyson H.J., Wright P.E. ZZ domain of CBP: An unusual zinc finger fold in a protein interaction module. J. Mol. Biol. 2004;343:1081–1093. doi: 10.1016/j.jmb.2004.08.087. 15476823. [DOI] [PubMed] [Google Scholar]
- 19.Mastushita-Sakai T., White-Grindley E., Samuelson J., Seidel C., Si K. Drosophila Orb2 targets genes involved in neuronal growth, synapse formation, and protein turnover. Proc. Natl. Acad. Sci. U. S. A. 2010;107:11987–11992. doi: 10.1073/pnas.1004433107. 20547833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khan M.R., Li L., Pérez-Sánchez C., Saraf A., Florens L., Slaughter B.D., Unruh J.R., Si K. Amyloidogenic oligomerization transforms Drosophila Orb2 from a translation repressor to an activator. Cell. 2015;163:1468–1483. doi: 10.1016/j.cell.2015.11.020. 26638074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gill J., Park Y., McGinnis J.P., Perez-Sanchez C., Blanchette M., Si K. Regulated intron removal integrates motivational state and experience. Cell. 2017;169:836–848.e15. doi: 10.1016/j.cell.2017.05.006. 28525754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.White-Grindley E., Li L., Mohammad Khan R., Ren F., Saraf A., Florens L., Si K. Contribution of Orb2A stability in regulated amyloid-like oligomerization of Drosophila Orb2. PLoS Biol. 2014;12 doi: 10.1371/journal.pbio.1001786. 24523662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li L., Sanchez C.P., Slaughter B.D., Zhao Y., Khan M.R., Unruh J.R., Rubinstein B., Si K. A putative biochemical engram of long-term memory. Curr. Biol. 2016;26:3143–3156. doi: 10.1016/j.cub.2016.09.054. 27818176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bajakian T.H., Cervantes S.A., Soria M.A., Beaugrand M., Kim J.Y., Service R.J., Siemer A.B. Metal binding properties of the N-terminus of the functional amyloid Orb2. Biomolecules. 2017;7:57. doi: 10.3390/biom7030057. 28763009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Joag H., Ghatpande V., Desai M., Sarkar M., Raina A., Shinde M., Chitale R., Deo A., Bose T., Majumdar A. A role of cellular translation regulation associated with toxic Huntingtin protein. Cell. Mol. Life Sci. 2020;77:3657–3670. doi: 10.1007/s00018-019-03392-y. 31796991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li M., Liu J., Ran X., Fang M., Shi J., Qin H., Goh J.M., Song J. Resurrecting abandoned proteins with pure water: CD and NMR studies of protein fragments solubilized in salt-free water. Biophys. J. 2006;91:4201–4209. doi: 10.1529/biophysj.106.093187. 16980357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Treviño M.Á., Pantoja-Uceda D., Menéndez M., Gomez M.V., Mompeán M., Laurents D.V. The singular NMR fingerprint of a polyproline II helical bundle. J. Am. Chem. Soc. 2018;140:16988–17000. doi: 10.1021/jacs.8b05261. 30430829. [DOI] [PubMed] [Google Scholar]
- 28.Pantoja-Uceda D., Oroz J., Fernández C., de Alba E., Giraldo R., Laurents D.V. Conformational priming of RepA-WH1 for functional amyloid conversion detected by NMR spectroscopy. Structure. 2020;28:336–347.e4. doi: 10.1016/j.str.2019.12.007. 31918960. [DOI] [PubMed] [Google Scholar]
- 29.Escobedo A., Topal B., Kunze M.B.A., Aranda J., Chiesa G., Mungianu D., Bernardo-Seisdedos G., Eftekharzadeh B., Gairí M., Pierattelli R., Felli I.C., Diercks T., Millet O., García J., Orozco M. Side chain to main chain hydrogen bonds stabilize a polyglutamine helix in a transcription factor. Nat. Commun. 2019;10 doi: 10.1038/s41467-019-09923-2. 31048691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reimer U., Scherer G., Drewello M., Kruber S., Schutkowski M., Fischer G. Side-chain effects on peptidyl-prolyl cis/trans isomerisation. J. Mol. Biol. 1998;279:449–460. doi: 10.1006/jmbi.1998.1770. 9642049. [DOI] [PubMed] [Google Scholar]
- 31.Spera S., Bax A. Empirical correlation between protein backbone conformation and Cα and Cβ 13C NMR chemical shifts. J. Am. Chem. Soc. 1991;113:5490–5492. doi: 10.1021/ja00014a071. [DOI] [Google Scholar]
- 32.Wishart D.S., Sykes B.D. The 13C chemical-shift index: A simple method for the identification of protein secondary structure using 13C chemical-shift data. J. Biomol. NMR. 1994;4:171–180. doi: 10.1007/BF00175245. 8019132. [DOI] [PubMed] [Google Scholar]
- 33.Charlier C., Cousin S.F., Ferrage F. Protein dynamics from nuclear magnetic relaxation. Chem. Soc. Rev. 2016;45:2410–2422. doi: 10.1039/c5cs00832h. 26932314. [DOI] [PubMed] [Google Scholar]
- 34.Wang Y., Jardetzky O. Probability-based protein secondary structure identification using combined NMR chemical-shift data. Prot. Sci. 2002;11:852–861. doi: 10.1110/ps.3180102. 11910028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chakrabartty A., Baldwin R.L. Stability of α-helices. Adv. Prot. Chem. 1995;46:141–176. doi: 10.1016/S0065-3233(08)60334-4. [DOI] [PubMed] [Google Scholar]
- 36.Ford L., Ling E., Kandel E.R., Fioriti L. CPEB3 inhibits translation of mRNA targets by localizing them to P bodies. Proc. Nat. Acad. Sci. U. S. A. 2019;116:18078–18087. doi: 10.1073/pnas.1815275116. 31416913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Conicella A.E., Zerze G.H., Mittal J., Fawzi N.L. ALS mutations disrupt phase separation mediated by α-helical structure in the TDP-43 low-complexity C-terminal domain. Structure. 2016;24:1537–1549. doi: 10.1016/j.str.2016.07.007. 27545621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fiumara F., Fioriti L., Kandel E.R., Hendrickson W.A. Essential role of coiled coils for aggregation and activity of Q/N-rich prions and PolyQ proteins. Cell. 2010;143:1121–1135. doi: 10.1016/j.cell.2010.11.042. 21183075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pelassa I., Corà D., Cesano F., Monje F.J., Montarolo P.G., Fiumara F. Association of polyalanine and polyglutamine coiled coils mediates expansion disease-related protein aggregation and dysfunction. Hum. Mol. Gen. 2014;23:3402–3420. doi: 10.1093/hmg/ddu049. 24497578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hervás R., del Carmen Fernández-Ramírez M., Galera-Prat A., Suzuki M., Yagai Y., Bruix M., Menéndez M., Laurents D.V., Carrión-Vázquez M. Divergent CPEB prion-like domains reveal different assembly mechanisms for a generic amyloid-like fold. bioRxiv. 2020 doi: 10.1101/2020.05.19.103804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ramírez de Mingo D., Pantoja-Uceda D., Hervás R., Carrión-Vázquez M., Laurents D.V. Preferred conformations in the intrinsically disordered region of human CPEB3 explain its role in memory consolidation. bioRxiv. 2020 doi: 10.1101/2020.05.12.091587. [DOI] [Google Scholar]
- 42.Vitalis A., Pappu R.V. Assessing the contribution of heterogeneous distributions of oligomers to aggregation mechanisms of polyglutamine peptides. Biophys. Chem. 2011;159:14–23. doi: 10.1016/j.bpc.2011.04.006. 21530061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Raveendra B.L., Siemer A.B., Puthanveettil S.V., Hendrickson W.A., Kandel E.R., McDermott A.E. Characterization of prion-like conformational changes of the neuronal isoform of Aplysia CPEB. Nat. Struct. Mol. Biol. 2013;20:495–501. doi: 10.1038/nsmb.2503. 23435382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ramírez de Mingo D., López-García P., Hervás R., Laurents D.V., Carrión-Vázquez M. Molecular determinants of liquid demixing and amyloidogenesis in human CPEB3. bioRxiv. 2020 doi: 10.1101/2020.06.02.129783. [DOI] [Google Scholar]
- 45.Auton M., Bolen W.D. Predicting the energetics of osmolyte-induced protein folding/unfolding. Proc. Natl. Acad. Sci. U. S. A. 2005;102:15065–15068. doi: 10.1073/pnas.0507053102. 16214887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pace C.N., Vajdos F., Fee L., Grimsley G., Gray T. How to measure and predict the molar absorption coefficient of a protein. Prot. Sci. 1995;4:2411–2423. doi: 10.1002/pro.5560041120. 8563639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sattler M., Schleucher J., Griesinger C. Heteronuclear multidimensional NMR experiments for the structure determination of proteins in solution employing pulsed field gradients. Prog. Nucl. Magn. Reson. Spectrom. 1999;34:93–158. doi: 10.1016/S0079-6565(98)00025-9. [DOI] [Google Scholar]
- 48.Pantoja-Uceda D., Santoro J. New 13C-detected experiments for the assignment of intrinsically disordered proteins. J. Biomol. NMR. 2014;59:43–50. doi: 10.1007/s10858-014-9827-1. 24699834. [DOI] [PubMed] [Google Scholar]
- 49.Sun Z.Y., Frueh D.P., Selenko P., Hoch J.C., Wagner G. Fast assignment of 15N-HSQC peaks using high-resolution 3D HNcocaNH experiments with non-uniform sampling. J. Biomol. NMR. 2005;33:43–50. doi: 10.1007/s10858-005-1284-4. 16222556. [DOI] [PubMed] [Google Scholar]
- 50.Pantoja-Uceda D., Santoro J. Direct correlation of consecutive C′-N groups in proteins: A method for the assignment of intrinsically disordered proteins. J. Biomol. NMR. 2013;57:57–63. doi: 10.1007/s10858-013-9765-3. 23929272. [DOI] [PubMed] [Google Scholar]
- 51.Vuister G.W., Bax A. Quantitative J correlation: A new approach for measuring homonuclear J(HN-Hα) coupling constants in 15N-enriched proteins. J. Am. Chem. Soc. 1993;115:7772–7777. doi: 10.1021/ja00070a024. [DOI] [Google Scholar]
- 52.Kjaergaard M., Poulsen F.M. Sequence correction of random coil chemical shifts: Correlation between neighbor correction factors and changes in the Ramachandran distribution. J. Biomol. NMR. 2011;50:157–165. doi: 10.1007/s10858-011-9508-2. 21604143. [DOI] [PubMed] [Google Scholar]
- 53.Kjaergaard M., Brander S., Poulsen F.M. Random coil chemical shift for intrinsically disordered proteins: effects of temperature and pH. J. Biomol. NMR. 2011;49:139–149. doi: 10.1007/s10858-011-9472-x. 21234644. [DOI] [PubMed] [Google Scholar]
- 54.Bai Y., Milne J.S., Mayne L., Englander S.W. Primary structure effects on peptide group hydrogen exchange. Proteins. 1993;17:75–86. doi: 10.1002/prot.340170110. 8234246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xue B., Dunbrack R.L., Williams R.W., Dunker A.K., Uversky V.N. PONDR-FIT: A meta-predictor of intrinsically disordered amino acids. Biochim. Biophys. Acta. 2010;1804:966–1010. doi: 10.1016/j.bbapap.2010.01.011. 20100603. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
NMR chemical shifts of the Orb2A PLD recorded at pH 4 and at pH 7 have been deposited in the BMRB data bank under file number 50274. The raw NMR data can be requested from D. V. Laurents at dlaurents@iqfr.csic.es. All other data presented and discussed are contained within the article and its supporting material.