Summary
Polycomb group (PcG) proteins exist in distinct multi-protein complexes and play a central role in silencing developmental genes, yet the underlying mechanisms remain elusive. Here, we show that deficiency of retinoblastoma binding protein 4 (RBBP4), a component of the Polycomb repressive complex 2 (PRC2), in embryonic stem cells (ESCs) leads to spontaneous differentiation into mesendodermal lineages. We further show that Rbbp4 and core PRC2 share an important number of common genomic targets, encoding regulators involved in early germ layer specification. Moreover, we find that Rbbp4 is absolutely essential for genomic targeting of PRC2 to a subset of developmental genes. Interestingly, we demonstrate that Rbbp4 is necessary for sustaining the expression of Oct4 and Sox2 and that the forced co-expression of Oct4 and Sox2 fully rescues the pluripotency of Rbbp4-null ESCs. Therefore, our study indicates that Rbbp4 links maintenance of the pluripotency regulatory network with repression of mesendoderm lineages.
Keywords: RBBP4, PRC2, polycomb, embryonic stem cells, self-renewal, pluripotency, mesendoderm, Oct4, Sox2
Graphical Abstract
Highlights
-
•
RBBP4 deficiency in ESCs leads to spontaneous differentiation into mesendodermal lineages
-
•
Rbbp4 binding sites in ESCs substantially overlap with PRC2 binding
-
•
Rbbp4 is absolutely essential for PRC2 chromatin occupancy
-
•
Rbbp4 is necessary for sustaining the expression levels of Oct4 and Sox2
Polycomb group (PcG) proteins play a critical role in silencing developmental genes, yet the underlying mechanisms remain elusive. Yikai et al. report that PcG protein Rbbp4 represses developmental gene expression by recruiting PRC2 to the genes' promoter regions, thereby maintaining stem cell self-renewal and identity.
Introduction
The polycomb group (PcG) proteins function as chromatin-based transcriptional repressors that are essential for the specification and maintenance of cell fates (Kuroda et al., 2020; Schuettengruber et al., 2017). They broadly assemble into two biochemically and functionally distinct chromatin-associated complexes, the polycomb repressive complexes 1 and 2 (PRC1 and PRC2). PRC1 mono-ubiquitylates histone H2A at position 119 (H2AK119ub1) using its E3 ubiquitin ligase subunits, RING1A/B. Multiple PRC1 sub-complexes have been identified, which are characterized by the incorporation of distinct PCGF homologs and versatile accessory partners and can be grouped into canonical and non-canonical PRC1 (cPRC1 and ncPRC1). The cPRC1 is composed of four core subunits: RING E3 ligase (RING1A/B), PCGF (Pcgf1–6), PHC (polyhomeotic homologs; PHC1/2/3), and CBX (polycomb; CBX2/4/6/7/8) (Di Croce and Helin, 2013; Simon and Kingston, 2009; Turner and Bracken, 2013). In ncPRC1 complexes, RYBP, or its paralog YAF2 replaces CBX and PHC subunits found in cPRC1s (Turner and Bracken, 2013).
PRC2 consists of four core proteins, SUZ12, EED, RBBP4/7, and either of the two histone H3K27 methyltransferases, EZH1 or EZH2, which mono-, di-, and tri-methylates histone H3 at lysine 27 (H3K27me1/2/3) (Margueron et al., 2008; Piunti and Shilatifard, 2016). In addition to the four core subunits, PRC2 interacts with several auxiliary components that fine-tune its enzymatic activity and/or modulate its recruitment to chromatin. The PRC2.1 contains one of the three PCL1–3 (Polycomb-like protein 1–3) paralogs, whereas PRC2.2 is characterized by the presence of AEBP2 together with JARID2 (Holoch and Margueron, 2017). Notably, it has long been thought that PRC2-mediated H3K27me3, recognized by a chromodomain-containing CBX protein subunit of cPRC1, is required for recruiting PRC1 at PcG repressed sites, providing a functional bridge between the two major PcG complexes. However, this view was challenged by the identification of ncPRC1 complexes, which lack H3K27me3-binding CBX subunits. Furthermore, recent studies have shown that the ncPRC1 complex, but not the cPRC1 complex, promotes H2AK119ub-dependent recruitment of PRC2, suggesting a critical role for ncPRC1 in PcG-mediated gene control (Blackledge et al., 2014).
Mice deficient for Suz12, Ezh2, or Eed exhibited embryonic lethality due to gastrulation defects (Faust et al., 1998; O'Carroll et al., 2001; Pasini et al., 2004), underscoring the importance of these core components of PRC2 in early embryogenesis. In line with this, embryonic stem cells (ESCs) lacking Ezh2, Eed, and Suz12, were deficient in somatic cell reprogramming by cell fusion (Pereira et al., 2010a). Interestingly, ESCs deficient for Suz12, Eed, or Ezh2 appear to be normal with little effect on morphology and self-renewal, indicating that PRC2 may be dispensable for overall maintenance of pluripotency (Chamberlain et al., 2008; Montgomery et al., 2005; Pasini et al., 2007). It is worth highlighting here that functional redundancy among members of the PRC2 complex could account for the mild to no phenotype of mutants lacking individual PRC2 subunits in ESCs.
Retinoblastoma binding protein 4 (RBBP4) and RBBP7 are a pair of WD40 motif containing proteins sharing a sequence identity of 92%, which often exist together in multiple chromatin modifying complexes, such as the PRC2 (Conway et al., 2018), the NuRD complex (Feng and Zhang, 2003), and Sin3/HDAC complex (Kuzmichev et al., 2002). WD40 domain proteins are involved in a variety of fundamental biological processes, in which WD40 domains, containing 40–60 residues with a defining tryptophan-aspartate (WD) dipeptide, serve as scaffolds for complex assembly or provide platforms to recruit diverse molecules that form functional complexes. A recent study indicated that Rbbp4 is essential for early embryonic development in mice (Miao et al., 2020). However, the exact cause of early embryonic lethality of Rbbp4 knockouts and its potential roles in maintenance of stem cells or pluripotency have not been studied.
In this report, we demonstrate that ablating Rbbp4 gene expression in ESCs results in a loss of the undifferentiated state and the initiation of differentiation along mesendodermal lineage. Mechanistically, we demonstrate that Rbbp4 plays an important role in guiding the PRC2 to the loci of developmental genes where they establish and maintain the repressive states of lineage-specific genes in ESCs. Our study therefore identifies RBBP4 as an essential chromatin factor for the maintenance of ESC pluripotency and shows that it functions within the PRC2 complex to repress mesendoderm specification.
Results
Rbbp4 is essential in the maintenance of self-renewal and pluripotency in ESCs
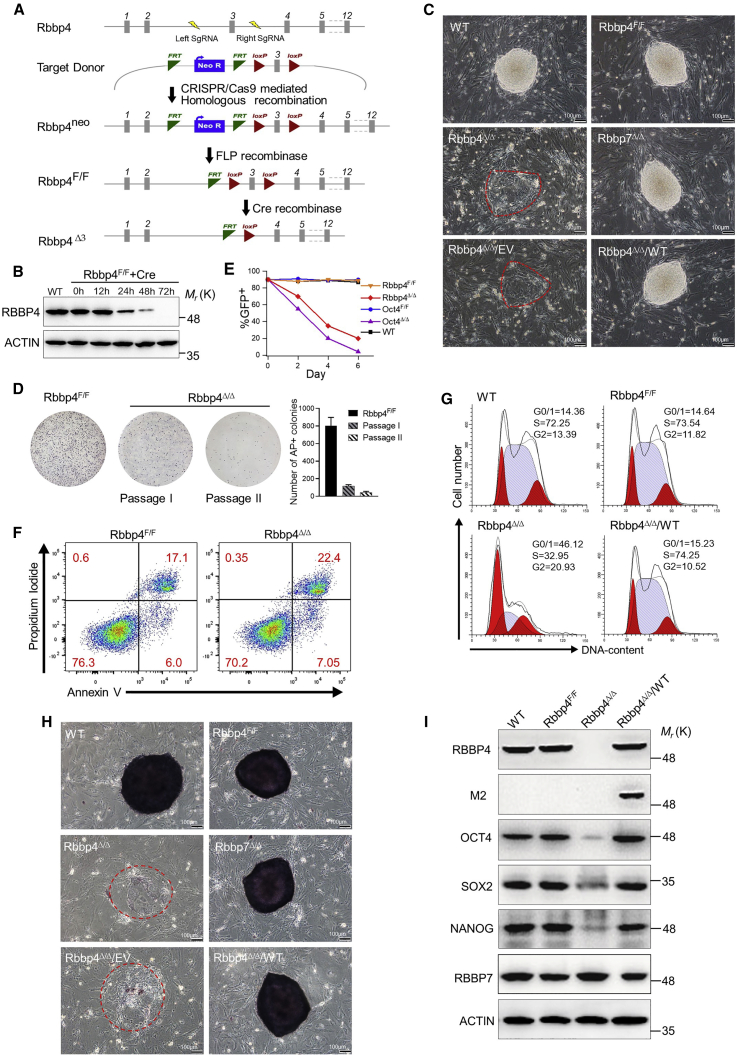
To circumvent the potential lethality that results from complete loss of Rbbp4 and to gain insight into the role of this gene in ESCs, we generated Rbbp4 conditional knockout mouse ESCs (Figures 1A and S1A–S1C), in which the third coding exon of both Rbbp4 alleles was flanked by parallel loxP sites, based on Cre-loxP system. In this Rbbp4 conditional floxed ESC line (Rbbp4F/F), Rbbp4 was completely removed 72 h after transfection with a plasmid encoding Cre, as evident in western blot (Figures 1B and S1D). Therefore, the cells were used for assay after 72 h of transfection throughout this study unless otherwise stated in the figure legends. Notably, Rbbp4F/F showed normal RBBP4 protein levels and formed robust colonies that were morphologically indistinguishable from wild-type ESCs. However, as shown in Figure 1C, in contrast to the Rbbp4F/F, ablation of Rbbp4 resulted in flat and spreading colonies without smooth edges. Importantly, ablation of Rbbp4 dramatically decreased secondary ES colony formation (Figure 1D). Consistently, in ESC competition assays, complete abolishment of Rbbp4 resulted in loss of self-renewal similar to deletion of Oct4 (Figure 1E). Rbbp4Δ/Δ ESCs displayed the same apoptotic rates as the Rbbp4F/F, as shown by annexin V staining (Figure 1F). Cell-cycle analysis revealed cells lacking Rbbp4 have a markedly extended G1 phase and a shortened S phase, suggesting that the impaired growth of Rbbp4Δ/Δ ESCs was due to an altered cell-cycle profile (Figure 1G). After alkaline phosphatase (AP) staining, Rbbp4F/F as well as wild-type ESC colonies stained bright and uniform, while the Rbbp4Δ/Δ colonies appeared dim, weak, and mosaic, suggesting loss of ESC self-renewal capability (Figure 1H). Consistent with the AP staining results, the expression of pluripotency genes Oct4, Sox2, and Nanog dramatically declined upon loss of Rbbp4 gene expression (Figure 1I). Importantly, lentivirus-mediated delivery of FLAG-tagged full-length Rbbp4 fully rescued the defective phenotypes associated with Rbbp4 deficiency. Remarkably, the complete loss of Rbbp7 (Figures S2A–S2E), which shares 92% of sequence identity with Rbbp4, caused no detectable defects in ESCs (Figures 1C and 1H). Together, these data indicate that Rbbp4 plays an important role in governing the pluripotent state in ESCs.
Figure 1.
Rbbp4 is essential in the maintenance of self-renewal and pluripotency in ESCs
(A) Schematic representation of the production for conditional inactivation of Rbbp4 in ESCs.
(B) Western blot showing RBBP4 levels in Rbbp4F/F-transfected Cre recombinase for different time points.
(C) Morphology of ESC colonies of indicated genotypes. Bright-field images of ESC colonies after 7 days of culture (grown from a single ESC). All ESC colony images were photographed at day 7 after seeding single-cell suspensions on feeder layers. Scale bar, 100 μm. The outlines of obviously defective colonies were circled in red. Rbbp4Δ/Δ ESCs expressing empty FLAG vector (Rbbp4Δ/Δ/EV) and vectors encoding wild-type RBBP4-FLAG (Rbbp4Δ/Δ/WT) are also shown.
(D) Secondary ES colony-replating assay. Bar graph shows the number of cells with AP-positive staining in the absence of RBBP4. Error bars represent means and STD from three 6 cm dishes.
(E) ESC growth competition assay. Oct4 knockout ESCs serve as a negative control.
(F) Representative fluorescence-activated cell sorting plots of annexin V and propidium iodide (PI) levels in Rbbp4F/F and Rbbp4Δ/Δ ESCs. Percentages of cells with different apoptosis marker levels are shown.
(G) Cell-cycle analysis of indicated ESCs. Percentages of cells in different phases are indicated. Representative histograms presented here show distribution of cells in sequential phases (G0/G1, red; S, light blue; and G2/M, red) of cell cycle.
(H) Alkaline phosphatase activity staining on each indicated ESC colony. Scale bar, 100 μm.
(I) Western blot analysis of OCT4, SOX2, NANOG, RBBP4, and RBBP7 protein expression in indicated ESCs; ACTIN served as a loading control.
See also Figures S1 and S2.
Loss of Rbbp4 results in aberrant expression of ESC core pluripotency factors and differentiation-associated genes
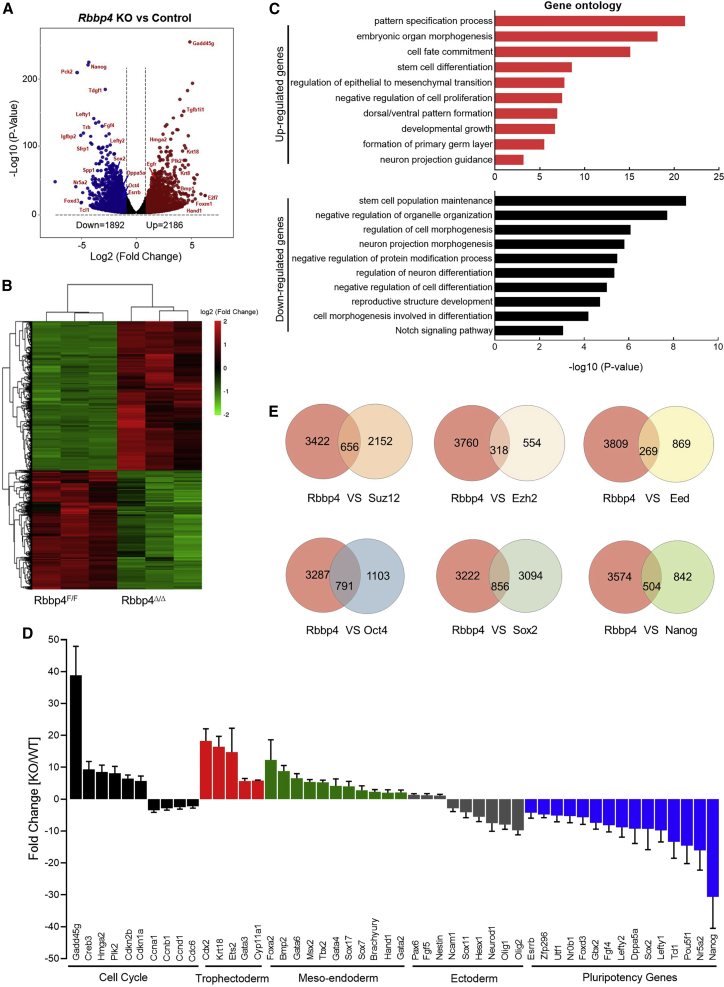
To explore the molecular basis of phenotypic alterations observed in Rbbp4-null ESCs and to rule out the possibility that loss of the undifferentiated state and self-renewal observed in Rbbp4Δ/Δ ESCs was due to a general impairment in cell proliferation, we performed gene expression analysis for Rbbp4F/F and Rbbp4Δ/Δ ESCs by RNA sequencing (RNA-seq) analysis. There were 4,078 genes that were differentially expressed by more than 2-fold upon Rbbp4 ablation (Figures 2A and 2B). About an equal number of genes were upregulated (2,186) or downregulated (1,892) in Rbbp4Δ/Δ ESCs. Gene ontology (GO) analysis showed that among genes downregulated in Rbbp4Δ/Δ ESCs were genes related to stem cell population maintenance, regulation of cell morphogenesis, and negative regulation of cell differentiation. Processes related to cell fate commitment, stem cell differentiation, pattern specification process, and tissue morphogenesis, were over-represented among the genes upregulated in the Rbbp4Δ/Δ ESCs (Figure 2C). By performing qRT-PCR analysis, we confirmed the differential expression of selected genes observed by RNA-seq analysis and showed that, while the expression of these genes was significantly reduced or increased in Rbbp4Δ/Δ ESCs (Figure 2D), their expression was largely unaffected in Rbbp7Δ/Δ ESCs (data not shown). Importantly, among Rbbp4Δ/Δ downregulated genes, our analysis revealed the pluripotency signature genes and transcription factors, including Oct4 (Pou5f1), Sox2, Nanog, Nr5a2, Dppa5a, and Esrrb (Figure 2D), which were shown to have critical roles in maintaining pluripotency in ESCs. In addition, qRT-PCR analysis of several lineage-specific markers demonstrated that loss of Rbbp4 led to the upregulation of mesendoderm markers (Foxa2, Gata6, Gata4, Sox17, Brachyury, Msx2, and Tbx2) and trophectoderm markers (Cdx2, Ets2, Gata3, and Krt18), whereas ectoderm markers (Pax6, Sox11, Neurod1, Olig1, and Olig2) were instead either unchanged or slightly decreased (Figure 2D). Consistent with our cell-cycle analysis (Figure 1G), we found a substantial number of genes involved in proliferation to be differentially expressed in Rbbp4Δ/Δ cells. These include p21WAF1/CIP1, a potent inducer of G1 arrest, and members of the cyclin family, key regulators in cell-cycle machinery (Figure 2D). Interestingly, deficiency of Rbbp4 in ESCs was associated with substantially reduced levels of the phosphorylated and hyperphosphorylated (ppRb) forms of the Rb protein at several residues (Figure S3A). As expected, a large number of the genes de-regulated in Rbbp4Δ/Δ cells was also observed in ESCs deficient for Suz12, Ezh2, and Eed (Figure 2E), consistent with the idea that they exist in the same complex. Moreover, transcriptional changes in Rbbp4Δ/Δ cells significantly overlapped with those seen in Oct4-, Sox2-, or Nanog-deficient ESCs, and those overlapping genes were enriched for pluripotency and cell differentiation. Altogether, the evidences collected so far clearly suggest that Rbbp4 has key roles in controlling the pluripotent state of ESCs.
Figure 2.
Loss of Rbbp4 results in aberrant expression of ESC core pluripotency factors and differentiation-associated genes
(A) Volcano plot showing the distribution of the differentially expressed (DE) genes with 2-fold changes upon Rbbp4 deletion. p < 0.05. Up- and downregulated genes are colored red and blue, respectively.
(B) Heatmap illustrating the RNA expression in Rbbp4F/F and Rbbp4Δ/Δ ESCs of RNA-seq analysis for 2-fold expression differentially expressed genes. False discovery rate < 0.05. Up- and downregulated genes are reported as red and green, respectively.
(C) Gene ontology (GO) enrichment analyses for biological processes associated with genes differentially expressed upon Rbbp4 deletion in ESCs. Analysis was carried out using Metascape (Zhou et al., 2019).
(D) Validation of RNA-seq data by qRT-PCR analysis. Relative mRNA levels of indicated cell-cycle-related genes, lineage-specific genes, and pluripotency-related genes in Rbbp4F/F after 3 days of Cre transfection were measured and data were normalized to β-actin relative to Rbbp4F/F. Data are pooled from three independent experiments and the error bars represent standard deviation of triplicate qPCR data.
(E) Venn diagrams (top) showing the overlap of the target genes between Rbbp4 and the core subunits of the PRC2 complex, respectively (Das et al., 2015; Pasini et al., 2007). Venn diagrams (bottom) showing the numbers of the regulated genes between Rbbp4 and pluripotency markers (Oct4, Sox2, and Nanog) (Ding et al., 2015; Loh et al., 2006).
See also Figure S3.
RBBP4 shares target genes with PRC2
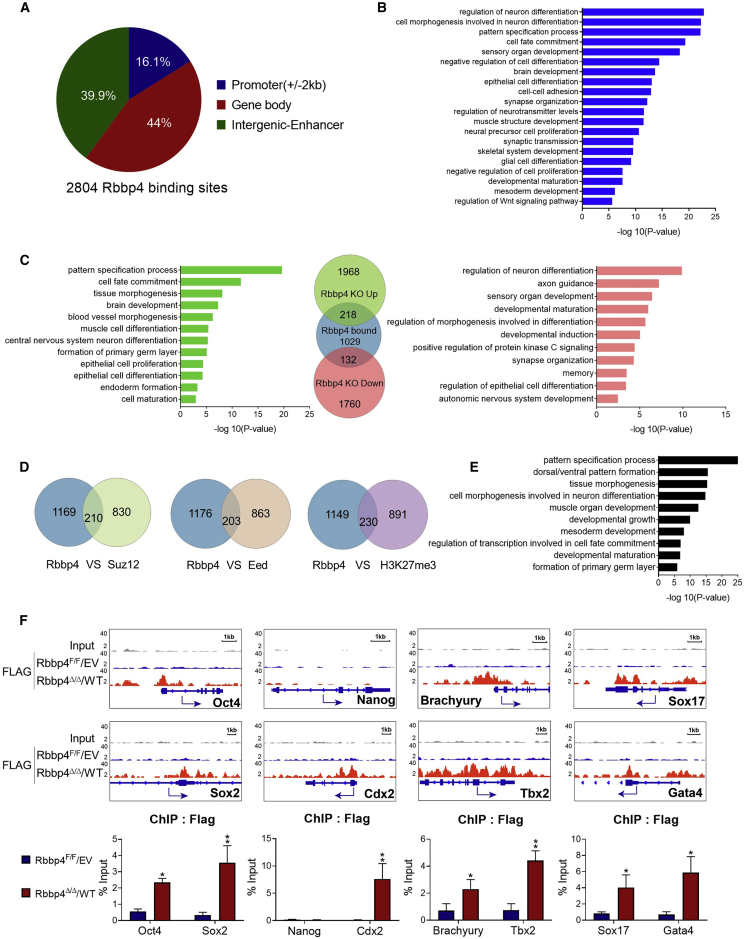
Given the rapid changes in gene expression patterns observed in Rbbp4Δ/Δ ESCs, we hypothesized that Rbbp4 may directly regulate both pluripotency and lineage-specific genes. We performed chromatin immunoprecipitation followed by high-throughput sequencing (ChIP-seq) analysis of RBBP4 binding. We found that the RBBP4 protein bound to 2,804 sites. A total of 16.1% of these sites were around gene promoters (defined as up to 2 kb upstream from the transcription start sites of the gene), 44% were within gene coding sequences, and 39.9% were intergenic-enhancer binding sites (Figure 3A). Thus, our analyses suggest that, on the genome, RBBP4 is mostly present at distal intergenic and intronic regions. Interestingly, GO analysis showed significant enrichment in genes involved in neuron development, pattern specification process, and regulation of cell differentiation (Figure 3B), strikingly resembling classical GO of PRC2 targets (Pereira et al., 2010b). Comparison of the de-regulated genes in Rbbp4Δ/Δ cells with the occupancy of RBBP4 in ESCs revealed that, among 2,186 genes that were upregulated in Rbbp4Δ/Δ, 218 genes were bound by RBBP4, whereas 132 of 1,892 downregulated genes were RBBP4 bound (Figure 3C). GO analysis of these overlapping genes again demonstrated a strong enrichment of terms involved in neuron differentiation, cell fate commitment, and pattern specification process. Since RBBP4 is physically associated with PRC2, as expected, we found that 210, 203, or 230 of Rbbp4 targets are also bound by SUZ12, EED, and H3K27me3, respectively (Figure 3D). Many of these genes were known critical regulators of pattern specification process, primary germ layer formation, cell fate commitment, and neuron differentiation (Figure 3E). Importantly, a detailed examination of RBBP4 ChIP-seq revealed that RBBP4 bound to the promoters of a group of genes associated with pluripotency or mesendodermal differentiation (Figure 3F).
Figure 3.
RBBP4 shares target genes with PRC2
(A) Pie chart showing the distribution of RBBP4 binding sites in mouse ESCs.
(B) GO enrichment analyses for genes that RBBP4 binds to. The biological processes of the top 20 are shown.
(C) Venn diagram showing the overlap between genes differentially expressed after Rbbp4 deletion and those occupied by RBBP4. GO analysis for biological processes associated with the overlapping genes.
(D) Venn diagram showing the overlap between genes that RBBP4 binds to and those occupied by SUZ12, EED, or H3K27me3.
(E) GO analysis of overlapping genes between RBBP4 binds and PRC2 targets.
(F) Genome browser tracks to show RBBP4 occupancy near lineage-specific markers for trophectoderm (Cdx2), mesoderm (Brachyury, Tbx2), and endoderm (Sox17, Gata4), Oct4 and Sox2 (up). ChIP-qPCR data showing binding of RBBP4-FLAG to representative RBBP4 target promoters (bottom).
Data are plotted as mean ± SD. (n = 3). ∗p < 0.05, ∗∗p < 0.01. See also Figure S4.
To identify the regions within the RBBP4 protein that are required for its chromatin targeting, we generated a set of FLAG-tagged Rbbp4 deletion mutants and introduced them into Rbbp4F/F ESCs (Figure S4A). Notably, all mutants were expressed at similar levels as evaluated by western blotting (Figure S4B). To test whether these mutants could retain recruitment to targets and to examine the ability of these mutants to maintain the expression of Rbbp4 target genes in the absence of endogenous Rbbp4, the cells were transduced with Cre recombinase and assayed 14 days later. ChIP-qPCR analyses demonstrated that the N-terminally deleted protein and the mutants with deletion of WD40 domains (individual or tandem), did not bind to targets (Figures S4C and S4D). Consistent with their failure to bind chromatin in ESCs, these mutants also lost the ability to restore target gene expression (Figure S4E). In contrast, the C terminus of RBBP4 was dispensable for binding and thus maintain these gene expressions as effectively as the wild type. To gain insight into the ability of these mutants to rescue self-renewal defect observed in Rbbp4Δ/Δ ESCs, we analyzed their ability to produce colonies after seeding on mouse embryonic fibroblasts (MEFs). As shown in Figures S4F–S4H, C-terminal deletion mutants fully rescued wild-type levels of growth in ESCs. However, RBBP4 lacking the WD40 domains or N-terminal virtually abolished its ability to restore the colony growth defect. Taken together, these results indicate that both N-terminal and WD40 domains are essential for the chromatin targeting of RBBP4 and show that these regions confer upon RBBP4 the capacity to control ESC properties.
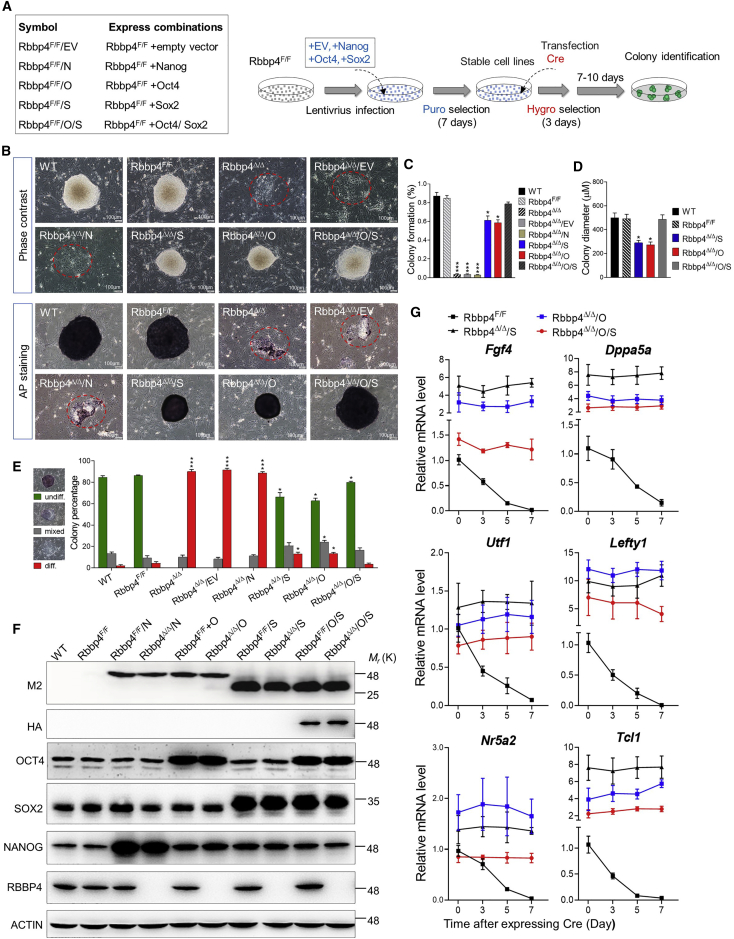
Rbbp4 governs pluripotency via maintaining the expression of Oct4 and Sox2
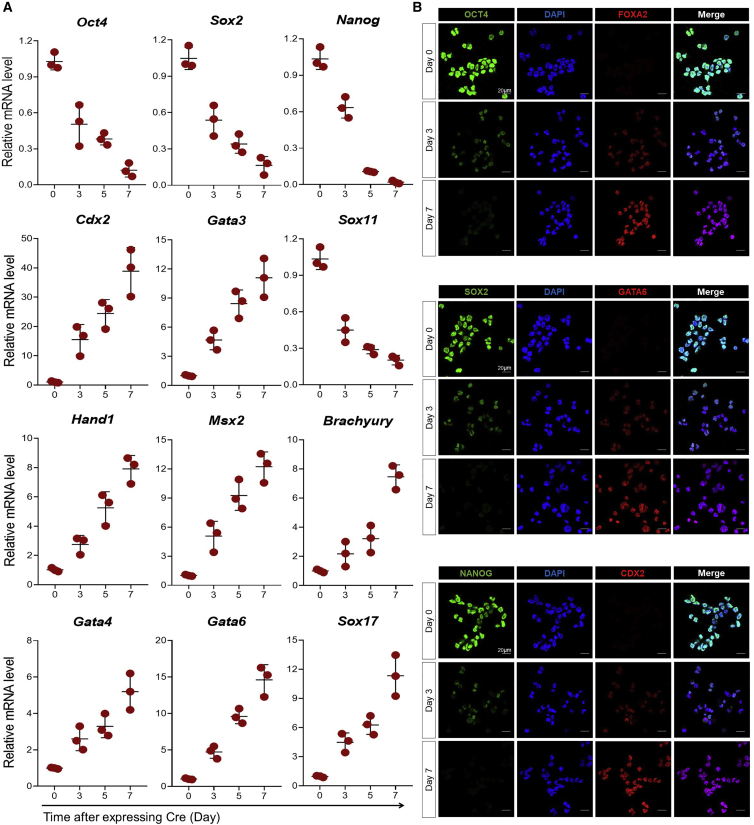
To evaluate transcriptional mediators responsible for loss of pluripotency and lineage specification upon Rbbp4 deficiency, we examined the temporal mRNA expression of pluripotency-related genes as well as three germ layer marker genes in Rbbp4F/F ESCs after transfection with Cre. As shown in Figure 4A, real-time qRT-PCR analysis revealed that Oct4, Sox2, and Nanog mRNA levels significantly decreased within 3 days after Rbbp4 ablation and declined steadily thereafter to baseline levels on day 7. In contrast, mesendoderm markers (Gata4, Gata6, Sox17, Hand1, Msx2, and Brachyury) and trophectoderm markers (Cdx2 and Gata3) were upregulated at the same time point that the mRNA of core pluripotency factors decreased. The precocious differentiation of Rbbp4Δ/Δ ESCs was also monitored by immunofluorescence microscopy to detect the expression profile of germ layer markers alongside pluripotency factors (Figure 4B). In Rbbp4F/F ESCs, OCT4, SOX2, and NANOG expression was high, whereas positivity for FOXA2, GATA6, and CDX2 was rarely observed, indicating the undifferentiated state of the cells. After transient transfection of a Cre recombinase expression plasmid, FOXA2, GATA6, and CDX2 expression gradually increased in a time-dependent manner, which was accompanied by the decreased expression of a panel of pluripotency genes.
Figure 4.
Rbbp4 knockout significantly reduces the expression of pluripotency-associated genes and promotes mesendodermal gene expression in ESCs
(A) Time-course analyses of pluripotency-associated genes and the lineage-specific markers for trophectoderm, mesoderm, and endoderm expression levels in Rbbp4F/F ESCs after transient expression of Cre recombinase. Expression is normalized by β-actin.
(B) Immunofluorescence showing co-staining of the pluripotency markers (OCT4, SOX2, and NANOG) and the lineage-specific markers (FOXA2, GATA6, and CDX2) at the indicated time points after expressing Cre recombinase in Rbbp4F/F ESCs, respectively. DAPI (blue), pluripotency marker genes (green), the lineage-specific markers (red), and the merge picture are shown. Magnification, 63×.
Data are represented as mean ± SD of three independent experiments.
We found RBBP4 binding enrichment at genes encoding critical components of the pluripotency network, such as Klf4, Sox2, and Oct4, whose expression is downregulated upon Rbbp4 ablation (Figures 3F, 4A, and S4C). These observations support the notion that Rbbp4 silences differentiation programs in ESCs through regulation of these genes. Therefore, it would be extremely interesting to know if the defects observed in Rbbp4Δ/Δ ESCs depend on lack of Oct4, Nanog, or Sox2 repression. To this end, we attempted to rescue Rbbp4Δ/Δ ESCs by introducing the transgenes of Oct4, Nanog, or Sox2. The Rbbp4-null rescue system utilized Rbbp4F/F ESCs, in which the floxed Rbbp4 alleles were excised by Cre recombinase (Figure 5A). Introducing Nanog (Rbbp4Δ/Δ/Nanog) did not significantly restore the self-renewal defect observed in Rbbp4Δ/Δ ESCs, but introducing either the Oct4 (Rbbp4Δ/Δ/Oct4) or Sox2 (Rbbp4Δ/Δ/Sox2) transgenes successfully partially rescued the propagation of Rbbp4Δ/Δ ESCs. Most importantly, forced (ectopic) expression of both Oct4 and Sox2 together (Rbbp4Δ/Δ/Oct4/Sox2) completely rescued the proliferation defect of Rbbp4-deficient ESCs (Figures 5B–5E). The Rbbp4Δ/Δ/Oct4, Rbbp4Δ/Δ/Sox2, or Rbbp4Δ/Δ/Oct4/Sox2 cells were able to form viable colonies on feeder cells and expand continuously for at least 50 passages without any significant change in colony morphology. It is worth noting that, although these three rescued lines exhibited similar expression levels of Oct4 and Sox2 (Figure 5F), the expression levels of other pluripotency-associated genes, such as Fgf4, Dppa5a, Utf1, Lefty1, Nr5a2, and Tcl1, in Rbbp4Δ/Δ/Oct4 or Rbbp4Δ/Δ/Sox2 were relatively higher than those in Rbbp4Δ/Δ/Oct4/Sox2 (Figure 5G).
Figure 5.
Rbbp4 governs pluripotency by maintaining the expression of Oct4 and Sox2
(A) Left: reference legend for cell lines used in Figure 5. Right: experimental diagrams of rescue assay.
(B) Representative images of the ESC colony of the indicated genotypes cultured for 7 days. Top, phase-contrast microscopy; bottom, AP staining.
(C) Percentage of isolated single ESCs of the indicated genotypes giving rise to macroscopic colonies.
(D) Bar graphs show the mean diameter of 30 random ESC colonies of the indicated genotypes.
(E) Quantitative analysis of colony formation assay in ESCs of indicated genotypes. AP-stained colonies were scored as undifferentiated (undiff.), mixed or differentiated (diff.).
(F) Western blot analysis of OCT4, SOX2, and NANOG protein levels after the overexpression of Oct4, Sox2, and Nanog in Rbbp4F/F ESCs and Rbbp4Δ/Δ ESCs.
(G) Time-course analysis of pluripotency-associated gene expression after expressing Cre in the corresponding cell lines by qRT-PCR.
Data in (C–E) represent the mean ± SD of three independent experiments. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001 (Student's t test) compared with the control.
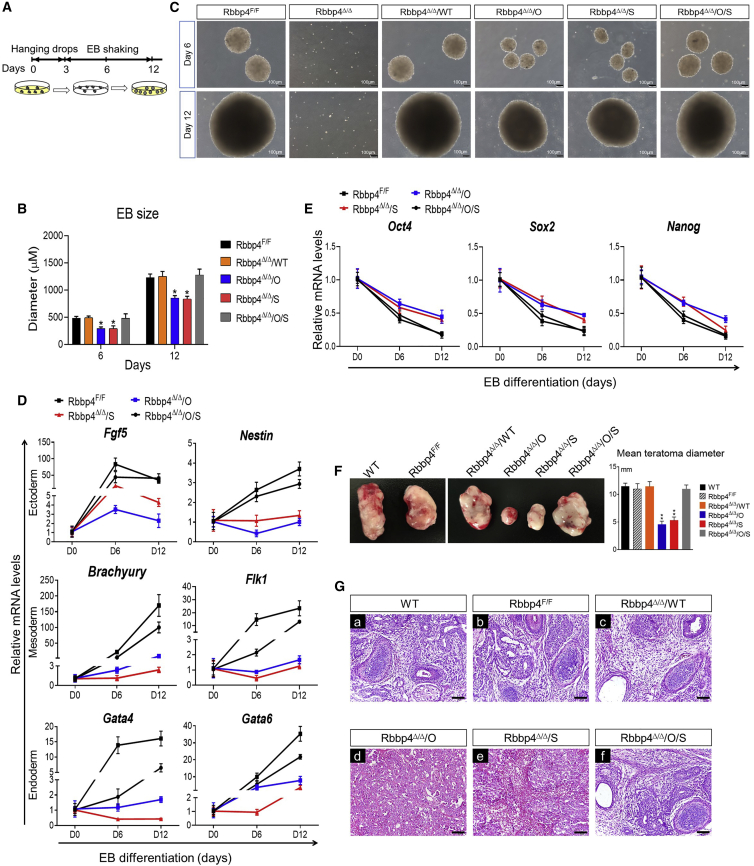
We next examined the potential of these rescued cells to undergo in vitro differentiation. Rbbp4Δ/Δ/Oct4, Rbbp4Δ/Δ/Sox2, or Rbbp4Δ/Δ/Oct4/Sox2 and control ESCs were tested for their capacity to form embryoid bodies (EBs) (Figure 6A). Rbbp4Δ/Δ ESCs failed to form EBs. However, as shown in Figures 6B and 6C, Rbbp4Δ/Δ/Oct4/Sox2 ESCs produce EBs comparable with those of the wild type in size. Rbbp4Δ/Δ/Oct4 or Rbbp4Δ/Δ/Sox2 had the capacity to form EBs, but they were greatly reduced in size than those of the wild type during the differentiation process. In addition, qRT-PCR analysis revealed that Rbbp4Δ/Δ/Oct4 or Rbbp4Δ/Δ/Sox2 EBs expressed reduced mRNA levels of marker genes for the three germ layers (endoderm, mesoderm, and ectoderm), whereas the expression levels of these germ layer markers in Rbbp4Δ/Δ/Oct4/Sox2 EBs were similar to that of wild type (Figure 6D). In contrast to the expression of these germ layer-specific genes, pluripotency-related genes, namely Oct4, Nanog, and Sox2, were sharply depleted during the course of wild-type and Rbbp4Δ/Δ/Oct4/Sox2 EB differentiation, while Rbbp4Δ/Δ/Oct4 or Rbbp4Δ/Δ/Sox2 EB exhibited similar trends, but of a lesser magnitude (Figure 6E). Consistent with these results, teratomas derived from wild-type and Rbbp4Δ/Δ/Oct4/Sox2 ESCs contained multiple tissue types from all three germ layers, whereas Rbbp4Δ/Δ/Oct4 or Rbbp4Δ/Δ/Sox2 teratomas lacked ectodermal, endodermal, and mesodermal structures (Figures 6F and 6G). The differentiation defects observed in Rbbp4Δ/Δ/Oct4 or Rbbp4Δ/Δ/Sox2 might be due to higher expression levels of a subset of pluripotency-associated genes.
Figure 6.
Overexpression of Sox2 and Oct4 maintains ESC self-renewal and pluripotency in the absence of Rbbp4 in vitro and in vivo
(A) Schematic illustration of the embryoid body (EB) formation procedure for differentiation of ESCs.
(B) The diameter statistics of EB. Thirty 30 EB diameters of each genotype were measured at each time point.
(C) Phase-contrast images of floating EB derived from indicated ESCs at day 6 and day 12. Scale bar, 100 μm.
(D) qRT-PCR analysis of lineage-specific markers at day 0, day 6, and day 12 during EB formation from the indicated ESC lines. Fgf5, Nestin (ectoderm), Brachyury, Flk1 (mesoderm), and Gata4, Gata6 (endoderm). Data are normalized to the expression levels in ESCs. Error bars represent ± SD (n = 3).
(E) qRT-PCR mRNA analysis of pluripotency markers during time-course differentiation of Rbbp4F/F, and rescue of Rbbp4 in Rbbp4Δ/Δ EBs. Samples were collected at different days.
(F) In vivo differentiation potential of formation teratomas of indicated ESCs. Left: the morphological characteristics of teratomas are shown. Right: average teratoma size at 4 weeks after injection in the indicated groups.
(G) Histological analysis of teratomas. (A–F) Histological sections of hematoxylin and eosin-stained teratomas. (A, B, C, F) Each teratoma contained three embryonic germ layer tissues. (D and E) Immature germ layer differentiation is shown. Scale bar, 50 μm.
Data in (B and D–F) represent mean ± SD obtained from three independent experiments. ∗p < 0.05 and ∗∗p < 0.01 (Student's t test) compared with the control.
RBBP4 guides PRC2 recruitment and H3K27 trimethylation at genomic loci
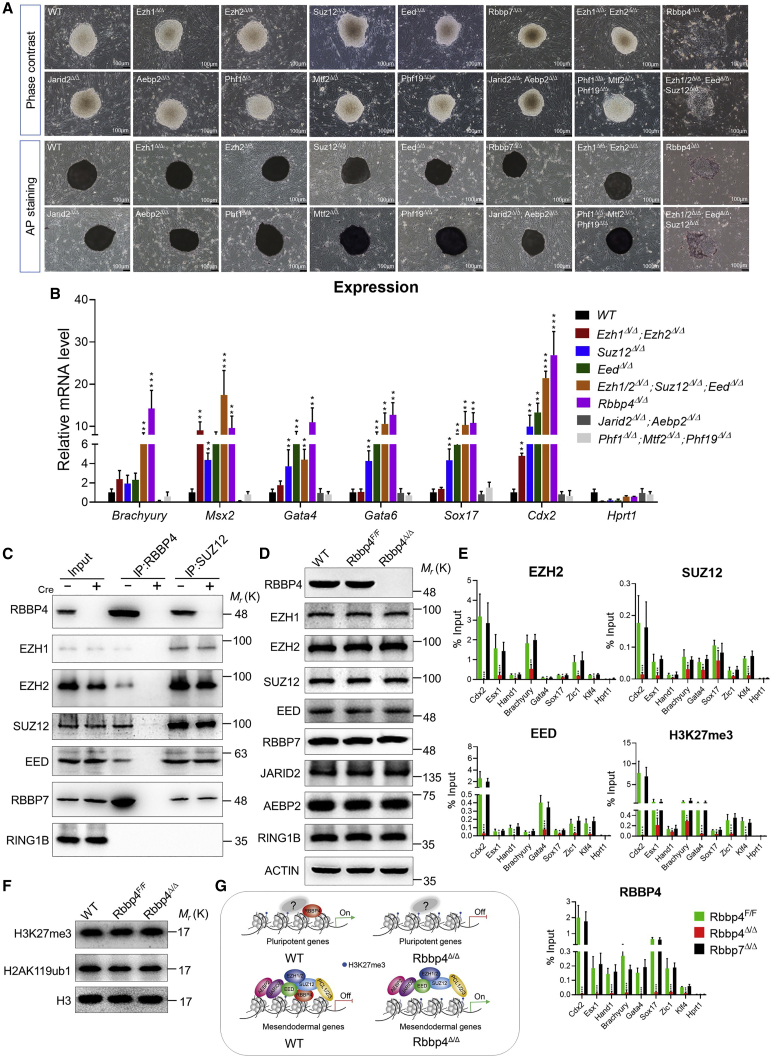
RBBP4 has been found to exist in a complex with PRC2 (Conway et al., 2018). ESCs lacking a single component of the PRC2 complex, such as Eed, Ezh2, or Suz12, show marginal disruption of self-renewal (Chamberlain et al., 2008). However, the functional redundancy among the members of the PRC2 complex has not yet been addressed. To characterize the ability of each PRC2 member to support the self-renewal of ESCs, we generated individual or combined PRC2-deficient ESC lines by using CRISPR-Cas9 technology (Figures S5A–S5J and S6A–S6I) and analyzed their ability to form colonies after seeding on mitotically inactivated MEFs. The ESCs deficient for each of the PRC2 subunits were viable and formed dome-shaped undifferentiated AP-positive colonies, which were morphologically indistinguishable from those formed by wild-type cells (Figure 7A). In addition, the lack of deficits in ESC proliferation in Ezh1, Jarid2, and Phf1, or their closest family members Ezh2, Aebp2, and Mtf2/Phf19, respectively, single knockout is not likely caused by each other's compensation because double or triple knockout ESCs (Ezh1Δ/Δ; Ezh2Δ/Δ, Jarid2Δ/Δ; Aebp2Δ/Δ, or Phf1Δ/Δ; Mtf2Δ/Δ; Phf19Δ/Δ) did not display significantly worse defects than single knockout ESCs. However, quadruple PRC2 knockout ESCs (Ezh1Δ/Δ; Ezh2Δ/Δ; EedΔ/Δ; Suz12Δ/Δ) exhibited reduction of both colony number and size with an increased proportion of partially and fully differentiated populations and reduced AP activity, suggestive of a degree of functional redundancy among core members of the PRC2 family (Figures 7A and S5K-S5M). Interestingly, the mesendoderm (Brachyury, Msx2, Gata4, Gata6, and Sox17) and trophectoderm genes (Cdx2) in Ezh1Δ/Δ; Ezh2Δ/Δ; EedΔ/Δ; Suz12Δ/Δ ESCs reached similar levels as those in Rbbp4Δ/Δ ESCs, suggesting that the induction of this lineage-specific gene expression is the result of loss of function of the PRC2 complex in which RBBP4 is a component. These lineage-specific genes also showed significant elevations in their expression levels in Suz12Δ/Δ, EedΔ/Δ, or Ezh1Δ/Δ; Ezh2Δ/Δ ESCs, although the magnitude of the elevation was not as prominent as that observed in Rbbp4Δ/Δ ESCs (Figure 7B). However, no such induction was evident in Jarid2Δ/Δ; Aebp2Δ/Δ or Phf1Δ/Δ; Mtf2Δ/Δ; Phf19Δ/Δ ESCs. The distinct phenotypes observed in Ezh1Δ/Δ; Ezh2Δ/Δ; EedΔ/Δ; Suz12Δ/Δ, Jarid2Δ/Δ; Aebp2Δ/Δ, or Phf1Δ/Δ; Mtf2Δ/Δ; Phf19Δ/Δ ESCs strongly suggest that additional recruitment mechanisms contribute to recruiting the PRC2 complex to its genomic targets.
Figure 7.
RBBP4 guides PRC2 recruitment and H3K27 trimethylation at genomic loci
(A) Bright-field images of an ESC colony of wild-type and indicated genotypes after 7 days of culture. AP staining images of the ESC colony that arose from the wild-type and indicated genotypes. Scale bar, 100 μm.
(B) qRT-PCR quantification of Rbbp4 targets in indicated cell lines normalized to β-actin. Hprt1 is represented as a negative control.
(C) Endogenous coimmunoprecipitations of RBBP4 and SUZ12 in Rbbp4F/F transfected with Cre (+) or control vector (−) for 72 h, followed by western blot analysis with the indicated antibodies against core subunits of PRC2.
(D) Western blot analyses using the indicated antibodies against PRC2 subunits on whole-cell lysates from wild-type (WT), Rbbp4F/F, and Rbbp4Δ/Δ ESCs.
(E) ChIP-qPCR analysis of EZH2, SUZ12, EED, H3K27me3, and RBBP4 binding at the indicated regions of pluripotent transcription factors and mesendodermal genes (normalized to input) in Rbbp4F/F, Rbbp7Δ/Δ, or Rbbp4F/F ESCs that were transfected with Cre after 3 days.
(F) Western blot for H3K27 tri-methylation and H2AK119 mono-ubiquitination on nuclear lysates from Rbbp4Δ/Δ and matched control ESCs. Histone 3 was used as a loading control.
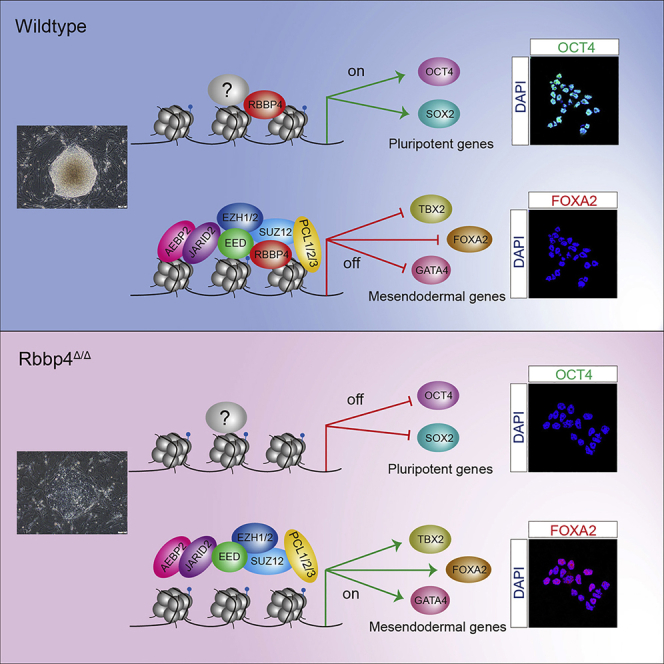
(G) Proposed model of Rbbp4-mediated pluripotency maintenance of ESCs.
Data in (B and E) represent the mean ± SD of three independent experiments. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001 (Student's t test) compared with the control. See also Figures S5 and S6.
To confirm the interaction between RBBP4 and PRC2 core members, we performed endogenous coimmunoprecipitation experiments in ESCs and demonstrated that RBBP4 coprecipitated RBBP7, EZH1, EZH2, EED, and SUZ12. Importantly, immunoprecipitation analysis for SUZ12 in Rbbp4Δ/Δ ESCs revealed that Rbbp4 ablation did not affect the association of core PRC2 components (Figure 7C). Furthermore, as shown in Figure 7D, Rbbp4 deletion did not appear to adversely affect the protein levels of the components of PRC2. We next examined a possible cooperation between RBBP4 and PRC2 at the chromatin level. Thus, we analyzed PRC2 at its target genes in Rbbp4Δ/Δ ESCs by utilizing ChIP-qPCR assays. These experiments showed an impairment of genomic EZH2, SUZ12, and EED binding at target loci that we examined in the absence of RBBP4 but not RBBP7, accompanied by a dramatic decrease in H3K27me3 (Figure 7E). The global H3K27me3 levels were similar in wild type (Figure 7F), Rbbp4F/F, and Rbbp4Δ/Δ ESCs, showing that the observed reduction of H3K27me3 at the PRC2 target promoters is due to changes in local EZH1/2 deposition. Collectively, these results demonstrate that RBBP4 is absolutely crucial for genomic loading of the entire PRC2 complex and for H3K27 methylation at its target genes in ESCs.
Discussion
RBBP4 is a ubiquitously expressed nuclear protein that is a member of a highly conserved subfamily of WD repeat-containing proteins (Qian et al., 1993), and targeted disruption of Rbbp4 leads to preimplantation embryonic lethality in mice (Miao et al., 2020), but the underlying molecular mechanisms are largely undetermined. Here, our data demonstrated that Rbbp4 maintains the pluripotent and self-renewal state of ESCs by promoting transcription of pluripotency factors and repressing the expression of lineage-specific genes. We also show that Rbbp4 targets the PRC2 complex to the promoters of lineage-specific genes to repress their expression in ESCs; thus, RBBP4 activity appears to be a key determinant of cell fate specification and lineage commitment in ESCs. Surprisingly, despite the striking similarity of the sequence shared by RBBP4 and RBBP7, and although both of them are often found within the same complexes, Rbbp4 but not Rbbp7 null mutants displayed severe defects, suggesting a specific role of Rbbp4 in sustaining ESC identity. The reason for such striking differences between Rbbp4 and Rbbp7 is not fully understood but it may be related to their chromatin binding ability in ESCs (Figure 7E).
RBBP4 tightly associates with EED, EZH1/2, and SUZ12 to form the core PRC2 complex
Although EZH1/2 catalyze H3K27me2/3, all the PRC2 core members are required for EZH1/2 to exert their catalytic activity. In addition, there are several substoichiometric accessory factors that associate with PRC2. In mammals, these include JARID2, AEBP2, PHF1, MTF2, and PHF19. Although these accessory components seem not particularly essential for PRC2 catalytic activity, they appear to modulate PRC2 enzymatic activity and/or its targeting to specific genomic loci. In this study, we demonstrated that Rbbp4-deficient ESCs, despite maintaining normal H3K27me3 levels, lost their normal undifferentiated colony morphologies and gave rise to flattened fibroblast-like cells. In addition, Rbbp4Δ/Δ cells failed to efficiently form colonies in secondary replating assays. Therefore, spontaneous differentiation induced by the ablation of Rbbp4 in ESCs resulted in the loss of self-renewal and pluripotency. Apart from Rbbp4, no other PRC2 members has been shown to play an essential role in the maintenance of self-renewal of ESCs (Chamberlain et al., 2008). Importantly, concomitant disruption of Ezh1/2, Eed, and Suz12 triggers spontaneous differentiation of ESCs toward primitive endoderm and mesoderm, a phenotype reminiscent of, but not as severe as, that seen in Rbbp4Δ/Δ ESCs. Notably, a considerable number of RBBP4 target regions was also occupied by EZH1/2, EED, and SUZ12, and was decorated with H3K27me3. These findings strongly implicate RBBP4 in recruitment of the PRC2 complex to target loci in ESCs. Indeed, EED, SUZ12, and EZH2, as well as the H3K27me3 mark, were absent or markedly reduced in Rbbp4Δ/Δ but not Rbbp7Δ/Δ ESCs. The most severe phenotype of Rbbp4Δ/Δ ESCs is accordance with the crucial importance of RBBP4 for genomic PRC2 targeting. As RBBP4 can associate with protein complexes other than PRC2 (Feng and Zhang, 2003; Kuzmichev et al., 2002), the severe phenotype in Rbbp4Δ/Δ ESCs can also be explained by the impaired function of these complexes in which RBBP4 is a common component. Notably, Rbbp4 was previously identified as being essential for the maintenance of human ESCs (O'Connor et al., 2011). It will be interesting to see whether a similar mechanism also exists in human ESCs. Our study establishes a critical role of RBBP4 in the recruitment of the PRC2 complex to genes essential for ESC differentiation along the mesendoderm lineage. Dissecting the potential contribution of JARID2, AEBP2, PHF1, MTF2, and PHF19 to RBBP4-guided PRC2 chromatin occupancy is warranted for future investigation.
The maintenance of self-renewal and pluripotency of ESCs depends mostly on three core transcription factors namely SOX2, OCT4, and NANOG, which induce genes necessary for sustaining the undifferentiated state and repress others involved in lineage commitment and terminal differentiation (Loh et al., 2006; Young, 2011). Interestingly, these three master transcription factors jointly sustain each other's transcription in autoregulatory and feedforward loops in ESCs (Boyer et al., 2005). Here, we show that knockout of Rbbp4 in ESCs dramatically reduces the expression of Oct4, Sox2, and Nanog. ChIP-seq and ChIP-qPCR demonstrated that the promoters of Oct4 and Sox2 were bound by RBBP4, suggesting that Rbbp4 positively regulates their expression. Oct4 and Sox2 are of interest as they have been shown to be critical for the pluripotency and self-renewal of ESCs. Importantly, forced expression of Oct4 or Sox2 but not Nanog could partially rescue the defect in Rbbp4-deficient ESCs. In addition, the phenotypes in Rbbp4Δ/Δ ESCs could be fully rescued by co-overexpression of both Oct4 and Sox2. However, several ChIP-seq studies have consistently failed to identify OCT4 and SOX2 as PRC2 target genes (Højfeldt et al., 2019; Tamburri et al., 2020), suggesting that Rbbp4 keeps the ESCs in a pluripotent state by maintaining the requisite levels of OCT4 and SOX2 in a PRC2-independent manner. In conclusion, our study strongly supports our model in which RBBP4 maintains ESCs in a pluripotent state by preventing mesendoderm specification and by regulating expression of Oct4 and Sox2 (Figure 7G).
RBBP4 was originally identified based on its interaction with the Rb tumor suppressor protein (Qian et al., 1993), which is essential for the maintenance of self-renewal and pluripotency in human ESCs (Chetty et al., 2013; Conklin et al., 2012; Conklin and Sage, 2009; Li et al., 2018b). The results of these studies in human ESCs together with our finding consolidate the notion that cell-cycle machinery and the maintenance of pluripotency are intricately connected to safeguard ESC identity (Pauklin and Vallier, 2013; Ruiz et al., 2011; Shcherbina et al., 2019). Interestingly, prevalent overexpression of Rbbp4 has been implicated in a variety of malignancies (Li et al., 2018a), suggesting that induction of carcinoma cell differentiation by directly targeting Rbbp4 might be a promising therapeutic approach for tumor treatment.
Experimental procedures
CRISPR-Cas9-mediated genome editing
The pSptCas9(BB)-2A-Puro(PX459) V2.0 vector was obtained from Addgene (no. 62988) and sgRNAs were designed using online CRISPR design tool (http://crispor.tefor.net). To generate stable knockout ESC lines, ESCs were transfected with a pair of Cas9 guides flanking the deletion region using Lipofectamine 2000 according to the manufacturer's instruction. Genomic DNA samples from individual ESC clones were PCR screened for the desired deletion and correct clones were validated by both qRT-PCR and western blot. To generate a conditional knockout of Rbbp4 in ESCs, ESCs were subjected to genomic engineering to flank the third coding exon of Rbbp4 (exon 3) with loxP sites in a parallel orientation using homology arms of approximately 2 kb and appropriate Cas9 guides. Correct Rbbp4F/F clones were validated by both qRT-PCR and western blot to confirm Rbbp4 removal by transient transfection of a Cre-expressing plasmid.
Statistics
Statistical analysis was carried out using GraphPad Prism 8.0 software. All experiments were carried out in at least three independent biological replicates for each group unless stated otherwise. All data are expressed as mean ± SD. Statistical significance is presented in the figures as ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and not significant (ns, p > 0.05) (Student’s t test).
Data and code availability
Sequencing data performed for this study are deposited in GEO database under the accession numbers GSE144155 (RNA-seq) and GSE155029 (ChIP-seq). Additional datasets used in this study are detailed in Table S4.
Author contributions
Conceptualization, J.Q.; methodology, T.S., C.W., S.L., Y.Z., and K.H.; formal analysis, C.W., T.S., Y.Z., L.D., and J.Q.; mechanistic investigations, C.W., S.L., L.D., Y.Z., Y.H., and J.Q.; writing – original draft, Y.H. and J.Q.; writing – review & editing, J.Q., Y.H., Y.X., and Q.J.; funding acquisition, J.Q.; supervision, J.Q.
Acknowledgments
We are indebted to Drs Zhenji Gan and Lin Liu for their helpful suggestions and for kindly providing reagents. We thank members of our laboratory for helpful discussion. This work was supported by grants from the National Natural Science Foundation of China (31970810 and 31671532) to J.Q.
Published: February 18, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.stemcr.2021.01.009.
Contributor Information
Yin Xia, Email: xia.yin@cuhk.edu.hk.
Qing Jiang, Email: qingj@nju.edu.cn.
Jinzhong Qin, Email: qinjz@nju.edu.cn.
Supplemental information
References
- Blackledge N.P., Farcas A.M., Kondo T., King H.W., McGouran J.F., Hanssen L.L., Ito S., Cooper S., Kondo K., Koseki Y. Variant PRC1 complex-dependent H2A ubiquitylation drives PRC2 recruitment and polycomb domain formation. Cell. 2014;157:1445–1459. doi: 10.1016/j.cell.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer L.A., Lee T.I., Cole M.F., Johnstone S.E., Levine S.S., Zucker J.P., Guenther M.G., Kumar R.M., Murray H.L., Jenner R.G. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamberlain S.J., Yee D., Magnuson T. Polycomb repressive complex 2 is dispensable for maintenance of embryonic stem cell pluripotency. Stem Cells. 2008;26:1496–1505. doi: 10.1634/stemcells.2008-0102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chetty S., Pagliuca F.W., Honore C., Kweudjeu A., Rezania A., Melton D.A. A simple tool to improve pluripotent stem cell differentiation. Nat. Methods. 2013;10:553–556. doi: 10.1038/nmeth.2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conklin J.F., Baker J., Sage J. The RB family is required for the self-renewal and survival of human embryonic stem cells. Nat. Commun. 2012;3:1244. doi: 10.1038/ncomms2254. [DOI] [PubMed] [Google Scholar]
- Conklin J.F., Sage J. Keeping an eye on retinoblastoma control of human embryonic stem cells. J. Cell. Biochem. 2009;108:1023–1030. doi: 10.1002/jcb.22342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway E., Jerman E., Healy E., Ito S., Holoch D., Oliviero G., Deevy O., Glancy E., Fitzpatrick D.J., Mucha M. A family of vertebrate-specific polycombs encoded by the LCOR/LCORL genes balance PRC2 subtype activities. Mol. Cell. 2018;70:408–421 e408. doi: 10.1016/j.molcel.2018.03.005. [DOI] [PubMed] [Google Scholar]
- Das P.P., Hendrix D.A., Apostolou E., Buchner A.H., Canver M.C., Beyaz S., Ljuboja D., Kuintzle R., Kim W., Karnik R. PRC2 is required to maintain expression of the maternal Gtl2-rian-Mirg locus by preventing de novo DNA methylation in mouse embryonic stem cells. Cell Rep. 2015;12:1456–1470. doi: 10.1016/j.celrep.2015.07.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Croce L., Helin K. Transcriptional regulation by Polycomb group proteins. Nat. Struct. Mol. Biol. 2013;20:1147–1155. doi: 10.1038/nsmb.2669. [DOI] [PubMed] [Google Scholar]
- Ding J., Huang X., Shao N., Zhou H., Lee D.F., Faiola F., Fidalgo M., Guallar D., Saunders A., Shliaha P.V. Tex10 coordinates epigenetic control of super-enhancer activity in pluripotency and reprogramming. Cell Stem Cell. 2015;16:653–668. doi: 10.1016/j.stem.2015.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faust C., Lawson K.A., Schork N.J., Thiel B., Magnuson T. The Polycomb-group gene eed is required for normal morphogenetic movements during gastrulation in the mouse embryo. Development. 1998;125:4495–4506. doi: 10.1242/dev.125.22.4495. [DOI] [PubMed] [Google Scholar]
- Feng Q., Zhang Y. The NuRD complex: linking histone modification to nucleosome remodeling. Curr. Top. Microbiol. Immunol. 2003;274:269–290. doi: 10.1007/978-3-642-55747-7_10. [DOI] [PubMed] [Google Scholar]
- Højfeldt J.W., Hedehus L., Laugesen A., Tatar T., Wiehle L., Helin K. Non-core subunits of the PRC2 complex are collectively required for its target-site specificity. Mol. Cell. 2019;76:423–436.e423. doi: 10.1016/j.molcel.2019.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holoch D., Margueron R. Mechanisms regulating PRC2 recruitment and enzymatic activity. Trends Biochem. Sci. 2017;42:531–542. doi: 10.1016/j.tibs.2017.04.003. [DOI] [PubMed] [Google Scholar]
- Kuroda M.I., Kang H., De S., Kassis J.A. Dynamic competition of polycomb and trithorax in transcriptional programming. Annu. Rev. Biochem. 2020;89:235–253. doi: 10.1146/annurev-biochem-120219-103641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmichev A., Zhang Y., Erdjument-Bromage H., Tempst P., Reinberg D. Role of the Sin3-histone deacetylase complex in growth regulation by the candidate tumor suppressor p33(ING1) Mol. Cell. Biol. 2002;22:835–848. doi: 10.1128/MCB.22.3.835-848.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D., Song H., Mei H., Fang E., Wang X., Yang F., Li H., Chen Y., Huang K., Zheng L. Armadillo repeat containing 12 promotes neuroblastoma progression through interaction with retinoblastoma binding protein 4. Nat. Commun. 2018;9:2829. doi: 10.1038/s41467-018-05286-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Narayanan C., Bian J., Sambo D., Brickler T., Zhang W., Chetty S. A transient DMSO treatment increases the differentiation potential of human pluripotent stem cells through the Rb family. PLoS one. 2018;13:e0208110. doi: 10.1371/journal.pone.0208110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh Y.H., Wu Q., Chew J.L., Vega V.B., Zhang W., Chen X., Bourque G., George J., Leong B., Liu J. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat. Genet. 2006;38:431–440. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- Margueron R., Li G., Sarma K., Blais A., Zavadil J., Woodcock C.L., Dynlacht B.D., Reinberg D. Ezh1 and Ezh2 maintain repressive chromatin through different mechanisms. Mol. Cell. 2008;32:503–518. doi: 10.1016/j.molcel.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao X., Sun T., Barletta H., Mager J., Cui W. Loss of RBBP4 results in defective inner cell mass, severe apoptosis, hyperacetylated histones and preimplantation lethality in mice. Biol. Reprod. 2020;103:13–23. doi: 10.1093/biolre/ioaa046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery N.D., Yee D., Chen A., Kalantry S., Chamberlain S.J., Otte A.P., Magnuson T. The murine polycomb group protein Eed is required for global histone H3 lysine-27 methylation. Curr. Biol. 2005;15:942–947. doi: 10.1016/j.cub.2005.04.051. [DOI] [PubMed] [Google Scholar]
- O'Carroll D., Erhardt S., Pagani M., Barton S.C., Surani M.A., Jenuwein T. The polycomb-group gene Ezh2 is required for early mouse development. Mol. Cell. Biol. 2001;21:4330–4336. doi: 10.1128/MCB.21.13.4330-4336.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor M.D., Wederell E., Robertson G., Delaney A., Morozova O., Poon S.S., Yap D., Fee J., Zhao Y., McDonald H. Retinoblastoma-binding proteins 4 and 9 are important for human pluripotent stem cell maintenance. Exp. Hematol. 2011;39:866–879.e861. doi: 10.1016/j.exphem.2011.05.008. [DOI] [PubMed] [Google Scholar]
- Pasini D., Bracken A.P., Hansen J.B., Capillo M., Helin K. The polycomb group protein Suz12 is required for embryonic stem cell differentiation. Mol. Cell. Biol. 2007;27:3769–3779. doi: 10.1128/MCB.01432-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasini D., Bracken A.P., Jensen M.R., Lazzerini Denchi E., Helin K. Suz12 is essential for mouse development and for EZH2 histone methyltransferase activity. EMBO J. 2004;23:4061–4071. doi: 10.1038/sj.emboj.7600402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauklin S., Vallier L. The cell-cycle state of stem cells determines cell fate propensity. Cell. 2013;155:135–147. doi: 10.1016/j.cell.2013.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira C.F., Piccolo F.M., Tsubouchi T., Sauer S., Ryan N.K., Bruno L., Landeira D., Santos J., Banito A., Gil J. ESCs require PRC2 to direct the successful reprogramming of differentiated cells toward pluripotency. Cell Stem Cell. 2010;6:547–556. doi: 10.1016/j.stem.2010.04.013. [DOI] [PubMed] [Google Scholar]
- Pereira J.D., Sansom S.N., Smith J., Dobenecker M.W., Tarakhovsky A., Livesey F.J. Ezh2, the histone methyltransferase of PRC2, regulates the balance between self-renewal and differentiation in the cerebral cortex. Proc. Natl. Acad. Sci. U S A. 2010;107:15957–15962. doi: 10.1073/pnas.1002530107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piunti A., Shilatifard A. Epigenetic balance of gene expression by Polycomb and COMPASS families. Science. 2016;352:aad9780. doi: 10.1126/science.aad9780. [DOI] [PubMed] [Google Scholar]
- Qian Y.W., Wang Y.C., Hollingsworth R.E., Jr., Jones D., Ling N., Lee E.Y. A retinoblastoma-binding protein related to a negative regulator of Ras in yeast. Nature. 1993;364:648–652. doi: 10.1038/364648a0. [DOI] [PubMed] [Google Scholar]
- Ruiz S., Panopoulos A.D., Herrerías A., Bissig K.D., Lutz M., Berggren W.T., Verma I.M., Izpisua Belmonte J.C. A high proliferation rate is required for cell reprogramming and maintenance of human embryonic stem cell identity. Curr. Biol. 2011;21:45–52. doi: 10.1016/j.cub.2010.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuettengruber B., Bourbon H.M., Di Croce L., Cavalli G. Genome regulation by polycomb and trithorax: 70 years and counting. Cell. 2017;171:34–57. doi: 10.1016/j.cell.2017.08.002. [DOI] [PubMed] [Google Scholar]
- Shcherbina A., Li J., Narayanan C., Greenleaf W., Kundaje A., Chetty S. Brief report: cell cycle dynamics of human pluripotent stem cells primed for differentiation. Stem Cells. 2019;37:1151–1157. doi: 10.1002/stem.3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon J.A., Kingston R.E. Mechanisms of polycomb gene silencing: knowns and unknowns. Nat. Rev. Mol. Cel. Biol. 2009;10:697–708. doi: 10.1038/nrm2763. [DOI] [PubMed] [Google Scholar]
- Tamburri S., Lavarone E., Fernández-Pérez D., Conway E., Zanotti M., Manganaro D., Pasini D. Histone H2AK119 mono-ubiquitination is essential for polycomb-mediated transcriptional repression. Mol. Cell. 2020;77:840–856.e845. doi: 10.1016/j.molcel.2019.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner S.A., Bracken A.P. A "complex" issue: deciphering the role of variant PRC1 in ESCs. Cell Stem Cell. 2013;12:145–146. doi: 10.1016/j.stem.2013.01.014. [DOI] [PubMed] [Google Scholar]
- Young R.A. Control of the embryonic stem cell state. Cell. 2011;144:940–954. doi: 10.1016/j.cell.2011.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Zhou B., Pache L., Chang M., Khodabakhshi A.H., Tanaseichuk O., Benner C., Chanda S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019;10:1523. doi: 10.1038/s41467-019-09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data performed for this study are deposited in GEO database under the accession numbers GSE144155 (RNA-seq) and GSE155029 (ChIP-seq). Additional datasets used in this study are detailed in Table S4.