Abstract
The COVID-19 pandemic has caused a significant burden since December 2019 that has negatively impacted the global economy owing to the fact that the SARS-CoV-2 virus is fast-transmitting and highly contagious. Efforts have been taken to minimize the impact through strict screening measures in country borders in order to isolate potential virus carriers. Effective fast-screening methods are thus needed to identify infected individuals. The standard diagnostic methods for screening SARS-CoV-2 virus have always been to perform nucleic acid-based and serological tests. However, with each having drawbacks on producing false results at very early or later stage after symptoms onset, supplementary techniques are needed to back up these tests. Surface-enhanced Raman spectroscopy (SERS) as a detection technique has continuously advanced throughout the years in terms of sensitivity and capability to detect ultralow concentration of analytes ranging from single molecule to pathogens, to present as a highly potential alternative to known sensing methods. SERS technology as a candidate for an alternative and supplementary diagnostic method for the viral envelope of SARS-CoV-2 virus is presented, comparing its pros and cons to the standard methods and what other aspects it could offer that the other methods are not capable of. Factors that contribute to the detection effectivity of SERS is also discussed to show the advantages and limitations of this technique. Despite its promising capabilities, challenges like sources of SARS-CoV-2 virus and its variations, reliable SERS spectra, mass production of SERS-active substrates, and compliance to regulations for wide-scale testing scenario are highlighted.
Keywords: COVID-19 pandemic, SARS-CoV-2 virus, Fast-screening, Surface-enhanced Raman spectroscopy, Viral envelope, Detection effectivity
1. Introduction
The highly transmissible coronavirus disease 2019 (COVID-19) pandemic caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has caused a worldwide emergency due to its rapid increase in cases. Symptoms could vary from the mildest cough to severe respiratory tract infections which usually manifest a few days after being infected; there are also reported cases of asymptomatic individuals that are virus carriers but does not display any signs of infection. Currently, the number of new cases reported globally is still rising, although the situation differs with every region – there is an uptrend in new cases in the American, European, Mediterranean, and African regions, but a decline in new cases and deaths are reported in the South-East Asian regions. As of mid-February 2021, the World Health Organization (WHO) records over 109.6 million cases and 2.4 million deaths worldwide.
Thus, the emergence of the SARS-CoV-2 virus indicates that people need more complete, more effective disease control and advance prevention deployment. For viral infections that have not yet been approved for cure, it is important to reduce and control the spread of infection by screening and diagnosing suspected carriers. For the SARS-CoV-2 virus, it is reported that carriers of the virus may be asymptomatic within 14 days (Hadi et al., 2020), therefore, poor surveillance may pose a huge risk to any population (Varotsos and Krapivin, 2020). Reportedly, there are various modes of SARS-CoV-2 virus transmission with the primary mode being through exposure to droplets expelled through coughing or talking, by the virus carrier. Prolonged exposure to symptomatic virus carriers poses a higher risk of transmission than a brief exposure to asymptomatic carriers. In contact with surfaces having the virus is also another mode of transmission aside from suspended aerosols in the air (Wiersinga et al., 2020). This shows that diagnostic procedures play a vital role in mitigating rapidly spreading viruses. Generally, several analytical aspects of nucleic acid-based and serological diagnostic tests should be considered (Giri et al., 2020; Ravi et al., 2020; Sheikhzadeh et al., 2020) including: accuracy, i.e., the ability of the tests to not produce false positives/negatives, reproducibility, i.e., the consistency of results after repeated tests, and specificity, i.e., the tests can correctly to identify target analyte/s.
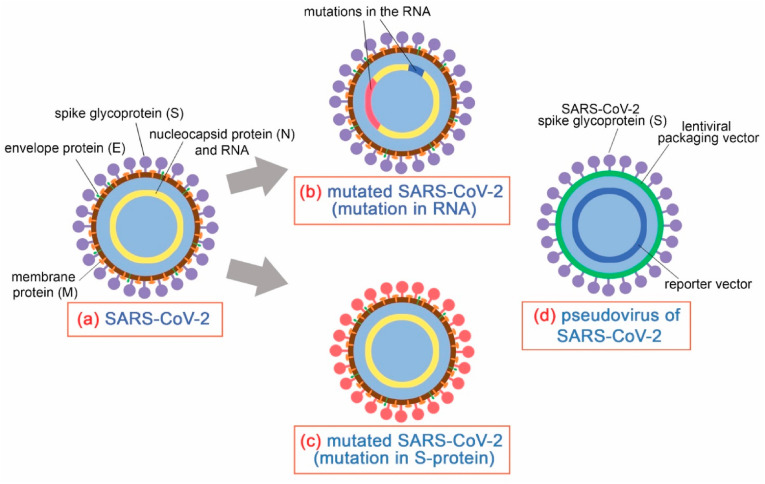
The SARS-CoV-2 virus is a coronavirus type which is characterized by the presence of proteins on their surfaces– spike (S), nucleocapsid (N), membrane (M), and envelope (E), and the genomic material is a single-stranded RNA, as illustrated in Fig. 1 (a). The S protein is responsible in mediating attachment and entry to the host cell and is thus the subject to most studies focusing on developing drugs against the virus. On the other hand, the N protein is responsible for the encapsulation of the genetic material, while the M and E proteins are in charge of morphogenesis to form the viral envelope. SARS-CoV-2 virus exhibits a diameter of around 60–140 nm with spikes having lengths ranging from 9–12 nm. The outbreak has caused the virus to adapt for infection of the intermediate hosts and human to human transmission, causing the virus to undergo several types of mutations, usually with an alteration in the genome of the single-stranded RNA inside the viral particle, as illustrated in Fig. 1(b). However, one notable mutation called D614G is characterized by alterations in the spike protein, as briefly described in Fig. 1(c), implying that this would cause a change in the pathogenesis and consequently, the virulence of the virus. In fact, it has been found that this particular mutation exhibits improved infectiousness and binding ability to the angiotensin-converting enzyme 2 (ACE2) of host cells. This particular mutation has so far been the only one noted to exhibit a change in the surface properties in reference to the original SARS-CoV-2 virus strain. Fig. 1(d) also shows the structure of the SARS-CoV-2 pseudovirus, which is much simpler than the actual virus – it mimics the actual virus for its spike glycoproteins, with the rest of its constituents being derived from other viruses and less infectious. It is suitable in studies focusing on drug development and virus detection particularly for COVID-19, as this could be used in facilities having lower biosafety levels due to the simplicity of its structure.
Fig. 1.
Structure of the (a) SARS-CoV-2 virus, (b) changes in RNA and (c) amino acid sequence of the S-protein leading to mutation, and (d) pseudovirus of SARS-CoV-2.
1.1. Currently applied diagnostic techniques
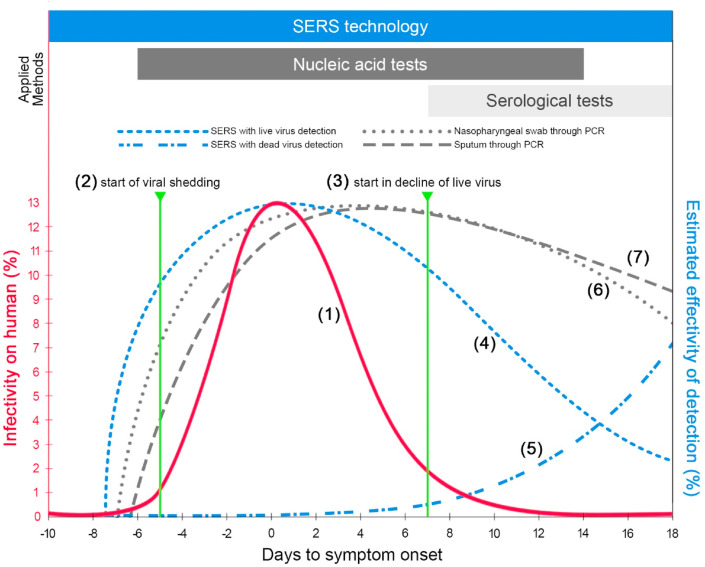
Currently, viral or bacterial infections are usually diagnosed through nucleic acid-based tests using polymerase chain reaction (PCR). Several tests through reverse transcriptase loop-mediated isothermal amplification (LAMP) and emerging methods using clustered regularly interspaced short palindromic repeats (CRISPR) are also employed (D'Cruz et al., 2020). Since the SARS-CoV-2 is a single-stranded RNA virus, reverse transcription PCR (RT-PCR) would be the most appropriate method because the RNA in the sample undergoes reverse transcription into DNA before exponentially amplified through heating and cooling to drive cycles of the process. The RT-PCR is thus the most widely used high-precision screening tool. It is reported to be sensitive (Poon et al., 2003; Tahamtan and Ardebili, 2020) with a limit of 39 gene copies per milliliter (Zhen et al., 2020); the DNA copies are then used to identify the presence of the target virus genome sequence. Consequently, the genetic information of the target virus is required prior to analysis. RT-LAMP technologies and variants of such (i.e. coupled with colorimetry) have been gaining attention as strong potential alternative detection methods as they are faster and cheaper due to the elimination of the RNA extraction step in conventional PCR (Dao Thi et al., 2020; Lalli et al., 2020; Ventura et al., 2020). However, as with any other nucleic acid-based tests, specific primers are still needed and in the event of virus mutation, these particular primers must be redeveloped to be able to detect mutated strains. The infectivity of the SARS-CoV-2 virus with respect to the timeline of infection is shown in Fig. 2 , plot (1); the curve indicates that the infectivity increases as it approaches the day of symptom onset and declines at almost the same rate of its increase.
Fig. 2.
General data of the infectivity on human, in %, is illustrated as the reference, plot (1). The start of viral shedding and the start in decline of live virus are respectively pointed, plots (2) and (3). The estimated effectivity of detection, in %, with respect to the timeline of infection (days to symptom onset) through SERS with live virus (plot (4)), and with dead virus (plot (5)) are compared with that through PCR by nasopharyngeal swab, plot (6) and sputum, plot (7). The stages of infection timeline corresponding to the diagnostic tools – SERS technology, nucleic acid-based test, and serological tests are also shown above based on their scope of applicability.
The nucleic acid-based method relies on viral genetic material. As long as a sufficient amount of virus particle can be collected from the sample, it can be extensively tested from 5 days before the onset of symptoms to 14 days after the onset of symptoms, as shown in Fig. 2, plot (1). As viral shedding starts 5 days before symptom onset, as shown in Fig. 2, plot (2), and peaking at −2 to 1 days relative to the day of symptom onset, nucleic acid-based tests such as RT-PCR are most effective within this timeframe that may make it accurate for early diagnosis, even during asymptomatic periods (Afzal, 2020; He et al., 2020). Another advantage of RT-PCR is its versatility in sample types. It has been found that nasopharyngeal swabs, sputum, stool and isolated viruses from the respiratory tract can all be used in this diagnostic method (Sethuraman et al., 2020).
The use of nucleic acid-based tests has its drawbacks – there is a need for sophisticated equipment and costly reagents (i.e., primers, enzymes, buffers, and polymerases) that must be replenished. In addition, these tests have shown to produce false positive/negative results due to the differences in viral loading in various samples (Sethuraman et al., 2020; van Kasteren et al., 2020). For instance, in a nasopharyngeal swab sample, it was found that viral RNA could be detected within the first week of symptoms, but the specific signals declined at around 6–7 days after the onset of symptoms, as shown in Fig. 2, plot (1) and (3). After which, seroconversion begins to peak with the start in the decline of live virus (Norman et al., 2020). In some cases, nasopharyngeal specimens give false-negatives (Sethuraman et al., 2020). Due to the variation of sample types, nucleic acid-based tests might give contradictory results (van Kasteren et al., 2020). Moreover, the process time of completing nucleic acid-based tests can be obtained 0.5 hours after sampling (Wu et al., 2020), and the throughput of detection equipment varies which may thus pose a challenge for monitoring and containing rapidly spreading viral infections.
In clinically suspected cases that are negative on nucleic acid-based tests, serological testing (i.e., antigen-based immunoassays) can be used as a confirmatory test to supplement the results, however, it is proven to be accurate only if the patients have developed immunity to the infected virus (Meng et al., 2020; Serrano et al., 2020). With the start in the decline of live virus, seroconversion peaks around 7–14 days after the onset of symptom, as shown in Fig. 2, the effectiveness and sensitivity of nucleic acid-based tests subsequently decline, which makes serology-based diagnosis most suitable for testing within this time frame (Jan Van Elslande et al., 2020). Currently, enzyme-linked immunosorbent assay (ELISA) and lateral flow immunoassay (LFIA) are employed as rapid diagnostic tests to detect the presence of antibodies (J. Van Elslande et al., 2020). As far as the SARS-CoV-2 virus is concerned, most of the antibodies produced are directed against its nucleocapsid protein as it is the most abundant protein expressed by the virus during infection. Another protein that is essential in the immunoassay-based detection of SARS-CoV-2 virus is the spike protein receptor-binding domain (S-RBD) (Yang et al., 2020), a spike glycoprotein which indicates the attachment to the host and eliciting neutralizing immune response. It has been reported that the simultaneous use of two antigens to detect IgM, IgA, and IgG will lead to more sensitive and accurate detection (Meng et al., 2020; Sethuraman et al., 2020).
Aside from the limited effectiveness of serological tests and a late window, as the nucleocapsid protein is the most conservative component in the virus classification, they are also prone to provide false positive results to detect other coronaviruses other than the SARS-CoV-2 virus, i.e., the antigen used in ELISA is most likely to react with antibodies of other coronaviruses (Wechselberger et al., 2020; Younes et al., 2020). Since this technology is only suitable for the late stage of the disease, the test results should not be used to screen for asymptomatic suspicious individuals, so it is not a suitable tool to control the spread of the virus; on the other hand, it is more suitable for confirming infected patients that show COVID-19 symptoms. Moreover, the antibody development of each infected patient is different. The 7–14 days after the occurrence of seroconversion is only the average of the collected data. Certain conditions may be contrary to this, leading to invalid tests and possibly providing false results (Stowell and Guarner, 2020).
In summary, the currently applied detection techniques are supplementary to each other to some extent, that is, nucleic acid-based detection can be applied in the early infection stage. As the patient develops immunity, the detection will eventually lead to a decline in effectiveness, thereafter, serological tests can be used effectively. Both tests are subject to timeframe restrictions, an appropriate method can be employed according to the specific conditions and stages of the disease. Nevertheless, having a diagnostic test that can cover the entire timeframe of the disease would not only eliminate the need to switch between timeframe-restricted tests, but also minimize the uncertainties from using diagnostic tests that may produce false positive results (Giri et al., 2020).
1.2. SERS and other techniques for virus detection
Aside from the aforementioned SARS-CoV-2 diagnostic tests, some studies have also attempted to explore alternative detection technologies. For examples, the use of field-effect transistor (FET), which uses graphene sheets conjugated with SARS-CoV-2 antibody to coat the transistor; then a nasopharyngeal swab sample is placed on the sensing area of the transistor, and the generated electricity signals are used to match and confirm the presence of viruses (Seo et al., 2020). By using antibodies to detect viruses, the technique combines electrical induction with immunoassays, which means that this method is only suitable for patients who have already been found to have antibodies, making the detection tool suitable for later-stage infections.
In another study, electrical signals were also used monitor the presence of SARS-CoV-2 virus wherein exhaled breath was used as the sample. The detection platform has multiple sensors composed of Au nanoparticles functionalized with ligands, and the volatile organic compounds (VOCs) in the exhaled breath will be adsorbed by these ligands. This is based on discovering pathogens and releasing them directly into the capture environment (Shan et al., 2020). With the gaseous sample as exhaled breath, unwanted particles may interfere with analyte collection in addition to the fact that there could be instances wherein the virus particles in VOCs from exhaled breath may not be at a detectable amount. Considered to be a rapid screening method, this technique may be used as a supplementary test at the early stage of infection.
The application of surface enhanced Raman spectroscopy (SERS) in the detection of viruses and bacteria has attracted the attention of medical experts due to the feasibility of biosensing (S. Luo et al., 2014). Since this technique only utilizes SERS-active substrate and a Raman spectrometer to detect the biochemical structures on the viral envelope, unlike a biochemical-based diagnosis, it does not require any other reagents, calibration or special treatment, so it is considered as a simple and feasible method that can be applied for a fast and accurate detection, in particular for fast-screening of SARS-CoV-2 virus, suitable for the pandemic (Asif et al., 2020). Raman spectroscopy involves inelastic scattering of light to identify the target analyte through the vibrational modes obtained from its components. However, Raman signals are typically weak that it could only be effective in characterizing materials in solid form and there would be a difficulty in detecting analytes in liquid as these are already diluted. A sample containing the analyte is placed on a SERS-active substrate before subjecting to a laser of a suitable wavelength, which will generate a unique SERS spectrum that serves as the “fingerprint” of the target molecule/species. Then, the Raman peak corresponding to the vibration pattern is associated with the analyte (Harper et al., 2013; Kho et al., 2015). The degree of Raman signal enhancement through a well synergized SERS-active substrate has been reported to be capable to detect single molecules at very low concentrations (S. C. Luo et al., 2014; Sivashanmugan et al., 2015).
SERS has proven its accuracy in detecting small molecules, such as environmental pollutants, pesticides, fertilizers, ions, and large molecules, such as proteins, antibodies, and nucleic acids (Chao et al., 2016). Further development allows SERS technology to successfully detect large molecules and other complex structure analytes, such as viruses and bacteria. SERS technology can even identify other pathogens based on the differences in the obtained spectra (Boardman et al., 2016). Table 1 summarizes the comparison among SERS technology and other commonly used diagnostic methods, including: analytes, acquisition time to obtain information, timeframe of effectivity, selectivity, and estimated cost per test (Afzal, 2020; Krüttgen et al., 2020; Sethuraman et al., 2020; J. Van Elslande et al., 2020). In detecting SARS-CoV-2 virus through antibodies, the test detects total antibodies that are mostly IgM and IgG. The self-pay price for this test is to $155 (including specimen collection fee) in the USA. In England, typical prices range from £100–150. Because testing in the absence of symptoms is not covered by Japan's national insurance, such individuals will need to pay about 40,000 yen ($373) per test. In Taiwan, the self-pay price for this test is to $250–400 (including specimen collection fee). The Korean government covers the cost for testing those with suspected symptoms or in recent contact with confirmed cases; anyone else can pay 150,000 KRW (~125 USD) to get tested, with fees reimbursable from the nation's single-payer health care (Lee and Lee, 2020; Taiwan Centers for Disease Control - COVID-19).
Table 1.
Comparison of SERS technology versus currently applied methods for SARS-CoV-2 detection.
| SERS | Nucleic acid-based tests | Serological tests | |
|---|---|---|---|
| Analyte/s | viral envelope and membrane proteins | viral RNA | antibody/antigen |
| Acquisition time | 5 mina | 15 min - 8 hr | 15–30 min |
| Timeframe of effectivity | no limit | early infection stage (6 days before to 14 days after symptom onset) | late infection stage (7 days after symptom onset) |
| Selectivity | depends upon the condition – some substrates have components with high affinity to the target, thus resulting to high selectivity | highly specific; targets viral RNA specific to a particular virus | low; can produce false positives with other same-category viruses |
| Estimated cost per test | 10–50 USD | 100–300 USDb | 25–100 USDb |
Approximate acquisition time based on SERS studies in general.
Reference: (Krouse and Abbott, 2020).
For the detection of SARS-CoV-2 virus, in particular, Fig. 2 shows the comparison of the estimated effectivity of detection, as well as its corresponding timeframe, utilizing SERS as a diagnostic method. SERS is capable of detecting the virus at any phase of the infection but the detectable analyte changes with the timeline – live (Fig. 2, plot (4)) and dead (Fig. 2, plot (5)) viruses are predominantly detected on the early and late stages of the infection, respectively. With this change in the virus integrity over time, the obtained SERS spectra is expected to attribute to the changes in the chemical composition of the viral envelope. As there is a slow decline in the amount of live virus after symptoms appear, and an increase in the amount of dead virus increases due to seroconversion, during the later stages of infection, both live and dead viruses could be detected through SERS at the same time. With a highly sensitive and selective SERS diagnosis, this could be a powerful tool in quantitative studies investigating the dynamics of live and dead viruses in a host over the course of an infection (Khan and Rehman, 2020; Tadesse et al., 2020).
1.3. General considerations in SERS technology as a detection tool
Aside from the advantages and limitations of SERS technology in comparison to other diagnostic methods, it is also important to emphasize that since a SERS-active substrate, the main component, is comprised of a plasmonic metal or a dielectric material (Lee et al., 2020; Sitjar et al., 2019); it needs not be used urgently as it is stable and thus could be stored for a long time, given that a proper storage condition and environment are followed. Processes involved in the fabrication of substrates are thus crucial in coming up with an effective SERS-active substrate.
Moreover, SERS is a versatile technique that is capable of detecting a wide range of analytes provided that the substrate and laser used for detection is suitable for the particular analyte/s. SERS can only detect vibrational modes related to the composition of analyte/s, and the characteristic Raman spectra generated are likely to infer the presence of a target analyte/s in a specimen (Shan et al., 2018; Yuan et al., 2017). It then follows that small-molecule analytes such as pesticides and ions exhibit less vibrational modes and thus having less characteristic peaks in their corresponding SERS spectra, compared to macromolecules and other large analytes such as bacteria and viruses, which possess various proteins on their surfaces, resulting to even more complex spectra which could pose a challenge in assigning peaks to the corresponding vibrational modes (Boardman et al., 2016; Galvan and Yu, 2018). In the case of a virus with an envelope, under the exposure of an appropriate substrate and laser wavelength, the SERS spectrum is presented in the form of characteristic peaks corresponding to the vibration modes of the biochemical components on the unique viral envelope (Dardir et al., 2020; Sivashanmugan et al., 2013).
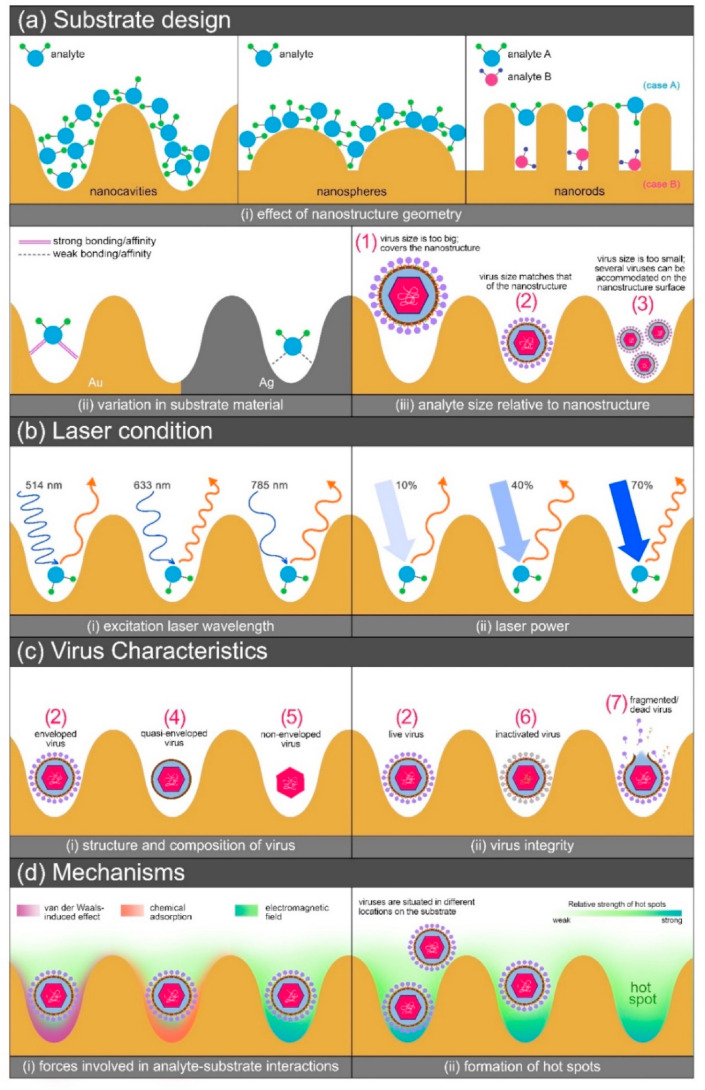
As shown in Fig. 3 , there are several factors that need to be considered in coming up with a SERS detection system specifically for analyte/virus detection; a detailed discussion of each contributing factor is discussed in the next section. In summary, the first three factors – substrate design, laser condition, and analyte characteristics, simultaneously impart their influence on the resulting SERS signal enhancement, providing a synergistic effect. Thus, an optimum condition for a particular application could be achieved in consideration of the combination of the three factors, resulting to the fourth factor, which is the proposed mechanism. Slight variations in the SERS spectra of the same analyte subjected to the same substrate and laser conditions should still be expected as uncontrollable cases such as variation in analyte orientation (Mühlig et al., 2017) and uneven distribution of analytes on the substrate would affect the reproducibility of the results.
Fig. 3.
Factors to consider in analyte detection through SERS: (a) Substrate design – (i) nanostructure geometry; (ii) substrate material; (iii) variation in the virus sizes, marks (1), (2), and (3) with respect to nanostructure size. (b) Laser condition – (i) wavelength and (ii) power. (c) Virus characteristics – (i) structure; (ii) virus integrity (d) Mechanisms – (i) substrate-analyte interactive forces and (ii) hot spot formation.
Particularly, a variety of nanostructures as SERS substrates have been fabricated and tested for bacteria and virus detection. For example, label-free detection of methicillin-resistant Staphylococcus aureus was demonstrated with the use of an aptamer-modified 2D hybrid graphene oxide SERS substrate; prominent peaks from the resulting spectra were attributed mostly to the vibrational modes produced by the lipids and proteins in the virus structure (Sinha et al., 2016).
3D plasmonic nanostructures have also shown to detect influenza virus strains, which possesses a spherical structure, and is predominantly composed of spike proteins (Sivashanmugan et al., 2013). In the said study, peaks with highest intensities corresponded to vibrational modes of adenine and tyrosine, which could be attributed to the tyrosine-histidine interactions that are responsible for the formation of the neuraminidase amino acid sequences on the viral envelope. These studies demonstrate that SERS technique is capable of detecting pathogens regardless of size – S. aureus being around 500–1000 nm, whereas influenza viruses ranging around 80–120 nm. Since viruses possess a variety of components on their membrane surface, each Raman-active component is expected to have at least a characteristic peak in the SERS spectrum, which may correspond to a particular vibrational mode such as what is shown in Table 2 with a variety of enveloped viruses.
Table 2.
Reported SERS substrates studied for virus detection and the corresponding vibrational modes attributed to viral components.
| Ref. | Substrate | Target virus | Vibrational mode/component | Peak assignment (cm−1) |
|---|---|---|---|---|
| Fan et al. (2010) | Klarite (Au on Si) | a variety of enveloped viruses: | S–S stretch | 540 |
| Tyrosine (skeletal) | 640 | |||
| Adenine | 720, 744 | |||
| norovirus MNV4 | Tyrosine | 844 | ||
| C–COO- stretch | 921, 937, 943 | |||
| Phenylalanine (symmetrical ring breathing) | 1001 | |||
| adenovirus MAD | Phenlyalanine (in-plane C–H bending) | 1018, 1022 | ||
| C–N stretch | 1047 | |||
| C–N and C–C stretch | 1129 | |||
| Shanmukh et al. (2006) | Ag nanorod | adenovirus | Guanine | ~643–656 |
| Adenine | ~719–730 | |||
| rhinovirus | Tyrosine | ~848–843 | ||
| Phenylalanine | ~1002 | |||
| HIV | CH2 deformation | ~1448–1454 | ||
| νaCOO− Trp | ~1523–1597 | |||
| respiratory syncytial virus | disulfide stretching | 527, 546 | ||
| C–N stretching | 1044 | |||
| CH2 deformation | 1456 | |||
| Zhang et al. (2019) | Au/Cu hollow nanocones of microbowls (HNCMB) | adenovirus type 5, coxsackievirus type 3 |
carbohydrates for solids | 1015 |
| C–N stretching | ~1041–1062 |
2. Design of SERS systems for detecting SARS-CoV-2 virus
SERS technique is gaining more attention for its potential in analyte detection, even at low concentrations, and cases wherein this technology is applicable would require systems that are not only accurate and sensitive but also selective, to be able to detect specific analytes out of a complex specimen (Mauriz, 2020). It exhibits potential in detecting SARS-CoV-2 as demonstrated by prior studies on SERS as a tool in detecting various viruses (Chen et al., 2020; Magdy et al., 2020). Specially developed SERS detection systems could even be used to perform semi-quantitative analysis of particular analytes present on a sample (Zhao et al., 2021). In order to accommodate the continuous demands from various applications, the technology should as well continuously advance and thus, major factors, as shown in Fig. 3, are suggested to be considered with respect to desired applications.
2.1. Major factors influencing SERS effects and their consequential uncertainties
Substrate design is crucial in SERS detection systems as parameters involved predominantly dictate the sensitivity and accuracy of the technique (S. C. Luo et al., 2014; Mauriz, 2020). In Fig. 3(a)-i, it can be noted that the geometry of the nanostructure influences the distribution of analytes on the substrate surface. For the cases of nanocavities, nanospheres, and nanorods, the cavities and/or gaps on the substrate serve as reservoirs where analytes would accumulate. If the analyte molecules are smaller than the cavities or gaps, as shown in the figures, the analytes are presumably enhanced by the nanostructured substrates. For the case of analytes located upon/between/among the top of the nanorods such as what is shown on the same figure with the analyte distribution on the nanorods, the contribution of the substrate to the SERS signal enhancement differs with the location of the analytes. Signals are stronger in case A (i.e., for analytes that exhibit bigger sizes than the gaps) compared to those in case B (smaller analytes) – the edges on the top of the nanorods induce stronger electromagnetic fields, meanwhile, analytes that are deep into the gaps between the nanorods experience weaker enhancement due to the lack of these edges (Bell et al., 2020; Lin et al., 2011; Zhao et al., 2020). Furthermore, a change in the orientation of an analyte molecule could also lead to a change in the SERS spectra since vibrational modes would then consequently change, which results to the appearance, disappearance, and/or shift in Raman peaks (Rzeznicka and Horino, 2018).
SERS-active substrates are most often fabricated with transition metals (Rh, Pd, Pt, Fe, Co, etc.) and plasmonic metals such as Au and Ag, which have the ability to induce localized surface plasmon resonance (LSPR), which is a result of the coupling of resonant oscillations of electrons and electromagnetic fields near the nanostructures; however, transition metals have also explored found to impart weaker SERS effects compared to plasmonic metals (Sharifi et al., 2020). In the material selection process, it is also important to consider the suitability of the substrate material in terms of its stability in an environment or condition where it is to be applied. Each plasmonic metal exhibit their own characteristic properties that are not found in other metals; for example, gold is a biocompatible material but provides a lower signal enhancement than silver, which on the other hand is less stable due to it being prone to oxidation (Liu et al., 2020). Studies have thus explored the use of both of these metals to take advantage of combining these properties, giving synergistic effects. The interaction of analyte with the substrate material also contributes to the total enhancement brought by the substrate, as illustrated on Fig. 3(a)-ii as an example. Strong affinities from analyte-substrate interactions result to higher enhancement due to the contributions made by the chemical mechanism (Pérez-Jiménez et al., 2020).
In Fig. 3(a)-iii, it is shown that a variety in the virus sizes would consequently result to a variety in the distribution of viruses on the nanostructures. In the illustration, nanocavities serve as the SERS-active substrate, and a virus particle that is bigger than the cavity itself, case (1), would not be able to maximize the effects brought by the cavity as it somehow covers the structure whereas in cases (2) and (3), the viruses are subjected to stronger plasmonic effect by the cavities as they are situated inside the cavity (Yao et al., 2014).
Aside from parameters involving the substrate design, the settings of the excitation laser used in conjunction with the substrate is equally important (Pilot et al., 2019; Starowicz et al., 2018). In an example shown in Fig. 3(b)-i, different laser wavelengths have varying effects on the resulting SERS spectra; if the laser wavelength used is roughly the same or near the plasmon resonance wavelength of the nanostructure, a stronger SERS signal could be obtained, which in the case in the figure, is at a laser wavelength of 633 nm. There is no absolute rule that the higher or lower the excitation laser wavelength is, the stronger is the resulting SERS signal – the best choice of laser depends on the combination of substrate, laser, and analyte, and thus could be obtained based on experimental results. Laser power should also be considered in performing SERS measurements (Yang et al., 2019; Zheng et al., 2014); a high laser power could be an advantage in some cases such as in the example in Fig. 3(b)-ii, as high power could induce more plasmonic resonance to occur on the substrate. However, high power could also degrade the analyte and thus, weak to no SERS or fake signals are obtained. It is therefore important to also determine the best laser power setting to maximize the SERS effects brought by the laser conditions.
Particularly, when viruses are taken as analytes to be detected through SERS, it is critical to know the classification of the virus in terms of structure; this way, when the resulting SERS spectra are analyzed for interpretation, it would be convenient to assign the peaks based on the composition of the virus. Fig. 3(c)-i illustrates the differences among the variety of viral structures – enveloped viruses, case (2), such as SARS-CoV-2 virus possess spike glycoproteins and membrane and envelope proteins on their surface, unlike quasi-enveloped, case (4), and non-enveloped, case (5), viruses which lack the surface proteins (Rivera-Serrano et al., 2019) and surface proteins and lipid membrane, respectively. Virus integrity should as well be considered as a factor since there is a difference in the biochemical composition among cases (2), (6), and (7) in Fig. 3(c)-ii. With this, a particular timeframe, such as in Fig. 2, in which the designed SERS-active substrate/s be used for virus detection, should be defined. In the case of SARS-CoV-2, live viruses, case (2), are expected to be detected at the early stage of infection until 7 days after the onset of symptoms, after which, fragmented/dead viruses, case (7), are most likely to be detected as seroconversion occurs at the later stage of infection (Norman et al., 2020). Therefore, SERS spectra would vary due to the variation in the viral components present on a specimen due to the variation in virus integrity, depending on the stage of infection.
As much as each of the abovementioned factors impart individual contributions to the total SERS effects, their synergistic contributions should not be disregarded. As an example, a gold nanostructure might perform the best with a virus analyte only if this factor is considered but when combined with a 633 nm laser that performed the best in a similar nanostructure that is made of silver, it is possible that the SERS effects in the case of the gold nanostructure and 633 nm laser might be weaker than that when silver is used. Therefore, for the SERS effects to be maximized, experiments should be meticulously designed with careful considerations of the factors mentioned.
2.2. Mechanism of SERS in the detection of viruses
Furthermore, as illustrated in Fig. 3(d)-i, analytes experience different types of adsorption on the substrate – CM effect which, as previously mentioned is due to the affinities between the analyte and substrate leading to possible charge transfers between these two systems; EM effect which is due to the formation of electromagnetic fields from the localized surface plasmon resonance produced by the plasmonic nanostructures upon exposure to incident radiation; and the van-der Waals-induced effect which is a relatively weak physical adsorption attributed to the electrical interactions between the analyte and substrate when in close proximity to each other.
An understanding of how signal enhancement takes place in SERS detection is essential in designing appropriate substrates for specific applications. The effect of SERS is dominated by two mechanisms - chemical (CM) and electromagnetic (EM) (S. C. Luo et al., 2014). CM mechanism is based on the interaction between the analyte molecule and the metal substrate, as there occurs a charge transfer between these components (Prakash, 2020; Trivedi et al., 2020). Chemisorption of the analyte causes a change in the electronic states of the molecules, causing the absorbance of the analyte to shift and when it is exposed to the incident radiation, its Raman cross section increases, leading to signal enhancement. On the other hand, EM mechanism relies on the localized surface plasmon resonance (LSPR) and the local electromagnetic field resulting from collective oscillations of conduction electrons of the plasmonic nanostructures (Sharma et al., 2012). The oscillation must be in resonance with the incident laser to produce a significant LSPR and consequently, higher signal enhancement. This leads to intensified Raman signals of the target analyte up to several orders of magnitude. However, LSPR can only have its maximum contribution to the SERS effect when the dimensions of the nanostructures are shorter than the excitation laser wavelength, this is why another influencing factor in designing SERS substrates is the geometry of the nanostructures (Ding et al., 2017). Both mechanisms work simultaneously but differ in the degree of contribution to the enhancement; EM is considered the predominant mechanism with a contribution to the overall enhancement factor of 104-108 whereas CM only contributes 10-103 (Shiohara et al., 2014; Su et al., 2019). Thus, research on substrate designs takes advantage of the EM effect by tuning the physical attributes of the nanostructures and material/s.
Physical factors play an important role in SERS technology – shape, size, and interstructure gaps are parameters that directly influence the capability of a SERS-active substrate to enhance the Raman signals. A variety of nanomorphologies were developed to extensively investigate the resulting electromagnetic fields produced with varying shapes; nanostructures in colloidal form (nanoparticles, nanostars, nanorods, nanocubes) and 3D form immobilized on glass or Si (nanocolumns, nanospheres, nanocavities, nanocones) (S. C. Luo et al., 2014). Aside from metals, dielectric materials in 2D form as sheets (graphene, graphene oxide, MoS2) were also found to provide SERS effects (Wolosiuk et al., 2014). Substrates in colloidal form were found to agglomerate uncontrollably, causing sites of enhancement to be distributed unevenly, resulting to inconsistent measurements throughout the substrate. Immobilized substrates fabricated through sophisticated methods were then developed to control nanostructure distribution in a uniform arrangement to address the problem of low reproducibility. Moreover, as much as the geometry of the nanostructures plays a vital role in the signal enhancement capability of the substrate, it is also important to consider that the gaps between these nanostructures are where hot spots (Rodriguez-Lorenzo and Alvarez-Puebla, 2014), localized regions of concentrated electromagnetic field where high signal enhancements occur, are formed, as shown in Fig. 3(d)-ii. However, despite these developments, having reproducible results while maintaining a highly sensitive and selective sensing platform is still a challenge in applying SERS technology.
A variety of materials in one substrate were studied upon to combine the properties of each of the individual components. In one study, bimetallic nanorods of Ag and Au were investigated to detect pesticides; Ag is known to provide higher signal enhancement than Au, while Au is chemically stable as it does not oxidize unlike Ag. Other dielectric materials such as ZrO2 and TiO2 have been incorporated to plasmonic nanostructures to create hybrid nanocomposites; these hybrids are said to exhibit synergistic effects, combining the strong plasmonic properties of the metallic component/s, and with the dielectric material providing sites for charge transfers (Sitjar et al., 2019; Sivashanmugan et al., 2015). The capture efficiency of the substrate could even increase if any of the components exhibit strong affinities to the target, improving the sensitivity of the substrate.
2.3. Labelling in SERS
What has been discussed so far only deals with label-free detection wherein the analyte of interest is directly detected with Raman spectroscopy through the SERS spectra of the analyte itself from its intrinsic molecular properties such as functional groups, molecular weight, charge, and its overall interaction with the components of the SERS substrate. However, as not all analytes are Raman-active and thus could not be detected through this direct detection method, further development in SERS technology brought the utilization of labels to address this challenge (Shan et al., 2018). SERS-active substrates are labelled with Raman reporters, Raman-active small molecules, usually that act as extrinsic tags, to facilitate the assignment of peaks in the SERS spectra (Kho et al., 2015). The suitable choice of Raman reporter in particular applications usually considers several factors including its affinity to the metallic component of the substrate and stability when subjected to harsh environment and strong laser powers.
In the detection of complex macromolecules of biological nature such as viruses, bacteria, and protein biomarkers, SERS spectra obtained through label-free detection are most often complicated and peaks coming from the analytes are hard to isolate. However, there are some exceptions to this case as some viruses were reported to be easily detectable even through label-free detection, and this could possibly be attributed to the structure and composition of the virus. Influenza strains were successfully detected on Au/Ag nanorods without the use of any Raman reporters. In the study, peaks found in the resulting SERS spectra were associated with the spike proteins found on the viral envelope – hemagglutinin and neuraminidase. In another case wherein the analyte was the Human Enterovirus 71 (EV71), collection of SERS signals was not possible due to the incompatibility of the analyte with the substrate. It should be considered that EV71 lacks the envelope and spike proteins, unlike the influenza strains as it is classified as a picornavirus, a nonenveloped virus, which is only characterized by a capsid on its surface. In cases, wherein the surface components are undetectable through label-free detection, it might be necessary to consider using SERS labels to obtain defined signals associated with the virus (Mauriz, 2020).
3. Challenges in using SERS technology as a diagnostic tool
Despite the advantages and convenience SERS technology may offer, there are still challenges being faced in the process of developing the technique. The factors discussed in the previous section tackled the specific parameters related to the design and fabrication of the SERS system, while in this section, important considerations when SERS is taken into a large-scale scenario are addressed. These major challenges are discussed given the current situation of the pandemic that needs urgent attention and so, alternative diagnostic methods such as SERS technology would prove to be significant.
3.1. Sources of SARS-CoV-2 virus and its variations and reliable SERS spectra
Experimental studies performed to detect SARS-CoV-2 virus through SERS technology would require the use of live virus to come up with its corresponding SERS spectra; as SARS-CoV-2 virus is highly infectious, a biosafety level 3 virology laboratory (BSL-3) is required. With the possible emergence of more SARS-CoV-2 mutations in the S-protein, or any other surface proteins, consequently, SERS spectra unique to SARS-CoV-2 virus strains would then diversify, requiring the need to update the SERS spectra database regularly in consideration of the new strains. However, as most of the mutations involve only the genetic material inside the viral particle, the resulting SERS spectra of the mutations would not deviate much as SERS is a surface-based technique; still, mutations with variations in the surface proteins should not be ruled out.
In spite, pseudovirus system could be used alternatively to imitate the actual SARS-CoV-2 virus. As SERS is able to analyze only the composition of the surface of an analyte, which possess all of the surface proteins of the SARS-CoV-2 virus would be most suitable for experimental cases without risking the possibility of an infection from the actual virus. The use of a pseudovirus to perform SERS experiments allows investigation to be carried out in lower biosafety level facilities. However, it should also be noted that there could still be a discrepancy on the amount of surface proteins present on the surface of the actual SARS-CoV-2 virus from that of the pseudovirus.
Moreover, in actual applications, there are several sources of specimen to be used for testing – it has been reported that samples could be obtained from nasopharyngeal swabs, saliva, and throat swabs. Viral loading could differ from samples taken at different sources from a single subject and this could pose false results in such way that the virus could be present in one location but could be absent in another. Another issue that needs to be addressed is that in actual cases, swab samples does not contain only the SARS-CoV-2 virus if present, there could be other viruses and biochemicals present in the sample that to some degree could cause a difficulty in the interpretation of the SERS spectra due to the interferences brought by these unwanted chemicals. Thus, it is desirable to have a SERS system that is selective or one that could distinguish between various other viruses, making multiplex detection of viruses possible (Ambartsumyan et al., 2020; Chen et al., 2020).
3.2. Mass production of SERS-active substrates
Most of the SERS studies done on a laboratory scale have shown potential to be produced in a larger scale as intended for their applications in the field of chemical sensing. With the recent event of the pandemic that calls for urgent solutions, quick but reliable mass testing is much needed especially in airports and large-scale events wherein the transmission of virus is most likely to occur. This consequently calls for testing supplies that could cater to a large number of people and to meet this demand, a mass production of the materials should be done.
SERS-active substrates produced for experimental studies could sometimes follow a complicated fabrication route that requires not only costly materials and sophisticated equipment but could also undergo processes that takes time to proceed, which defeats the purpose of mass production that should be quick but cost-effective. Thus, a process flow in a laboratory scale that could easily be transferred for mass production is highly desirable. With mass testing as the intended objective, SERS substrates should be designed with a device or a platform that could easily process the sample from the test subject.
3.3. Compliance to regulations for urgent use
A recent study showed that at least 35% of people are asymptomatic, revealing an increased risk of rapid community spread and the need for widespread testing (“COVID-19 Pandemic Planning Scenarios,” 2020). With the quick spread of COVID-19, the FDA has begun to issue Emergency Use Authorizations (EUA) to several diagnostic tests for COVID-19. In emergency cases such as the COVID-19 pandemic wherein urgent strategies are needed, medical devices could be utilized to address these issues through a short-term EUA or a regular application of in-vitro diagnostic (IVD) device (“FDA MOU 225-14-017,” 2020). The challenge posed in both cases is the requirement of performing a minimum number of screenings using cultured or actual samples from test subjects; however, the regulation depends upon countries. And so, for SERS technology to be applied as a potential diagnostic tool in the case of a pandemic, the previously mentioned challenges – source of the virus and mass production of the substrates should be addressed to be able to comply with the regulations that would prepare and approve the use of the device.
Centers for Disease Control (CDC) of each country is called to detect and investigate diseases as well as assist the nation in implementing disease prevention tactics and health policies. The FDA primarily serves as a regulatory agency for medical devices, and acquiring the EUA for diagnostic tools (“Emergency Use Authorizations for Medical Devices,” 2020) to give emergency approval to COVID-19 diagnostic tests allows protocols for a wide range of activities related to the monitoring, diagnostics and treatment of COVID-19 to be implemented urgently. Similar to the EUA approval procedure by the FDA, the Emergency Use Listing (EUL) by the WHO validates in vitro diagnostics (IVD) used for the detection of COVID-19, focusing on IVDs most likely to be used in countries with limited resources for testing. While the EUA is meant to provide accelerated approval for all IVDs that meet requirements, the EUL prioritizes simpler products to support countries most in need. (“COVID-19 Puts the WHO’s EUL) to Work,” 2020).
4. Conclusion and perspective
The current diagnostic tools for SARS-CoV-2 virus and its variants have always been the convention but with the advancement of method, more techniques are being developed to address the limitations brought by these standard methods, such as their applicability to only limited timeframes. In this article, we introduce the development of SERS technology, which makes it a complementary choice suitable for conventional methods. It should be emphasized that in order to produce a SERS-active substrate specifically for SARS-CoV-2 virus detection, its substrate design is a critical factor, because not any SERS substrate has the ability to detect viruses. With the use of a corresponding SARS-CoV-2 pseudovirus, there is no need to perform SERS experiments in the BSL-3 laboratory; although convenient, this is only to test the effectiveness of SERS-active substrates in detecting viruses. Identifying positive and negative cases through the SERS system will be able to meet EUA regulations and subsequent IVD applications. It is foreseeable that with the further development of high-throughput manufacturing technology and modular design, SERS-active substrates dedicated to virus detection can be produced on a large scale to meet the high demand during the outbreak.
Author statement
Writing original draft preparation, Jaya Sitjar, Han Lee, and Jiunn-Der Liao ; writing review and editing, Jiunn-Der Liao, Huey-Pin Tsai, Jen-Ren Wang, and Ping-Yen Liu . All authors have read and agree to the published version of the manuscript.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
This work was supported by the Ministry of Science and Technology of Taiwan (grant numbers: MOST 109-2224-E-006-008, 109-2327-B-006-005, and 109-2811-E-006-531-MY2), The BSL-3 Virology Laboratory of National Cheng Kung University Hospital, Tainan, Taiwan (with authorization), and Center for Drug Evaluation (CDE) of Taiwan (case number: 109IDX08030). The authors declare no conflict of interest.
References
- Afzal A. J. Adv. Res. 2020;26:149–159. doi: 10.1016/j.jare.2020.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambartsumyan O., Gribanyov D., Kukushkin V., Kopylov A., Zavyalova E. Int. J. Mol. Sci. 2020;21:1–15. doi: 10.3390/ijms21093373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asif M., Ajmal M., Ashraf G., Muhammad N., Aziz A., Iftikhar T., Wang J., Liu H. Curr. Opin. Electrochem. 2020;23:174–184. doi: 10.1016/j.coelec.2020.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell S.E.J., Charron G., Cortés E., Kneipp J., de la Chapelle M.L., Langer J., Procházka M., Tran V., Schlücker S. Angew. Chem. Int. Ed. 2020;59:5454–5462. doi: 10.1002/anie.201908154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boardman A.K., Wong W.S., Premasiri W.R., Ziegler L.D., Lee J.C., Miljkovic M., Klapperich C.M., Sharon A., Sauer-Budge A.F. Anal. Chem. 2016;88:8026–8035. doi: 10.1021/acs.analchem.6b01273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao J., Cao W., Su S., Weng L., Song S., Fan C., Wang L. J. Mater. Chem. B. 2016;4:1757–1769. doi: 10.1039/c5tb02135a. [DOI] [PubMed] [Google Scholar]
- Chen H., Das A., Bi L., Choi N., Moon J.-I., Wu Y., Park S., Choo J. 2020. Nanoscale 21560–21570. [DOI] [PubMed] [Google Scholar]
- COVID-19 pandemic planning scenarios. 2020. https://www.cdc.gov/coronavirus/2019-ncov/hcp/planning-scenarios.html [WWW Document]
- D'Cruz R.J., Currier A.W., Sampson V.B. Front. Cell Dev. Biol. 2020;8:1–11. doi: 10.3389/fcell.2020.00468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dao Thi V.L., Herbst K., Boerner K., Meurer M., Kremer L.P.M., Kirrmaier D., Freistaedter A., Papagiannidis D., Galmozzi C., Stanifer M.L., Boulant S., Klein S., Chlanda P., Khalid D., Miranda I.B., Schnitzler P., Kräusslich H.G., Knop M., Anders S. Sci. Transl. Med. 2020;12 doi: 10.1126/scitranslmed.abc7075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dardir K., Wang H., Martin B.E., Atzampou M., Brooke C.B., Fabris L. J. Phys. Chem. C. 2020;124:3211–3217. [Google Scholar]
- Ding S.Y., You E.M., Tian Z.Q., Moskovits M. Chem. Soc. Rev. 2017;46:4042–4076. doi: 10.1039/c7cs00238f. [DOI] [PubMed] [Google Scholar]
- Emergency Use Authorizations for Medical Devices 2020. https://www.fda.gov/medical-devices/emergency-situations-medical-devices/emergency-use-authorizations-medical-devices [WWW Document]
- Fan C., Hu Z., Riley L.K., Purdy G.A., Mustapha A., Lin M. J. Food Sci. 2010;75:302–307. doi: 10.1111/j.1750-3841.2010.01619.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FDA MOU 225-14-017 2020. https://www.fda.gov/about-fda/domestic-mous/mou-225-14-017 [WWW Document] accessed 12.4.20.
- Galvan D.D., Yu Q. Adv. Healthc. Mater. 2018;7:1–27. doi: 10.1002/adhm.201701335. [DOI] [PubMed] [Google Scholar]
- Giri B., Pandey S., Shrestha R., Pokharel K., Ligler F.S., Neupane B.B. Anal. Bioanal. Chem. 2020;413(1):35–48. doi: 10.1007/s00216-020-02889-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadi A.G., Kadhom M., Hairunisa N., Yousif E., Mohammed S.A. ABiointerface Res. Appl. Chem. 2020;10:7234–7242. [Google Scholar]
- Harper M.M., McKeating K.S., Faulds K. Phys. Chem. Chem. Phys. 2013;15:5312. doi: 10.1039/c2cp43859c. [DOI] [PubMed] [Google Scholar]
- He X., Lau E.H.Y., Wu P., Deng X., Wang J., Hao X., Lau Y.C., Wong J.Y., Guan Y., Tan X., Mo X., Chen Y., Liao B., Chen W., Hu F., Zhang Q., Zhong M., Wu Y., Zhao L., Zhang F., Cowling B.J., Li F., Leung G.M. Nat. Med. 2020;26:672–675. doi: 10.1038/s41591-020-0869-5. [DOI] [PubMed] [Google Scholar]
- Khan R.S., Rehman I.U. Expert Rev. Mol. Diagn. 2020:1–3. doi: 10.1080/14737159.2020.1784008. 00. [DOI] [PubMed] [Google Scholar]
- Kho, K.W., Dinish, U.S., Olivo, M., 2015. Elsevier Ltd.
- Krouse S., Abbott B. What kind of covid test should I get? Answers on cost, accuracy and more. Wall St. J. 2020 https://www.wsj.com/articles/covid-19-tests-answers-on-cost-accuracy-and-turnaround-time-11599134378 [WWW Document] [Google Scholar]
- Krüttgen A., Cornelissen C.G., Dreher M., Hornef M., Imöhl M., Kleines M. J. Clin. Virol. 2020;128:104394. doi: 10.1016/j.jcv.2020.104394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalli, M.A., Langmade, S.J., Chen, X., Fronick, C.C., Sawyer, C.S., Burcea, L.C., Wilkinson, M.N., Fulton, R.S., Heinz, M., Buchser, W.J., Head, R.D., Mitra, R.D., Milbrandt, J., 2020. medRxiv 1–34.
- Lee D., Lee J. Transp. Res. Interdiscip. Perspect. 2020;5:100111. doi: 10.1016/j.trip.2020.100111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H., Yang J.W., Liao J. Der, Sitjar J., Liu B.H., Sivashanmugan K., Fu W.E., Chen G.D. Coatings. 2020;10 [Google Scholar]
- Lin Y.Y., Liao J. Der, Ju Y.H., Chang C.W., Shiau A.L. Nanotechnology. 2011;22 doi: 10.1088/0957-4484/22/18/185308. [DOI] [PubMed] [Google Scholar]
- Liu Y., Zhu J., Weng G., Li J., Zhao J. Microchim. Acta. 2020;187 doi: 10.1007/s00604-020-04460-y. [DOI] [PubMed] [Google Scholar]
- Luo S.C., Sivashanmugan K., Liao J. Der, Yao C.K., Peng H.C. Biosens. Bioelectron. 2014;61:232–240. doi: 10.1016/j.bios.2014.05.013. [DOI] [PubMed] [Google Scholar]
- Magdy C., Issam F., Amir P., Adel S., El Gohary S. MedCrave Online J. Appl. Bionics Biomech. 2020;4:86–91. [Google Scholar]
- Mauriz E. Sensors. 2020;20:1–27. [Google Scholar]
- Meng X., Shi L., Yao L., Zhang Y., Cui L. Colloids Surfaces A Physicochem. Eng. Asp. 2020:124658. [Google Scholar]
- Mühlig A., Cialla-May D., Popp J. J. Phys. Chem. C. 2017;121:2323–2332. [Google Scholar]
- Norman M., Gilboa T., Ogata A.F., Maley A.M., Cohen L., Busch E.L., Lazarovits R., Mao C.P., Cai Y., Zhang J., Feldman J.E., Hauser B.M., Caradonna T.M., Chen B., Schmidt A.G., Alter G., Charles R.C., Ryan E.T., Walt D.R. Nat. Biomed. Eng. h. 2020;4(12):1180–1187. doi: 10.1038/s41551-020-00611-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Jiménez A.I., Lyu D., Lu Z., Liu G., Ren B. Chem. Sci. 2020;11:4563–4577. doi: 10.1039/d0sc00809e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilot R., Signorini R., Durante C., Orian L., Bhamidipati M., Fabris L. Biosensors. 2019;9 doi: 10.3390/bios9020057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon L.L.M., Chan K.H., Wong O.K., Yam W.C., Yuen K.Y., Guan Y., Lo Y.M.D., Peiris J.S.M. J. Clin. Virol. 2003;28:233–238. doi: 10.1016/j.jcv.2003.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash O. J. Chem. Phys. 2020;153 doi: 10.1063/5.0022880. [DOI] [PubMed] [Google Scholar]
- Ravi N., Cortade D.L., Ng E., Wang S.X. 2020. Diagnostics for SARS-CoV-2 Detection: A Comprehensive Review of the FDA-EUA COVID-19 Testing Landscape. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera-Serrano E.E., González-López O., Das A., Lemon S.M. Elife. 2019;8:1–24. doi: 10.7554/eLife.43983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Lorenzo L., Alvarez-Puebla R.A. Woodhead Publishing Limited; 2014. Nanosensors for Chemical and Biological Applications. [Google Scholar]
- Rzeznicka I., Horino H. 2018. Raman Spectroscopy. [Google Scholar]
- Seo G., Lee G., Kim M.J., Baek S.H., Choi M., Ku K.B., Lee C.S., Jun S., Park D., Kim H.G., Kim S.J., Lee J.O., Kim B.T., Park E.C., Il Kim S. RACS Nano. 2020;14:5135–5142. doi: 10.1021/acsnano.0c02823. [DOI] [PubMed] [Google Scholar]
- Serrano M.M., Rodríguez D.N., Palop N.T., Arenas R.O., Córdoba M.M., Mochón M.D.O., Cardona C.G. J. Clin. Virol. 2020;129:104529. doi: 10.1016/j.jcv.2020.104529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethuraman N., Jeremiah S.S., Ryo A. JAMA, J. Am. Med. Assoc. 2020;323:2249–2251. doi: 10.1001/jama.2020.8259. [DOI] [PubMed] [Google Scholar]
- Shan B., Broza Y.Y., Li Wenjuan, Wang Y., Wu S., Liu Z., Wang Jiong, Gui S., Wang L., Zhang Z., Liu W., Zhou S., Jin W., Zhang Qianyu, Hu D., Lin L., Zhang Qiujun, Li Wenyu, Wang Jinquan, Liu H., Pan Y., Haick H. ACS Nano. 2020 doi: 10.1021/acsnano.0c05657. [DOI] [PubMed] [Google Scholar]
- Shan B., Pu Y., Chen Y., Liao M., Li M. Coord. Chem. Rev. 2018;371:11–37. [Google Scholar]
- Shanmukh S., Jones L., Driskell J., Zhao Y., Dluhy R., Tripp R.A. Nano Lett. 2006;6:2630–2636. doi: 10.1021/nl061666f. [DOI] [PubMed] [Google Scholar]
- Sharifi M., Hosseinali S.H., Hossein Alizadeh R., Hasan A., Attar F., Salihi A., Shekha M.S., Amen K.M., Aziz F.M., Saboury A.A., Akhtari K., Taghizadeh A., Hooshmand N., El-Sayed M.A., Falahati M. Talanta. 2020;212:120782. doi: 10.1016/j.talanta.2020.120782. [DOI] [PubMed] [Google Scholar]
- Sharma B., Frontiera R.R., Henry A.-I., Ringe E., Van Duyne R.P. Mater. Today. 2012;15:16–25. doi: 10.1016/S1369-7021(12)70017-2. [DOI] [Google Scholar]
- Sheikhzadeh E., Eissa S., Ismail A., Zourob M. Diagnostic techniques for COVID-19 and new developments. Talanta. 2020;220 doi: 10.1016/j.talanta.2020.121392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiohara A., Wang Y., Liz-Marzán L.M. J. Photochem. Photobiol. C Photochem. Rev. 2014;21:2–25. [Google Scholar]
- Sinha S.S., Jones S., Pramanik A., Ray P.C. Acc. Chem. Res. 2016;49:2725–2735. doi: 10.1021/acs.accounts.6b00384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitjar J., Liao J.-D., Lee H., Liu B.H., Fu W. Nanomaterials. 2019;9:664. doi: 10.3390/nano9050664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivashanmugan K., Liao J. Der, Liu B.H., Yao C.K., Luo S.C. Sensor. Actuator. B Chem. 2015;207:430–436. [Google Scholar]
- Sivashanmugan K., Liao J. Der, You J.W., Wu C.L. Sensor. Actuator. B Chem. 2013;181:361–367. [Google Scholar]
- Starowicz Z., Wojnarowska-Nowak R., Ozga P., Sheregii E.M. Colloid Polym. Sci. 2018;296:1029–1037. doi: 10.1007/s00396-018-4308-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stowell S., Guarner J. Clin. Infect. Dis. 2020;71:1935–1936. doi: 10.1093/cid/ciaa510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Y., Shi Y., Wang P., Du J., Raschke M.B., Pang L. Beilstein J. Nanotechnol. 2019;10:549–556. doi: 10.3762/bjnano.10.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadesse L.F., Safir F., Ho C.S., Hasbach X., Khuri-Yakub B.P., Jeffrey S.S., Saleh A.A.E., Dionne J. J. Chem. Phys. 2020;152 doi: 10.1063/1.5142767. [DOI] [PubMed] [Google Scholar]
- Tahamtan A., Ardebili A. Expert Rev. Mol. Diagn. 2020;20:453–454. doi: 10.1080/14737159.2020.1757437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taiwan Centers for Disease Control - COVID-19 2020. https://www.cdc.gov.tw/Bulletin/Detail/tN4ppuNAO444e1D4bFgfkA?typeid=9 [WWW Document]
- Trivedi D.J., Barrow B., Schatz G.C. J. Chem. Phys. 2020;153 doi: 10.1063/5.0023359. [DOI] [PubMed] [Google Scholar]
- Van Elslande Jan, Decru B., Jonckheere S., Van Wijngaerden E., Houben E., Vandecandelaere P., Indevuyst C., Depypere M., Desmet S., André E., Van Ranst M., Lagrou K., Vermeersch P. Clin. Microbiol. Infect. 2020;26(11):1557.e1–1557.e7. doi: 10.1016/j.cmi.2020.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Elslande J., Houben E., Depypere M., Brackenier A., Desmet S., André E., Van Ranst M., Lagrou K., Vermeersch P. Clin. Microbiol. Infect. 2020;26:1082–1087. doi: 10.1016/j.cmi.2020.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Kasteren P.B., van der Veer B., van den Brink S., Wijsman L., de Jonge J., van den Brandt A., Molenkamp R., Reusken C.B.E.M., Meijer A. J. Clin. Virol. 2020;128:104412. doi: 10.1016/j.jcv.2020.104412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varotsos C.A., Krapivin V.F. Costas. Saf. Sci. 2020;132 doi: 10.1016/j.ssci.2020.104962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura B. Della, Cennamo M., Minopoli A., Campanile R., Censi S.B., Terracciano D., Portella G., Velotta R. ACS Sens. 2020;5:3043–3048. doi: 10.1021/acssensors.0c01742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wechselberger C., Susanne S., Doppler S. Performance evaluation of serological assays to determine the immunoglobulin status in SARS-CoV-2 infected patients. J. Clin. Virol. 2020;131 doi: 10.1016/j.jcv.2020.104589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiersinga W.J., Rhodes A., Cheng A.C., Peacock S.J., Prescott H.C. JAMA, J. Am. Med. Assoc. 2020;324:782–793. doi: 10.1001/jama.2020.12839. [DOI] [PubMed] [Google Scholar]
- Wolosiuk A., Tognalli G., Mart E.D., Granada M., Fuertes M.C., Troiani H., Bilmes S.A., Fainstein A., Soler-illia G.J.A.A. ACS Appl. Mater. Interfaces. 2014;6(7):5263–5272. doi: 10.1021/am500631f. [DOI] [PubMed] [Google Scholar]
- Wu Z. gang, Zheng H. yun, Gu J., Li F., Lv R., long Deng, yun Y., Xu W., zhou Tong, qmg Y. Curr. Med. Sci. 2020;40:614–617. doi: 10.1007/s11596-020-2224-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Jingyun, Wang W., Chen Z., Lu S., Yang F., Bi Z., Bao L., Mo F., Li X., Huang Y., Hong W., Yang Y., Zhao Y., Ye F., Lin S., Deng W., Chen H., Lei H., Zhang Z., Luo M., Gao H., Zheng Y., Gong Y., Jiang X., Xu Y., Lv Q., Li D., Wang M., Li F., Wang S., Wang Guanpeng, Yu P., Qu Y., Yang L., Deng H., Tong A., Li J., Wang Z., Yang Jinliang, Shen G., Zhao Z., Li Y., Luo J., Liu H., Yu W., Yang M., Xu J., Wang J., Li H., Wang H., Kuang D., Lin P., Hu Z., Guo W., Cheng W., He Y., Song X., Chen C., Xue Z., Yao S., Chen L., Ma X., Chen S., Gou M., Huang W., Wang Y., Fan C., Tian Z., Shi M., Wang F.S., Dai L., Wu M., Li G., Wang Guangyu, Peng Y., Qian Z., Huang C., Lau J.Y.N., Yang Z., Wei Y., Cen X., Peng X., Qin C., Zhang K., Lu G., Wei X. Nature. 2020;586:572–577. doi: 10.1038/s41586-020-2599-8. [DOI] [PubMed] [Google Scholar]
- Yang L., Peng Y., Yang Y., Liu J., Huang H., Yu B., Zhao J., Lu Y., Huang Z., Li Z., Lombardi J.R. Adv. Sci. 2019;6:1–13. doi: 10.1002/advs.201900310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao C.K., Liao J. Der, Lin C.H., Yang Y.S., Yu S.H., Yang J.W. Sensor. Actuator. B Chem. 2014;191:219–226. [Google Scholar]
- Younes N., Al-Sadeq D.W., Al-Jighefee H., Younes S., Al-Jamal O., Daas H.I., Yassine H.M., Nasrallah G.K. Viruses. 2020;12:1–27. doi: 10.3390/v12060582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y., Panwar N., Yap S.H.K., Wu Q., Zeng S., Xu J., Tjin S.C., Song J., Qu J., Yong K.T. Coord. Chem. Rev. 2017;337:1–33. [Google Scholar]
- Zhang Xingang, Zhang Xiaolei, Luo C., Liu Z., Chen Y., Dong S., Jiang C., Yang S., Wang F., Xiao X. Small. 2019;15:1–8. doi: 10.1002/smll.201805516. [DOI] [PubMed] [Google Scholar]
- Zhao F., Wang W., Zhong H., Yang F., Fu W., Ling Y., Zhang Z. R Talanta. 2021;221:121465. doi: 10.1016/j.talanta.2020.121465. [DOI] [PubMed] [Google Scholar]
- Zhao X., Wen J., Zhu A., Cheng M., Zhu Q., Zhang X., Wang Y., Zhang Y. Nanomaterials. 2020;10:1–13. doi: 10.3390/nano10091667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhen W., Manji R., Smith E., Berry G.J. J. Clin. Microbiol. 2020;58:1–8. doi: 10.1128/JCM.00743-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X.S., Hu P., Zhong J.H., Zong C., Wang X., Liu B.J., Ren B. J. Phys. Chem. C. 2014;118:3750–3757. [Google Scholar]