Summary
The three peripheral sensory neuron (SN) subtypes, nociceptors, mechanoreceptors, and proprioceptors, localize to dorsal root ganglia and convey sensations such as pain, temperature, pressure, and limb movement/position. Despite previous reports, to date no protocol is available allowing the generation of all three SN subtypes at high efficiency and purity from human pluripotent stem cells (hPSCs). We describe a chemically defined differentiation protocol that generates all three SN subtypes from the same starting population, as well as methods to enrich for each individual subtype. The protocol yields high efficiency and purity cultures that are electrically active and respond to specific stimuli. We describe their molecular character and maturity stage and provide evidence for their use as an axotomy model; we show disease phenotypes in hPSCs derived from patients with familial dysautonomia. Our protocol will allow the modeling of human disorders affecting SNs, the search for treatments, and the study of human development.
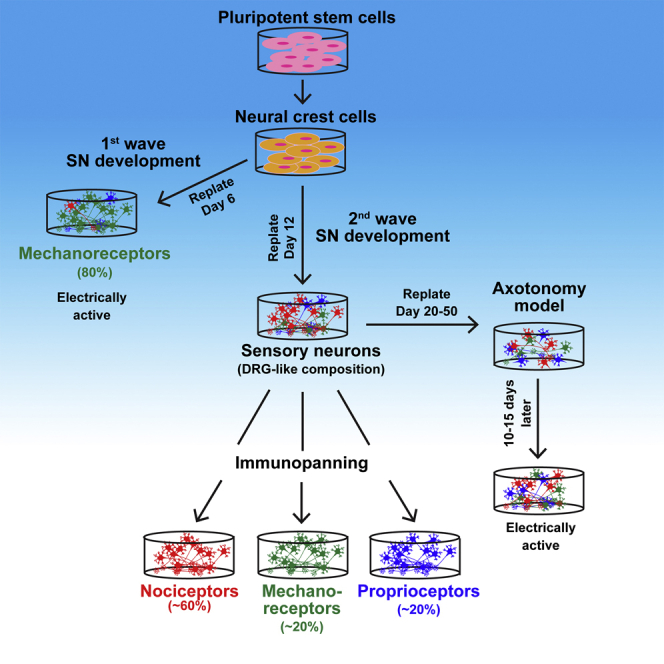
Keywords: pluripotent stem cells, induced pluripotent stem cells, neural crest, sensory neurons, nociceptors, mechanoreceptors, proprioceptors, dorsal root ganglia, axotonomy, immunopanning
Graphical Abstract
Highlights
-
•
Efficient differentiation protocol for functional, DRG-like peripheral sensory neurons
-
•
Enrichment of nociceptors, mechanoreceptors, and proprioceptors via immunopanning
-
•
Generation of mechanoreceptors via the first wave of SN development
-
•
Disease phenotypes in iPSC-derived SNs from familial dysautonomia and axotomy model
In this article, Zeltner and colleagues describe an efficient protocol to differentiate functional peripheral sensory neurons that resemble the human dorsal root ganglia. Methods are provided to isolate nociceptors, mechanoreceptors, and proprioceptors individually using immunopanning or by catching progeny from developmental waves, i.e., mechanoreceptors. This protocol was used as a model to study axotomy and familial dysautonomia using patients' induced pluripotent stem cells.
Introduction
Peripheral sensory neurons (SNs) localize to the dorsal root ganglia (DRG) that run parallel to the spinal cord and innervate peripheral limbs, tissues, and organs. There are three main subtypes of SNs with specialized functions and innervation targets. (1) Nociceptors respond to noxious heat and chemical stimuli and convey pain sensations. They are typically of small-to-medium diameter, either thinly myelinated or unmyelinated, and marked by the neurotrophin receptor TRKA (Lallemend and Ernfors, 2012). (2) Mechanoreceptors are large-diameter, myelinated neurons that express TRKB/TRKC and convey touch sensations. A subset has an extremely low threshold for activation (Poole et al., 2014). (3) Proprioceptors are large-diameter TRKC+ neurons that sense limb movement and position (Lallemend and Ernfors, 2012).
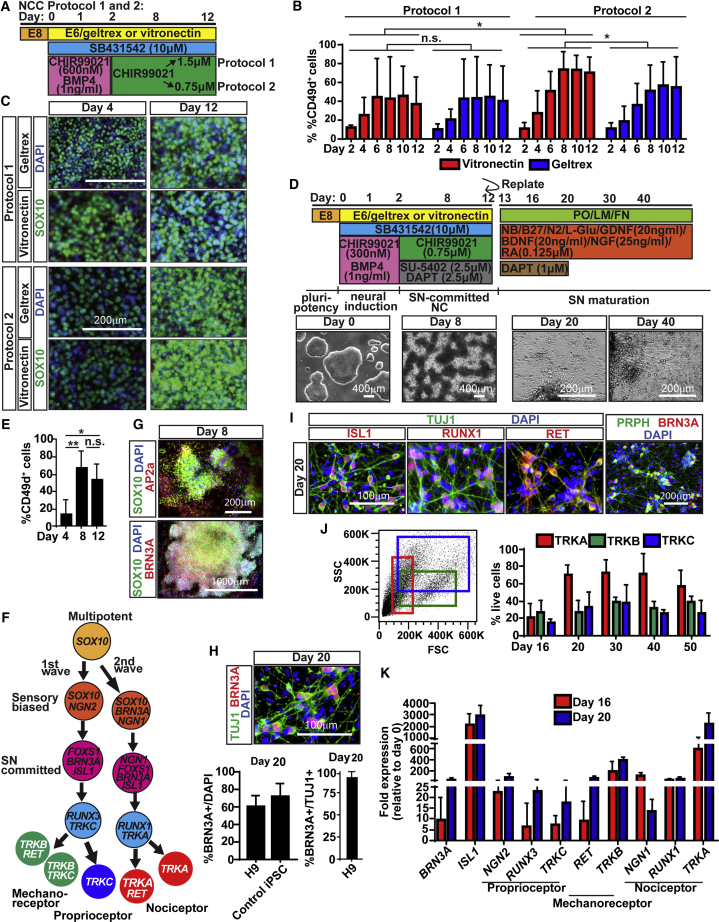
DRG SNs originate from migrating neural crest cells (NCCs), which originate from the neural plate border and are identified by markers such as SNAIL2, ETS1, and SOX10 (Pla and Monsoro-Burq, 2018). SN specification originates from two main distinct NCC migration waves (Figure 1F), mostly defined in mice. First, mechanoreceptors and proprioceptors are specified by sensory-biased NCCs expressing NEUROGENIN2 (NGN2). RUNX3 expression is then upregulated, promoting TRKC expression in proprioceptors, or downregulated, causing TRKB, TRKC, and RET expression in mechanoreceptors. A second wave is characterized by the expression of NEUROGENIN1 (NGN1) and RUNX1 and gives rise to TRKA-expressing nociceptors (Marmigère and Ernfors, 2007). Neurotrophins bind to specific TRK receptors to promote SN specification. Nerve growth factor (NGF), brain-derived neurotrophic factor (BDNF), and neurotrophin-3 (NT3) bind to TRKA, TRKB, and TRKC, respectively (Huang and Reichardt, 2001). Subtypes of mechanoreceptors and nociceptors express the neurotrophin receptor RET, which is activated by glial cell line-derived neurotrophic factor (GDNF) (Donnelly et al., 2018). Despite these differences, all SN subtypes express the transcription factors BRN3A and ISLET1 (ISL1) (Lallemend and Ernfors, 2012; Sun et al., 2008).
Figure 1.
Generation and characterization of hESC-derived SNs
(A) Timeline of NCC differentiation and conditions tested.
(B) Flow-cytometry quantification of CD49d+ NCCs. ∗p < 0.05 using two-way ANOVA; n.s., not significant.
(C) IF of NCCs for SOX10 and DAPI.
(D) Timeline of SN differentiation protocol (top). Representative bright-field (BF) images for each indicated day (bottom).
(E) Flow-cytometry quantification of SN-biased NCCs stained for CD49d. ∗p < 0.01, ∗∗p < 0.001 using one-way ANOVA; n.s., not significant.
(F) SN gene expression summary.
(G) AP2a, BRN3A, and DAPI expression by SN-biased NCCs generated.
(H) Quantification of SN generation efficiency. Day-20 SNs were stained for TUJ1, BRN3A, and DAPI (top). Number of BRN3A+/all (DAPI+) cells and BRN3A+/neurons (TUJ1+) were quantified based on IF (bottom).
(I) Day-20 SNs express markers specific of peripheral neurons and SN subtypes (refer to F).
(J) SN subtype quantification. The unstained cell population that corresponds to the expected subtype size was gated on the SSC/FSC plot and then quantified based on its corresponding TRK receptor. Example: small cells were gated in the red gate, and of those ~70% stained positive for TRKA at day 20. DAPI was used to exclude dead cells.
(K) SNs express general (BRN3A, ISL1) and subtype-specific markers based on qRT-PCR.
In all experiments n > 4, and bar graphs show mean ± SD. See also Figure S1.
To date, most of the studies of DRG neurons have been carried out in animal models. Thus, it is important to establish human models of this system to further our knowledge of DRG and SNs in humans and study these cells' dysfunction in disease. Pluripotent stem cell (PSCs, including embryonic stem cells [ESCs] and induced pluripotent stem cells [iPSCs]) technology is a powerful tool to study SN diseases, critically assess animal model-derived data, and validate potential drug compounds. There are several reports describing the differentiation of SNs from human PSCs (hPSCs) (Table S1) (Alshawaf et al., 2018; Boisvert et al., 2015; Chambers et al., 2012; Goldstein et al., 2010; Namer et al., 2019; Pomp et al., 2005; Schrenk-Siemens et al., 2015; Young et al., 2014). The protocols vary widely in their efficiency and the SN subtypes that they generate, but they share similarities, most notably using some combination of NGF, BDNF, and NT3 growth factors.
Here, we describe a versatile method to derive SNs from hPSCs in feeder-free and chemically defined conditions. We characterize each developmental stage (from the neural plate border to SNs) and effectively quantify each SN subtype. Our protocol generates SN subtypes at the same proportions seen in the DRG (Ernsberger, 2009). Furthermore, we can bias culture conditions toward the generation of nociceptors or mechanoreceptors. Additionally, we describe immunopanning as a gentle isolation technique of specific SN subtypes. Finally, we find that our SNs can be replated at a mature state; they regrow their axons and form functional synapses, suggesting that our protocol can be used as a model for axotomy. Thus, this protocol can be adapted to study various ailments of the peripheral nervous system (PNS).
Results
A method for SN generation mimicking the DRG neuronal composition
Since all DRG SNs are derived from NCCs, we first aimed to generate a protocol to derive NCCs efficiently. The WNT pathway is a key regulator of NCC development in vivo (Ji et al., 2019) and in generating NCCs from hPSCs (Menendez et al., 2011; Mica et al., 2013). Based on a recent study (Tchieu et al., 2017), we tested differentiation initiation timing, CHIR99021 concentrations (= WNT activation), and surface coatings (Figure 1A). We found that (1) initiating the differentiation the day of seeding (rather than 24 h later) and (2) lowering the concentration of CHIR99021 and using vitronectin (rather than Geltrex) resulted in dense NC “ridges” (Figure S1A, arrows), ∼80% NCCs, measured by expression of CD49D (Figure 1B, which correlates with SOX10 [Fattahi et al., 2016]), and SOX10 robust expression (Figure 1C). To confirm NC identity, we showed a decrease of the pluripotency gene POU5F1(OCT4) by day 4 (Figure S1B) and high expression of the NCC-related genes SOX10 and P75NTR (Figure S1B) (Simões-Costa and Bronner, 2015) with a peak at day 6 (Figure S1B). To increase SN differentiation efficiency and speed, we used fibroblast growth factor (FGF) receptor/vascular endothelial growth factor receptor inhibition (SU-5402) and Notch inhibition (DAPT), as previously shown (Chambers et al., 2012) (Figure 1D). To ensure that the cells properly passed through the NCC stage (Figure S1C), we showed OCT4 downregulation within the first 4 days and upregulation of genes expressed during neural plate border formation (GATA2/3 and TFAP2A) (de Crozé et al., 2011) and in specification of premigratory NCCs (PAX3, ETS1, and SNAIL2) (Théveneau et al., 2007) (Figures S1D and S1E). We further found expression of CD49D in ∼70% of cells by day 8 (Figure 1E). Similar to in vivo studies, we found expression of TFAP2A, SOX10, FOXS1, and the SN marker BRN3A (Figures 1F, 1G, and S1F), suggesting the presence of SN-committed NCCs. Our cultures showed low expression of contaminating endoderm (SOX17/FOXA2), mesoderm (TBXT), and central nervous system (CNS) progenitors (PAX6/OTX2, Figure S1G). However, we found expression of the placode progenitor SIX1 and EYA1 (Zou et al., 2004) (Figure S1G). Placode and neural crest lineages are intimately associated during development (Lallemend and Ernfors, 2012), and placode derivation from hPSCs has some similarities with our protocol (Tchieu et al., 2017; Zimmer et al., 2018). Together, these data suggest that we are progressing through the proper developmental stages of NCCs, and that FGF and Notch signaling inhibition pushes the differentiation toward a SN fate.
Next, we sought to push the NCCs toward SNs by replating them on day 12 in a combination of SN-favoring growth factors (Figure 1D) (Chambers et al., 2012). Addition of DAPT (days 12–20) reduced the number of SOX10+ progenitor cells and enriched the purity of the culture (Figure S1H). By day 20, SNs formed defined clusters reminiscent of ganglia and showed axonal outgrowth (Figure 1D, bottom). Sixty to seventy percent of all cells expressed the pan-SN marker BRN3A (Figure 1H). Additionally, almost all (∼99%) of the TUJ1+ neurons expressed BRN3A, indicating little contamination of other neuronal cell types (Figure 1H). This was confirmed by low expression of markers of autonomic (ASCL1), motor (HB9 and OLIG2), CNS (TBR1), and enteric neurons (EDNRB) (Figures S1I and S1J). However, we found that the remaining 30%–40% of the cells in the cultures have a character of myofibroblasts (α-smooth muscle actin, desmin) and Schwann cells (MPZ, MBP) (Figures S1K and S1J), similar to previous reports (Dionisi et al., 2020).
PSC differentiations are notoriously variable. We sought to define the variability in our protocol and found that sequential culture of human ESCs (hESCs) decreased NCC and SN differentiation efficiency over time, affecting about one in every five experiments (Figure S2A). This limitation was overcome using the CryoPause method (Figure S2B) (Wong et al., 2017). Furthermore, we found that cells can be frozen at the NCC stage (day 12) without compromising further differentiation after thawing (Figures S2E–S2G). This highlights the ease of working with this protocol and allows researchers to generate large, well-characterized batches of cells for their analysis without the compromise of potentially failed experiments.
Molecular characterization and analysis of SN subtypes in our cultures revealed robust expression of ISL1, BRN3A, Peripherin (PRPH), and TUJ1 (Figure 1I), as well as RUNX1+ nociceptor-fated SNs and mechanoreceptor RET+ SNs similar to in vivo studies (Figure 1I). We used fluorescence-activated cell sorting (FACS) analysis and gating on specific cell populations in side scatter/forward scatter (SSC/FSC) plots according to the expected cell size followed by sorting for their associated TRK receptor. For example, to detect the portion of nociceptors in our cultures, we gated on the small-cell-size population (Figure 1J, red gate, left) and then selected TRKA+ cells to be ∼70% starting at day 20 (∼50% of total cells) (Figure 1J, right, red bars). Accordingly, ∼30% of SNs (∼34% total cells) were a combination of TRKB+/C+ mechanoreceptors (due to this double staining, the numbers add up to more than 100%) and TRKC+ proprioceptors (Figure 1J). This ratio correlates with previous studies showing the ratio of TRKA+, TRKB+, and TRKC+ neurons in the human DRG is approximately 2:1:1 (Ernsberger, 2009), suggesting that our model resembles the DRG neuronal composition. Finally, qRT-PCR analysis on days 16 and 20 confirmed the presence of each SN subtype (Figure 1K). In sum, our data show that we are generating SNs, which go through the proper developmental stages.
Late-stage molecular and functional characterization of hESC-derived SNs
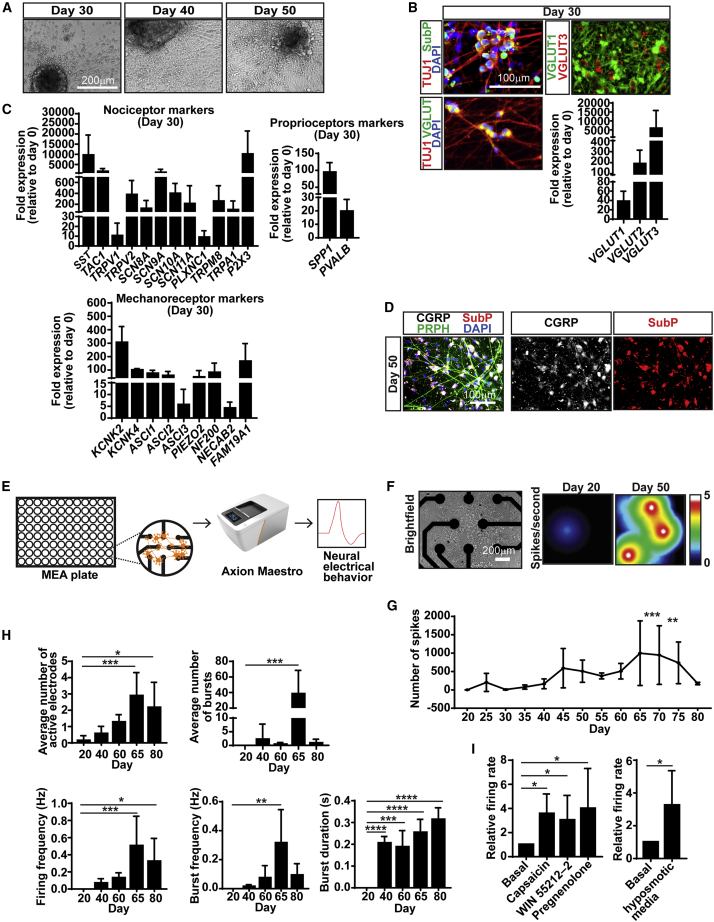
Next, we functionally characterized and assessed our SNs’ maturity via late-stage marker expression and electrophysiology. Beginning on day 20, we observed formation of large clusters with dense, radiating axons (Figure 2A). General markers indicating maturation of SNs were present at day 30 (VGLUT1/2/3, Figure 2B). Nociceptors expressed SST, PLXNC1, and Substance P (TAC1) (Figures 2B–2D). Also, we found expression of the members of the transient receptor (TRP) family TRPV1 and TRPV2 (expressed in medium-to-large-diameter nociceptors), the temperature-sensitive receptor TRPM8, and the cold-activated receptor TRPA1 (Figure 2C). Finally, expression of SCN8A-11A genes (encoding Na+ channels Nav1.6–9) and the ATP-activated receptor P2X3 was noted. Similarly, mechanoreceptors expressed the mechanically activated K+ channels TREK-1 (KCNK2) and TRAAK (KCNK4), the acid-sensing ion channels (ASIC1-3) expressed in the Meissner corpuscles and Merkel cells, NEFH/NF200 (expressed in myelinated A-β fiber neurons), PIEZO2 (expressed in C-low-threshold mechanoreceptors), NECAB2, and FAM19A1 (expressed in low-threshold mechanoreceptors). Proprioceptors expressed SPP1 and Parvalbumin (PVALB) (Figure 2C). We confirmed protein expression of the nociceptor-related marker calcitonin-gene related protein(CGRP) and Substance P on day 50 by immunofluorescence (Figure 2D).
Figure 2.
Characterization of late-stage SNs
(A) Representative BF images of SNs.
(B) SNs express general mature markers, by IF and qRT-PCR.
(C and D) SNs express subtype-specific mature markers, by qRT-PCR (C) and IF (D).
(E) Workflow of experiments using a multielectrode array (MEA).
(F) Representative image of SNs on MEA plates (left) and heatmap generated by spike firing on days 20 and 50 (right).
(G) Functional profile of SNs. NCCs were replated on MEA plates on day 12 and continued to differentiate. For each measurement day, 5 min were recorded and averaged over six wells. Statistical analysis was done by comparing with day 20.
(H) Quantification of average number of active electrodes, number and duration of bursts, and firing and burst frequency of SNs.
(I) SN activity is modulated by nociceptor and mechanoreceptor activation. SNs were incubated with nociceptor agonists (left) or hypo-osmotic medium (right) followed by 5-min recordings.
Data are normalized to untreated control (basal). n > 4 in (A) to (D), n = 3 in (F) to (I). Graphs show mean ± SD. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001 using one-way ANOVA. See also Figure S3.
Next, we asked whether the SNs were functional. Using a multielectrode array (MEA, Figure 2E), we found spontaneous neural activity starting on day 25 and peaking on day 65 (Figures 2F and 2G). We also found that by day 65 the number of bursts and active electrodes peaked (Nehme et al., 2018), and firing and burst frequency was the highest. However, the duration of the bursts increased up to day 80 and correlated with a decrease in burst but not in firing frequency, suggesting firing of longer but less frequent bursts (Figure 2H). We did not see synchronous network activity. It is unclear whether this is because our SNs are not mature enough or whether PNS neurons do not form networks in vivo and in vitro (Alshawaf et al., 2018; Namer et al., 2019). We then sought to assess specific modulation of the SN subtypes. We found that nociceptor agonists capsaicin (Caterina et al., 1997), WIN 55212-2, and pregnenolone (Quallo et al., 2017) increased the firing rate (Figure 2I, left). Additionally, incubation of SNs with hypo-osmotic medium (which mimics pressure) increased the firing rate, suggesting that the mechanoreceptors are functional (Figure 2I, right).
Long-term culture of SNs (50 days or more) causes neuronal detachment from the wells (Figure S3A, arrows). Thus, we asked whether replating cells a second time would be suitable for long-term cultures. We found that SNs can be replated any time between day 16 and day 50 and form a dense network 20 days post replating (Figure S3B). To investigate whether this might be used as an axotomy model, we needed to answer two questions: (1) are the neurons 20 days post replating truly postmitotic SNs or were they replenished from remaining SOX10+ progenitor cells? and (2) are the replated neurons electrically active? Our cultures have very few SOX10+ cells at days 30, 40, and 50 (Figure S3C). However, we found that in the normal course of differentiation (when replating at day 12), BRN3A peaks 10–20 days post replating and is downregulated in mature SNs (Figure S3D). Thus, we hypothesized that if new SNs were being generated from SOX10+ cells at the day-50 replating, they would show high BRN3A levels 20 days later (day 70). This, however, was not the case (Figure S3B, bottom lane). Also, SNs replated at day 30 are electrically active 25 days later (Figure S3G). In sum, we show the efficient generation of functional SNs that can be maintained for long-term studies.
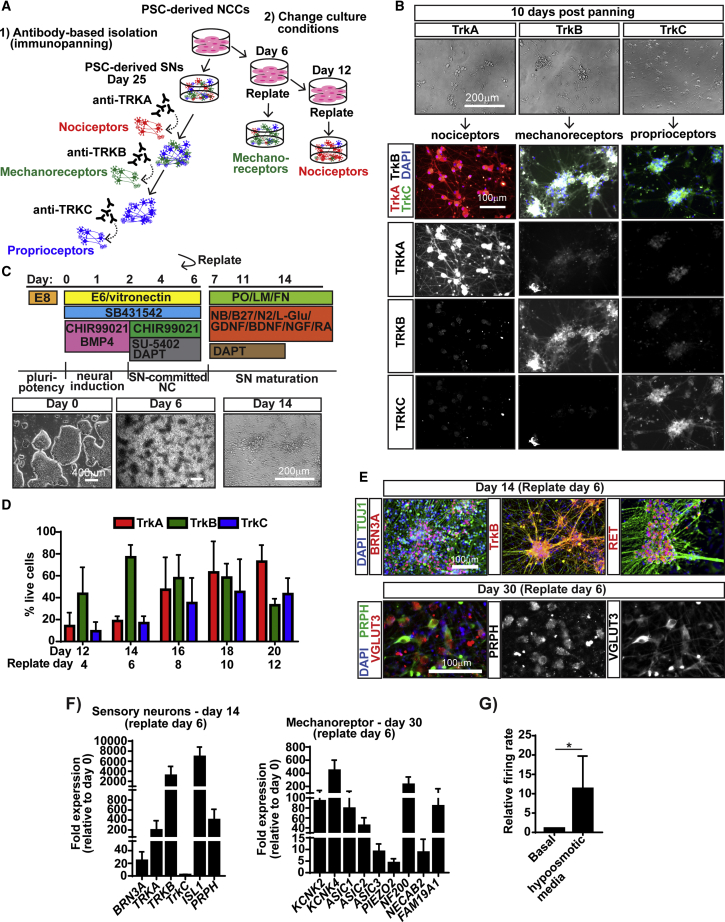
Strategies for SN-subtype enrichment
Contrary to previous reports (Alshawaf et al., 2018; Chambers et al., 2012; Schrenk-Siemens et al., 2015), our protocol generates all three SN subtypes (Figure 1J). Thus, we searched for methods to enrich distinct SN subtypes from our bulk cultures. We first tried using NGF, BDNF, and NT3 to promote differentiation of nociceptors, mechanoreceptors, and proprioceptors, respectively (Bibel and Barde, 2000) (Figure S4A). FACS analysis on day 20, however, indicated no subtype enrichment (Figure S4B). We next tried to use FACS to isolate SN subtypes (Figure S4C). Unfortunately, the sorted cells failed to survive, a known phenomenon. Finally, we adapted immunopanning (Sloan et al., 2017), a gentle antibody-based purification technique, to segregate the different SN subtypes (Figure 3A, left). This method allows the binding of specific cells from a mix to a dish precoated with antibodies against cell-surface proteins. The cells of interest attach to the antibody and the following wash and dissociation steps are much gentler compared with FACS. We showed that TRKA+ nociceptors, TRKB+ mechanoreceptors, and TRKC+ proprioceptors could be isolated from bulk day-25 SN cultures (Figures 3B and S4F). We also found isolated TRKB+/C+ SNs, so future improvement of this method will need to address its efficiency. Thus, we further searched for a technically easier approach to enrich for specific SN subtypes by changing the culture conditions in our protocol (Figure 3A, right). SNs develop in vivo in two waves marked by NGN2 followed by NGN1 (Ma et al., 1999). In our SN protocol, we found TUJ1+ neurons at day 8 (Figure S4D) compared with the control NCC protocol. This coincided with NGN2 expression early (day 3) followed by NGN1 expression later (day 8, Figure S4E), suggesting that both SN development waves occur. Based on these results, we tested whether early replating (which pushes neuron differentiation) would promote NGN2-mediated mechanoreceptor and proprioceptor enrichment (Figures 1F and 3C). We replated the SN-specified NCCs at days 4, 6, 8, 10, and 12 and assessed TRK expression by FACS 8 days later (Figure 3D). We found a dramatic enrichment (80%) of TRKB+ mechanoreceptors when replating was done at day 6, supporting our hypothesis that we catch the first wave of SN development. However, we were not able to generate TRKC+ proprioceptors at a higher efficiency than in the original bulk protocol (∼40%) (Figure 3D). The mechanoreceptor-enriched cultures expressed pan-SN markers (BRN3A, TUJ1, PRPH, VGLUT3, and ISL1, Figures 3E, 3F, and S4G) as well as mechanoreceptor-specific markers TRKB, RET (Figure 3E), KCKN4, and FAM19A1 (Figure 3F), and responded to hypo-osmotic pressure (Figure 3G) at a higher level compared with bulk cultures (Figure 2I). In sum, we show two methods (immunopanning and catching the first SN development wave) to enrich for specific SN subtypes from our cultures.
Figure 3.
Approaches for SN-subtype enrichment
(A) Schematics of proposed enrichment methods.
(B) Isolation of SN subtypes by immunopanning. Cells on day 25 were immunopanned using anti-TRKA, -TRKB, or -TRKC antibodies. Cells were cultured for 10 days (representative BF images, top) followed by IF for TRKA, TRKB, and TRKC (bottom).
(C) Timeline of culture conditions to promote mechanoreceptor enrichment (top). Representative BF images of each day (below).
(D) Flow-cytometry quantification of neurons expressing TRK receptors. The days when cells were replated and analyzed are indicated.
(E and F) SNs differentiated from SN-biased NCC replated on day 6 express mechanoreceptor markers TRKB, RET, BRN3A, TUJ1, VGLUT3, and PRPH by IF (E) and qRT-PCR (F).
(G) SN activity is modulated by hypo-osmotic medium. SNs were incubated for 1 min prior to data acquisition.
Data are normalized to untreated control (basal). In all experiments n > 4. Graphs show mean ± SD. ∗p < 0.05 using t test with Welch's correction. See also Figure S4.
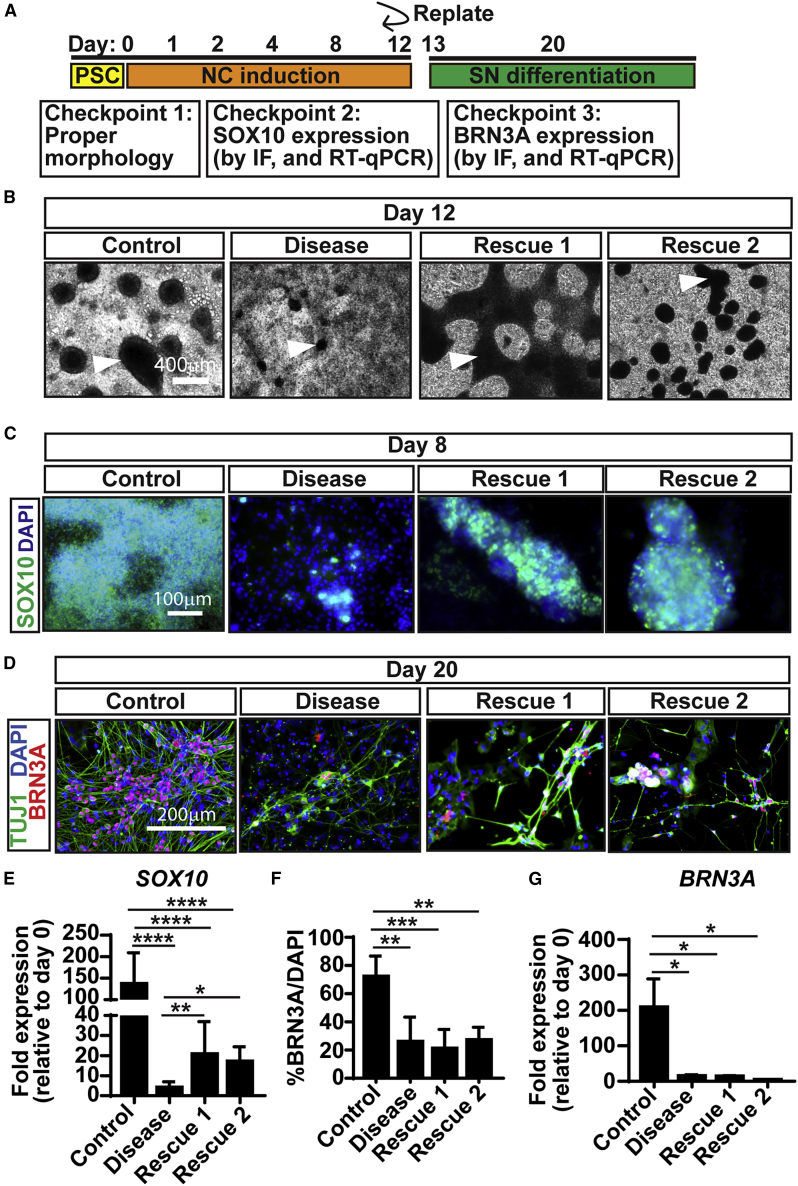
Efficient SN differentiation of patient-derived iPSCs
For successful disease modeling, drug screening, and future cell-replacement approaches it is imperative that differentiation protocols work appropriately with iPSCs. Thus, here we focused on familial dysautonomia (FD), a devastating disorder caused by a mutation in the ELP1 gene that decreases the numbers of SNs (Norcliffe-Kaufmann et al., 2017). We differentiated previously characterized (Zeltner et al., 2016) healthy control iPSCs (Control), FD patient-derived iPSCs (Disease), and FD iPSCs where the ELP1 mutation was rescued using CRISPR/Cas9 (Rescue 1 and Rescue 2, i.e., two clones) into SNs (Table S2). To confirm that the differentiation was occurring properly, we tested a series of quality control checkpoints (Figure 4A). Checkpoint 1: proper NCC generation is indicated by dense ridges (arrow) in Control and Rescue 1/2, but is diminished in Disease (Figure 4B). Checkpoint 2: these NCCs express high levels of SOX10 in Control, are somewhat diminished in Rescue 1/2, and drastically reduced in Disease (Figures 4C and 4E). Checkpoint 3: control iPSCs generated SNs properly (∼75% BRN3A+ SNs) while Disease and Rescue 1/2 were deficient (∼25%, Figures 4D and 4F). Overall, rescuing ELP1 increased the number of NCCs but did not rescue the SN phenotype, suggesting that ELP1 is important for NCC development, but another mechanism is responsible for the SN defect in FD, agreeing with previous results (Zeltner et al., 2016). Together, these results show that our protocol (1) can be utilized for both hESCs and iPSCs and (2) is capable of recapitulating disease phenotypes observed in FD.
Figure 4.
Differentiation of SNs from iPSCs
(A) Timeline of differentiation highlighting quality control checkpoints to ensure a successful differentiation.
(B) Checkpoint 1. Representative BF images of NCCs (arrowheads) derived from healthy (control), FD (disease), and FD ELP1-rescue (Rescue 1 and Rescue 2) iPSCs.
(C and E) Checkpoint 2. SOX10 expression by IF (C) and qRT-PCR (E) on day 8.
(D, F and G) Checkpoint 3. BRN3A expression by IF (D) and its image quantification (F) and qRT-PCR (G) on day 20.
In all experiments n = 3. Graphs show mean ± SD. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001 using one-way ANOVA.
Discussion
Here we describe an easy, efficient, and versatile method to derive SNs from hPSCs (Table S1). Our differentiation is a chemically defined monolayer culture, which bypasses the need to generate neurospheres (Alshawaf et al., 2018; Boisvert et al., 2015; Schrenk-Siemens et al., 2015; Young et al., 2014), with only one replating step (day 12) (Alshawaf et al., 2018; Boisvert et al., 2015; Young et al., 2014). We provide culturing aids to reduce variability by using the CryoPause method (Wong et al., 2017) as well as a mid-differentiation freezing option (Figure S2). By day 20, we reach an efficiency of 60%–70% of all cells being BRN3A+ and ∼99% of those being TUJ1+ SNs (Figure 1H). It has been shown that H9 cells are prone to differentiate into SNs. Interestingly, SN differentiation from control iPSCs was more efficient (∼70%) compared with H9 cells, suggesting that the efficiency observed is due to our protocol and not any intrinsic mechanism in H9 cells.
By day 20, our SN cultures are composed of all three SN subtypes—nociceptors, mechanoreceptors, and proprioceptors (Figure 1J)—at a ratio of approximately 2:1:1, which correlates with the relative SN distribution in the human DRG in vivo (Ernsberger, 2009). Accordingly, between days 30 and 50, the expression level of TRKA declines, which mimics what happens in vivo (Figure 1J) (Ernsberger, 2009). Thus, we are providing an in vitro model to study the neuronal interactions within ganglia. We report expression of SN progenitor (Figures 1I and 1K) and mature markers (Figures 2B–2D) of each subtype. For example, we found high expression of RET (co-expressed in 50% of TRKA+ human SNs) (Figure 1K) and TRPA1 (expressed in TRPV1+ TRKA+ human SNs) (Figure 2B), as well as the Na channels SCN8A-11A, agreeing with a previous report in human DRG (Rostock et al., 2018). Lastly, our SNs are functionally active and respond to specific stimuli (nociceptive agonists or hypo-osmotic pressure) (Figures 2F–2I).
Previous reports have shown generation of SN subtypes (Alshawaf et al., 2018), albeit at a lower efficiency (up to 11% of BRN3A+/ISL1+ cells), and the percentage of TRKA+, TRKB+, and TRKC+ SNs range from 17% to 25%. Another report (Schrenk-Siemens et al., 2015) showed directed differentiation of hPSCs to mechanoreceptors reaching up to ∼28% when combined with mitotic blockers and ∼18% upon lentivirus-mediated overexpression of NGN2, which may blur results if used for modeling SN disorders. Our bulk SN protocol reaches better efficiencies. Additionally, our efforts to develop methods for the enrichment of each SN subtype provides the possibility to generate nociceptors at ∼90%, proprioceptors at ∼38% (by FACS/immunopanning, Figure S4C), and mechanoreceptors at ∼75% (by day-6 replating). In our molecular characterization of these mechanoreceptors, we found expression of NECAB2, FAM19A1, KCNK2, KCNK4, and PIEZO2 (Figure 3F), similar to previous reports (Alshawaf et al., 2018; Nickolls et al., 2020). We also found the ASIC family of genes, which have a modulatory role in mechanotransduction (Delmas et al., 2011), as well as NF200 expression. These genes were found to be expressed in mouse DRG, but human DRG gene expression might be different. For example, evidence from human DRG shows that NF200 is expressed in nearly all SNs (Rostock et al., 2018). Future transcriptomics analysis of our hPSC-derived DRG-like culture might provide more clarity regarding these species' differences.
Lastly, a recent study showed a method to generate proprioceptors (70% of SNs) from SOX10+ NCCs using high concentrations of NT3 and low concentrations of NGF, GDNF, and BDNF (Dionisi et al., 2020). The authors also see increases of NGN1 and NGN2 expression. It is possible that by modifying the growth factor concentrations in our system we could enrich our culture with TRKC+ proprioceptors.
We show that the SNs generated by our protocol can be replated in late stages of differentiation and still be viable and functional (Figure S4). Our initial hypothesis was that these SNs were differentiated from SOX10+ progenitor cells. In this scenario, we would see BRN3A expression within 30 days of replating based on our qRT-PCR analysis (Figure S4E). However, because we do not see BRN3A expression 20 days post replating (Figure S4F), we hypothesize that we are replating fully differentiated SNs, which are also functional by electrophysiology analysis (Figure S4G). This suggests that our proposed protocol could be used as a model to study peripheral axotomy in vitro.
In conclusion, we developed a versatile protocol for the generation of functional SNs that mimic the proportions of subtypes in the DRG. Several methods can be employed to isolate each of the specific SN subtypes, nociceptors, mechanoreceptors, and proprioceptors from these cultures. This protocol can be adapted for a myriad of research interests ranging from studying the human DRG to investigating PNS diseases such as FD using patient-derived iPSCs that affect either all SN subtypes or each of the individual subtypes. Our work can be used to further advance numerous fields interested in studying the functions of, and diseases associated with, human SNs.
Experimental procedures
Reagents, companies, and catalog numbers are listed in Table S3. All antibodies are listed in Table S4.
hPSC and iPSC maintenance
Human embryonic stem cells (WA-09, WiCell) were grown at 37°C/5% CO2 and fed daily with Essential 8 Medium (E8) + Supplement on dishes coated with vitronectin (5 μg/mL, 1 h at room temperature). For splitting, H9 colonies were washed with PBS and treated with 0.5 mM EDTA in PBS with 3.08 M NaCl for 2 min at 37°C. Cells were resuspended in E8 + Supplement and split at a ratio of 1:10. Control and FD iPSCs were previously characterized (Zeltner et al., 2016) and maintained under the same conditions.
Neural crest and sensory neuron culture conditions
Prior to differentiation, plates were coated with Geltrex at 1:100 dilution and stored at 4°C overnight, or vitronectin. The next day, hPSCs were harvested using EDTA for 15 min and plated at a density of 200,000 cells/cm2. On day 0 (day of plating) to day 1 of the differentiation, cultures were fed with Essential 6 Medium (E6) containing 10 μM SB431542, 1 ng/mL bone morphogenetic protein 4, 300 nM CHIR99021, and 10 μM Y-27632. On day 2, NCC D2+ medium was made with E6 containing 10 μM SB431542 and 1.5 μM or 0.75 μM CHIR99021. For SN induction, cells were maintained in D2-12 medium (E6 + 10 μM SB431542, 0.75 μM CHIR99021, 2.5 μM SU5402, and 2.5 μM DAPT). Cells were fed every 48 h between day 2 and day 12.
On day 12, cells were replated at a density of 250,000 cells/cm2 onto plates coated with 15 μg/mL poly-L-ornithine hydrobromide, 2 μg/mL mouse laminin-1, and 2 μg/mL human fibronectin (PO/LM/FN). Cells were dissociated with Accutase for 20 min, washed with PBS, and resuspended in SN medium: neurobasal medium containing N2, B-27, 2 mM L-glutamine, 20 ng/mL GDNF, 20 ng/mL BDNF, 25 ng/mL NGF, 600 ng/mL laminin-1 and fibronectin, 1 μM DAPT, and 0.125 μM retinoic acid. NT-3 (20 ng/mL) was added where indicated. Cells were fed every 2–3 days through day 20. On day 20, DAPT was removed and cells were fed every 3–4 days.
Flow cytometry
Cells were dissociated with Accutase for 20 min and washed in Flow buffer (DMEM, 2% fetal bovine serum [FBS], and 1 mM L-glutamine). Cells were centrifuged at 200 × g for 4 min and resuspended in cold PBS, counted, diluted to a concentration of 1 × 106 cells/100 μL, and incubated for 20 min (CD49D) or 1 h (TRKA, TRKB, TRKC) with conjugated antibody. Cells were then washed twice in Flow buffer and incubated with DAPI (1:1,000) in 300 μL of Flow buffer for 5 min. Cells were filtered and analyzed using a Cytoflex S (Beckman). Analysis was carried out using FlowJo.
Electrophysiology
Experiments were performed using a Maestro Pro (Axion Biosystems) MEA system. CytoView MEA 96 plates containing eight embedded electrodes per well were coated with PO/LM/FN and seeded with NCCs (day 6 or 12) or SNs (day 30). Repeated recordings were made every 5 days at 37°C with a sampling frequency of 12.5 kHz for 5 min. Recordings from at least six wells were averaged per reading. Bursts were detected using Inter-Spike Interval. Capsaicin, WIN 55212, and pregnenolone were titrated and incubated for 1 min before data acquisition. Hypo-osmotic medium was obtained by mixing SN medium with sterile water in a 45:55 ratio.
Immunopanning
Three 10-cm Petri dishes were coated with 40 μL of anti-mouse immunoglobulin G (IgG) or anti-goat IgG in 13 mL of 50 mM Tris-HCl (pH 9.5) at 4°C overnight. The dishes were washed three times with PBS and incubated with 6.6 μg of TRKA, TRKB, or TRKC antibody in 5 mL of 0.2% BSA + 10 μg DNAse in PBS for at least 2 h at room temperature. Day-25 SNs were washed with PBS, incubated with Accutase for 45 min at 37°C, collected in PBS, and centrifuged at 200 × g for 4 min. The pellet was resuspended in Panning buffer (20% FBS, 10 μg of DNAse, and 10 μM Y-27632 in PBS) using a p1000 micropipette and passed through a 0.22-μm filter, after which the cells were counted. The TRKA panning dish was washed three times with PBS. Cells were added and incubated for 15 min at room temperature and transferred to the next panning dish (TRKB), previously washed three times with PBS. This was repeated for the final panning dish. Positive selection plates were carefully washed with PBS about seven times. The dish was then incubated with 5 mL of dissociation buffer (1 mL of Accutase in 14 mL of 1× Earle’s balanced salt solution) at 37°C for 5 min. Cells were dislodged with 30% FBS in 50:50 neurobasal medium/DMEM, collected, and centrifuged at 200 × g for 4 min. The pellet was resuspended in SN differentiation medium + 10 μM Y-27632 and plated in 96-well plates, or concentrated in a 10-μL drop and plated in dried 24-well plates coated with PO/LM/FN. The medium was replaced the following day.
Author contributions
N.Z. and K.S.-D. conceived and designed the experiments; K.S.-D., J.R.S., and H.U. conducted experiments; K.S.-D., J.R.S., and N.Z. analyzed and interpreted data; J.R.S., K.S.-D., and N.Z. wrote the manuscript; N.Z. provided mentoring and financial and administrative support, and approved the final version of the manuscript.
Conflicts of interests
The authors declare no competing interests. This work is linked to the patent “Compositions and methods for making sensory neurons,” serial number 63/1279, held by the University of Georgia.
Acknowledgments
We wish to thank Dr. Steven Sloan for his feedback in the optimization of the immunopanning protocol. We also thank Dr. Lorenz Studer for his support at the initial stages of this work. This work was funded by Faculty Start-up from the University of Georgia to N.Z.
Published: February 4, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.stemcr.2021.01.001.
Supplemental information
References
- Alshawaf A.J., Viventi S., Qiu W., D'Abaco G., Nayagam B., Erlichster M., Chana G., Everall I., Ivanusic J., Skafidas E., Dottori M. Phenotypic and functional characterization of peripheral sensory neurons derived from human embryonic stem cells. Sci. Rep. 2018;8:603. doi: 10.1038/s41598-017-19093-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibel M., Barde Y.A. Neurotrophins: key regulators of cell fate and cell shape in the vertebrate nervous system. Genes Dev. 2000;14:2919–2937. doi: 10.1101/gad.841400. [DOI] [PubMed] [Google Scholar]
- Boisvert E.M., Engle S.J., Hallowell S.E., Liu P., Wang Z.W., Li X.J. The specification and maturation of nociceptive neurons from human embryonic stem cells. Sci. Rep. 2015;5:16821. doi: 10.1038/srep16821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caterina M.J., Schumacher M.A., Tominaga M., Rosen T.A., Levine J.D., Julius D. The capsaicin receptor: a heat-activated ion channel in the pain pathway. Nature. 1997;389:816–824. doi: 10.1038/39807. [DOI] [PubMed] [Google Scholar]
- Chambers S.M., Qi Y., Mica Y., Lee G., Zhang X.J., Niu L., Bilsland J., Cao L., Stevens E., Whiting P. Combined small-molecule inhibition accelerates developmental timing and converts human pluripotent stem cells into nociceptors. Nat. Biotechnol. 2012;30:715–720. doi: 10.1038/nbt.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Crozé N., Maczkowiak F., Monsoro-Burq A.H. Reiterative AP2a activity controls sequential steps in the neural crest gene regulatory network. Proc. Natl. Acad. Sci. U S A. 2011;108:155–160. doi: 10.1073/pnas.1010740107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmas P., Hao J., Rodat-Despoix L. Molecular mechanisms of mechanotransduction in mammalian sensory neurons. Nat. Rev. Neurosci. 2011;12:139–153. doi: 10.1038/nrn2993. [DOI] [PubMed] [Google Scholar]
- Dionisi C., Rai M., Chazalon M., Schiffmann S.N., Pandolfo M. Primary proprioceptive neurons from human induced pluripotent stem cells: a cell model for afferent ataxias. Sci. Rep. 2020;10:7752. doi: 10.1038/s41598-020-64831-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly C.R., Gabreski N.A., Suh E.B., Chowdhury M., Pierchala B.A. Non-canonical Ret signaling augments p75-mediated cell death in developing sympathetic neurons. J. Cell Biol. 2018;217:3237–3253. doi: 10.1083/jcb.201703120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernsberger U. Role of neurotrophin signalling in the differentiation of neurons from dorsal root ganglia and sympathetic ganglia. Cell Tissue Res. 2009;336:349–384. doi: 10.1007/s00441-009-0784-z. [DOI] [PubMed] [Google Scholar]
- Fattahi F., Steinbeck J.A., Kriks S., Tchieu J., Zimmer B., Kishinevsky S., Zeltner N., Mica Y., El-Nachef W., Zhao H. Deriving human ENS lineages for cell therapy and drug discovery in Hirschsprung disease. Nature. 2016;531:105–109. doi: 10.1038/nature16951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein R.S., Pomp O., Brokhman I., Ziegler L. Generation of neural crest cells and peripheral sensory neurons from human embryonic stem cells. Methods Mol. Biol. 2010;584:283–300. doi: 10.1007/978-1-60761-369-5_15. [DOI] [PubMed] [Google Scholar]
- Huang E.J., Reichardt L.F. Neurotrophins: roles in neuronal development and function. Annu. Rev. Neurosci. 2001;24:677–736. doi: 10.1146/annurev.neuro.24.1.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Y., Hao H., Reynolds K., McMahon M., Zhou C.J. Wnt signaling in neural crest ontogenesis and oncogenesis. Cells. 2019;8:1173. doi: 10.3390/cells8101173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lallemend F., Ernfors P. Molecular interactions underlying the specification of sensory neurons. Trends Neurosci. 2012;35:373–381. doi: 10.1016/j.tins.2012.03.006. [DOI] [PubMed] [Google Scholar]
- Ma Q., Fode C., Guillemot F., Anderson D.J. Neurogenin1 and neurogenin2 control two distinct waves of neurogenesis in developing dorsal root ganglia. Genes Dev. 1999;13:1717–1728. doi: 10.1101/gad.13.13.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marmigère F., Ernfors P. Specification and connectivity of neuronal subtypes in the sensory lineage. Nat. Rev. Neurosci. 2007;8:114–127. doi: 10.1038/nrn2057. [DOI] [PubMed] [Google Scholar]
- Menendez L., Yatskievych T.A., Antin P.B., Dalton S. Wnt signaling and a Smad pathway blockade direct the differentiation of human pluripotent stem cells to multipotent neural crest cells. Proc. Natl. Acad. Sci. U S A. 2011;108:19240–19245. doi: 10.1073/pnas.1113746108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mica Y., Lee G., Chambers S.M., Tomishima M.J., Studer L. Modeling neural crest induction, melanocyte specification, and disease-related pigmentation defects in hESCs and patient-specific iPSCs. Cell Rep. 2013;3:1140–1152. doi: 10.1016/j.celrep.2013.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namer B., Schmidt D., Eberhardt E., Maroni M., Dorfmeister E., Kleggetveit I.P., Kaluza L., Meents J., Gerlach A., Lin Z. Pain relief in a neuropathy patient by lacosamide: proof of principle of clinical translation from patient-specific iPS cell-derived nociceptors. EBioMedicine. 2019;39:401–408. doi: 10.1016/j.ebiom.2018.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nehme R., Zuccaro E., Ghosh S.D., Li C., Sherwood J.L., Pietilainen O., Barrett L.E., Limone F., Worringer K.A., Kommineni S. Combining NGN2 programming with developmental patterning generates human excitatory neurons with NMDAR-mediated synaptic transmission. Cell Rep. 2018;23:2509–2523. doi: 10.1016/j.celrep.2018.04.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickolls A.R., Lee M.M., Espinoza D.F., Szczot M., Lam R.M., Wang Q., Beers J., Zou J., Nguyen M.Q., Solinski H.J. Transcriptional programming of human mechanosensory neuron subtypes from pluripotent stem cells. Cell Rep. 2020;30:932–946.e7. doi: 10.1016/j.celrep.2019.12.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norcliffe-Kaufmann L., Slaugenhaupt S.A., Kaufmann H. Familial dysautonomia: history, genotype, phenotype and translational research. Prog. Neurobiol. 2017;152:131–148. doi: 10.1016/j.pneurobio.2016.06.003. [DOI] [PubMed] [Google Scholar]
- Pla P., Monsoro-Burq A.H. The neural border: induction, specification and maturation of the territory that generates neural crest cells. Dev. Biol. 2018;444:S36–S46. doi: 10.1016/j.ydbio.2018.05.018. [DOI] [PubMed] [Google Scholar]
- Pomp O., Brokhman I., Ben-Dor I., Reubinoff B., Goldstein R.S. Generation of peripheral sensory and sympathetic neurons and neural crest cells from human embryonic stem cells. Stem Cells. 2005;23:923–930. doi: 10.1634/stemcells.2005-0038. [DOI] [PubMed] [Google Scholar]
- Poole K., Herget R., Lapatsina L., Ngo H.D., Lewin G.R. Tuning Piezo ion channels to detect molecular-scale movements relevant for fine touch. Nat. Commun. 2014;5:3520. doi: 10.1038/ncomms4520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quallo T., Alkhatib O., Gentry C., Andersson D.A., Bevan S. G protein βγ subunits inhibit TRPM3 ion channels in sensory neurons. ELife. 2017;6:e26138. doi: 10.7554/eLife.26138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rostock C., Schrenk-Siemens K., Pohle J., Siemens J. Human vs. mouse nociceptors - similarities and differences. Neuroscience. 2018;387:13–27. doi: 10.1016/j.neuroscience.2017.11.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrenk-Siemens K., Wende H., Prato V., Song K., Rostock C., Loewer A., Utikal J., Lewin G.R., Lechner S.G., Siemens J. PIEZO2 is required for mechanotransduction in human stem cell-derived touch receptors. Nat. Neurosci. 2015;18:10–16. doi: 10.1038/nn.3894. [DOI] [PubMed] [Google Scholar]
- Simões-Costa M., Bronner M.E. Establishing neural crest identity: a gene regulatory recipe. Development. 2015;142:242–257. doi: 10.1242/dev.105445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan S.A., Darmanis S., Huber N., Khan T.A., Birey F., Caneda C., Reimer R., Quake S.R., Barres B.A., Paşca S.P. Human astrocyte maturation captured in 3D cerebral cortical spheroids derived from pluripotent stem cells. Neuron. 2017;95:779–790.e6. doi: 10.1016/j.neuron.2017.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Dykes I.M., Liang X., Eng S.R., Evans S.M., Turner E.E. A central role for Islet1 in sensory neuron development linking sensory and spinal gene regulatory programs. Nat. Neurosci. 2008;11:1283–1293. doi: 10.1038/nn.2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchieu J., Zimmer B., Fattahi F., Amin S., Zeltner N., Chen S., Studer L. A modular platform for differentiation of human PSCs into all major ectodermal lineages. Cell Stem Cell. 2017;21:399–410.e7. doi: 10.1016/j.stem.2017.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Théveneau E., Duband J.L., Altabef M. Ets-1 confers cranial features on neural crest delamination. PLoS One. 2007;2:e1142. doi: 10.1371/journal.pone.0001142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong K.G., Ryan S.D., Ramnarine K., Rosen S.A., Mann S.E., Kulick A., De Stanchina E., Müller F.J., Kacmarczyk T.J., Zhang C. CryoPause: a new method to immediately initiate experiments after cryopreservation of pluripotent stem cells. Stem Cell Reports. 2017;9:355–365. doi: 10.1016/j.stemcr.2017.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young G.T., Gutteridge A., Fox H., Wilbrey A.L., Cao L., Cho L.T., Brown A.R., Benn C.L., Kammonen L.R., Friedman J.H. Characterizing human stem cell–derived sensory neurons at the single-cell level reveals their ion channel expression and utility in pain research. Mol. Ther. 2014;22:1530–1543. doi: 10.1038/mt.2014.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeltner N., Fattahi F., Dubois N.C., Saurat N., Lafaille F., Shang L., Zimmer B., Tchieu J., Soliman M.A., Lee G. Capturing the biology of disease severity in a PSC-based model of familial dysautonomia. Nat. Med. 2016;22:1421–1427. doi: 10.1038/nm.4220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmer B., Ewaleifoh O., Harschnitz O., Lee Y.S., Peneau C., McAlpine J.L., Liu B., Tchieu J., Steinbeck J.A., Lafaille F. Human iPSC-derived trigeminal neurons lack constitutive TLR3-dependent immunity that protects cortical neurons from HSV-1 infection. Proc. Natl. Acad. Sci. U S A. 2018;115:E8775–E8782. doi: 10.1073/pnas.1809853115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou D., Silvius D., Fritzsch B., Xu P.X. Eya1 and Six1 are essential for early steps of sensory neurogenesis in mammalian cranial placodes. Development. 2004;131:5561–5572. doi: 10.1242/dev.01437. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.