ABSTRACT
Numerous neutralizing antibodies that target SARS-CoV-2 have been reported, and most directly block binding of the viral Spike receptor-binding domain (RBD) to angiotensin-converting enzyme II (ACE2). Here, we deliberately exploit non-neutralizing RBD antibodies, showing they can dramatically assist in neutralization when linked to neutralizing binders. We identified antigen-binding fragments (Fabs) by phage display that bind RBD, but do not block ACE2 or neutralize virus as IgGs. When these non-neutralizing Fabs were assembled into bispecific VH/Fab IgGs with a neutralizing VH domain, we observed a ~ 25-fold potency improvement in neutralizing SARS-CoV-2 compared to the mono-specific bi-valent VH-Fc alone or the cocktail of the VH-Fc and IgG. This effect was epitope-dependent, reflecting the unique geometry of the bispecific antibody toward Spike. Our results show that a bispecific antibody that combines both neutralizing and non-neutralizing epitopes on Spike-RBD is a promising and rapid engineering strategy to improve the potency of SARS-CoV-2 antibodies.
KEYWORDS: Bispecific, neutralizing antibody, knob-in-hole, COVID-19, SARS-CoV-2
Introduction:
SARS-CoV-2 has emerged as a global health concern and effective therapeutics are necessary to curb the COVID-19 pandemic. Many potential therapeutic options for treating COVID-19 have been explored, including small molecules,1 convalescent patient sera,2 decoy receptors,3–6 neutralizing antibodies,7–18 and other protein scaffolds.19–21 In particular, antibodies are advantageous due to their specific and potent binding, demonstrated pharmacokinetics, and ability to be recombinantly produced and manufactured at scale. SARS-CoV-2 antibodies have been derived from several sources, including B-cells of convalescent patients7,12,13,17 and people with prior coronavirus infections,14,15 animal immunization,10,18 and synthetic libraries or de novo design.8,9,11,16,19 Most of the antibodies reported to date potently target the receptor-binding domain (RBD) in the trimeric Spike protein on the surface of SARS-CoV-2,7,13,22–24 which is highly immunogenic and is the key protein that mediates cellular entry via interaction with the host angiotensin-converting enzyme II (ACE2) receptor.25 However, given the widespread global impact of this pandemic and limitations in biologic manufacturing capacities, means to further increase the potency of these antibodies and thereby decrease the dose required will be critical in meeting the global demand for therapeutics.26 Additionally, testing different scaffolds and targeting mechanisms against coronavirus could lead to a better understanding of the most effective modalities and ultimately lead to a more resilient therapeutic arsenal against viral infections.
Following the identification of an initial candidate antibody, various methods are used to improve its affinity and potency, but each approach has both advantages and drawbacks. Affinity maturation using mutagenesis or library display is a powerful tool to improve candidate antibodies and can screen large sequence spaces.27 However, this process is labor-intensive and may result in an antibody sequence with altered biophysical or pharmacokinetic properties that require additional optimization. A parallel strategy to improve potency is to target multiple epitopes, either by engineering bispecific or multi-specific molecules or by combining multiple antibodies into a cocktail.21,28 Targeting multiple epitopes has the added benefit of decreasing the likelihood of viral escape and resistance,18,29 and has shown promise as a powerful viral immunotherapy against viruses, such as influenza30 and HIV.31 Indeed, several cocktails12,29 and engineered multi-specific binders19,32 have been shown to be effective against SARS-CoV-2. Recently, our lab demonstrated the benefits of linking multiple neutralizing epitopes on the SARS-CoV-2 Spike using bi-paratopic binders derived from variable heavy (VH) domains.8 By linking multiple neutralizing VH together in tandem, we were able to improve antibody potency through avidity.
Here, we explored whether linking non-neutralizing binders to neutralizing binders in a bispecific scaffold could be used as a means to rapidly improve neutralization potency. Using phage display, we identified Fabs that bind RBD but do not block ACE2 binding and then assembled them in a knob-in-hole (KIH) bispecific IgG scaffold with VH binders that block ACE2. These VH/Fab bispecifics have the additional advantage of avoiding the light-chain mispairing problem common to bispecific IgGs that include Fabs on both arms.33 Remarkably, the resulting VH/Fab bispecifics are ~20 to 25-fold more potent in neutralizing both pseudotyped and authentic SARS-CoV-2 virus than the mono-specific bi-valent VH-Fc or IgG alone or as a cocktail. This effect is epitope-dependent, illustrating the unique geometry that bispecific VH/Fab IgGs could capture on the trimeric Spike protein. Our findings highlight how targeting multiple epitopes within a single therapeutic molecule, both neutralizing and non-neutralizing, can confer significant gains in efficacy, and could potentially be generalized to other therapeutic targets to rapidly enhance antibody potency.
Results
Identification and characterization of Fabs against SARS-CoV-2 Spike
We recently reported the identification and engineering of human VH binders against SARS-CoV-2 Spike from an in-house VH-phage library, using a masked phage selection strategy to enrich binders to Spike-RBD that compete with ACE2. From this process, we identified VH domains against two epitopes (sites A and B) that bind within the ACE2 binding site of SARS-CoV-2 Spike. In the bi-valent VH-Fc format, both site A and site B binders block binding of ACE2 to Spike and neutralize pseudotyped and authentic SARS-CoV-2. VH domains that bind outside of the ACE2 binding site were not identified with this selection campaign.8
Here, we used an in-house Fab-phage library to identify unbiased Fab binders that recognize Spike-RBD. Briefly, for each round of selection, the Fab-phage pool was pre-cleared with biotinylated Fc immobilized on streptavidin (SA)-coated magnetic beads before incubating with SA-beads conjugated with biotinylated Spike-RBD-Fc (Figure 1a). After 3–4 rounds of selection, significant enrichment was observed for Fab-phage that bound Spike-RBD-Fc over Fc alone. Individual phage clones were isolated and phage ELISA was used to characterize binding to Spike-RBD-Fc alone and in complex with ACE2-Fc. We hypothesized that Fab-phage that can bind similarly to Spike-RBD-Fc alone or when masked with ACE2-Fc would bind an epitope outside of the ACE2 binding site, and would therefore occupy a unique epitope from the VH. From here, we identified over 200 unique Fab-phage sequences that bound Spike-RBD-Fc, a majority of which did not bind at the Spike-ACE2 interface (Figure 1b). We characterized a subset of these and identified two lead Fabs, C01 and D01, which bound Spike-RBD-Fc and the trimeric Spike full ectodomain (Secto) with high affinity (Figure 1c-d, Table 1). Conversion of these Fabs into a traditional bivalent IgG scaffold further improved affinity to Secto to single-digit nanomolar KD (figure 1f-g). The increased affinity of the IgG compared to the Fab is driven by the avidity of the two binding arms. Due to the challenges of modeling the interaction between a bi-valent binder and a conformationally dynamic, trimeric Spike, we have reported affinities as apparent KDs (KD,app) derived from a 1:1 binding model of the data.
Figure 1.
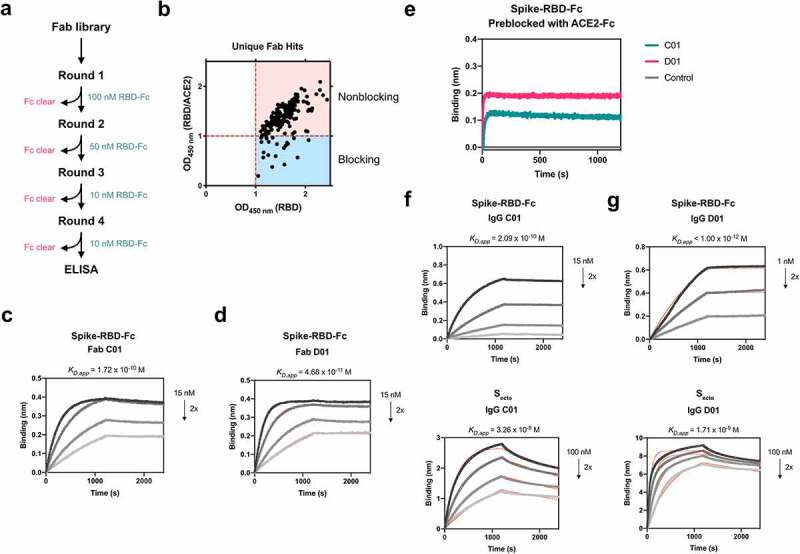
Fabs identified by phage display bind Spike RBD and Secto with high affinity outside of the ACE2 binding site
(a) Schematic of phage display used to isolate binders to Spike RBD-Fc from an in-house Fab-phage library. (b) Phage ELISA used to characterize binders shows that a majority of binders isolated did not bind similarly to Spike RBD in complex with ACE2 as to RBD alone. Multipoint BLI measurements of (c) Fab C01 and (d) Fab D01 on Spike RBD-Fc demonstrate high affinity binding. (e) Sequential epitope binning BLI demonstrates when Spike RBD-Fc is pre-saturated with ACE2-Fc, both Fabs C01 and D01 can still bind, indicating a non-overlapping epitope with ACE2-Fc. Multipoint BLI measurements of (f) IgG C01 and (g) IgG D01 show that conversion of Fab to IgG increases affinity to both Spike RBD-Fc (top) and trimeric Secto (bottom).
Table 1.
In vitro binding affinities of antibodies against SARS-CoV-2 Spike
| Antibody | KD,app (nM) |
|
|---|---|---|
| Spike-RBD-Fc | Secto | |
| Fab C01 | 0.172 | 29.9 |
| Fab D01 | 0.047 | 92.0 |
| IgG C01 | 0.209 | 3.26 |
| IgG D01 | <0.001a | 1.71 |
| VH-Fc A01 | <0.001a | 4.09 |
| VH-Fc B01 | 0.056 | 0.313 |
| Bis1 (VH A01/Fab C01) | <0.001a | 0.603 |
| Bis2 (VH B01/Fab C01) | <0.001a | 0.611 |
| Bis3 (VH A01/Fab D01) | <0.001a | 0.395 |
| Bis4 (VH B01/Fab D01) | <0.001a | 0.127 |
akoff was below the limit of detection (<1 x 10–7 sec−1) and could not be fit.
We proceeded to characterize the epitopes of Fab C01 and Fab D01 by sequential bio-layer interferometry (BLI), where one binder is incubated with the antigen until saturation, followed by incubation with the second binder. Robust binding of the second binder in the presence of the first suggests that the two binders occupy non-overlapping epitopes on the same antigen. Both Fab C01 and Fab D01 do not overlap with ACE2-Fc, indicating that they bind outside of the ACE2 binding site (Figure 1e). Interestingly, while Fab C01 and D01 compete for binding on Spike-RBD, on Secto their epitopes do not appear to overlap (Supplementary Figure 1a). Additionally, we observe that pre-saturation with Fab D01 blocks binding of Fab C01 on Secto, while pre-saturation with Fab C01 does not block Fab D01 (Supplementary Figure 1b-c). These data suggest that, although C01 and D01 have overlapping epitopes on isolated Spike-RBD, these Fabs have different binding mechanisms and could be influenced by the dynamics and accessibility of the RBDs in the context of the Spike trimer. Therefore, we assign site C (for Fab C01) and site D (for Fab D01) to unique epitopes on Secto though they overlap on Spike-RBD. Additionally, we compared the epitopes of Fab C01 and Fab D01 to a previously described antibody CR302214 and found that Fab D01 competes with CR3022 while Fab C01 does not (Supplementary Figure 1d). CR3022 has been shown to recognize the RBD outside of the ACE2 binding site at an epitope only accessible when the RBD is in the “up” conformation.14 Fab D01 appears to bind an overlapping epitope, but with a higher affinity to Spike-RBD-Fc compared to CR3022 (KD = 115 nM). Thus, from these studies, we have identified binders against four unique epitopes on Spike-RBD: site A (VH A01), site B (VH B01), site C (Fab C01), and site D (Fab D01).
Generation of bispecific KIH IgGs using Fab and VH against SARS-CoV-2 Spike
We wondered whether combining VH and Fab binders directed at different Spike epitopes into a bispecific antibody scaffold could improve potency. One of the most straightforward and successful methods to make bispecific IgG-type binders is the classic KIH strategy, which was pioneered by Paul Carter and colleagues at Genentech.34 In this approach, a complementary set of mutations in the CH3 domains of the Fc promotes the heterodimerization between a “knob” Fc and a “hole” Fc. A KIH bispecific IgG modality has not been reported for COVID-19 to date, but has been previously developed for other viral infections, such as HIV, and showed improved potency compared to the mono-specific parental binders.35 Therefore, generating KIH bispecifics against Spike could present a novel, effective modality against SARS-CoV-2.
When the KIH strategy is applied for two Fab arms, the individual half IgGs must be separately expressed and purified before they can be assembled into a bispecific IgG molecule due to the presence of two different light chains. This process is required to circumvent the problem of light chains and heavy chain mispairing that can occur when the antibodies are co-expressed in the same cell. While many strategies have been developed to try and address this issue, including using common light chains, additional purifications, or scaffold engineering, they all require additional labor-intensive steps.36–38 However, since the VH/Fab bispecific molecules contain only one light chain in the Fab arm, co-expression of the VH-Fc knob with Fab-Fc hole should generate the proper VH/Fab bispecific IgG (Figure 2a).
Figure 2.
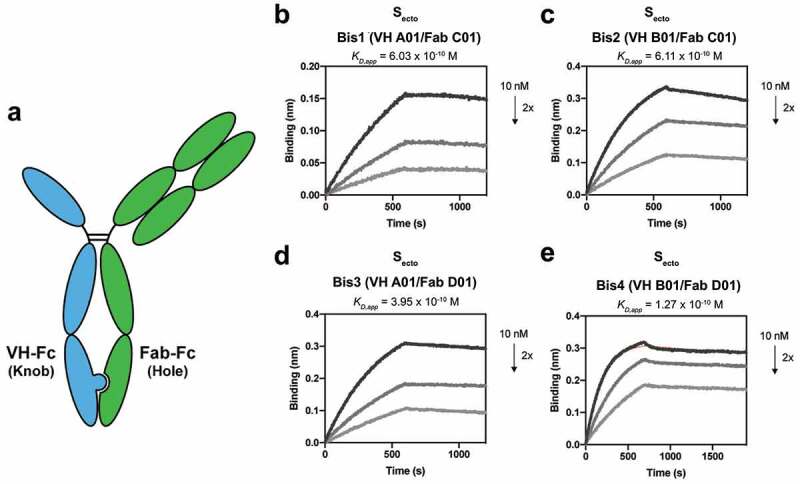
Bispecific VH/Fab IgGs bind with high affinity to trimeric Secto. (a) Cartoon schematic of the bispecific VH/Fab IgG antibody scaffold utilized in this study. (b-e) Multipoint BLI measurements (10 nM, 5 nM, and 2.5 nM) of the indicated bispecific antibody on Secto. (b) Bis1 (VH A01/FabC01) (c) Bis2 (VH B01/FabC01) (d) Bis3 (VH A01/FabD01) (e) Bis4 (VH B01/FabD01)
Using this strategy, we generated four different VH/Fab bispecifics to explore combinations of targeting sites A and B with sites C and D; Bis1 is VH(A01)/Fab(C01) fusion, Bis2 is VH(B01)/Fab(C01), Bis3 is VH(A01)/Fab(D01), and Bis4 is VH(B01)/Fab(D01). For each bispecific, the VH was cloned into the “knob” Fc, and the heavy chain of the Fab was cloned into the “hole” Fc. All four bispecific IgGs were successfully expressed and purified and shown by gel to be bispecific, containing both a VH-Fc and Fab-Fc arm on each molecule (Supplementary Figure 2). All four bispecifics bound Secto with higher affinity than their parental mono-specific counterparts (Figure 2b-e, Table 1). In particular, Bis3 and Bis4, which both include Fab(D01), bound Secto with highest affinity with KD,app of 395 pM and 127 pM, respectively. Reversing the orientation and immobilizing the bispecific IgG on the biosensor and probing with soluble Secto did not significantly change the measured affinity (Supplementary Figure 3).
Bispecific KIH IgGs show enhanced neutralization against pseudotyped and authentic SARS-CoV-2 compared to mono-specific binders
We then tested the neutralization potency of these VH/Fab bispecifics using HIV-1-derived lentivirus pseudotyped with SARS-CoV-2 Spike to compare their potencies to the mono-specific bi-valent VH-Fc and IgGs. Spike pseudotypes were generated and used in neutralization assays with HEK293T cells expressing ACE2 on the surface; the assays were performed using established vectors and protocols.39 VH-Fc A01 and VH-Fc B01 neutralized Spike pseudotypes with half-maximal inhibitory concentration (IC50) of 2.86 nM (0.23 μg/mL) and 2.01 nM (0.16 μg/mL), respectively. In contrast, we found that IgG C01 and IgG D01 did not neutralize pseudovirus at the concentrations tested (up to 100 nM). This is likely because these two IgGs do not target the ACE2 binding site, and there are likely epitope and geometry-specific mechanisms that determine whether and to what extent non-blocking binders neutralize the virus. Indeed, anti-Spike antibodies that do not compete with ACE2 binding, including CR3022, have been reported, but they have mixed and variable success in neutralizing SARS-CoV-2, depending on the specific antibody and its epitope.14,15,17,40–42
Next, we tested the neutralization profile of the bispecific VH/Fab IgGs. To our surprise, we found that bispecifics that use the non-neutralizing Fab D01 neutralized significantly more potently than the parental mono-specific binders. Bis3 and Bis4 neutralized with IC50s of 0.128 nM (0.015 μg/mL) and 0.107 nM (0.012 μg/mL), respectively, which is ~20-fold more potent than the VH-Fc alone (Figure 3c-d, Table 2). This indicates that a bispecific antibody, even if one of the arms is non-neutralizing, can show enhanced potency compared to the mono-specific counterparts. In contrast, the bispecific antibodies that included Fab C01 (Bis1, Bis2) did not show such improved potency and neutralized with similar IC50s to the parental VH-Fc (Figure 3a-b). This suggests that the enhanced potency provided by a non-neutralizing binder is epitope-specific. Interestingly, we observed no significant correlation between IC50 and binding affinity against Secto of the bispecifics and VH-Fcs (R2 = 0.06) (Supplementary Figure 4). This suggests that an increase in neutralization potency of anti-Spike binders is not highly predictive from affinity alone and that the specific mechanism and geometry of epitopes targeted by the bispecific IgG scaffold likely play an important role in increasing potency. Additionally, we find that a cocktail of the mono-specific antibodies (IgG + VH-Fc) does not improve potency (Supplementary Figure 5), which suggests that a bispecific IgG, where two epitopes are targeted by a single agent, has unique mechanisms and advantages.
Figure 3.
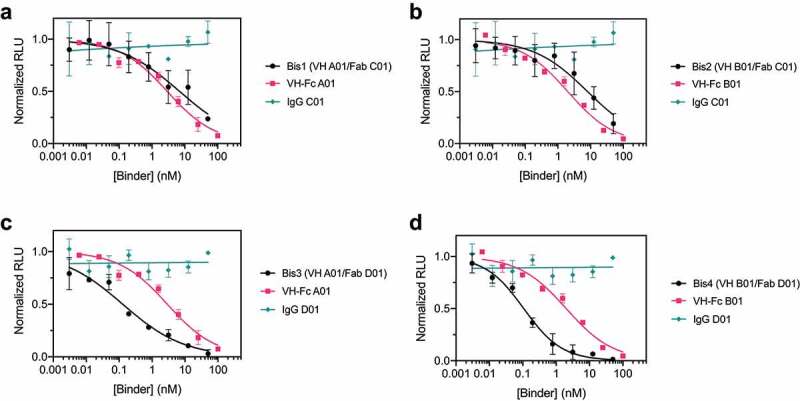
Bispecific VH/Fab IgGs are more potent in neutralizing SARS-CoV-2 pseudovirus than the mono-specific counterparts
Pseudovirus neutralization curves for (a) Bis1 (VH A01/Fab C01) compared to VH-Fc A01 and IgG C01, (b) Bis2 (VH B01/Fab C01) compared to VH-Fc B01 and IgG C01, (c) Bis3 (VH A01/Fab D01) compared to VH-Fc A01 and IgG D01, (d) Bis4 (VH B01/Fab D01) compared to VH-Fc A01 and IgG D01. Data represent the average and standard deviation of three independent experiments and were fit to a non-linear regression using Prism 8 software to obtain IC50 values
Table 2.
SARS-CoV-2 Pseudovirus Neutralization IC50
| Antibody | IC50 |
|
|---|---|---|
| nM (95% CI) | μg/mL (95% CI) | |
| VH-Fc A01 | 2.86 (1.63–5.03) | 0.23 (0.13–0.40) |
| VH-Fc B01 | 2.01 (1.22–3.34) | 0.16 (0.10–0.27) |
| IgG C01 | ≫100 | ≫15 |
| IgG D01 | ≫100 | ≫15 |
| VH-Fc A01 + IgG C01 | 6.63 (4.57–9.58) | 0.75 (0.52–1.09) |
| VH-Fc B01 + IgG C01 | 5.13 (3.48–7.57) | 0.58 (0.39–0.86) |
| VH-Fc A01 + IgG D01 | 2.25 (1.66–3.04) | 0.26 (0.19–0.35) |
| VH-Fc B01 + IgG D01 | 3.23 (2.34–4.42) | 0.37 (0.27–0.50) |
| Bis1 (VH A01/Fab C01) | 6.87 (3.41–17.07) | 0.78 (0.39–1.94) |
| Bis2 (VH B01/Fab C01) | 8.07 (4.63–16.00) | 0.92 (0.53–1.82) |
| Bis3 (VH A01/Fab D01) | 0.128 (0.080–0.201) | 0.015 (0.009–0.023) |
| Bis4 (VH B01/Fab D01) | 0.107 (0.080–0.143) | 0.012 (0.009–0.016) |
Lastly, we examined the neutralization profile of the bispecific IgGs and VH-Fc on authentic SARS-CoV-2 virus with Vero E6 cells as the host cell. Each binder was assayed for its ability to decrease the cytopathic effect (CPE) of Vero E6 cells caused by SARS-CoV-2 infection. Consistent with the pseudovirus neutralization results, bispecific IgGs Bis3 and Bis4 neutralized authentic SARS-CoV-2 significantly more potently (~25-fold) than the VH-Fcs (Figure 4, Table 3). The IC50s of VH-Fc A01 and VH-Fc B01 against authentic SARS-CoV-2 were 25–30 nM. These values are similar to a previously reported IC50 of VH-Fc B01 of 33.5 nM, which used qPCR to read out intracellular viral RNA.8 The neutralization IC50s of Bis3 and Bis4 against authentic SARS-CoV-2 was 1.00 nM (0.11 μg/mL) and 1.19 nM (0.14 μg/mL), respectively. Interestingly, Bis1 and Bis2, which did not show a significant improvement over the VH-Fcs in the pseudovirus neutralization assays, were about 2.5 to 3-fold more potent than the VH-Fcs when assayed on the authentic virus. Taken together, we find that bispecific IgGs, particularly Bis3 and Bis4, are significantly more potent in neutralizing both pseudotyped and authentic SARS-CoV-2 virus than their mono-specific counterparts.
Figure 4.
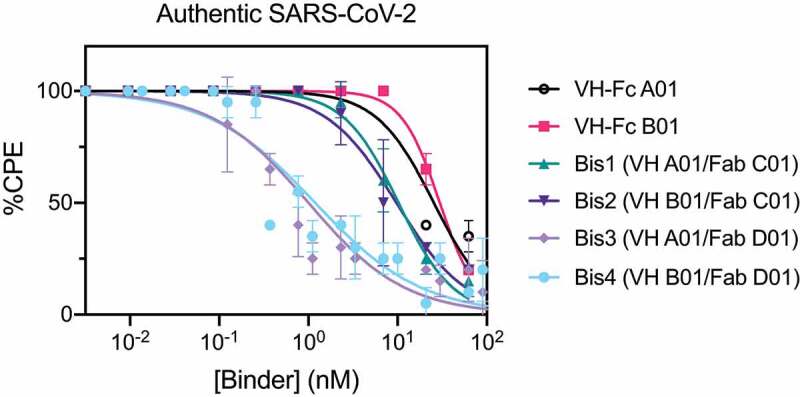
Bispecific VH/Fab IgGs neutralize authentic SARS-CoV-2 virus more potently than the mono-specific VH-Fcs
Authentic SARS-CoV-2 virus neutralization curves of VH-Fc A01, VH-Fc B01, Bis1 (VH A01/Fab C01), Bis2 (VH B01/Fab C01), Bis3 (VH A01/Fab D01), Bis4 (VH B01/Fab D01). Vero E6 cells were incubated with the virus and threefold dilution of binder and assessed for cytopathic effect (CPE) 7 d after infection. Data represent the average and standard deviation of two independent experiments. Data were fit to a non-linear regression using Prism 8 software to obtain IC50 values.
Table 3.
Authentic SARS-CoV-2 Virus Neutralization IC50
| Antibody | IC50 |
|
|---|---|---|
| nM (95% CI) | μg/mL (95% CI) | |
| VH-Fc A01 | 25.5 (18.8–36.0) | 2.04 (1.50–2.88) |
| VH-Fc B01 | 29.8 (26.5–33.5) | 2.38 (2.12–2.68) |
| Bis1 (VH A01/Fab C01) | 10.3 (8.5–12.4) | 1.16 (0.95–1.40) |
| Bis2 (VH B01/Fab C01) | 9.70 (6.80–14.2) | 1.10 (0.77–1.61) |
| Bis3 (VH A01/Fab D01) | 1.00 (0.67–1.54) | 0.11 (0.08–0.17) |
| Bis4 (VH B01/Fab D01) | 1.19 (0.80–1.80) | 0.14 (0.09–0.20) |
Structural modeling of the epitopes targeted by bispecific IgG Bis4 (VH B01/Fab D01)
Bis4, which uses the combination of VH B01 and Fab D01, was one of the most potent molecules we tested in this study. The epitope of VH B01 was determined previously by cryo-EM.8 In the absence of a high-resolution structure of Fab D01 bound to Spike, we used the structure of CR302212 as a surrogate to model how the two arms on Bis4 could engage Spike. We find that in the context of the same RBD, the VH and Fab bind at separate, non-overlapping epitopes (Supplementary Figure 6a-b). The CR3022 epitope is fully exposed only in the RBD “up” conformation, while the VH B01 epitope is largely accessible regardless of the RBD conformation. VH B01 and Fab CR3022 point away from each other at an angle of 133 degrees, with a distance of 134.5 Å between the C-termini of the VH and the heavy chain of the Fab. Although the exact epitope and angle of engagement of Fab D01 may differ from CR3022, it appears challenging for both arms of Bis4 to engage the same RBD. However, it could be possible for the Bis4 VH arm to bind one RBD, and the Fab arm to bind a neighboring RBD in the context of the Spike trimer when one or more RBDs are in the “up” conformation (Supplementary Figure 6c). We hypothesize that simultaneous engagement of two RBDs on Spike by Bis4 could explain its potent neutralization mechanism, although there could be other unexplored mechanisms at play. Determining the exact epitope and binding mechanism of Fab D01 would shed further light on how these bispecific IgGs engage Spike and neutralize SARS-CoV-2.
Discussion
Here, we report the generation of bispecific IgG antibodies against SARS-CoV-2 Spike that combine neutralizing and non-neutralizing binders against different epitopes on the Spike-RBD as a promising protein engineering strategy to rapidly improve the potency of antibody therapeutics for COVID-19. We show that certain combinations of these binders in a bispecific IgG scaffold are significantly more potent in neutralizing pseudotyped and authentic SARS-CoV-2 virus than the mono-specific bi-valent counterparts and may enable efficacy increases not predicted by affinity alone. Additionally, we show that non-blocking, non-neutralizing epitopes can provide an unexpected benefit and boost the potency of molecules when combined in a bispecific format with neutralizing epitopes. Combing neutralizing and non-neutralizing epitopes can be a useful approach to rescue binders that would have been deprioritized due to their lack of neutralization. Exploring the use of this strategy in parallel with other affinity maturation campaigns on some of the most potent antibodies reported against SARS-CoV-2 to date could enable the generation of even more potent neutralizing binders. This can decrease the effective dose necessary for therapeutic effect, thus lessen the burden on manufacturing capacity and enable the wider distribution of these treatments.
In aggregate, we have produced binders against four distinct epitopes within the RBD of Spike. This is somewhat remarkable given the small size of the RBD (206 amino acids, ~23 kDa). Binders to many epitopes on Spike have been reported,15,17,19,41–43 showing that this antigen is highly immunogenic both in vivo and in vitro. It will be interesting to map them relative to those we have found. Our results show that the combination via a KIH bispecific scaffold is particularly useful in improving potency through avidity and multi-epitope targeting and could be less labor and time-intensive than affinity maturation.
Our study also highlights how different epitopes on Spike differ in their neutralization potency and their engineering potential. To what extent an antibody neutralizes SARS-CoV-2 is likely influenced by its binding mechanism, affinity, and scaffold, and is made more complex by the oligomeric and dynamic nature of the target antigen Spike. It is well established that the RBDs of Spike proteins of the coronavirus family are capable of adopting various conformations and that the trimer can undergo large conformational changes.44 These structural dynamics likely affect the accessibility and orientation of the different binding epitopes, which can then influence the potency of binders and how they behave when engineered into multi-specific modalities. This may help explain why bispecifics that use Fab D01 are superior to bispecifics that use Fab C01. Additionally, even binders that overlap in epitope, such as Fab D01 and CR3022, may differ in their properties. Fab D01 has a much higher affinity to Spike than CR3022 and was non-neutralizing. CR3022 was also non-neutralizing (IC50 ≫ 400 μg/mL),14 although another recent study contradicts this observation,40 and showed that CR3022 can neutralize SARS-CoV-2, possibly through the destabilization of the Spike trimer. Which combinations of binders will synergize and what antibody scaffolds are optimal for efficacy remain unclear, and this uncertainty provides wide latitude for protein engineers to explore how these different factors affect antibody potency. A deeper biophysical understanding of Spike dynamics will also be important for the rational engineering of potent biologics to this therapeutically important target. We believe these principles for the design of bispecific and bi-paratopic binders combining neutralizing and non-neutralizing binders can apply to other therapeutic targets even in the absence of high-resolution structures.
Materials & Methods
Cloning
Spike-RBD-Fc and trimeric Spike ectodomain (Secto), and ACE2-Fc were produced as biotinylated proteins as previously described.45 Fabs were subcloned from the Fab-phagemid into an E. coli expression vector pBL347. VH-Fc was cloned into a pFUSE (InvivoGen) vector with a human IgG1 Fc domain as previously described.8 The heavy chain of the IgG was cloned from the Fab plasmid into a pFUSE (InvivoGen) vector with a human IgG1 Fc domain. The light chain of the IgG was cloned from the Fab plasmid into the same vector but lacking the Fc domain. All constructs were sequence verified by Sanger sequencing.
Protein Expression and Purification
Fabs were expressed in E. coli C43(DE3) Pro + using an optimized autoinduction medium and purified by protein A affinity chromatography.46 VH-Fc, IgGs, and bispecifics were expressed in Expi293 BirA cells using transient transfection (Expifectamine, Thermo Fisher Scientific). After transfection for 3–5 d, media was harvested, and VH-Fc and IgGs were purified using protein A affinity chromatography. Bispecific antibodies were purified by Ni-NTA affinity chromatography. The bispecifics were then buffer exchanged into phosphate-buffered saline (PBS) containing 20% glycerol, concentrated, and flash frozen for storage. All other proteins were buffer exchanged into PBS by spin concentration and stored in aliquots at −80°C. The purity and integrity of all proteins were assessed by SDS-PAGE.
Fab-phage selection
Phage selections were done according to previously established protocols.46 Selections were performed using biotinylated antigens captured with SA-coated magnetic beads (Promega). In each round, the phage pool was first cleared by incubation with beads loaded with Fc domain only. The unbound phage was then incubated with beads loaded with Spike-RBD-Fc. After washing, the bound phage was eluted by the addition of 2 μg/mL of TEV protease. In total, four rounds of selection were performed with decreasing amounts of Spike-RBD-Fc. All steps were done in PBS buffer + 0.02% Tween-20 + 0.2% bovine serum albumin (PBSTB). Individual phage clones from the third and fourth rounds of selections were analyzed by phage ELISA.
Phage ELISA
For each phage clone, four different conditions were tested – Direct: Spike-RBD-Fc, Competition: Spike-RBD-Fc with an equal concentration of Spike-RBD-Fc in solution, Negative selection: ACE2-Fc/Spike-RBD-Fc complex, and Control: Fc. 384-well Nunc Maxisorp flat-bottom clear plates (Thermo Fisher Scientific) were coated with 0.5 μg/mL of NeutrAvidin in PBS overnight at 4°C and subsequently blocked with PBSTB for 1 h at room temperature. Plates were washed 3X with PBS containing 0.05% Tween-20 (PBST) and were washed similarly between each of the steps. 20 nM of biotinylated Spike-RBD-Fc, ACE2-Fc/Spike-RBD-Fc complex, or Fc diluted in PBSTB was captured on the NeutrAvidin-coated wells for 30 min, then blocked with PBSTB + 10 μM biotin for 30 min. Phage supernatant diluted 1:5 in PBSTB was added for 20 min. For the competition samples, the phage supernatant was diluted into PBSTB with 20 nM Spike-RBD-Fc. Bound phage was detected by incubation with anti-M13-horseradish peroxidase conjugate (Sino Biologics catalog number 11973-MM05-H, 1:5000) for 30 min, followed by the addition of TMB substrate (VWR International). The reaction was quenched with the addition of 1 M phosphoric acid and the absorbance at 450 nm was measured using a Tecan M200 Pro spectrophotometer.
Bio-layer Interferometry
BLI measurements were made using an Octet RED384 (ForteBio) instrument. Spike-RBD-Fc or Secto were immobilized on an SA biosensor and loaded until a 0.4 nm signal was achieved. After blocking with 10 μM biotin, purified binders in solution were used as the analyte. PBSTB was used for all buffers. Data were analyzed using the ForteBio Octet analysis software and kinetic parameters were determined using a 1:1 monovalent binding model.
Pseudovirus generation
HEK293T-ACE2 cells were a gift from Arun Wiita’s laboratory at the University of California, San Francisco. Cells were cultured in D10 media (DMEM + 1% Pen/Strep + 10% heat-inactivated fetal bovine serum). Plasmids to generate pseudotyped HIV-1-derived lentivirus were a gift from Peter Kim’s lab at Stanford University and pseudovirus displaying SARS-CoV-2 Spike was prepared as previously described.39 Briefly, plasmids at the designated concentrations were added to OptiMEM media with FuGENE HD Transfection Reagent (Promega) at a 3:1 FuGENE: DNA ratio, incubated for 30 min and subsequently transfected into HEK-293 T cells. After 24 h, the supernatant was removed and replaced with D10 culture media. The virus was propagated for an additional 48 h, and the supernatant was harvested and filtered. The virus was stored in flash-frozen aliquots at −80°C and thawed before use.
HEK-ACE2 were seeded at 10,000 cells/well on 96-well white plates (Corning, cat. 354620). After 24 h, pseudovirus stocks were titered via a two-fold dilution series in D10 media, and 40 μL were added to cells. After 60 h, infection and intracellular luciferase signal was determined using Bright-Glo™ Luciferase Assay (Promega), and the dilution achieving maximal luminescent signal within the linear range, ~3-5 x 105 luminescence units, was chosen as the working concentration for neutralization assays.
Pseudovirus neutralization assay
HEK-ACE2 were seeded at 10,000 cells/well in 40 μL of D10 on 96-well white plates (Corning, cat. 354620) 24 h before infection. To determine IC50 for pseudovirus neutralization, dose series of each VH binder were prepared at 3x concentration in D10 media, and 50 μL was aliquoted into each well in 96-well plate format. Next, 50 μL of the virus was added to each well, except no virus control wells, and the virus and blocker solution was allowed to incubate for 1 h at 37°C. After pre-incubation, 80 μL of the virus and blocker inoculum was transferred to HEK-ACE2. After 60 h of infection at 37°C, intracellular luciferase signal was measured using the Bright-Glo™ Luciferase Assay. Luminescence was normalized to the no binder control and plotted using Prism 8 software. Non-linear four-parameter regression was used to determine the IC50.
Authentic SARS-CoV-2 virus neutralization assay
Authentic SARS-CoV-2 virus was handled and neutralization assays were conducted using biosafety level 3 containment with approved protocols. SARS-CoV-2 isolate USA-WA1/2020 (NR-52281) was obtained from BEI resources47 and expanded with a minimal passage in Vero E6 cells. SARS-CoV-2 at 103 TCID50/ml was incubated with threefold serially diluted binder at 37°C for 1 h before infection of Vero E6 cell monolayer in 96-well-plates. Virus/binder mixtures were added to 10 replicate wells at 100 μL per well. The plates were incubated for 7 d at 37°C with 5% CO2 until clear CPE developed. The experiment was repeated twice. Wells with clear CPE were counted positive and the percentage of positive wells for each concentration of binder was plotted and analyzed using Prism 8 software. Non-linear four-parameter regression was used to determine the IC50.
Supplementary Material
Acknowledgments
We thank members of the Wells Lab, particularly those working on COVID-19 projects for their efforts and contributions. We would specifically like to thank Dr. Jie Zhou, Dr. Jeff Glasgow, and Dr. James Byrnes for assistance in phage selection and Fab cloning and purification and Dr. Kaitlin Schaefer for assistance in Spike purification. We also thank Dr. Matthew Nix (in Dr. Arun Wiita’s Lab, UCSF) for guidance on pseudovirus neutralization assays and the laboratory of Dr. Peter Kim (Stanford University) for providing plasmids for pseudovirus production. We also thank Dr. Ian Wilson (Scripps Research) for providing plasmids for CR3022. The following reagent was deposited by the Centers for Disease Control and Prevention and obtained through BEI Resources, NIAID, NIH: SARS-Related Coronavirus 2, Isolate USA-WA1/2020, NR-52281.
J.A.W. is supported by generous grants from NCI (R35 GM122451-01); Chan-Zuckerberg Biohub, UCSF Program for Breakthrough Biomedical Research (PBBR), Fast Grants from Emergent Ventures at the Mercatus Center at the George Mason University (#2154), The Harry and Dianna Hind Professorship in Pharmaceutical Sciences, and funding from The Harrington Discovery Institute (GA33116). S.A.L. is a Merck Fellow of the Helen Hay Whitney Foundation. N.J.R., I.L., and S.K.E. are supported by the National Science Foundation. X.X.Z. is a Merck Fellow of the Damon Runyon Cancer Research Foundation, DRG-2297-17.
Funding Statement
This work was supported by the Damon Runyon Cancer Research Foundation [DRG-2297-17]; Helen Hay Whitney Foundation; National Cancer Institute [R35 GM122451-01]; National Science Foundation; The Harrington Discovery Institute [GA33116]; Chan-Zuckerberg Biohub; UCSF Program for Breakthrough Biomedical Research; Fast Grants from Emergent Ventures at the Mercatus Center at George Mason University [2154]; The Harry and Dianna Hind Professorship in Pharmaceutical Sciences.
Abbreviations
| ACE2 | Angiotensin-converting enzyme II |
| CPE | Cytopathic effect |
| KIH | Knob-in-hole |
| RBD | Receptor binding domain |
| SA | Streptavidin |
| Secto | Spike ectodomain |
| VH | Variable heavy |
Author Contributions:
S.A.L., J.A.G., and K.P. co-wrote the manuscript with input from all authors. J.A.G. designed and characterized the bispecific antibodies. K.P., S.A.L., S.K.E., J.L., and C.J.B. conducted the phage selections and/or characterized the Fab and IgG binders. S.A.L., P.S., and N.J.R. designed and conducted the pseudovirus neutralization assays. J.J. and G.S. conducted and/or supervised the authentic virus neutralization assays. I.L. assisted in protein purification. S.A.L., X.X.Z., K.K.L., and J.A.W. supervised the project.
Disclosure of Potential Conflicts of Interest:
S.A.L, J.A.G, K.P., C.J.B., J.A.W., and the Regents of The University of California have filed provisional patent applications on the VH constructs (US. Application No. 63/139,484) and bispecific VH/Fab constructs (US. Application No. 63/139,487) described in this study.
Data availability statement:
All data supporting the findings of this study are available from the corresponding author upon request.
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website.
References
- 1.Grein J, Ohmagari N, Shin D, Diaz G, Asperges E, Castagna A, Feldt T, Green G, Green ML, Lescure FX, et al. Compassionate use of remdesivir for patients with severe Covid-19. N Engl J Med. 2020;382:2327–10. doi: 10.1056/NEJMoa2007016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Joyner MJ, Senefeld JW, Klassen SA, Mills JR, Johnson PW, Theel ES, Wiggins CC, Bruno KA, Klompas AM, Kunze KL, et al. Effect of convalescent plasma on mortality among hospitalized patients with COVID-19: initial three-2 month experience. medRxiv. 2020. doi: 10.1101/2020.08.12.20169359. [DOI] [Google Scholar]
- 3.Chan KK, Dorosky D, Sharma P, Abbasi SA, Dye JM, Kranz DM, Herbert AS, Procko E.. Engineering human ACE2 to optimize binding to the spike protein of SARS coronavirus 2. Science. 2020;1265:eabc0870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Monteil V, Kwon H, Prado P, Hagelkruys A, Wimmer R, Stahl M, Leopoldi A, Garreta E. Inhibition of SARS-CoV-2 infections in engineered human tissues using clinical-grade soluble human ACE2. Cell. 2020;181:905–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Glasgow A, Glasgow J, Limonta D, Solomon P, Lui I, Zhang Y, Nix MA, Rettko NJ, Lim SA, Zha S, et al. Engineered ACE2 receptor traps potently neutralize SARS-CoV-2. Proc Natl Acad Sci USA. 2020;117:28046–55. doi: 10.1073/pnas.2016093117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guo L, Bi W, Wang X, Xu W, Yan R, Zhang Y, Zhao K, Li Y, Zhang M, Bao X, et al. Engineered trimeric ACE2 binds and locks “three-up” spike protein to potently inhibit SARS-CoVs and mutants. bioRxiv. 2020. doi: 10.1101/2020.08.31.274704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Robbiani DF, Gaebler C, Muecksch F, Lorenzi JCC, Wang Z, Cho A, Agudelo M, Barnes CO, Gazumyan A, Finkin S, et al. Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature. 2020;584(7821):437–42. doi: 10.1038/s41586-020-2456-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bracken CJ, Lim SA, Solomon P, Rettko NJ, Nguyen DP, Zha BS, Schaefer K, Byrnes JR, Zhou J, Lui I, et al. Bi-paratopic and multivalent VH domains block ACE2 binding and neutralize SARS-CoV-2. Nat Chem Biol. 2020. doi: 10.1038/s41589-020-00679-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang B, Asarnow D, Lee W-H, Huang C-W, Faust B, Ng PML, Ngoh EZX, Bohn M, Bulkley D, Pizzorno A, et al. Bivalent binding of a fully human IgG to the SARS-CoV-2 spike proteins reveals mechanisms of potent neutralization. bioRxiv. 2020. doi: 10.1101/2020.07.14.203414. [DOI] [Google Scholar]
- 10.Lv Z, Deng Y-Q, Ye Q, Cao L, Sun C-Y, Fan C, Huang W, Sun S, Sun Y, Zhu L, et al. Structural basis for neutralization of SARS-CoV-2 and SARS-CoV by a potent therapeutic antibody. Science. 2020;1509:eabc5881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schoof M, Faust B, Saunders RA, Sangwan S, Rezelj V, Hoppe N, Boone M, Billesbølle CB, Puchades C, Azumaya CM, et al. An ultra-potent synthetic nanobody neutralizes SARS-CoV-2 by locking Spike into an inactive conformation. bioRxiv. 2020. doi: 10.1101/2020.08.08.238469. [DOI] [Google Scholar]
- 12.Liu L, Wang P, Nair MS, Yu J, Rapp M, Wang Q, Luo Y, Chan JFW, Sahi V, Figueroa A, et al. Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. Nature. 2020;584:450–56. doi: 10.1038/s41586-020-2571-7. [DOI] [PubMed] [Google Scholar]
- 13.Cao Y, Su B, Guo X, Sun W, Deng Y, Bao L, Zhu Q, Zhang X, Zheng Y, Geng C, et al. Potent Neutralizing Antibodies against SARS-CoV-2 Identified by High-Throughput Single-Cell Sequencing of Convalescent Patients’ B Cells. Cell. 2020;182:73–84. doi: 10.1016/j.cell.2020.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yuan M, Wu NC, Zhu X, Lee -C-CD, So RTY, Lv H, Mok CKP, Wilson IA. A highly conserved cryptic epitope in the receptor-binding domains of SARS-CoV-2 and SARS-CoV. Science. 2020;368:630–33. doi: 10.1126/science.abb7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pinto D, Park YJ, Beltramello M, Walls AC, Tortorici MA, Bianchi S, Jaconi S, Culap K, Zatta F, De Marco A, et al. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature. 2020;583:290–95. doi: 10.1038/s41586-020-2349-y. [DOI] [PubMed] [Google Scholar]
- 16.Miersch S, Ustav M, Li Z, Case JB, Ganaie S, Matusali G, Colavita F, Lapa D, Capobianchi MR, Novelli G, et al. Synthetic antibodies neutralize SARS-CoV-2 infection of mammalian cells. bioRxiv. 2020. doi: 10.1101/2020.06.05.137349. [DOI] [Google Scholar]
- 17.Noy-Porat T, Makdasi E, Alcalay R, Mechaly A, Levy Y, Bercovich-Kinori A, Zauberman A, Tamir H, Yahalom-Ronen Y, Israeli M, et al. A panel of human neutralizing mAbs targeting SARS-CoV-2 spike at multiple epitopes. Nat Commun. 2020;11:1–7. doi: 10.1038/s41467-020-18159-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hansen J, Baum A, Pascal KE, Russo V, Giordano S, Wloga E, Fulton BO, Yan Y, Koon K, Patel K, et al. Studies in humanized mice and convalescent humans yield a SARS-CoV-2 antibody cocktail. Science. 2020;1014:eabd0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Walser M, Rothenberger S, Hurdiss DL, Schlegel A, Calabro V, Villemagne D, Paladino M, Hospodarsch T, Neculcea A, Zürcher A, et al. Highly potent anti-SARS-CoV-2 multi-DARPin therapeutic candidates. bioRxiv. 2020. doi: 10.1101/2020.08.25.256339. [DOI] [Google Scholar]
- 20.Cao L, Goreshnik I, Coventry B, Case JB, Miller L, Kozodoy L, Chen RE, Carter L, Walls L, Park Y-J, et al. De novo design of picomolar SARS-CoV-2 miniprotein inhibitors. Science. 2020;370:426–31. doi: 10.1126/science.abd9909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miao X, Luo Y, Huang X, Lee SMY, Yuan Z, Tang Y, Chen L, Wang C, Wu F, Xu Y, et al. A novel biparatopic hybrid antibody-ACE2 fusion that blocks SARS-CoV-2 infection: implications for therapy. MAbs. 2020;12:e1804241. doi: 10.1080/19420862.2020.1804241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Byrnes JR, Zhou XX, Lui I, Elledge SK, Glasgow JE, Lim SA, Loudermilk RP, Chiu CY, Wang TT, Wilson MR, et al. Competitive SARS-CoV-2 serology reveals most antibodies targeting the spike receptor-binding domain compete for ACE2 binding. mSphere. 2020;5:e00802–20. doi: 10.1128/mSphere.00802-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huo J, Le Bas A, Ruza RR, Duyvesteyn HME, Mikolajek H, Malinauskas T, Tan TK, Rijal P, Dumoux M, Ward PN, et al. Neutralizing nanobodies bind SARS-CoV-2 spike RBD and block interaction with ACE2. Nat Struct Mol Biol. 2020;27:846–54. doi: 10.1038/s41594-020-0469-6. [DOI] [PubMed] [Google Scholar]
- 24.Shi R, Shan C, Duan X, Chen Z, Liu P, Song J, Song T, Bi X, Han C, Wu L, et al. A human neutralizing antibody targets the receptor binding site of SARS-CoV-2. Nature. 2020;584:120–24. doi: 10.1038/s41586-020-2381-y. [DOI] [PubMed] [Google Scholar]
- 25.Wrapp D, Wang N, Corbett KS, Goldsmith JA, Hsieh CL, Abiona O, Graham BS, McLellan JS. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–63. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sharma I, Wosińska M, Kroetsch A, Sullivan H, McClellan M. COVID-19. Manufacturing for monoclonal antibodies. Duke Margolis Center for Health Policy. 2020. 1–6. Retrieved from https://hdl.handle.net/10161/21332.. [Google Scholar]
- 27.Daugherty PS, Chen G, Olsen MJ, Iverson BL, Georgiou G. Antibody affinity maturation using bacterial surface display. Protein Eng. 1998;11:825–32. doi: 10.1093/protein/11.9.825. [DOI] [PubMed] [Google Scholar]
- 28.Logtenberg T. Antibody cocktails: next-generation biopharmaceuticals with improved potency. Trends Biotechnol. 2007;25:390–94. doi: 10.1016/j.tibtech.2007.07.005. [DOI] [PubMed] [Google Scholar]
- 29.Baum A, Fulton BO, Wloga E, Copin R, Pascal KE, Russo V, Giordano S, Lanza K, Negron N, Ni M, Baum A, Fulton BO, Wloga E, Copin R, Pascal KE, Russo V, Giordano S, Lanza K, Negron N, Ni M, et al . Antibody cocktail to SARS-CoV-2 spike protein prevents rapid mutational escape seen with individual antibodies. Science. 2020;369:1014–18. doi: 10.1126/science.abd0831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zanin M, Keck Z-Y, Rainey GJ, Lam C-YK, Boon ACM, Rubrum A, Darnell D, Wong -S-S, Griffin Y, Xia J, et al. An anti-H5N1 influenza virus FcDART antibody is a highly efficacious therapeutic agent and prophylactic against h5n1 influenza virus infection. J Virol. 2015;89:4549–61. doi: 10.1128/JVI.00078-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Asokan M, Rudicell RS, Louder M, McKee K, O’Dell S, Stewart-Jones G, Wang K, Xu L, Chen X, Choe M, et al. Bispecific antibodies targeting different epitopes on the HIV-1 envelope exhibit broad and potent neutralization. J Virol. 2015;89:12501–12. doi: 10.1128/JVI.02097-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dong J, Huang B, Jia Z, Wang B, Gallolu Kankanamalage S, Titong A, Liu Y. Development of multi-specific humanized llama antibodies blocking SARS-CoV-2/ACE2 interaction with high affinity and avidity. Emerg Microbes Infect. 2020;9:1034–36. doi: 10.1080/22221751.2020.1768806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Joshi KK, Phung W, Han G, Yin Y, Kim I, Sandoval W, Carter PJ. Elucidating heavy/light chain pairing preferences to facilitate the assembly of bispecific IgG in single cells. MAbs. 2019;11:1254–65. doi: 10.1080/19420862.2019.1640549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ridgway JBB, Presta LG, Carter P. “Knobs-into-holes” engineering of antibody C(H)3 domains for heavy chain heterodimerization. Protein Eng. 1996;9:617–21. doi: 10.1093/protein/9.7.617. [DOI] [PubMed] [Google Scholar]
- 35.Davis-Gardner ME, Alfant B, Weber JA, Gardner MR, Farzan M, Koup RA, Goff SP. A bispecific antibody that simultaneously recognizes the V2-and V3-glycan epitopes of the HIV-1 envelope glycoprotein is broader and more potent than its parental antibodies. MBio. 2020;11:1–16. doi: 10.1128/mBio.03080-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Merchant AM, Zhu Z, Yuan JQ, Goddard A, Adams CW, Presta LG, Carter P. An efficient route to human bispecific IgG. Nat Biotechnol. 1998;16:677–81. doi: 10.1038/nbt0798-677. [DOI] [PubMed] [Google Scholar]
- 37.Klein C, Sustmann C, Thomas M, Stubenrauch K, Croasdale R, Schanzer J, Brinkmann U, Kettenberger H, Regular JT, Schaefer W. Progress in overcoming the chain association issue in bispecific heterodimeric IgG antibodies. MAbs. 2012;4:653–63. doi: 10.4161/mabs.21379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bönisch M, Sellmann C, Maresch D, Halbig C, Becker S, Toleikis L, Hock B, Novel RF. CH1:CL interfaces that enhance correct light chain pairing in heterodimeric bispecific antibodies. Protein Eng Des Sel. 2017;30:685–96. doi: 10.1093/protein/gzx044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Crawford KHD, Eguia R, Dingens AS, Loes AN, Malone KD, Wolf CR, Chu HY, Tortorici MA, Veesler D, Murphy M, et al. Protocol and reagents for pseudotyping lentiviral particles with SARS-CoV-2 spike protein for neutralization assays. Viruses. 2020;12:513. doi: 10.3390/v12050513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huo J, Zhao Y, Ren J, Zhou D, Duyvesteyn HME, Ginn HM, Carrique L, Malinauskas T, Ruza RR, Shah PNM, et al. Neutralization of SARS-CoV-2 by destruction of the prefusion spike. Cell Host Microbe. 2020;28:1–10. doi: 10.1016/j.chom.2020.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chi X, Yan R, Zhang J, Zhang G, Zhang Y, Hao M, Zhang Z, Fan P, Dong Y, Yang Y, et al. A potent neutralizing human antibody reveals the N-terminal domain of the Spike protein of SARS-CoV-2 as a site of vulnerability. bioRxiv. 2020. doi: 10.1101/2020.05.08.083964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brouwer PJM, Caniels TG, van der Straten K, Snitselaar JL, Aldon Y, Bangaru S, Torres JL, Okba NMA, Claireaux M, Kerster G, et al. Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. Science. 2020;369:643–50. doi: 10.1126/science.abc5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang C, Li W, Drabek D, Okba NMA. A panel of human neutralizing mAbs targeting SARS-CoV-2 spike at multiple epitopes. Nat Commun. 2020;11:2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ke Z, Oton J, Qu K, Cortese M, Zila V, McKeane L, Nakane T, Zivanov J, Neufeldt CJ, Cerikan B, et al. Structures and distributions of SARS-CoV-2 spike proteins on intact virions. Nature. 2020;588:498–502. doi: 10.1038/s41586-020-2665-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lui I, Zhou XX, Lim SA, Elledge SK, Solomon P, Rettko NJ, Zha BS, Kirkemo LL, Gramespacher JA, Liu J, et al. Trimeric SARS-CoV-2 Spike interacts with dimeric ACE2 with limited intra-Spike avidity. bioRxiv. 2020. doi: 10.1101/2020.05.21.109157. [DOI] [Google Scholar]
- 46.Hornsby M, Paduch M, Miersch S, Sääf A, Matsuguchi T, Lee B, Wypisniak K, Doak A, King D, Usatyuk S; Hornsby M, Paduch M, Miersch S, Sääf A, Matsuguchi T, Lee B, Wypisniak K, Doak A, King D, Usatyuk S, et al . A high through-put platform for recombinant antibodies to folded proteins. Mol Cell Proteomics. 2015;14(10):2833–47. doi: 10.1074/mcp.O115.052209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rogers TF, Zhao F, Huang D, Beutler N, Burns A, He W, Limbo O, Smith C, Song G, Woehl J, et al. Isolation of potent SARS-CoV-2 neutralizing antibodies and protection from disease in a small animal model. Science. 2020;7520:eabc7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data supporting the findings of this study are available from the corresponding author upon request.


