Summary
Chondrodysplasias are hereditary diseases caused by mutations in the components of growth cartilage. Although the unfolded protein response (UPR) has been identified as a key disease mechanism in mouse models, no suitable in vitro system has been reported to analyze the pathology in humans. Here, we developed a three-dimensional culture protocol to differentiate hypertrophic chondrocytes from induced pluripotent stem cells (iPSCs) and examine the phenotype caused by MATN3 and COL10A1 mutations. Intracellular MATN3 or COL10 retention resulted in increased ER stress markers and ER size in most mutants, but activation of the UPR was dependent on the mutation. Transcriptome analysis confirmed a UPR with wide-ranging changes in bone homeostasis, extracellular matrix composition, and lipid metabolism in the MATN3 T120M mutant, which further showed altered cellular morphology in iPSC-derived growth-plate-like structures in vivo. We then applied our in vitro model to drug testing, whereby trimethylamine N-oxide led to a reduction of ER stress and intracellular MATN3.
Keywords: iPSC, chondrodysplasia, MATN3, COL10A1, unfolded protein response
Highlights
-
•
A new induction method enables hypertrophic chondrocyte differentiation from iPSCs
-
•
Chondrodysplasia mutants show intracellular MATN3 or COL10 accumulation
-
•
Some, but not all, mutants have ER stress with an unfolded protein response
-
•
This induction system is applicable to transcriptomic analysis and drug development
Chondrodysplasias are highly heterogeneous genetic cartilage diseases. Here, Toguchida and colleagues establish a new induction system to differentiate hypertrophic chondrocytes from iPSCs and analyze these disorders in vitro. They found that different chondrodysplasia mutants, despite mutations being in the same gene, showed varying phenotypes and transcriptomic changes. This system provides an initial platform for further investigation and drug development.
Introduction
Chondrodysplasias are hereditary cartilage disorders, which often manifest by early childhood as mild to severe skeletal abnormalities due to mutations in the components of growth cartilage. In multiple epiphyseal dysplasia (MED; OMIM #607078) and metaphyseal chondrodysplasia type Schmid (MCDS; OMIM #156500), short-limbed dwarfism and deformities of the hips or knees are commonly observed (Czarny-Ratajczak et al., 2001; Mäkitie et al., 2005), but large variations in the skeletal phenotype and disease severity between patients have made these diseases difficult to research and treat. To overcome the obstacles posed by this heterogeneity and deepen our understanding of chondrodysplasias, it is imperative to obtain relevant patient samples and establish accurate disease models.
However, as it is ethically questionable to obtain samples from patients' growth plates, much of our understanding of chondrodysplasia disease mechanisms has come from studies of animal models. Mutations in the MATN3 vWFa and COL10A1 NC1 domains, which respectively cause MED and MCDS, have been reported to disrupt folding and oligomerization in vitro (Cotterill et al., 2005; Wilson et al., 2005), suggesting a gain-of-function effect that has been further supported by Matn3 or Col10a1 knockout mice having no significant skeletal dysplasia (Kwan et al., 1997; van der Weyden et al., 2006). In contrast, model mice with the p.V194D mutation in Matn3, known to cause MED in humans, showed short-limbed dwarfism and structural disruption of the growth plate with decreased chondrocyte proliferation and increased apoptosis (Leighton et al., 2007). Similarly, model mice with the MCDS-causing p.N617K mutation in Col10a1 also showed short-limbed dwarfism, but the growth plate had an extended hypertrophic zone without decrease in chondrocyte proliferation (Rajpar et al., 2009). In both models, ER stress was detected in growth plate chondrocytes as a result of intracellular accumulation of MATN3 or COL10, indicating that the unfolded protein response (UPR) is a key event in these diseases. However, these results were obtained using homozygous mice, while most MED- and MCDS-causing mutations are heterozygous with autosomal dominant inheritance in humans (Mortier et al., 2019). Of the heterozygous mouse models, only the COL10A1 Y632X heterozygotes demonstrated a clear phenotype (Forouhan et al., 2018), but contrary to this model, where the mutant Y632X mRNA was stable, human cartilage cells with an identical mutation showed no mRNA transcripts from the mutant allele (Chan et al., 1998), suggesting that haploinsufficiency may be causative in humans. Due to these species differences, models that more closely reflect the pathology in humans are required.
Recently, disease-specific induced pluripotent stem cells (iPSCs) have emerged as a powerful tool to further our understanding of human hereditary diseases and screen for candidate drugs. For example, the clinical phenotype of type II collagenopathies, a subgroup of chondrodysplasias, has been recapitulated in vitro using patient-derived iPSCs (Okada et al., 2015). Therefore, in this study, we aimed to apply this approach to MED and MCDS. As these two chondrodysplasias mainly or partly affect hypertrophic chondrocytes, we first developed a three-dimensional (3D) culture protocol to robustly derive such late-stage chondrocytes. Then, using both patient-derived and artificially mutated iPSC lines with heterozygous MATN3 or COL10A1 mutations, we applied our protocol to the in vitro recapitulation of MED and MCDS phenotypes. Comparison with isogenic controls enabled us to demonstrate a phenotype in some mutants that is similar to previous observations in model mice, including ER stress and a UPR caused by the intracellular accumulation of the affected protein, indicating the usefulness of our model for the analysis of diseases affecting the growth plate. However, evidence of ER stress and the UPR was weaker in other mutants, suggesting that each mutation, even in the same causative gene, may have a different impact on the phenotype. Our system provides an initial platform for further investigation and drug development of growth plate diseases.
Results
Differentiation of iPSCs into hypertrophic chondrocytes through the sclerotome
To model chondrodysplasias in vitro, we first developed a protocol of differentiating hypertrophic chondrocytes from the wild-type 414C2 iPSC line (Okita et al., 2011) in serum-free conditions. We combined previously reported protocols of sclerotome induction (Matsuda et al., 2020) and chondrogenic induction (Umeda et al., 2012) with slight modifications. After sclerotome induction (SI), hypertrophic chondrocyte induction (HI) was performed in 3D culture for up to 70 days, starting with a 14-day-period of standard chondrogenic induction (Umeda et al., 2012) before adding the thyroid hormone T3, which has been reported to promote hypertrophic maturation (Mueller and Tuan, 2008) (Figure 1A).
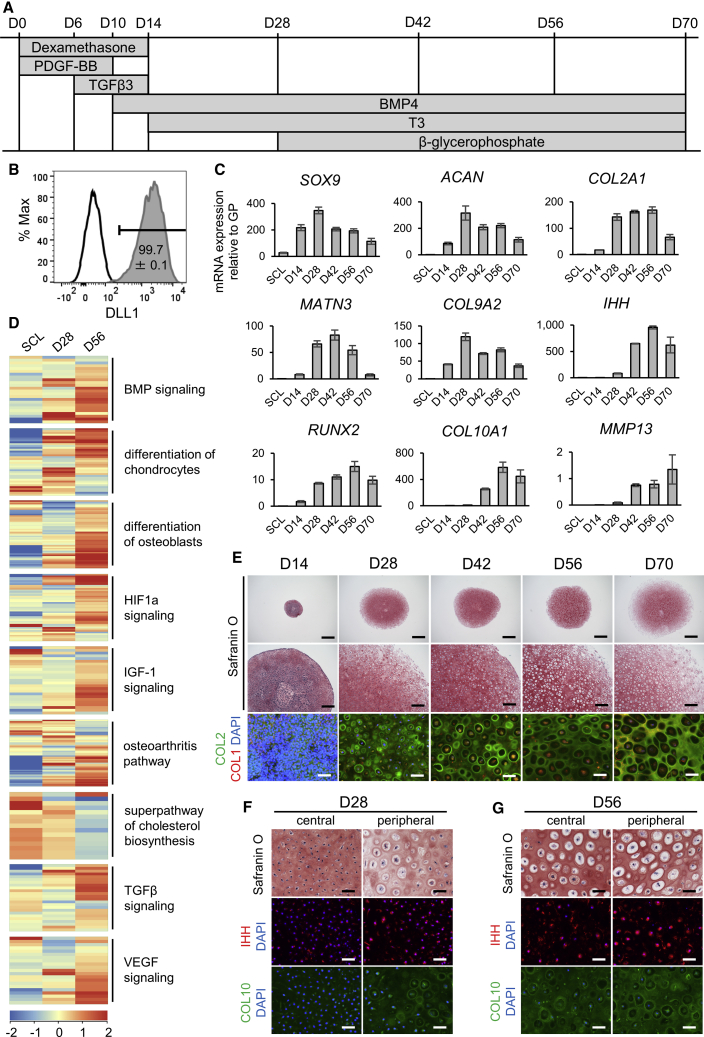
Figure 1.
Differentiation of hypertrophic chondrocytes from iPSCs
(A) Protocol of hypertrophic chondrocyte induction (HI) in 3D pellet culture from sclerotome (SCL) cells (D0).
(B) Representative result of DLL1-positive cells (compared with isotype control) on day 2 of sclerotome induction (SI), with the mean and SEM (standard error of the mean) of n = 4 independent experiments displayed.
(C) mRNA expression of chondrocyte markers of different stages over time from SCL on day −1 to day 70 of HI by qPCR (top row, early markers; middle row, proliferating to prehypertrophic markers; bottom row, hypertrophic markers). Values are shown as mean ± SEM (n = 4 independent experiments), relative to the mean of six pieces of a human distal femoral growth plate (GP).
(D) Heatmap of averaged normalized intensity values of gene expression in chondrocyte hypertrophy-related pathways from IPA in SCL, day-28, and day-56 samples (n = 3 independent experiments).
(E) Safranin O staining (top and middle rows) and COL2 (green) with COL1 (red) immunostaining (bottom row) of pellets from days 14–70 of HI. Scale bars, 1 mm (top row), 200 μm (middle row), 50 μm (bottom row).
(F and G) Safranin O and IHH or COL10 immunostaining on day 28 (F) and day 56 (G) of HI in the central or peripheral area of each pellet. Scale bars, 50 μm.
All results shown are from experiments using the 414C2 wild-type iPSC line. For (E), (F), and (G), similar results were obtained in n = 3 independent experiments. See also Figure S1 and Table S1.
This protocol required no cell sorting, as fluorescence-activated cell sorting (FACS) analysis at the presomitic mesoderm (PSM) stage during SI showed that almost 100% of cells were positive for the PSM marker DLL1 (Figure 1B). The expression of chondrocyte markers was detected from day 14 of HI, peaking on day 28 and declining thereafter, while hypertrophic markers mostly appeared on day 42, with an expression similar to or greater than the human distal femoral growth plate (Figure 1C). Transcriptome and Ingenuity Pathway Analysis (IPA) of day-56 pellets compared with day-28 pellets showed gene expression changes in several chondrocyte hypertrophy-related pathways, including increases in bone morphogenetic protein (BMP) and insulin-like growth factor-1 (IGF-1) signaling (Karl et al., 2014; Wang et al., 1999) and decreases in cholesterol biosynthesis (Tsushima et al., 2018) (Figure 1D and Table S1). Apart from these pathways, both chondrocyte and osteoblast differentiation-related gene expression were increased as the chondrocytes matured from sclerotome until day 56.
The pellet dramatically increased in size from 1 mm on day 14 to 4–5 mm on day 28, stabilizing thereafter (Figure 1E). The cartilage matrix stained with Safranin O by day 14, COL2 by day 28, and COL1 by day 70. On day 28, cells resembled proliferating chondrocytes in the pellet interior, with only a thin layer of cells with a hypertrophic morphology in the periphery of the pellet, but by day 56 a hypertrophic morphology was detected throughout the pellet. Prehypertrophic marker IHH and hypertrophic marker COL10 were present at a higher level in the peripheral area than the central area on day 28, further increasing in both areas by day 56 (Figures 1F and 1G). The expression of chondrocyte markers and the change in chondrocyte morphology during HI were similar in the 1231A3 iPSC line (Nakagawa et al., 2014) (Figures S1A–S1E). Markers of other tissues, such as adipose and ligament, were not expressed at high levels throughout the induction, but markers of bone, including IBSP and SP7, increased as chondrocytes moved toward hypertrophy (Figure S1F). Together with COL1 staining on day 70, this suggests some cells may also have differentiated into the osteoprogenitor lineage in the later stages.
Patient analysis and establishment of COL10A1 and MATN3 mutant iPSC lines
iPSC lines were established from one MED patient with a previously reported heterozygous MATN3 c.359C>T (p.T120M) mutation (Jackson et al., 2004), one MCDS patient with a novel heterozygous COL10A1 c.1841_1841delT (p.L614Rfs∗8) mutation, and one MCDS patient with a previously reported heterozygous COL10A1 c.53G>A (p.G18E) mutation (Ikegawa et al., 1997). Karyotype analysis showed no chromosomal abnormalities in the two clones from each patient (Figures S2A, S3A, and S4A). Each clone showed normal morphology, the presence of pluripotency markers, and the ability to differentiate into all three germ layers (Figures S2B–S2E, S3B–S3E, and S4B–S4E).
Radiological findings in the MED patient included bowing of the femora with genu varum, as well as mild platyspondyly and scoliosis (Figure S2F). The patient’s height at age 9 years was 2.2 SD (standard deviations) below normal. The MATN3 T120M mutation was corrected in the rescued clone (Figure S2G). In addition to the MATN3 T120M mutation, the SNP MATN3 c.659T>C (p.V220A), which has been reported in both MED patients and normal controls (Kim et al., 2011), was also detected in the MED patient’s healthy allele (Figure S2H). To further analyze the pathology of MATN3 mutations, we created a heterozygous MATN3 c.626G>C (p.R209P) mutation in 414C2 iPSCs (Figure S2I). This mutation has been previously reported in MED, causing genu valgum but no dwarfism (Kim et al., 2011).
MCDS patient #1, who had the COL10A1 L614Rfs∗8 mutation, showed a typical MCDS phenotype by age 2 years, with radiological findings including metaphyseal flaring and coxa vara (Figure S3F). The mutation was corrected in the rescued clone (Figure S3G). Despite the early stop codon resulting from the frameshift mutation, nonsense-mediated decay (NMD) was not detected (Figure S3H). MCDS patient #2, with the COL10A1 G18E mutation, has been previously described with radiological findings including widening of the physes, bowing of the femora, and coxa vara (Ikegawa et al., 1997). The mutation was corrected in the rescued clone (Figure S4F). Another heterozygous COL10A1 mutant with the c.1798T>C (p.S600P) mutation was created using 414C2 iPSCs (Figure S4G). This mutation has been reported in an MCDS patient with short-limbed dwarfism, coxa vara, and metaphyseal abnormalities (Gregory et al., 2000). Sequencing of cDNA reverse transcribed from RNA showed that mutant RNA is present in all mutants (Figures S2J, S2K, S3I, S4H, and S4I).
COL10A1 and MATN3 mutants differentiate into hypertrophic chondrocytes
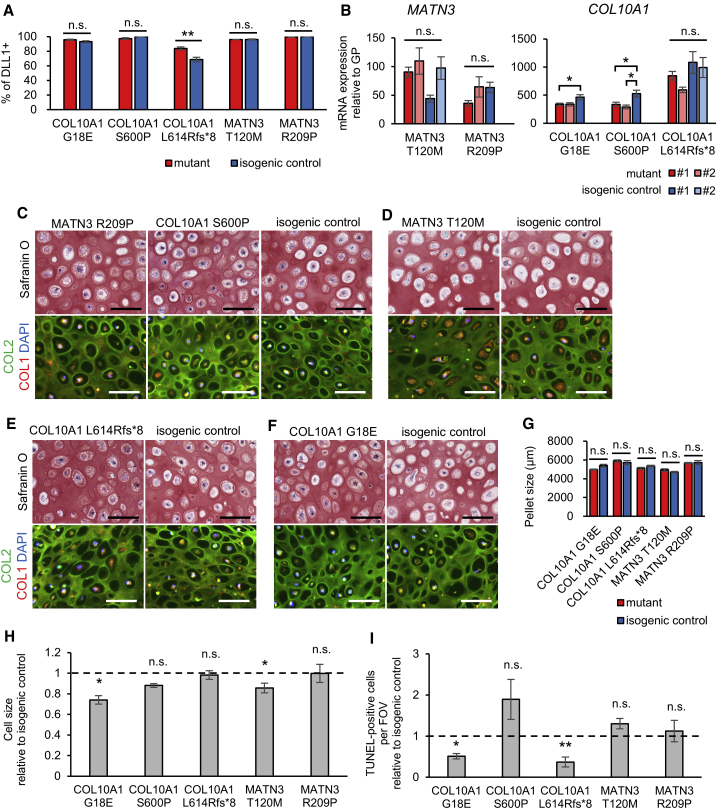
We next assessed the ability of our mutant iPSC lines to differentiate into hypertrophic chondrocytes. At the PSM stage, mutants showed an equal or higher percentage of DLL1-positive cells (Figure 2A). On day 56 of HI, the expression of chondrocyte markers from various stages was, with some clonal variation, similar in mutants and isogenic controls (Figure S5A). However, some mutants showed a lower expression of IHH, which is part of the IHH-PTHrP feedback loop that regulates chondrocyte differentiation in the growth plate (Kobayashi et al., 2002), suggesting a possible disruption in the signaling that regulates chondrocyte maturation. MATN3 expression in MATN3 mutants was unchanged, but COL10A1 expression decreased in the COL10A1 S600P mutant (Figure 2B). Cartilage matrix production was not disrupted in mutants, as Safranin O, COL2, and COL1 staining, as well as the pellet size, were similar to that in isogenic controls (Figures 2C–2G and S5B). Despite the changes in IHH and COL10A1 expression, the cell morphology was not different between mutants and controls, with both showing a hypertrophic morphology. The cell size showed no consistent changes for COL10A1 or MATN3 mutants and, surprisingly, cell death was unchanged or actually decreased in mutants (Figures 2H and 2I).
Figure 2.
COL10A1 and MATN3 mutants show no disruption of chondrogenic differentiation
(A) Percentage of DLL1-positive cells on day 2 of SI.
(B) mRNA expression of the heterozygously mutated gene by qPCR, with values from a second clone also shown for each mutant and isogenic control (number of independent experiments shown in Table S2). Values are relative to the mean of six pieces of the human distal femoral growth plate. Statistical analysis by ANOVA and post hoc Tukey’s HSD.
(C–F) Safranin O staining (top panels) or COL2 (green) and COL1 (red) immunostaining (bottom panels) of mutants (left panels) and their isogenic controls (right panels). Similar results were obtained in n = 4 independent experiments. Scale bars, 100 μm.
(G) Pellet size of each mutant and its isogenic control. Three technical replicates per independent experiment were measured.
(H) Cell size of each mutant relative to the isogenic control, quantified from the inverse image of COL2 fluorescence.
(I) TUNEL-positive cells per FOV (field of view) relative to the isogenic control of each mutant.
All results except (A) are from day 56 of HI. Values are expressed as mean ± SEM. Dotted lines indicate the value = 1 of the isogenic controls in (H) and (I). Except where stated otherwise, the results are from n = 4 independent experiments, and statistical analysis was performed using unpaired two-sided t tests. (n.s., no significant difference; ∗p < 0.05, ∗∗p < 0.01). See also Figures S2–S5.
COL10 or MATN3 is retained intracellularly in mutants
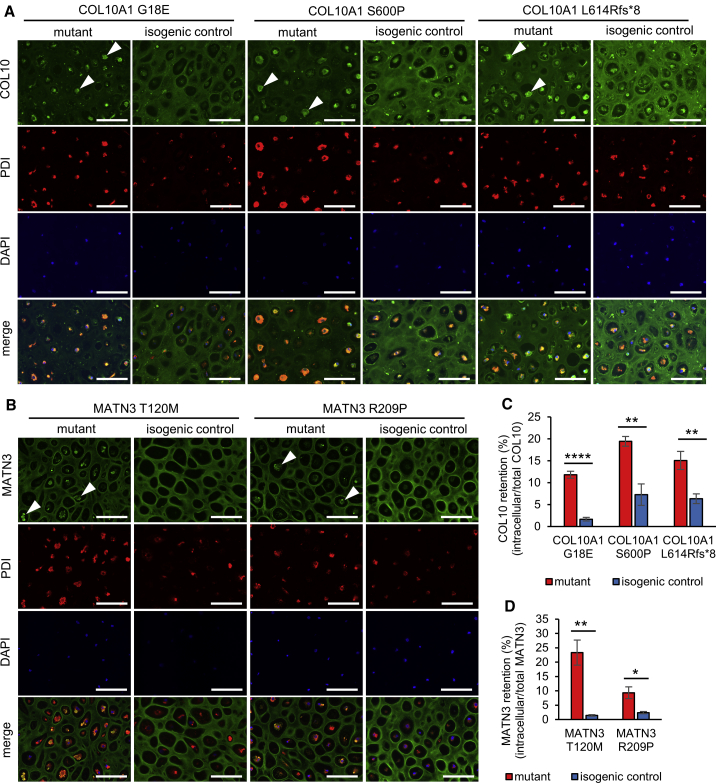
Since MCDS and MED model mice accumulate the mutated protein intracellularly, we examined whether this is also the case in humans. Indeed, immunostaining of the protein on day 56 of HI revealed the presence of intracellular aggregates in both COL10A1 and MATN3 mutants (Figures 3A, 3B, S5C, and S5D). These aggregates co-stained with the ER marker PDI, showing that they are retained within the ER. At the same time, extracellular COL10 or MATN3 was decreased in mutants. All mutants showed a significant increase of COL10 or MATN3 retention, which differed depending on the mutation and was particularly elevated in the MATN3 T120M mutant (Figures 3C and 3D).
Figure 3.
COL10A1 and MATN3 mutants retain COL10 or MATN3 within the ER
(A and B) Immunostaining of COL10 (A) or MATN3 (B) and PDI on day 56 of HI. Arrowheads indicate intracellular aggregates in mutants. Similar results were obtained in n = 4 independent experiments. Scale bars, 100 μm.
(C and D) Intracellular retention quantified from fluorescence intensity of COL10 (C) or MATN3 (D) co-staining with PDI (intracellular) divided by total fluorescence intensity in n = 4 independent experiments on day 56 of HI. Values are expressed as mean ± SEM. (n.s., no significant difference; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗∗p < 0.0001 by unpaired two-sided t test).
See also Figure S5.
ER expansion, ER stress, and a UPR are detected in some but not all mutants
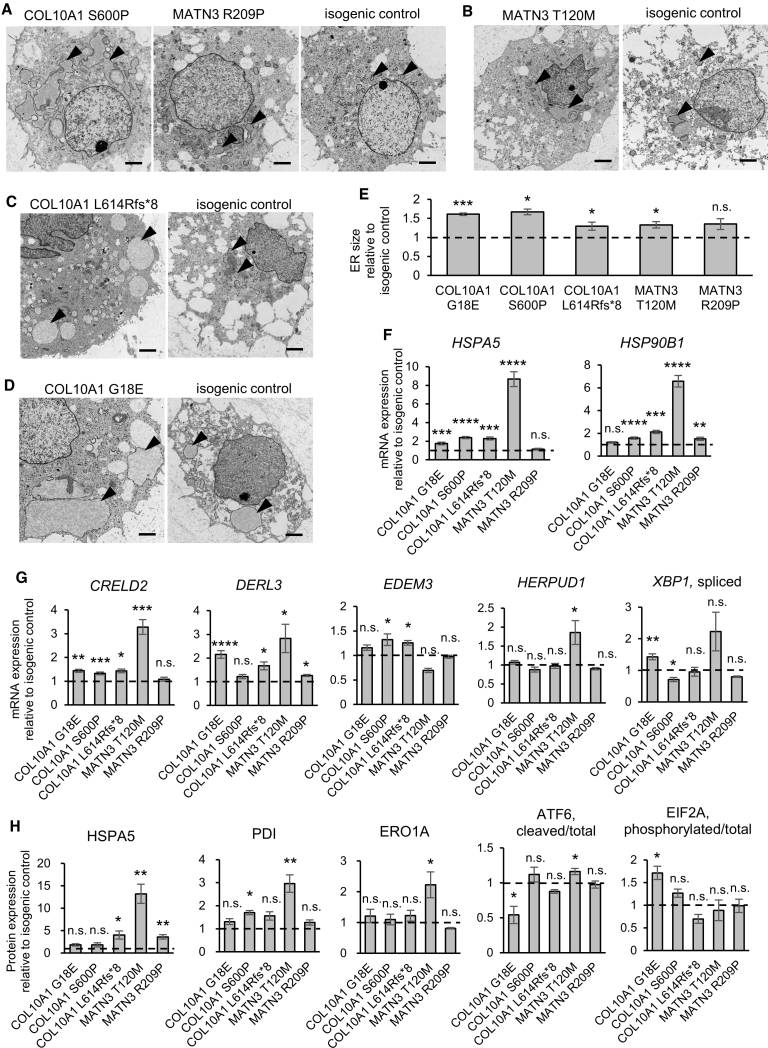
To analyze the impact of intracellular COL10 or MATN3 accumulation in human cartilage, we next used transmission electron microscopy to assess the ER, which was found to be mildly to severely enlarged in mutants (Figures 4A–4D). Quantification of the PDI-positive areas confirmed these results, with all but the MATN3 R209P mutant showing a significant increase in ER size (Figure 4E). We next examined ER stress and UPR markers at the mRNA and protein level, revealing significant increases in one or more markers in every mutant (Figures 4F–4H and S5E). The largest changes were seen in the MATN3 T120M mutant, which showed high expression of the chaperones HSPA5, HSP90B1, and PDI, as well as increased ATF6 cleavage and upregulation of downstream UPR genes CRELD2, DERL3, HERPUD1, and ERO1A. Activation of the ATF6 branch was also reported in MATN3 V194D model mice (Pirog et al., 2019), but this was not observed in our MATN3 R209P mutant, which had little to no increase in ER stress and UPR markers. In the case of COL10A1 mutants, COL10A1 G18E showed an increase in spliced XBP1 and phosphorylated EIF2A, as well as greater expression of CRELD2 and DERL3, suggesting the activation of PERK and IRE1 branches. COL10A1 N617K model mice have also shown IRE1 and PERK activation (Cameron et al., 2011), but the activation of these branches was not clear in our COL10A1 S600P and L614Rfs∗8 mutants.
Figure 4.
Some, but not all, COL10A1 and MATN3 mutants show an increased ER size, ER stress, and a UPR
(A–D) Transmission electron microscopy of mutants (left panels) and their isogenic controls (right panels). Arrowheads indicate the ER. Scale bars, 2 μm.
(E) ER size quantified from the area of PDI fluorescence from n = 4 independent experiments.
(F and G) mRNA expression of ER stress markers (F) and UPR markers (G) by qPCR (number of independent experiments shown in Table S2).
(H) Protein expression of ER stress and UPR markers by Simple Western (HSPA5, PDI, ERO1A) or western blotting (ATF6, EIF2A) from n = 4 independent experiments.
All results are from day 56 of HI and are expressed as mean ± SEM, relative to the respective isogenic control (dotted lines). Statistical analysis was performed by unpaired two-sided t test (n.s., no significant difference; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001). See also Figure S5.
Zonal disorganization in growth-plate-like structures
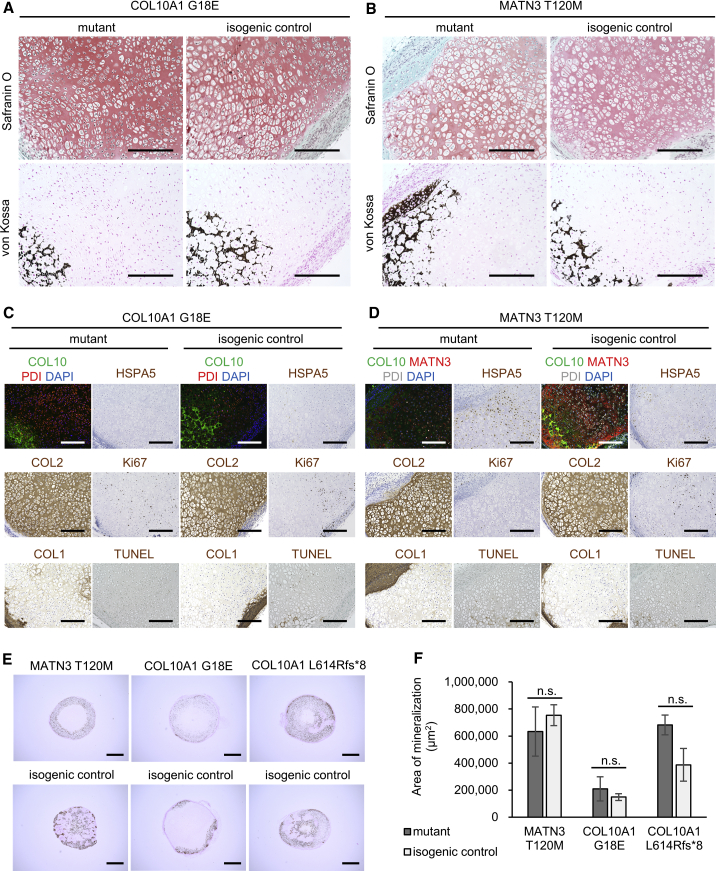
Since cell morphology and chondrocyte maturation markers in mutants showed little or no change in our in vitro system, we next assessed whether an in vivo environment may allow the detection of further differences. iPSC-derived sclerotome cells, which served as the chondroprogenitors in our in vitro model, were transplanted into immunodeficient mice to allow spontaneous development into tissues including cartilage and self-organizing growth-plate-like structures (previously described in Matsuda et al., 2020 and Loh et al., 2016). Histological examination of the tissues 56 days after transplantation revealed growth-plate-like structures both in mutants and isogenic controls, with similar Safranin O and von Kossa staining (Figures 5A and 5B).
Figure 5.
COL10A1 and MATN3 mutants show zonal disorganization without change in mineralization
(A and B) Histology of growth-plate-like structures from tissue collected on day 56 after transplantation into immunodeficient mice. (Top) Safranin O staining; (bottom) von Kossa staining. Similar results were observed in two or more growth-plate-like structures for each clone. Scale bars, 200 μm.
(C and D) Immunofluorescence, immunohistochemistry, and TUNEL staining of the growth-plate-like structures in (A) and (B). Scale bars, 200 μm.
(E) von Kossa staining of mutant (top) or isogenic control (bottom) pellets transplanted on day 28 of HI and collected after 20 days in vivo. Similar results were obtained in six pellets transplanted into three different mice (two pellets per mouse). Scale bars, 1 mm.
(F) Quantification of von Kossa-positive areas. Values are expressed as mean ± SEM. Results are calculated as n = 3 from three different mice with two pellets each. (n.s., no significant difference by unpaired two-sided t test).
However, in the COL10A1 G18E mutant, chondrocyte morphology changed little throughout the structure, and no distinct zonal accumulation of hypertrophic chondrocytes could be observed ahead of the mineralized areas, reflecting the morphological changes in hypertrophic chondrocytes to a smaller, more proliferating chondrocyte-like morphology observed in mouse models (Rajpar et al., 2009). In the MATN3 T120M mutant, cells with a hypertrophic morphology were observed throughout the growth-plate-like structure, which lacked columnar chondrocytes similarly to mouse models (Leighton et al., 2007). In both mutants, COL2 and COL1 production was unaffected, confirming the results of the in vitro observations, while COL10 expression was variable (Figures 5C and 5D). Ki67 mostly stained in areas where cells had a proliferating chondrocyte-like morphology, while TUNEL staining was mostly observed in the hypertrophic-like areas, without a notable increase. HSPA5 stained weakly in both the COL10A1 G18E mutant and isogenic control, but more intensely in the MATN3 T120M mutant compared with the isogenic control, confirming the severe ER stress of the latter observed in vitro.
To examine whether COL10A1 and MATN3 mutations affect the amount of mineralization, we transplanted cartilage pellets created using our in vitro system into immunodeficient mice on day 28 of HI. Within 20 days, mineralization was detected in all mutants and isogenic controls (Figure 5E), but the area of mineralization was not different between the isogenic pairs (Figure 5F).
Application of the in vitro model to transcriptomics and drug testing
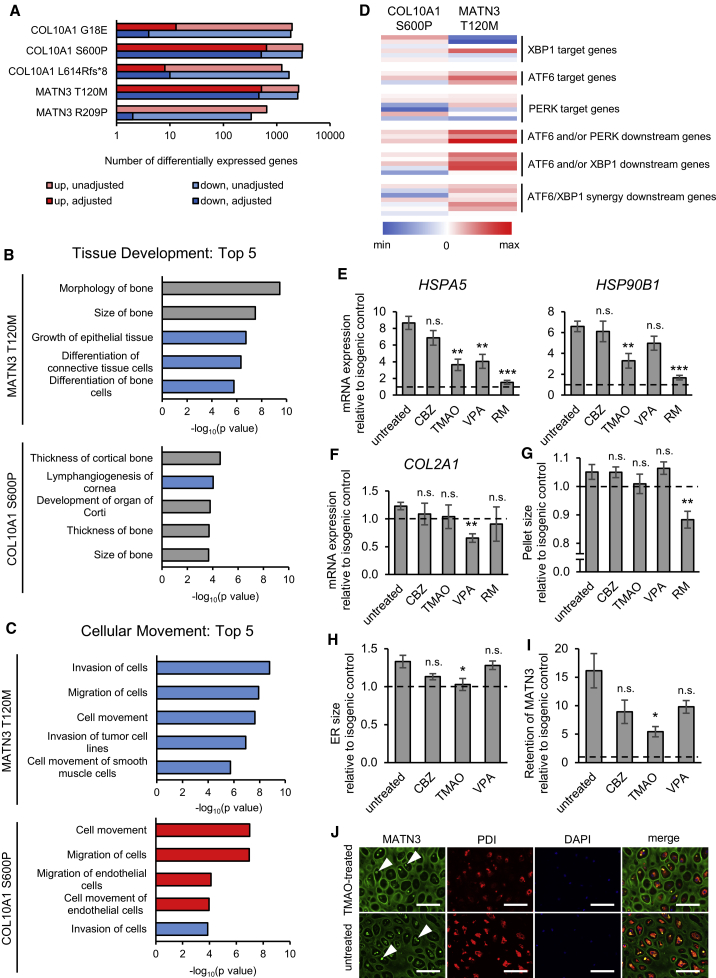
Due to the differences in ER stress levels and UPR activation in each mutant, we next asked whether our in vitro model could be used to further examine the effects of each MATN3 and COL10A1 mutation on the transcriptome. Microarray analysis revealed large differences in the amount of differentially expressed genes (DEGs) in each mutant. The COL10A1 G18E, COL10A1 L614Rfs∗8, and MATN3 R209P mutants showed only few DEGs with false discovery rate (FDR)-adjusted p value of <0.05, suggesting that these mutations are mostly tolerated in vitro (Figure 6A). However, the COL10A1 S600P and MATN3 T120M mutations led to wide-ranging changes in multiple signaling pathways and cellular functions (Figures 6B, 6C, and S6A–S6D; Table S1).
Figure 6.
iPSC-derived hypertrophic chondrocytes allow drug testing and further exploration of chondrodysplasia pathology
(A) Number of differentially expressed genes by microarray analysis using FDR-adjusted or unadjusted p value of <0.05 by moderated t test in each mutant compared with its isogenic control.
(B and C) Top five Tissue Development (B) and Cellular Movement (C) terms in IPA Diseases and Functions for the MATN3 T120M and COL10A1 S600P mutants versus their isogenic controls by microarray analysis. Genes with adjusted p value of <0.05 by moderated t test were used for IPA. Red bars, positive activation Z score; blue bars, negative activation Z score; gray bars, zero or unavailable activation Z score.
(D) Heatmap of the log2 fold change of UPR genes from different branches in the COL10A1 S600P and MATN3 T120M mutants by microarray analysis.
(E and F) mRNA expression of ER stress markers (E) and chondrocyte marker COL2A1 (F) by qPCR in the MATN3 T120M mutant with and without drug treatment.
(G) Pellet size of the MATN3 T120M mutant with and without treatment.
(H) ER size of the MATN3 T120M mutant with and without treatment, quantified from the area of PDI fluorescence.
(I) Intracellular retention of MATN3 in the MATN3 T120M mutant with and without treatment, quantified from fluorescence intensity of MATN3 co-staining with PDI (intracellular) divided by total fluorescence intensity.
(J) Immunostaining of MATN3 and PDI in the MATN3 T120M mutant with and without treatment. Arrowheads indicate intracellular aggregates. Similar results were obtained in n = 4 independent experiments. Scale bars, 100 μm.
All results are from day 56 of HI. The number of independent experiments is n = 3 in (A) to (D) and n = 4 in (E) to (I). In (E) to (I), values are expressed as mean ± SEM, relative to the isogenic control (dotted lines), and statistical analysis was performed by unpaired two-sided t test of treated compared with untreated samples (n.s., no significant difference; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001). As experiments were performed at the same time, data of untreated mutants and isogenic controls in (E) to (J) are the same as in Figures 2G, 3D, 4E, 4F, and S5A. CBZ, carbamazepine; TMAO, trimethylamine N-oxide; VPA, valproic acid; RM, rapamycin. See also Figure S6 and Table S1.
For both mutants, top terms in the IPA Tissue Development category were related to bone size, thickness, and morphology, exemplified by the common upregulation of CXXC5, GPNMB, and CHAD, known to be involved in bone homeostasis and osteoblast differentiation (Figure 6B) (Frara et al., 2016; Hessle et al., 2013; Kim et al., 2015). However, while both mutants showed changes in the Cellular Movement category, many were in the opposite direction (Figure 6C). In the COL10A1 S600P mutant, the integrins ITGA8, ITGA11, ITGB2, and ITGB3 were downregulated, with ILK and integrin signaling among the most significantly affected pathways (Figure S6B). The MATN3 T120M mutant had significant changes in several extracellular matrix (ECM) proteins, with decreased TNC, LAMA4, and POSTN, and increased COMP expression. Interestingly, both mutants showed a negative change in angiogenesis-related terms in the Organismal Development category, with a common downregulation of VEGFC and an upregulation of thrombospondins THBS2 or THBS3 in the COL10A1 S600P and MATN3 T120M mutants, respectively (Figure S6C). In the Small Molecule Biochemistry category, MATN3 T120M led to upregulated sterol synthesis through increases in INSIG1, SQLE, HMGCR, and SREBF2, while the changes in the COL10A1 S600P mutant included amino acid uptake and glucose metabolism (Figure S6D).
Together, these results indicate that these mutations result in both common and unique changes in cell-matrix interactions, metabolism, matrix composition, and bone homeostasis that can be detected using our system. This is despite the weaker ER stress and the unclear UPR activation in the COL10A1 S600P mutant, which had increases in the ER stress response genes CREB3, SERP1, and CRYAB, but in only few of the canonical UPR genes (Figure 6D, details in Table S1). In the MATN3 T120M mutant, however, the UPR was one of the most significantly affected pathways, mostly through the activation of ATF6 target and downstream genes that have been reported in the MATN3 V194D model mice (Pirog et al., 2019) (Figures S6A and 6D).
Therefore, we next utilized the MATN3 T120M mutant to examine the possibility of our system to be used as a drug-testing platform. As autophagy inducers and chemical chaperones have been shown to improve the phenotype in models of skeletal dysplasias (Forouhan et al., 2018; Kawai et al., 2019; Okada et al., 2015; Posey et al., 2014), we assessed their effect on this mutant on day 56 of HI after 28 days of treatment. Both HSPA5 and HSP90B1 were significantly reduced using trimethylamine N-oxide (TMAO) and rapamycin (RM), neither of which affected COL2A1 expression, but RM led to a significant reduction of pellet size, indicating a possible disruption of ECM production (Figures 6E–6G). TMAO was also successful in reducing the ER size and intracellular MATN3 retention in this mutant, demonstrating a marked alleviation of its phenotype in our system (Figures 6H–6J).
Discussion
In this study, we developed a robust, serum-free protocol to induce hypertrophic chondrocytes from iPSCs to model chondrodysplasias in a 3D culture system. This protocol mimics the process of chondrocyte differentiation in the growth plate, with cells characteristic of hypertrophic chondrocytes in their morphology as well as their gene expression being obtained after 56 days in 3D culture. Genes that are highly upregulated in the physis but not articular cartilage, such as MATN3 and COL9A2 (Paradise et al., 2018), were expressed in the course of induction, supporting the validity of our model to research disorders of the growth plate.
Using this system, we found that MATN3 and COL10A1 mutants had intracellular accumulation of the affected protein and an increased expression of ER stress markers, the severity of which depended on the mutation. Strong activation of UPR genes was detected only in the MATN3 T120M mutant, indicating heterogeneity in the phenotype even when mutations are in the same disease-causing gene. The patient with the MATN3 T120M mutation had a short stature (−2.2 SD) and a severe phenotype in the radiological findings, with skeletal abnormalities reminiscent of spondyloepimetaphyseal dysplasia, matrilin 3 type (SEMD-MATN3; OMIM #608728), which is caused by homozygous MATN3 mutations (Borochowitz et al., 2004). Since the SNP rs187943382, causing MATN3 V220A, was found in the healthy allele of MATN3 in this patient, we cannot exclude that it could exacerbate the pathology caused by T120M, as both are within the von Willebrand factor type A (vWFa) domain and may act in synergy to worsen the misfolding. Because V220A has also been detected in healthy controls (Kim et al., 2011), and the rescue of the MATN3 T120M mutant showed no intracellular accumulation of MATN3, we propose that this SNP may only worsen existing MED pathology without causing it on its own. In contrast to the MATN3 T120M mutant, the MATN3 R209P mutant had only few transcriptomic changes and little ER stress. The height of the R209P patient was in the 51st percentile (Kim et al., 2011), indicating that this mutation may result in fewer structural changes in MATN3 and a milder phenotype than other mutations, which caused very short statures. These observations indicate that our system is capable of recapitulating the differences in phenotype severity of MED patients with mutations in MATN3.
However, the genotype-phenotype relationship in the COL10A1 mutants was less clear. Despite the different types and locations of the mutations in COL10A1, the MCDS patients all had a similar phenotype typical of this chondrodysplasia. One of the patients had a previously unreported COL10A1 L614Rfs∗8 frameshift mutation in the NC1 domain, which did not lead to NMD despite the early stop codon. This suggests that unlike the Y632X mutation (Chan et al., 1998), this mutation will not result in haploinsufficiency and likely acts in a gain-of-function manner similar to that of substitution mutations in the same domain. The COL10A1 G18E mutation, however, likely leads to impaired signal peptide cleavage and failure to release the COL10A1 chains into the ER lumen (Chan et al., 2001). Our results also show an enlargement of the ER and a mild increase of UPR markers in this mutant, possibly causing a phenotype with similar molecular mechanisms to mutations in the NC1 domain. Nevertheless, neither L614Rfs∗8 nor G18E led to substantial changes in the transcriptome, raising the possibility that their phenotype was not completely recapitulated in our system or may require environmental factors such as mechanical stresses to fully manifest.
Indeed, in both MATN3 and COL10A1 mutants, changes in chondrocyte maturation markers were small, and changes in cell morphology required an in vivo environment to appear. Together with the expression of COL2A1 remaining high on day 56, this suggests that our in vitro system may still lack some of the required regulatory network. However, a distinct advantage of our in vitro model over the in vivo sclerotome transplantation is the stability of results across different iPSC lines, allowing for the quantification and detection of mild changes as well as drug testing. In contrast, the formation of in vivo growth-plate-like structures was rare and spontaneous, with the variability in their size and orientation precluding the quantification of the length of the proliferating and hypertrophic zones, the latter of which has been reported to extend in Col10a1 mutant mice (Rajpar et al., 2009). Therefore, improving on the stability of the in vivo model or developing an in vitro model that includes both proliferating and hypertrophic chondrocytes in separate zones, similarly to a growth plate in vivo, may greatly expand the capabilities of chondrodysplasia-specific iPSCs to precisely recapitulate the disease phenotype.
Despite these limitations, the MATN3 T120M and COL10A1 S600P mutants displayed wide-ranging transcriptomic changes in our in vitro model. Since the COL10A1 S600P mutant, contrary to mouse models of MCDS (Wang et al., 2018), showed almost no signs of activation of any of the UPR branches, this raises the question of whether the UPR is always a key disease mechanism of MCDS in humans. COL10A1 N617K model mice had little to no phenotype when heterozygous (Rajpar et al., 2009), and while COL10A1 Y632X mutant mRNA was stable in model mice (Forouhan et al., 2018), no Y632X mutant mRNA transcripts were found in humans (Chan et al., 1998), so that species differences may exist in the exact effects of each mutation. In our COL10A1 S600P mutant, changes in genes involved in cell-matrix signaling and bone homeostasis were detected, but the molecular mechanisms are unclear. As CREB3, CREB3L1, and the vesicular trafficking genes ARF1, ARF4, and MIA3 were upregulated, the mild ER stress in this mutant may result in altered protein secretion and ECM composition, which may be further affected by the increase in the ECM modulators TIMP1 and ADAM15. Interestingly, ADAM15 has been found to have inhibitory effects on apoptosis in osteoarthritic chondrocytes (Böhm et al., 2010), but whether apoptosis is a disease mechanism in MCDS is unclear (Cameron et al., 2015). Since we did not observe an increase in TUNEL-positive cells in vitro or in vivo, the ER stress in the human heterozygous COL10A1 and MATN3 mutants is, at least at the tested time point, not strong enough to induce the apoptotic pathway and may instead trigger the adaptive response.
While there was little evidence of a canonical UPR driving the pathology in the COL10A1 S600P mutant, many of the most significantly upregulated genes from the microarray analysis of the MATN3 T120M mutant were related to ER stress and the UPR, including PDIA4, HSP90B1, MANF, HSPA5, HYOU1, and CRELD2. Upregulation of MANF and CRELD2 has also been reported in MATN3 V194D mouse and in vitro models due to inappropriate disulfide bond formation in mutant MATN3 (Hartley et al., 2013), suggesting that the molecular mechanism by which the T120M mutation disrupts folding may be similar. In our model, the MATN3 T120M mutant mainly activated ATF6 target and downstream genes, resulting in expression changes related to functions such as bone homeostasis and lipid metabolism, as well as ECM proteins. As cholesterol production regulates hedgehog signaling in differentiating chondrocytes (Tsushima et al., 2018), the increase of genes related to cholesterol biosynthesis and the decrease of IHH expression in MATN3 T120M is one possible mechanism by which cartilage maturation may be altered in this mutant, but whether these changes are pathological or adaptive remains to be established. Since the UPR is likely a key disease trigger, and treatment with the chemical chaperone TMAO was able to decrease intracellular MATN3 retention as well as ER stress markers, chaperones provide a promising therapeutic avenue for MED caused by this mutation.
Taken together, our results show that our system of hypertrophic chondrocyte differentiation from iPSCs serves as an initial platform for the study of human chondrodysplasias and drug development in vitro. Surprisingly mild phenotypes in MATN3 R209P, COL10A1 G18E, and COL10A1 L614Rfs∗8 compared with MATN3 T120M and COL10A1 S600P mutants in our model indicate that the effects on cell behavior caused by mutations even in the same gene are highly diverse. With further improvements of our model to more completely recapitulate MED and MCDS phenotypes in vitro, we expect it to become a valuable tool to better assess rare patient mutations, shed light on the genotype-phenotype relationships, and perform drug screening for different chondrodysplasia mutations in the future.
Experimental procedures
Establishment of iPSC lines and isogenic controls
Patient iPSC lines were established from skin fibroblasts (MCDS patient #1) or peripheral blood mononuclear cells (MCDS patient #2 and MED patient) as previously described (Okita et al., 2011). Gene-corrected rescues were created using the CRISPR/Cas9 system. Further details are available in Supplemental information, including guide RNAs and repair templates in Table S3. All experiments with human subjects were performed with written informed consent and approved by the Ethics Committee of the Department of Medicine and Graduate School of Medicine, Kyoto University, the Ethics Committee of the Shiga Medical Center for Children, and the Ethical Committee of RIKEN Yokohama Institute.
Sclerotome and hypertrophic chondrocyte induction
All iPSCs were maintained feeder-free on dishes coated with iMatrix-511 silk (Nippi) in StemFit AK02N (Ajinomoto) with 50 U of penicillin and 50 μg/mL streptomycin (Gibco). Cells were passaged at a density of 1.1 × 103 to 3.2 × 103 cells/cm2 5 days before induction. SI was performed for 6 days as previously described, after which cells were detached and resuspended in CDMi base medium containing 100 nM SAG (Calbio), 600 nM LDN193189 (Stemgent), and 10 μM Y-27632 (Wako) (Matsuda et al., 2020). Cells were seeded into low-attachment 96-well plates (SUMILON) at 2.5 × 105 cells/well and, on the next day, i.e., day 0 of HI, induced as previously described for the first 14 days (Umeda et al., 2012). In brief, on day 0, the medium was changed to basal chondrogenic medium supplemented with 40 ng/mL PDGF-BB (R&D) and 0.1 μM dexamethasone (Wako). From day 6, 10 ng/mL TGFβ3 (R&D) was additionally supplemented. From day 10, PDGF-BB was removed and 50 ng/mL BMP4 (R&D) was added.
Then, from day 14, dexamethasone and TGFβ3 were removed, and 10 nM triiodothyronine (T3) (Sigma) was added. From day 28, 10 mM β-glycerophosphate (Sigma) was added. For the drug-treated groups, 20 μM carbamazepine (CBZ) (Sigma), 50 mM trimethylamine N-oxide (TMAO) (Sigma), 200 μM valproic acid (VPA) (Sigma), or 10 nM rapamycin (RM) (MedChem Express) was added from days 28 to 56 of HI. The CDMi and basal chondrogenic medium compositions and other information about induction are detailed in Supplemental information, including key reagents in Table S6. All independent experiments in this study refer to pellets from separate differentiations.
Animal experiments
For observation of growth-plate-like structures, cells were detached on day 6 of SI and resuspended in CDMi containing 100 nM SAG, 600 nM LDN193189, and 10 μM Y-27632 at a concentration of up to 1 × 108 cells/mL. This cell suspension was mixed 1:1 with Matrigel (BD) and 100 μL was injected subcutaneously into a minimum of six male immunodeficient NOD/ShiJic-scid Jcl (NOD-SCID) mice (CLEA Japan) as previously described (Matsuda et al., 2020). The transplanted tissue was collected after 56 days. For quantification of mineralization of cartilage, pellets were transplanted subcutaneously into three male NOD-SCID mice on day 28 of HI. Pellets were collected after 20 days. All animal experiments were approved by the UCSF Institutional Animal Care and Use Committee and performed in accordance with the Regulations on Animal Experimentation at Kyoto University.
Expression analysis and flow cytometry
After RNA extraction using the RNeasy Micro or Mini Kit (QIAGEN), quantitative PCR (qPCR) was performed with the Thunderbird SYBR qPCR Mix (Toyobo). Microarray analysis was performed with the Human Gene 1.0ST Array (Affymetrix). Protein expression was analyzed by western blotting and Simple Western (ProteinSimple). Flow cytometry was performed using FACS Aria II (BD) as previously described (Matsuda et al., 2020) with minor modifications. More details are provided in Supplemental information, including primer information in Table S5.
Histological analysis
In vitro pellets and in vivo tissue were fixed in 4% paraformaldehyde for 2 days before paraffin embedding. Staining protocols are detailed in Supplemental information.
Data and code availability
The microarray data are available in the GEO database under the accession numbers GEO: GSE148728 and GEO: GSE157955.
Author contributions
Y.P., S.K., and J.T. designed the study. Y.P. developed the HI protocol, performed the gene editing and disease modeling, analyzed all experiments, and drafted the manuscript. S.N. and M.N. assisted in the in vitro and in vivo experiments, respectively. M.W. assisted in data analysis. S.T. supervised the establishment of patient iPSC lines. C.A. and Y.Y. devised the SI and transplantation protocols. J.-Y.X. and Z.W. performed the mutation search by NGS. K.F., M.T., T.F., and S.I. provided patient samples for the establishment of iPSC lines. J.T. supervised the study and revised the manuscript, which was further reviewed by all authors.
Conflicts of interest
The authors declare no competing interests.
Acknowledgments
We thank J. Ma and T. Nakashima for their assistance in the in vitro and in vivo experiments, respectively, H. Maekawa for providing human growth plate samples, and H. Yoshitomi, Y. Jin, and T. Takarada for comments and discussion. Preparation of tissue slides and staining was supported by the Center for Anatomical, Pathological and Forensic Medical Research, Graduate School of Medicine, Kyoto University, and Applied Medical Research Laboratory. Karyotyping was performed by Chromocenter. This study was supported by a grant-in-aid for the Acceleration Program for Intractable Disease Research Utilizing Disease Specific iPS Cells (AMED) for S.I. and J.T. and the Centers for Clinical Application Research on Specific Disease/Organ (type B) grants (AMED) for J.T.
Published: February 25, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.stemcr.2021.01.014.
Supplemental information
References
- Böhm B., Hess S., Krause K., Schirner A., Ewald W., Aigner T., Burkhardt H. ADAM15 exerts an antiapoptotic effect on osteoarthritic chondrocytes via up-regulation of the X-linked inhibitor of apoptosis. Arthritis Rheum. 2010;62:1372–1382. doi: 10.1002/art.27387. [DOI] [PubMed] [Google Scholar]
- Borochowitz Z.U., Scheffer D., Adir V., Dagoneau N., Munnich A., Cormier-Daire V. Spondylo-epi-metaphyseal dysplasia (SEMD) matrilin 3 type: homozygote matrilin 3 mutation in a novel form of SEMD. J. Med. Genet. 2004;41:366–372. doi: 10.1136/jmg.2003.013342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron T.L., Bell K.M., Gresshoff I.L., Sampurno L., Mullan L., Ermann J., Glimcher L.H., Boot-Handford R.P., Bateman J.F. XBP1-independent UPR pathways suppress C/EBP-β mediated chondrocyte differentiation in ER-stress related skeletal disease. PLoS Genet. 2015;11:e1005505. doi: 10.1371/journal.pgen.1005505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron T.L., Bell K.M., Tatarczuch L., Mackie E.J., Rajpar M.H., McDermott B.T., Boot-Handford R.P., Bateman J.F. Transcriptional profiling of chondrodysplasia growth plate cartilage reveals adaptive ER-stress networks that allow survival but disrupt hypertrophy. PLoS One. 2011;6:e24600. doi: 10.1371/journal.pone.0024600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan D., Ho M.S., Cheah K.S. Aberrant signal peptide cleavage of collagen X in Schmid metaphyseal chondrodysplasia. Implications for the molecular basis of the disease. J. Biol. Chem. 2001;276:7992–7997. doi: 10.1074/jbc.M003361200. [DOI] [PubMed] [Google Scholar]
- Chan D., Weng Y.M., Graham H.K., Sillence D.O., Bateman J.F. A nonsense mutation in the carboxyl-terminal domain of type X collagen causes haploinsufficiency in schmid metaphyseal chondrodysplasia. J. Clin. Invest. 1998;101:1490–1499. doi: 10.1172/JCI1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotterill S.L., Jackson G.C., Leighton M.P., Wagener R., Mäkitie O., Cole W.G., Briggs M.D. Multiple epiphyseal dysplasia mutations in MATN3 cause misfolding of the A-domain and prevent secretion of mutant matrilin-3. Hum. Mutat. 2005;26:557–565. doi: 10.1002/humu.20263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czarny-Ratajczak M., Lohiniva J., Rogala P., Kozlowski K., Perälä M., Carter L., Spector T.D., Kolodziej L., Seppänen U., Glazar R. A mutation in COL9A1 causes multiple epiphyseal dysplasia: further evidence for locus heterogeneity. Am. J. Hum. Genet. 2001;69:969–980. doi: 10.1086/324023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forouhan M., Sonntag S., Boot-Handford R.P. Carbamazepine reduces disease severity in a mouse model of metaphyseal chondrodysplasia type Schmid caused by a premature stop codon (Y632X) in the Col10a1 gene. Hum. Mol. Genet. 2018;27:3840–3853. doi: 10.1093/hmg/ddy253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frara N., Abdelmagid S.M., Sondag G.R., Moussa F.M., Yingling V.R., Owen T.A., Popoff S.N., Barbe M.F., Safadi F.F. Transgenic expression of osteoactivin/gpnmb enhances bone formation in vivo and osteoprogenitor differentiation ex vivo. J. Cell. Physiol. 2016;231:72–83. doi: 10.1002/jcp.25020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory C.A., Zabel B., Grant M.E., Boot-Handford R.P., Wallis G.A. Equal expression of type X collagen mRNA from mutant and wild type COL10A1 alleles in growth plate cartilage from a patient with metaphyseal chondrodysplasia type Schmid. J. Med. Genet. 2000;37:627–629. doi: 10.1136/jmg.37.8.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley C.L., Edwards S., Mullan L., Bell P.A., Fresquet M., Boot-Handford R.P., Briggs M.D. Armet/Manf and Creld2 are components of a specialized ER stress response provoked by inappropriate formation of disulphide bonds: implications for genetic skeletal diseases. Hum. Mol. Genet. 2013;22:5262–5275. doi: 10.1093/hmg/ddt383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hessle L., Stordalen G.A., Wenglén C., Petzold C., Tanner K.E., Brorson S.H., Baekkevold E.S., Önnerfjord P., Reinholt F.P., Heinegård D. The skeletal phenotype of chondroadherin deficient mice. PLoS One. 2013;8:e63080. doi: 10.1371/journal.pone.0063080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikegawa S., Nakamura K., Nagano A., Haga N., Nakamura Y. Mutations in the N-terminal globular domain of the type X collagen gene (COL10A1) in patients with Schmid metaphyseal chondrodysplasia. Hum. Mutat. 1997;9:131–135. doi: 10.1002/(SICI)1098-1004(1997)9:2<131::AID-HUMU5>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Jackson G.C., Barker F.S., Jakkula E., Czarny-Ratajczak M., Mäkitie O., Cole W.G., Wright M.J., Smithson S.F., Suri M., Rogala P. Missense mutations in the beta strands of the single A-domain of matrilin-3 result in multiple epiphyseal dysplasia. J. Med. Genet. 2004;41:52–59. doi: 10.1136/jmg.2003.011429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karl A., Olbrich N., Pfeifer C., Berner A., Zellner J., Kujat R., Angele P., Nerlich M., Mueller M.B. Thyroid hormone-induced hypertrophy in mesenchymal stem cell chondrogenesis is mediated by bone morphogenetic protein-4. Tissue Eng. Part A. 2014;20:178–188. doi: 10.1089/ten.tea.2013.0023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai S., Yoshitomi H., Sunaga J., Alev C., Nagata S., Nishio M., Hada M., Koyama Y., Uemura M., Sekiguchi K. In vitro bone-like nodules generated from patient-derived iPSCs recapitulate pathological bone phenotypes. Nat. Biomed. Eng. 2019;3:558–570. doi: 10.1038/s41551-019-0410-7. [DOI] [PubMed] [Google Scholar]
- Kim H.Y., Yoon J.Y., Yun J.H., Cho K.W., Lee S.H., Rhee Y.M., Jung H.S., Lim H.J., Lee H., Choi J. CXXC5 is a negative-feedback regulator of the Wnt/β-catenin pathway involved in osteoblast differentiation. Cell Death Differ. 2015;22:912–920. doi: 10.1038/cdd.2014.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim O.H., Park H., Seong M.W., Cho T.J., Nishimura G., Superti-Furga A., Unger S., Ikegawa S., Choi I.H., Song H.R. Revisit of multiple epiphyseal dysplasia: ethnic difference in genotypes and comparison of radiographic features linked to the COMP and MATN3 genes. Am. J. Med. Genet. A. 2011;155A:2669–2680. doi: 10.1002/ajmg.a.34246. [DOI] [PubMed] [Google Scholar]
- Kobayashi T., Chung U.I., Schipani E., Starbuck M., Karsenty G., Katagiri T., Goad D.L., Lanske B., Kronenberg H.M. PTHrP and Indian hedgehog control differentiation of growth plate chondrocytes at multiple steps. Development. 2002;129:2977–2986. doi: 10.1242/dev.129.12.2977. [DOI] [PubMed] [Google Scholar]
- Kwan K.M., Pang M.K., Zhou S., Cowan S.K., Kong R.Y., Pfordte T., Olsen B.R., Sillence D.O., Tam P.P., Cheah K.S. Abnormal compartmentalization of cartilage matrix components in mice lacking collagen X: implications for function. J. Cell Biol. 1997;136:459–471. doi: 10.1083/jcb.136.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leighton M.P., Nundlall S., Starborg T., Meadows R.S., Suleman F., Knowles L., Wagener R., Thornton D.J., Kadler K.E., Boot-Handford R.P. Decreased chondrocyte proliferation and dysregulated apoptosis in the cartilage growth plate are key features of a murine model of epiphyseal dysplasia caused by a matn3 mutation. Hum. Mol. Genet. 2007;16:1728–1741. doi: 10.1093/hmg/ddm121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh K.M., Chen A., Koh P.W., Deng T.Z., Sinha R., Tsai J.M., Barkal A.A., Shen K.Y., Jain R., Morganti R.M. Mapping the pairwise choices leading from pluripotency to human bone, heart, and other mesoderm cell types. Cell. 2016;166:451–467. doi: 10.1016/j.cell.2016.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mäkitie O., Susic M., Ward L., Barclay C., Glorieux F.H., Cole W.G. Schmid type of metaphyseal chondrodysplasia and COL10A1 mutations—findings in 10 patients. Am. J. Med. Genet. A. 2005;137A:241–248. doi: 10.1002/ajmg.a.30855. [DOI] [PubMed] [Google Scholar]
- Matsuda M., Yamanaka Y., Uemura M., Osawa M., Saito M.K., Nagahashi A., Nishio M., Guo L., Ikegawa S., Sakurai S. Recapitulating the human segmentation clock with pluripotent stem cells. Nature. 2020;580:124–129. doi: 10.1038/s41586-020-2144-9. [DOI] [PubMed] [Google Scholar]
- Mortier G.R., Cohn D.H., Cormier-Daire V., Hall C., Krakow D., Mundlos S., Nishimura G., Robertson S., Sangiorgi L., Savarirayan R. Nosology and classification of genetic skeletal disorders: 2019 revision. Am. J. Med. Genet. A. 2019;179:2393–2419. doi: 10.1002/ajmg.a.61366. [DOI] [PubMed] [Google Scholar]
- Mueller M.B., Tuan R.S. Functional characterization of hypertrophy in chondrogenesis of human mesenchymal stem cells. Arthritis Rheum. 2008;58:1377–1388. doi: 10.1002/art.23370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa M., Taniguchi Y., Senda S., Takizawa N., Ichisaka T., Asano K., Morizane A., Doi D., Takahashi J., Nishizawa M. A novel efficient feeder-free culture system for the derivation of human induced pluripotent stem cells. Sci. Rep. 2014;4:3594. doi: 10.1038/srep03594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada M., Ikegawa S., Morioka M., Yamashita A., Saito A., Sawai H., Murotsuki J., Ohashi H., Okamoto T., Nishimura G. Modeling type II collagenopathy skeletal dysplasia by directed conversion and induced pluripotent stem cells. Hum. Mol. Genet. 2015;24:299–313. doi: 10.1093/hmg/ddu444. [DOI] [PubMed] [Google Scholar]
- Okita K., Matsumura Y., Sato Y., Okada A., Morizane A., Okamoto S., Hong H., Nakagawa M., Tanabe K., Tezuka K. A more efficient method to generate integration-free human iPS cells. Nat. Methods. 2011;8:409–412. doi: 10.1038/nmeth.1591. [DOI] [PubMed] [Google Scholar]
- Paradise C.R., Galeano-Garces C., Galeano-Garces D., Dudakovic A., Milbrandt T.A., Saris D.B.F., Krych A.J., Karperien M., Ferguson G.B., Evseenko D. Molecular characterization of physis tissue by RNA sequencing. Gene. 2018;668:87–96. doi: 10.1016/j.gene.2018.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirog K.A., Dennis E.P., Hartley C.L., Jackson R.M., Soul J., Schwartz J.M., Bateman J.F., Boot-Handford R.P., Briggs M.D. XBP1 signalling is essential for alleviating mutant protein aggregation in ER-stress related skeletal disease. PLoS Genet. 2019;15:e1008215. doi: 10.1371/journal.pgen.1008215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posey K.L., Coustry F., Veerisetty A.C., Liu P., Alcorn J.L., Hecht J.T. Chondrocyte-specific pathology during skeletal growth and therapeutics in a murine model of pseudoachondroplasia. J. Bone Miner. Res. 2014;29:1258–1268. doi: 10.1002/jbmr.2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajpar M.H., McDermott B., Kung L., Eardley R., Knowles L., Heeran M., Thornton D.J., Wilson R., Bateman J.F., Poulsom R. Targeted induction of endoplasmic reticulum stress induces cartilage pathology. PLoS Genet. 2009;5:e1000691. doi: 10.1371/journal.pgen.1000691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsushima H., Tang Y.J., Puviindran V., Hsu S.C., Nadesan P., Yu C., Zhang H., Mirando A.J., Hilton M.J., Alman B.A. Intracellular biosynthesis of lipids and cholesterol by Scap and Insig in mesenchymal cells regulates long bone growth and chondrocyte homeostasis. Development. 2018;145:dev162396. doi: 10.1242/dev.162396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umeda K., Zhao J., Simmons P., Stanley E., Elefanty A., Nakayama N. Human chondrogenic paraxial mesoderm, directed specification and prospective isolation from pluripotent stem cells. Sci. Rep. 2012;2:455. doi: 10.1038/srep00455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Weyden L., Wei L., Luo J., Yang X., Birk D.E., Adams D.J., Bradley A., Chen Q. Functional knockout of the matrilin-3 gene causes premature chondrocyte maturation to hypertrophy and increases bone mineral density and osteoarthritis. Am. J. Pathol. 2006;169:515–527. doi: 10.2353/ajpath.2006.050981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Tan Z., Niu B., Tsang K.Y., Tai A., Chan W.C.W., Lo R.L.K., Leung K.K.H., Dung N.W.F., Itoh N. Inhibiting the integrated stress response pathway prevents aberrant chondrocyte differentiation thereby alleviating chondrodysplasia. eLife. 2018;7:e37673. doi: 10.7554/eLife.37673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Zhou J., Bondy C.A. Igf1 promotes longitudinal bone growth by insulin-like actions augmenting chondrocyte hypertrophy. FASEB J. 1999;13:1985–1990. doi: 10.1096/fasebj.13.14.1985. [DOI] [PubMed] [Google Scholar]
- Wilson R., Freddi S., Chan D., Cheah K.S., Bateman J.F. Misfolding of collagen X chains harboring Schmid metaphyseal chondrodysplasia mutations results in aberrant disulfide bond formation, intracellular retention, and activation of the unfolded protein response. J. Biol. Chem. 2005;280:15544–15552. doi: 10.1074/jbc.M410758200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The microarray data are available in the GEO database under the accession numbers GEO: GSE148728 and GEO: GSE157955.