Summary
Human induced pluripotent stem cell-derived cardiomyocytes (hiPSC-CMs) are a powerful platform for biomedical research. However, they are immature, which is a barrier to modeling adult-onset cardiovascular disease. Here, we sought to develop a simple method that could drive cultured hiPSC-CMs toward maturity across a number of phenotypes, with the aim of utilizing mature hiPSC-CMs to model human cardiovascular disease. hiPSC-CMs were cultured in fatty acid-based medium and plated on micropatterned surfaces. These cells display many characteristics of adult human cardiomyocytes, including elongated cell morphology, sarcomeric maturity, and increased myofibril contractile force. In addition, mature hiPSC-CMs develop pathological hypertrophy, with associated myofibril relaxation defects, in response to either a pro-hypertrophic agent or genetic mutations. The more mature hiPSC-CMs produced by these methods could serve as a useful in vitro platform for characterizing cardiovascular disease.
Keywords: hypertrophic cardiomyopathy, cardiomyocyte maturation, disease modeling, hiPSC-CM, hiPSC-CM maturation, HIF1α
Graphical Abstract
Highlights
-
•
Standard (glucose) cultured hiPSC-CMs demonstrate a blunted hypertrophic response
-
•
A maturation method induces hiPSC-CM maturation and suppresses HIF1A expression
-
•
Mature hiPSC-CMs demonstrate improved sarcomeric morphology and contractility
-
•
Mature hiPSC-CMs respond to agonist- or mutation-induced hypertrophy
In this article, Song and colleagues show that a combination of fatty acid medium and micropatterned surfaces induces maturation in human induced pluripotent stem cell-derived cardiomyocytes. Matured cells display improved sarcomere morphology, metabolic maturation, and contractility. These cells also show increased sensitivity to hypertrophic stimuli, including hypertrophic agonist and genetic mutations, representing an ideal system to model cardiovascular disease.
Introduction
Human induced pluripotent stem cell-derived cardiomyocytes (hiPSC-CMs) represent a powerful tool to study cardiovascular physiology and disease, offering several unique benefits. Significant differences exist between the hearts of humans and those of the small animals most often used for modeling cardiovascular disease, including heart rate, cell size, multinucleation frequency, and myosin heavy chain expression (Mills and Hudson, 2019). Therefore, the capability to study cardiovascular disease in a human cell system is critical. hiPSC-CMs offer several additional benefits, including scalability, generation of patient-specific cells, and rapid creation of knockout or transgenic lines via gene editing. However, hiPSC-CMs also have a significant drawback: they are functionally immature, typically resembling fetal or neonatal cardiomyocytes in terms of cell size and morphology, gene expression, myofibril contractility, and metabolic activity (Mills and Hudson, 2019; Pioner et al., 2016; Tan and Ye, 2018). Although certain forms of cardiovascular disease do affect infants, it is most commonly an adult disease. The functional immaturity of hiPSC-CMs is thus a barrier to modeling and studying most forms of cardiovascular disease. Improving hiPSC-CM maturity is a topic of acute interest, with numerous studies reporting a plethora of methods to achieve this aim, including, for example, prolonged culture (Kamakura et al., 2013), utilization of fatty acid medium (Correia et al., 2017), culturing on nanopatterned surfaces (Carson et al., 2016), engineered heart tissue (EHT) (Weinberger et al., 2017), electromechanical EHT stimulation (Ronaldson-Bouchard et al., 2018), and 3D microtissues composed of multiple types of cells (Giacomelli et al., 2020).
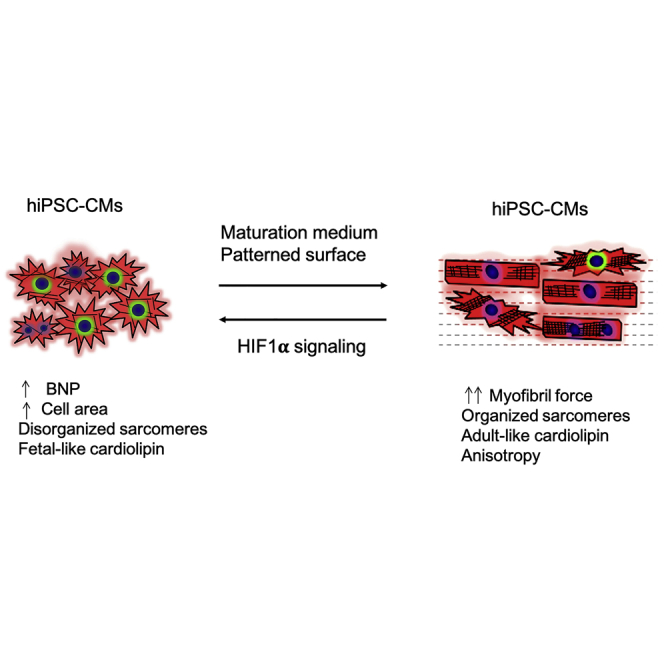
Here, we sought to develop a method to produce structurally and functionally mature hiPSC-CMs that requires little specialized equipment and can be scaled up easily. To this end, we combined and optimized existing approaches to induce hiPSC-CM maturation. First, we attempted to facilitate metabolic maturation by culturing cells in medium where fatty acids represent the primary energy source. In addition, cells were plated on micropatterned surfaces designed to promote elongation and anisotropy. hiPSC-CMs were maintained in standard glucose-based medium (RPMI 1640, GLUC), with maturation medium (MM), on patterned surfaces in standard medium, (GLPAT), or with maturation medium and patterning (MPAT). Compared with GLUC, both MM and, especially, MPAT hiPSC-CMs displayed a number of structural and functional improvements indicative of maturation, including increased expression of fatty acid-oxidizing genes, sarcomeric organization, and increased myofibril force generation. GLPAT cells displayed only modest improvements. In addition, MPAT cells displayed a robust, reversible response to the hypertrophic agonist phenylephrine (PE), which was not observed in GLUC cells. hiPSC-CMs derived from patients with Danon disease cultured in MPAT also displayed a robust, spontaneous hypertrophic response. Finally, expression of hypoxia-inducible factor 1α (HIF1A) was suppressed in MPAT-cultured cells, and restoration of its expression induced significant cellular hypertrophy. Our results indicate that the combinatorial approach employed in MPAT culture produces hiPSC-CMs that demonstrate both adult-like myofibril mechanics and an adult-like hypertrophic response, which will facilitate further study of adult cardiovascular disease with hiPSC-CMs.
Results
MPAT Improves Both hiPSC-CM Morphology and Sarcomeric Organization
To investigate whether hiPSC-CMs could be shifted toward more adult-like morphology and behavior, a combinatorial approach was employed (Figure S1A). First, instead of using medium where glucose is the main energy source (GLUC), our cells were cultured in medium where glucose was substituted with a combination of galactose, oleic acid, and palmitic acid (MM), which improved cellular morphology, contractility, ATP content, and metabolic behavior in previous studies (Correia et al., 2017; Hu et al., 2018). Although both GLUC and MM each contain the B-27 supplement, which contains fatty acids, the total fatty acid content in MM is ∼150 μM versus ∼144 nM in GLUC (Brewer and Cotman, 1989). As an additional step to improve hiPSC-CM maturity, MM-cultured hiPSC-CMs were plated onto plastic coverslips that had been micropatterned with grooves via 20-μm lapping paper, to induce cell elongation and anisotropic alignment (MPAT). As an additional control, we also plated cells onto patterned surfaces but continued to culture them in GLUC medium (GLPAT). When plated onto patterned surfaces, hiPSC-CMs displayed coordinated uniaxial contraction along the direction of patterning, which was not observed under other conditions (Videos S1, S2, S3, and S4).
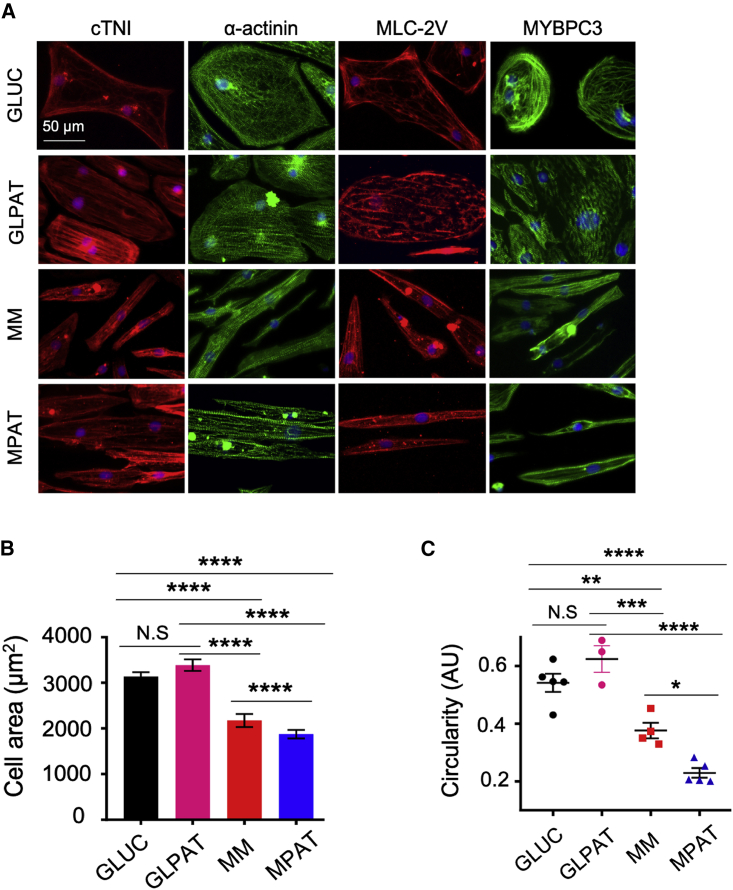
We then assessed whether either patterning or MM induced changes in cellular morphology or sarcomere organization by immunofluorescence microscopy. Cells cultured under each condition expressed the sarcomeric proteins cardiac troponin I, α-actinin, ventricular myosin light chain 2 (MLC-2V), and myosin-binding protein C3 (Figure 1A). In MM and MPAT cells, sarcomeres showed greater levels of organization, with Z lines oriented perpendicular to the long axis of the cells, whereas in GLUC, sarcomeres wrapped around the cells in a circular fashion or were oriented chaotically throughout the cytosol. GLPAT-cultured cells showed some degree of alignment, but with a relatively round morphology. To assess differences in cell morphology quantitatively, we measured cell area and circularity. Cells cultured with MM or MPAT were approximately 30% smaller than cells cultured in GLUC, consistent with previous reports (Correia et al., 2017) (Figures 1B, 1C, S1B, and S1C). MM cells showed significantly lower circularity than GLUC cells, which was further reduced in MPAT cells, indicating that MPAT cells were more elongated than MM cells, given their similar size (Figures 1C and S1C–S1E). GLPAT cells were both less elongated and larger than MM or MPAT cells (Figures 1B, 1C, and S1B–S1D). Whereas the average cell area of GLUC cells was comparable to that of adult mouse ventricular cardiomyocytes (AMVCMs), MM and MPAT cells were both significantly smaller, and all hiPSC-CM groups demonstrated greater variability of cell area compared with AMVCMs (Figure S1B). All hiPSC-CM groups were smaller than adult human cardiomyocytes (10,000–14,000 μm2) (Tan and Ye, 2018). As the heart develops, average sarcomere length increases from approximately 1.6 to 2.2 μm (Tan and Ye, 2018). Sarcomere length was significantly increased in MPAT hiPSC-CMs relative to the other culture groups assessed (Figure S1F), potentially indicating the greatest degree of sarcomeric maturity in this group. More than 75% of hiPSC-CMs stained positive for MLC-2V (Figure S1G).
Figure 1.
Morphological Improvements with Maturation Methods in CUSO-2 hiPSC-CMs
(A) Representative images of staining for various sarcomeric proteins in hiPSC-CMs cultured under each condition. cTNI, cardiac troponin I; MLC-2V, myosin light chain, ventricular isoform; MYBPC3, myosin-binding protein C3.
(B) Average cell areas in hiPSC-CMs cultured under each condition.
(C) Cellular circularity, defined by circularity = 4 × π × area/perimeter2 in hiPSC-CMs cultured under each condition. A lower circularity is indicative of more elongated objects. Data were pooled from three replicates from two inductions (GLPAT) or three or more (all other conditions) inductions, shown as mean ± SEM. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001; Kruskal-Wallis test followed by Dunn's post test (B) or one-way ANOVA with Tukey's post test (C).
See also Figure S1.
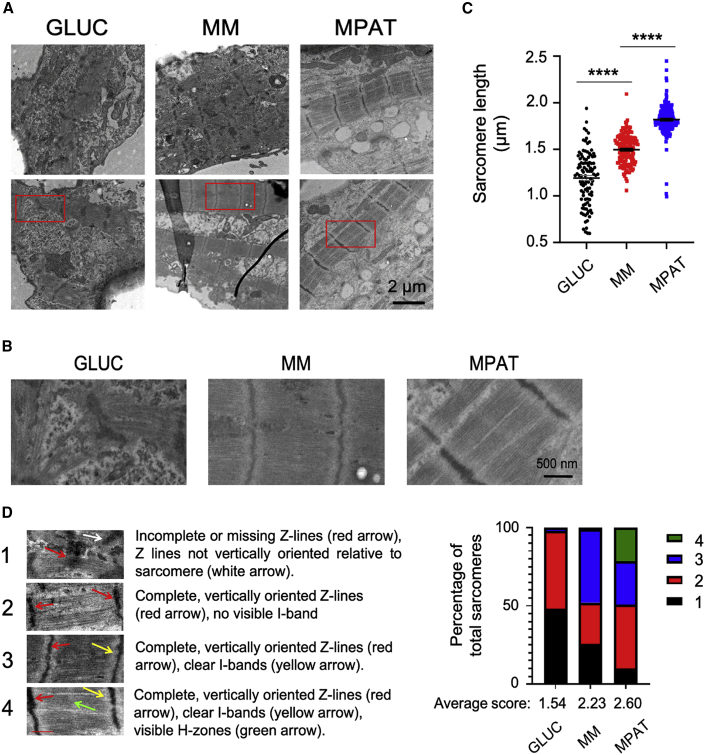
To further investigate sarcomeric morphology we used electron microscopy. When cultured in standard glucose medium, hiPSC-CMs displayed chaotically aligned, disorganized sarcomeres with Z lines of varying thickness, which often did not pass through the entirety of sarcomeres, consistent with developing cardiomyocytes in the fetal heart (Zhang and Pasumarthi, 2007). By contrast, MM cells displayed more diverse sarcomere morphology: some sarcomeres were disordered, as seen in GLUC cells, whereas others were highly regular and organized, with I bands, but lacking H zones. MPAT cells consistently displayed organized sarcomeres in which both I bands and H zones could be observed (Figures 2A and 2B), as is typically seen in more developed cardiomyocytes in vivo, or in more mature hiPSC-CMs (Lundy et al., 2013; Zhang and Pasumarthi, 2007). To confirm these findings quantitatively, we measured sarcomere length in our electron microscopy images and developed a scoring metric for sarcomere organization. MPAT hiPSC-CMs demonstrated the longest average sarcomere length and the greatest proportion of highly organized sarcomeres of any culture condition assessed, while GLUC hiPSC-CMs had the shortest sarcomeres and the greatest proportion of disorganized sarcomeres (Figures 2C and 2D). Our results indicate that the MPAT maturation method produced hiPSC-CMs with the greatest maturity as evidenced by sarcomeric morphology and structure relative to other conditions.
Figure 2.
Sarcomere Morphology Is Improved in CUSO-2 hiPSC-CMs with Maturity Methods
(A and B) (A) Low-magnification and (B) high-magnification election microscopy images of hiPSC-CMs cultured under each condition.
(C) Sarcomere length (SL) in hiPSC-CMs cultured under each condition. SLs were measured between Z lines.
(D) Representative images and criteria for sarcomere organization, and distribution of sarcomere organization of hiPSC-CMs cultured under each indicated culture condition, as well as average organization score for each group. 1 is least organized, 4 is most organized. SLs and organization were assessed on electron microscopy images from two inductions.
Each dot represents an individual SL measurement, data shown as mean ± SEM. ∗∗∗∗p < 0.0001, one-way ANOVA with Tukey's post test.
MPAT Improves Myofibril Force Generation
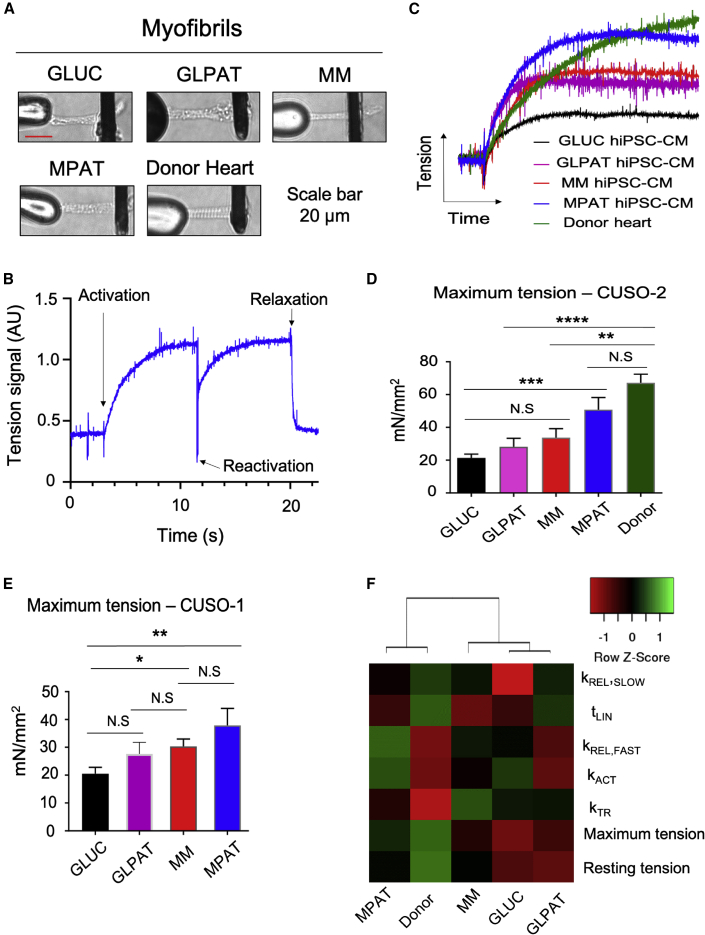
As MPAT improved hiPSC-CM sarcomeric morphology, we investigated whether sarcomeric function was correspondingly improved. To compare sarcomeric function of hiPSC-CMs to adult cardiac tissue, we isolated myofibrils from hiPSC-CMs cultured under each condition as well as from left ventricular tissue from a 35-year-old male donor heart for mechanics analysis (Figures 3A and 3B). Remarkably, relative to GLUC, myofibrils from CUSO-2 MPAT hiPSC-CMs demonstrated a nearly 150% increase in maximum tension generation, approaching the level of force generation measured in myofibrils from the donor heart. In the CUSO-1 line, MPAT myofibril maximum tension generation was increased approximately 85% relative to GLUC (Figures 3C–3E and Table S1). GLPAT and MM myofibrils displayed an intermediate level of tension generation for both cell lines. Activation and relaxation kinetics were similar between all CUSO-2 hiPSC-CM-derived and donor heart-derived myofibrils. We also conducted clustering analysis on myofibril mechanical parameters in CUSO-2 hiPSC-CMs and human myofibrils and found that MPAT myofibrils clustered most closely with human adult myofibrils, potentially indicating increased maturity (Figure 3F). To explore potential mechanisms causing increased MPAT force generation, we examined sarcomeric protein expression. We found that our cells expressed mostly MYH7 (Figure S2A), and that the expression of most sarcomeric proteins was quite similar across various culture conditions. However, while MM and GLUC cells expressed both ventricular and atrial forms of myosin light chain (MLC-2V and MLC-2A), MPAT cells expressed primarily MLC-2V (Figures S2B and S2C). Low expression of slow skeletal troponin I and significant expression of myosin IIB, typically expressed in the fetal heart (Ma and Adelstein, 2012; Saggin et al., 1989), were detected in our samples. From our results at this stage, it became apparent that MPAT produced hiPSC-CMs to the greatest degree of maturity in terms of force generation, cell morphology, or sarcomeric organization. For the remainder of the experiments, we focused on comparing GLUC, MM, and MPAT, or GLUC and MPAT.
Figure 3.
Myofibril Mechanics in hiPSC-CMs and Human Heart Tissue
(A) Representative images of CUSO-2 myofibrils isolated from each group of cells and from a human donor heart, mounted between stretcher (left) and force probes (right).
(B) Representative myofibril activation/relaxation trace from CUSO-2 MPAT myofibril.
(C) Myofibril activation traces from each indicated CUSO-2 culture group and from a human donor heart. Greater trace height indicates greater force generation.
(D and E) Average maximum tension generation of myofibrils isolated from CUSO-2 hiPSC-CMs cultured as indicated and from a human donor heart (D) or from CUSO-1 hiPSC-CMs (E).
(F) Heatmap of CUSO-2 hiPSC-CM myofibril mechanics properties. Unsupervised hierarchical clustering was performed on columns, by centroid linkage, indicating that MPAT cells cluster most closely with human adult donor myofibrils. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001, one-way ANOVA with Tukey's post test on log2-transformed data. Myofibrils were isolated from hiPSC-CMs from four inductions (CUSO-2 GLUC, MM, MPAT; CUSO-1 GLUC, MM), two inductions (CUSO-1 GLPAT, MPAT), or a single induction (CUSO-2 GLPAT), while 13 myofibrils were isolated from the donor heart.
Data shown as mean ± SEM. See also Figures S2 and S4 and Table S1.
MPAT and MM Induce a Gene Expression Program Controlling Fatty Acid Oxidation
To determine the mechanisms by which our methods induced cardiomyocyte maturation, RNA sequencing (RNA-seq) was performed on hiPSC-CMs cultured under each condition. Numerous genes displayed differential expression between the GLUC and either the MM or the MPAT condition, with a total of approximately 1,000 genes showing either a 1.5-fold increase or a decrease in expression (Figures S3A and S3B). Most genes (733 upregulated and 529 downregulated) differentially regulated between either GLUC and MM or GLUC and MPAT were in common between these comparisons, with a smaller number differentially expressed between MM and MPAT. We also performed gene ontology (GO) analysis using PANTHER and KEGG pathway analysis on differentially expressed genes. GO terms enriched in genes upregulated in MM or MPAT relative to GLUC hiPSC-CMs were frequently related to cardiac development or differentiation or fatty acid metabolism (Table S2, MM versus GLUC and MPAT versus GLUC). We also observed a general upregulation of expression of numerous genes involved in fatty acid metabolism in MPAT hiPSC-CMs (Figure S3C). GO and KEGG terms enriched in downregulated genes in MM versus GLUC corresponded to DNA synthesis and cell division, many of which were further downregulated in MPAT cells compared with MM (Table S2). This could indicate that culture in fatty acid medium arrests hiPSC-CM cell division, which is consistent with a previous study in EHT (Mills et al., 2017). KEGG terms enriched in upregulated genes in MM or MPAT versus GLUC frequently involved fatty acid metabolism (Table S2). Interestingly, one KEGG term enriched in upregulated genes in MPAT versus MM, the PPAR signaling pathway, is important to mitochondrial function and cardiac energy metabolism (Table S2). We also examined expression of several genes involved in fatty acid transport/oxidation using qPCR, and found several that were significantly upregulated in MPAT versus GLUC (Figure S4A), with a trend toward higher expression in MPAT than in MM.
We then analyzed metabolic changes in maturing hiPSC-CMs using metabolite screening. We measured 136 polar metabolites in high-throughput profiling, of which 51 were significantly altered across our three conditions (Figure S3D). Metabolites were examined on a candidate basis, focusing on glucose and long-chain fatty acid metabolism. Surprisingly, glucose levels in hiPSC-CMs cultured under each condition were similar. However, many metabolites of the glycolytic pathway were significantly reduced in MPAT hiPSC-CMs relative to GLUC, while intracellular levels of several fatty acids, including oleic, myristoleic, and several medium- or long-chain acylated fatty acids, were much higher in MM and MPAT cells (Figures S3D, S4B, and S4C). We also observed significant suppression of the glycolytic enzyme GAPDH in MM and especially MPAT cells, further supporting the notion of suppressed glycolysis in these cells (Figures S2B and S2C). Together with our RNA-seq data, these results indicate that MPAT induces a shift from glycolysis to fatty acid oxidation as a source of energy production, as is seen in the developing heart. Developmentally, this shift to fatty acid oxidation is accompanied by an increase in activity of the carnitine palmitoyltransferase (CPT) system, which imports fatty acids into the mitochondria. As we observed increased CPT gene expression, we tested CPT activity in hiPSC-CMs and undifferentiated hiPSCs. CPT1 activity was increased in all hiPSC-CM groups relative to hiPSCs and was further increased in MM/MPAT groups (Figure S4D). CPT2 activity was similar between all hiPSC-CM groups but was significantly increased only in MPAT hiPSC-CMs relative to hiPSCs. As CPT1 activity is considered to be rate limiting in long-chain fatty acid oxidation (Noland, 2015), enhanced CPT1 activity is consistent with increased long-chain fatty acid use.
Cardiolipin (CL) is the critical phospholipid component of mitochondrial membranes and is essential for mitochondrial function (Shen et al., 2015). Each CL molecule has four fatty acid side chains, which vary in a tissue-specific manner. In the developing heart, CL content changes, resulting in increased CL with side chains of 72 total carbon length (72C-CL; MW 1,448–1,456), such that tetralineolyl CL (MW 1,448) represents the dominant species in the adult heart (Schlame et al., 2005). Considering the metabolic improvements we observed in our MPAT cells, we investigated whether CL remodeling was occurring in our more mature cells, using adult human heart as control. Induction of hiPSCs into hiPSC-CMs caused significant CL remodeling, in particular a decrease in 68C- and 70C-CL and an increase in 72C- and 74C-CL. With maturation methods, a further loss in 70C-CL and an increase in 72C-CL were observed (Figures S4E and S4F), shifting the CL profile toward that of adult human hearts.
MPAT and MM Methods Allow an Adult-like Hypertrophic Response in hiPSC-CMs
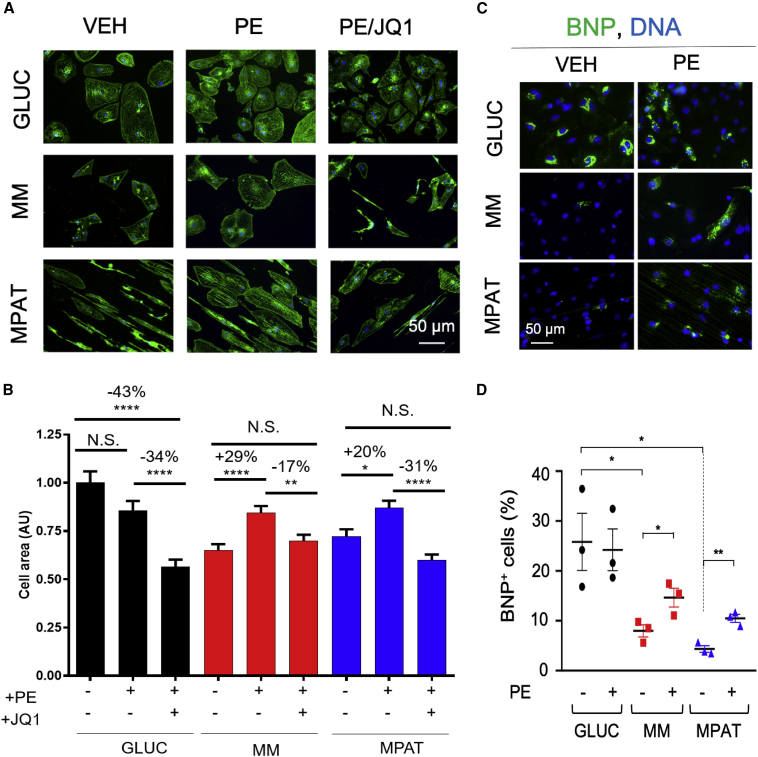
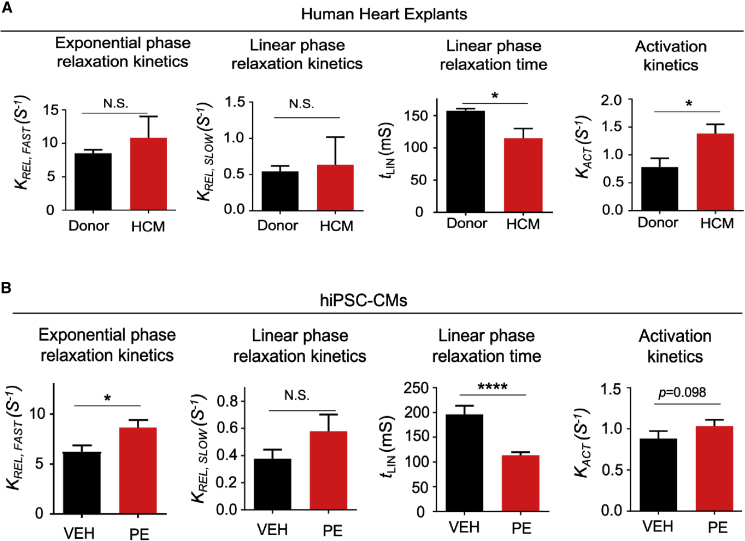
We next investigated the efficacy of maturation methods for studying hypertrophic remodeling. Significant differences in the magnitude of in vitro hypertrophic response have been observed in neonatal versus adult cardiac myocytes. In neonatal cardiomyocytes, agents such as the α-adrenergic receptor agonist PE induce a 50%–100% increase in cell area (Anand et al., 2013; Miller et al., 2009), whereas the response in adult cardiomyocytes is typically only 10%–30% (Bupha-Intr et al., 2012; Miller et al., 2009). Therefore, we investigated the response of hiPSC-CMs to PE. The BET-bromodomain inhibitor JQ1 is known to inhibit cardiomyocyte hypertrophy both in vivo and in vitro (Anand et al., 2013; Duan et al., 2017; Spiltoir et al., 2013); thus we also tested whether JQ1 could block the effects of PE in our system. Surprisingly, PE treatment had no effect on cell area of GLUC hiPSC-CMs, yet JQ1 treatment nonetheless profoundly reduced cell area. However, in MM and MPAT hiPSC-CMs, PE treatment induced a strong hypertrophic response, causing a 29% or 20% increase in cell area in MM or MPAT cells, respectively, which could be blocked by JQ1 (Figures 4A and 4B). To compare hiPSC-CMs directly to adult cells, we also treated isolated AMVCMS with PE and JQ1. In these cells, PE induced a 16% increase in cell area, which JQ1 reversed (Figures S5A and S5B), comparable to the response of MPAT hiPSC-CMs. To validate these findings, we also investigated the expression of NPPB (B-type natriuretic peptide, BNP), a robust marker of hiPSC-CM hypertrophy (Carlson et al., 2013), by qPCR and immunofluorescence. By qPCR, MM and MPAT cells displayed a 5- to 6-fold reduction in BNP expression relative to GLUC cells at baseline (Figure S5C). Moreover, more than 20% of GLUC cells stained positive for BNP, which was not increased by PE treatment, while BNP staining in MM and MPAT cells was much lower at baseline and was increased approximately 2-fold by PE treatment (Figures 4C and 4D). Taken together, these results suggest that prolonged glucose culture may be sufficient to induce hiPSC-CM hypertrophy.
Figure 4.
Culture in Maturation Medium Is Necessary to Induce a Hypertrophic Response in CUSO-2 hiPSC-CMs
(A) Representative images of hiPSC-CMs cultured under each indicated condition, fixed, and stained for α-actinin (green) and Hoechst 33342 (blue).
(B) Quantification of cell area in cardiomyocytes treated with vehicle, 10 μM PE, or 10 μM PE and 1 μM JQ1 for 48 h and then fixed and stained; there were 600 or more hiPSC-CMs from four inductions per group.
(C and D) (C) Representative images and (D) quantification of BNP/pro-NT-BNP-positive CUSO-2 hiPSC-CMs treated with or without PE, from three hiPSC-CM inductions. BNP/pro-NT-BNP signal is green, Hoechst 33342 is blue.
Data shown as mean ± SEM. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗∗p < 0.0001, Kruskal-Wallis test followed by Dunn's post test (B), or one-way ANOVA with Tukey's post test (D). See also Figure S5.
PE Treatment of MPAT hiPSC-CMs Induces Myofibril Relaxation Changes as Observed in Hypertrophic Cardiomyopathy
Previous reports have indicated that myofibrils isolated from hearts of humans with hypertrophic cardiomyopathy (HCM) display profound differences in mechanical behavior (Belus et al., 2008). We examined whether myofibrils isolated from hypertrophic hiPSC-CMs might demonstrate similar myofibril mechanical perturbations. We first assessed myofibril mechanics from human donor and HCM hearts in our tissue bank. Myofibrils were isolated from four donor hearts and four hearts from patients who underwent heart transplant due to HCM with a left ventricular posterior wall thickness of >12 mm as assessed by echocardiography. All myofibril mechanical parameters from donor and HCM hearts are listed in Table S3. Relaxation of myofibrils is biphasic, with a slow, linear phase and a fast, exponential phase (Stehle et al., 2002). Compared with donor hearts, HCM heart-derived myofibrils showed a shortened linear-phase relaxation time and faster activation kinetics (Figure 5A), similar to previous findings in a human MYH7 mutant HCM heart (Belus et al., 2008).
Figure 5.
Phenylephrine Treatment Induces Changes in Myofibril Relaxation that Are Similar to Those Observed in Hypertrophic Cardiomyopathy
(A) Myofibril mechanics data from human donor and idiopathic hypertrophic cardiomyopathy (HCM) hearts. Myofibrils were isolated from four donor and four HCM hearts, with at least eight myofibrils isolated per heart.
(B) Myofibril mechanics data from CUSO-2 MPAT hiPSC-CMs treated with either vehicle or 10 μM PE for 48 h. There were 39–44 myofibrils isolated per condition, pooled from three inductions, and presented as the mean ± SEM. ∗p < 0.05, ∗∗∗∗p < 0.0001, t test on log2-transformed data.
To compare with hypertrophic human hearts, we isolated myofibrils from MPAT hiPSC-CMs, as these displayed the most adult-like myofibril behavior. Hypertrophy was again induced via treatment with PE. As an additional comparison, we also isolated myofibrils from vehicle- or PE-treated AMVCMs. Remarkably, we found that myofibril relaxation in PE-treated hiPSC-CMs and in HCM hearts was similarly altered: the exponential-phase relaxation constant was increased, while the duration of linear-phase relaxation was shortened (indicating faster relaxation) (Figures 5B and S6A). As JQ1 treatment blocks the induction of PE-induced hypertrophy, we tested whether it could also reverse the PE-induced changes in myofibril relaxation in hiPSC-CMs but found that it had no significant effects (Figure S6B). In AMVCMs, PE induced relaxation changes similar to those in hiPSC-CMs, but no changes in activation kinetics could be detected (Figure S6C). Together, these results indicate that the MPAT hiPSC-CM platform is a suitable in vitro model for HCM, allowing parallel assessment of changes in cell area, hypertrophic gene expression, and myofibril mechanical behavior.
Modeling Danon Cardiomyopathy Using MPAT hiPSC-CMs
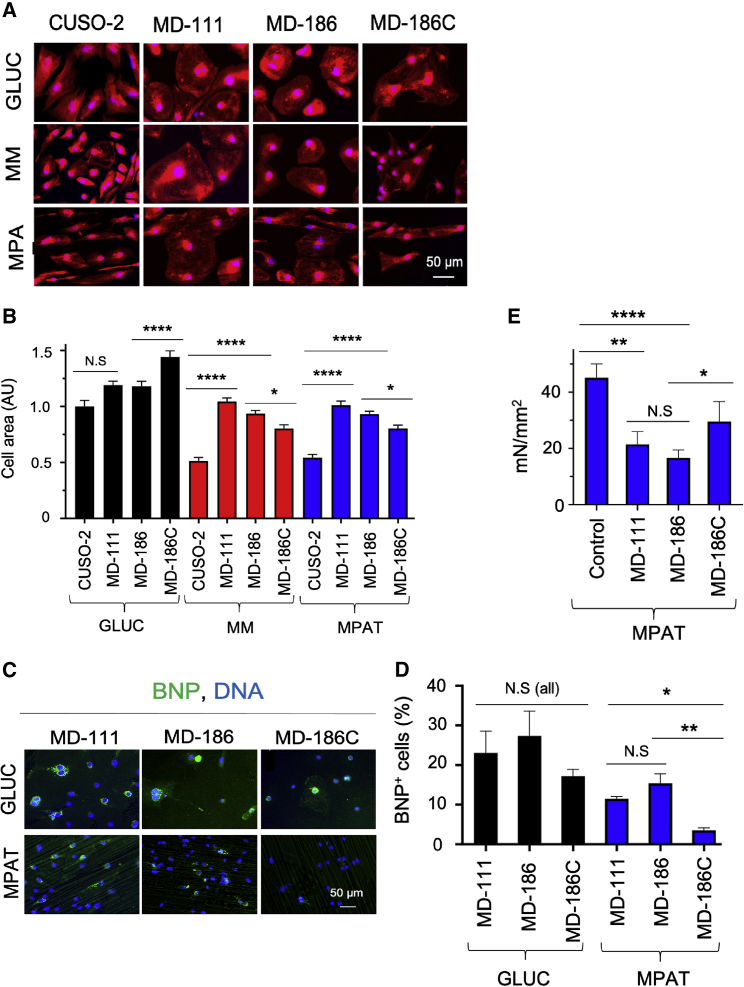
We next investigated whether we could characterize cardiac hypertrophy in patient-specific cells. In a previous study (Chi et al., 2019), we generated multiple lines of hiPSC-CMs from Danon disease patients carrying mutations in the LAMP2 gene that lead to pathological cardiac remodeling. We focused on hiPSC-CMs from two patients, MD-186 and MD-111, who at time of transplant had moderate and severe cardiac hypertrophy, respectively. The MD-186C line, in which the LAMP2 disease-causing mutation in patient MD-186 was corrected, was used as an isogenic control, in addition to the non-isogenic control CUSO-2 line. For these experiments, we assessed both cell area and expression of BNP to assess cardiac hypertrophy. We found that when cultured in GLUC medium, both sets of Danon hiPSC-CMs displayed quite large cell areas. In GLUC, the control CUSO-2 line was smaller than either Danon line, while the corrected MD-186C line was even larger. However, when cultured in MPAT, the Danon cell lines were significantly larger than CUSO-2, while MD-186C was significantly smaller than MD-186 (Figures 6A and 6B). Similarly, each Danon line demonstrated significantly more BNP staining than did MD-186C when cultured in MPAT, but not GLUC (Figures 6C and 6D). Similar to our findings with PE treatment, we observed significant induction of hypertrophy in Danon lines only when cultured in MM or MPAT and not GLUC. Our previous work indicates that myofibrils isolated from the hearts of Danon patients display reduced maximum tension relative to control donor heart myofibrils (Chi et al., 2019). To examine whether this phenomenon occurred in a cell-based model of Danon disease, we isolated myofibrils from MD-186, MD-111, and MD-186C hiPSC-CMs cultured under MPAT. Interestingly, Danon hiPSC-CM myofibrils demonstrated reduced tension generation relative to either wild-type or mutation-corrected myofibrils (Figure 6E and Table S3), similar to patient hearts.
Figure 6.
Culture in Maturation Medium Allows Characterization of Hypertrophy in Danon hiPSC-CMs
(A and B) (A) Representative images of and (B) quantification of cell area in hiPSC-CMs derived from control (CUSO-2), Danon (MD-111, MD-D186), or Danon-corrected (MD-186C) lines, cultured under the indicated conditions, fixed, and stained with CellMask orange (red) and Hoechst 33342 (blue).
(C and D) (C) Representative images of and (D) quantification of BNP/pro-NT-BNP-positive hiPSC-CMs of Danon and D186C cell lines, cultured under indicated conditions, fixed, and stained.
(E) Maximum myofibril tension generation from myofibrils from Danon, D186C, and control lines. Here, “control” indicates the maximum tension generation of pooled CUSO-1/CUSO-2 MPAT myofibrils. There were 6–22 myofibrils per Danon group and 56 myofibrils in the “control” group. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗∗p < 0.0001, Kruskal-Wallis test followed by Dunn's post test (B), one-way ANOVA with Tukey's post test (D and E), or t test for two-group comparisons (E). Log2-transformed data were used for tests in (E). Data were collected from three or more biological replicates from two or more inductions per experiment, shown as mean ± SEM. Cell areas were measured on 200 or more cells per group.
See also Table S3.
Sensitivity of MPAT-Cultured hiPSC-CMs to Hypertrophic Signaling Is Dependent on Suppression of Hypoxia-Inducible Factor 1α
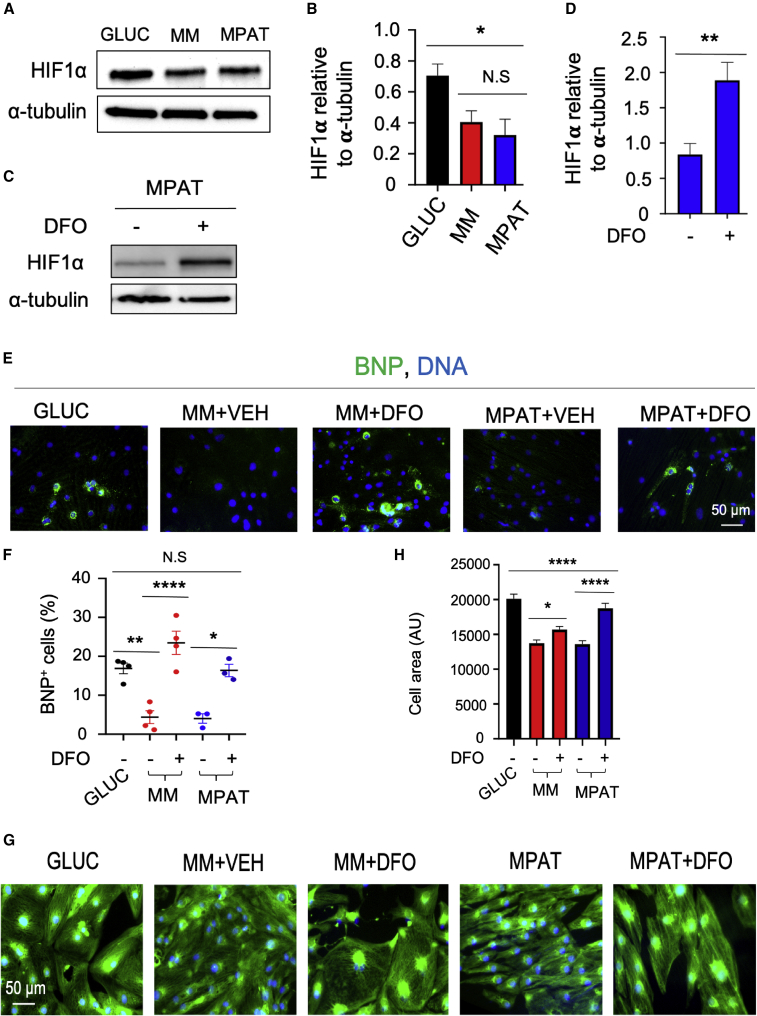
We next sought to investigate the mechanisms by which culture under the MM or MPAT conditions suppresses hypertrophic signaling. A previous report indicated that hiPSC-CM culture in glucose medium can induce expression of HIF1A, while culture in fatty acids suppresses this (Hu et al., 2018). It is also known that hypoxia can induce expression of the hypertrophic marker BNP in a HIF-1α-dependent mechanism (Casals et al., 2009) and, furthermore, that activation or increased expression of HIF1A can induce hypertrophy in cultured cardiomyocytes (Chu et al., 2012), while HIF-1α can block this process (Kumar et al., 2018). Therefore, we speculated that modulation of HIF1A expression might be responsible for the basal cardiomyocyte hypertrophy we observed in GLUC-cultured cells and the suppression of this phenomenon in MM/MPAT culture. We examined HIF1A expression under each culture condition and found that HIF-1α levels were significantly lower in MPAT cells compared with GLUC (Figures 7A and 7B). We then investigated the effects of restoring HIF-1α levels in MM/MPAT cells by treating with desferrioxamine (DFO), an iron chelator that can induce HIF1A expression in hiPSC-CMs (Hoes et al., 2018). Treatment of cells with 30 mM DFO for 96 h significantly increased HIF1A expression (Figures 7C and 7D). Remarkably, in multiple hiPSC lines, we found that DFO treatment significantly increased both BNP levels and cell area in MM/MPAT cells, to an extent relatively comparable to that of GLUC culture (Figures 7E–7H and S7). Taken together, these results support the notion that suppression of HIF1A expression by culture under MM or MPAT conditions is at least partially responsible of the responsiveness of these cells to hypertrophic signaling and the lack thereof of GLUC-cultured cells.
Figure 7.
Regulation of Hypertrophic Signaling by HIF-1α in CUSO-1 hiPSC-CMs
(A) Representative western blot of hiPSC-CMs cultured under indicated conditions and blotted for HIF-1α and α-tubulin.
(B) Quantification of HIF-1α levels in hiPSC-CMs cultured under indicated conditions, as assessed by western blot, and normalized to α-tubulin. N = 5 lysates.
(C) Representative western blot of hiPSC-CMs cultured under MPAT conditions and treated with DMSO vehicle or 30 mM desferrioxamine (DFO) for 96 h.
(D) Quantification of HIF-1α levels in hiPSC-CMs cultured under indicated conditions, as assessed by western blot, and normalized to α-tubulin. N = 10–11 lysates.
(E–H) hiPSC-CMs were cultured as indicated and treated with either vehicle or DFO for 96 h. Cells were fixed and stained for BNP/pro-NT-BNP (green, E) or using CellMask green for cell area (green, G), and for nuclei (Hoechst 33342, blue). (F) Quantification of BNP-positive cells; (H) quantification of cell area. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗∗p < 0.0001. Three hiPSC-CM inductions were used per experiment, data are shown as mean ± SEM. Cell areas were measured on 450 or more cells per group. (B and F) One-way ANOVA with Tukey's post test; (D) t test, (H) Kruskal-Wallis test followed by Dunn's post test.
See also Figure S7.
Discussion
Here, we present a relatively simple method to produce more mature hiPSC-CMs by culturing cells in medium containing fatty acids as a significant energy source and plating these cells on micropatterned growth surfaces. Under these conditions, cells demonstrate elongation (Figure 1), enhanced sarcomeric maturity (Figure 2), and metabolic and CL maturation (Figures S3 and S4). Force generation of myofibrils isolated from these cells is also comparable to that from adult human ventricular tissue (Figure 3). Notably, MPAT hiPSC-CMs also respond well to hypertrophic stimulation, producing an adult-like hypertrophic response, and demonstrate alterations in myofibril relaxation similar to those of human HCM tissue (Figures 4, 5, 6, and 7).
Compared with many maturation approaches, our approach may have potential applications due to its simplicity. Fatty acid medium can be prepared at low cost, while patterned surfaces require only a few tools to produce. This culturing setup can therefore be introduced into a new research environment quite quickly. The combination of these methods induces a more mature cardiac phenotype across a variety of parameters, from cell morphology to contractility and hypertrophic response. For example, most of the improvements in myofibril mechanics occurred in MPAT compared with other groups, whereas metabolic changes were driven primarily by the switch from GLUC to MM, while other phenotypes, such as sarcomeric morphology, were sequentially improved by each maturation step. This indicates that combinatorial approaches may be optimal for producing mature hiPSC-CMs. We believe that these methods have a number of potential applications, from drug screening to interrogating the effects of genetic or pharmacological perturbations in a human cell system. The approaches used here could also be combined with methods such as cell pacing, growth hormone treatment, or use of altered extracellular matrix composition or stiffness to promote further maturation.
The maturity-inducing effects of micro- and nanopatterned substrates on hiPSC-CMs are well reported (Pioner et al., 2016), but the mechanisms driving this remain undefined. A previous report indicated that a 700- to 1,000-nm nanopatterned groove size is optimal to induce hiPSC-CM maturity (Carson et al., 2016). Although the micropatterned grooves described here are much larger, they clearly exert positive effects on maturity as well. Our results indicate that patterned surfaces improved cellular morphology, sarcomere organization, and myofibril force generation, but we also observed a trend toward improved CL remodeling and increased fatty acid gene expression in MPAT versus MM cells. Immature hiPSC-CMs and fetal cardiomyocytes typically have an atrial cell-like identity, with dominant expression of the atrial isoforms of myosin light and heavy chains, MYL7 encoding MLC-2A and MYH6, respectively (Bedada et al., 2016; Tan and Ye, 2018). We observed dominant expression of MYH7 in all hiPSC-CM cultures but increased relative expression of MYL2 encoding MLC-2V to MYL7 only in MPAT hiPSC-CMs (Figure S2). The activation constants (KACT) we measured in hiPSC-CM myofibrils under most conditions (0.95–1.32, 1.9 for CUSO-1 GLPAT) are lower than previously reported values in human atrial tissue myofibrils (3.73) (Piroddi et al., 2007) or in embryonic stem cell derived-CM myofibrils expressing either mostly MYH7 or mixed MYH6/7 (1.70 or 2.44, respectively) (Weber et al., 2016). Together, these findings are indicative of sarcomeric maturity and potentially a more ventricular-like identity in our MPAT cells. As forced expression of MYL2 results in increased contractility in isolated cardiomyocytes (Pawloski-Dahm et al., 1998), this could be a mechanism underlying the increased myofibril force generation observed in MPAT cells.
Previous reports have indicated that prolonged (>100 days) culture of hiPSC-CMs induces a more adult-like phenotype, as indicated by improvements in sarcomeric ultrastructure, cell morphology, and contractility (Kamakura et al., 2013; Lundy et al., 2013). Yet in our hands, hiPSC-CMs cultured in glucose-containing medium had no hypertrophic response to PE and there was little to modest induction of hypertrophy in Danon hiPSC-CMs (Figures 4, 6, and 7). The antihypertrophic agent JQ1 also reduced cell area significantly beyond baseline in GLUC hiPSC-CMs, which did not occur in MM or MPAT cells (Figures 4A and 4B). A recent report indicated that 14 days of culture in 22 mM glucose was sufficient to induce cardiac hypertrophy and dysfunction in hiPSC-CMs (Ng et al., 2018). Although our GLUC medium contains only 10.9 mM glucose, prolonged culture under these conditions may be sufficient to induce a hypertrophic phenotype. hiPSC-CMs are increasingly used to model various types of cardiomyopathies, and these studies, along with our findings, indicate that a careful choice of conditions for long-term culture is essential. Mechanistically, a previous study indicated that repression of HIF1A expression improves metabolic maturity of hiPSC-CMs (Hu et al., 2018). We found that HIF-1α suppression also regulates the capability of hiPSC-CMs to initiate a hypertrophic response. However, it is possible that modulation of additional signaling pathways may play a role in regulating this response as well.
Although we observed similar changes in relaxation in PE-treated hiPSC-CMs and human HCM hearts, PE treatment did not dramatically increase myofibril activation kinetics. This could be due to differences between myofibrils from human heart and those from hiPSC-CMs, as hiPSC-CMs show higher baseline activation kinetics (Tables S1 and S3). However, 48 h of PE treatment is also unlikely to fully recapitulate the remodeling that occurs in HCM, which is mutation driven and occurs over years. A recent report demonstrated that 48 h of PE treatment on adult rat cardiac myofibrils did not affect the kinetics of myofibril activation or relaxation (Woulfe et al., 2019). Here, we demonstrated that 72 h of PE treatment on AMVCMs resulted in a slight (∼10 ms) reduction in linear-phase relaxation time as opposed to an 80-ms reduction in hiPSC-CMs (Figures 5 and S6, Table S3). It is possible that these divergent effects are due to differences between the PE responses of mouse, rat, and human, or variations in treatment times. However, the kinetics of human and rodent myofibrils are also dissimilar: in rodent, activation and relaxation constants are much larger, and linear-phase relaxation time is much shorter (∼50 ms in rodents versus 150–200 ms in humans). The capability of PE to further speed up this already short linear phase may therefore be abrogated, leading to modest effects in rodents.
In totality, our findings suggest that matured hiPSC-CMs are an appropriate in vitro model for conducting translational research of human myofibril mechanics in response to pathological stresses. The platform we describe here could also be of particular use when combined with CRISPR-based editing strategies to create cardiomyopathy patient-specific cells, allowing simultaneous assessment of cellular morphology, myofibril mechanics, metabolic behavior, and hypertrophic remodeling. A limitation of the hiPSC-CM-based platform is the potential presence of phenotypic variability between inductions and lines. For example, when we compared induction-matched or all-GLUC with GLPAT CUSO-1 hiPSC-CMs, we observed similar but not exactly the same changes in cell circularity and area and myofibril maximum tension (Table S1). To avoid results being affected by interinduction and interline variability, it may be appropriate to conduct paired experiments from multiple inductions of various hiPSC lines.
Experimental Procedures
Human and Animal Subject Information
The portion of this study related to human subjects was reviewed and approved by the Colorado Multiple Institutions Review Board (COMIRB 06-0452 and 01-568). All research involving animals complied with protocols approved by the Institutional Animal Care and Use Committee of the University of Colorado.
hiPSC-CM Differentiation and Culture
hiPSCs were induced into cardiomyocytes via the Wnt signaling modulation method (Lian et al., 2012). Twenty to twenty-five days post induction, cardiomyocytes were enriched using lactate medium (Tohyama et al., 2013). At approximately day 40 post induction, hiPSC-CMs were replated (onto fibronectin-coated patterned surfaces, flat cell culture dishes, or flat plastic coverslips) (Figure S1A). The four experimental conditions were GLUC, cultured on flat surfaces in RPMI-1640/B-27; GLPAT, cultured on patterned surfaces in RPMI-1640/B-27; MM, cultured on flat surfaces in maturation medium consisting of glucose-free RPMI, 50 μM palmitic/100 μM oleic acid, 10 mM galactose, and B-27 diluted 1:50, based on a previous report (Correia et al., 2017); and MPAT, cultured on patterned surfaces in maturation medium. Experiments were generally performed 65–75 days post induction, while certain CUSO-1 myofibril experiments were performed 90–100 days post differentiation.
Cell Lines Used
All hiPSC lines were reprogrammed from skin fibroblasts. The five hiPSC lines used in this study were CUSO-1 and CUSO-2, which were derived from healthy male donors; MD-186 and MD-111, which were derived from male Danon disease patients; and MD-186C, in which CRISPR-Cas9 was used to correct the Danon-causing mutation in the LAMP2 gene in MD-186 hiPSCs, described in Chi et al. (2019).
Patterned Surface Preparation
Patterned surfaces were prepared from clear plastic coverslips using 20-μm lapping paper, incubated in 100% ethanol followed by UV sterilization.
Myofibril Mechanics
hiPSC-CMs were lysed, and myofibrils were mounted between two glass instruments. Contraction was induced via introduction of calcium, followed by rapid calcium withdrawal via a pipette switching technique (Woulfe et al., 2019).
Immunostaining
Cells were fixed with either 4% paraformaldehyde or 50% methanol/50% acetone, permeabilized, blocked, and stained. Imaging was conducted on the EVOS FL Cell Imaging System (Thermo Fisher Scientific, Waltham, MA.).
Immunoblotting
Cells were lysed in ice-cold cell lysis buffer, loaded onto Tris-HCl gels, and transferred overnight onto polyvinylidene fluoride membranes. Membranes were then blocked, incubated with antibodies, and developed.
RNA Extraction and RNA-seq
Cells were lysed using Trizol followed by RNA extraction and precipitation from the aqueous phase and removal of genomic DNA. RNA-seq was performed by the BGI Group (Shenzhen, China).
Detailed methods are described in the Supplemental experimental procedures.
Data and Code Availability
The accession number for the RNA-seq is GEO: GSE143608.
Author Contributions
W.E.K. and K.S. conceptualized this project. W.E.K., C.C., B.B., Y.C., Y.Z., and P.L. performed experiments. Y.H.L., K.C.W., M.Y.J., Y.D., and L.A.W. assisted with myofibril experiments and analysis. J.A.R., B.C.B., and A.D.A. performed metabolic screening. G.C.S. and K.C.C. performed cardiolipin and CPT assays. H.X. analyzed RNA-seq data. M.R.G.T., A.V.A., J.C.C., M.R.B., and P.M.B. are responsible for the tissue bank containing human heart samples. A.V.A. assisted with analysis of HCM function. W.E.K., M.Y.J., T.A.M., P.M.B., and K.S. analyzed data and prepared the manuscript.
Conflicts of Interest
An application for a patent has been filed by the University of Colorado.
Acknowledgments
The authors thank Andrew Riching for advice. W.E.K. was supported by a postdoctoral fellowship from the University of Colorado Consortium for Fibrosis Research & Translation, NIH Cardiology training grant T32HL007822, an American Heart Association postdoctoral fellowship (19POST34380250), and an NIH/CCTSI CO-Pilot Mentored Faculty Award. K.S. was supported by funds from the Boettcher Foundation, American Heart Association, University of Colorado Department of Medicine Outstanding Early Career Scholar Program, Gates Frontiers Fund, and NIH HL133230. Y.H.L. (16POST30960017) and T.A.M. (16SFRN31400013) received support from the American Heart Association. T.A.M. was also supported by NIH HL150225, HL127240, DK119594, and HL116848.
Published: February 25, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.stemcr.2021.01.018.
Supplemental Information
Table S2-1: Gene ontology (GO) on differentially expressed genes. RNA-seq data were filtered to include only the top 12,500 expressed genes for all groups, then GO analysis was performed on genes displaying >1.5-fold regulation between designated groups. Table S2-2: KEGG pathway analysis on differentially expressed genes. RNA-seq data were filtered to include only the top 12,500 expressed genes for all groups, then KEGG pathway analysis was performed on genes displaying >1.5-fold regulation between designated groups.
References
- Anand P., Brown J.D., Lin C.Y., Qi J., Zhang R., Artero P.C., Alaiti M.A., Bullard J., Alazem K., Margulies K.B. BET bromodomains mediate transcriptional pause release in heart failure. Cell. 2013;154:569–582. doi: 10.1016/j.cell.2013.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedada F.B., Wheelwright M., Metzger J.M. Maturation status of sarcomere structure and function in human iPSC-derived cardiac myocytes. Biochim. Biophys. Acta. 2016;1863:1829–1838. doi: 10.1016/j.bbamcr.2015.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belus A., Piroddi N., Scellini B., Tesi C., D'Amati G., Girolami F., Yacoub M., Cecchi F., Olivotto I., Poggesi C. The familial hypertrophic cardiomyopathy-associated myosin mutation R403Q accelerates tension generation and relaxation of human cardiac myofibrils. J. Physiol. 2008;586:3639–3644. doi: 10.1113/jphysiol.2008.155952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer G.J., Cotman C.W. Survival and growth of hippocampal neurons in defined medium at low density: advantages of a sandwich culture technique or low oxygen. Brain Res. 1989;494:65–74. doi: 10.1016/0006-8993(89)90144-3. [DOI] [PubMed] [Google Scholar]
- Bupha-Intr T., Haizlip K.M., Janssen P.M. Role of endothelin in the induction of cardiac hypertrophy in vitro. PLoS One. 2012;7:e43179. doi: 10.1371/journal.pone.0043179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson C., Koonce C., Aoyama N., Einhorn S., Fiene S., Thompson A., Swanson B., Anson B., Kattman S. Phenotypic screening with human iPS cell-derived cardiomyocytes: HTS-compatible assays for interrogating cardiac hypertrophy. J. Biomol. Screen. 2013;18:1203–1211. doi: 10.1177/1087057113500812. [DOI] [PubMed] [Google Scholar]
- Carson D., Hnilova M., Yang X., Nemeth C.L., Tsui J.H., Smith A.S., Jiao A., Regnier M., Murry C.E., Tamerler C. Nanotopography-induced structural anisotropy and sarcomere development in human cardiomyocytes derived from induced pluripotent stem cells. ACS Appl. Mater. Interfaces. 2016;8:21923–21932. doi: 10.1021/acsami.5b11671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casals G., Ros J., Sionis A., Davidson M.M., Morales-Ruiz M., Jimenez W. Hypoxia induces B-type natriuretic peptide release in cell lines derived from human cardiomyocytes. Am. J. Physiol. Heart Circ. Physiol. 2009;297:H550–H555. doi: 10.1152/ajpheart.00250.2009. [DOI] [PubMed] [Google Scholar]
- Chi C., Leonard A., Knight W.E., Beussman K.M., Zhao Y., Cao Y., Londono P., Aune E., Trembley M.A., Small E.M. LAMP-2B regulates human cardiomyocyte function by mediating autophagosome-lysosome fusion. Proc. Natl. Acad. Sci. U S A. 2019;116:556–565. doi: 10.1073/pnas.1808618116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu W., Wan L., Zhao D., Qu X., Cai F., Huo R., Wang N., Zhu J., Zhang C., Zheng F. Mild hypoxia-induced cardiomyocyte hypertrophy via up-regulation of HIF-1alpha-mediated TRPC signalling. J. Cell Mol. Med. 2012;16:2022–2034. doi: 10.1111/j.1582-4934.2011.01497.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correia C., Koshkin A., Duarte P., Hu D., Teixeira A., Domian I., Serra M., Alves P.M. Distinct carbon sources affect structural and functional maturation of cardiomyocytes derived from human pluripotent stem cells. Sci. Rep. 2017;7:8590. doi: 10.1038/s41598-017-08713-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan Q., McMahon S., Anand P., Shah H., Thomas S., Salunga H.T., Huang Y., Zhang R., Sahadevan A., Lemieux M.E. BET bromodomain inhibition suppresses innate inflammatory and profibrotic transcriptional networks in heart failure. Sci. Transl. Med. 2017;9:eaah5084. doi: 10.1126/scitranslmed.aah5084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giacomelli E., Meraviglia V., Campostrini G., Cochrane A., Cao X., van Helden R.W.J., Krotenberg Garcia A., Mircea M., Kostidis S., Davis R.P. Human-iPSC-Derived cardiac stromal cells enhance maturation in 3D cardiac microtissues and reveal non-cardiomyocyte contributions to heart disease. Cell Stem Cell. 2020;26:862–879.e11. doi: 10.1016/j.stem.2020.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoes M.F., Grote Beverborg N., Kijlstra J.D., Kuipers J., Swinkels D.W., Giepmans B.N.G., Rodenburg R.J., van Veldhuisen D.J., de Boer R.A., van der Meer P. Iron deficiency impairs contractility of human cardiomyocytes through decreased mitochondrial function. Eur. J. Heart Fail. 2018;20:910–919. doi: 10.1002/ejhf.1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu D., Linders A., Yamak A., Correia C., Kijlstra J.D., Garakani A., Xiao L., Milan D.J., van der Meer P., Serra M. Metabolic maturation of human pluripotent stem cell-derived cardiomyocytes by inhibition of HIF1alpha and LDHA. Circ. Res. 2018;123:1066–1079. doi: 10.1161/CIRCRESAHA.118.313249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamakura T., Makiyama T., Sasaki K., Yoshida Y., Wuriyanghai Y., Chen J., Hattori T., Ohno S., Kita T., Horie M. Ultrastructural maturation of human-induced pluripotent stem cell-derived cardiomyocytes in a long-term culture. Circ. J. 2013;77:1307–1314. doi: 10.1253/circj.cj-12-0987. [DOI] [PubMed] [Google Scholar]
- Kumar S., Wang G., Liu W., Ding W., Dong M., Zheng N., Ye H., Liu J. Hypoxia-induced mitogenic factor promotes cardiac hypertrophy via calcium-dependent and hypoxia-inducible factor-1alpha mechanisms. Hypertension. 2018;72:331–342. doi: 10.1161/HYPERTENSIONAHA.118.10845. [DOI] [PubMed] [Google Scholar]
- Lian X., Hsiao C., Wilson G., Zhu K., Hazeltine L.B., Azarin S.M., Raval K.K., Zhang J., Kamp T.J., Palecek S.P. Robust cardiomyocyte differentiation from human pluripotent stem cells via temporal modulation of canonical Wnt signaling. Proc. Natl. Acad. Sci. U S A. 2012;109:E1848–E1857. doi: 10.1073/pnas.1200250109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundy S.D., Zhu W.Z., Regnier M., Laflamme M.A. Structural and functional maturation of cardiomyocytes derived from human pluripotent stem cells. Stem Cells Dev. 2013;22:1991–2002. doi: 10.1089/scd.2012.0490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X., Adelstein R.S. In vivo studies on nonmuscle myosin II expression and function in heart development. Front. Biosci. (Landmark Ed.) 2012;17:545–555. doi: 10.2741/3942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller C.L., Oikawa M., Cai Y., Wojtovich A.P., Nagel D.J., Xu X., Xu H., Florio V., Rybalkin S.D., Beavo J.A. Role of Ca2+/calmodulin-stimulated cyclic nucleotide phosphodiesterase 1 in mediating cardiomyocyte hypertrophy. Circ. Res. 2009;105:956–964. doi: 10.1161/CIRCRESAHA.109.198515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills R.J., Hudson J.E. Bioengineering adult human heart tissue: how close are we? APL Bioeng. 2019;3:010901. doi: 10.1063/1.5070106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills R.J., Titmarsh D.M., Koenig X., Parker B.L., Ryall J.G., Quaife-Ryan G.A., Voges H.K., Hodson M.P., Ferguson C., Drowley L. Functional screening in human cardiac organoids reveals a metabolic mechanism for cardiomyocyte cell cycle arrest. Proc. Natl. Acad. Sci. U S A. 2017;114:E8372–E8381. doi: 10.1073/pnas.1707316114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng K.M., Lau Y.M., Dhandhania V., Cai Z.J., Lee Y.K., Lai W.H., Tse H.F., Siu C.W. Empagliflozin ammeliorates high glucose induced-cardiac dysfuntion in human iPSC-derived cardiomyocytes. Sci. Rep. 2018;8:14872. doi: 10.1038/s41598-018-33293-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noland R.C. Exercise and regulation of lipid metabolism. Prog. Mol. Biol. Transl. Sci. 2015;135:39–74. doi: 10.1016/bs.pmbts.2015.06.017. [DOI] [PubMed] [Google Scholar]
- Pawloski-Dahm C.M., Song G., Kirkpatrick D.L., Palermo J., Gulick J., Dorn G.W., 2nd, Robbins J., Walsh R.A. Effects of total replacement of atrial myosin light chain-2 with the ventricular isoform in atrial myocytes of transgenic mice. Circulation. 1998;97:1508–1513. doi: 10.1161/01.cir.97.15.1508. [DOI] [PubMed] [Google Scholar]
- Pioner J.M., Racca A.W., Klaiman J.M., Yang K.C., Guan X., Pabon L., Muskheli V., Zaunbrecher R., Macadangdang J., Jeong M.Y. Isolation and mechanical measurements of myofibrils from human induced pluripotent stem cell-derived cardiomyocytes. Stem Cell Reports. 2016;6:885–896. doi: 10.1016/j.stemcr.2016.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piroddi N., Belus A., Scellini B., Tesi C., Giunti G., Cerbai E., Mugelli A., Poggesi C. Tension generation and relaxation in single myofibrils from human atrial and ventricular myocardium. Pflugers Arch. 2007;454:63–73. doi: 10.1007/s00424-006-0181-3. [DOI] [PubMed] [Google Scholar]
- Ronaldson-Bouchard K., Ma S.P., Yeager K., Chen T., Song L., Sirabella D., Morikawa K., Teles D., Yazawa M., Vunjak-Novakovic G. Advanced maturation of human cardiac tissue grown from pluripotent stem cells. Nature. 2018;556:239–243. doi: 10.1038/s41586-018-0016-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saggin L., Gorza L., Ausoni S., Schiaffino S. Troponin I switching in the developing heart. J. Biol. Chem. 1989;264:16299–16302. [PubMed] [Google Scholar]
- Schlame M., Ren M., Xu Y., Greenberg M.L., Haller I. Molecular symmetry in mitochondrial cardiolipins. Chem. Phys. Lipids. 2005;138:38–49. doi: 10.1016/j.chemphyslip.2005.08.002. [DOI] [PubMed] [Google Scholar]
- Shen Z., Ye C., McCain K., Greenberg M.L. The role of cardiolipin in cardiovascular health. Biomed. Res. Int. 2015;2015:891707. doi: 10.1155/2015/891707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiltoir J.I., Stratton M.S., Cavasin M.A., Demos-Davies K., Reid B.G., Qi J., Bradner J.E., McKinsey T.A. BET acetyl-lysine binding proteins control pathological cardiac hypertrophy. J. Mol. Cell Cardiol. 2013;63:175–179. doi: 10.1016/j.yjmcc.2013.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stehle R., Kruger M., Pfitzer G. Force kinetics and individual sarcomere dynamics in cardiac myofibrils after rapid ca(2+) changes. Biophys. J. 2002;83:2152–2161. doi: 10.1016/S0006-3495(02)73975-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan S.H., Ye L. Maturation of pluripotent stem cell-derived cardiomyocytes: a critical step for drug development and cell therapy. J. Cardiovasc. Transl. Res. 2018;11:375–392. doi: 10.1007/s12265-018-9801-5. [DOI] [PubMed] [Google Scholar]
- Tohyama S., Hattori F., Sano M., Hishiki T., Nagahata Y., Matsuura T., Hashimoto H., Suzuki T., Yamashita H., Satoh Y. Distinct metabolic flow enables large-scale purification of mouse and human pluripotent stem cell-derived cardiomyocytes. Cell Stem Cell. 2013;12:127–137. doi: 10.1016/j.stem.2012.09.013. [DOI] [PubMed] [Google Scholar]
- Weber N., Schwanke K., Greten S., Wendland M., Iorga B., Fischer M., Geers-Knorr C., Hegermann J., Wrede C., Fiedler J. Stiff matrix induces switch to pure beta-cardiac myosin heavy chain expression in human ESC-derived cardiomyocytes. Basic Res. Cardiol. 2016;111:68. doi: 10.1007/s00395-016-0587-9. [DOI] [PubMed] [Google Scholar]
- Weinberger F., Mannhardt I., Eschenhagen T. Engineering cardiac muscle tissue: a maturating field of research. Circ. Res. 2017;120:1487–1500. doi: 10.1161/CIRCRESAHA.117.310738. [DOI] [PubMed] [Google Scholar]
- Woulfe K.C., Ferrara C., Pioner J.M., Mahaffey J.H., Coppini R., Scellini B., Ferrantini C., Piroddi N., Tesi C., Poggesi C. A novel method of isolating myofibrils from primary cardiomyocyte culture suitable for myofibril mechanical study. Front. Cardiovasc. Med. 2019;6:12. doi: 10.3389/fcvm.2019.00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F., Pasumarthi K.B. Ultrastructural and immunocharacterization of undifferentiated myocardial cells in the developing mouse heart. J. Cell Mol. Med. 2007;11:552–560. doi: 10.1111/j.1582-4934.2007.00044.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S2-1: Gene ontology (GO) on differentially expressed genes. RNA-seq data were filtered to include only the top 12,500 expressed genes for all groups, then GO analysis was performed on genes displaying >1.5-fold regulation between designated groups. Table S2-2: KEGG pathway analysis on differentially expressed genes. RNA-seq data were filtered to include only the top 12,500 expressed genes for all groups, then KEGG pathway analysis was performed on genes displaying >1.5-fold regulation between designated groups.
Data Availability Statement
The accession number for the RNA-seq is GEO: GSE143608.