Abstract
Adult echinostomes having 37 collar spines collected from the intestine of Pitalah ducks in Aceh Province, Indonesia in 2018 were morphologically and molecularly determined to be Echinostoma miyagawai Ishii, 1932 (Digenea: Echinostomatidae). Among 20 ducks examined, 7 (35.0%) were found to be infected with this echinostome, and the number of flukes collected was 48 in total with average 6.9 (1–17) worms per duck. The adult flukes were 7.2 (6.1–8.5) mm in length and 1.2 (1.0–1.4) mm in width (pre-ovarian or testicular level) and characterized by having a head collar armed with 37 collar spines (dorsal spines arranged in 2 alternating rows), including 5 end group spines, and variable morphology of the testes, irregularly or deeply lobed (3–5 lobes) at times with horizontal extension. The eggs within the worm uterus were 93 (79–105) μm long and 62 (56–70) μm wide. These morphological features were consistent with both E. miyagawai and Echinostoma robustum, for which synonymy to each other has been raised. Sequencing of 2 mitochondrial genes, cox1 and nad1, revealed high homology with E. miyagawai (98.6–100% for cox1 and 99.0–99.8% for nad1) and also with E. robustum (99.3–99.8% for nad1) deposited in GenBank. We accepted the synonymy between the 2 species and diagnosed our flukes as E. miyagawai (syn. E. robustum) with redescription of its morphology. Further studies are required to determine the biological characteristics of E. miyagawai in Aceh Province, Indonesia, including the intermediate host and larval stage information.
Keywords: Echinostoma miyagawai; Echinostoma robustum; ITS; cox1, nad1; Pitalah duck; Indonesia
INTRODUCTION
The 37-collar-spined Echinostoma spp. (family Echinostomatidae) or ‘Echinostoma revolutum group’ are taxonomically diverse consisting of at least 26 species, including 16 valid and 10 validity-retained species, worldwide [1]. E. revolutum is the type, and there are several closely related species which include Echinostoma miyagawai Ishii, 1932 (syn. Echinostoma friedi Toledo et al., 2000) and Echinostoma robustum Yamaguti, 1935 [1].
E. miyagawai was originally described from ducks (Anas platyrhynchos and Anas boschas domesticus) and fowls (Gallus gallus domesticus) in Japan [2]. Human experimental infection with this fluke was successful in Zhejiang Province, China [3]. Beaver [4] synonymized this species with E. revolutum because of morphological similarities, and Yamaguti [5] followed this synonymy. However, Russian researchers, including Bashkirova [6] and Skrjabin and Bashkirova [7], did not accept this synonymy, and Kosupko [8,9] validated both species through morphological and ecological approaches. On the other hand, Kanev [10] denied the validity of E. miyagawai and synonymized it with Echinostoma echinatum. Later, Kostadinova et al. [11,12] re-validated E. miyagawai on the basis of its unique cercarial chaetotaxy and morphometric data of larvae and adults obtained in Europe (Bulgaria) in comparison with E. revolutum. Fried and Graczyk [13] and Toledo et al. [14] acknowledged the taxonomic validity of E. miyagawai. Faltýnková et al. [15] redescribed the adult stage of E. miyagawai based on specimens collected from ducks in Central and Western Europe (Czech Republic, Poland, Bulgaria, and Spain) and synonymized Echinostoma friedi Toledo et al., 2000 with E. miyagawai. The first intermediate host is freshwater snails, including Planorbis planorbis, Anisus vortex, Radix peregra, Galba corvus, and Gyraulus chinensis [15], and the second intermediate host (experimental) is lymnaeid snails, including Lymnaea stagnalis and L. truncatula [11].
Echinostoma robustum was originally described from 2 avian species in Taiwan [16]. Thereafter, the existence of this fluke has been reported from Asia, Europe, and North and South Americas [1]. Bezubik K. in 1956 (reference not traceable) supported Dubinina who first considered this species as a synonym of E. revolutum but Rayski and Fahmy [17] acknowledged this as a distinct species, which was agreed by Bashkirova [6], Skrjabin and Bashkirova [7], and Yamaguti [5,18]. However, Huffman and Fried [19], Kanev [10], Kostadinova and Gibson [20], Fried and Graczyk [13], and Toledo et al. [14] did not recognize this species among the ‘E. revolutum’ group. Thereafter, E. robustum had not drawn much taxonomic attention until Detwiler et al. [21,22] reported the cercariae of this species from USA and Brazil based on molecular data of cox1, nad1, and ITS loci. Subsequently, Mohanta et al. [23] reported adult flukes of E. robustum collected from ducks in Bangladesh and suggested a synonymy of E. miyagawai (syn. E. friedi) with E. robustum, although they did not actually synonymize the 2 species.
We performed a survey of intestinal flukes in ducks from Aceh Province, Indonesia and collected a number of adult echinostomes having 37 collar spines. They were morphologically and molecularly analyzed to determine the species. Our final diagnosis was E. miyagawai (syn. E. robustum, E. friedi) based on various morphological features with review of related literature as well as unique molecular data. The adult morphology of E. miyagawai was redescribed, and reasons for the synonymy between E. miyagawai and E. robustum were discussed.
MATERIALS AND METHODS
Worm collection
In March and October 2018, 2 times’ small surveys were undertaken in Aceh Province, Indonesia to detect intestinal flukes infecting domestic Pitalah ducks (Anas sp.) (n=20) reared in 2 localities, Aceh Besar and Matang Glumpang Counties. The ducks were brought to the laboratory of Permatahati Mazas Foundation, Banda Aceh, the capital of Aceh Province and killed by cervical dislocation or bleeding, and their intestinal tract, including ceca, were removed. The intestinal tract was longitudinally opened with a pair of scissors in saline, and helminths were collected from the sediment under stereomicroscopy. Some of the fluke specimens were fixed in 10% neutral formalin under a cover slip pressure for morphological studies. Other specimens were stored in 70–80% ethanol for molecular studies. The experimental protocol followed the Institutional Guidelines for Animal Care and User Committee, Institute of Parasitic Diseases, Korea Association of Health Promotion, Seoul, Korea.
Morphological analyses
The formalin-fixed specimens were stained with Semichon’s acetocarmine, dehydrated with graded series of ethanol, cleared in carbol-xylene, and mounted in Canada balsam. Because of deteriorated (shrunken) morphology of intrauterine eggs during the process of dehydration and clearing, some specimens stained with acetocarmine were not processed for dehydration but directly cleared in glycerin-alcohol (70%) and mounted in glycerin jelly. Fourteen specimens (10 were balsam-mounted and 4 were glycerin jelly-mounted) were used for morphological observations, measurements (Table 1), and description. The egg size (n=40) was measured in 4 glycerin-jelly mounted specimens (10 eggs from each specimen). The comparison of our specimens with E. miyagawai and other 37-collar-spined Echinostoma spp. was based on morphological characters given by previous authors [10,15,24,25]. Photomicrographs of the worms were taken with a digital camera (eXcope XCAM1080, Tokyo, Japan) on an Olympus BX43 light microscope (Tokyo, Japan). Measurements were taken from digital images with the aid of LAS v4.12 image analysis software (Leica, Wetzlar, Germany). The measurements are in μm unless stated otherwise.
Table 1.
Measurements of Echinostoma miyagawai adults in comparison with other 37-collar-spined Echinostoma species (unit: μm)
| Species | E. miyagawai (this study) (n=14) | E. miyagawai [15] (n=13) | E. revolutum [15] (n=16) | E. paraulum [15] (n=10) | ||||
|---|---|---|---|---|---|---|---|---|
|
|
|
|
|
|
||||
| Organ | Mean | Range | Mean | Range | Mean | Range | Mean | Range |
| Body length (BL) | 7,186 | 6,186–8,536 | 9,990 | 9,163–11,014 | 10,531 | 9,454–11,846 | 6,345 | 5,600–6,862 |
|
| ||||||||
| Body width 1 (BW1) | 843 | 868–1,274 | 1,159 | 1,029–1,272 | 1,298 | 1,182–1,454 | 1,043 | 923–1,108 |
|
| ||||||||
| Body width 2 (BW2) | 1,064 | 667–1,053 | 1,026 | 898–1,178 | 1,618 | 1,303–1,815 | 1,308 | 1,138–1,446 |
|
| ||||||||
| Body width 3 (BW3) | 1,220 | 1,012–1,416 | 1,305 | 1,197–1,459 | 1,815 | 1,454–2,092 | 1,514 | 1,231–1,662 |
|
| ||||||||
| Head collar length (CL) | 485 | 395–599 | 434 | 393–505 | 392 | 351–424 | 488 | 432–553 |
|
| ||||||||
| Head collar width (CW) | 614 | 528–725 | 656 | 598–692 | 607 | 552–652 | 729 | 644–781 |
|
| ||||||||
| Oral sucker length (OSL) | 267 | 227–307 | 305 | 262–337 | 316 | 261–358 | 287 | 265–318 |
|
| ||||||||
| Oral sucker width (OSW) | 301 | 233–412 | 293 | 262–355 | 282 | 246–303 | 335 | 280–371 |
|
| ||||||||
| Angle spine length (ASL) | 78 | 71–83 | 70 | 52–93 | 74 | 58–92 | 100 | 73–124 |
|
| ||||||||
| Angle spine with (ASW) | 21 | 19–25 | 22 | 17–29 | 17 | 11–24 | 21 | 15–28 |
|
| ||||||||
| Lateral spine length (LSL) | 81 | 75–91 | 76 | 67–87 | 79 | 64–91 | 101 | 85–120 |
|
| ||||||||
| Lateral spine width (LSW) | 21 | 18–24 | 23 | 20–26 | 17 | 12–21 | 21 | 17–25 |
|
| ||||||||
| Dorsal spine length (DSL) | 76 | 69–79 | 68 | 58–75 | 85 | 71–94 | 99 | 90–105 |
|
| ||||||||
| Dorsal spine width (DSW) | 21 | 18–24 | 21 | 19–26 | 18 | 13–23 | 21 | 19–23 |
|
| ||||||||
| Prepharynx length (PL) | 18 | 4–41 | 10 | 0–37 | 41 | 7–99 | 45 | 15–68 |
|
| ||||||||
| Pharynx length (PHL) | 273 | 230–292 | 299 | 262–337 | 237 | 209–258 | 324 | 273–379 |
|
| ||||||||
| Pharynx width (PHW) | 277 | 197–335 | 250 | 187–355 | 219 | 194–258 | 238 | 212–280 |
|
| ||||||||
| Esophagus length (ESL) | 659 | 569–790 | 585 | 542–655 | 408 | 318–569 | 408 | 265–546 |
|
| ||||||||
| Cirrus sac length (CSL) | 491 | 410–710 | 506 | 468–598 | 552 | 410–735 | 413 | 318–531 |
|
| ||||||||
| Cirrus sac width (CSW) | 262 | 242–292 | 300 | 281–355 | 316 | 243–403 | 281 | 205–326 |
|
| ||||||||
| Seminal vesicle length (SVL) | 244 | 205–330 | - | - | 336 | 224–440 | 238 | 152–303 |
|
| ||||||||
| Seminal vesicle width (SVW) | 133 | 82–183 | - | - | 146 | 60–212 | 134 | 114–144 |
|
| ||||||||
| Ventral sucker length (VSL) | 658 | 594–830 | 706 | 655–748 | 873 | 796–1,038 | 667 | 569–766 |
|
| ||||||||
| Ventral sucker width (VSW) | 648 | 544–910 | 738 | 655–785 | 893 | 796–1,061 | 731 | 705–766 |
|
| ||||||||
| Ovary length (OVL) | 281 | 198–342 | 283 | 243–355 | 349 | 288–394 | 150 | 114–182 |
|
| ||||||||
| Ovary width (OVW) | 410 | 290–620 | 397 | 337–449 | 416 | 291–493 | 246 | 212–288 |
|
| ||||||||
| Mehlis’ gland length (MEL) | 631 | 413–765 | 386 | 337–468 | 399 | 326–455 | 153 | 106–190 |
|
| ||||||||
| Mehlis’ gland width (MEW) | 233 | 135–295 | 373 | 318–411 | 734 | 582–849 | 310 | 258–341 |
|
| ||||||||
| Ant. testis length (ATL) | 534 | 403–653 | 564 | 430–692 | 788 | 629–932 | 384 | 341–455 |
|
| ||||||||
| Ant. testis width (ATW) | 728 | 588–982 | 409 | 337–505 | 597 | 425–750 | 377 | 326–447 |
|
| ||||||||
| Post. testis length (PTL) | 564 | 477–674 | 562 | 449–655 | 879 | 627–1,061 | 406 | 296–515 |
|
| ||||||||
| Post. testis width (PTW) | 708 | 593–787 | 411 | 337–524 | 579 | 395–705 | 415 | 288–531 |
|
| ||||||||
| Egg length (EL) | 93 | 79–105 | 95 | 94–96 | 114 | 108–125 | 113 | 104–122 |
|
| ||||||||
| Egg width (EW) | 62 | 56–70 | 60 | 59–60 | 65 | 57–75 | 62 | 53–70 |
|
| ||||||||
| Forebody length (FORE) | 1,121 | 912–1,269 | 1,408 | 1,253–1,533 | 1,369 | 1,200–1,662 | 1,218 | 985–1,600 |
|
| ||||||||
| ODIV | 982 | 808–1,131 | 1,181 | 1,103–1,324 | 1,027 | 923–1,262 | 1,074 | 923–1,354 |
|
| ||||||||
| OVAR | 3,923 | 3,536–4,311 | 4,856 | 4,507–5,348 | 4,461 | 3,969–5,046 | 3,043 | 2,739–3,385 |
|
| ||||||||
| TEND | 2,497 | 2,245–2,795 | 3,081 | 2,824–3,329 | 3,653 | 3,000–4,308 | 2,059 | 1,262–2,431 |
|
| ||||||||
| OSW/PHW | 1.09 | 0.88–1.30 | 0.97 | 0.78–1.11 | 1.29 | 1.15–1.54 | 1.41 | 1.24–1.58 |
|
| ||||||||
| BW1/BL (%) | 11.7 | 9.6–14.0 | 11.6 | 11.2–12.5 | 12.4 | 11.2–14.3 | 16.5 | 15.0–19.8 |
|
| ||||||||
| BW2/BL (%) | 14.8 | 13.1–17.0 | 10.3 | 9.7–10.9 | 15.4 | 13.8–17.4 | 20.7 | 18.9–24.7 |
|
| ||||||||
| BW3/BL (%) | 17.1 | 14.2–20.9 | 13.1 | 12.4–13.6 | 17.3 | 15.2–19.5 | 23.9 | 21.5–26.9 |
|
| ||||||||
| FORE/BL (%) | 15.6 | 14.0–17.2 | 14.1 | 13.4–14.9 | 13.0 | 10.9–14.8 | 19.3 | 15.5–26.3 |
|
| ||||||||
| CW/BW3 (%) | 50.6 | 43.1–58.4 | 50.4 | 46.2–56.9 | 33.6 | 28.3–38.3 | 48.4 | 42.0–57.3 |
|
| ||||||||
| OVID/BL (%) | 13.7 | 12.3–14.7 | 11.8 | 11.2–12.5 | 9.8 | 8.3–11.2 | 17.0 | 14.5–23.7 |
|
| ||||||||
| OVAR/BL (%) | 54.7 | 50.3–57.7 | 48.6 | 47.7–49.7 | 42.5 | 39.1–46.2 | 48.1 | 43.5–57.0 |
|
| ||||||||
| TEND/BL (%) | 34.8 | 31.2–36.7 | 30.9 | 30.1–31.9 | 34.6 | 31.4–37.4 | 32.3 | 22.0–35.4 |
ODIV, distance from anterior extremity to intestinal bifurcation; OVAR, distance from the posterior margin of ventral sucker to ovary; TEND, length of post-testicular region [15].
Molecular analyses (cox1 and nad1)
For molecular analyses, fluke specimens preserved in 70–80% ethanol were used. Genomic DNA was extracted using the Spin-Column Protocol of DNeasy® Blood & Tissue kit (QIAGEN, Hilden, Germany). PCR was conducted using specific primers designed to amplify the mitochondrial cox1 and nad1 [26] genes in echinostomes. The primers for cox1 were JB3 and JB13, and those for nad1 were JB11 and JB12 [26]. Sequencing of the PCR products was performed by Macrogen Inc. (Seoul, Korea). For evaluation of the genetic identity of the samples, the basic local alignment search tool (BLAST; http://blast.ncbi.nlm.nih.gov/Blast.cgi) was used. Our obtained sequences were aligned with the reference sequences of cox1 and nad1 for 37-collar-spined Echinostoma species in GenBank using the Geneious® program version 10.2.6 (Biometers Ltd., Auckland, New Zealand). Phylogenetic information was assessed via maximum-likelihood (ML) analyses using the MEGA v6 program applying Tamura-Nei model of nucleotide substitution with 1,000 bootstrap replications [27].
RESULTS
Echinostoma miyagawai Ishii, 1932
[syn. Echinostoma robustum Yamaguti, 1935; Echinostoma friedi Toledo et al., 2000]
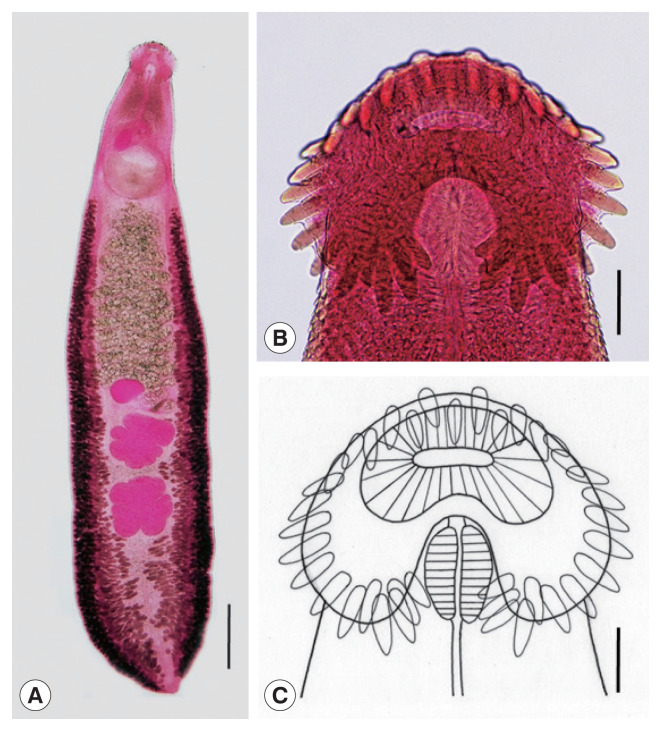
Adult: Body dorsoventrally flattened, robust, muscular, elongated leaf-like with slightly attenuated anterior and blunt posterior ends, 7.2 (6.2–8.5) mm in length and 1.2 (1.0–1.4) mm in width at pre-ovarian or testicular region (n=14) (Table 1; Fig. 1A–C). Body slightly constricted at level of posterior third of ventral sucker. Tegument beset with triangular, scale-like spines, less dense posteriorly, extending to level of pharynx dorsally, level of posterior margin of ventral sucker laterally, and level of testes ventrally. Forebody relatively short representing about 16% of whole body length. Anterior end with characteristic features of an echinostome, equipped with an oral sucker and a head collar. Oral sucker small, muscular, spherical, subterminal, and about 1/3 of the size of ventral sucker. Head collar well-developed, prominent, and muscular armed with collar spines (Fig. 1B, C). Collar spines 37 in total number, with the formula of 5-6-6-3-6-6-5, including angle (corner) spines 5+5, lateral spines 6+6, dorsal spines 6+3+6; dorsal spines arranged in 2 alternating rows (Fig. 1B, C). Collar spines large, conspicuous but not sharply pointed. Prepharynx very short or absent; pharynx muscular, elongate-oval. Esophagus long, at times meandering; intestinal bifurcation from anterior extremity at about 14% of total body length; ceca blind, narrow, overlapped by vitelline follicles, and terminate near the posterior extremity. Ventral sucker large, subspherical, muscular, located at end of first one-fourth of body. Cirrus sac elongate-oval, with muscular walls, and located dorsally between intestinal bifurcation and middle level of ventral sucker, containing oval seminal vesicle, well-developed pars prostatica, coiled ejaculatory duct, and muscular cirrus with smooth unarmed surface. Genital atrium and genital pore median, just posterior to intestinal bifurcation, receiving female (metraterm) and male reproductive (ejaculatory) ducts. Uterus intercecal, long and slender, with numerous transverse coils between ventral sucker and ovary, containing a large number of eggs. Metraterm short, weakly muscular, connected to genital pore. Ovary slightly elongate-oval, median or slightly submedian, almost equatorial, between uterus and Mehlis’ gland. Mehlis’ gland transversely oval, median, connected with ovary and uterine tubule. Uterine seminal receptacle present, ventral to Mehlis’ gland. Laurer’s canal absent. Vitelline follicles extensive, extending laterally forming 2 lateral groups, from the level slightly posterior to ventral sucker to near posterior extremity; the 2 groups of vitellaria merge (7/14; 50.0%) at posterior field of posterior testes but in some specimens (7/14; 50.0%) they do not actually merge. Two testes tandem, irregularly lobed (3–5 lobes), frequently with deep indentations (9/14; 64.3%), transversely extended (9/14; 64.3%), located in posterior field of body, more or less separated from each other. Eggs numerous, yellowish, oval, immature, operculate with a small, inconspicuous operculum and a small abopercular thickening or wrinkling at abopercular end, and 93 (79–105) μm long and 62 (56–70) μm wide (n=40). Excretory vesicle Y-shaped, bifurcates just posterior to posterior testis; excretory pore terminal.
Fig. 1.
Echinostoma miyagawai from a Pitalah duck in Aceh Province, Indonesia. (A) An adult worm showing its characteristic head collar with collar spines, oral and ventral suckers, uterus with eggs, cirrus sac, ovary, Mehlis’ gland, testes, and vitelline follicles. Acetocarmine-stained. Ventral view. Scale bar=0.72 mm. (B) Head collar equipped with 37 collar spines. Acetocarmine-stained. Ventral view. Scale bar=100 μm. (C) Line drawing of the head collar with 37 collar spines. Scale bar=100 μm.
Taxonomic summary
Type host: Pitalah duck (Anas sp.) (natural infection), one of Indonesia’s native ducks.
Site of infection: Small intestine, cecum, and rectum.
Locality: Aceh Besar and Matang Glumpang Counties, Aceh Province, Indonesia.
Deposition of specimens: The specimens are deposited in the Parasite Museum, Institute of Parasitic Diseases, Korea Association of Health Promotion, Seoul, Republic of Korea (no. 2020-0021-01~24). Voucher specimens are also deposited in Meguro Parasitological Museum, Tokyo, Japan (MPM Coll. No. 21712).
Molecular results
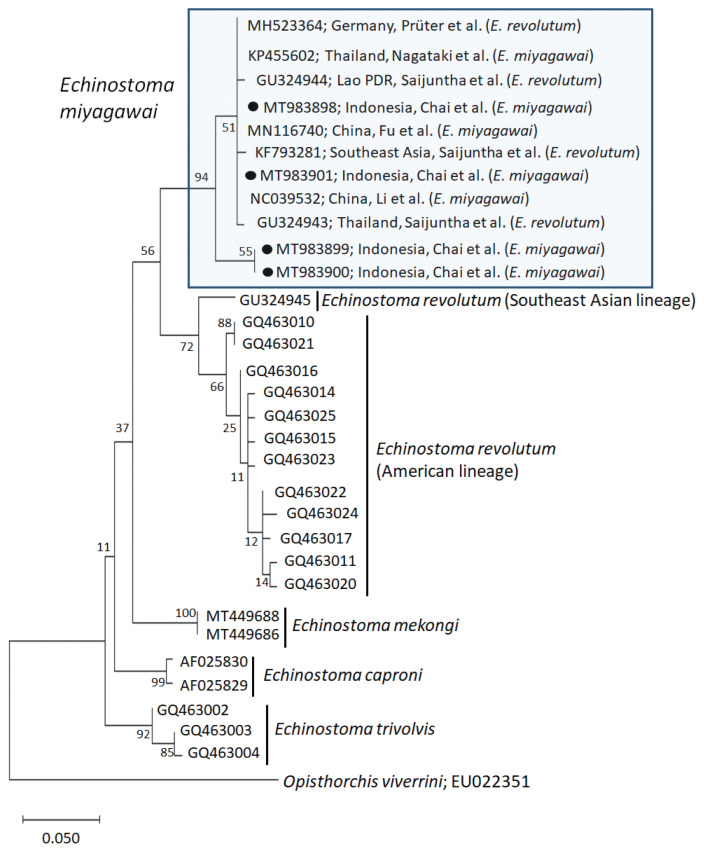
A phylogenetic tree based on cox1 (197 bp after trimming) sequences was constructed using the ML analyses. The tree comprised of 5 species of 37-collar-spined Echinostoma group, including E. revolutum Southeast Asian and American lineages, E. miyagawai, E. mekongi, E. caproni, and E. trivolvis. The pairwise alignment of our 4 isolates (GenBank accession nos. MT983898-MT983901) was 99.0–100%, and they were strongly clustered with E. miyagawai reported in GenBank, i.e., 99.0–100% with MN116740 of Fu et al. [28] and NC039532 of Li et al. [29], and 98.6–100% with KP455602 of Nagataki et al. [30] (Table 2; Fig. 2). The sequence similarity of our samples with 3 isolates by Saijuntha et al. [31], originally named as E. revolutum but suggested to be E. miyagawai by Heneberg [32] was 99.0–100%. Our samples appeared to be a sister group with E. revolutum. However, the sequences of our samples showed only 90.4–91.6% similarity with the Southeast Asian lineage of E. revolutum (GU324945; reported originally under the name Echinoparyphium recurvatum by Saijuntha et al. [31] and amended as E. revolutum (Southeast Asian lineage) by Nagataki et al. [30]) and 92.6% similarity with the American lineage of E. revolutum (GQ463016; Detwiler et al. [21]) (Table 2).
Table 2.
Sequence comparison of our samples with other 37-collar-spined Echinostoma spp. based on cox1 and nad1 region
| cox1 | nad1 | ||
|---|---|---|---|
|
|
|
||
| Our samples (n=4) 99.0–100 |
Our samples (n=4) 99.5–100 |
||
| E. miyagawai (MN116740; China; Fu et al.) | 99.0–100 | E. miyagawai (KP065635; Czech Republic; Georgieva et al.) | 99.3–99.8 |
|
| |||
| E. miyagawai (NC039532; China; Li et al.) | 99.0–100 | E. robustum (LC224091; Bangladesh; Mohanta et al.) | 99.3–99.8 |
|
| |||
| E. miyagawai (KP455602; Thailand; Nagataki et al.) | 98.6–100 | E. miyagawai (KP455620; Thailand; Nagataki et al.) | 99.0–99.6 |
|
| |||
| E. miyagawai (GU324943; Thailand; Saijuntha et al.) | 98.8–100 | E. friedi (AJ564379; Spain; Marcilla et al.) | 99.3–99.8 |
|
| |||
| E. revolutum American lineage (GQ463016; USA; Detwiler et al.) | 92.6–93.2 | E. revolutum (AF026287; Australia; Morgan and Blair) | 98.5–99.0 |
|
| |||
| E. revolutum Southeast Asian lineage (GU324945; Thailand; Saijuntha et al.) | 90.4–91.6 | E. miyagawai (KY436400; New Zealand; Georgieva et al.) | 97.3–97.8 |
|
| |||
| E. robustum (GQ463053; USA; Detwiler et al.) | 94.3–94.8 | ||
|
| |||
| E. robustum (GQ463055; Brazil; Detwiler et al.) | 90.5–91.0 | ||
|
| |||
| E. revolutum American lineage (GQ463061; USA; Detwiler et al.) | 88.8 | ||
Fig. 2.
A phylogenetic tree of Echinostoma miyagawai in comparison with other 37-collar-spined ‘E. revolutum group’ constructed based on 197 bp of mitochondrial cox1 sequences. ●, our specimens.
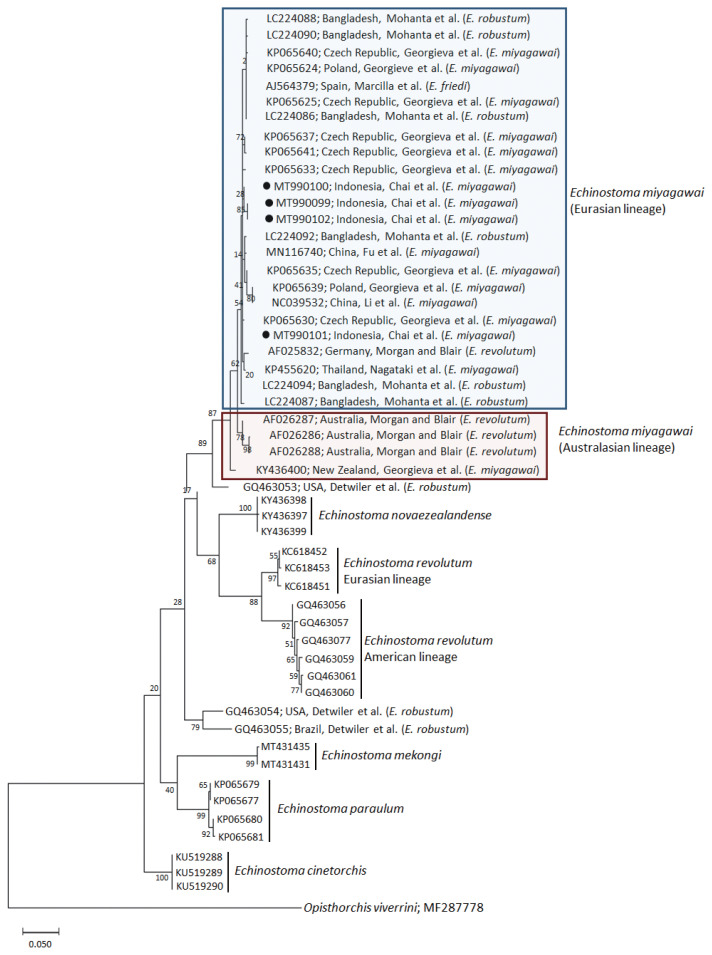
A phylogenetic tree based on nad1 (401 bp after trimming) sequences constructed using the ML method is shown in Fig. 3. The tree composed of sequences of 6 different species of 37-collar-spined Echinostoma group available in GenBank, including E. miyagawai Eurasian and Australasian lineages, E. novaezealandense, E. revolutum Eurasian and American lineages, E. mekongi, E. paraulum, and E. cinetorchis. The pairwise alignment of our 4 isolates (GenBank accession nos. MT990099-MT990102) was 99.5–100%, and they were strongly clustered with the Eurasian lineage of E. miyagawai and also with E. robustum and E. friedi (Table 2; Fig. 3). The sequence similarities of our samples with E. miyagawai (KP065635) of Georgieva et al. [33] and E. miyagawai (KY455621) of Nagataki et al. [30] were 99.3–99.8% and 99.0–99.6%, respectively. The similarity of our samples with E. robustum (LC224091) of Mohanta et al. [34] and E. friedi of Marcilla et al. (unpublished) was also very high, both 99.3–99.8% (Table 2). We propose to include all these isolates among the Eurasian lineage of E. miyagawai [30] (Fig. 3). On the other hand, juvenile E. miyagawai worms (KY436400) of Georgieva et al. [24] found from New Zealand revealed lower similarity with our samples, 97.3–97.8% (Table 2; Fig. 3). However, the nad1 sequence of this isolate showed high homology with the 3 isolates (AF026286-AF026288) from Australia by Morgan and Blair [26]. We propose to put these 4 isolates together and designate them as the Australasian lineage of E. miyagawai.
Fig. 3.
A phylogenetic tree of Echinostoma miyagawai in comparison with other 37-collar-spined ‘E. revolutum group’ constructed based on 401 bp of mitochondrial nad1 sequences. ●, our specimens.
By nad1 analysis, a North American (USA) isolate of E. robustum (GQ463053) by Detwiler et al. [21] showed only 94.3–94.8% similarity with E. miyagawai, including our samples. Two other American isolates (USA and Brazil) of E. robustum (GQ463054-GQ463055) also revealed lower similarity (90.5–91.0%) with our samples. These 2 clones may represent 2 separate species different from E. miyagawai (E. robustum). Moreover, the sequences of E. revolutum American lineage (GQ463061) reported by Detwiler et al. [21] showed only 88.8% similarity compared with our samples (Table 2).
DISCUSSION
Mohanta et al. [23] assigned their worms from ducks in Bangladesh as E. robustum and suggested the synonymy of E. miyagawai, E. robustum, and E. friedi based on both morphological and molecular results. They stated that their worms had a comparatively long forebody, comprising of 16.4–20.9% of the fluke length, with a distinctive constriction at the level of the ventral sucker, elongate-oval cirrus sac extending from the level of intestinal bifurcation to the middle of the ventral sucker, and median, tandem, and markedly separated testes as common features for E. miyagawai, E. robustum, and E. friedi [23]. Moreover, based on nad1 Bayesian tree, their 11 isolates (Erob-nad1-G1~G11) belonged to the same phylogenetic clade with E. miyagawai of Georgieva et al. [33] (KP065625 and KP065640 from Czech Republic), E. miyagawai of Nagataki et al. [30] (KP455622, KP455623, and KP455624 from Thailand), E. friedi of Marcilla et al., unpublished (AJ564379 from Spain), and another isolate from Germany (AF025832; Morgan and Blair [26] under the name E. revolutum) [23]. However, they [23] designated all these isolates as E. robustum Eurasian clade, and also assigned 2 isolates by Detwiler et al. [21,22] (GQ463054 and GQ463055 from USA and Brazil, respectively) as E. robustum American clade. This was like synonymizing E. miyagawai and E. friedi with E. robustum. However, prior to this, E. miyagawai was morphologically redescribed by Faltýnková et al. [15], and E. friedi was synonymized with E. miyagawai.
Against the synonymy raised by Mohanta et al. [23], Heneberg [32] argued that E. miyagawai is a valid species and the available evidence does not allow proposing the synonymization of E. miyagawai with E. robustum/friedi. Mohanta et al. [34] rebutted against this and insisted that E. robustum, E. miyagawai, and E. friedi are very similar in morphology, and their characteristics overlap each other. For example, the testes of E. miyagawai were considered to be irregular on the surface and not distinctly lobated, which had been one of the differential characters from the testes of E. robustum [34]. However, Faltýnková et al. [15] showed that E. miyagawai adults also had indented or lobulated testes. Minor morphological differences may occur depending on the final host species, fluke age, and fixation methods; thus, there is no strong evidence for that E. robustum and E. miyagawai are notably different species [34].
Based on the results of our study, we agree with Mohanta et al. [23,34] in that E. miyagawai, E. robustum, and E. friedi are conspecific. However, we assigned our flukes as E. miyagawai synonymizing E. robustum and E. friedi with E. miyagawai, considering the rule of priority in scientific nomenclature; E. miyagawai was named in 1932 [2] which precedes E. robustum which was reported in 1935 [16].
In our study, we found several interesting and significant findings which strongly support this synonymy. Among our specimens, there were specimens which have mixed morphological features for both E. miyagawai and E. robustum, and also E. friedi. For example, in the morphology of testes, specimens with shallow lobed testes with no distinct indentation (E. miyagawai; Ishii [2]) and those with deeply indented and 3–5 lobed testes (E. robustum; Yamaguti [16]) were both found. These 2 features of testes were also shown in E. friedi [35]. Another differential character between E. miyagawai and E. robustum is transversely extended testes (often) in E. robustum and no such extension of testes in E. miyagawai [23,34]. In E. friedi, some specimens collected from naturally infected rats revealed transversely extended testes (usually tri-lobed) [35]. Among our specimens, about 2/3 (64.3%) had transversely extended testes, while the others revealed almost globular testes with only slight lobulations. In addition, regarding the confluence of 2 lateral groups of vitelline follicles beyond the posterior testicular area, E. miyagawai has either confluent [2,36] or non-confluent vitellaria behind the posterior testis [15], and those of E. robustum or E. friedi are either non-confluent [16,23,36] or confluent behind the posterior testis in most specimens [35]. Our specimens revealed both features, merging of vitellaria in some specimens (50%) and not strictly merging in other specimens (50%). Therefore, merging of vitellaria behind the posterior testis does not seem to be a specific character for these echinostomes.
In molecular analysis of E. miyagawai and E. revolutum (Southeast Asian and American lineages) based on cox1 ML tree, our 4 samples were strongly clustered with E. miyagawai of Nagataki et al. [30], and Fu et al. [28] and Li et al. [29] from Thailand and China, respectively. In addition, previously misidentified isolates as E. revolutum, including 3 isolates (KF793281, GU324943, and GU324944) from Southeast Asia (Thailand and Lao PDR) by Saijuntha et al. [31] and 1 (MH523364) from Germany by Prüter et al. [37] appeared to be conspecific with E. miyagawai in this study as suggested by Heneberg [32]. In our cox1 tree, E. miyagawai and E. revolutum (Southeast Asian and American lineages) were closely related forming a sister group. However, they revealed only 90.4–92.6% homology between the 2 species supporting their distinct positions.
In molecular analysis of E. miyagawai, E. robustum, and E. friedi based on nad1 neighbor-joining tree, Georgieva et al. [33] established a seemingly unique clade under the name E. robustum/friedi (GQ463054 and GQ463055 from USA and Brazil; Detwiler et al. [21,22]) separately from E. miyagawai clade, including the isolates obtained by themselves (KP065624-KP065641 from Central Europe), those reported by Morgan and Blair [26] under the name E. revolutum (AF025832 from Germany and AF026286-AF026288 from Australia), those reported by Detwiler et al. [21,22] under the name E. robustum/friedi (GQ463053 from USA), and those deposited by Marcilla et al. (unpublished) under the name E. friedi (AJ564379 from Spain). Further, Nagataki et al. [30] classified these isolates into 3 different clades, including E. robustum (GQ463053-GQ463055), E. miyagawai Australian lineage (AF026286-AF026288), and E. miyagawai Eurasian lineage (AF025832, KP065624-KP065641, and AJ564379). However, Mohanta et al. [23] recognized only 2 lineages of E. robustum, Eurasian and American, based on nad1 sequences and transferred E. miyagawai isolates (AF025832, KP065622-KP065625, KP065640, and AJ564379) to Eurasian E. robustum, including 11 of their own isolates (Erob-nad1-G1~G11). However, Heneberg [32] disagreed this transfer of E. miyagawai isolates to E. robustum clade and claimed that E. miyagawai is a distinct species.
In our study, we strongly support the claim of Heneberg [32] in that E. miyagawai is a distinct species morphologically and molecularly. In addition, our study agrees also with Mohanta et al. [23] in that E. miyagawai, E. robustum, and E. friedi are conspecific from the morphological as well as molecular points of view. Thus, we determined our samples from Pitalah ducks in Indonesia as E. miyagawai and synonymized E. robustum and E. friedi with E. miyagawai. Studies are required on the biological characteristics of E. miyagawai, including the intermediate and definitive hosts, in Aceh Province, Indonesia.
The status of American isolates of E. robustum (GQ463053 and GQ463054 from USA and GQ463055 from Brazil) reported by Detwiler et al. [21,22] remains to be further clarified. Regarding these 3 isolates together with the German isolate (AF025832) designated as E. revolutum by Morgan and Blair [26,38] and the Spanish isolate (AJ564379) deposited in GenBank as E. friedi by Marcilla et al. (unpublished), Georgieva et al. [39] suggested that they are a complex structure comprising of at least 3 species. In our study, the German (AF025832) and Spanish (AJ564379) isolates could be included within the Eurasian clade of E. miyagawai [30]; however, it seems inappropriate to include the 3 American isolates under the name E. robustum within the same clade (see Fig. 3). The 3 isolates revealed less than 95% similarity with our samples (E. miyagawai). Therefore, we strongly suggest that the 3 American isolates could be 2 different species distinguished from E. miyagawai (syn. E. robustum, E. friedi) (see Fig. 3). It was stated by McDonald [36] that E. robustum occurs frequently in North America and Eurasia. However, morphological descriptions for E. robustum American isolates have not been available, and further studies are required to clarify this point.
ACKNOWLEDGMENTS
We are grateful to the staff of Permatahati Mazas Foundation, Banda Aceh, Aceh Province, Indonesia who helped this study. We also thank the staff of Korea Association of Health Promotion, Seoul, Korea who participated in International Project on “Health Promotion Support for Elementary School Children in Indonesia (2018–2020)”.
Footnotes
CONFLICT OF INTEREST
We have no conflict of interest related to this study.
REFERENCES
- 1.Chai JY, Cho J, Chang T, Jung BK, Sohn WM. Taxonomy of Echinostoma revolutum and 37-collar-spined Echinostoma spp.: a historical review. Korean J Parasitol. 2020;58:343–371. doi: 10.3347/kjp.2020.58.4.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ishii N. Studies on bird trematodes. I. Bird trematodes in Japan. II. Four new bird trematodes. Jpn J Exp Med. 1932;11:91–100. [Google Scholar]
- 3.Mao SP. Protozoan and helminth parasites of humans in mainland China. Int J Parasitol. 1991;21:347–351. doi: 10.1016/0020-7519(91)90037-8. [DOI] [PubMed] [Google Scholar]
- 4.Beaver PC. Experimental studies on Echinostoma revolutum (Froelich), a fluke from birds and mammals. Illin Biol Monogr. 1937;15:1–96. [Google Scholar]
- 5.Yamaguti S. Synopsis of Digenetic Trematodes of Vertebrates. I. Tokyo, Japan: Keigaku Publishing Co; 1971. pp. 1–1074. [Google Scholar]
- 6.Bashkirova EI. Family echinostomatidae dietz, 1909. In: Skrjabin KI, editor. Trematodes and Animals and Man. Moscow, Russia: USSR Academy of Sciences; 1947. pp. 310–391. (English translated version) [Google Scholar]
- 7.Skrjabin KI, Bashkirova EI. Family echinostomatidae dietz, 1909. In: Skrjabin KI, editor. Trematodes and Animals and Man. Moscow-Leningrad, Russia: 1956. pp. 51–930. (in Russian) [Google Scholar]
- 8.Kosupko GA. Morphological peculiarity of cercariae of Echinostoma revolutum and Echinostoma miyagawai . Trudy vsesoyuznogo Instituta Helminthologii. 1969;15:159–165. (in Russian). [Google Scholar]
- 9.Kosupko GA. PhD Thesis. Moscow, Russia: 1972. Studies on the morphological and biological peculiarity of Echinostoma revolutum (von. Froelich, 1802) and Echinostoma miyagawai Ishii, 1932 (Trematoda: Echinostomatidae) on experimental material; pp. 1–258. (in Russian) [Google Scholar]
- 10.Kanev I. Life-cycle, delimitation and redescription of Echinostoma revolutum (Froelich, 1802) (Trematoda: Echinostomatidae) Syst Parasitol. 1994;28:125–144. doi: 10.1007/BF00009591. [DOI] [Google Scholar]
- 11.Kostadinova A, Gibson DI, Biserkow V, Chipev N. Re-validation of Echinostoma miyagawai Ishii, 1932 (Digenea: Echinostomatidae) on the basis of the experimental completion of its life-cycle. Syst Parasitol. 2000;45:81–108. doi: 10.1023/A:1006241610689. [DOI] [PubMed] [Google Scholar]
- 12.Kostadinova A, Gibson DI, Biserkow V, Chipev N. A quantitative approach to the evaluation of the morphological variability of two echinostomes, Echinostoma miyagawai Ishii, 1932 and E. revolutum (Froelich, 1802), from Europe. Syst Parasitol. 2000;45:1–15. doi: 10.1023/A:1006232612469. [DOI] [PubMed] [Google Scholar]
- 13.Fried B, Graczyk TK. Recent advances in the biology of Echinostoma species in the ‘revolutum’ group. Adv Parasitol. 2004;58:139–195. doi: 10.1016/S0065-308X(04)58003-X. [DOI] [PubMed] [Google Scholar]
- 14.Toledo R, Esteban JG, Fried B. Chapter 3. Recent advances in the biology of echinostomes. Adv Parasitol. 2009;69:147–204. doi: 10.1016/S0065-308X(09)69003-5. [DOI] [PubMed] [Google Scholar]
- 15.Faltýnková A, Georgieva S, Soldánová M, Kostadinova A. A re-assessment of species diversity within the ‘revolutum’ group of Echinostoma Rudolphi, 1809 (Digenea: Echinostomatidae) in Europe. Syst Parasitol. 2015;90:1–25. doi: 10.1007/s11230-014-9530-3. [DOI] [PubMed] [Google Scholar]
- 16.Yamaguti S. Studies on the helminth fauna of Japan. Part 5. Trematodes of birds, III. Jpn J Zool. 1935;6:159–182. [Google Scholar]
- 17.Rayski C, Fahmy MAM. Investigation on some trematodes of birds from the East Scotland. Z Parazitenkd. 1962;22:186–195. doi: 10.1007/BF00260005. [DOI] [PubMed] [Google Scholar]
- 18.Yamaguti S. Systema Helminthum. Vol. I. The Digenetic Trematodes of Vertebrates (Part I) New York, USA: Interscience Publishers Inc; 1958. pp. 1–979. [Google Scholar]
- 19.Huffman JE, Fried B. Echinostoma and echinostomiasis. Adv Parasitol. 1990;29:215–269. doi: 10.1016/s0065-308x(08)60107-4. [DOI] [PubMed] [Google Scholar]
- 20.Kostadinova A, Gibson DI. The systematic of the echinostomes. In: Fried B, Graczyk TK, editors. Echinostomes as Experimental Models for Biological Research. Dordrecht, The Netherlands: Kluwer Academic Publishers; 2000. pp. 31–57. [Google Scholar]
- 21.Detwiler JT, Bos DH, Minchella DJ. Revealing the secret lives of cryptic species: examining the phylogenetic relationships of echinostome parasites in North America. Mol Phylogenet Evol. 2010;55:611–620. doi: 10.1016/j.ympev.2010.01.004. [DOI] [PubMed] [Google Scholar]
- 22.Detwiler JT, Zajac AM, Minchella DJ, Belden LK. Revealing cryptic parasite diversity in a definitive host: echinostomes in muskrats. J Parasitol. 2012;98:1148–1155. doi: 10.1645/GE-3117.1. [DOI] [PubMed] [Google Scholar]
- 23.Mohanta UK, Watanabe T, Anisuzzaman, Ohari Y, Itagaki T. Characterization of Echinostoma revolutum and Echinostoma robustum from ducks in Bangladesh based on morphology, nuclear ribosomal ITS2 and mitochondrial nad1 sequences. Parasitol Int. 2019;69:1–7. doi: 10.1016/j.parint.2018.11.002. [DOI] [PubMed] [Google Scholar]
- 24.Georgieva S, Blasco-Costa I, Kostadinova A. Molecular characterization of four echinostomes (Digenea: Echinostomatidae) from birds in New Zealand, with descriptions of Echinostome novaezealandense n. sp. and Echinoparyphium poulini n.sp. Syst Parasitol. 2017;94:477–497. doi: 10.1007/s11230-017-9712-x. [DOI] [PubMed] [Google Scholar]
- 25.Cho J, Jung BK, Chang T, Sohn WM, Sinuon M, Chai JY. Echinostoma mekongi n. sp. (Digenea: Echinostomatidae) from Riparian people along the Mekong River in Cambodia. Korean J Parasitol. 2020;58:431–443. doi: 10.3347/kjp.2020.58.4.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morgan JA, Blair D. Relative merits of nuclear ribosomal internal transcribed spacers and mitochondrial CO1 and ND1 genes for distinguishing among Echinostoma species (Trematoda) Parasitology. 1998;116:289–297. doi: 10.1017/s0031182097002217. [DOI] [PubMed] [Google Scholar]
- 27.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fu YT, Jin YC, Li F, Liu GH. Characterization of the complete mitochondrial genome of the echinostome Echinostoma miyagawai and phylogenetic implications. Parasitol Res. 2019;118:3091–3097. doi: 10.1007/s00436-019-06417-4. [DOI] [PubMed] [Google Scholar]
- 29.Li Y, Qiu YY, Zeng MH, Diao PW, Chang QC, Gao Y, Zhang Y, Wang CR. The complete mitochondrial genome of Echinostoma miyagawai: Comparisons with closely related species and phylogenetic implications. Infect Genet Evol. 2019;75:103961. doi: 10.1016/j.meegid.2019.103961. [DOI] [PubMed] [Google Scholar]
- 30.Nagataki M, Tantrawatpan C, Agatsuma T, Sugiura T, Duenngai K, Sithithaworn P, Andrews RH, Petney TN, Saijuntha W. Mitochondrial DNA sequences of 37 collar-spined echinostomes (Digenea: Echinostomatidae) in Thailand and Lao PDR reveals presence of two species: Echinostoma revolutum and E. miyagawai . Infect Genet Evol. 2015;35:56–62. doi: 10.1016/j.meegid.2015.07.022. [DOI] [PubMed] [Google Scholar]
- 31.Saijuntha W, Sithithaworn P, Duenngai K, Kiatsopit N, Andrews RH, Petney TN. Genetic variation and relationships of four species of medically important echinostomes (Trematoda: Echinostomatidae) in South-East Asia. Infect Genet Evol. 2011;11:375–381. doi: 10.1016/j.meegid.2010.11.009. [DOI] [PubMed] [Google Scholar]
- 32.Heneberg P. Taxonomic comments on the validity of Echinostoma miyagawai Ishii, 1932 (Trematoda: Echinostomatidae) Parasitol Int. 2020;74:101931. doi: 10.1016/j.parint.2019.101931. [DOI] [PubMed] [Google Scholar]
- 33.Georgieva S, Faltýnková A, Brown R, Blasco-Costa I, Soldánová M, Sitko J, Scholz T, Kostadinova A. Echinostoma ‘revolutum’ (Digenea: Echinostomatidae) species complex revisited: species delimitation based on novel molecular and morphological data gathered in Europe. Parasit Vectors. 2014;7:520. doi: 10.1186/s13071-014-0520-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mohanta UK, Watanabe T, Anisuzzaman, Ohari Y, Itagaki T. A rebuttal letter to Letter to the Editor by P. Heneberg on “Taxonomic comments on the validity of Echinostoma miyagawai Ishii, 1932 (Trematoda: Echinostomatidae)”. Parasitol Int. 2020;74:101971. doi: 10.1016/j.parint.2019.101971. [DOI] [PubMed] [Google Scholar]
- 35.Toledo R, Muñoz-Antolí C, Esteban JG. The life-cycle of Echinostoma friedi n. sp. (Trematoda: Echinostomatidae) in Spain and a discussion on the relationships within the ‘revolutum’ group based on cercarial chaetotaxy. Syst Parasitol. 2000;45:199–217. doi: 10.1023/a:1006385902664. [DOI] [PubMed] [Google Scholar]
- 36.McDonald ME. Key to Trematodes Reported in Waterfowl. Washington DC, USA: US Department of the Interior, Fish and Wildlife Service; 1981. pp. 1–156. [Google Scholar]
- 37.Prüter H, Franz M, Auls S, Czirják GA, Greben O, Greenwood AD, Lisitsyna O, Syrota Y, Sitko J, Krone O. Chronic lead intoxication decreases intestinal helminth species richness and infection intensity in mallards (Anas platyrhynchos) Sci Total Environ. 2018;644:151–160. doi: 10.1016/j.scitotenv.2018.06.297. [DOI] [PubMed] [Google Scholar]
- 38.Morgan JA, Blair D. Mitochondrial ND1 gene sequences used to identify echinostome isolates from Australia and New Zealand. Int J Parasitol. 1998;28:493–502. doi: 10.1016/s0020-7519(97)00204-x. [DOI] [PubMed] [Google Scholar]
- 39.Georgieva S, Selbach C, Faltýnková A, Soldánová M, Sures B, Skirnisson K, Kostadinova A. New cryptic species of the ‘revolutum’ group of Echinostoma (Digenea: Echinostomatidae) revealed by molecular and morphological data. Parasit Vectors. 2013;6:64. doi: 10.1186/1756-3305-6-64. [DOI] [PMC free article] [PubMed] [Google Scholar]