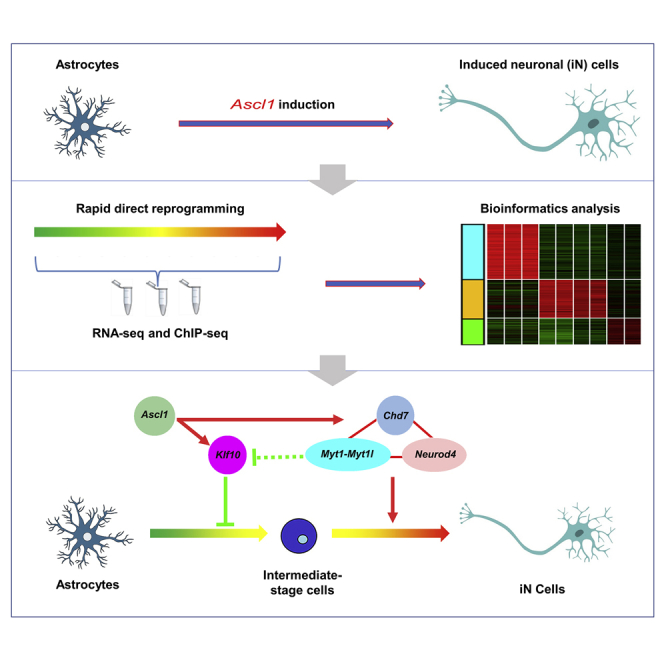
Summary
Direct neuronal reprogramming potentially provides valuable sources for cell-based therapies. Proneural gene Ascl1 converts astrocytes into induced neuronal (iN) cells efficiently both in vitro and in vivo. However, the underlying mechanisms are largely unknown. By combining RNA sequencing and chromatin immunoprecipitation followed by high-throughput sequencing, we found that the expression of 1,501 genes was markedly changed during the early stages of Ascl1-induced astrocyte-to-neuron conversion and that the regulatory regions of 107 differentially expressed genes were directly bound by ASCL1. Among Ascl1's direct targets, Klf10 regulates the neuritogenesis of iN cells at the early stage, Myt1 and Myt1l are critical for the electrophysiological maturation of iN cells, and Neurod4 and Chd7 are required for the efficient conversion of astrocytes into neurons. Together, this study provides more insights into understanding the molecular mechanisms underlying Ascl1-mediated astrocyte-to-neuron conversion and will be of value for the application of direct neuronal reprogramming.
Keywords: direct neuronal reprogramming, astrocyte-to-neuron conversion, transcription factor Ascl1, transcription factor Klf10, transcription factor Myt1, transcription factor Myt1l, chromo-helicase-DNA-binding protein 7 (Chd7), induced neuronal (iN) cells, transcription factor Neurod4, molecular mechanisms
Graphical Abstract
Highlights
-
•
RNA-seq and ChIP-seq were used to study Ascl1-induced astrocyte-to-neuron conversion
-
•
Early Klf10 regulates neuritogenesis and electrophysiological properties of iN cells
-
•
Myt1 and Myt1l are critical for the electrophysiological maturation of iN cells
-
•
Neurod4 and Chd7 are required for efficient conversion of astrocytes to neurons
Rao et al. studied molecular mechanisms underlying Ascl1-induced conversion of astrocytes to neurons. Among 107 direct targets of Ascl1, early expression of Klf10 regulates the neuritogenesis and the electrophysiological properties of induced neuronal (iN) cells, Myt1 and family member Myt1l are critical for the electrophysiological maturation of iN cells, and Neurod4 and Chd7 are required for efficient direct neuronal conversion.
Introduction
The central nervous system of mammals has a limited capacity to regenerate when neurons are injured or lost in traumatic or neurodegenerative diseases. Together with stem cell-derived neuronal products, direct neuronal reprogramming offers promising alternatives to achieve neuronal repair (Colasante et al., 2019; Masserdotti et al., 2016; Tsunemoto et al., 2015; Xu et al., 2015). Mounting evidence shows that non-neuronal cells such as fibroblasts and astrocytes can be directly converted into neurons and a number of neuronal subtypes (Addis et al., 2011; Berninger et al., 2007; Caiazzo et al., 2011; Colasante et al., 2015; Guo et al., 2014; Heinrich et al., 2010; Heins et al., 2002; Kim et al., 2011; Li et al., 2019; Liu et al., 2013, 2015; Niu et al., 2013; Pfisterer et al., 2011; Son et al., 2011; Torper et al., 2013; Vadodaria et al., 2016; Vierbuchen et al., 2010; Xu et al., 2016). However, the molecular mechanisms underlying direct neuronal reprogramming remain poorly understood.
The past several years has seen much progress in our understanding of how fibroblasts are converted into neurons. It has been reported a hierarchical mechanism operates in the direct reprogramming of fibroblasts into neurons mediated by the transcription factors (TFs) Ascl1, Brn2, and Myt1l. Ascl1 acts as an “on-target” pioneer factor by occupying most cognate genomic sites in both the opened and closed chromatin in fibroblasts (Wapinski et al., 2013). Further study showed that Ascl1 opens closed chromatin at its target sites within 12 h, and induces rapid chromatin remodeling and nucleosome phasing that precedes neuronal maturation in direct reprogramming of fibroblasts to neurons (Wapinski et al., 2017). By performing single-cell RNA sequencing (RNA-seq), Treutlein et al. (2016) revealed that the direct reprogramming of fibroblasts to neurons contains two stages: the initiation stage when Ascl1 induces neuronal and myocyte fates and the maturation stage when Brn2 and Myt1l promote reprogrammed fibroblasts to permanently acquire neuronal identity. Mall et al. (2017) further showed that the TF Myt1l represses multiple somatic cell lineage programs to establish and maintain neuronal identity. Besides TFs, polypyrimidine tract-binding proteins, microRNAs, and epigenetic regulators also actively participate in the conversion of fibroblasts into neurons (He et al., 2018; Hu et al., 2018; Lee et al., 2018; Lu and Yoo, 2018; Luo et al., 2019; Zhang et al., 2016).
During direct reprogramming of astrocytes into neurons, it has been reported that Neurog2 and Ascl1 rapidly induced distinct transcriptional programs with only a small subset of target genes in common at 24 h after induction, including Insm1, NeuroD4, Prox1, and Sox11. Among these downstream TFs, only NeuroD4 is sufficient to induce a small fraction of neuronal cells (1%–3%) from cerebral cortex astrocytes at postnatal day 6–7 (P6–7), and together with Insm1 they induced a glutamatergic neuronal phenotype (Masserdotti et al., 2015). However, how the downstream factors of proneural genes contribute to the astrocyte-to-neuron conversion remains largely unknown. Here, we combined RNA-seq, chromatin immunoprecipitation followed by high-throughput sequencing (ChIP-seq), and analysis of the TF regulation network to dissect the mechanisms underlying Ascl1-induced astrocyte-to-neuron conversion. We found that Ascl1 induced rapid and global transcriptional changes by directly binding to its target genes. Among these direct target genes, the TFs Klf10, Myt1, and Neurod4, and chromatin remodeling factor Chd7 played important roles in the direct reprogramming of astrocytes into neurons.
Results
Ascl1 Induces Rapid and Global Transcriptional Changes
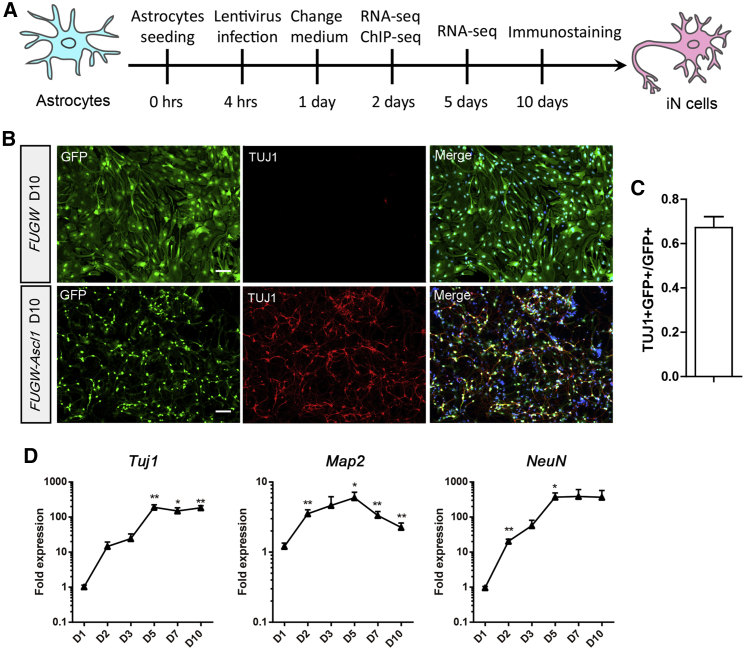
To investigate the molecular mechanisms underlying Ascl1-induced astrocyte-to-neuron conversion, we infected the cultured dorsal midbrain astrocytes with FUGW-Ascl1 or control viruses and performed the assays within 10 days post infection (DPI) (Figure 1A). We assessed the purity of the starter astrocytes by performing immunostaining. The results showed that the majority of the cells were stained positive for GFAP (83.3% ± 1.6%, n = 3 independent experiments) while few cells were stained positive for TUJ1 (0.8% ± 0.2%, n = 3 independent experiments) and NG2 (0.5% ± 0.2%, n = 3 independent experiments). The TUJ1+ cells did not exhibit neuronal morphology. Notably, in Marius Wernig's work on direct conversion of fibroblasts to functional neurons by defined factors (Vierbuchen et al., 2010), it has been reported that rare TUJ1-positive cells with fibroblast-like morphology exist in starting mouse embryonic fibroblasts (MEFs), indicating weak expression of TUJ1 in non-neuronal cells. Conversely, SOX2 (a neural stem cell marker)-positive cells could hardly be detected in the astrocyte cultures (data not shown). At 10 DPI when infected with the control lentivirus FUGW expressing GFP only, the astrocytes maintained the glial morphology and did not express the neuronal marker TUJ1. In contrast, most of the astrocytes infected with FUGW-Ascl1 adopted a neuronal fate and expressed TUJ1 (67.2% ± 5.1%, n = 3 independent experiments, 5701 GFP+ cells counted) and exhibited characteristic neuronal morphology (Figures 1B and 1C). We then examined the expression of the neuronal markers Tuj1, Map2, and NeuN by performing quantitative RT-PCR (qRT-PCR) at various time points during the direct reprogramming. We found that the expression of neuronal markers was markedly increased at 2 DPI and reached peak levels at 5 DPI (Figure 1D).
Figure 1.
Ascl1 Converts Mouse Dorsal Midbrain Astrocytes into Neurons
(A) Schematic representation of the overall experimental design of this study.
(B) Cultured astrocytes infected with lentivirus Ascl1-FUGW expressed TUJ1 at 10 DPI and displayed characteristic neuronal morphology.
(C) Histogram showing the efficiency of direct reprogramming.
(D) qRT-PCR results showing the expression of neuronal markers in iN cells at different time points during direct reprogramming.
Data are presented as mean ± SEM, n = 3 independent experiments. ∗p < 0.05, ∗∗p < 0.01. Scale bars, 100 μm. See also Table S8.
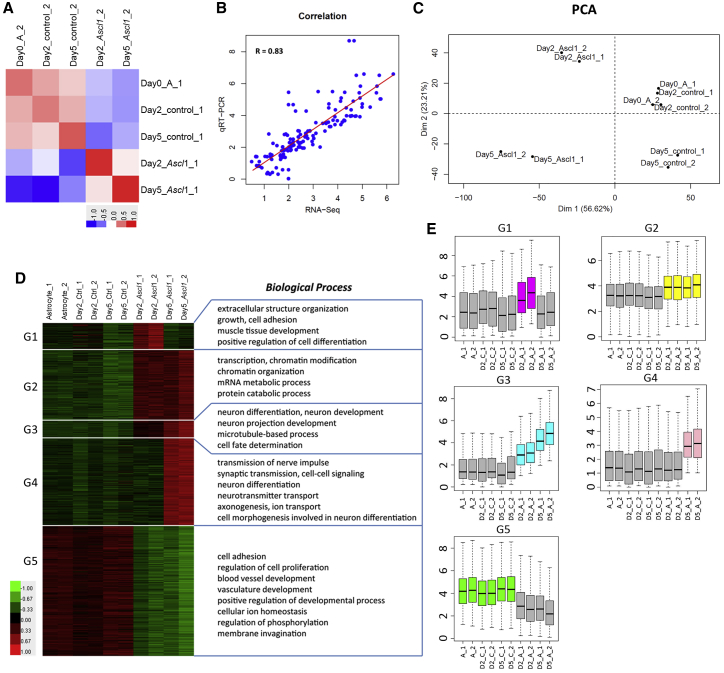
To reveal the genome-wide transcriptional changes during astrocyte-to-neuron conversion, we performed RNA-seq assays at 2 DPI and 5 DPI with five groups of samples including day0-A (astrocytes control without virus infection at day 0), day2-Ctrl, and day5-Ctrl (astrocytes infected with FUGW at day 2 and day 5), day2-Ascl1 and day5-Ascl1 (astrocytes infected with FUGW-Ascl1 at day 2 and day 5). The results showed that the two biological replicates in each group correlated very well (Figure 2A). Correlation analysis confirmed that the normalized RNA-seq tag counts of these genes were consistent with their expression levels that were detected by qRT-PCR (Figure 2B). Principal component analysis (PCA) separated the Ascl1-infected samples from controls in the principal component 1 (PC1) dimension and revealed that day2-Ascl1 was an intermediate state across the direct reprogramming (Figures 2C, S1A, and S1B). We found that the expression of 1,501 genes was markedly changed (fold change >1.5, p < 0.05) in FUGW-Ascl1-infected astrocytes compared with that in control astrocytes during direct reprogramming, and these differentially expressed genes (DEGs) could be clustered into five groups by their expression levels at different time points. The expression of genes in group 1 was upregulated only at day 2 (G1), the genes in group 2 were upregulated at day 2 and their expression was maintained at day 5 (G2), group 3 was composed of genes whose expression was sequentially upregulated from day 2 to day 5 (G3), and group 4 consisted of genes upregulated only at day 5 (G4), whereas the genes in group 5 were downregulated at both day 2 and day 5 (G5) (Figures 2D and 2E). Furthermore, functional enrichment analysis revealed that the genes in each group were associated with distinct biological processes (Figure 2D). The list of 1,501 DEGs can be found in Table S1. We have compared the results with previous works (Masserdotti et al., 2015; Wapinski et al., 2013), and the comparisons can be found in Tables S2–S4 and Figure S2.
Figure 2.
Ascl1 Induces Rapid and Global Transcriptional Changes
(A) Correlation analysis of RNA-seq biological replicates.
(B) Correlation analysis of RNA-seq and qRT-PCR results.
(C) Principal component analysis (PCA) of RNA-seq samples.
(D) Heatmaps show the expression of differentially expressed genes during the direct reprogramming processes by RNA-seq across different time points (n = 2 biological replicates). Top gene ontology terms are shown on the right side.
(E) Box plots show average expression levels of genes in the five groups that are Astrocyte-1/2 (A_1/2), day2-control-1/2 (D2_C_1/2), day5-control-1/2 (D5_C_1/2), day2-Ascl1-induced-1/2 (D2_A_1/2), and day5-Ascl1-induced-1/2 (D5_A_1/2), respectively. The y axis represents the normalized expression of log2(FPKM [fragments per kilobase of transcript per million mapped reads] + 1).
Genome-wide Binding Sites of ASCL1 in Astrocytes
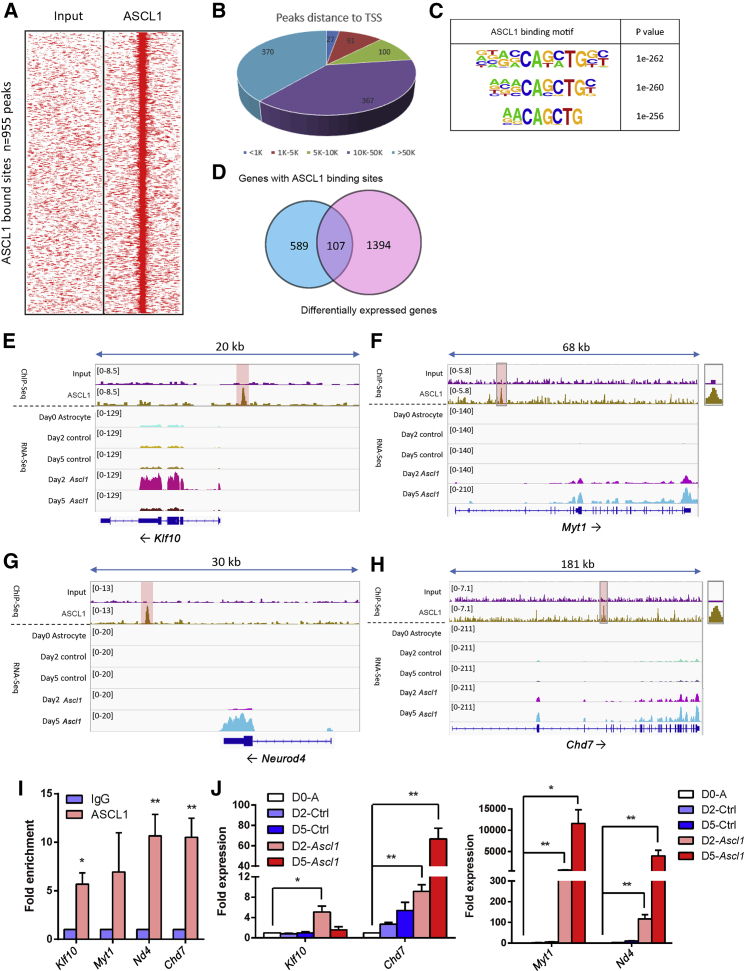
To search for the genome-wide binding sites of ASCL1 in astrocytes, we performed ChIP-seq for ASCL1 in astrocytes at day 2 after virus infection. The results showed that 955 ASCL1-binding peaks were identified with at least 3-fold enrichment (Figure 3A), and the ASCL1 bound sites were mainly located in the distal regions of target genes (Figure 3B and Table S5). The genes with enriched binding peaks within 50 kb upstream or downstream of transcriptional start sites (TSSs) were defined as ASCL1 direct binding targets. We found that 696 genes were directly bound by ASCL1 (Table S6). De novo motif discovery analysis was performed to identify ASCL1 binding motifs within its binding regions. Among the top-ranking motifs, the canonical E-box motif CANNTG, which is associated with ASCL1 binding in fibroblasts (Wapinski et al., 2013) and neural stem cells (Raposo et al., 2015), was highly enriched across the binding sites (Figure 3C). Combining the results of ChIP-seq with RNA-seq, we found that 107 of 1,501 DEGs were directly bound by ASCL1 during direct reprogramming (Figure 3D and Table S7).
Figure 3.
Genome-wide Binding Sites of ASCL1 in Astrocytes during Direct Reprogramming Mediated by Ascl1
(A) Heatmaps representing genome-wide occupancy profiles of ASCL1 in astrocytes 2 days after transduction of Ascl1.
(B) Genome-wide distributions of ASCL1 binding peaks. The peak distances to TSSs are shown.
(C) ASCL1 binding motifs and the p value for each motif.
(D) Of the 1,501 differentially expressed genes from RNA-seq (Table S1), 107 overlapped with those genes with ASCL1 bound within 50 kb (Tables S5 and S6).
(E–H) Enrichment of ASCL1 in the regulatory regions of Klf10 (E), Myt1 (F), Neurod4 (G), and Chd7 (H) in ChIP-seq, and their tag counts in RNA-seq. The scale bars indicating the enrichment density of sequencing tags are listed on the left side of each enrichment plot.
(I) ChIP-qPCR validation of ASCL1 enrichment at the regulatory regions of target genes at 2 DPI.
(J) qRT-PCR shows the fold expression of target genes. Genes in day0-A (D0-A), day2-Ctrl (D2-Ctrl), day5-Ctrl (D5-Ctrl), day2-Ascl1 (D2-Ascl1), and day5-Ascl1 (D5-Ascl1) are shown in white, slate blue, blue, pink, and red, respectively.
Data are presented as mean ± SEM, n = 3 independent experiments. ∗p < 0.05, ∗∗p < 0.01. See also Figure S1; Tables S5 and S6–S8.
We applied the same criteria to analyze the ChIP-seq results, which were collected in fibroblasts reprogrammed for 2 days (Wapinski et al., 2013), and identified 12,321 ASCL1-binding peaks and 6,115 possible binding targets. Among these possible binding targets, 2,644 genes were distributed on either PC1 or PC2 dimension, which classified the direct neuronal reprogramming process to distinct stages. We defined the 2,644 genes as ASCL1 direct downstream targets in this system. Notably, 92 genes out of the 107 ASCL1 direct downstream targets in astrocytes induced for 2 days (this study) were also identified as ASCL1 downstream targets in fibroblasts reprogrammed for 2 days (Wapinski et al., 2013) (Table S7).
To unravel the connection of ASCL1 and the downstream targets, we analyzed the Connection Specificity Index (CSI) of TFs in five DEG groups and generated a TF co-expression network (Ascl1 negatively correlated TF group was removed in Figure S1C). We connected Ascl1 and its direct downstream targets through deep yellow arrows. Being closer to ASCL1, the target gene displayed more significant differential expression, and we made the arrow bold. We then reconnected and reclustered the TFs based on the detailed CSI coefficient, and the correlation between every two TFs was shown (Figure S1C). TFs Klf10, Myt1, and Neurod4 were chosen as representative DEGs of day2 upregulated only (G1), day2-day5 upregulated sequentially (G3), and day5 upregulated only (G4) to investigate what roles they might play during the Ascl1-induced astrocyte-to-neuron conversion (Figures 3E–3G and S1C). Furthermore, among the 1,501 DEGs identified during Ascl1-induced astrocyte-to-neuron conversion, 61 epigenetic factors were differentially expressed (fold change >1.5, p < 0.05), and an ATP-dependent chromatin remodeler chromo-helicase-DNA-binding protein 7 (Chd7) was the most significant DEG. Meanwhile, the expression profile of Chd7 highly correlated with that of Ascl1 (Figure S3). Thus, we also studied the function of the epigenetic factor Chd7 (Figure 3H). We have confirmed the binding of ASCL1 on the regulatory regions of Klf10, Myt1, Neurod4, and Chd7 by ChIP-PCR at 2 DPI and the expression of these genes at 2 DPI and 5 DPI by qRT-PCR (Figures 3I and 3J).
Klf10 Regulates Neuritogenesis and the Electrophysiological Properties of Induced Neuronal Cells
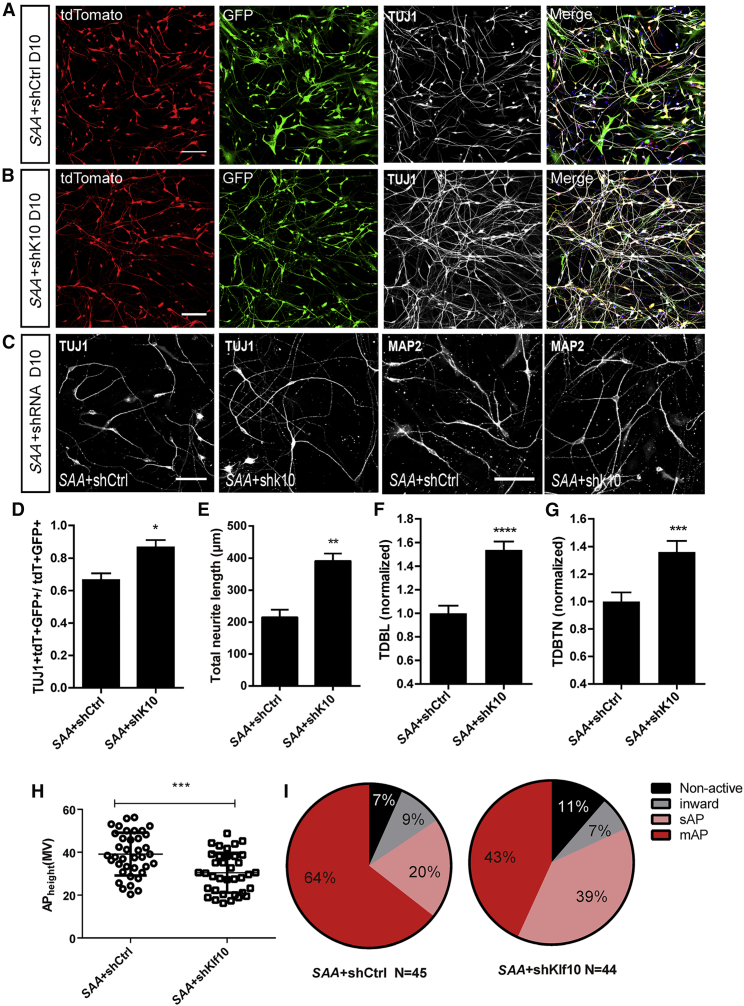
The expression of TF Klf10 was upregulated only at day 2 during direct reprogramming (Figure 3J). To investigate whether Klf10 plays critical roles in initiating the astrocyte-to-neuron conversion, we designed specific short hairpin RNAs (shRNAs) against Klf10 (Figure S4A) to reduce its expression. A previous work reported that phosphomutant ASCL1 can enhance neuronal induction activity in Xenopus embryos and improve neuronal transdifferentiation efficiency (Ali et al., 2014). Therefore, we mutated all six serine-proline sites (SP) to alanine-proline (SA) in ASCL1 to generate S-A ASCL1 (SAA) and used SAA as a substitute for ASCL1 to induce astrocyte-to-neuron conversion. Astrocytes were co-infected with FUW-SAA-IRES-tdTomato and pLKD-shKlf10-GFP or pLKD-shCtrl-GFP lentiviruses. We found that the morphology of SAA virus-infected astrocytes was transformed quicker upon Klf10 knockdown compared with that of control (data not shown). The cellular phenotypes of SAA and shRNA virus co-infected cells (tdTomato+GFP+) were analyzed at 10 DPI (Figures 4A and 4B). Notably, more TUJ1-positive neuronal cells were generated (86.6% ± 4.4% versus 66.5 ± 4.2, n = 4, 1,207–1,370 tdTomato+GFP+ cells counted), and the total neurite length (TUJ1+) was increased (391.3 ± 23.08 μm versus 215.1 ± 23.53 μm, n = 3, 61–70 tdTomato+GFP+ cells counted) upon Klf10 knockdown (Figures 4C–4E). Moreover, the dendrite complexity was also enhanced upon Klf10 knockdown, as measured by increased total dendritic branch length (MAP2+) (1.54 ± 0.07 versus 1.00 ± 0.07, n = 3, 60 tdTomato+GFP+ cells measured) and total dendritic branch tip number (MAP2+) (1.36 ± 0.08 versus 1.00 ± 0.07, n = 3, 60 tdTomato+GFP+ cells measured) (Figures 4C, 4F, and 4G).
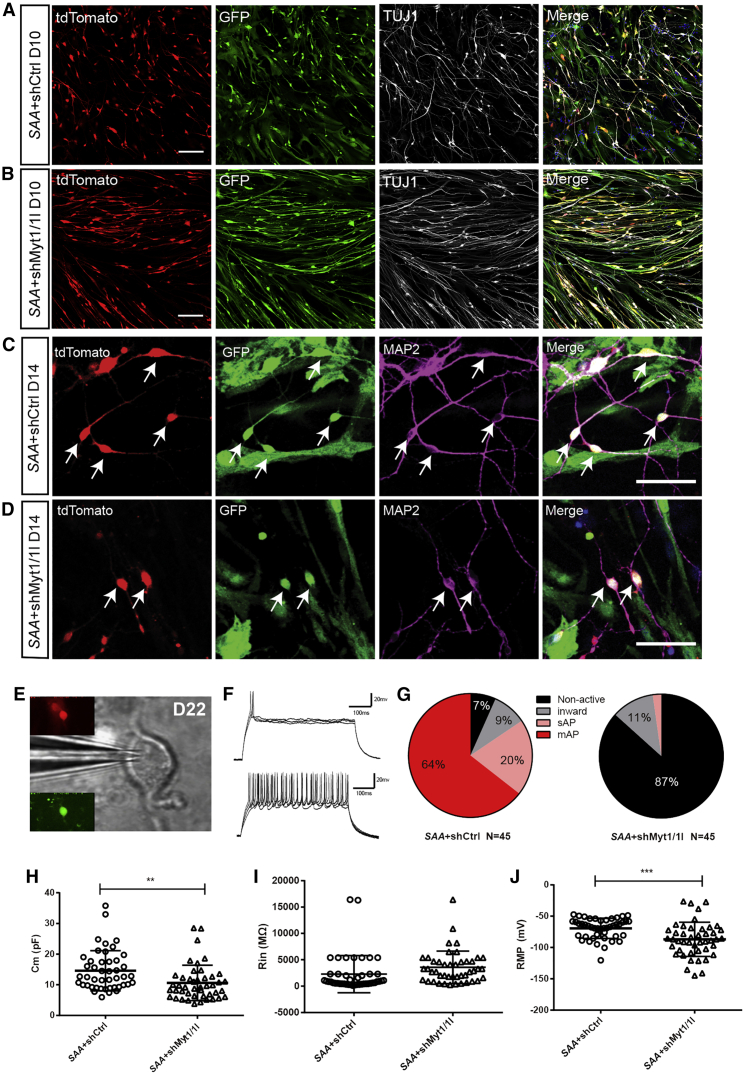
Figure 4.
Knockdown of Klf10 Promotes Neurite Outgrowth and Reduces Electrophysiological Activity of iN Cells
(A and B) Micrographs of cells co-infected with FUW-SAA-IRES-tdTomato and shRNA at 10 DPI.
(C) Enlarged images showing the immunostaining of TUJ1 and MAP2 in iN cells at 10 DPI.
(D) Quantification of TUJ1-positive cells among FUW-SAA-IRES-tdTomato and shRNA co-infected cells.
(E) Statistical results of the total neurite length of iN cells.
(F and G) Total dendrite branch length (TDBL; F) and total dendrite branch tip number (TDBTN; G) were measured and analyzed.
(H) Action potential height of iN cells when Klf10 was knocked down.
(I) Percentages of iN cells with four different degrees of membrane excitability (non-active, inward, sAP, or mAPs) when co-infected with FUW-SAA-IRES-tdTomato and shRNA against Klf10 or control shRNA.
Data are presented as mean ± SEM, n = 3 independent experiments. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. Scale bars, 100 μm. See also Figure S4 and Table S9.
To evaluate whether early Klf10 expression controls other aspects of neuronal differentiation, we performed electrophysiological recording and qRT-PCR. The recording results showed that upon Klf10 knockdown, the membrane properties of induced neuronal (iN) cells, such as membrane capacitance (Cm), input resistance (Rin), and resting membrane potential (RMP), remained largely unchanged (data not shown). Conversely, the height of action potential (AP) was markedly reduced (Figure 4H). As we reported previously, we categorized iN cells into four groups based on their current and voltage response patterns: non-active cells (“non-active”), cells exhibiting inward current without AP (“inward”), single AP (“sAP”), and multiple APs (“mAPs”) (Liu et al., 2015). When Klf10 was knocked down, the percentage of iN cells firing mAPs was markedly reduced whereas the percentage of non-active cells and iN cells firing sAP increased (Figure 4I). Meanwhile, the qRT-PCR results showed that the mRNA expression of synaptic proteins such as Synapsin I and Homer1 was not markedly changed when Klf10 was knocked down. Interestingly, the mRNA expression of sodium channel gene Scn1a but not sodium channel genes Scn2b and Scn8a increased when Klf10 was knocked down (data not shown). These results indicate that early Klf10 expression controls the neuritogenesis and the electrophysiological properties of iN cells.
Myt1 and Myt1l Are Critical for the Electrophysiological Maturation of iN Cells
A recent study reported that pan-neuron-specific TF Myt1-like (Myt1l) exerts a pro-neuronal function by direct repression of many different somatic lineage programs except the neuronal program during direct reprogramming of fibroblasts to neurons (Mall et al., 2017). Besides Myt1l, there are two other Myt family members, Myt1 and Myt3 (also known as St18) (Yee and Yu, 1998). Interestingly, during astrocyte-to-neuron conversion, the expression of Myt1 was increased enormously both at day 2 and day 5, whereas the expression of Myt1l and Myt3 was upregulated only at day 5 by 116-fold and 26-fold, respectively (Figure S4B). We investigated their functions by knocking down Myt1, Myt1l, or Myt3 individually or Myt1 and My1l simultaneously (Figure S4C). The results showed that individual knockdown of Myt1, Myt1l, or Myt3 did not affect the morphology and reprogramming efficiency of iN cells (data not shown). Interestingly, double knockdown of Myt1 and My1l resulted in iN cells with thicker neurites and stretched-out morphology (Figures 5A and 5B), although the number of induced TUJ1-positive cells remained largely unchanged compared with that of the control (70.1% ± 2.9% versus 56.4% ± 5.85%, n = 3, 1,020–1,036 tdTomato+ GFP+ cells counted). We also performed immunostaining with antibodies of more mature markers such as MAP2 and SYNAPSIN I. The results showed that compared with the scramble control, iN cells induced by FUW-SAA-IRES-tdTomato together with pLKD-shMyt1/1L-GFP expressed MAP2 (Figures 5C and 5D) and SYNAPSIN I (data not shown) as well.
Figure 5.
Knockdown of Myt1 and Myt1l Inhibits the Electrophysiological Maturation of iN Cells
(A and B) TUJ1 immunostaining of cells co-infected with FUW-SAA-IRES-tdTomato and shMyt1/1L (B) or shCtrl (A) at 10 DPI.
(C and D) MAP2 immunostaining of cells co-infected with FUW-SAA-IRES-tdTomato and shMyt1/1L (D) or shCtrl (C) at 14 DPI.
(E) Differential interference contrast image of whole-cell recording from an iN cell (green and red fluorescence) at 22 DPI.
(F) Representations of single action potentials (sAP) or multiple action potentials (mAPs) generated by iN cells from control group when recorded in current-clamp mode.
(G) Percentages of iN cells with four different degrees of membrane excitability (non-active, inward, sAP, or mAPs) when co-infected with FUW-SAA-IRES-tdTomato and shRNA against Myt1 and Myt1l or control shRNA.
(H–J) Cm (H), Rin (I), and RMP (J) of iN cells when both Myt1 and Mytl1 were knocked down.
Data are presented as mean ± SEM, n = 3 independent experiments. ∗p < 0.05, ∗∗p < 0.01. Scale bars, 100 μm. See also Figure S4 and Table S9.
We further examined the electrophysiological properties of iN cells by performing whole-cell recording upon Myt1 and/or Myt1l knockdown. FUW-SAA-IRES-tdTomato and pLKD-shMyt1/shMyt1l-GFP lentivirus co-infected cells (tdTomato+ GFP+) were recorded at 22 DPI (Figure 5E). Knockdown of Myt1 or Myt1l individually resulted in a decreased percentage of iN cells firing mAPs, increased percentage of iN cells firing sAP, and generation of non-active iN cells compared with that in cells infected with only FUW-SAA-IRES-tdTomato (Figure S4D). More notably, upon knockdown of both Myt1 and Myt1l, the majority of iN cells were non-active (Figures 5F and 5G). Moreover, upon Myt1 and Myt1l double knockdown, the iN cells had lower Cm (shMyt1/1L 10.6 ± 0.9 pF, shCtrl 14.6 ± 1.0 pF, n = 45), tended to have higher Rin (shMyt1/1L 3575 ± 458.4 MΩ, shCtrl 2278 ± 526.8 MΩ, n = 45) and had more negative RMP (shMyt1/1L −86.9 ± 4.1 mV, shCtrl −69.4 ± 2.4 mV, n = 45) compared with that of control (Figures 5H–5J). Taking these results together, Myt1 and Myt1l were critical for the electrophysiological maturation of iN cells.
Neurod4 Can Partially Substitute Ascl1 to Induce iN Cells
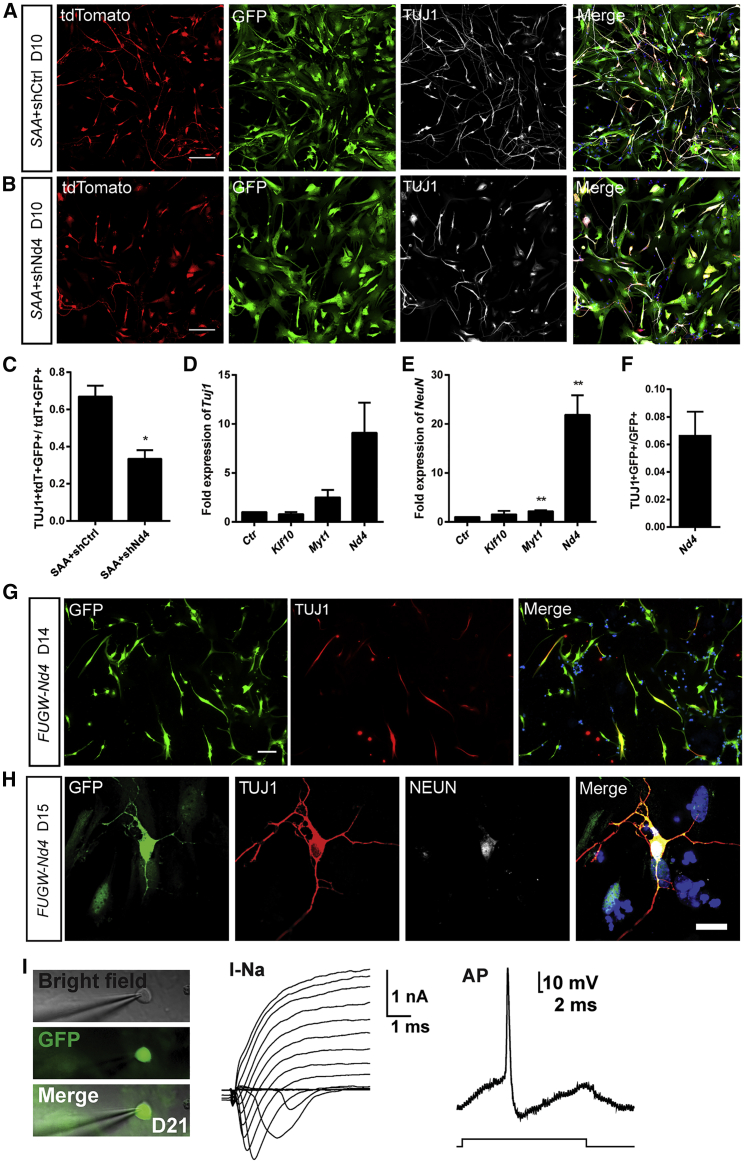
Neurod4 is a TF that contributes to the neuronal differentiation program (Guillemot, 2007). During astrocyte-to-neuron conversion, the expression of Neurod4 was markedly increased (Figure 3J). Consistent with a previous report (Masserdotti et al., 2015), the conversion efficiency of astrocytes into neurons markedly decreased upon Neurod4 knockdown (33.4% ± 4.6% versus 66.9% ± 5.9%, n = 3, 876–1,062 tdTomato+ GFP+ cells counted) (Figures S5A and 6A–6C). To explore whether the downstream TFs of Ascl1 are sufficient to induce neuronal reprogramming, we infected the astrocytes with FUGW-Klf10, FUGW-Myt1, or FUGW-Neurod4 and examined the expression of Tuj1 and NeuN at 10 DPI. The results showed that Klf10 and Myt1 induced the expression of Tuj1 by 0.8-fold and 5.1-fold and the expression NeuN by 1.5-fold and 1.9-fold, respectively, compared with that of control while Neurod4 induced the expression of Tuj1 by 9.1-fold and the expression of NeuN by 21.9-fold (Figures 6D and 6E). This was corroborated with the generation of TUJ1-positive neuronal cells in astrocytes infected with FUGW-Neurod4, and the conversion efficiency was about 6% (308 GFP+ cells counted) (Figures 6F and 6G).
Figure 6.
Neurod4 Can Partially Substitute Ascl1 to Induce a Small Fraction of TUJ1-Positive Neuronal Cells
(A and B) Micrographs of cells infected with FUW-SAA-IRES-tdTomato and shNeurod4 (shNd4; B) or shCtrl (A) at 10 DPI.
(C) Statistical results of direct reprogramming efficiency upon Neurod4 knockdown.
(D and E) Expression of Tuj1 (D) and NeuN (E) after overexpression of Klf10, Myt1, or Neurod4 (Nd4).
(F) Efficiency of TUJ1-positive cells induced by Neurod4.
(G) Representative images of cells infected with FUGW-Neurod4 at 14 DPI.
(H) TUJ1 and NEUN immunostaining of cells infected with FUGW-Neurod4 at 15 DPI.
(I) Patch-clamp recordings of FUGW-Neurod4 infected cells at 21 DPI.
Data are presented as mean ± SEM, n = 2–3 independent experiments. ∗p < 0.05, ∗∗p < 0.01. Scale bars, 100 μm. See also Figure S5; Tables S8 and S9.
To further investigate whether the downstream TF Neurod4 can induce a genuine astrocyte-to-neuron conversion, we performed more immunostaining with antibodies of mature neuronal markers. The results showed that Neurod4 could convert astrocytes into neuron-like cells expressing not only TUJ1 but also NEUN and SYNAPSIN I (Figure 6H and data not shown). We also performed patch-clamp recordings on neuron-like cells induced by Neurod4, the results of which showed that these neuron-like cells were able to produce inward currents in response to voltage steps and to fire APs in response to current injections (n = 4/16, Figure 6I). These results suggest that Neurod4 can induce a genuine astrocyte-to-neuron switch and are in line with the observation that Neurod4 is sufficient to induce a small but consistent fraction of TUJ1+ neuronal cells (1%–3%) from astrocytes and generate APs upon receiving an injection of suprathreshold current pulses (Masserdotti et al., 2015).
Knockdown of Chd7 Reduces Astrocyte-to-Neuron Conversion Efficiency
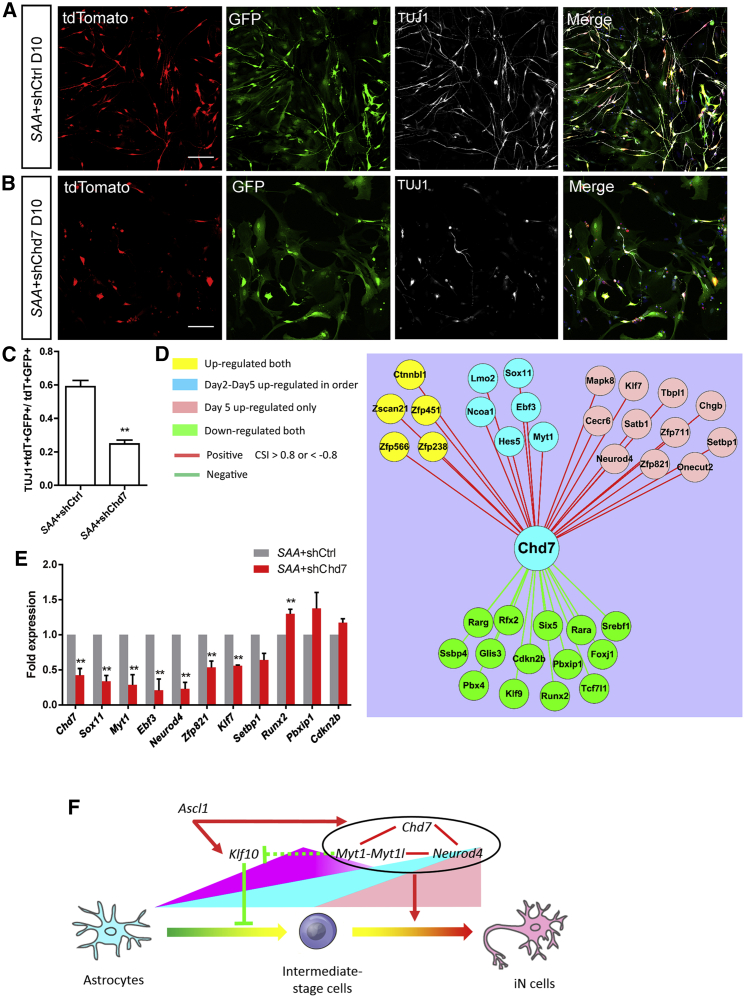
Together with TFs, epigenetic regulators are actively involved in the direct conversion of fibroblasts into neurons (He et al., 2018; Lee et al., 2018; Luo et al., 2019). Notably, we found the expression of Chd7, a chromatin remodeling factor, to be markedly increased during the reprogramming process of astrocytes into neurons (Figure 3J). Therefore, we characterized the function of Chd7 in Ascl1-induced astrocyte-to-neuron conversion by shRNA-mediated knockdown (Figure S5B). The results showed that the conversion efficiency markedly decreased upon Chd7 knockdown (24.7% ± 2.3%, n = 3, 786 tdTomato+GFP+ cells counted) compared with that of the control (59.0% ± 3.7%, n = 3, 2,516 tdTomato+GFP+ cells counted) (Figures 7A–7C, S5C, and S5D). To investigate how Chd7 affects the reprogramming, we constructed a network of Chd7 correlated TFs based on their expression correlation CSI (CSI > 0.8) (Figure 7D). The TFs included those that were upregulated at day 2 and day 5 (in yellow), upregulated continuously at day 2 and day 5 (in blue), upregulated only at day 5 (in brown), and downregulated at both day 2 and day 5 (in green) (Figure 7D). In line with the CSI analysis, qRT-PCR results showed that the expression of Chd7 positively correlated genes was markedly reduced upon Chd7 knockdown (Figure 7E). Interestingly, these decreased genes, such as Sox11, Myt1, Ebf3, Neurod4, and Zfp821, are related to neuronal development and differentiation, which may explain why knockdown of Chd7 reduced the efficiency of direct reprogramming. These results indicate that besides neurogenic TFs, epigenetic regulator Chd7 is also indispensable for the astrocyte-to-neuron conversion. Combining the results of Klf10, Myt1, Mytl1, Neurod4, and Chd7 during Ascl1-mediated astrocyte-to-neuron conversion, we found that different downstream targets of Ascl1 may play distinct roles: Klf10 was transiently expressed at 2 DPI, and its early expression controlled the neuritogenesis and the electrophysiological properties of iN cells; the expression of Myt1 and Chd7 was continually increased at 2 DPI and 5 DPI, and they were positive driving forces for iN cells to proceed with the reprogramming process and acquire mature neuronal properties; the expression of Neurod4 was upregulated largely at 5 DPI and promoted the maturation of iN cells (Figure 7F).
Figure 7.
Knockdown of Chd7 Reduces the Efficiency of Direct Reprogramming
(A and B) Representative images of cells co-infected with FUW-SAA-IRES-tdTomato and shCtrl (A) or shChd7 (B) at 10 DPI.
(C) Statistical results of direct reprogramming efficiency upon Chd7 knockdown.
(D) Co-expression network of Chd7 correlated transcription factors. The positive correlations between these genes are marked in red and the negative correlations in green.
(E) qRT-PCR expression analysis of Chd7 and its positive or negative correlated transcription factors.
(F) Schematic illustration of distinct roles played by Klf10, Myt1, Mytl1, Neurod4, and Chd7 during Ascl1-mediated astrocyte-to-neuron conversion.
Data are presented as mean ± SEM, n = 3 independent experiments. ∗p < 0.05, ∗∗p < 0.01. Scale bars, 100 μm. See also Figure S5; Tables S8 and S9.
Discussion
In this study we found that Ascl1 induced rapid and global transcriptional changes at 2 DPI and 5 DPI by performing RNA-seq during astrocyte-to-neuron conversion. Meanwhile, ChIP-seq performed at 2 DPI showed that the regulatory regions of 696 genes were directly bound by ASCL1. Combining the results of RNA-seq and ChIP-seq, we found that ASCL1 directly bound to the regulatory regions of 107 DEGs. Among these downstream genes of ASCL1, we found that the early expression of Klf10 controlled the neuritogenesis and the electrophysiological properties of iN cells, Myt1 and Myt family member Myt1l were critical for the electrophysiological maturation of iN cells, and Neurod4 and Chd7 were required for the efficient conversion of astrocytes to neurons.
The similarity between this study and that of Wapinski et al. (2013) is that 2 days after Ascl1 induction, both systems entered intermediate stages (Figures S2A and S2B) and showed similar transcriptome identities (Figure S2C). Genes upregulated on day 2, such as TFs Sox11, Hes5, and Hes6, were mostly related to neuronal differentiation and relevant biological processes. Notably, Klf10 was upregulated in both systems as well (Figure S2C). PC1 dimension reflected the neuronal maturation procedure (Figure S2A). Meanwhile, hierarchical clustering analysis revealed that Day5_Ascl1_1/2 clustered closer to BAM_22d of Wapinski et al. (2013) (Figure S2B), indicating a faster reprogramming process of our system. However, in PC2, our samples (Day5_Ascl1_1/2) showed differences compared with BAM_13d and BAM_22d (Figure S2D), indicating that induction time plays important roles in direct reprogramming.
Comparative analysis showed the correlation between our system and the reprogramming method applied in Masserdotti et al., 2015 (Figure S2E) as well. Pearson correlation coefficient assays with RNA-seq and microarray data showed that, at 4 h after ASCL1ERT2 activation, there was no significant change in transcriptome, corroborating with the fact that there were only a few DEGs at 4 h (data not shown). At 24 h after ASCL1ERT2 activation, astrocytes were transformed into another stage, similar to our intermediate state (day2-Ascl1). At 48 h after ASCL1ERT2 activation, the astrocytes had a tendency toward later reprogramming stages (day2-Ascl1 to day5-Ascl1) (Figure S2E). Notably, poor reprogramming was observed in the sample 2 Ascl1-48h-2 (Figure S2E).
Moreover, we found the expression of 630 overlapping genes was markedly changed during Ascl1-mediated conversion of astrocytes and MEFs into neurons, and the regulatory regions of 629 genes were bound by ASCL1 in both astrocytes and MEFs. For example, the expression of Klf10 and Sox11 were augmented during Ascl1-mediated conversion and their regulatory regions were bound by ASCL1 in both astrocytes and MEFs (data not shown). Wapinski et al. (2013) identified Zfp238 as a key downstream target of Ascl1 that can partially substitute for Ascl1 during direct reprogramming of fibroblasts into neurons. However, in this study we found that the regulatory regions of Zfp238 were not bound by ASCL1 and that Ascl1 largely could not induce the expression of Zfp238 in astrocytes (data not shown). Instead, we found that another ASCL1 downstream target Neurod4 could convert astrocytes into TUJ1-positive neuronal cells, which is consistent with a previous study (Masserdotti et al., 2015). In MEFs, Neurod4 alone was incapable of eliciting neuronal conversion, while together with Insm1 they generated iN cells at 14 DPI (Masserdotti et al., 2015). The direct binding of ASCL1 to large amounts of overlapping genes in astrocytes and fibroblasts reveals a highly similar occupancy of ASCL1 even in distantly related cell types. Conversely, a modest fraction of overlapping genes was differentially expressed during the conversion of astrocytes and fibroblasts to neurons, demonstrating that Ascl1 regulates the expression of downstream genes in cell context-dependent fashion. Furthermore, it has been reported that Ascl1 converts Müller glia cells from the retina and astrocytes from the neocortex and cerebellum into different neuronal subtypes (Chouchane et al., 2017; Guimaraes et al., 2018; Pollak et al., 2013). In Table S4, Pvalb was upregulated in iN cells generated from neocortex astrocytes but not in iN cells generated from dorsal midbrain astrocytes (this study), indicating different neuronal subtypes. This warrants further studies to reveal the identities of iN cells converted from astrocytes that are located in different regions.
It is unknown whether Klf10 plays physiological functions in the nervous system (Subramaniam et al., 2010). In this study, we found that the expression of Klf10 was upregulated at day 2 but downregulated at day 5 during direct reprogramming of astrocytes into neurons. Interestingly, upon Klf10 knockdown, more TUJ1-positive neuronal cells were generated with longer neurite length and more complex dendrites but with reduced electrophysiological activity. Moreover, the expression of Klf10 was also upregulated during direct reprogramming of fibroblasts to neurons at day 2 (Treutlein et al., 2016), and we found that the efficiency of fibroblast-to-neuron conversion was increased upon Klf10 knockdown as well (data not shown). These results show that Klf10 may play a similar role during the direct reprogramming from different donor cells at the early stages.
Although Ascl1 alone is sufficient to convert MEFs into iN cells in optimized culture conditions, endogenous Myt1l is induced during the conversion and exogenous Myt1l considerably increases the efficiency of conversion and the functional maturation of the iN cells (Chanda et al., 2014; Vierbuchen et al., 2010). We have previously shown that Ascl1 alone is sufficient to convert midbrain astrocytes into functional, synapse-forming neurons in vitro (Liu et al., 2015). In this study, we found that the expression of Myt1 was increased more remarkably compared with that of Myt1l and Myt3 during Ascl1-induced astrocyte-to-neuron conversion. Meanwhile, ASCL1 directly bound to the regulatory region of Myt1. This is corroborated by the observation that Myt1 is a direct target of ASCL1 at the onset of neuronal differentiation (Vasconcelos et al., 2016). However, knockdown of Myt1, Myt1l, or Myt3 individually did not affect the conversion efficiency of astrocytes into iN cells induced by Ascl1 (data not shown). Moreover, double knockdown of Myt1 and Myt1l largely did not affect the conversion efficiency of astrocytes into iN cells, although these cells had thicker neurites, stretched-out morphology, and inactive electrophysiological properties. This warrants investigating further whether the Myt1 family members play similar roles also for the iN cells converted from fibroblasts to obtain functional neuronal properties.
Wapinski et al. (2013) found that Ascl1 acts as a pioneer factor at neurogenic loci marked by a close chromatin state to direct conversion of fibroblasts into neurons, and this is in agreement with the observation that Ascl1 coordinately regulates gene expression and the chromatin landscape during neurogenesis (Raposo et al., 2015). Enhanced chromatin accessibility has also been observed in TF Neurog2, microRNAs, and CRISPR/cas9-mediated fibroblast-to-neuron conversion (Abernathy et al., 2017; Black et al., 2016; Smith et al., 2016). Moreover, epigenetic factor ten-eleven translocation 3 (Tet3), which regulates DNA demethylation, and a histone H3 lysine 4 methylase KMT2B contribute to the direct conversion of fibroblast into functional neurons (Barbagiovanni et al., 2018; Zhang et al., 2016). Conversely, incomplete MyoD-induced transdifferentiation of human fibroblasts is associated with chromatin remodeling deficiencies (Manandhar et al., 2017). It has been reported that the chromatin remodeling factor Chd7 regulates adult neurogenesis via activation of SoxC TFs and is indispensable for normal cerebellar development (Feng et al., 2013, 2017). Here, we found the efficiency of astrocyte-to-neuron conversion to be markedly decreased upon Chd7 knockdown. Moreover, CSI results showed that Chd7 is required for expression of the genes Sox11, Myt1, Ebf3, Neurod4, and Zfp821 that are related to neuronal development and differentiation. It will be of interest to investigate how Chd7 is recruited to specific targets and remodels the chromatin structure. Indeed one way to improve direct neuronal reprogramming is to understand the possible barriers in the context of a higher-order chromatin landscape (Gascon et al., 2017; Guo and Morris, 2017; Ninkovic and Gotz, 2018; Riemens et al., 2018).
Experimental Procedures
Astrocyte Culture
Astrocytes were cultured as previously described with some modifications (McCarthy and de Vellis, 1980).
Viral Production
Lentiviruses were produced from HEK293FT cells that were transiently transfected with lentiviral, viral envelope-typing, and VSVG-pseudo-typing plasmids (Tiscornia et al., 2006).
Immunostaining
Immunostaining on cultured cells was performed essentially as previously described except that the primary antibodies were incubated for overnight (Vierbuchen et al., 2010).
RNA Sequencing
Total RNA was isolated with TRIzol reagent. Libraries were prepared, and sequencing was performed in 100-bp paired-end format on the Illumina HiSeq 2000 system.
Chromatin Immunoprecipitation Followed by Sequencing
ChIP-seq was carried out in astrocytes 2 days after FUGW-Ascl1 infection as described previously (Jin et al., 2009). Approximately 9–12 × 106 cells were used for each ChIP-seq experiment, and sequencing reads were generated on HiSeq 2000 Illumina platforms.
Gene Knockdown
To acutely knock down Klf10, Myt1, Neurod4, and Chd7, we cloned one or two shRNAs specifically against targeting sequence and one control scramble shRNA sequence into the lentiviral vector pLKD (OBiO Technology [Shanghai]). The shRNA sequences can be found in Table S9.
Electrophysiological Recording
Whole-cell voltage-clamp or current-clamp recording was performed as described previously (Lu et al., 2007).
Data and Code Availability
The accession number for the RNA-seq data is GEO: GSE132674. The accession number for the ChIP-seq data is GEO: GSE132671.
Author contributions
L.C., N.J., and Z.R. designed the project. Z.R. and S.L. performed the experiments. R.W. and L.M. analyzed the RNA-seq and ChIP-seq. S.H. and Y.S. conducted electrophysiological experiments. J.Y. carried out the lentivirus production. Z.R. and L.C. wrote the manuscript.
Acknowledgments
We thank Dr. Xiang Yu for technical assistance and Dr. Jie He for comments on the project. This work was supported by the National Key Research and Development Program of China (grant no. 2018YFA0108003), the National Natural Science Foundation of China (nos. 31871037, 32070976, and 31371096), Guangxi First-class Discipline Project for Basic Medicine Sciences (no. GXFCDP-BMS-2018) (L.C.), and Fundamental Research Funds for the Central Universities Under Grant No. XJS201211 (Z.R.).
Published: February 11, 2021
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.stemcr.2021.01.006.
Contributor Information
Naihe Jing, Email: njing@sibcb.ac.cn.
Leping Cheng, Email: lpcheng@gxmu.edu.cn.
Supplemental Information
References
- Abernathy D.G., Kim W.K., McCoy M.J., Lake A.M., Ouwenga R., Lee S.W., Xing X., Li D., Lee H.J., Heuckeroth R.O. MicroRNAs induce a permissive chromatin environment that enables neuronal subtype-specific reprogramming of adult human fibroblasts. Cell Stem Cell. 2017;21:332–348. doi: 10.1016/j.stem.2017.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Addis R.C., Hsu F.C., Wright R.L., Dichter M.A., Coulter D.A., Gearhart J.D. Efficient conversion of astrocytes to functional midbrain dopaminergic neurons using a single polycistronic vector. PLoS One. 2011;6:e28719. doi: 10.1371/journal.pone.0028719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali F.R., Cheng K., Kirwan P., Metcalfe S., Livesey F.J., Barker R.A., Philpott A. The phosphorylation status of Ascl1 is a key determinant of neuronal differentiation and maturation in vivo and in vitro. Development. 2014;141:2216–2224. doi: 10.1242/dev.106377. [DOI] [PubMed] [Google Scholar]
- Barbagiovanni G., Germain P.L., Zech M., Atashpaz S., Lo Riso P., D'Antonio-Chronowska A., Tenderini E., Caiazzo M., Boesch S., Jech R. KMT2B is selectively required for neuronal transdifferentiation, and its loss exposes dystonia candidate genes. Cell Rep. 2018;25:988–1001. doi: 10.1016/j.celrep.2018.09.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berninger B., Costa M.R., Koch U., Schroeder T., Sutor B., Grothe B., Gotz M. Functional properties of neurons derived from in vitro reprogrammed postnatal astroglia. J. Neurosci. 2007;27:8654–8664. doi: 10.1523/JNEUROSCI.1615-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black J.B., Adler A.F., Wang H.G., D'Ippolito A.M., Hutchinson H.A., Reddy T.E., Pitt G.S., Leong K.W., Gersbach C.A. Targeted epigenetic remodeling of endogenous loci by CRISPR/Cas9-based transcriptional activators directly converts fibroblasts to neuronal cells. Cell Stem Cell. 2016;19:406–414. doi: 10.1016/j.stem.2016.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caiazzo M., Dell'Anno M.T., Dvoretskova E., Lazarevic D., Taverna S., Leo D., Sotnikova T.D., Menegon A., Roncaglia P., Colciago G. Direct generation of functional dopaminergic neurons from mouse and human fibroblasts. Nature. 2011;476:224–227. doi: 10.1038/nature10284. [DOI] [PubMed] [Google Scholar]
- Chanda S., Ang C.E., Davila J., Pak C., Mall M., Lee Q.Y., Ahlenius H., Jung S.W., Sudhof T.C., Wernig M. Generation of induced neuronal cells by the single reprogramming factor ASCL1. Stem Cell Rep. 2014;3:282–296. doi: 10.1016/j.stemcr.2014.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chouchane M., Melo de Farias A.R., Moura D.M.S., Hilscher M.M., Schroeder T., Leao R.N., Costa M.R. Lineage reprogramming of astroglial cells from different origins into distinct neuronal subtypes. Stem Cell Reports. 2017;9:162–176. doi: 10.1016/j.stemcr.2017.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colasante G., Lignani G., Rubio A., Medrihan L., Yekhlef L., Sessa A., Massimino L., Giannelli S.G., Sacchetti S., Caiazzo M. Rapid conversion of fibroblasts into functional forebrain GABAergic interneurons by direct genetic reprogramming. Cell Stem Cell. 2015;17:719–734. doi: 10.1016/j.stem.2015.09.002. [DOI] [PubMed] [Google Scholar]
- Colasante G., Rubio A., Massimino L., Broccoli V. Direct neuronal reprogramming reveals unknown functions for known transcription factors. Front. Neurosci. 2019;13:283. doi: 10.3389/fnins.2019.00283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng W., Kawauchi D., Korkel-Qu H., Deng H., Serger E., Sieber L., Lieberman J.A., Jimeno-Gonzalez S., Lambo S., Hanna B.S. Chd7 is indispensable for mammalian brain development through activation of a neuronal differentiation programme. Nat. Commun. 2017;8:14758. doi: 10.1038/ncomms14758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng W., Khan M.A., Bellvis P., Zhu Z., Bernhardt O., Herold-Mende C., Liu H.K. The chromatin remodeler CHD7 regulates adult neurogenesis via activation of SoxC transcription factors. Cell Stem Cell. 2013;13:62–72. doi: 10.1016/j.stem.2013.05.002. [DOI] [PubMed] [Google Scholar]
- Gascon S., Masserdotti G., Russo G.L., Gotz M. Direct neuronal reprogramming: achievements, hurdles, and new roads to success. Cell Stem Cell. 2017;21:18–34. doi: 10.1016/j.stem.2017.06.011. [DOI] [PubMed] [Google Scholar]
- Guillemot F. Spatial and temporal specification of neural fates by transcription factor codes. Development. 2007;134:3771–3780. doi: 10.1242/dev.006379. [DOI] [PubMed] [Google Scholar]
- Guimaraes R.P.M., Landeira B.S., Coelho D.M., Golbert D.C.F., Silveira M.S., Linden R., de Melo Reis R.A., Costa M.R. Evidence of Muller glia conversion into retina ganglion cells using neurogenin2. Front. Cell. Neurosci. 2018;12:410. doi: 10.3389/fncel.2018.00410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo C., Morris S.A. Engineering cell identity: establishing new gene regulatory and chromatin landscapes. Curr. Opin. Genet. Dev. 2017;46:50–57. doi: 10.1016/j.gde.2017.06.011. [DOI] [PubMed] [Google Scholar]
- Guo Z., Zhang L., Wu Z., Chen Y., Wang F., Chen G. In vivo direct reprogramming of reactive glial cells into functional neurons after brain injury and in an Alzheimer's disease model. Cell Stem Cell. 2014;14:188–202. doi: 10.1016/j.stem.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He B., Deng T., Zhu I., Furusawa T., Zhang S., Tang W., Postnikov Y., Ambs S., Li C.C., Livak F. Binding of HMGN proteins to cell specific enhancers stabilizes cell identity. Nat. Commun. 2018;9:5240. doi: 10.1038/s41467-018-07687-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrich C., Blum R., Gascon S., Masserdotti G., Tripathi P., Sanchez R., Tiedt S., Schroeder T., Gotz M., Berninger B. Directing astroglia from the cerebral cortex into subtype specific functional neurons. PLoS Biol. 2010;8:e1000373. doi: 10.1371/journal.pbio.1000373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heins N., Malatesta P., Cecconi F., Nakafuku M., Tucker K.L., Hack M.A., Chapouton P., Barde Y.A., Gotz M. Glial cells generate neurons: the role of the transcription factor Pax6. Nat. Neurosci. 2002;5:308–315. doi: 10.1038/nn828. [DOI] [PubMed] [Google Scholar]
- Hu J., Qian H., Xue Y., Fu X.D. PTB/nPTB: master regulators of neuronal fate in mammals. Biophys. Rep. 2018;4:204–214. doi: 10.1007/s41048-018-0066-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Z., Liu L., Bian W., Chen Y., Xu G., Cheng L., Jing N. Different transcription factors regulate nestin gene expression during P19 cell neural differentiation and central nervous system development. J. Biol. Chem. 2009;284:8160–8173. doi: 10.1074/jbc.M805632200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Su S.C., Wang H., Cheng A.W., Cassady J.P., Lodato M.A., Lengner C.J., Chung C.Y., Dawlaty M.M., Tsai L.H. Functional integration of dopaminergic neurons directly converted from mouse fibroblasts. Cell Stem Cell. 2011;9:413–419. doi: 10.1016/j.stem.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.W., Oh Y.M., Lu Y.L., Kim W.K., Yoo A.S. MicroRNAs overcome cell fate barrier by reducing EZH2-controlled REST stability during neuronal conversion of human adult fibroblasts. Dev. Cell. 2018;46:73–84. doi: 10.1016/j.devcel.2018.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Shi Y., Yao X., Wang X., Shen L., Rao Z., Yuan J., Liu Y., Zhou Z., Zhang Z. Conversion of astrocytes and fibroblasts into functional noradrenergic neurons. Cell Rep. 2019;28:682–697. doi: 10.1016/j.celrep.2019.06.042. [DOI] [PubMed] [Google Scholar]
- Liu M.L., Zang T., Zou Y.H., Chang J.C., Gibson J.R., Huber K.M., Zhang C.L. Small molecules enable neurogenin 2 to efficiently convert human fibroblasts into cholinergic neurons. Nat. Commun. 2013;4:2183. doi: 10.1038/ncomms3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Miao Q., Yuan J., Han S., Zhang P., Li S., Rao Z., Zhao W., Ye Q., Geng J. Ascl1 converts dorsal midbrain astrocytes into functional neurons in vivo. J. Neurosci. 2015;35:9336–9355. doi: 10.1523/JNEUROSCI.3975-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J.T., Li C.Y., Zhao J.P., Poo M.M., Zhang X.H. Spike-timing-dependent plasticity of neocortical excitatory synapses on inhibitory interneurons depends on target cell type. J. Neurosci. 2007;27:9711–9720. doi: 10.1523/JNEUROSCI.2513-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y.L., Yoo A.S. Mechanistic insights into microRNA-induced neuronal reprogramming of human adult fibroblasts. Front. Neurosci. 2018;12:522. doi: 10.3389/fnins.2018.00522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo C., Lee Q.Y., Wapinski O., Castanon R., Nery J.R., Mall M., Kareta M.S., Cullen S.M., Goodell M.A., Chang H.Y. Global DNA methylation remodeling during direct reprogramming of fibroblasts to neurons. eLife. 2019;8:e40197. doi: 10.7554/eLife.40197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mall M., Kareta M.S., Chanda S., Ahlenius H., Perotti N., Zhou B., Grieder S.D., Ge X., Drake S., Euong Ang C. Myt1l safeguards neuronal identity by actively repressing many non-neuronal fates. Nature. 2017;544:245–249. doi: 10.1038/nature21722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manandhar D., Song L., Kabadi A., Kwon J.B., Edsall L.E., Ehrlich M., Tsumagari K., Gersbach C.A., Crawford G.E., Gordan R. Incomplete MyoD-induced transdifferentiation is associated with chromatin remodeling deficiencies. Nucleic Acids Res. 2017;45:11684–11699. doi: 10.1093/nar/gkx773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masserdotti G., Gascon S., Gotz M. Direct neuronal reprogramming: learning from and for development. Development. 2016;143:2494–2510. doi: 10.1242/dev.092163. [DOI] [PubMed] [Google Scholar]
- Masserdotti G., Gillotin S., Sutor B., Drechsel D., Irmler M., Jorgensen H.F., Sass S., Theis F.J., Beckers J., Berninger B. Transcriptional mechanisms of proneural factors and REST in regulating neuronal reprogramming of astrocytes. Cell Stem Cell. 2015;17:74–88. doi: 10.1016/j.stem.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy K.D., de Vellis J. Preparation of separate astroglial and oligodendroglial cell cultures from rat cerebral tissue. J. Cell Biol. 1980;85:890–902. doi: 10.1083/jcb.85.3.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ninkovic J., Gotz M. Understanding direct neuronal reprogramming-from pioneer factors to 3D chromatin. Curr. Opin. Genet. Dev. 2018;52:65–69. doi: 10.1016/j.gde.2018.05.011. [DOI] [PubMed] [Google Scholar]
- Niu W., Zang T., Zou Y., Fang S., Smith D.K., Bachoo R., Zhang C.L. In vivo reprogramming of astrocytes to neuroblasts in the adult brain. Nat. Cell Biol. 2013;15:1164–1175. doi: 10.1038/ncb2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfisterer U., Kirkeby A., Torper O., Wood J., Nelander J., Dufour A., Bjorklund A., Lindvall O., Jakobsson J., Parmar M. Direct conversion of human fibroblasts to dopaminergic neurons. Proc. Natl. Acad. Sci. U S A. 2011;108:10343–10348. doi: 10.1073/pnas.1105135108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollak J., Wilken M.S., Ueki Y., Cox K.E., Sullivan J.M., Taylor R.J., Levine E.M., Reh T.A. ASCL1 reprograms mouse Muller glia into neurogenic retinal progenitors. Development. 2013;140:2619–2631. doi: 10.1242/dev.091355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raposo A., Vasconcelos F.F., Drechsel D., Marie C., Johnston C., Dolle D., Bithell A., Gillotin S., van den Berg D.L.C., Ettwiller L. Ascl1 coordinately regulates gene expression and the chromatin landscape during neurogenesis. Cell Rep. 2015;10:1544–1556. doi: 10.1016/j.celrep.2015.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riemens R.J.M., van den Hove D.L.A., Esteller M., Delgado-Morales R. Directing neuronal cell fate in vitro: achievements and challenges. Prog. Neurobiol. 2018;168:42–68. doi: 10.1016/j.pneurobio.2018.04.003. [DOI] [PubMed] [Google Scholar]
- Smith D.K., Yang J., Liu M.L., Zhang C.L. Small molecules modulate chromatin accessibility to promote NEUROG2-mediated fibroblast-to-neuron reprogramming. Stem Cell Reports. 2016;7:955–969. doi: 10.1016/j.stemcr.2016.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son E.Y., Ichida J.K., Wainger B.J., Toma J.S., Rafuse V.F., Woolf C.J., Eggan K. Conversion of mouse and human fibroblasts into functional spinal motor neurons. Cell Stem Cell. 2011;9:205–218. doi: 10.1016/j.stem.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramaniam M., Hawse J.R., Rajamannan N.M., Ingle J.N., Spelsberg T.C. Functional role of KLF10 in multiple disease processes. Biofactors. 2010;36:8–18. doi: 10.1002/biof.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiscornia G., Singer O., Verma I.M. Production and purification of lentiviral vectors. Nat. Protoc. 2006;1:241–245. doi: 10.1038/nprot.2006.37. [DOI] [PubMed] [Google Scholar]
- Torper O., Pfisterer U., Wolf D.A., Pereira M., Lau S., Jakobsson J., Bjorklund A., Grealish S., Parmar M. Generation of induced neurons via direct conversion in vivo. Proc. Natl. Acad. Sci. U S A. 2013;110:7038–7043. doi: 10.1073/pnas.1303829110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treutlein B., Lee Q.Y., Camp J.G., Mall M., Koh W., Shariati S.A., Sim S., Neff N.F., Skotheim J.M., Wernig M. Dissecting direct reprogramming from fibroblast to neuron using single-cell RNA-seq. Nature. 2016;534:391–395. doi: 10.1038/nature18323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsunemoto R.K., Eade K.T., Blanchard J.W., Baldwin K.K. Forward engineering neuronal diversity using direct reprogramming. EMBO J. 2015;34:1445–1455. doi: 10.15252/embj.201591402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vadodaria K.C., Mertens J., Paquola A., Bardy C., Li X., Jappelli R., Fung L., Marchetto M.C., Hamm M., Gorris M. Generation of functional human serotonergic neurons from fibroblasts. Mol. Psychiatry. 2016;21:49–61. doi: 10.1038/mp.2015.161. [DOI] [PubMed] [Google Scholar]
- Vasconcelos F.F., Sessa A., Laranjeira C., Raposo A., Teixeira V., Hagey D.W., Tomaz D.M., Muhr J., Broccoli V., Castro D.S. MyT1 counteracts the neural progenitor program to promote vertebrate neurogenesis. Cell Rep. 2016;17:469–483. doi: 10.1016/j.celrep.2016.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierbuchen T., Ostermeier A., Pang Z.P., Kokubu Y., Sudhof T.C., Wernig M. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010;463:1035–1041. doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wapinski O.L., Lee Q.Y., Chen A.C., Li R., Corces M.R., Ang C.E., Treutlein B., Xiang C., Baubet V., Suchy F.P. Rapid chromatin switch in the direct reprogramming of fibroblasts to neurons. Cell Rep. 2017;20:3236–3247. doi: 10.1016/j.celrep.2017.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wapinski O.L., Vierbuchen T., Qu K., Lee Q.Y., Chanda S., Fuentes D.R., Giresi P.G., Ng Y.H., Marro S., Neff N.F. Hierarchical mechanisms for direct reprogramming of fibroblasts to neurons. Cell. 2013;155:621–635. doi: 10.1016/j.cell.2013.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Du Y., Deng H. Direct lineage reprogramming: strategies, mechanisms, and applications. Cell Stem Cell. 2015;16:119–134. doi: 10.1016/j.stem.2015.01.013. [DOI] [PubMed] [Google Scholar]
- Xu Z., Jiang H., Zhong P., Yan Z., Chen S., Feng J. Direct conversion of human fibroblasts to induced serotonergic neurons. Mol. Psychiatry. 2016;21:62–70. doi: 10.1038/mp.2015.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yee K.S., Yu V.C. Isolation and characterization of a novel member of the neural zinc finger factor/myelin transcription factor family with transcriptional repression activity. J. Biol. Chem. 1998;273:5366–5374. doi: 10.1074/jbc.273.9.5366. [DOI] [PubMed] [Google Scholar]
- Zhang J., Chen S., Zhang D., Shi Z., Li H., Zhao T., Hu B., Zhou Q., Jiao J. Tet3-mediated DNA demethylation contributes to the direct conversion of fibroblast to functional neuron. Cell Rep. 2016;17:2326–2339. doi: 10.1016/j.celrep.2016.10.081. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession number for the RNA-seq data is GEO: GSE132674. The accession number for the ChIP-seq data is GEO: GSE132671.