Abstract
Claudin-11, a tight junction protein, is indispensable in the formation of the radial component of myelin. Here, we report de novo stop-loss variants in the gene encoding claudin-11, CLDN11, in three unrelated individuals presenting with an early-onset spastic movement disorder, expressive speech disorder and eye abnormalities including hypermetropia. Brain MRI showed a myelin deficit with a discrepancy between T1-weighted and T2-weighted images and some progress in myelination especially involving the central and peripheral white matter. Exome sequencing identified heterozygous stop-loss variants c.622T>C, p.(*208Glnext*39) in two individuals and c.622T>G, p.(*208Gluext*39) in one individual, all occurring de novo. At the RNA level, the variant c.622T>C did not lead to a loss of expression in fibroblasts, indicating this transcript is not subject to nonsense-mediated decay and most likely translated into an extended protein. Extended claudin-11 is predicted to form an alpha helix not incorporated into the cytoplasmic membrane, possibly perturbing its interaction with intracellular proteins. Our observations suggest that stop-loss variants in CLDN11 expand the genetically heterogeneous spectrum of hypomyelinating leukodystrophies.
Keywords: CLDN11, hypomyelinating leukodystrophy, tight junction, exome, stop-loss
Riedhammer et al. present three unrelated individuals with a hypomyelinating leukodystrophy harbouring de novo stoploss variants in CLDN11 identified by exome sequencing. The individuals share a phenotype consisting of an early-onset spastic movement disorder, speech delay and eye abnormalities including hypermetropia.
Introduction
Impaired development of myelin is the hallmark of hypomyelinating leukodystrophies, a group of genetically heterogeneous disorders (Pouwels et al., 2014). MRI led to the delineation of distinct disease entities around 15 years ago. As a result of the development of new methods of molecular genetic testing, the number of genetically defined leukodystrophies has increased in recent years (van der Knaap et al., 2019). Since the association of variants in PLP1 with Pelizaeus-Merzbacher disease [PMD (MIM 312080)], more than 10 different hypomyelinating leukodystrophies with more or less characteristic clinical features, such as 4H leukodystrophy, have been described and received a sequential ‘HLD-’ number in OMIM (Vanderver et al., 2015). Increasing knowledge on the diversity of white matter pathology underlying leukodystrophies confirms that all its constituents determine white matter integrity and function. In the past decade, it became evident that not only variants in genes encoding structural oligodendrocyte proteins may be involved in hypomyelination leukodystrophies, but also variants in genes coding for transcription factors controlling expression of structural genes as well as variants in other genes with cellular roles less obviously linked with oligodendrocyte function (van der Knaap and Bugiani, 2018).
The correct formation of myelin plays a pivotal role in proper function of the CNS. The myelin sheath is assembled in multiple layers of lipid-rich membranes encasing the axon. This sheath is crucial for saltatory conduction along axons allowing the emergence of action potentials only at the nodes of Ranvier (Denninger et al., 2015). The principal component of myelin integrity in the CNS is proteolipid protein (PLP), a lipid-associated tetraspan protein that is widely expressed in oligodendrocytes (Luders et al., 2019). Tight junctions are further important elements of the myelin sheath. They structure the radial component of myelin and act as diffusion barriers. Claudin-11 (CLDN11, 3q26.2), a tetraspan protein like PLP and a member of the claudin tight junction family, is an indispensable part of the radial component of myelin (Denninger et al., 2015).
Here, we describe three unrelated individuals with a hypomyelinating leukodystrophy, in whom exome sequencing detected heterozygous de novo stop-loss variants in CLDN11.
Materials and methods
Consent
Written informed consent for sequencing and publication in a scientific journal was obtained by the legal guardians of all three individuals according to the Declaration of Helsinki and this study has been approved by the ethical committees of the respective institutions in Munich, Warsaw and Vancouver.
Next-generation sequencing
Trio exome sequencing (exome sequencing of index and parents) was performed in Individuals 1 and 3. Proband-only exome sequencing with subsequent targeted-sequencing of the CLDN11 stop-loss variant in the parents was performed in Individual 2 (see Supplementary material for further details).
RNA sequencing
RNA from fibroblasts was sequenced as described in Kremer et al. (2017) but as strand-specific mRNA according to the TruSeq Stranded mRNA Sample Prep Guide on a NovaSeq 6000 platform (Illumina) (Kremer et al., 2017). Next, RNA sequencing data were analysed to detect differentially expressed genes compared to 156 control fibroblasts (Supplementary material).
Protein visualization and modelling
The protein sequence of extended claudin-11 was visualized using the free web-based tool Protter (http://wlab.ethz.ch/protter/start/) (Omasits et al., 2014). Modelling of tertiary structure of added amino acids was done with PEP-FOLD and cytoplasmic localization of added amino acids was predicted with TMHMM (all for variant c.622T>C) (Krogh et al., 2001; Thevenet et al., 2012; Shen et al., 2014).
Cranial MRI interpretation
MRIs were analysed visually and described with respect to myelin specific changes. For standardized evaluation of myelination, T2-weighted images were scored with a semiquantitative myelination score with a maximum score of 36 characterizing complete myelination (Plecko et al., 2003).
Data availability
All data can be shared. Results from molecular genetic analysis and RNA sequencing are available at the authors’ institutions in Munich, Warsaw and Vancouver.
Results
Clinical synopsis
All three affected individuals were of non-Finnish European descent.
In Individual 1 (female), spasticity predominantly of the lower limbs and truncal hypotonia became apparent in the first year of life. Walking independently never exceeded a few metres and she had knee and ankle contractures. She presented with nystagmus, a hyperopic astigmatism of +2 dpt and strabismus, surgically treated at the age of 3 years.
Individual 2 (male) showed first signs of spasticity of all extremities with truncal hypotonia at the age of 4 months. Over time he developed muscular rigidity in the trunk, shoulder girdle and in the upper extremities. Walking without help became possible only over a couple months at the age of 5 years. He showed oral hypotonia with severe drooling and presented with nystagmus, early-onset strabismus (apparent already at age 4 months), which was multiply surgically treated, and hypermetropia of +4.5/+4.25 dpt (at age 8 years).
In Individual 3 (male), a spastic movement disorder with increased tone of the extremities (pronounced in the lower extremities), arching of neck, and plagiocephalus, became apparent at age 11 months. Walking without help was never achieved. He had oral hypotonia with constant drooling and presented with strabism with exotropic eye alignment, a fine nystagmus, decreased visual acuity near and far and a hyperopic astigmatism +3.25 dpt OD and +2.75 dpt OS.
All showed a pronounced delay in speech development, reached the ability to speak in sentences, but were all severely dysarthric. IQ was not tested in any of them, but intellectual disability seems to be mild at most. Hearing was unaffected in all and no involvement of internal organs occurred in any.
Electrophysiology revealed disturbed sensory evoked potentials in individual 1 (not performed in Individuals 2 and 3). Peripheral nerve conduction studies were normal in all three.
Broad diagnostic work-up included genetic and metabolic tests in all individuals, which remained without any indicative findings despite repeatedly elevated blood lactate in Individual 3 (2.3–6.1 mmol/l).
Table 1 provides geno- and phenotypic characteristics of the three individuals. Extended case reports are provided as case reports in the Supplementary material. See also Supplementary Video 1 for Individual 1.
Table 1.
Phenotype and genotype of the individuals
| Individual 1 (German) | Individual 2 (Polish) | Individual 3 (Canadian) | |
|---|---|---|---|
| Genetic and proband data | |||
| Variant in CLDN11, chromosomal position (NC_000012.11, hg19) | chr3:g.170150542T>C | chr3:g.170150542T>G | chr3:g.170150542T>C |
| Variant in CLDN11, cDNA and protein position (NM_005602.5, NP_005593.2) |
c.622T>C p.(*208Glnext*39) |
c.622T>G p.(*208Gluext*39) |
c.622T>C p.(*208Glnext*39) |
| Inheritance | De novo | De novo | De novo |
| Gender | Female | Male | Male |
| Ethnicity | Non-Finnish European (German; family from Germany) | Non-Finnish European (Polish; family from Poland) | Non-Finnish European (Canadian; maternal family from former Yugoslavia and Scotland, paternal family from Russia, Norway and Germany) |
| Age at last examination | 13 years | 8 years | 6.5 years |
| Pyramid signs | |||
| Babinski (yes/no) | + | + | + |
| Lower extremity tendon reflexes | ++ | +++ | ++ |
| Axial/trunk muscle tone | Hypotonic | Rigid | Normal |
| Lower extremity muscle tone | High | High | High |
| Upper extremity muscle tone | High | High / rigid | Normal-high |
| Motor developmental milestones | |||
| Sitting without support | Age 4.5 years | Age 3.5 years | Age 4 years |
| Walking without assistance | Few metres (at home) | Few metres age 5 years, lost ability to walk age 5.5 years | Never learned, stand shortly unsupported age 6 years |
| GMFCS level | III | III | III |
| Skeletal findings | |||
| Scoliosis | − | − | − |
| Contractures | + | + | + |
| Other | Flat feet | Abnormal shoulder belt, short neck, concave chest | − |
| Mental development and speech | |||
| Intelligence | Low-normal range | Low-normal range | Low-normal range |
| Autism | No | No | No |
| First words | 2.2 years | 4 years | 2 years |
| Speech receptive | Good | Good | Good |
| Speech expressive | Simple sentences | Sentences up to 6 words | Sentences up to 6 words |
| Dysarticulation | Severe | Severe | Severe |
| Drooling | + | + | + |
| Visual system | |||
| Nystagmus | + | + | + |
| Head nodding | − | − | + |
| Refraction error | Hypermetropia (+2.0 dpt) and astigmatism | Hypermetropia (+4.5/+4.25 dpt) and astigmatism | Hypermetropia (+3.25 dpt/+2.75 dpt) and astigmatism |
| Fundus | Normal | Normal | Normal (age 14 months) pale right optic disc (age 4 years) |
| Cataract | − | − | − |
| Strabism | + | + | + |
| Epilepsy | |||
| EEG | Normal | Unspecific changes | Normal |
| Seizures | Single seizure at age 3 years, currently seizure-free without medication | No seizures | No seizures |
+/++/+++/− = present (with graduation)/absent; GMFCS = gross motor function classification system.
Neuroimaging
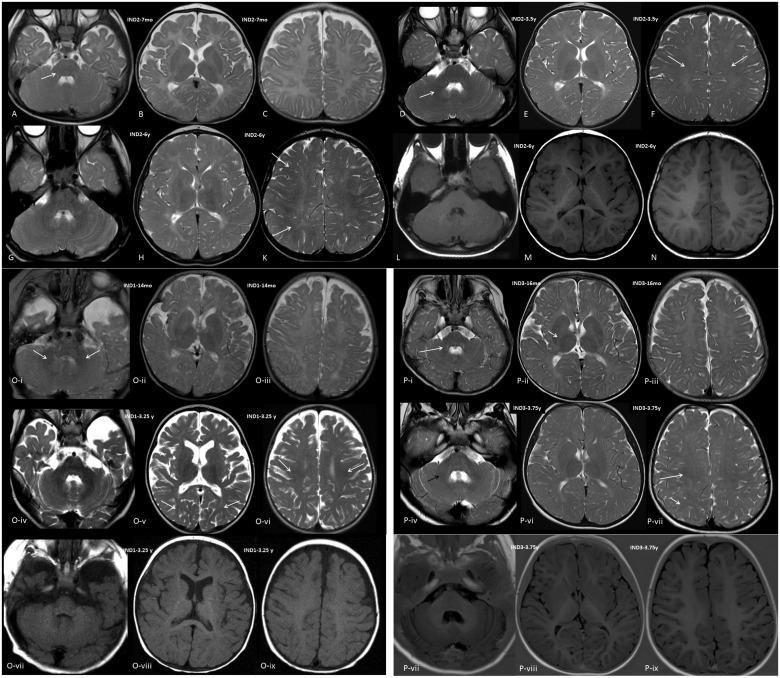
In Individual 1, MRI of the brain was performed at the age of 14 months and 3.25 years, and showed clear characteristics of hypomyelination with a discrepancy between T1-weighted and T2-weighted images but with some progress in myelination over time. T1-weighted images showed signal hyperintensity near to normal for age with respect to structures involved. T2-weighted images, however, did not demonstrate relevant myelination of white matter structures (signal hypointensity) at 14 months; at 3.25 years, the cerebellar white matter had a hypointense signal, and mild hypointensity was seen in the corpus callosum, the posterior limb of the internal capsule (PLIC), as well as the frontal and occipital central white matter; myelination scores were 7 and 13, respectively. Individual 1 is the only individual showing some atrophy of the telencephalon on follow-up MRI.
In Individual 2, MRI scans were done at 7 months, 3.5 years, 4.3 years, and 6 years, which showed hypomyelination with characteristics similar to Individual 1. Over the years, some progress in myelination could be seen especially in central and even peripheral white matter, but not in the PLIC or periventricular white matter. Myelination scores were 4, 8, 10 and 14.
In Individual 3, MRIs were performed at age 16 months and 3.75 years and showed hypomyelination comparable to Individuals 1 and 2, also with some progress in myelination especially involving the central and peripheral white matter. Myelination scores were 7 and 16, respectively (Fig. 1).
Figure 1.
Cranial MRI images. Images shown are of Individual 2 (A–N), Individual 1 (O) and Individual 3 (P). (A–C) T2-weighted axial images of Individual 2 at the age of 7 months, (D–F) T2-weighted axial images at age 3.5 years, (G–K) T2-weighted axial images, at age 6 years, and (L–N) T1-weighted axial images at age 6 years at the level of the cerebellum (A, D, G and L), diencephalon (B, E, H and M) and centrum semiovale (C, F, K and N). There is no myelin equivalent at 7 months on T2-weighted images except for a mildly lower signal in the cerebellar peduncles (arrow); at 3.5 years, central white matter shows some lower signal (arrows), as well as the central cerebellar white matter (short arrow); at 6 years, lower signal is seen in the more peripheral frontal and occipital white matter (arrows); T1-weighted images at 6 years appear near normal, U-fibres are not clearly myelinated, however, and signal is slightly less hyperintense than normal. [O(i–iii)] T2-weighted axial images of Individual 1 at the age of 14 months, (iv–vi) T2-weighted axial images at age 3.25 years, (vii–ix) T1-weighted images at age 3.25 years at the level of the cerebellum (i, iv and vii), diencephalon (ii, v and viii) and centrum semiovale (iii, vi and ix). There is no myelin equivalent at 14 months on T2-weighted images except for a lower signal in the cerebellar peduncles (arrows) and cerebellar white matter, central white matter may show some minimally lower signal; at 3.25 years, central white matter shows lower signal (arrows), as well as the more peripheral occipital white matter (short arrows), the cerebellar white matter is clearly of low signal. T1-weighted images at 3.25 years show hyperintensity of white matter with signal; however, less hyperintense than normal; U-fibres are not clearly myelinated. There is evidence of some global atrophy of the telencephalon (somewhat reduced white matter volume and enlarged sulci). [P(i–iii)] T2-weighted axial images of Individual 3 at the age of 16 months, (iv–vi) T2-weighted axial images and (vii–ix) Inversion recovery axial at age 3.75 years, at the level of the cerebellum (i, iv and vii), diencephalon (ii, v and viii) and centrum semiovale (iii, vi and ix). There is no myelin equivalent at 16 months on T2-weighted except for a lower signal in the cerebellar peduncles and cerebellar white matter (arrow), and minimally in the posterior limb of the internal capsule (PLIC; short arrow); at 3.75 years, central white matter shows lower signal (arrow), as does the PLIC and the more peripheral occipital white matter (short arrow), the cerebellar white matter is clearly of low signal (black arrow); inversion recovery images at 3.75 years appear near normal, U-fibres are not clearly myelinated; however, as evident through the somewhat blurry differentiation between cortex and white matter; and signal is somewhat less hyperintense than normal. IND = individual; mo = months; y = years.
Molecular genetic analysis and protein prediction
No disease-causing variant(s) in known disease-associated genes could be identified by exome sequencing in Individuals 1–3.
Trio exome sequencing of Individual 1 at 12 years of age revealed a heterozygous de novo stop-loss variant in CLDN11 NM_005602.5:c.622T>C, p.(*208Glnext*39).
Proband-only exome sequencing of Individual 2 at 7 years of age revealed a heterozygous stop-loss variant in CLDN11 NM_005602.5:c.622T>G, p.(*208Gluext*39). Subsequent sequencing of the parents confirmed a de novo status of the variant.
Individual 3 was enrolled in the TIDEX study and trio exome sequencing at 3 years of age revealed a heterozygous de novo stop-loss variant in CLDN11 NM_005602.5:c.622T>C, p.(*208Glnext*39), the same as in Individual 1 (Tarailo-Graovac et al., 2016).
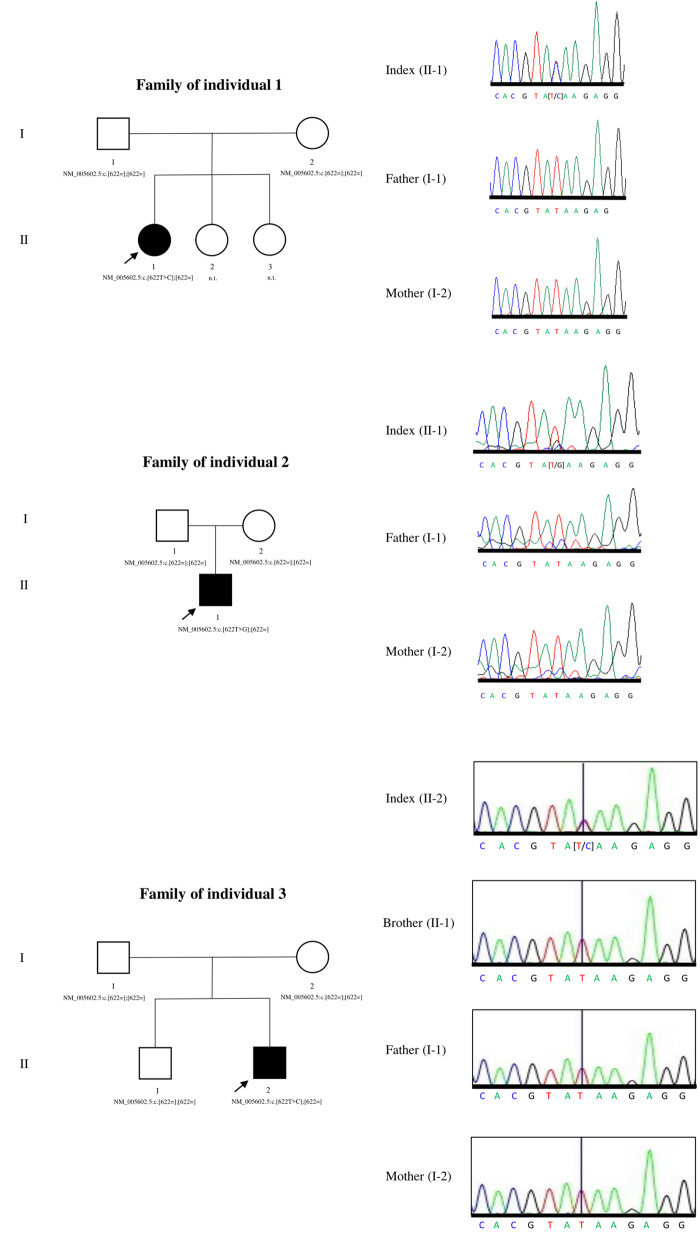
Sanger sequencing of index individuals, their parents and the unaffected brother of Individual 3 confirmed the identified variants and their de novo status.
Both variants c.622T>C, p.(*208Glnext*39) and c.622T>G, p.(*208Gluext*39) are not listed in more than 20 000 in-house exomes (Munich Exome Server) or in over 140 000 individuals of the Genome Aggregation Database (gnomAD v2.1.1; https://gnomad.broadinstitute.org/). The gnomAD does not list any stop-loss variants in CLDN11 (in v.2.1.1, or in v.3).
Figure 2 features pedigrees of the three families and electropherograms of the stop-loss variant region of the three individuals and their parents (also of the unaffected brother of Individual 3).
Figure 2.
Pedigrees of the three families and electropherograms of the stop-loss variant region of the three individuals and their parents (also of the unaffected brother of Individual 3). Squares denote male individuals, circles denote female individuals.
RNA sequencing of cultivated skin fibroblasts (passage 3) of Individual 3 showed that the stop-loss variant has heterozygous expression on RNA level (Supplementary Fig. 1).
CLDN11 transcript expression levels are not significantly different from 156 control fibroblasts (Kremer et al., 2017). Significantly downregulated genes (adjusted P < 0.05, fold change 0.5, 0.51, and 0.68, respectively) were ESYT1, ZNF160, and DPM1 (Supplementary Table 1). The ESYT1 expression level may be explained by the presence of a rare, heterozygous nonsense variant in ESYT1 NM_015292.2:c.2956C>T, p.(Arg986*) in the individual’s exome (inherited from the unaffected mother). DPM1 is associated with an autosomal-recessive disease [congenital disorder of glycosylation, type Ie (MIM 608799)]. There is no direct interaction between CLDN11, DPM1 and ZNF160 (https://genemania.org/). PLP1 is not expressed in fibroblasts (https://www.gtexportal.org/home/).
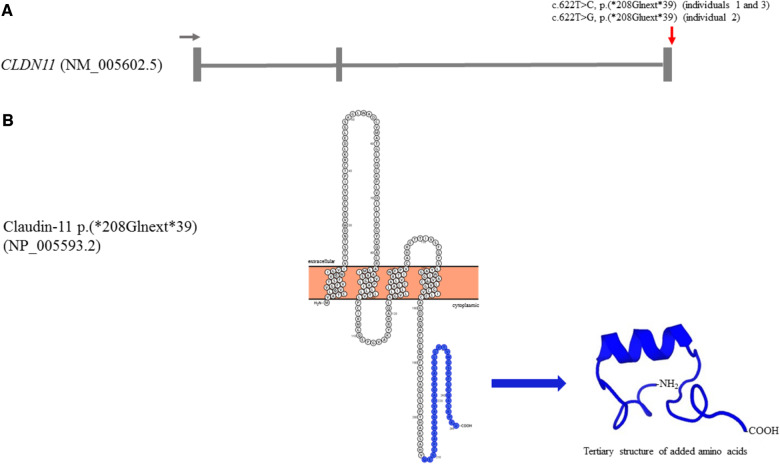
At the protein level, the de novo stop-loss variants are predicted to lead to an extension of 39 amino acids to the cytoplasmic C-terminus of claudin-11 and the added sequences (analysed for c.622T>C and c.622T>G) are unique as they do not have any significant similarity to any RefSeq protein sequence (Altschul et al., 2005; Heinemann and Schuetz, 2019). The extension is predicted to add a domain with an alpha helix to the cytoplasmic tail of claudin-11 (computed by PEP-FOLD for variant c.622T>C) not incorporated into the cytoplasmic membrane (computed by TMHMM for variant c.622T>C; see ‘Materials and methods’ section). Figure 3 illustrates the gene structure of CLDN11 and protein structure of extended claudin-11.
Figure 3.
Gene structure and protein sequence/structure. (A) Gene structure of CLDN11 featuring three exons (transcript NM_005602.5). Position of stop-loss variants is indicated by a red arrow (5′–3′ direction indicated by a grey arrow). (B) Protein sequence and tertiary structure of extended claudin-11. Added amino acids are depicted in blue in protein sequence [designed using the free web-based tool Protter (http://wlab.ethz.ch/protter/start/)]. Modelling of tertiary structure of added amino acids added was done with PEP-FOLD (Thevenet et al., 2012; Shen et al., 2014). Predicted tertiary structure of 39 added amino acid showing an alpha helix. All pertinent to variant NM_005602.5:c.622T>C, p.(*208Glnext*39) in CLDN11 (individuals 1 and 3).
Discussion
We present three unrelated individuals with hypomyelinating leukodystrophy, harbouring de novo stop-loss variants in CLDN11 identified by exome sequencing, sharing strikingly overlapping genotypic and phenotypic characteristics.
Clinically, all three individuals had an early-onset phenotype, subsequently developed a spastic-rigid disorder without ataxia or dystonia, combined with a pronounced deficit in oral motor skills. Intellectual abilities were only mildly impaired.
Interestingly, all individuals had hypermetropia which has not been described in any other hypomyelinating leukodystrophy. This seems to be a noteworthy finding and might be a specific clinical sign of this hypomyelinating leukodystrophy, as is a high myopia in 4H leukodystrophy (Wolf et al., 2005). However, it is certainly too early to call this an essential sign, since the prevalence of hyperopia in the general population is at least 6% at age 8–9 years (Donnelly et al., 2005).
On cranial MRI, there was a discrepancy between T1-weighted and T2-weighted images, as described for hypomyelinating leukodystrophies, with lessening/absence of T2-hypointensity without the significant lessening of T1-hyperintensity seen in non-hypomyelinating leukodystrophies (Pouwels et al., 2014). Signal intensity on T1-weighted and inversion recovery images was higher than isointensity, but lower than considered normal for age, which indicated the presence of some myelin, and, thus, a moderate rather than severe myelin deficit. On T2-weighted images, some progress in myelination with respect to central and peripheral white matter occurred which is not seen in classical PMD, for example (Plecko et al., 2003). When clustering features of MRI in hypomyelinating disorders, Steenweg et al. (2010) did not describe such a feature as consistent in any disorder of their analysis. This feature, therefore, may help to draw attention to CLDN11.
CLDN11 encodes claudin-11, which has been identified as an indispensable protein in the formation of the radial component of myelin (Denninger et al., 2015). After PLP (50–60% of myelin protein in CNS) and myelin basic protein (MBP, 40%), claudin-11 is the third most frequent protein in CNS myelin (7%) (Bronstein et al., 1996, 1997). Knockout of Cldn11 in mice leads to mild neurological deficits including body tremors and hindlimb weakness after birth, behavioural abnormalities and a reduction of nerve conduction. Interestingly, the absence of claudin-11does not result in defective myelin formation underlining the role of claudin-11in the physical properties but not integrity of myelin (Gow et al., 1999; Devaux and Gow, 2008; Maheras et al., 2018).
There is an association between CLDN11 and PLP1; when both orthologues are absent in mice, a severe movement disorder develops with disruption of myelin formation and strong reduction in nerve conduction velocities. In contrast, knockout of either one of the genes does not lead to a substantial deficit in myelin. This might be because of compensatory effects as a knockout of one of the genes leads to upregulation of the other (Boison and Stoffel, 1994; Klugmann et al., 1997; Gow et al., 1999; Chow et al., 2005). Interestingly, MBP, the second most abundant protein in CNS myelin after PLP is not differentially expressed when either or both Plp1 or Cldn11 are knocked out in mice, pointing towards the specific reciprocal regulation of Plp1 and Cldn11 (Chow et al., 2005).
We identified the stop-loss variant c.622T>C, p.(*208Glnext*39) in two of the individuals and the stop-loss variant c.622T>G, p.(*208Gluext*39) in one individual in CLDN11 (NM_005602.5). The genotypic data with three unrelated individuals with de novo stop-loss variants in CLDN11 is very convincing since mutations of stop codons are rare events in human genetic disease [only 0.27% of 159 705 variants reported in the Human Gene Mutation Database (HGMD® Professional 2019.4)]. The variant c.622T>C, p.(*208Glnext*39) did not lead to a loss of CLDN11 expression on RNA level in fibroblasts indicating this transcript is not subject to nonsense-mediated decay and most likely translated into an extended protein. As PLP1 is not expressed in fibroblasts, our RNA sequencing data did not allow us to demonstrate a potential compensatory upregulation of PLP1 expression in the presence of mutated CLDN11.
Both proteins claudin-11 and PLP1 are tetraspan integral membrane proteins of the myelin sheath (Devaux et al., 2010). There is evidence that claudin-11and PLP1 form complexes with integrins and further associated proteins in oligodendrocytes important for myelin proliferation and migration (Tiwari-Woodruff et al., 2001; Gudz et al., 2002). Importantly, claudin-11-associated protein 1 (OAP-1/tetraspanin 3; gene TSPAN3) forms a complex with claudin-11 and β1-integrin. For this, OAP-1 and claudin-11 interact via the C-terminus of claudin-11, which is heavily altered by the stop-loss variants identified in the three individuals (Bronstein et al., 2000). Therefore, one possible mechanism could be that interaction of claudin-11 with OAP-1 (and β1-integrin and eventually PLP1) is disturbed by the added amino acids impairing myelinogenesis.
In conclusion, we describe three unrelated individuals with an autosomal dominant hypomyelinating leukodystrophy and a shared genotype of heterozygous de novo stop-loss variants in CLDN11. The consistent clinical phenotype and an MRI-pattern differing in part from classical PMD and other hypomyelinating disorders, may give guidance to the clinician.
Supplementary Material
Acknowledgements
We are grateful to the affected individuals and their families for their participation in this study. We acknowledge the clinicians and laboratory scientists involved in the management of these individuals and for providing their medical reports. We acknowledge the following individuals for their contributions: Dr Michael Sargent, Department of Radiology at BC Children's Hospital for performing brain MRI investigation; Mrs Xiaohua Han for Sanger sequencing; Mrs Dora Pak and Mrs Evelyn Lomba for research management support; Mrs Michelle Higginson for DNA extraction, sample handling, and technical data; Mrs Meriam Waqas for data management; Prof Wyeth Wasserman (bioinformatic analysis), Dr Britt Drögemöller and Dr Colin Ross (Sanger analysis). Some of the individuals listed in this publication have been connected via GeneMatcher (Sobreira et al., 2015).
Funding
T.B.H. was supported by the German Bundesministerium für Bildung und Forschung (BMBF) through the Juniorverbund in der Systemmedizin “mitOmics” (FKZ 01ZX1405C to T.B.H.), the intramural fortüne program (#2435-0-0) and by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) – Projektnummer (418081722). Funding to C.v.K. was provided by the Canadian Institutes of Health Research (grant number #301221), National Ataxia Foundation. Informatics infrastructure supported by Genome British Columbia and Genome Canada (ABC4DE Project). C.v.K. is a recipient of the Michael Smith Foundation for Health Foundation Research Scholar Award and a Foundation Metakids salary award. R.v.d.L. is supported by a Rubicon fellowship from the Netherlands Organization for Scientific Research (NWO and ZONMW, 452172015). R.K. was funded by the German Network for Mitochondrial Disorders (mitoNET 01GM1906B).
Competing interests
The authors report no competing interests.
Supplementary material
Supplementary material is available at Brain online.
References
- Altschul SF, Wootton JC, Gertz EM, Agarwala R, Morgulis A, Schaffer AA, et al. Protein database searches using compositionally adjusted substitution matrices. Febs J 2005; 272: 5101–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boison D, Stoffel W. Disruption of the compacted myelin sheath of axons of the central nervous system in proteolipid protein-deficient mice. Proc Natl Acad Sci USA 1994; 91: 11709–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bronstein JM, Chen K, Tiwari-Woodruff S, Kornblum HI. Developmental expression of OSP/claudin-11. J Neurosci Res 2000; 60: 284–90. [DOI] [PubMed] [Google Scholar]
- Bronstein JM, Micevych PE, Chen K. Oligodendrocyte-specific protein (OSP) is a major component of CNS myelin. J Neurosci Res 1997; 50: 713–20. [DOI] [PubMed] [Google Scholar]
- Bronstein JM, Popper P, Micevych PE, Farber DB. Isolation and characterization of a novel oligodendrocyte-specific protein. Neurology 1996; 47: 772–8. [DOI] [PubMed] [Google Scholar]
- Chow E, Mottahedeh J, Prins M, Ridder W, Nusinowitz S, Bronstein JM. Disrupted compaction of CNS myelin in an OSP/Claudin-11 and PLP/DM20 double knockout mouse. Mol Cell Neurosci 2005; 29: 405–13. [DOI] [PubMed] [Google Scholar]
- Denninger AR, Breglio A, Maheras KJ, LeDuc G, Cristiglio V, Deme B, et al. Claudin-11 tight junctions in myelin are a barrier to diffusion and lack strong adhesive properties. Biophys J 2015; 109: 1387–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaux J, Fykkolodziej B, Gow A. Claudin proteins and neuronal function. Curr Top Membr 2010; 65: 229–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaux J, Gow A. Tight junctions potentiate the insulative properties of small CNS myelinated axons. J Cell Biol 2008; 183: 909–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly UM, Stewart NM, Hollinger M. Prevalence and outcomes of childhood visual disorders. Ophthalmic Epidemiol 2005; 12: 243–50. [DOI] [PubMed] [Google Scholar]
- Gow A, Southwood CM, Li JS, Pariali M, Riordan GP, Brodie SE, et al. CNS myelin and sertoli cell tight junction strands are absent in Osp/claudin-11 null mice. Cell 1999; 99: 649–59. [DOI] [PubMed] [Google Scholar]
- Gudz TI, Schneider TE, Haas TA, Macklin WB. Myelin proteolipid protein forms a complex with integrins and may participate in integrin receptor signaling in oligodendrocytes. J Neurosci 2002; 22: 7398–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinemann U, Schuetz A. Structural features of tight-junction proteins. Int J Mol Sci 2019; 20: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klugmann M, Schwab MH, Puhlhofer A, Schneider A, Zimmermann F, Griffiths IR, et al. Assembly of CNS myelin in the absence of proteolipid protein. Neuron 1997; 18: 59–70. [DOI] [PubMed] [Google Scholar]
- Kremer LS, Bader DM, Mertes C, Kopajtich R, Pichler G, Iuso A, et al. Genetic diagnosis of Mendelian disorders via RNA sequencing. Nat Commun 2017; 8: 15824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 2001; 305: 567–80. [DOI] [PubMed] [Google Scholar]
- Luders KA, Nessler S, Kusch K, Patzig J, Jung RB, Mobius W, et al. Maintenance of high proteolipid protein level in adult central nervous system myelin is required to preserve the integrity of myelin and axons. Glia 2019; 67: 634–49. [DOI] [PubMed] [Google Scholar]
- Maheras KJ, Peppi M, Ghoddoussi F, Galloway MP, Perrine SA, Gow A. Absence of claudin 11 in CNS myelin perturbs behavior and neurotransmitter levels in mice. Sci Rep 2018; 8: 3798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omasits U, Ahrens CH, Muller S, Wollscheid B. Protter: interactive protein feature visualization and integration with experimental proteomic data. Bioinformatics 2014; 30: 884–6. [DOI] [PubMed] [Google Scholar]
- Plecko B, Stockler-Ipsiroglu S, Gruber S, Mlynarik V, Moser E, Simbrunner J, et al. Degree of hypomyelination and magnetic resonance spectroscopy findings in patients with Pelizaeus Merzbacher phenotype. Neuropediatrics 2003; 34: 127–36. [DOI] [PubMed] [Google Scholar]
- Pouwels PJ, Vanderver A, Bernard G, Wolf NI, Dreha-Kulczewksi SF, Deoni SC, et al. Hypomyelinating leukodystrophies: translational research progress and prospects. Ann Neurol 2014; 76: 5–19. [DOI] [PubMed] [Google Scholar]
- Shen Y, Maupetit J, Derreumaux P, Tuffery P. Improved PEP-FOLD approach for peptide and miniprotein prediction. J Chem Theory Comput 2014; 10: 4745–58. [DOI] [PubMed] [Google Scholar]
- Sobreira N, Schiettecatte F, Valle D, Hamosh A. GeneMatcher: a matching tool for connecting investigators with an interest in the same gene. Hum Mutat 2015; 36: 928--30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steenweg ME, Vanderver A, Blaser S, Bizzi A, de Koning TJ, Mancini GM, et al. Magnetic resonance imaging pattern recognition in hypomyelinating disorders. Brain 2010; 133: 2971–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarailo-Graovac M, Shyr C, Ross CJ, Horvath GA, Salvarinova R, Ye XC, et al. Exome sequencing and the management of neurometabolic disorders. N Engl J Med 2016; 374: 2246–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thevenet P, Shen Y, Maupetit J, Guyon F, Derreumaux P, Tuffery P. PEP-FOLD: an updated de novo prediction server for both linear and disulfide bonded cyclic peptides. Nucleic Acids Res 2012; 40: W288–93. (Web Server issue). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari-Woodruff SK, Buznikov AG, Vu TQ, Micevych PE, Chen K, Kornblum HI, et al. OSP/claudin-11 forms a complex with a novel member of the tetraspanin super family and beta1 integrin and regulates proliferation and migration of oligodendrocytes. J Cell Biol 2001; 153: 295–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Knaap MS, Bugiani M. Leukodystrophies - much more than just diseases of myelin. Nat Rev Neurol 2018; 14: 747–8. [DOI] [PubMed] [Google Scholar]
- van der Knaap MS, Schiffmann R, Mochel F, Wolf NI. Diagnosis, prognosis, and treatment of leukodystrophies. Lancet Neurol 2019; 18: 962–72. [DOI] [PubMed] [Google Scholar]
- Vanderver A, Prust M, Tonduti D, Mochel F, Hussey HM, Helman G, et al. Case definition and classification of leukodystrophies and leukoencephalopathies. Mol Genet Metab 2015; 114: 494–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf NI, Harting I, Boltshauser E, Wiegand G, Koch MJ, Schmitt-Mechelke T, et al. Leukoencephalopathy with ataxia, hypodontia, and hypomyelination. Neurology 2005; 64: 1461–4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data can be shared. Results from molecular genetic analysis and RNA sequencing are available at the authors’ institutions in Munich, Warsaw and Vancouver.