Abstract
Fast, accurate, and reliable diagnostic tests are critical for controlling the spread of the coronavirus disease 2019 (COVID-19) associated with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection. The current gold standard for testing is real-time PCR; however, during the current pandemic, supplies of testing kits and reagents have been limited. We report the validation of a rapid (30 minutes), user-friendly, and accurate microchip real-time PCR assay for detection of SARS-CoV-2 from nasopharyngeal swab RNA extracts. Microchips preloaded with COVID-19 primers and probes for the N gene accommodate 1.2-μL reaction volumes, lowering the required reagents by 10-fold compared with tube-based real-time PCR. We validated our assay using contrived reference samples and 21 clinical samples from patients in Canada, determining a limit of detection of 1 copy per reaction. The microchip real-time PCR provides a significantly lower resource alternative to the Centers for Disease Control and Prevention–approved real-time RT-PCR assays with comparable sensitivity, showing 100% positive and negative predictive agreement of clinical samples.
In late 2019, an outbreak of the novel human severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), known as coronary virus disease 2019 (COVID-19) began in Wuhan in the Hubei province of China1 and has rapidly spread around the globe. As of February 26, 2021, >131 million cases and 2,850,521 deaths have been reported worldwide, resulting in an enormous economic impact (World Health Organization, https://covid19.who.int). Rapid, sensitive, and cost-effective diagnostics can play an important role in the containment of COVID-19 and in bringing society from pandemic to normalcy. Throughout the current pandemic, test kits and reagents required for manufacturing such test kits have been limiting, slowing the rapid expansion of clinical testing.2 These limitations have encouraged the search for alternative protocols that use reagents in a more cost-effective manner, are user-friendly, and preserve the sensitivity and speed of conventional tests for early-stage detection.3 Multiple laboratories around the world are beginning to offer diverse diagnostic solutions; however, quantitative RT-PCR (RT-qPCR) remains the current gold standard test used in clinical diagnostics laboratories.4, 5, 6, 7, 8, 9, 10
The current COVID-19 RT-qPCR test detects the virus by single-plex amplification of one or two segments of the N, ORF1b, E, or RdRp genes, whereas other assays have been developed to take advantage of multiplex amplification, amplifying multiple genes in a single reaction.4, 5, 6, 7, 8, 9, 10 Regardless of the method, amid the pandemic, most of these assays require supply-limited reagents in high volumes and significant technical labor for preparing complex reagent mixtures, resulting in high-cost assays with potential for human error.11 , 12
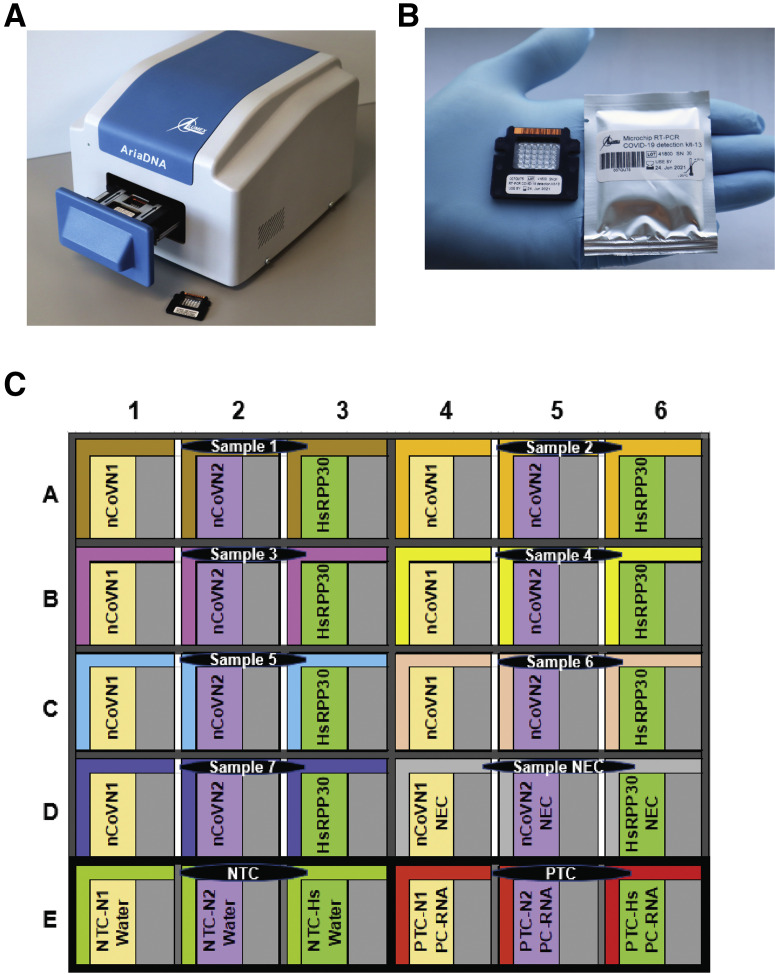
Microchip real-time PCR, an alternative to conventional real-time PCR, has been proven to be a user-friendly technology that can provide reliable, sensitive, and specific results in less time.13, 14, 15, 16, 17, 18 These advantages are attributed to the miniaturized reaction volumes, where the chip's high surface area to volume ratio offers high heat transfer efficiency.13 This technology is expected to meet the current standards of existing detection assays [Centers for Disease Control and Prevention (CDC), https://www.cdc.gov/flu/professionals/diagnosis/molecular-assays.htm, last accessed January 14, 2021], while giving accurate results in <30 minutes and addressing the current supply-limiting situations by lowering the cost, labor, and reagent consumption. This report presents the clinical validation of a microchip-based real-time PCR system that consists of a disposable microchip with 30 microreactors (6 columns × 5 rows) where each well can accommodate a miniature TaqMan chemistry-based reaction of 1.2 μL (Figure 1 ). The protocol uses a single-plex assay based on one-step RT-qPCR reactions targeting the SARS-CoV-2–specific N gene. The microchip is preloaded with lyophilized CDC-recommended N1 and N2 primers and probes for detecting two regions of the N gene, along with Homo sapiens ribonuclease P/MRP subunit p30 (HsRPP30) as a human specimen control. The accompanying lightweight AriaDNA microchip-based PCR analyzer offers user-friendly, rapid real-time monitoring and analysis of PCR reactions while using significantly fewer resources than tube-based RT-PCR. Therefore, this robust and easy-to-operate system can be applied for COVID-19 testing in both clinical diagnostics laboratories and point-of-care scenarios.
Figure 1.
A: AriaDNA analyzer. B: Microchip for coronavirus disease 2019 detection with lyophilized reagents in the microwells displayed along with its packaging. C: Layout of the microchip. Each sample and its associated control reactions are loaded into three wells, nCoVN1 (novel coronavirus N1 primer/probe], nCoVN2 (novel coronavirus N2 primer/probe), and Homo sapiens ribonuclease P/MRP subunit p30 (HsRPP30) (human sample control primer/probe) at 1.2 μL each. Three controls are loaded per chip. NEC, negative extraction control; NTC, negative template control; PTC, positive template control.
Materials and Methods
Procurement of Reagents
The CDC emergency authorization uses the 2019-nCoV CDC EUA Kit (catalog number 10006770; Integrated DNA Technology Inc., Coralville, IA), which includes primers and probes. As per their kit, two primer-probe sets, N1 and N2, were used to detect regions of the N gene of SARS-CoV-2, and a third primer-probe set was used to detect HsRPP30, a housekeeping gene of humans as a sample control. UltraPlex 1-Step ToughMix (4X) (catalog number 95166-01K; Quantabio, Gaithersburg, MD) was used as the enzymatic premix for real-time PCR on the microchip.
For limit of detection (LOD) determination, heat-inactivated SARS-related coronavirus 2, isolate USA-WA1/2020 (catalog number NR-52347; BEI Resources, Manassas, VA) was procured. As a positive control in all experiments, in vitro transcribed Armored RNA Quant SARS-CoV-2 (1 × 1011 copies/mL; catalog number 52030; Asuragen, Austin, TX) was used as a SARS-CoV-2 control, and Armored RNA Quant RNase P (1 × 1011 copies/mL; catalog number 52031; Asuragen) was used as an alternative template to the human housekeeping gene HsRPP30.
Preparation of Disposable Pre-Filled Microchips
Empty microchips with 30 microwells (6 columns × 5 rows) were manufactured from aluminum sheets by metal stamping technology and coated with surface modifiers (Figure 1B). This coating imparts hydrophilicity to the microwells of 1.2 μL capacity and hydrophobicity to the upper surface of the microchips. The empty microchips were then filled with a 1.2 μL solution of primers and probes of N1, N2, and HsRPP30 along with stabilizing agents using an OT2 robotic workstation (Opentrons, Brooklyn, NY) in a predefined layout (Figure 1C). The prefilled microchips were then lyophilized by Lumex Instruments Canada (Mission, BC, Canada) using a SJIA-10N Lyophilizer (Ningbo Shuangjia Instrument Co. Ltd, Zhejiang,China), and each microchip was individually packaged in a moisture-free package (Figure 1B). These microchips were supplied to the Unrau Laboratory in the Molecular Biology and Biochemistry Department of Simon Fraser University (Burnaby, BC, Canada) for sample validation and testing.
Viral RNA Preparation
The LOD measurements were performed using qPCR control RNA from heat-inactivated SARS-related coronavirus 2, isolate USA-WA1/2020. As described by the provider, nucleic acid was extracted using a QIAamp Viral RNA Mini Kit (catalog number 52906; Qiagen, Hilden, Germany) and vialed in TE buffer (10 mmol/L Tris hydrochloride, 1 mmol/L EDTA, pH 8.0). Samples were verified by RT-PCR amplification of the ORF1ab gene, and the viral genome copy number was determined using the BioRad QX200 Droplet Digital PCR System (BioRad, Hercules, CA). The resultant sample of approximately 50,000 copies/μL was diluted in RNA Storage Solution (catalog number AM7001; Thermo Fisher Scientific, Waltham, MA) and used in measuring LOD experiments with approximately 1500, 150, 15, 4, 2, 1.5, 1, 0.72, 0.5, 0.25, 0.15, and 0.015 copies per reaction.
Human Clinical Sample Collection and Preparation
Nasopharyngeal (NP) swabs of 8 patients with positive COVID-19 test results and 13 patients with negative COVID-19 test results were collected and tested by a clinical team at St. Paul's Hospital in Vancouver, BC, Canada, between March 30 and May 5, 2020. Because of a shortage of supplies early during the pandemic, samples were collected with a combination of a COPAN UTM collection kit and BD universal viral transport system.19 Viral RNA was extracted from 500 μL NP swab media using the MagNA Pure 96 DNA and Viral NA LV extraction kit (catalog number 06374891001; Roche, Indianapolis, IN) on the MagNA Pure Compact Instrument before eluting in 50 μL.
St. Paul's Hospital Real-Time RT-qPCR Assays
RT-PCR (20 μL reactions) was performed on the extracted samples described above using the LightCycler Multiplex RNA virus Master kit (catalog number 07083173001; Roche) with LightMix Modular SARS-CoV-2 (COVID-19) primers targeting the E gene (catalog number 53-0776-10; TIB MolBiol, Berlin, Germany). An EAV Extraction Control (catalog number 66-0909-96) was added during the extraction and was detected by RT-PCR using LightMix Modular EAV RNA extraction Control primers20 (catalog number 66-0909-96; Roche). RT-PCR was performed on a LightCycler 480 II instrument (Roche) as per company guidelines.
Microchip-Based RT-qPCR
Real-time qPCR was performed on a microchip-based PCR analyzer (Lumex Instruments Canada) (Figure 1A) using AriaDNA software version 1.4b.129 (AriaDNA Software, Valencia, Spain) to control the instrument and obtain PCR results. For LOD determinations, RT-qPCR assays were performed in a mixture of two parts double distilled water (ddH2O), one part viral RNA (to a final expected concentration of 1500, 150, 15, 4, 2, 1.5, 1, 0.72, 0.5, 0.25, 0.15, and 0.015 copies per reaction of 1.2 μL), and one part UltraPlex 1-Step ToughMix, a 4× concentrated master mix for one-step RT-qPCR that contains deoxyadenosine triphosphate, deoxycytidine triphosphate, deoxyguanosine triphosphate, deoxythymidine triphosphate, deoxyuridine triphosphate, magnesium, qScript XLT reverse transcriptase, RNase inhibitor protein, and AccuStart II hot-start Taq DNA polymerase. For testing patient clinical samples, reactions were performed in a mixture of three parts viral RNA extracted from NP swabs and one part 4× UltraPlex 1-Step ToughMix. Each microchip was loaded with the prepared reaction samples alongside two negative template controls (one part ddH2O or RNA Storage Solution, two parts ddH2O and one part 4× UltraPlex 1-Step ToughMix) and a positive template control (one part equivolume in vitro transcribed SARS-Cov-2 nuclear RNA (1 × 105 copies/μL) and HsRPP30 RNA (1 × 105 copies/μL), two parts ddH2O and one-part 4X UltraPlex 1-Step ToughMix). Each sample and control were loaded into three wells that contained SARS-CoV-2 targets N1 and N2 and human sample control HsRPP30 primer probes with 1.2 μL per well. Reactions were performed on the AriaDNA PCR analyzer as two-step PCR cycling, eliminating the standard extension step at 72°C. Slow thermal settings were applied, including a reverse transcription step at 50°C for 900 seconds, followed by a denaturing step at 95°C for 120 seconds and 45 cycles of 95°C for 3 seconds followed by extension and signal recording at 55°C for 30 seconds. Ct values were determined as a second derivative maximum (SDM) once fluorescence passed an autoset SDM threshold. PCR curves that represented a dependence of fluorescent signal (S) versus cycle (C) were used by the AriaDNA software to obtain its first (dS/dC) and second (d2S/d2C) derivatives. A cycle corresponding to a maximum of the second derivative was obtained.21 , 22 This value is termed the SDM value and reported as Ct. The SDM values are reported only when an amplitude of the fluorescent signal and an amplitude of its first derivative are both above the preset thresholds. Those threshold values can be adjusted within AriaDNA software and were set at 150 and 50 arbitrary units, respectively. No edge effect was detected in the microwells because the thermal conductivity of the microchip material is high and the entire chip sits on a uniformly heated Peltier element. The equilibration of temperature across the microchip is rapid, and temperature differences across the chip are <0.2°C.
Assay Duration Optimization
A comparison was performed at Lumex Instruments Canada's Research and Development Laboratory between the slow thermal cycling settings at 50°C for 900 seconds, 95°C for 120 seconds, 95°C for 3 seconds, and 55°C for 30 seconds and fast thermal cycling settings at 50°C for 300 seconds, 95°C for 120 seconds, 95°C for 1 second, and 55°C for 20 seconds. Four replicates were run each day for 5 consecutive days using 25 copies/μL and 2500 copies/μL of in vitro transcribed Armored RNA Quant SARS-CoV-2.
Results
LOD Determination
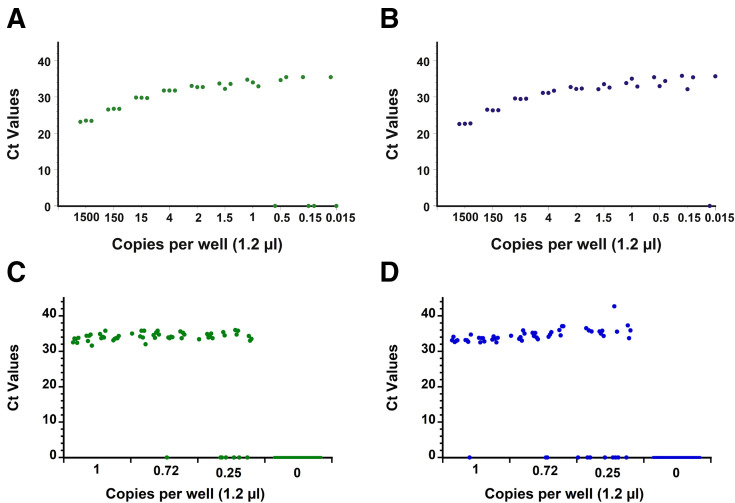
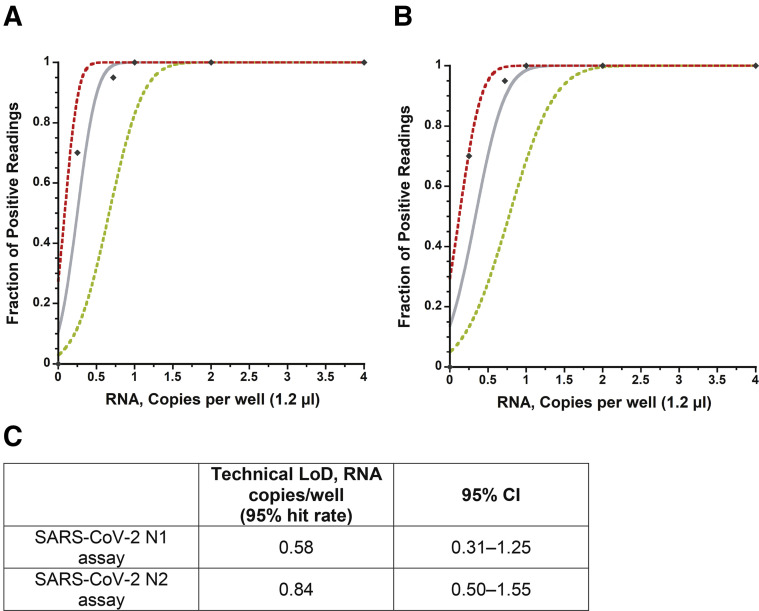
The capability of the RT-qPCR assay on the AriaDNA PCR analyzer was assessed by measuring the LOD using serially diluted extracted viral RNA from heat-inactivated SARS-related coronavirus 2 with a known titer (BEI Resources) as validated by droplet digital PCR (see Materials and Methods). Initially, a preliminary LOD was determined by measuring a 10-fold serial dilution in triplicate of expected concentrations of 1500 to 0.015 copies per well and two-fold dilutions from 4 to 0.5 copies per well, resulting in the following samples: 1500, 150, 15, 4, 2, 1.5, 1, 0.5, 0.15, and 0.015 copies per well. The N1 and N2 primer sets could detect as low as 1 copy per reaction with 100% reproducibility, whereas below these expected concentrations, results were stochastic in nature for both primer sets (Figure 2, A and B, Supplemental Figure S1 and Supplemental Tables S1 and S2). Twenty viral RNA replicates of approximately 1, 0.72, or 0.25 expected copies per reaction and 36 replicates that contained no template (water) were then tested for both the N1 and N2 primer sets. As before, results become stochastic below 1 copy per well, confirming with 95% confidence a LOD of at least 1 copy per reaction on the AriaDNA analyzer (Figure 2, C and D, Supplemental Figure S2 and Supplemental Table S3). A Probit analysis (Figure 3 ) found a confidence limit of 0.58 and 0.84 copies per well for N1 and N2 primers, respectively, suggesting that the reported droplet digital PCR determined concentration of the acquired viral RNA was in fact slightly lower than what it was on the AriaDNA device.
Figure 2.
Limit of detection (LoD) determination with extracted cultured severe acute respiratory syndrome coronavirus 2 RNA. Ct values of 10-fold and 2-fold serial dilutions of extracted cultured viral RNA obtained by microchip quantitative RT-PCR assay and measured on the AriaDNA analyzer with N1 (green dots) (A) and N2 (blue dots) (B) primers/probes. Ct values of 20 positive replicates per concentration of viral RNA (1 copy per well, 0.72 copies per well, and 0.25 copies per well) and 12 double distilled water negative controls per viral concentration, Ct values are determined as described in the Materials and Methods, and a Ct of zero indicates that the fluorescence signal did not cross the threshold limit within 45 cycles. n = 3 (A and B); n = 36 (C and D).
Figure 3.
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) detection of novel coronarvirus N1 (nCoVN1) primer/probe target (A) and novel coronavirus N2 (nCoV-N2) primer/probe target (B). The x axis shows input RNA copies per well, and the y axis shows positive results across all parallel reactions performed. Diamonds are experimental data points resulting from replicate testing at given concentrations (x axis) (20 replicate reactions per datum point for RNA concentrations up to 1.0 copies per well and 3 replicates for RNA concentrations >1.0 copies per well). The inner bold line represents a Probit curve, with outer dotted lines representing 95% CIs. C: Limit of detection (LOD) calculated from the Probit analysis.
Clinical Patient Samples
Using the microchip RT-qPCR assay on the AriaDNA analyzer, extracted RNA from 21 NP swab samples collected from 8 patients with positive COVID-19 test results and 13 patients with negative COVID-19 test results were tested. By CDC standards (Supplemental Table S4), of the 21 patient samples, 8 tested positive for COVID-19 infection, 12 tested negative, and 1 had an invalid sample (Table 1 , Supplemental Figure S3). There was 100% positive and negative agreement relative to the conventional CDC RT-qPCR assay performed at St. Paul's Hospital. The invalid sample, identified as negative by hospital testing, similarly had no amplification of COVID-19 target on the microchip RT-qPCR assay; however, the corresponding lack of the HsRPP30 sample control detection deemed this sample invalid by CDC standards. It is likely that this result reflects an error in patient sampling because this has been reported as a probable cause in many false-negative COVID-19 diagnostic test results.19 In this case, a new sample would have to be collected from the patient and retested, which was not possible because of the wide sample collection timeframe and subsequent transfer of the sample to the university testing laboratory. Furthermore, a blind replication experiment was performed with the available samples. Randomly, four positive and three negative patient samples were selected, relabeled, and shuffled by a third party to keep the sample identity secret from the tester. The tester ran the samples on a microchip with no prior knowledge of the samples, correctly identifying all seven samples as per previous RT-qPCR assays on the AriaDNA PCR analyzer and conventional RT-qPCR assay (Supplemental Table S5).
Table 1.
Conventional versus Microchip RT-qPCR of Patient Nasopharyngeal Swab Samples∗
| Patient No. | Conventional RT-qPCR (St. Paul's Hospital) |
Microchip RT-qPCR |
|||||
|---|---|---|---|---|---|---|---|
| Ct nCoVE | Ct EAV | Status | Ct nCoVN1 | Ct nCoVN2 | Ct HsRPP30 |
Status | |
| 1 | 30.45 | 31.55 | Positive | 36.65 | 36.03 | 25.39 | Positive |
| 2 | — | 31.16 | Negative | — | — | 29.54 | Negative |
| 3 | 27.14 | 31.34 | Positive | 30.88 | 32.79 | 26.34 | Positive |
| 4 | — | 31.24 | Negative | — | — | 27.63 | Negative |
| 5 | 30.33 | 30.76 | Positive | 32.79 | 33.86 | 32.79 | Positive |
| 6 | 26.32 | 31.79 | Positive | 29.06 | 29.90 | 26.27 | Positive |
| 7 | — | 31.08 | Negative | — | — | 25.84 | Negative |
| 8 | — | 33.34 | Negative | — | — | 26.92 | Negative |
| 9 | 21.07 | 31.31 | Positive | 22.75 | 23.68 | 24.98 | Positive |
| 10 | 25.54 | 30.16 | Positive | 26.18 | 28.84 | 21.79 | Positive |
| 11 | 28.35 | 30.23 | Positive | 33.66 | 32.73 | 27.31 | Positive |
| 12 | 28.64 | 30.37 | Positive | 34.66 | 33.92 | — | Positive |
| 13 | — | 31.05 | Negative | — | — | 26.44 | Negative |
| 14 | — | 31.20 | Negative | — | — | — | Invalid |
| 15 | — | 31.73 | Negative | — | — | 25.63 | Negative |
| 16 | — | 31.24 | Negative | — | — | 26.32 | Negative |
| 17 | — | 31.74 | Negative | — | — | 25.60 | Negative |
| 18 | — | 31.15 | Negative | — | — | 22.60 | Negative |
| 19 | — | 31.24 | Negative | — | — | 26.49 | Negative |
| 20 | — | 31.84 | Negative | — | — | 24.30 | Negative |
| 21 | — | 31.59 | Negative | — | — | 23.53 | Negative |
EAV, extraction control; HsRPP30, Homo sapiens ribonuclease P/MRP subunit p30; nCoVE, novel coronavirus E gene; nCoVN1, novel coronavirus N1; nCoVN2, novel coronavirus N2; —, signal did not cross the threshold limit within 45 cycles, so a Ct value was not determined.
Ct values for 8 patients with positive and 13 patients with negative nasopharyngeal samples with nCoVE and EAV primers/probes using conventional quantitative RT-PCR versus nCoVN1 (N gene), nCoV-N2 (N gene), and HsRPP30 (sample control) primers/probes using microchip quantitative RT-PCR.
Assay Duration Optimization
To take advantage of the high heat transfer efficiency the system has to offer, a comparison was performed between the slow thermal cycling settings used in this study (51 minutes for 45 cycles) and a fast-thermal cycling (30 minutes for 45 cycles) to potentially speed up testing assays. Four replicates were run each day for 5 consecutive days using 25 copies/μL and 2500 copies/μL of synthetic Armored RNA Quant SARS-CoV-2 RNA. The dCt and CV data of Ct values obtained for N1, N2, and HsRPP30 targets at slow- and fast-thermal cycling parameters suggest that assays can be performed in as quickly as 30 minutes with 45 shorter cycles, satisfying the rapid tests criteria (CDC, https://www.cdc.gov/flu/professionals/diagnosis/molecular-assays.htm) (Supplemental Table S6).
Discussion
The current golden standard for COVID-19 diagnostic testing is the RT-qPCR assay, which is a robust technology that is hindered by expensive instrumentation, relatively slow turnaround times, high cost, and low reagent availability, making it restrictive to clinical and public health laboratories amid the current pandemic. In this report, we validated a microchip RT-qPCR technology for detection of SARS-CoV-2 in clinical samples. The microchip kit miniaturizes the reaction volumes by 10-fold, resulting in lower reagent consumption and faster assay times (as low as 30 minutes vs approximately 70 minutes), while maintaining the same gold standard in sensitivity as the higher-volume techniques. Because the kit comes preloaded with SARS-CoV-2 primers and probes, it may reduce operator-associated errors, improving the reliability of analysis in remote settings. The reported assay reliably has a limit of detection of approximately 1 viral copy per reaction, with mean Ct values that correlate with dilutions that range from approximately 1500 to 1 expected copies per reaction. Notably, the validated assay shows comparable accuracy to that of a clinically validated RT-qPCR assay for the tested 21 COVID-19 NP patient samples.
Testing accuracy has been reported to be partially dependent on how the patient samples were collected, with sputum being the most accurate, followed by NP swabs and saliva, and lastly oropharyngeal swabs.23, 24, 25, 26 In addition, NP swabs may increase the viral exposure of the health care worker in situations in which insufficient personal protective equipment is available, increasing the risk of transmission.27 However, NP samples are currently widely used for diagnostics and have been reported to be essential for COVID-19 management, including risk assessment of transmission and decision-making regarding quarantine of patients.23, 24, 25, 26 This study confirms that NP samples can be used to reliably detect SARS-CoV-2 by microchip analysis.
Because the numbers of infections and transmissions continue to increase, there is an urgent need for rapid and inexpensive diagnostic tests. Available internationally, the low-energy (100 W), compact, lightweight system and rapid processing presented here can not only benefit clinical diagnostic laboratories but also enable point-of-care testing in remote locations, clinics, and airports while maintaining a similar detection sensitivity as observed with traditional RT-qPCR. Although further testing of additional clinical samples and sample types may be needed before this assay can be widely deployed in clinical and public health settings, these preliminary results demonstrate a promising versatile technology that can be easily configured and mobilized to detect infections of current and future emerging viruses, overcoming current bottlenecks and ensuring a faster response in the future.
Acknowledgments
We thank Dr. Rajwant Gill, Dr. Irina Gelimson, and Steven Hao for research and development, quality control, and technical support concerning the production of lyophilized microchips.
Footnotes
Supported by Canadian Institutes of Health Research grant 202002OV1-440215-COVCACA-54157 (P.J.U.).
R.C. and I.Y. contributed equally to this work.
Disclosures: S.G. and M.S. are employed by Lumex Instruments Canada.
Supplemental material for this article can be found at http://doi.org/10.1016/j.jmoldx.2021.02.009.
Supplemental Data
Fluorescent quantitative RT-PCR curves of limit of detection determination. Ten-fold serial dilution and two-fold serial dilution in triplicates of extracted viral RNA. N1 (green) (A) and N2 (blue) (B) primers/probes were used. cp, copies; RFU, relative fluorescence unit; rxn, reaction.
Fluorescent quantitative RT-PCR curves of 20 viral RNA replicates of 1 (A), 0.72 (B), and 0.25 (C) copies per reaction and 12 negative replicates of double distilled water (ddH2O) with N1 (green) and N2 (blue) primer/probes (0 copies per reaction) for each of the 20 replicates of extracted viral RNA (D, E, and F). cp, copies; RFU, relative fluorescence unit; rxn, reaction.
Fluorescent quantitative RT-PCR curves of 8 coronavirus disease 2019 (COVID-19)–positive patient samples and 13 COVID-19–negative patient samples. Samples were tested for severe acute respiratory syndrome coronavirus 2 using two primer/probe sets for N gene detection: N1 primer/probe (green) (A), N2 (blue) primer/probe (B), and Homo sapiens ribonuclease P/MRP subunit p30 (HsRPP30) (red) primers/probes for human sample control (C). RFU, relative fluorescence unit.
References
- 1.Wang C., Horby P.W., Hayden F.G., Gao G.F. A novel coronavirus outbreak of global health concern. Lancet. 2020;395:470–473. doi: 10.1016/S0140-6736(20)30185-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Babiker A., Myers C.W., Hill C.E., Guarner J. SARS-CoV-2 testing. Am J Clin Pathol. 2020;153:706–708. doi: 10.1093/ajcp/aqaa052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., Chen H.D., Chen J., Luo Y., Guo H., Jiang R.D., Liu M.Q., Chen Y., Shen X.-R., Wang X., Zheng X.S., Zhao K., Chen Q.J., Deng F., Liu L.L., Yan B., Zhan F.X., Wang Y.Y., Xiao G.F., Shi Z.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang Y., Odiwuor N., Xiong J., Sun L., Nyaruaba R.O., Wei H., Tanner N.A. Rapid molecular detection of SARS-CoV-2 (COVID-19) virus RNA using colorimetric LAMP. medRxiv. 2020 doi: 10.1101/2020.02.26.20028373. [DOI] [Google Scholar]
- 5.Lamb L.E., Bartolone S.N., Ward E., Chancellor M.B. Rapid detection of novel coronavirus (COVID-19) by reverse transcription-loop-mediated isothermal amplification. medRxiv. 2020 doi: 10.1101/2020.02.19.20025155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.El-Tholoth M., Bau H.H., Song J. 2020. A single and two-stage, closed-tube, molecular test for the 2019 novel coronavirus (COVID-19) at home, clinic, and points of entry. ChemRxiv. [DOI] [Google Scholar]
- 7.Yu L., Wu S., Hao X., Dong X., Mao L., Pelechano V., Chen W.-H., Yin X. Rapid detection of COVID-19 coronavirus using a reverse transcriptional loop-mediated isothermal amplification (RT-LAMP) diagnostic platform. Clin Chem. 2020;66:975–977. doi: 10.1093/clinchem/hvaa102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rabe B.A., Cepko C. SARS-CoV-2 detection using an isothermal amplification reaction and a rapid, inexpensive protocol for sample inactivation and purification. PNAS. 2020;117:24450–24458. doi: 10.1073/pnas.2011221117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bustin S.A., Nolan T. RT-qPCR Testing of SARS-CoV-2: A Primer. Int J Mol Sci. 2020;21:3004. doi: 10.3390/ijms21083004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Udugama B., Kadhiresan P., Kozlowski H.N., Malekjahani A., Osborne M., Li V.Y.C., Chen H., Mubareka S., Gubbay J.B., Chan W.C.W. Diagnosing COVID-19: the disease and tools for detection. ACS Nano. 2020;14:3822–3835. doi: 10.1021/acsnano.0c02624. [DOI] [PubMed] [Google Scholar]
- 11.Petrillo S., Carrà G., Bottino P., Zanotto E., De Santis M.C., Margaria J.P., Giorgio A., Mandili G., Martini M., Cavallo R., Barberio D., Altruda F. A novel multiplex qRT-PCR assay to detect SARS-CoV-2 infection: high sensitivity and increased testing capacity. Microorganisms. 2020;8:1064. doi: 10.3390/microorganisms8071064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kumar R., Nagpal S., Kaushik S., Mendiratta S. COVID-19 diagnostic approaches: different roads to the same destination. Virusdisease. 2020;31:97–105. doi: 10.1007/s13337-020-00599-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tong R., Zhang L., Song Q., Hu C., Chen X., Lou K., Gong X., Gao Y., Wen W. A fully portable microchip real-time polymerase chain reaction for rapid detection of pathogen. Electrophoresis. 2019;40:1699–1707. doi: 10.1002/elps.201900090. [DOI] [PubMed] [Google Scholar]
- 14.Gill R., Gill S., Slyadnev M., Stroganov A. Identification and quantitation of cashmere (pashmina) fiber and wool using novel microchip based real-time PCR technology. J Textile Sci Technol. 2018;4:141–150. [Google Scholar]
- 15.Nikitin M.M., Statsyuk N.V., Frantsuzov P.A., Dzhavakhiya V.G., Golikov A.G. Matrix approach to the simultaneous detection of multiple potato pathogens by real-time PCR. J Appl Microbiol. 2018;124:797–809. doi: 10.1111/jam.13686. [DOI] [PubMed] [Google Scholar]
- 16.Nikitin M.M., Statsyuk N.V., Frantsuzov P.A., Pridannikov M.V., Golikov A.G. Rapid and simple detection of two potato cyst nematode species by real-time multiplex PCR using preserved microarray-based test systems. Russ J Nematol. 2017;25:51–60. [Google Scholar]
- 17.Bogdanov K.V., Nikulina T.S., Lomaia E.G., Slyadnev M.N., Zaritskey A.Y. Identification of oncogene mutations in leukemia patients using microchip-based PCR analysis. Russ J Bioorg Chem. 2017;43:544–551. [Google Scholar]
- 18.Abdulina D.R., Purish L.M., Iutynska G.A., Nikitin M.M., Golikov A.G. Test-Systems for monitoring of corrosion-relevant sulfate-reducting bacteria using real-time PCR assay. Biotechnol Acta. 2016;9:48–54. [Google Scholar]
- 19.Kinloch N.N., Ritchie G., Brumme C.J., Dong W., Dong W., Lawson T., Jones R.B., Montaner J.S.G., Leung V., Romney M.G., Stefanovic A., Matic N., Lowe C.F., Brumme Z.L. Suboptimal biological sampling as a probable cause of false-negative COVID-19 diagnostic test results. J Infect Dis. 2020;222:899–902. doi: 10.1093/infdis/jiaa370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K., Bleicker T., Brünink S., Schneider J., Schmidt M.L., Mulders D.G., Haagmans B.L., van der Veer B., van den Brink S., Wijsman L., Goderski G., Romette J.-L., Ellis J., Zambon M., Peiris M., Goossens H., Reusken C., Koopmans M.P., Drosten C. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25:23–30. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luu-The V., Paquet N., Calvo E., Cumps J. Improved real-time RT-PCR method for high-throughput measurements using second derivative calculation and double correction. BioTechniques. 2005;38:287–293. doi: 10.2144/05382RR05. [DOI] [PubMed] [Google Scholar]
- 22.Zhao S., Fernald R.D. Comprehensive algorithm for quantitative real-time polymerase chain reaction. J Comput Biol. 2005;12:1047–1064. doi: 10.1089/cmb.2005.12.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang Y., Yang M., Shen C., Wang F., Yuan J., Li J., Zhang M., Wang Z., Xing L., Wei J., Peng L., Wong G., Zheng H., Liao M., Feng K., Li J., Yang Q., Zhao J., Zhang Z., Liu L., Liu Y. Comparative sensitivity of different respiratory specimen types for molecular diagnosis and monitoring of SARS-CoV-2 shedding. Innovation. 2020;1:100061. doi: 10.1016/j.xinn.2020.100061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang H., Liu Q., Hu J., Zhou M., Yu M., Li K., Xu D., Xiao Y., Yang J., Lu Y., Wang F., Yin P., Xu S. Nasopharyngeal swabs are more sensitive than oropharyngeal swabs for COVID-19 diagnosis and monitoring the SARS-CoV-2 load. Front Med (Lausanne) 2020;7:334. doi: 10.3389/fmed.2020.00334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iwasaki S., Fujisawa S., Nakakubo S., Kamada K., Yamashita Y., Fukumoto T., Sato K., Oguri S., Taki K., Senjo H., Sugita J., Hayasaka K., Konno S., Nishida M., Teshima T. Comparison of SARS-CoV-2 detection in nasopharyngeal swab and saliva. J Infect. 2020;81:e145–e147. doi: 10.1016/j.jinf.2020.05.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wyllie A.L., Fournier J., Casanovas-Massana A., Campbell M., Tokuyama M., Vijayakumar P. Saliva or nasopharyngeal swab specimens for detection of SARS-CoV-2. N Engl J Med. 2020;383:1283–1286. doi: 10.1056/NEJMc2016359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tu Y.P., Jennings R., Hart B., Cangelosi G.A., Wood R.C., Wehber K., Verma P., Vojta D., Berke E.M. Swabs collected by patients or health care workers for SARS-CoV-2 testing. N Engl J Med. 2020;83:494–499. doi: 10.1056/NEJMc2016321. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fluorescent quantitative RT-PCR curves of limit of detection determination. Ten-fold serial dilution and two-fold serial dilution in triplicates of extracted viral RNA. N1 (green) (A) and N2 (blue) (B) primers/probes were used. cp, copies; RFU, relative fluorescence unit; rxn, reaction.
Fluorescent quantitative RT-PCR curves of 20 viral RNA replicates of 1 (A), 0.72 (B), and 0.25 (C) copies per reaction and 12 negative replicates of double distilled water (ddH2O) with N1 (green) and N2 (blue) primer/probes (0 copies per reaction) for each of the 20 replicates of extracted viral RNA (D, E, and F). cp, copies; RFU, relative fluorescence unit; rxn, reaction.
Fluorescent quantitative RT-PCR curves of 8 coronavirus disease 2019 (COVID-19)–positive patient samples and 13 COVID-19–negative patient samples. Samples were tested for severe acute respiratory syndrome coronavirus 2 using two primer/probe sets for N gene detection: N1 primer/probe (green) (A), N2 (blue) primer/probe (B), and Homo sapiens ribonuclease P/MRP subunit p30 (HsRPP30) (red) primers/probes for human sample control (C). RFU, relative fluorescence unit.