Summary
One cause of human male infertility is a scarcity of spermatogonial stem cells (SSCs) in testes with Sertoli cells that neither produce adequate amounts of GDNF nor form the Sertoli-Sertoli junctions that form the blood-testis barrier (BTB). These patients raise the issue of whether a pool of SSCs, depleted due to inadequate GDNF stimulation, will expand if normal signaling is restored. Here, we reduce adult mouse SSC numbers by 90% using a chemical-genetic approach that reversibly inhibits GDNF signaling. Signal resumption causes all remaining SSCs to replicate immediately, but they primarily form differentiating progenitor spermatogonia. Subsequently, self-renewing replication restores SSC numbers. Testicular GDNF levels are not increased during restoration. However, SSC replication decreases as numbers of SSCs and progenitors increase, suggesting important regulatory interactions among these cells. Finally, sequential loss of SSCs and then pachytene spermatocytes causes dissolution of the BTB, thereby recapitulating another important characteristic of some infertile men.
Keywords: spermatogonial stem cell, progenitor spermatogonia, GDNF, Ret, GFRα1, blood-testis barrier, Sertoli cell, infertility
Graphical Abstract
Highlights
-
•
SSCs are rapidly lost when GDNF signaling is inhibited
-
•
If signaling resumes the depleted SSC pool is restored by self-renewing replication
-
•
Testicular GDNF levels are not increased during restoration
-
•
SSC replication decreases as stem cell numbers increase
Wright and colleagues investigate the restoration of adult spermatogonial stem cells (SSCs) following 9 days of inhibited GDNF signaling. Immediately after signaling resumes, SSC replication generates progenitors. Then, self-renewing replication restores SSC numbers. Testicular GDNF levels are not increased during restoration. However, the fraction of replicating SSCs decreases as their numbers increase, suggesting that interactions among them regulate restoration.
Introduction
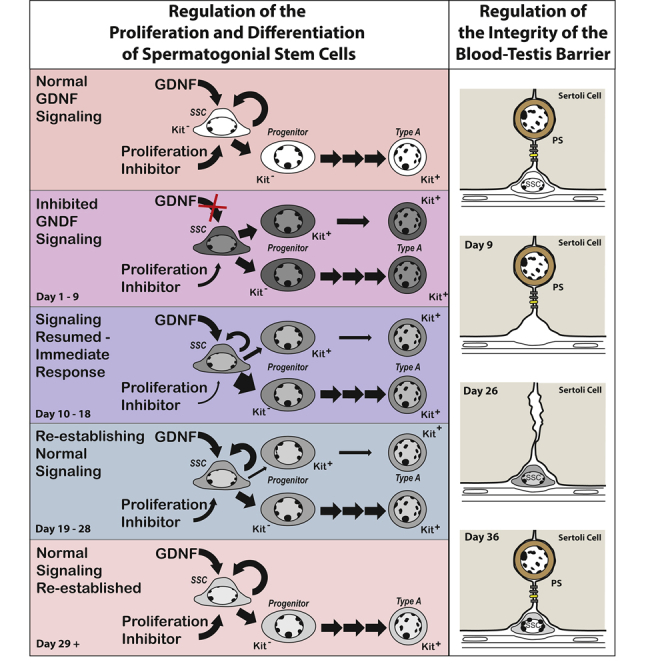
Many studies have demonstrated that glial cell line-derived neurotrophic factor (GDNF), a product of Sertoli cells and in some species, peritubular myoid cells, is essential for establishing and maintaining the pool of spermatogonial stem cells (SSCs), the foundational spermatogenic cells. Neonatal mice that are heterozygous null for GDNF fail to produce normal numbers of SSCs, and those stem cells are eventually lost (Meng et al., 2000). Juvenile mice that undergo Cre-mediated excision of the gene encoding the ligand binding subunit of the GDNF receptor, GFRα1, lose SSCs within 3 weeks (Sada et al., 2012). We have shown that GDNF is essential for maintaining SSCs in the adult mouse testis by use of a chemical-genetic approach that rapidly but reversibly inhibits GDNF signaling (Savitt et al., 2012). This approach combined mice with a V805A mutation in Ret, the kinase subunit of the GDNF receptor, and daily injections of 1NA-PP1, a bulky ATP competitive inhibitor that only binds the mutant kinase. We demonstrated that inhibition of GDNF signaling for 9 days caused loss of some SSCs, while inhibition for 11 to 30 days resulted in quantitative stem cell loss (Parker et al., 2014; Savitt et al., 2012). Decreased replication and the irreversible differentiation of SSCs into progenitor spermatogonia and then fully mature type A1 spermatogonia deplete the stem cell pool (Parker et al., 2014).
Over time, a seminiferous epithelium without stem cells loses increasingly mature spermatogenic cells, a process called maturation depletion. Eventually, only Sertoli cells remain, a condition called Sertoli cell-only (SCO) syndrome. Importantly, this syndrome is one cause of infertility of men. While all tubules are SCO in some of these men, tubules of others contain small segments with a full complement of spermatogenic cells and, thus, SSCs (Schlegel, 2004). The similarity in testicular histology of mouse testes deprived of GDNF signaling to men with SCO syndrome led us to hypothesize that Sertoli cells in these human testes produce abnormally low levels of GDNF. Our recent studies support this hypothesis (Singh et al., 2017). In addition, our detailed transcriptional analysis of highly regressed SCO human tubules detected all SSC markers, although at markedly reduced levels (Paduch et al., 2019). Thus, these tubules appear to contain a few unproductive stem cells.
An important question arose from our clinical and basic studies; if inadequate stimulation by GDNF causes loss of most but not all SSCs, could the stem cell pool be rebuilt if GDNF stimulation was restored to normal? Experiments described herein answer this question by evaluating the restoration of SSCs and their progenitor spermatogonia in Ret(V805A) mice that were treated for 9 days with 1NA-PP1. An affirmative answer might seem obvious, but it is not a forgone conclusion. SSCs are phenotypically heterogeneous both in their replicative activity and in their expression of receptors for critical paracrine factors, such as GDNF (Kubota, 2019). It is entirely possible that inhibition of GDNF signaling selects a subset of SSCs that are more resistant to differentiation but have a reduced capacity to rebuild the stem cell pool when GDNF signaling is restored. Alternatively, inhibition might result in epigenetic changes that alter a stem cell's self-renewing capacity (Farlik et al., 2015).
We also sought to determine if following 9 days of inhibited GDNF signaling, self-renewing stem cell replication was primarily responsible for restoring SSC numbers and consequently the entire spermatogenic lineage. We posited that processes that regulate numbers of SSCs in normal mature testis also regulate restoration. A long-standing model proposes that SSCs are a subset of solitary, highly undifferentiated spermatogonia, which upon replication generates either a new stem cell or differentiating progenitor spermatogonia. It is further proposed that niche factors, such as GDNF and FGF2, regulate which cell types are generated (de Rooij, 2001, 2017). However, two recent studies, which used busulfan to kill some SSCs, suggest that this classical model does not explain SSC restoration. One study concluded that an efficient restorative process requires fragmentation of chains of progenitor spermatogonia, and their subsequent dedifferentiation into SSCs (Carrieri et al., 2017). The second concluded that progenitors in short chains and SSCs functionally interconvert and that busulfan-induced damage to the testis increases GDNF concentration, which rebalances this interconversion toward stemness (La et al., 2018).
In our previous studies we treated mice for 9 days with 1NA-PP1; 19 days later about half of the seminiferous tubules were SCO (Parker et al., 2014). An important issue that arose was whether this result reflected a 50% loss of functional SSCs or whether a much larger percentage of SSCs were lost, but their numbers were partially restored after GDNF signaling resumed. We resolve this issue by evaluating the numbers of transplantable SSCs that remain in a testis immediately following 9 days of inhibition of GDNF signaling.
We also investigated the extent to which replication drives SSC restoration, as well as that of progenitor spermatogonia. Those studies required that we examine intact seminiferous tubules, enumerate both SSCs and progenitor spermatogonia, and also determine the percentage of cells that were replicating. In those studies, we defined SSCs as expressing a stem cell marker and existing as solitary, single (As) cells. We acknowledge that the pool of As spermatogonia contains both SSCs and the most undifferentiated of their immediate progeny, progenitor spermatogonia. However, these progenitors may de-differentiate into functional SSCs (Carrieri et al., 2017; Nakagawa et al., 2010). We defined progenitors as two or more spermatogonia that express the same stem cell marker and have plasma membranes in direct physical contact. Cells in pairs (Apr) cells are considered less differentiated than cells in chains of A aligned (Aal) spermatogonia (de Rooij, 2001).
Our studies were founded on the proposition that investigation of the loss and restoration of SSCs in 1NA-PP1-treated Ret(V805A) mice might shed light on the ontogeny of human SCO syndrome and eventually suggest therapies for men diagnosed with this syndrome who nonetheless retain some SCCs. For this proposition to be valid, the restoration of SSCs that follows a temporary inhibition of GDNF signaling must occur in a tissue that develops the same architectural deficit observed in human SCO tubules: the failure of adjacent Sertoli cells to form the blood-testis barrier, a series of adhesive junctions essential for fertility (Mruk and Cheng, 2015). We note that the presence of pachytene spermatocytes was recently reported to be required for barrier formation (Li et al., 2018). It follows that, if our mouse model recapitulates human SCO syndrome, loss of SSCs and then eventually of pachytene spermatocytes should cause dissolution of this barrier. Our last experiment tests this proposition.
The above considerations led us to test the following hypothesis: inhibition of GDNF signaling causes a substantial loss of SSCs and, with the resumption of signaling, self-renewing replication restores stem cell numbers. We predict that the loss of differentiated spermatogenic cells following loss of SSCs results in dissolution of the blood-testis barrier, as occurs in human SCO testes.
Results
Nine Days of Inhibited GDNF Signaling Results in a Substantial Loss of SSCs, but Their Numbers are Significantly Restored Once Signaling Resumes
This study had three parts, which were conducted simultaneously: (1) we assayed the effect of 9 days of inhibited GDNF signaling on numbers of transplantable SSCs, (2) we confirmed that the effect of inhibiting GDNF signaling recapitulated previous results, and (3) we evaluated both the loss and restoration of SSCs by enumerating spermatogonia that expressed a highly restricted SSC marker, ID4-eGFP (Helsel et al., 2017; Parker et al., 2014; Savitt et al., 2012). Ret(V805A) mice were transgenic for both LacZ and Id4-eGFP. (In subsequent studies mice were not transgenic.)
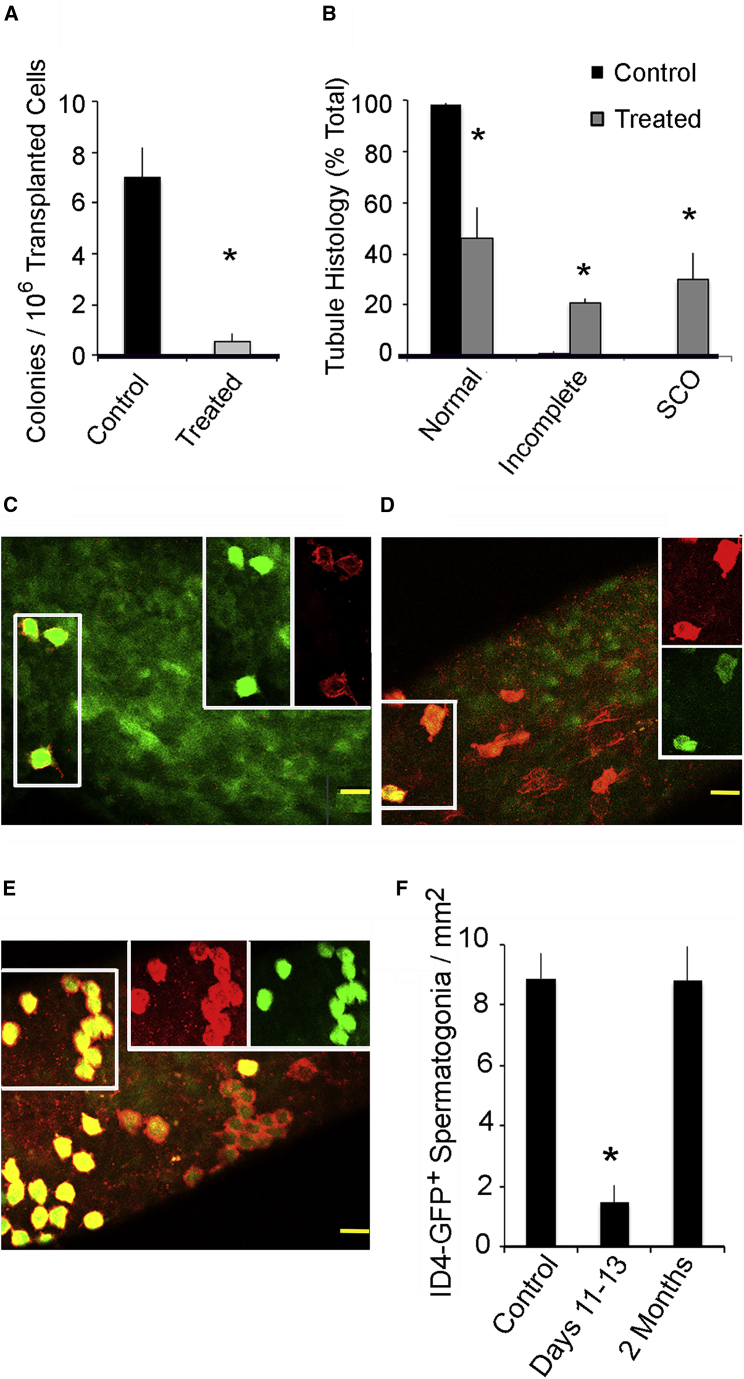
Part 1: single germ cell suspensions were prepared 2 to 3 days after the last injection with 1NA-PP1 or vehicle and ~800,000 cells from treated and control testes were transplanted into 17 and 16 germ cell-deficient testes, respectively. Two months later we enumerated colonies of LacZ+ spermatogenic cells that encircled a seminiferous tubule. Transplants from control mice generated 10 times more LacZ+ colonies than germ cells from treated animals (Figure 1A). Thus, inhibition of GDNF signaling for 9 days causes 90% loss of transplantable SSCs.
Figure 1.
Loss of SSCs during 9 Days of Inhibited GDNF Signaling and Their Partial Restoration 2 Months after Signaling Resumes
Adult, Ret V805A+/+, Rosa 26+/−, ID4-eGFP+/− mice were injected for 9 days with vehicle (control) or with 1NA-PP1 (treated). Quantitative data are presented as mean + SEM. An asterisk indicates that treated mice differ from controls (p < 0.05).
(A) The effects of inhibition of GDNF signaling on numbers of transplantable SSCs. Two to 4 days after the last injection, germ cells from each animal were transplanted into testes of germ cell-deficient mice. Germ cells from treated and control mice were transplanted into 17 and 16 testes, respectively, and LacZ+ germ cell colonies per testis enumerated 2 months later. Data are presented as number of colonies per 106 transplanted cells per testis.
(B) Fraction of tubule cross-sections in control and treated animals were classified as exhibiting normal spermatogenesis, incomplete spermatogenesis, or Sertoli cell-only (SCO) syndrome. Testes were collected 2 months after the last injection, 1 μm testis cross-sections were prepared and each tubule classified. Data (n = 3/group) are presented as fraction of tubule cross-sections per classification per testis.
(C–E). Confocal micrographs of GFRα1+ (red), ID4-eGFP+ (green) spermatogonia in tubules of control and treated mice. The two boxes on the right side of an image separate red and green channels of the left hand box. Scale bars, 40 μm. (C) Image from a control animal. (D) Image from an animal that was analyzed 2–4 days after the last injection 1NaPP1. Similar images were obtained for 75% of the surface of tubules collected 2 months after treatment. (E) The remaining 25% contained clusters of GFRα1+, ID4-eGFP+ spermatogonia.
(F) Numbers of GFRα1+, ID4-eGFP+ spermatogonia/mm2 of tubule surface for control (n = 5) and treated mice analyzed 2–4 days or 2 months (n = 3/group) after the last injection.
Part 2: we sacrificed three control and three treated animals 2 months after the last injection of vehicle or 1NA-PP1-HCl, prepared their testes for histological analysis, and determined the percentage of tubules that exhibited normal spermatogenesis, incomplete spermatogenesis (missing one to three generations of germ cells), or SCO syndrome (Parker et al., 2014; Savitt et al., 2012). Consistent with previous studies, normal or incomplete spermatogenesis was observed in 49% and 22% of tubules, respectively, of treated animals (Figure 1B). The rest were SCO (Parker et al., 2014; Savitt et al., 2012).
Part 3: we collected seminiferous tubules from five control mice, and from mice sacrificed 2–4 days or 2 months after treatment (n = 3/group). To distinguish As from Apr and Aal spermatogonia, tubules were processed for GFRα1 immunocytochemistry. Microscopic analysis revealed that all Id4-eGFP+ spermatogonia co-expressed GFRα1 and all existed as As spermatogonia in controls, in tubules collected 2–4 days after treatment and in about 75% of the surface of tubules collected 2 months later (Figures 1C and 1D). In the remaining areas, Id4-eGFP+ cells were in clusters (Figure 1E).
Morphometric analysis revealed that 2–4 days after treatment, the density of Id4-eGFP+ spermatogonia was reduced to 16% of control, while at 2 months cell densities were increased to 92% (Figure 1F). However, the clustering of some cells suggests that restoration was incomplete.
Self-Renewing Replication Restores Numbers of SSCs after GDNF Signaling Resumes
As discussed in the Introduction, three different mechanisms potentially explain the restoration of the stem cell pool that follows resumption of GDNF signaling. Our primary goal for the next four studies was to determine if self-renewing replication restored SSCs. A secondary goal was to determine if replication of both SSCs and progenitor spermatogonia restored progenitors. To meet these goals, we compared a direct measurement of increased cell numbers with an estimate of new cells formed by replication. The need for quantitation precluded lineage tracing, which generates only qualitative data unless a high percentage of SSCs are marked at a defined time (Lord and Oatley, 2018). Results of one study suggest that 2 weeks of induction may be necessary to mark most SSCs (Sharma et al., 2019), thereby excluding analysis of restoration initiation.
Study 1 tested an implicit assumption in our definition of progenitor spermatogonia: GFRα1+ spermatogonia whose plasma membranes make direct physical contact function in a coordinated manner. Study 2 defined changes in numbers and replication of SSCs and progenitor spermatogonia during and after 9 days of inhibited GDNF signaling. We identified replicating cells by their incorporation of the thymidine analog 5-ethynyl-2′deoxyuridine (EdU), which was injected 24 h before tissue collection. Study 3 defined the fraction of cells in S phase during a 24-h period that were labeled with EdU. Determining labeling efficiency is necessary for EdU’s half-life in serum is about 30 min and cells entering S phase hours after EdU injection might not be labeled (Cheraghali et al., 1995). Study 4 determined the average length of the cell cycle of SSCs. Finally, we used the results of the last three studies to estimate yield of new cells from replication after GDNF signaling resumption, and we compared this yield with the actual increase in cell number.
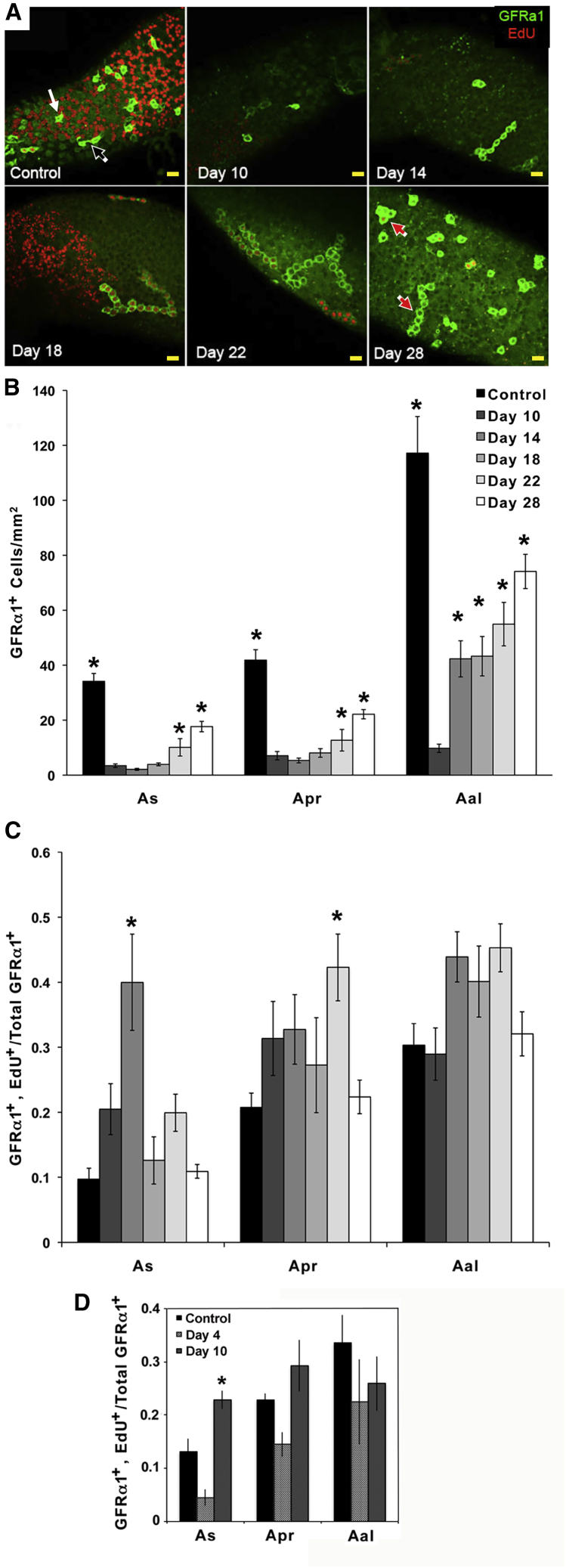
Study 1: we used cell replication to test if four GFRα1+ cells in a chain functioned coordinately. Mice were injected with EdU, tubules were collected 24 h later, and EdU+ and GFRα1+ cells in these tubules identified. GFRα1+, EdU+, and GFRα1+, EdU− cells were observed in the same microscopic field (Figure S1A). (This we observed in all experiments.) However, for 72% of all chains of four Aal cells, when at least one cell was EdU+, so, too, were the other three (Figure S1B). Thus, spermatogonia that contact one another coordinate their replication.
Study 2: Ret(V805A) mice were treated for 9 days with 1NA-PP1 or vehicle, and tissues collected 1 day later (day 10 of the experiment) or on days 14, 18, 22, and 28. Here, we used GFRα1 as the stem cell marker. Figure 2A presents confocal images of GFRα1+ As, Apr, and Aal spermatogonia on seminiferous tubules of control and treated mice. Few GFRα1+ cells were observed on day 10, but their numbers increased after GDNF signaling resumed. Unexpectedly on days 18 and 22, many of the chains of GFRα1+ Aal spermatogonia were substantially longer than in control animals, and average chain length was significantly increased (Figure S1C). Qualitatively, similar trends were observed when we used a different marker, ZBTB16 (Figure S2) (Buaas et al., 2004).
Figure 2.
The Effects of Loss and Resumption of GDNF Signaling on Densities of GFRα1+ As, Apr, and Aal Spermatogonia and on the Fraction of Cells that Incorporated EdU
(A) Representative 1.8 μm thick optical sections of GFRα1+ As, Apr, and Aal spermatogonia (green) on intact seminiferous tubules. All mice were injected with EdU (red) 24 h before sample collection. Mice were injected with vehicle (Control) or with 1NA-PP1 for 9 days and samples collected on days 10, 14, 18, 22, and 28 of the experiment. White, black, and red arrows identify representative GFRα1+ As, Apr, and Aal spermatogonia, respectively. Scale bars, 40 μm.
(B) Numbers of GFRα1+ As, Apr, and Aal spermatogonia/mm2 of seminiferous tubule surface in control mice and in mice treated with 1NA-PP1 for 9 days and analyzed on days 10, 14, 18, 22, and 28. Data are expressed as mean ± SEM (n = 8/group). An asterisk over a bar identifies that group as significantly different from treated animals on day 10. The inserted legend also applies to (C).
(C) Fractions of GFRα1+ As, Apr, and Aal spermatogonia that incorporated EdU in controls and in mice that were treated with 1NA-PP1 for 9 days and analyzed on days 10–28. An asterisk over a bar indicates a significant difference from control.
(D). Fraction of total GFRα1+ As, Apr, and Aal spermatogonia that incorporated EdU in controls and animals treated for 3 or 9 days with 1NA-PP1. EdU was injected 24 h before sample collection (days 4 or 10). Data are expressed as mean ± SEM (n = 5/group). An asterisk denotes a significant difference between days 4 and 10.
To define the time course of recovery, we enumerated GFRα1+ As, Apr, and Aal spermatogonia (Figure 2B). (From here on, we refer to these cells in the text, but not in figure legends, as As, Apr, and Aal cells.) On day 10, the densities of As and Apr cells were reduced to 8% and 10% of controls, respectively. Numbers of As and Apr cells remained low on day 14 and then increased to 53% of control by day 28. Numbers of Aal cells also were reduced to 8% of controls on day 10, but increased by 4-fold by day 14, and further to 63% of control on day 28. This more rapid recovery suggested that restoration was initially biased toward progenitor spermatogonia.
To define how cell replication changed during recovery, we evaluated the fraction of GFRα1+ cells that incorporated EdU (Figures 2A and 2C). As cells exhibited the greatest response to resumption of GDNF signaling; cell replication was four times higher than in controls on day 14, but returned to control levels by day 18. Replication of Apr cells was only greater than controls on day 22. Replication of Aal cells did not change significantly.
Our observation that replication of As cells was not decreased after 9 days of inhibited GDNF signaling was unexpected, since it is decreased after 3 days (Parker et al., 2014). To confirm and expand our new result, we compared cell replication after 3 and 9 days of inhibited GDNF signaling (n = 5/group). Results (Figure 2D) showed that a significantly higher fraction of As cells replicated after 9 days than after 3 days, while replication by controls was at an intermediate level.
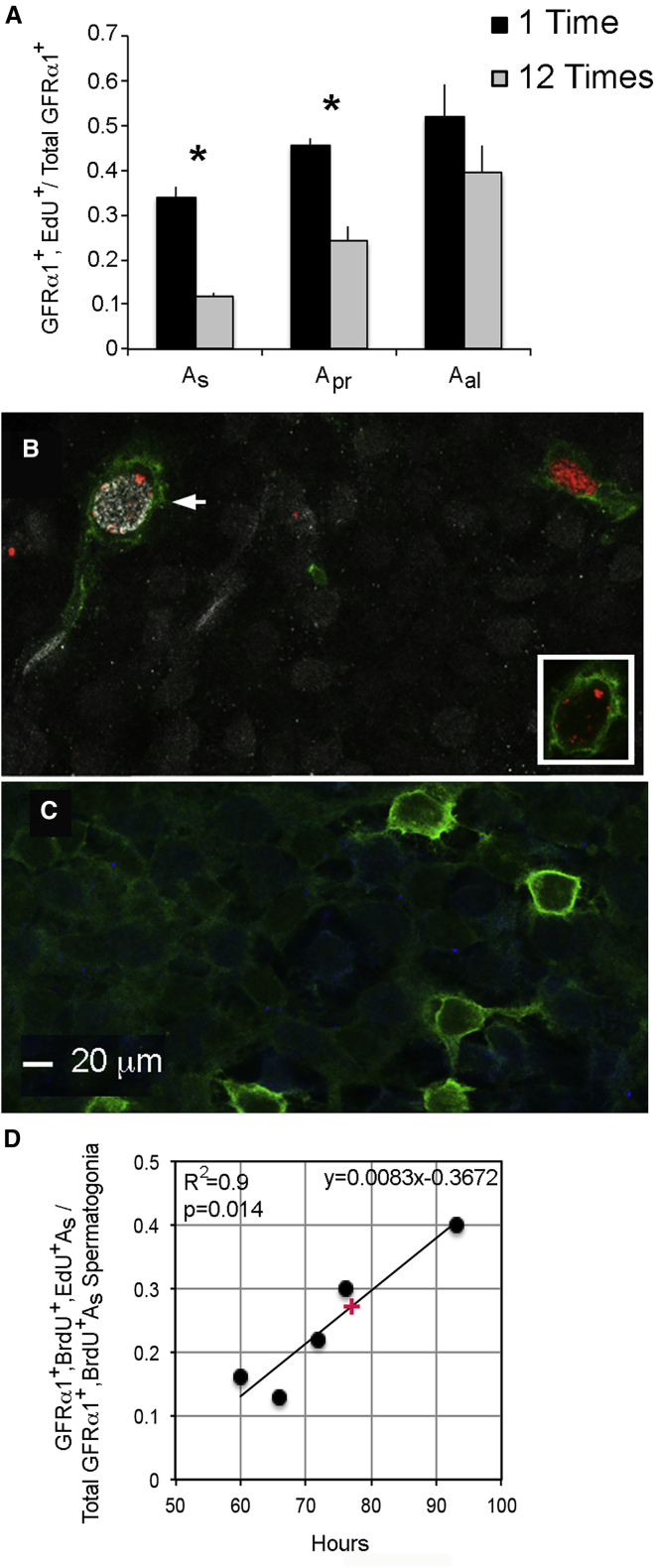
Study 3: to determine labeling efficiency, we injected mice once or every other hour for 24 h with EdU (n = 3/group), collected samples 24 h after the first injection, and determined the fraction of cells labeled with EdU after 1 or 12 injections (Figure 3A). Labeling efficiency (fraction of cells labeled after 1 injection/fraction labeled after 12) was 0.35, 0.53, and 0.76 for As, Apr, and Aal cells, respectively.
Figure 3.
Determinations of the Efficiency of Labeling GFRα1+ As, Apr, and Aal Spermatogonia with EdU and of the Average Length of the Cell Cycle of GFRα1+ As Spermatogonia
(A) Fractions of GFRα1+ As, Apr, and Aal spermatogonia that were EdU+ after 1 or 12 injections (n = 3/group). Tissues were collected 24 h after the first injection. Data are presented as mean + SEM. An asterisk denotes a significant difference between 1 and 12 injections.
(B) A GFRα1+, BrdU+, EdU+ As spermatogonium from an animal injected with BrdU (white), with EdU (red) 93 h later, and tubules collected 2 h later. The same cell is shown in the insert, but only signals for GFRα1 and EdU are shown. The BrdU−, EdU+ cell in the upper right hand corner shows that anti-BrdU does not cross-react with EdU.
(C) GFRα1+ As spermatogonia in a control mouse that was injected neither with BrdU nor EdU but processed for both. Scale bar, 20 μm.
(D) Determination of the average length of the cell cycle of GFRα1+ As spermatogonia. Mice were injected with BrdU and then 60, 66, 72, 76, or 93 h later with EdU (n = 1 animal per time point). GFRα1+, BrdU+, EdU+ As spermatogonia and total GFRα1+, BrdU+ As spermatogonia were enumerated and the number of the first divided by the number of the second. The resulting fractions were plotted against hours between injections. We defined average cell-cycle length, 76.5 h, for GFRα1+ As spermatogonia from the midpoint on the line (see red +).
Study 4: we used a classical approach to determine the average length of the cell cycle of As cells, sequential injection of animals with two different thymidine analogs and identification of the time interval producing dual-labeled cells. While Kanatsu-Shinohara et al. (2016) reported that the length of the cell cycle of CDH1+ spermatogonia was 93.6 h, we performed this experiment because our mice have a different genetic background, SSCs are heterogeneous, cycle lengths of other stem cells can vary, and CDH1 expression spans As to type A1 spermatogonia (Bragado Alonso et al., 2014; Kubota, 2019; Tokuda et al., 2007). We injected mice first with 5′-bromo-2′deoxyuridine (BrdU) and then 60, 66, 72, 76, and 93 h later with EdU and collected tubules 2 h after the second injection (n = 1/time point). Those times spanned the reported lengths of the cell cycle of mouse CDH1+ spermatogonia and of rat As spermatogonia (Huckins, 1971; Kanatsu-Shinohara et al., 2016). Dual-labeled GFRα1+ As cells were observed at all five time points (Figure 3B). We detected no apparent labeling in negative control tubules (Figure 3C). To establish the average cell-cycle length, we enumerated GFRα1+, BrdU+, EdU+, As cells and total GFRα1+, BrdU+ As cells at each time point, divided the former number by the latter, and plotted the resulting fractions against hours between injections (Figure 3D). This analysis revealed a linear, positive correlation between the fraction of cells that were dual-labeled and the time between injections. To estimate the average length of the cell cycle, we calculated the midpoint between the minimum and maximum fraction of double-labeled cells, and extrapolated from that midpoint an average cell cycle of 76.5 h, or 3.2 days.
Comparing cell yield with increased cell numbers: we used the data from Figure 2 to calculate cell densities and fraction of EdU-labeled As, Apr, and Aal cells for each 3.2-day interval after GDNF signaling resumed (Table S1). We assumed that numbers of replicating cells equaled cell yield; yield = cells/mm2 × (fraction of EdU+ cells/labeling efficiency). Table 1 compares total cell yield with total increase in cell density from days 10 to 28. Replication of As cells explains both the increase in their number and 53% of the increase of Apr cells. Replication of Apr cells accounts for 53% of the increase in Aal cells, while Aal cell replication explains the remainder. Yield from replication of As, Apr, plus Aal cells also exceed the sum of their increased numbers. Finally, Table S1 demonstrates that all remaining As cells replicate within 4 days of resumed GDNF signaling. These results support the conclusions that self-renewing replication restores numbers of SSCs and that replication of SSCs and progenitors restores progenitors.
Table 1.
Calculated Yield of New Cells from Replication of GFRα1+ As, Apr, and Aal Spermatogonia from Days 10 to 28 versus Measured Increase in the Numbers of Those Cells
| Cell Type | Calculated Yielda | Measured Increasea |
|---|---|---|
| GFRα1+ As sermatogoniab | 22.29 | 14.25 |
| GFRα1+ Apr spermatogonia | 37.74 | 15.09 |
| GFRα1+ Aal spermatogonia | 140.95 | 64.36 |
| Totalc | 200.98 | 93.7 |
Data are presented as numbers of new cells/mm2 of tubule surface.
Predicted yield from replication of GFRα1+ As spermatogonia exceeds the measured increase in their numbers.
The sum of the predicted yields from the replication of all three cell types exceeds the sum of the measured increases in their cell numbers.
When GDNF Signaling Resumes, Restoration is Initially Biased toward Formation of Progenitors
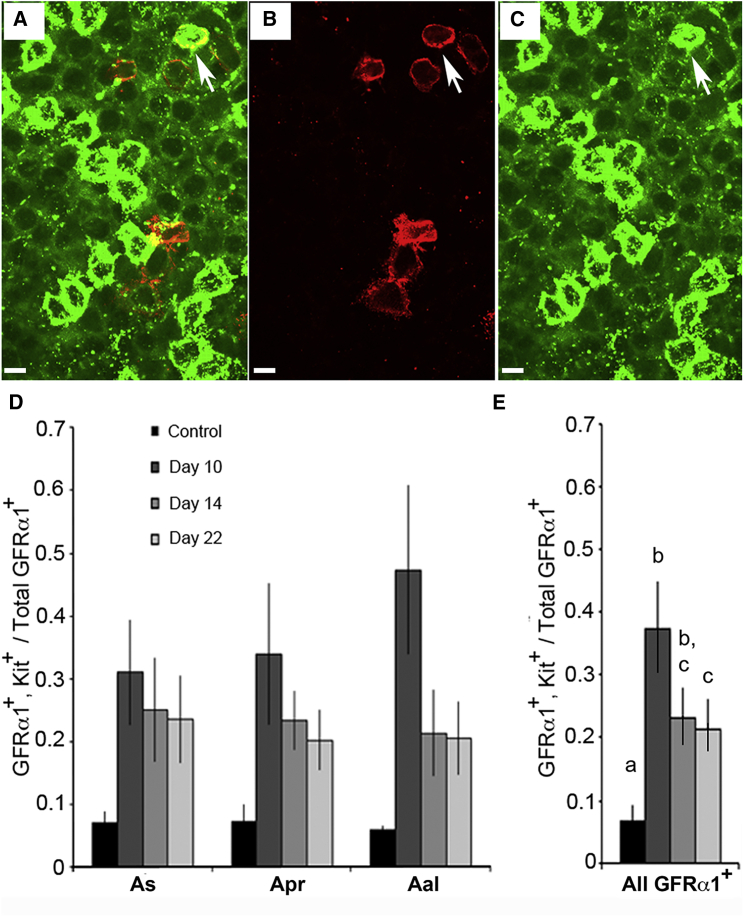
Aal cells are the first to increase in number after GDNF signaling resumes, suggesting that the restorative process is initially biased toward formation of progenitors. We wondered if these cells were poised to become fully differentiated type A1 spermatogonia and thus expressed Kit. We treated mice for 9 days with 1NA-PP1, collected samples on days 10, 14, and 22, and enumerated cells expressing GFRα1 or GFRα1 plus Kit (Figures 4A–4C). Results show a significant overall effect of 1NA-PP1 treatment on the fraction of GFRα1+ cells that co-expressed Kit (Figure 4D). When counts for all three cell types were summed, the fraction of cells that co-expressed Kit at day 10 was 5.5-fold higher than in controls. By day 22, this fraction was reduced but still greater than control (Figure 4E).
Figure 4.
After 9 Days of Inhibited GDNF Signaling, a Significant Fraction of GFRα1+ Spermatogonia Also Express the Differentiation Marker, Kit, but This Fraction Decreases Once GDNF Signaling Resumes
Spermatogonia co-expressing GFRα1 (red) and/or Kit (green) were identified by immunocytochemistry and confocal microscopy. Optical sections are 2 μm thick. Scale bars, 20 μm.
(A) Image with merged red and green channels. The arrow points to a cell expressing both proteins.
(B and C). Separate red (B) and green (C) channels.
(D). Fraction of GFRα1+ As, Apr, and Aal spermatogonia that co-expressed Kit. Mice were treated for 9 days with vehicle (control) or 1NA-PP1, and tubules analyzed on days 10, 14, 18, and 22. Data (mean ± SEM; n = 9–11 mice/group) are presented for each cell type. ANOVA shows an overall significant effect of treatment.
(E). Fraction of all GFRα1+ spermatogonia that co-expressed Kit. Data were summed across cell types, and each mean compared with the other means. Different letters over bars denote significant differences between means.
During Restoration, Increased SSC Replication is Not Associated with Increased GDNF Expression but with Numbers of SSCs and Progenitors
The marked spike on day 14 in As cell replication suggested exposure to increased mitogenic stimuli. A likely candidate was GDNF, because changes in GDNF signaling affect replication of GFRα1+ As, spermatogonia within 2 days (Parker et al., 2014) and testicular content of GDNF mRNA is elevated after toxicant-induced death of SSCs (Carrieri et al., 2017; Zohni et al., 2012). Thus, we measured testicular GDNF protein and mRNA levels in controls after inhibition and following resumption of GDNF signaling. Neither increased, nor did transcripts encoding four other regulators of SSCs, FGF2, FGF8, CSF1, and CXCL12 (Figure S3) (DeFalco et al., 2015; Hasegawa and Saga, 2014; Oatley et al., 2009; Yang et al., 2013). Next, we used RNA sequencing to search for potential increases in expression of other growth factors. Analysis of control testes and testes collected at day 14 (n = 3/group) identified 15,721 transcripts with Ensemble IDs. Principal-component analysis showed that testicular transcriptomes of the two groups were highly similar (Figure S4). However, expression of 144 transcripts differed (false discovery rate < 0.05). None encoded a growth factor.
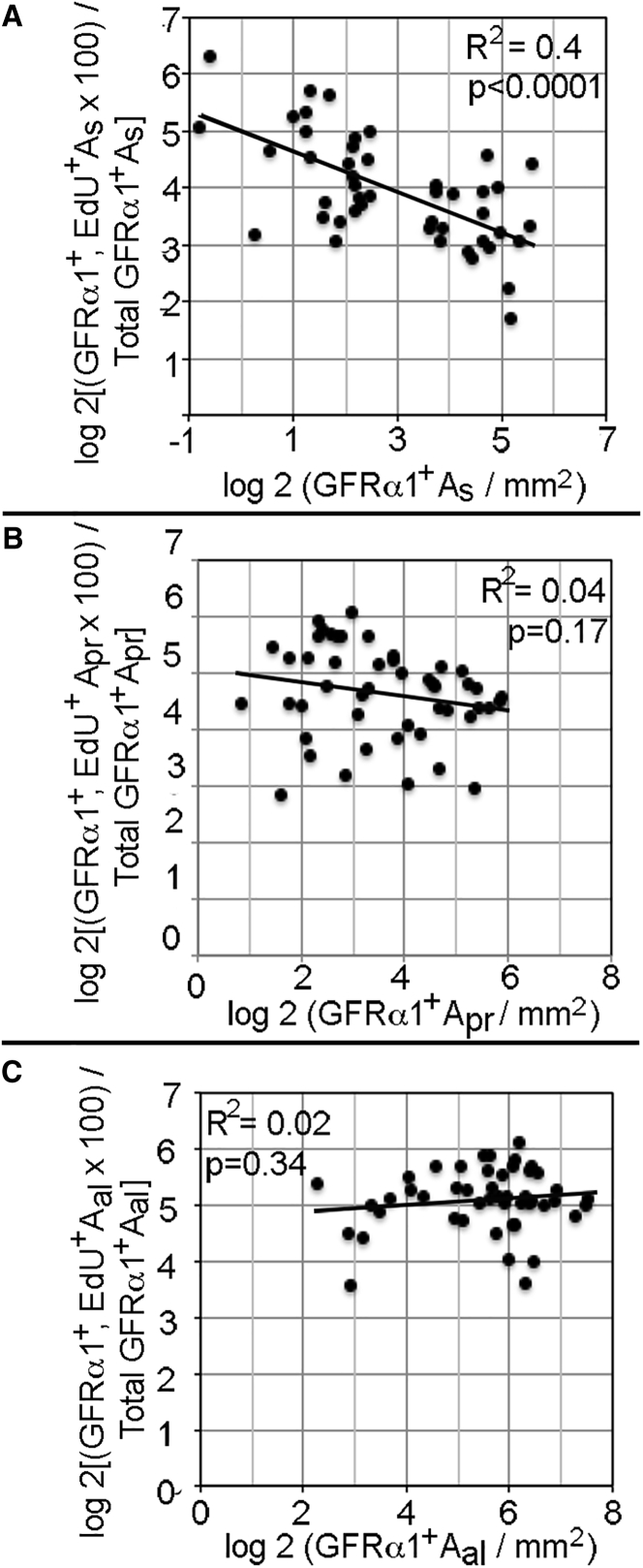
The lack of increased growth factor expression led us to look for any evidence of regulated replication of SSCs and progenitors. As SSCs appear to interact in vitro, we asked if data presented in Figures 2B and 2C revealed relationships in vivo between the densities of As, Apr, and Aal cells and their replications (Ebata et al., 2011). We pooled and log transformed data from individual animals and plotted the fraction of As, Apr, or Aal cells that incorporated EdU against their densities and the densities of the two other cell types. This analysis revealed highly significant negative correlations between the fraction of As cells that replicated and densities of As, Apr, and Aal cells. However, replication of Apr or Aal cells were not correlated with densities of the three cell types (Figures 5 and S5). These results suggest that As, Apr, and Aal cells, either directly or via another cell type, regulate replication of As spermatogonia.
Figure 5.
The Replication of GFRα1+ As Spermatogonia Decreases as Their Density Increases
For each control and treated animal (Figures 2B and 2C), densities of GFRα1+ As, Apr, and Aal spermatogonia and the EdU+ fractions of these cells were log 2 transformed and replication plotted against density. R2 and p values for each regression line are shown.
Loss of SSCs Followed by Loss of Pachytene Spermatocytes Results in Dissolution of the Blood-Testis Barrier
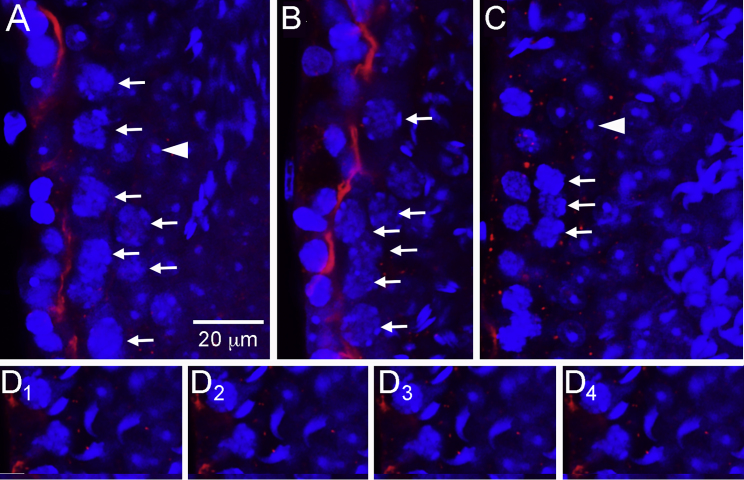
In the Introduction, we proposed that 1NaPP1-treated Ret(V805A) mice model the human SCO syndrome. An important caveat is that loss of SSCs in these mice results in the same architectural deficit seen in human SCO syndrome, the failure of Sertoli cells to form a blood-testis barrier (Camatini et al., 1981). To evaluate this possibility, we focused on claudin 11 because it is not concentrated at appropriate sites in the human SCO testes, and because claudin 11 is required for Sertoli cells to form the barrier (Mazaud-Guittot et al., 2010; Stammler et al., 2016). We focused on tubules of treated mice that contained or lacked pachytene spermatocytes because of the report that these cells stimulate barrier formation (Li et al., 2018). Mice were treated with vehicle or with 1NA-PP1 for 9 days and tubules were collected 32 days later. Claudin 11 was detected by immunocytochemistry, and nuclei were stained with DAPI. In all tubules of control mice, optical cross-sections of seminiferous tubules revealed a ring of claudin 11 just above spermatogonia and early preleptotene spermatocytes, the site of the barrier (Figure 6A) (Mruk and Cheng, 2015). Identical results were obtained with tubules of treated animals that contained a full complement of spermatogenic cells or that contained pachytene spermatocytes but lacked round spermatids (Figure 6B). However, claudin 11 was not detected at the presumptive site of the barrier in tubules with very few pachytene spermatocytes but with round spermatids (Figure 6C). Neither could we reconstruct normal claudin 11 localization from serial optical sections (Figure 6D). Thus, the restoration of SSCs and the subsequent rebuilding of the seminiferous epithelium occur in a tissue that undergoes dissolution of its blood-testis barrier.
Figure 6.
Loss of Pachytene Spermatocytes that Follows Loss of SSCs Results in Dissolution of the Blood-Testis Barrier
Mice were injected with vehicle or 1NA-PP1 for 9 days and testes collected 32 days later. Whole seminiferous tubules were immunostained for claudin 11 (red) and with DAPI (blue) and imaged by confocal microscopy. Thicknesses of the optical sections for the red and blue channels were 2.2 and 0.9 μm, respectively. An arrow points to each of the pachytene spermatocytes in (A–C); arrowheads point to a nucleolus of one of the round spermatids in (A and C). Scale bar, 20 μm (all panels).
(A) Optical section through a middle of a seminiferous tubule of a control animal. This tubule contains a full complement of spermatogenic cells. Identical results were obtained from tubules of treated animals that contained a full complement of germ cells. In all such tubules, claudin 11 is concentrated at the site of the blood-testis barrier.
(B) Optical section through the middle of a seminiferous tubule of a treated animal. This tubule contains many pachytene spermatocytes but no round spermatids. Claudin 11 localization is normal.
(C) Optical section through the middle of a seminiferous tubule of a treated animal. This tubule contains few pachytene spermatocytes but many round spermatids. Claudin 11 is present in small puncta, not at the presumptive site of the blood-testis barrier. This animal was also the source of the tubule shown in (B).
(D) Optical sections separated by 4 μm through a tubule, lacking pachytene spermatocytes, in a treated animal.
Discussion
Our principal goal was to test the hypothesis that inhibition of GDNF signaling caused a substantial loss of SSCs from the adult testis, which, upon resumption of signaling, were restored by self-renewing replication. We predicted that this loss of SSCs and the resulting maturation depletion of the seminiferous epithelium caused dissolution of the blood-testis barrier. Our data support both our hypothesis and our prediction. The loss of this barrier demonstrates that our mouse model recapitulates an important characteristic of human SCO syndrome.
Most SSCs are Lost during 9 Days of Inhibited GDNF Signaling, but Are Partially Restored When it Resumes
We demonstrated that 9 days of inhibited GDNF signaling results in a 90% decrease in transplantable SSCs. We noted a similar decrease when we defined SSCs as As spermatogonia that expressed Id4-eGFP and/or GFRα1. This loss should be considered in the light of the report that adult mouse testes contain two different subpopulations of SSCs; one expresses GFRα1, the other does not (Garbuzov et al., 2018). As Ret expression by GFRα1+ SSCs is 16.5-fold higher than by GFRα1− cells, the later should be immune to 1NA-PP1 treatment. However, as the report also indicated that individual SSCs rapidly interconvert between GFRα1− and GFRα1+ states, all SSCs in our studies should be affected by 9 days of treatment.
Our data show that, after resumption of GDNF signaling, Aal cells are the first to be restored and about 25% express Kit. Thus, the restorative process is biased initially toward regeneration of progenitor spermatogonia, some of which appear poised to become type A spermatogonia. However, after this initial phase, the SSC pool is substantially rebuilt.
Our analysis indicates that the total yield from replication of As, Apr, and Aal cells exceeds the sum of the increases in their numbers. We propose that this apparent excess cell production is due to loss of GFRα1 expression during differentiation of Aal cells. It is useful to view these results from the perspective of the simplest model for As, Apr, and Aal spermatogonia (de Rooij, 2001). This model posits that replication of As spermatogonia generates either a new As spermatogonia or two Apr spermatogonia. Replication of Apr spermatogonia generates Aal cells, while the replication of Aal cells increases chain length. When viewed from this perspective, our data indicate that, following resumption of GDNF signaling, self-renewing replication of As spermatogonia restores all SSCs and the most immature, As, progenitor spermatogonia. However, replication of As cells accounts for only 53% of the restored Apr cells. The remainder might be generated when two cells in a pair simultaneously replicate and the new chain of four cells splits into two pairs. Nonetheless, while we cannot fully explain how all Apr cells are restored, our quantitative analysis supports the conclusion that, following the resumption of GDNF signaling, self-renewing replication of GFRα+ As spermatogonia, acting as SSCs, rebuilds the stem cell pool. This process differs markedly from those proposed to restore SSCs following busulfan-induced testicular damage (Carrieri et al., 2017; La et al., 2018).
The Regulators of SSC Restoration
The rebuilding of the stem cell pool after GDNF signaling resumption raises the issue of how this process is regulated. The fact that all remaining GFRα1+ As spermatogonia replicated within 3–4 days of resumed signaling suggested that the concentration of an important regulator of SSC proliferation was altered. GDNF was a likely candidate because multiple studies report increased GDNF expression by Sertoli cells following busulfan-induced spermatogonial death (Anand et al., 2016; Zohni et al., 2012). We, therefore, were surprised that testicular levels of GDNF protein and mRNA were not increased in mice that had lost 90% of SSCs. The likely reason for our apparently unique result is that SSCs are lost to differentiation, while in the other studies SSCs die (Choi et al., 2004). Sertoli cells express Toll-like receptors and apoptosis of germ cells generates ligands for those receptors. It has been proposed that the binding of those ligands to their cognate receptors stimulates Sertoli cell GDNF expression, Wnt signaling, and pro-inflammatory pathways (Anand et al., 2016; Zhang et al., 2013; Zohni et al., 2012).
We detected no significant changes in expression of any other known growth factor or cytokine regulator of SSC replication. However, replication of GFRα1+ As spermatogonia decreased as their densities and those of GFRα1+ Apr and Aal spermatogonia increased, indicating that restoration is regulated. This regulation is specific for replications of Apr and Aal spermatogonia do not correlate with cell densities. One possible explanation for our data is that these three cell types secrete an inhibitor of SSC proliferation, whose testicular concentration increases as SSCs and progenitor spermatogonia are restored. Consistent with our proposal is the fact that SSCs express a number of cytokines and adhesion molecules as well as their cognate receptors (Green et al., 2018; Guo et al., 2017). It is also supported by the report that in vitro the clustering of SSCs into mounds suppresses their stem cell activity (Ebata et al., 2011). However, it is possible that interactions that regulate SSC replication are indirect, requiring the action of intermediaries, such as Sertoli or peritubular myoid cells, which secrete the proliferation inhibitor. Identifying both this inhibitor and its source are important future objectives.
Loss of SSCs Followed by Loss of Pachytene Spermatocytes Results in Dissolution of the Blood-Testis Barrier
Human SCO syndrome is characterized by the inability of adjacent Sertoli cells to form a functional barrier (Camatini et al., 1981). An important cause of this deficit is the Sertoli cell's inability to concentrate an essential barrier component, claudin 11, at the correct site (Mazaud-Guittot et al., 2010; Stammler et al., 2016). Therefore, it is noteworthy that SCO mouse tubules neither properly concentrate claudin-11 nor form a barrier (Li et al., 2018). We observed that claudin 11 was mislocalized in tubules with reduced numbers of pachytene spermatocytes but with round spermatids, while claudin 11 was correctly localized in tubules with pachytene spermatocytes but without round spermatids.
Conclusion and Relevance to Human SCO Syndrome
Our results demonstrate that inhibition of GDNF signaling for 9 days results in loss of 90% of SSCs. Upon the resumption of this signaling, numbers of these stem cells and of their immediate progeny, progenitor spermatogonia, are substantially restored. This restoration is initially biased toward formation of progenitor spermatogonia, some of which express Kit, and thus are primed to become fully differentiated type A1 spermatogonia. However, within 2 months of renewed signaling, SSC restoration is sufficient to support normal, complete spermatogenesis in about 50% of tubule cross-sections. This restoration is driven by self-renewing stem cell replication. In fact, immediately after GDNF signaling resumes, all remaining GFRα1+ As spermatogonia replicate. This event is not associated with elevated expression of GDNF or of four other growth factors. However, our data suggest that SSC replication is regulated directly or indirectly by feedback from other SSCs or from progenitor spermatogonia. Finally, our results demonstrate that loss of SSCs and subsequent maturation depletion of pachytene spermatocytes causes dissolution of the blood-testis barrier. Thus, our mouse model recapitulates important characteristics of human SCO testes: loss of SSCs due to inadequate GDNF signaling and dissolution of the blood-testis barrier.
Our results raise hope for the development of new therapies for the subset of infertile men with SCO syndrome, whose testes contain some SSCs. It is possible that increasing their testicular GDNF levels will stimulate replication of the few SSCs in their testis, thereby increasing stem cell numbers and seeding spermatogenesis in previously germ cell-deficient areas of tubule.
Experimental Procedures
Animals
The Johns Hopkins University and University of Pittsburgh Medical School Animal Care the Use Committees approved relevant animal protocols. All mice used to study the response to the inhibition and resumption of GDNF signaling were homozygous for Ret V805A+/+, had a C57BL/6J genetic background, and were aged 90–120 days (Savitt et al., 2012). Mice used to estimate numbers of transplantable SSCs were generated by crossing Ret V805A+/+ with B6129S7-Gt (Rosa)+/− 26Sor/J mice, Id4-eGFP+/− mice (Jackson Laboratory, Bar Harbor, ME). Offspring carrying both transgenes were backcrossed seven times to Ret(V805A) mice. We used C57BL/6J mice (Jackson Laboratory) to determine EdU-labeling efficiency and cell-cycle length. Supplemental Experimental Procedures describe protocols for injection of 1NaPP1, EdU, and BrdU, as well as primers and conditions for detection of transgenes.
Synthesis of 1NA-PP1-HCl
We synthesized 1NA-PP1-HCl as described previously (Savitt et al., 2012). For brevity we call it 1NA-PP1 throughout the paper.
Transplantation of Germ Cells into Germ Cell-Deficient Testes
Two to 4 days after the last injection of 1NA-PP1 or vehicle, a single-cell suspension was generated from each testis of treated and control animals, and approximately 800,000 viable cells were transplanted into the seminiferous tubules of germ cell-deficient, busulfan-treated C57BL/6 mice, as described previously (Medrano et al., 2014). Recipients were analyzed 2 months later by incubating fixed, decapsulated testes in 5-bromo-4-chloro-3-indolyl-b-D-galactopyranoside (X-gal) and counting X-Gal+ spermatogenic colonies. Very few incomplete colonies or patches were observed.
Immunocytochemistry, Histology, Microscopy, and Image Analysis
We used previously described methods to detect GFRα1, ZBTB16, Kit, and EdU (Parker et al., 2014; Savitt et al., 2012). Antibodies are listed in Table S2. Whole mounts of seminiferous tubules were imaged using Zeiss LSM700 or LSM710 confocal microscopes. ZBTB16+ cells were imaged as described previously (Savitt et al., 2012). Supplemental Experimental Procedures describe how we detected claudin 11 and BrdU. One-micron, plastic-embedded testis sections were prepared as described (Savitt et al., 2012).
Quantification of Specific Transcripts and of GDNF Protein in Mouse Testes
Testis GDNF protein and growth factor mRNA concentrations were assayed as described previously (Parker et al., 2014; Savitt et al., 2012; Singh et al., 2017). Transcript and GDNF protein levels were normalized for 18S rRNA and total protein, respectively (Bradford, 1976). Supplemental Experimental Procedures provide full protocols.
Statistical Analyses
Data were analyzed by two-tailed t tests, ANOVA, or correlation using GraphPad Prism 6.0 (La Jolla, CA). Post hoc comparisons were performed with the Holm-Side's multiple comparison test. Statistically significant differences were defined as p ≤ 0.05.
Data and Code Availability
The RNA sequencing data were uploaded to the GEO (GEO: GSE111487).
Author Contributions
N.P. performed most experiments and wrote the first manuscript draft A.L. analyzed cell differentiation. M.S. performed stem cell transplants. K.E.O. supervised M.S. and edited the manuscript drafts. J.M.O. provided Id4-eGFP mice and edited the manuscript drafts. C.Z. and F.U.R. synthesized 1NaPP1-HCl. K.S. directed 1NaPP1-HCl synthesis and discussed the results. W.W.W. directed the research, performed and analyzed the experiments, and wrote all but the first manuscript draft.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This research was supported by the Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Centers for Translational Research in Reproduction and Infertility Program (NCTRI) grant 1R01HD074542. RNA sequencing was performed by Dr. Jen Grenier, Cornell University Sequencing Core, Center for Reproductive Genomics. The center is supported in part by a grant from the NCTRI, P50HD076210. We thank Pamela Wright for editorial and graphical assistance and Janet Folmer for histology. We thank Dr. Manju Sharma and Dr. Bob Braun for providing their protocol for detecting BrdU+, EdU+ cells.
Published: February 25, 2021
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.stemcr.2021.01.015.
Supplemental Information
References
- Anand S., Bhartiya D., Sriraman K., Mallick A. Underlying mechanisms that restore spermatogenesis on transplanting healthy niche cells in busulphan treated mouse testis. Stem Cell Rev. Rep. 2016;12:682–697. doi: 10.1007/s12015-016-9685-1. [DOI] [PubMed] [Google Scholar]
- Bradford M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Bragado Alonso S., Schulze-Steikow M., Calegari F. Cell cycle activity of neural precursors in the diseased mammalian brain. Front. Neurosci. 2014;8:39. doi: 10.3389/fnins.2014.00039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buaas F.W., Kirsh A.L., Sharma M., McLean D.J., Morris J.L., Griswold M.D., de Rooij D.G., Braun R.E. Plzf is required in adult male germ cells for stem cell self-renewal. Nat. Genet. 2004;36:647–652. doi: 10.1038/ng1366. [DOI] [PubMed] [Google Scholar]
- Camatini M., Franchi E., Decurtis I. Permeability to lanthanum of blood testis barrier in human germinal aplasia. Anat. Rec. 1981;200:293–297. doi: 10.1002/ar.1092000308. [DOI] [PubMed] [Google Scholar]
- Carrieri C., Comazzetto S., Grover A., Morgan M., Buness A., Nerlov C., O'Carroll D. A transit-amplifying population underpins the efficient regenerative capacity of the testis. J. Exp. Med. 2017;214:1631–1641. doi: 10.1084/jem.20161371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheraghali A.M., Kumar R., Knaus E.E., Wiebe L.I. Pharmacokinetics and bioavailability of 5-ethyl-2'-deoxyuridine and its novel (5R,6R)-5-bromo-6-ethoxy-5,6-dihydro prodrugs in mice. Drug Metab. Dispos. 1995;23:223–226. [PubMed] [Google Scholar]
- Choi Y.J., Ok D.W., Kwon D.N., Chung J.I., Kim H.C., Yeo S.M., Kim T., Seo H.G., Kim J.H. Murine male germ cell apoptosis induced by busulfan treatment correlates with loss of c-kit-expression in a Fas/FasL- and p53-independent manner. FEBS Lett. 2004;575:41–51. doi: 10.1016/j.febslet.2004.08.034. [DOI] [PubMed] [Google Scholar]
- de Rooij D.G. Proliferation and differentiation of spermatogonial stem cells. Reproduction. 2001;121:347–354. doi: 10.1530/rep.0.1210347. [DOI] [PubMed] [Google Scholar]
- de Rooij D.G. The nature and dynamics of spermatogonial stem cells. Development. 2017;144:3022–3030. doi: 10.1242/dev.146571. [DOI] [PubMed] [Google Scholar]
- DeFalco T., Potter S.J., Williams A.V., Waller B., Kan M.J., Capel B. Macrophages contribute to the spermatogonial niche in the adult testis. Cell Rep. 2015;12:1107–1119. doi: 10.1016/j.celrep.2015.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebata K.T., Yeh J.R., Zhang X., Nagano M.C. Soluble growth factors stimulate spermatogonial stem cell divisions that maintain a stem cell pool and produce progenitors in vitro. Exp. Cell Res. 2011;317:1319–1329. doi: 10.1016/j.yexcr.2011.03.013. [DOI] [PubMed] [Google Scholar]
- Farlik M., Sheffield N.C., Nuzzo A., Datlinger P., Schonegger A., Klughammer J., Bock C. Single-cell DNA methylome sequencing and bioinformatic inference of epigenomic cell-state dynamics. Cell Rep. 2015;10:1386–1397. doi: 10.1016/j.celrep.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbuzov A., Pech M.F., Hasegawa K., Sukhwani M., Zhang R.J., Orwig K.E., Artandi S.E. Purification of GFRalpha1+ and GFRalpha1− spermatogonial stem cells reveals a niche-dependent mechanism for fate determination. Stem Cell Reports. 2018;10:553–567. doi: 10.1016/j.stemcr.2017.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green C.D., Ma Q., Manske G.L., Shami A.N., Zheng X., Marini S., Moritz L., Sultan C., Gurczynski S.J., Moore B.B. A comprehensive roadmap of murine spermatogenesis defined by single-cell RNA-seq. Dev. Cell. 2018;46:651–667 e610. doi: 10.1016/j.devcel.2018.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J., Grow E.J., Yi C., Mlcochova H., Maher G.J., Lindskog C., Murphy P.J., Wike C.L., Carrell D.T., Goriely A. Chromatin and single-cell RNA-seq profiling reveal dynamic signaling and metabolic transitions during human spermatogonial stem cell development. Cell Stem Cell. 2017;21:533–546 e536. doi: 10.1016/j.stem.2017.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa K., Saga Y. FGF8-FGFR1 signaling acts as a niche factor for maintaining undifferentiated spermatogonia in the mouse. Biol. Reprod. 2014;91:145. doi: 10.1095/biolreprod.114.121012. [DOI] [PubMed] [Google Scholar]
- Helsel A.R., Yang Q.E., Oatley M.J., Lord T., Sablitzky F., Oatley J.M. ID4 levels dictate the stem cell state in mouse spermatogonia. Development. 2017;144:624–634. doi: 10.1242/dev.146928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huckins C. The spermatogonial stem cell population in adult rats II. A radioautographic analysis of their cell cycle properties. Cell Tissue Kinet. 1971;4:313–334. doi: 10.1111/j.1365-2184.1971.tb01543.x. [DOI] [PubMed] [Google Scholar]
- Kanatsu-Shinohara M., Naoki H., Shinohara T. Nonrandom germline transmission of mouse spermatogonial stem cells. Dev. Cell. 2016;38:248–261. doi: 10.1016/j.devcel.2016.07.011. [DOI] [PubMed] [Google Scholar]
- Kubota H. Heterogeneity of spermatogonial stem cells. In: Birbrair A., editor. Stem Cells Heterogeneity in Different Organs. Springer International Publishing; 2019. pp. 225–242. [Google Scholar]
- La H.M., Makela J.A., Chan A.L., Rossello F.J., Nefzger C.M., Legrand J.M.D., De Seram M., Polo J.M., Hobbs R.M. Identification of dynamic undifferentiated cell states within the male germline. Nat. Commun. 2018;9:2819. doi: 10.1038/s41467-018-04827-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X.Y., Zhang Y., Wang X.X., Jin C., Wang Y.Q., Sun T.C., Li J., Tang J.X., Batool A., Deng S.L. Regulation of blood-testis barrier assembly in vivo by germ cells. FASEB J. 2018;32:1653–1664. doi: 10.1096/fj.201700681R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord T., Oatley J.M. Functional assessment of spermatogonial stem cell purity in experimental cell populations. Stem Cell Res. 2018;29:129–133. doi: 10.1016/j.scr.2018.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazaud-Guittot S., Meugnier E., Pesenti S., Wu X., Vidal H., Gow A., Le Magueresse-Battistoni B. Claudin 11 deficiency in mice results in loss of the Sertoli cell epithelial phenotype in the testis. Biol. Reprod. 2010;82:202–213. doi: 10.1095/biolreprod.109.078907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medrano J.V., Martinez-Arroyo A.M., Sukhwani M., Noguera I., Quinonero A., Martinez-Jabaloyas J.M., Pellicer A., Remohi J., Orwig K.E., Simon C. Germ cell transplantation into mouse testes procedure. Fertil. sterility. 2014;102 doi: 10.1016/j.fertnstert.2014.07.669. e11–12. [DOI] [PubMed] [Google Scholar]
- Meng X., Lindahl M., Hyvonen M.E., Parvinen M., de Rooij D.G., Hess M.W., Raatikainen-Ahokas A., Sainio K., Rauvala H., Lakso M. Regulation of cell fate decision of undifferentiated spermatogonia by GDNF. Science. 2000;287:1489–1493. doi: 10.1126/science.287.5457.1489. [DOI] [PubMed] [Google Scholar]
- Mruk D.D., Cheng C.Y. The mammalian blood-testis barrier: its biology and regulation. Endocr. Rev. 2015;36:564–591. doi: 10.1210/er.2014-1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T., Sharma M., Nabeshima Y.I., Braun R.E., Yoshida S. Functional hierarchy and reversibility within the murine spermatogenic stem cell compartment. Science. 2010;328:62–67. doi: 10.1126/science.1182868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oatley J.M., Oatley M.J., Avarbock M.R., Tobias J.W., Brinster R.L. Colony stimulating factor 1 is an extrinsic stimulator of mouse spermatogonial stem cell self-renewal. Development. 2009;136:1191–1199. doi: 10.1242/dev.032243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paduch D.A., Hilz S., Grimson A., Schlegel P.N., Jedlicka A.E., Wright W.W. Aberrant gene expression by Sertoli cells in infertile men with Sertoli cell-only syndrome. PLoS One. 2019;14:e0216586. doi: 10.1371/journal.pone.0216586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker N., Falk H., Singh D., Fidaleo A., Smith B., Lopez M.S., Shokat K.M., Wright W.W. Responses to glial cell line-derived neurotrophic factor change in mice as spermatogonial stem cells form progenitor spermatogonia which replicate and give rise to more differentiated progeny. Biol. Reprod. 2014;91:92. doi: 10.1095/biolreprod.114.119099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sada A., Hasegawa K., Pin P.H., Saga Y. NANOS2 acts downstream of glial cell line-derived neurotrophic factor signaling to suppress differentiation of spermatogonial stem cells. Stem Cells. 2012;30:280–291. doi: 10.1002/stem.790. [DOI] [PubMed] [Google Scholar]
- Savitt J., Singh D., Zhang C., Chen L.C., Folmer J., Shokat K.M., Wright W.W. The in vivo response of stem and other undifferentiated spermatogonia to the reversible inhibition of glial cell line-derived neurotrophic factor signaling in the adult. Stem Cells. 2012;30:732–740. doi: 10.1002/stem.1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlegel P.N. Causes of azoospermia and their management. Reprod. Fertil. Dev. 2004;16:561–572. doi: 10.10371/RD03087. [DOI] [PubMed] [Google Scholar]
- Sharma M., Srivastava A., Fairfield H.E., Bergstrom D., Flynn W.F., Braun R.E. Identification of EOMES-expressing spermatogonial stem cells and their regulation by PLZF. ife. 2019;8:e43352. doi: 10.7554/eLife.43352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh D., Paduch D.A., Schlegel P.N., Orwig K.E., Mielnik A., Bolyakov A., Wright W.W. The production of glial cell line-derived neurotrophic factor by human sertoli cells is substantially reduced in Sertoli cell-only testes. Hum. Reprod. 2017;32:1108–1117. doi: 10.1093/humrep/dex061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stammler A., Luftner B.U., Kliesch S., Weidner W., Bergmann M., Middendorff R., Konrad L. Highly conserved testicular localization of claudin-11 in normal and impaired spermatogenesis. PLoS One. 2016;11:e0160349. doi: 10.1371/journal.pone.0160349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokuda M., Kadokawa Y., Kurahashi H., Marunouchi T. CDH1 is a specific marker for undifferentiated spermatogonia in mouse testes. Biol. Reprod. 2007;76:130–141. doi: 10.1095/biolreprod.106.053181. [DOI] [PubMed] [Google Scholar]
- Yang Q.E., Kim D., Kaucher A., Oatley M.J., Oatley J.M. CXCL12-CXCR4 signaling is required for the maintenance of mouse spermatogonial stem cells. J. Cell Sci. 2013;126:1009–1020. doi: 10.1242/jcs.119826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Wang T., Deng T., Xiong W., Lui P., Li N., Chen Y., Han D. Damaged spermatogenic cells induce inflammatory gene expression in mouse Sertoli cells through the activation of Toll-like receptors 2 and 4. Mol. Cell. Endocrinol. 2013;365:162–173. doi: 10.1016/j.mce.2012.10.016. [DOI] [PubMed] [Google Scholar]
- Zohni K., Zhang X., Tan S.L., Chan P., Nagano M.C. The efficiency of male fertility restoration is dependent on the recovery kinetics of spermatogonial stem cells after cytotoxic treatment with busulfan in mice. Hum. Reprod. 2012;27:44–53. doi: 10.1093/humrep/der357. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA sequencing data were uploaded to the GEO (GEO: GSE111487).