Abstract
Genotype-first approach allows to systematically identify carriers of pathogenic variants in BRCA1/2 genes conferring a high risk of familial breast and ovarian cancer. Participants of the Estonian biobank have expressed support for the disclosure of clinically significant findings. With an Estonian biobank cohort, we applied a genotype-first approach, contacted carriers, and offered return of results with genetic counseling. We evaluated participants’ responses to and the clinical utility of the reporting of actionable genetic findings. Twenty-two of 40 contacted carriers of 17 pathogenic BRCA1/2 variants responded and chose to receive results. Eight of these 22 participants qualified for high-risk assessment based on National Comprehensive Cancer Network criteria. Twenty of 21 counseled participants appreciated being contacted. Relatives of 10 participants underwent cascade screening. Five of 16 eligible female BRCA1/2 variant carriers chose to undergo risk-reducing surgery, and 10 adhered to surveillance recommendations over the 30-month follow-up period. We recommend the return of results to population-based biobank participants; this approach could be viewed as a model for population-wide genetic testing. The genotype-first approach permits the identification of individuals at high risk who would not be identified by application of an approach based on personal and family histories only.
Subject terms: Genetic counselling, Population screening, Cancer genomics
Introduction
The rapidly increasing volume of genomic data at the population-scale imposes an intensified need for best practices to guide and standardize the way how clinically significant, but potentially unexpected, genetic findings from population-based research cohorts are handled. The human genomics community needs to address questions such as how to anticipate and manage such genetic events, and how to communicate such unexpected findings.
Several biobanks in Australia, Northern Europe, and the United States have applied a genotype-first approach, in which individuals carrying clinically significant variants are recontacted and offered returns of results [1–4]. Being irrespective of family history and the presence of medical indication, this approach varies greatly from common practice in clinical settings, where considerations regarding genetic analysis are based on personal or familial histories. Evidence for participants’ reactions to unexpected results is limited, but suggests that people tend to appreciate being given actionable information about their health risks [1, 3, 5–8].
Although the general consensus supports the return of clinically significant findings in clinical and research settings, numerous challenges must still be faced, especially in the research context [3, 6, 8]. One such challenge involves the provision of genomic risk information to apparently healthy individuals in a manner that improves disease risk prediction, prevention, diagnosis, and therapy. Procedural guidelines are currently limited, largely due to a dearth of conclusive studies on the clinical and psychological impacts of such disclosure.
The unique legislative setting in Estonia permits the recontacting of Estonian Biobank (EstBB) participants for specific research and prospective studies. The Estonian Human Genes Research Act [9] and the broad consent given by all biobank participants allow us to conduct valuable research on the impact of the communication of potentially unexpected genomic findings to research participants.
Given its high prevalence in the EstBB dataset, carrier screening for autosomal dominant hereditary breast and ovarian cancer (HBOC; MIM #604370, #612555) was selected for this study. In the general population, about 5–10% of all breast cancer and 10–15% of all ovarian cancer cases can be attributed to variants in the BRCA1 (MIM #113705) and BRCA2 (MIM #600185) genes, which can explain about half of breast/ovarian cancer aggregation in families [10–12]. The prevalence of clinically significant genetic variants in BRCA1/2 is about 1/400 in the general population, but can vary significantly depending on the characteristics of specific study cohorts [13]. Women with an inherited BRCA1 pathogenic variant have a 65–72% lifetime risk of breast cancer development, and a 37–62% risk of ovarian cancer development; BRCA2 pathogenic variant carriers are expected to have lifetime risks of 45–85% for breast cancer and 11–23% for ovarian cancer by the age of 80 years [14–16].
In Estonia, guidelines for the identification of high-risk individuals are based largely on those of the National Comprehensive Cancer Network (NCCN); BRCA1/2-associated HBOC is suspected in individuals with personal or family histories [17]. The main approach of identifying risk variants in multiplex families with high frequencies of breast and ovarian cancer is common in clinical contexts, but has been shown to miss a large percentage of high-risk individuals [18]. In this study, we applied a genotype-first method with data from the population-based EstBB to systematically identify individuals at high risk of HBOC, regardless of phenotypic heterogeneity, incomplete penetrance, or lack of family history.
Materials and methods
Study cohort
The EstBB is a population-based biobank managed by the Institute of Genomics at the University of Tartu [19]. It currently contains data from more than 200,000 participants, representing almost 20% of Estonia’s adult population. All participants have provided broad written consent, which encompasses the provision of samples for future research use and the collection of electronic health records from national registries and databases. EstBB participants can opt out of future recontact regarding participation in additional research projects or the receipt of research results.
Data for this study are from a sub-cohort of 17,679 EstBB participants recruited between 2002 and 2011. At the time of analysis, sequencing data were available from a genome sequencing (GS) and an exome sequencing (ES) dataset for 4594 individuals. Array-based genotype data (Illumina Infinium HumanCoreExome and OmniExpress-12 beadchip microarrays) were available for the remaining 13,085 individuals. Sequenced reads were aligned to the GRCh37 human reference genome assembly. After filtering and exclusion of duplicates between the two datasets, the final GS and ES sets contained 2240 and 2354 unique samples, respectively. Sequenced variants were annotated with the Variant Effect Predictor [20] (version 87; Gencode v19 on GRCh37.p13) and ANNOVAR [21]. GS and ES data preparation is described in detail in the Supplementary Materials and Methods.
For long-range phasing, we used high-coverage (30×) GS and array-based genomic data from 2240 to 13,085 EstBB participants, respectively. The GS panel was phased using the read aware phasing model of SHAPEIT v2.r837 [22], with the read-quality and base-quality parameters set to 20. Genotype data were phased with SHAPEIT v2.r837 using the default parameters. The GS and array-based data were merged, leaving only overlapping single nucleotide variants for further analysis. We calculated pairs of individuals based on shared identity by descent using PLINK v1.9 [23], with the min parameter set to 0.4. Haplotypes of calculated pairs were then visualized using R software [24], and the graphs generated were assessed visually. All cases of putative alternative allele carriage were analysed further by Sanger sequencing and no discrepancies were found.
Evaluation of variant pathogenicity
We extracted all BRCA1/2 coding variants from the GS/ES data, and cross-referenced those with likely pathogenic (LP) or known pathogenic (KP) classification and minor allele frequencies <0.5% with the ClinVar [25], VariantValidator [26] and dbSNP 153 databases [27]. For estimation of the pathogenicity of all LP/KP variants, several in silico prediction algorithms were used. Further details of variant detection and evaluation of variant pathogenicity are provided in the Supplementary Materials and Methods.
We compiled a list of carriers of 17 LP/KP BRCA1/2 variants [28] and validated each by Sanger sequencing. For confirmation, independent blood samples collected at participants’ first visits were tested by an ISO-certified (ISO 15189:2012, ISO 9001:2015) diagnostic laboratory (Asper BioGene Ltd, Tartu, Estonia).
Return of results procedure
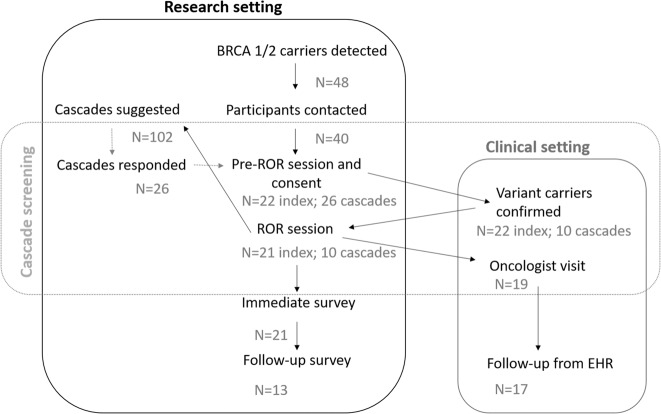
The procedural framework for the communication of unexpected genetic findings with high clinical relevance to EstBB participants was developed with participants’ rights and interests at the forefront. A previously developed procedure for the return of results and provision of clinical support [5] was implemented with a few modifications (Fig. 1). This framework is consistent with the precepts of Estonian Human Genes Research Act [9], and the protocol for this study was approved by the Research Ethics Committee of the University of Tartu.
Fig. 1. Framework for the return of results.
Most steps were conducted in the research setting. Female LP/KP variant carriers were referred to a collaborating oncologist.
The framework includes procedures for the contacting of variant carriers without immediate disclosure of their genetic status: the sending of an initial contact letter (Supplementary Material S1.1), acquisition of project-specific informed consent, independent validation of the finding in a certified diagnostic laboratory, disclosure of genetic risk accompanied by genetic counseling, and collaboration with an oncologist for the development of a personalized participant surveillance plan. Framework included cascade screening to identify additional high-risk individuals in carriers’ families. Information letters were provided to index individuals with a strong recommendation to hand them out to the respective first- and second-degree family members invited to cascade screen (Supplementary Material S1.2).
Follow-up
Data on participants’ responses to the receipt of results about clinically significant findings were gathered using immediate and long-term feedback surveys, developed based on findings from analogous previous studies [5, 29–31]. The first survey, administered immediately after the disclosure of genetic results, included questions about participants’ satisfaction, understanding, and psychological responses [30]. The second survey, mailed to participants 6 months later, included questions about decision regret [31], perceived personal control and coping [32], psychological adjustment [30], communication, support, and reported health behavior and healthcare utilization. Cascade screening participants were asked to fill in a survey immediately after the disclosure of clinically significant findings.
To facilitate the provision of effective clinical support for high-risk variant carriers identified in a research setting, all female participants carrying LP/KP BRCA1/2 variants were referred directly to a collaborating clinical oncologist. These participants’ ongoing medical management was reviewed via electronic health records from the collaborating hospital and national imaging and e-health databases. Depending on the dates of participants’ first visits, follow-up periods ranged from 12 to 30 months (January 2017–July 2019).
Results
Identified BRCA1/2 genetic variants
Forty-eight individuals in the study cohort were identified as carriers of 17 LP/KP BRCA1/2 variants (Table 1). Eighteen carriers were identified from the GS cohort, 19 were identified from the ES cohort, and additional 11 carriers were identified through long-range phasing of genotyping array data. Ten of the 17 variants were in BRCA1 (in 35 participants) and seven were in BRCA2 (in 13 participants). Fifteen (88%) of the variants had been classified in ClinVar previously: 14 KP variants had 3* status and one KP variant had 2* status. One LP variant had pending status in the Breast Cancer Information Core database (not actively curated) [33]. Based on GS and ES data, the prevalence of high-risk BRCA1/2 variants was 0.80% (1/124), first degree relatives excluded. For extended details refer to Supplementary Materials and Methods.
Table 1.
Identified HBOC-associated genetic variants.
| Gene | LP/KP variant | rsID | cDNA positiona | Protein position | GS/ESb | LRPc | AC_ESTd |
|---|---|---|---|---|---|---|---|
| BRCA1 | KP | rs80356898 | c.1687C>T | p.(Gln563*) | 1 | 0 | 1 |
| BRCA1 | KP | rs28897672 | c.181T>G | p.(Cys61Gly) | 1 | 0 | 1 |
| BRCA1 | KP | rs80357282 | c.1840A>T | p.(Lys614*) | 1 | 0 | 1 |
| BRCA1 | LP | Novel | c.2178delT | p.(Leu727Glufs*9) | 1 | 0 | 1 |
| BRCA1 | KP | rs80357711 | c.4035delA | p.(Glu1346Lysfs*20) | 8 | 8 | 16 |
| BRCA1 | KP | rs80357508 | c.4065_4068delTCAA | p.(Asn1355Lysfs*10) | 1 | 0 | 1 |
| BRCA1 | KP | rs80357305 | c.4258C>T | p.(Gln1420*) | 1 | 0 | 1 |
| BRCA1 | KP | rs80356860 | c.5117G>A | p.(Gly1706Glu) | 1 | 0 | 1 |
| BRCA1 | KP | rs80357906 | c.5266dupC | p.(Gln1756Profs*74) | 6 | 5 | 11 |
| BRCA1 | LP | rs483353102 | c.5534_5539delinsCCAGTGCCAGGACAGCAGG | p.(Tyr1845Serfs*39) | 1 | 0 | 1 |
| BRCA2 | KP | rs80358622 | c.37G>T | p.(Glu13*) | 1 | 0 | 1 |
| BRCA2 | KP | rs397515636 | c.3975_3978dupTGCT | p.(Ala1327Cysfs*4) | 1 | 0 | 1 |
| BRCA2 | KP | rs886040543 | c.469_470insT | p.(Lys157Ilefs*26) | 1 | 0 | 1 |
| BRCA2 | KP | rs80359584 | c.6405_6409delCTTAA | p.(Asn2135Lysfs*3) | 1 | 0 | 1 |
| BRCA2 | KP | rs80359112 | c.8572C>T | p.(Gln2858*) | 3 | 3 | 6 |
| BRCA2 | KP | rs1555288494 | c.9097_9098insT | p.(Thr3033Ilefs*11) | 2 | 0 | 2 |
| BRCA2 | KP | rs876661242 | c.9381G>A | p.(Trp3127*) | 1 | 0 | 1 |
HBOC hereditary breast and ovarian cancer, LP likely pathogenic, KP known pathogenic, GS genome sequencing, ES exome sequencing, LRP long-range phasing.
aProvided for BRCA1 ENST00000357654 (NM_007294.3) and BRCA2 ENST00000544455.1 (NM_000059.3).
bn = 4594 individuals.
cn = 13,085 individuals.
dHigh-risk variant carrier numbers in the Estonian Biobank subset (n = 17,679).
The 17 BRCA1/2 variants comprised nine frameshift variants, six nonsense variants (including a novel predicted loss-of-function variant), and two missense variants. Thirteen (76%) variants were singletons. The most frequent KP variant was the known Eastern European founder variant BRCA1 c.4035delA (NM_007294.3) [34], which accounted for 33% (n = 16/35) of BRCA1 variant carriers. BRCA1 c.5266dupC (NM_007294.3) was the second most prevalent KP variant [34], common throughout Eurasia [35], accounting for 23% (n = 11/35) of BRCA1 variant carriers. The most frequently detected KP variant in BRCA2 was c.8572C>T (NM_000059.3), seen in six (12.5%) carriers.
The novel, likely deleterious, BRCA1 variant c.2178delT (NM_007294.3; p.(Pro727Glnfs*9)) causes a frameshift and creates a premature stop codon. The change is predicted to cause loss of normal protein function through nonsense-mediated decay of an mRNA due to the presence of a stop codon within the first ~90% of the coding region. The variant is not present in ClinVar or dbSNP. As loss-of-function variants in BRCA1 are generally pathogenic [36], we included c.2178delT p.(Pro727Glnfs*9) in the dataset as an LP variant. According to data from the Estonian Causes of Death Registry, the carrier of this variant was diagnosed with breast cancer at the age of 45 years and died of the disease (coded as C50.9, diagnosis was classified according to the International Classification of Diseases, 10th revision (ICD-10)) at the age of 49 years.
Participants with identified high-risk variants
Eight of the 48 participants with detected LP/KP BRCA1/2 variants could not be recontacted due to changes in residency status or death (n = 8). The cause of death for all deceased participants was cancer (Table S2).
Of the 40 contacted participants, 22 attended the first visit (55% response rate). One participant (a 77-year-old woman) declined participation. For the 17 nonrespondents (8 male, 9 female; mean age, 48 [range, 32–83] years), no information was available about the receipt of invitation letters or reason for nonresponse. One nonrespondent had been diagnosed previously with breast cancer at the ages of 57 and 77 years.
Of the 22 respondents (8 male, 14 female; mean age, 47.6 [range, 25–75] years), 18 had biological children (Table 2). Four (18%) respondents had previously received HBOC-related cancer diagnoses, and 18 (82%) respondents had first- or second-degree relatives with breast, ovarian, prostate, pancreatic, and/or endometrial cancer. One participant (ID.19) provided consent, but chose not to attend the second visit and did not receive genetic risk information. Her clinical history includes mild intellectual disability and she was under regular surveillance for anxiety. The sister of ID.19 was contacted through another breast cancer project and tested clinically positive for HBOC.
Table 2.
Characteristics of study participants and their relatives.
| ID | Sex | Age | Gene with LP/KP variant | Personal ca.dx.and age at dx | Family historya | 1st degree relatives BC/OC/PC/PAC and age at dx | 2nd degree relatives BC/OC/PC/PAC and age at dx | Cascades attended/ suggested | BRCA1/2 variant positive cascades |
|---|---|---|---|---|---|---|---|---|---|
| 1 | F | 34 | BRCA1 | No | High | Mother (OC, 50) | No | 3 of 5 | 1 |
| 2 | F | 47 | BRCA2 | No | Moderate | Mother (BC, 48) | Grandfather (PC, 77) | 2 of 5 | 2 |
| 3 | M | 66 | BRCA1 | PC, 59 | High | Mother (OC, 54) | 3 aunts and grandmother (OC) | 0 of 5 | NA |
| 4 | F | 52 | BRCA1 | No | Moderate | No | Grandmother (BC) | 2 of 5 | 1 |
| 5 | F | 39 | BRCA1 | No | High | Mother (OC, 55) | Grandmother (BC, 55), aunt (OC, 40) | 4 of 7 | 1 |
| 6 | M | 40 | BRCA1 | No | Moderate | Mother (BC, 54) | No | 0 of 3 | NA |
| 7 | F | 59 | BRCA1 | No | Low | No | No | 5 of 8 | 2 |
| 8 | F | 75 | BRCA1 | BC, 75 | High | Daughter (OC, 42) | No | 0 of 3 | NA |
| 9 | F | 75 | BRCA2 | BC, 53 | Moderate | Brother (PAC, 59) | No | 3 of 8 | 0 |
| 10 | F | 33 | BRCA1 | No | High | Mother (BC, 38) | No | 1 of 7 | 1 |
| 11 | M | 75 | BRCA1 | PC, 64 | Moderate | Sister (BC, 70) | No | 0 of 4 | NA |
| 12 | F | 46 | BRCA1 | No | Moderate | No | Grandmother (BC, 55) | 1 of 2 | 1 |
| 13 | M | 42 | BRCA1 | No | High | Mother (OC), father (PC, 63) | No | 0 of 1 | NA |
| 14 | F | 31 | BRCA1 | No | Moderate | No | 3 aunts (BC, 50s) | 0 of 4 | NA |
| 15 | F | 42 | BRCA1 | No | Moderate | No | Aunt (BC, 50s) | 0 of 5 | NA |
| 16 | M | 25 | BRCA1 | No | Low | No | No | 0 of 6 | NA |
| 17 | M | 58 | BRCA1 | No | Moderate | No | Aunt (BC, 50s) | 4 of 5 | 1 |
| 18 | F | 55 | BRCA1 | No | High | Sister (BC, 45) | No | 0 of 8 | NA |
| 19 | F | 29 | BRCA2 | No | High | Mother (BC, 50), father (PC, 58) | Grandmother (BC, 40s), grandfather (PC, 60s) | NAb | NA |
| 20 | M | 43 | BRCA2 | No | Low | No | No | 1 of 6 | 0 |
| 21 | M | 47 | BRCA2 | No | Moderate | Father (PC, 72) | No | 0 of 2 | NA |
| 22 | F | 31 | BRCA2 | No | Limited | No | No | 0 of 1 | NA |
LP likely pathogenic, KP known pathogenic, BC breast cancer, OC ovarian cancer, PC prostate cancer, PAC pancreatic cancer.
aPer the National Comprehensive Cancer Network guidelines, high risk = one individual with two or more primary BCs, a first- or second-degree relative diagnosed with BC at ≤45 years of age, two or more relatives with BC, male BC, or a relative with OC; moderate risk = some HBOC-related diagnoses, but insufficient for high-risk qualification; low risk = no HBOC-related diagnosis reported.
bID19 choose not to attend the second visit and did not receive genetic risk information, and was thus lost to follow-up.
Of the 102 first- and second-degree relatives for whom cascade screening was recommended, 26 relatives of 10 index participants (13 male, 13 female; mean age, 41 [range, 19–76] years) responded and attended (25.5% response rate; Table S3). No information is available about whether the non-responding relatives (55 female, 47 male) were informed or about their decisions to not participate. Nine cascade screening respondents were offspring, eight were siblings, and two were mothers of the index participants; the other seven individuals were second-degree relatives. Cascade screening identified ten individuals as carriers of the family BRCA1/2 variant. Four cascade-screened individuals had previously received different cancer diagnoses, being early-onset breast cancer in two cases. The response to cascade screening was higher through invitation letters given by female index participants (62% of female index participants had at least one cascade participant respond compared to 25% for male index participants).
Response to disclosure
All 21 participants who received results completed the first survey, and 13 of these participants completed the second survey (62% response rate; Table S4). The response rate was higher among women (85%) than among men (25%).
Twenty of 21 index participants reported that they were glad to have been contacted about the genetic findings; only one participant was unsure of how she felt about being contacted (Table 3, Q1). All participants considered the information provided to be understandable, interesting, informative, and valuable (Table 3, Q2–Q5). At 6 months, all 13 respondents reported that they were coping with the genetic information received and had no regret regarding their decision to receive it (Table 3, Q21–Q26).
Table 3.
Responses to surveys administered immediately and 6 months after disclosure of genetic risk information.
| Theme | Item | Statement | Mean | Range | |
|---|---|---|---|---|---|
| Immediate response | Decision regreta | Q1 | I am glad that the Biobank contacted me regarding the genetic finding | 4.8 | 3–5 |
| Perceived impacta | Q2 | Information provided was understandable | 4.9b | 4–5 | |
| Q3 | Information provided was interesting | 4.7 | 4–5 | ||
| Q4 | Information provided was informative | 4.9 | 4–5 | ||
| Q5 | Information provided was valuable | 4.9 | 4–5 | ||
| Q6 | I understand the potential impact of the finding on my close relatives | 4.9 | 4–5 | ||
| Q7 | I can explain the potential impact of this genetic finding on health risks to my family members | 4.7 | 4–5 | ||
| Q8 | I know who to turn to regarding health concerns or for counseling | 4.8 | 4–5 | ||
| Emotional responsec | Q9 | I feel calm | 3.3 | 1–4 | |
| Q10 | I am tense | 1.8b | 1–4 | ||
| Q11 | I feel upset | 1.8b | 1–4 | ||
| Q12 | I am relaxed | 3.5 | 1–4 | ||
| Q13 | I feel content | 3.3 | 1–4 | ||
| Q14 | I am worried | 2.2b | 1–4 | ||
| 6-month follow-up | Emotional responsec | Q15 | I feel calm | 3.5 | 1–4 |
| Q16 | I am tense | 1.5d | 1–2 | ||
| Q17 | I feel upset | 1.7d | 1–2 | ||
| Q18 | I am relaxed | 3.3d | 2–4 | ||
| Q19 | I feel content | 2.9d | 1–4 | ||
| Q20 | I am worried | 1.8d | 1–3 | ||
| Coping and decision regreta | Q21 | I am able to cope with having this condition in my family | 4.8 | 4–5 | |
| Q22 | It was the right decision | 4.8 | 4–5 | ||
| Q23 | I regret the choice that was made | 1.3d | 1–2 | ||
| Q24 | I would go for the same choice if I had to do it over again | 4.8d | 4–5 | ||
| Q25 | The choice did me a lot of harm | 1.3d | 1–2 | ||
| Q26 | The decision was a wise one | 4.7d | 4–5 | ||
| Perceived impacta | Q27 | I now have better access to healthcare/specialists | 3.5 | 1–5 | |
| Q28 | I feel that my treatment and/or condition has improved | 2.8 | 1–4 | ||
| Q29 | The information received has somehow changed my life | 3.7 | 1–5 |
aResponses given on a five-point Likert scale: 5, agree; 4, slightly agree; 3, difficult to say; 2, slightly disagree; 1, disagree.
bTwenty of 21 participants responded.
cResponses given on a four-point scale: 4, very much; 3, moderately; 2, somewhat; 1, not at all.
dTwelve of 13 participants responded.
Immediately after receiving risk information with counseling, participants tended to feel content, calm, and relaxed (Table 3, Q9, Q12, and Q13), and only slightly or not at all upset, worried, or tense (Table 3, Q10, Q11, and Q14). However, the responses to these questions varied to a great extent. Overall, four individuals reported feeling tense and three reported feeling worried; one participant (ID.9) reported feeling tense, worried, and upset. Average response scores for these items were similar 6 months later (Table 3, Q15–Q20), although participants tended to feel less worried, upset, and tense than at the time of the first survey, with only one participant reporting feeling worried. Of the six individuals who reported feeling tense, worried, or upset immediately after receiving risk information, five responded to the 6-month survey and none of them reported having those negative feelings any longer. All 16 responding cascade screening participants reported that they were glad to have been contacted regarding the genetic finding in the family.
Medical impact of result disclosure
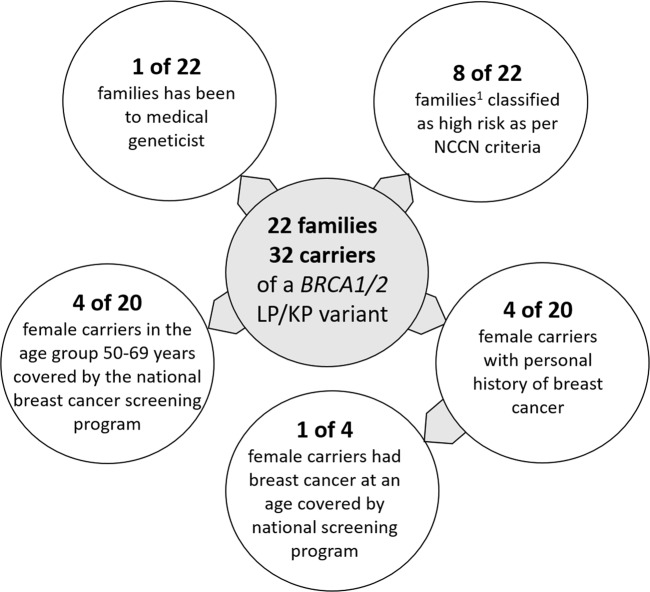
Currently, the clinical management of study participants have been dependent on the personal or family history of HBOC-related cancers or national breast cancer population screening program (Table S5). One of the 22 families studied reported familial risk management by a clinical geneticist prior to the study. Based on their personal and family histories, 8 (36.5%) of the 22 index participants qualified for HBOC genetic assessment, according to the NCCN criteria (Fig. 2). The other 14 participants did not qualify for HBOC assessment according to the current guidelines because they had insufficient family histories or very limited information available. Four of 20 female carriers belonged to the age group covered by the national breast cancer screening program (Fig. 2). Of the four women with histories of breast cancer, only one belonged to that age group at the time of diagnosis.
Fig. 2. Added value of the genotype-first approach to the identification of high-risk individuals.
The number of high-risk individuals who have been to a medical geneticist is limited. Majority of the families do not meet the NCCN criteria to be classified as having high risk for HBOC. Only limited number of female carriers had breast cancer diagnosis at an age group covered by the national breast cancer screening program. 1Self-reported family history of hereditary breast and ovarian cancer-related cancers indicative of high risk.
All 19 women with BRCA1/2 variants were referred to an oncologist for the development of personal surveillance plans. Two of these women were already seeing an oncologist due to previous cancer diagnoses. Over the 12–30-month follow-up period, 10 (59%) of these 19 women followed the clinical surveillance plan according to the HBOC guidelines (Table 4). Such adherence was not associated with participants’ age, whether they had children, family histories, or residence (rural or urban; proximity to a hospital).
Table 4.
Medical adherence among female index and cascade individuals with LP/KP BRCA1/2 variants.
| ID | Age | BSO/USO and surgery year | Mastectomy and surgery year | Regular surveillance | Ca.dx. and age at dx. | Family history | 1st degree relatives BC/OC/PC/PCA dx. and age at dx. |
|---|---|---|---|---|---|---|---|
| 1 | 34 | Yes (USO, 2017) | – | Yes | No | High | Mother (OC, 50) |
| 2 | 47 | Yes (BSO, 2017) | – | Yes | No | Moderate | Mother (BC, 48) |
| 4 | 52 | Yes (BSO, 2017) | – | Yes | No | Moderate | No |
| 5 | 39 | Yes (BSO, 2017) | – | Yes | No | High | Mother (OC, 55) |
| 7 | 59 | No | – | No | No | Low | No |
| 8 | 75 | No | Yes (UL, 2018)a | Yes | BC, 75 | High | Daughter (OC, 42) |
| 9 | 75 | N/Ab (BSO, 1989) | Yes (UL, 2002)b | N/A | BC, 53 | Moderate | Brother (PAC, 59) |
| 10 | 33 | No | – | Yes | No | High | Mother (BC, 38) |
| 12 | 46 | Yes (BSO, 2017) | – | No | No | Moderate | No |
| 14 | 31 | No | – | No | No | Moderate | No |
| 15 | 42 | No | – | No | No | Moderate | No |
| 18 | 55 | No | – | No | No | High | Sister (BC, 45) |
| 19c | 29 | N/A | N/A | N/A | No | High | Mother (BC, 50), father (PC, 58) |
| 22 | 31 | No | – | Yes | No | Limited | No |
| 2.1 | 76 | N/Ab (BSO, 1990) | Yes (UL, 1990)b | No | BC, 49 | Moderate | No |
| 10.1 | 58 | No | Yes (UL, 1999)b | Yes | BC, 39 | High | No |
| 12.1d | 21 | N/A | N/A | N/A | No | High | No |
| 4.2 | 29 | No | – | Yes | No | Moderate | No |
| 5.2 | 38 | No | – | No | No | High | Mother (OC, 59) |
| 17.2 | 34 | Yes (BSO, 2018) | Partial breast sector resection, 2018 | Yes | No | Moderate | No |
BSO bilateral salpingo-oophorectomy, USO unilateral salpingo-oophorectomy, BC breast cancer, OC ovarian cancer, PC prostate cancer, PAC pancreatic cancer, UL unilateral mastectomy.
aRisk-reducing surgery performed during this study due to previous diagnosis.
bRisk-reducing surgeries performed due to previous diagnoses before this study.
cThis individual chose not to attend the second visit and did not receive genetic risk information.
dThis individual was below the recommended age limit for medical surveillance.
The NCCN guidelines for the management of individuals with high HBOC risk include recommendations for risk reduction surgery, depending on the subject’s age. Three of the 19 women with BRCA1/2 variants had previously undergone unilateral mastectomy due to breast cancer. This includes two women with bilateral salpingo-oophorectomy (BSO) performed as part of empirical recurrence risk management. A 21-year-old woman (ID12.1) was considered to be too young for regular clinical surveillance or preventive surgery. During the follow-up period, 5 (31.3%) of 15, eligible for preventive surgery, women chose to undergo preventive BSO with an average age of 43.6 (age range, 34–52) years. Additionally, from therapeutic interventions, one participant underwent unilateral salpingo-oophorectomy when a nonmalignant lesion was detected. Another participant underwent partial breast sector resection due to a finding after the study-related oncologist visit. No woman chose to undergo prophylactic mastectomy following the return of results. The eight women who did not choose to undergo risk-reducing surgeries during the follow-up period were aged 29–59 (mean, 40) years.
Discussion
This study highlights the significant potential of population-based genomic studies for personal risk evaluation and population-wide risk-based management at the national healthcare level. It provides critical evidence for the application of genotype-first screening to improve long-term outcomes for high-risk individuals in the population, many of whom are unaware of their genetic risk. Most importantly, they were not captured by current clinical practice, which emphasizes the importance of the population-based genomic screening.
Potential of existing datasets
Population-based biobanks provide an excellent resource to study the frequency and penetrance of clinically significant genetic variants in unselected cohorts. A genotype-first approach to the analysis of unselected data yields an unbiased estimate of HBOC-related cancer prevalence in the general population, rather than solely in multiplex families. Intensive monitoring and early intervention can improve outcomes in carriers with no positive family history [18]. The information on HBOC-related LP/KP variants affects the therapy of choice (e.g., surgery approach, the use of PARP inhibitors in advanced cancer, etc.) [37].
The prevalence of LP/KP BRCA1/2 variants in our study cohort was 0.80% (1/124), previous prevalence data for BRCA1 (7.6%) and BRCA2 (12.5%) was on clinical cohort [34] of women with breast cancer diagnoses or predictive cases. The actual prevalence of BRCA1/2 pathogenic variants is uncertain, as estimates are based largely on data from clinical cohorts, rather than general populations expected to be cancer free. The approach used in this study led to the identification of previously unknown carriers of LP/KP variants in BRCA1/2, irrespective of personal or family history.
Population biobanks are also a good resource for the identification of novel genetic variants that may be clinically significant. One novel, presumably pathogenic, BRCA1 variant, c.2178delT p.(Pro727Glnfs*9), was identified in this study. The broad consent that biobank participants provide, the accessibility of biological sample collection, and the availability of long-term follow-up data in biobank datasets enable researchers to access and analyze information, including that from biobank participants who are deceased or cannot be contacted. The availability of multiple in silico pathogenicity evaluation scores and medical diagnoses enable estimation of the pathogenicity of novel variants, even when databases such as gnomAD, dbSNP, and ClinVar have no available data. Such variants may be population specific, yet still clinically relevant.
This study also revealed some limitations related to the approaching of high-risk individuals in the biobank setting. These limiting factors include difficulties with contact, as participants may have been contacted last more than a decade previously. Another challenge involved the composition of an invitation letter that respected participants’ right to not receive information while being sufficiently informative to allow participants to decide whether they were interested in participating in the present study. These factors could have contributed to the 55% response rate. The response rate to cascade screening (25.5%) was probably impacted, at least in part, by index individuals’ gateway roles. The response to cascade screening was higher through female than through male index participants, indicating the need for improved communication with gender-specific heritable disorders [38, 39]. The response rates in previous EstBB studies with similar approach had response rates of 89% [5] and 78% [2] among index and 57% [2] among cascade individuals. Illustrating that even small changes to the approach such as flexibility of time and location for receiving results or the specificity of the invite letter can influence the response rates. In other studies as reviewed in Menko et al., the uptake for presymptomatic DNA testing for HBOC in at-risk family members is similar to what we observed, reportedly ranging from 15 to 57% [40].
The contacting of family members for cascade screening is one way to maximize the impact of a population biobank. Further investigation is needed to determine the best way to increase response rates for optimization of this cascade approach.
Risk-based screening
As in many other countries, the national breast cancer screening program in Estonia targets women in a limited age group (50–69 years), thereby excluding some women who are potentially at high risk. In the present cohort, two women had received breast cancer diagnoses by the age of 50 years. Despite the broad access to medical services in Estonia, the medical system consistently misses individuals with greater cancer risk. In our cohort, only 1 of 22 participating families had previously visited a clinical geneticist. These findings suggest that the majority of high-risk variant carriers are unaware of their genetic predisposition to HBOC-related cancers and are not under optimal surveillance.
The efficiency of breast cancer risk assessment based on family history depends on family size, family members’ ages, the sex distribution of high-risk genetic variants, and the availability of detailed individual knowledge of cancer in the family. For instance, our cohort contained three families with multiple diagnoses of cancer, but the index individuals had no information about primary disease locations. Additionally, the majority of family histories did not fulfill the current criteria for high risk [17].
Genetic predisposition to cancer is usually suspected when the disease is diagnosed at a relatively young age. This pattern is largely true in familial cases seen in clinical settings, although the application of a genotype-first approach to an unselected population reveals great variability in the onset and prevalence of cancer. Our results indicate that age and family history alone are poor discriminators of breast cancer risk. We thus propose that existing genomic data be examined whenever possible during risk assessment.
Risk-based management
The guidelines for women with suspected hereditary predisposition to HBOC recommend earlier and more frequent screening than regular age dependent screening and optional preventive surgery [41]. In our cohort, 59% of female carriers followed the clinical surveillance recommendations during the follow-up period. Women with high-risk BRCA1/2 variants tend to prefer surveillance for breast cancer. The uptake of risk-reducing surgeries in our cohort was lesser than reported previously, with 31% of our participants choosing to undergo BSO compared with the 46% reported by Rowley et al. [1]. and no participant choosing to undergo bilateral mastectomy. Unfortunately there is no general registry for genetic disorders in Estonia, including hereditary cancer syndromes. However, the uptake of prophylactic mastectomy among patients is low. The uptake of risk-reducing procedures likely depends on different populations and cohorts, average age and other contextual factors including availability and reimbursement, clinical guidelines and traditions; these factors were currently not studied.
A possible limiting factor for long-term follow-up surveillance is that we obtained clinical follow-up data on individuals who participated in cascade screening from the collaborating oncologist and participating central hospital. Thus, these data may not reflect participants’ complete medical histories, including potential consultation with other oncologists and the use of surveillance services in the private sector. Thus, the actual rate of medical adherence may be somewhat higher than revealed by our analyses.
Population-scale impacts of screening
Our analysis demonstrated that EstBB participants and their family members who participated in cascade screening appreciated receiving their screening results. This finding is likely generalizable to the general population.
Population biobank data provide information on the potential impact of population screening for high-risk individuals using tools currently offered to a limited group of individuals with significant personal or family histories indicative of genetic predisposition. The results of analogous genotype-first projects inform us about perceptions regarding the receipt of unexpected genetic results. Long-term follow-up data on high-risk variant carriers in the general population will aid assessment of the clinical utility of population screening. Läll et al. suggests polygenic risk together with germline variant testing could be an efficient complimentary tool for risk stratification in clinical practice for better screening and prevention [42].
At the end of 2019, more than 200,000 EstBB participants had been recruited. Array-based genotype data for all of these participants will become available by June 2020, and will be implemented as an integral part of the national personalized medicine initiative. Based on the 0.82% estimated prevalence of LP/KP variants from this study, more than 1000 EstBB participants could be BRCA1/2 variant carriers. Considering a reduction in the detection rate due to the long-range phasing nature of array-based data and a response rate of ~50%, the application of a genotype-first approach to HBOC and breast cancer screening in EstBB could impact up to 1600 potential BRCA1/2 carriers among EstBB participants alone, demonstrating the importance of this pilot evaluation of this type of approach.
Supplementary information
Acknowledgements
We express our sincere thanks to the biobank participants and their relatives for participating in the study.
Funding
This research was supported by the European Union through the European Regional Development Fund (project no. 2014-2020.4.01.15-0012 GENTRANSMED), European Union Horizon 2020 (grant no. 810645, no. 654248), Estonian Research Council (PUT736 to NT, PUT PRG555 to NT, IUT20-60, PUT1660 to TE, PUTJD817 to MK, MOBERA15, RITA1/01-42-03).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Liis Leitsalu, Marili Palover
Supplementary information
The online version of this article (10.1038/s41431-020-00760-2) contains supplementary material, which is available to authorized users.
References
- 1.Rowley SM, Mascarenhas L, Devereux L, Li N, Amarasinghe KC, Zethoven M, et al. Population-based genetic testing of asymptomatic women for breast and ovarian cancer susceptibility. Genet Med. 2018;21:913–22. doi: 10.1038/s41436-018-0277-0. [DOI] [PubMed] [Google Scholar]
- 2.Alver M, Palover M, Saar A, Läll K, Zekavat SM, Tõnisson N, et al. Recall by genotype and cascade screening for familial hypercholesterolemia in a population-based biobank from Estonia. Genet Med. 2019;21:1173–80. doi: 10.1038/s41436-018-0311-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Haukkala A, Kujala E, Alha P, Salomaa V, Koskinen S, Swan H, et al. The return of unexpected research results in a biobank study and referral to health care for heritable long QT syndrome. Public Health Genomics. 2013;16:241–50. doi: 10.1159/000354105. [DOI] [PubMed] [Google Scholar]
- 4.Manickam K, Buchanan AH, Schwartz MLB, Hallquist MLG, Williams JL, Rahm AK, et al. Exome sequencing-based screening for BRCA1/2 expected pathogenic variants among adult biobank participants. JAMA Netw Open. 2018;1:e182140. doi: 10.1001/jamanetworkopen.2018.2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Leitsalu L, Alavere H, Jacquemont S, Kolk A, Maillard AM, Reigo A, et al. Reporting incidental findings of genomic disorder-associated copy number variants to unselected biobank participants. Per Med. 2016;13:303–14. doi: 10.2217/pme-2016-0009. [DOI] [PubMed] [Google Scholar]
- 6.Vornanen M, Aktan-Collan K, Hallowell N, Konttinen H, Haukkala A. Lay perspectives on receiving different types of genomic secondary findings: a qualitative vignette study. J Genet Couns. 2018;28:343–54. doi: 10.1007/s10897-018-0288-7. [DOI] [PubMed] [Google Scholar]
- 7.Francke U, Dijamco C, Kiefer AK, Eriksson N, Moiseff B, Tung JY, et al. Dealing with the unexpected: consumer responses to direct-access BRCA mutation testing. PeerJ. 2013;1:e8. doi: 10.7717/peerj.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Budin-ljøsne I, Mascalzoni D, Soini S, Machado H, Kaye J, Bentzen HB, et al. Feedback of individual genetic results to research participants. Biopreserv Biobank. 2016;14:241–8. doi: 10.1089/bio.2015.0115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Riigikogu. Human Genes Research Act. 2000. https://www.riigiteataja.ee/en/eli/508042019001/consolide.
- 10.Szabo C, King M. Inherited breast and ovarian cancer. Hum Mol Genet. 1995;4:1811–7. doi: 10.1093/hmg/4.suppl_1.1811. [DOI] [PubMed] [Google Scholar]
- 11.Miki Y, Swensen J, Shattuck-Eidens D, Futreal PA, Harshman K, Tavtigian S, et al. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science. 1994;266:66–71. doi: 10.1126/science.7545954. [DOI] [PubMed] [Google Scholar]
- 12.Wooster R, Bignell G, Lancaster J, Swift S, Seal S, Mangion J, et al. Identification of the breast cancer susceptibility gene BRCA2. Nature. 1995;378:789–92. doi: 10.1038/378789a0. [DOI] [PubMed] [Google Scholar]
- 13.McClain MR, Palomaki GE, Nathanson KL, Haddow JE. Adjusting the estimated proportion of breast cancer cases associated with BRCA1 and BRCA2 mutations: public health implications. Genet Med. 2005;7:28–33. doi: 10.1097/01.GIM.0000151155.36470.FF. [DOI] [PubMed] [Google Scholar]
- 14.Hartmann LC, Lindor NM. The role of risk-reducing surgery in hereditary breast and ovarian cancer. Obstet Gynecol Surv. 2016;71:598–9. doi: 10.1097/01.ogx.0000499510.12280.54. [DOI] [PubMed] [Google Scholar]
- 15.Kuchenbaecker KB, Hopper JL, Barnes DR, Phillips KA, Mooij TM, Roos-Blom MJ, et al. Risks of breast, ovarian, and contralateral breast cancer for BRCA1 and BRCA2 mutation carriers. JAMA. 2017;317:2402–16. doi: 10.1001/jama.2017.7112. [DOI] [PubMed] [Google Scholar]
- 16.Balmaña J, Díez O, Rubio IT, Cardoso F. BRCA in breast cancer: ESMO clinical practice guidelines. Ann Oncol. 2011;22:31–34. doi: 10.1093/annonc/mdr373. [DOI] [PubMed] [Google Scholar]
- 17.National Comprehensive Cancer Network (NCCN). Breast Cancer Risk Reduction. NCCN Guidelines Version 1.2020.
- 18.Gabai-Kapara E, Lahad A, Kaufman B, Friedman E, Segev S, Renbaum P, et al. Population-based screening for breast and ovarian cancer risk due to BRCA1 and BRCA2. Proc Natl Acad Sci. 2014;111:14205–10. doi: 10.1073/pnas.1415979111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leitsalu L, Haller T, Esko T, Tammesoo M-L, Alavere H, Snieder H, et al. Cohort Profile: Estonian Biobank of the Estonian Genome Center, University of Tartu. Int J Epidemiol. 2015;44:1137–47. doi: 10.1093/ije/dyt268. [DOI] [PubMed] [Google Scholar]
- 20.McLaren W, Gil L, Hunt SE, Riat HS, Ritchie GRS, Thormann A, et al. The ensembl variant effect predictor. Genome Biol. 2016;17:1–14. doi: 10.1186/s13059-016-0974-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang H, Wang K. Genomic variant annotation and prioritization with ANNOVAR and wANNOVAR. Nat Protoc. 2015;10:1556–66. doi: 10.1038/nprot.2015.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Delaneau O, Howie B, Cox AJ, Zagury JF, Marchini J. Haplotype estimation using sequencing reads. Am J Hum Genet. 2013;93:687–96. doi: 10.1016/j.ajhg.2013.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.R Core Team. R: A language and environment for statistical computing. 2018. https://www.r-project.org/.
- 25.Landrum MJ, Lee JM, Benson M, Brown GR, Chao C, Chitipiralla S, et al. ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 2018;46:D1062–67. doi: 10.1093/nar/gkx1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Freeman PJ, Hart RK, Gretton LJ, Brookes AJ, Dalgleish R. VariantValidator: accurate validation, mapping, and formatting of sequence variation descriptions. Hum Mutat. 2018;39:61–68. doi: 10.1002/humu.23348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sherry S, Ward M, Kholodov M, Baker J, Phan L, Smigielski E, et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–11. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–24. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gray SW, Martins Y, Feuerman LZ, Bernhardt BA, Biesecker BB, Christensen KD, et al. Social and behavioral research in genomic sequencing: approaches from the Clinical Sequencing Exploratory Research Consortium and Measures Working Group. Genet Med. 2014;16:727–35. doi: 10.1038/gim.2014.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marteau TM, Bekker H. The development of a six-item short-form of the state scale of the Spielberger State—Trait Anxiety Inventory (STAI) Br J Clin Psychol. 1992;31:301–6. doi: 10.1111/j.2044-8260.1992.tb00997.x. [DOI] [PubMed] [Google Scholar]
- 31.Brehaut JC, O’Connor AM, Wood TJ, Hack TF, Siminoff L, Gordon E, et al. Validation of a decision regret scale. Med Decis Mak. 2003;23:281–92. doi: 10.1177/0272989X03256005. [DOI] [PubMed] [Google Scholar]
- 32.Berkenstadt M, Shiloh S, Barkai G, Katznelson MBM, Goldman B. Perceived personal control (PPC): a new concept in measuring outcome of genetic counseling. Am J Med Genet. 1999;82:53–59. doi: 10.1002/(SICI)1096-8628(19990101)82:1<53::AID-AJMG11>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 33.Szabo C, Masiello A, Ryan JF, Brody LC. The breast cancer information core: database design, structure, and scope. Hum Mutat. 2000;16:123–31. doi: 10.1002/1098-1004(200008)16:2<123::AID-HUMU4>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 34.Tamboom K, Kaasik K, Aršavskaja J, Tekkel M, Lilleorg A, Padrik P, et al. BRCA1 mutations in women with familial or early-onset breast cancer and BRCA2 mutations in familial cancer in Estonia. Hered Cancer Clin Pr. 2010;8:4. doi: 10.1186/1897-4287-8-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hamel N, Feng BJ, Foretova L, Stoppa-Lyonnet D, Narod SA, Imyanitov E, et al. On the origin and diffusion of BRCA1 c.5266dupC (5382insC) in European populations. Eur J Hum Genet. 2011;19:300–6. doi: 10.1038/ejhg.2010.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Borg Å, Haile RW, Malone KE, Capanu M, Diep A, Törngren T, et al. Characterization of BRCA1 and BRCA2 deleterious mutations and variants of unknown clinical significance in unilateral and bilateral breast cancer: The WECARE study. Hum Mutat 2010;31. 10.1002/humu.21202. [DOI] [PMC free article] [PubMed]
- 37.Tung NM, Garber JE. BRCA1/2 testing: therapeutic implications for breast cancer management. Br J Cancer. 2018;119:141–52. doi: 10.1038/s41416-018-0127-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rauscher EA, Dean M, Campbell-Salome G, Barbour JB. “How do we rally around the one who was positive?” Familial uncertainty management in the context of men managing BRCA-related cancer risks. Soc Sci Med. 2019;242:112592. doi: 10.1016/j.socscimed.2019.112592. [DOI] [PubMed] [Google Scholar]
- 39.Menko FH, Jeanson KN, Bleiker EMA, Van Tiggelen CWM, Hogervorst FBL, Jacqueline A, et al. The uptake of predictive DNA testing in 40 families with a pathogenic BRCA1/BRCA2 variant. An evaluation of the proband-mediated procedure. Eur J Hum Genet. 2020. 10.1038/s41431-020-0618-8. [DOI] [PMC free article] [PubMed]
- 40.Menko FH, ter Stege JA, van der Kolk LE, Jeanson KN, Schats W, Moha DA, et al. The uptake of presymptomatic genetic testing in hereditary breast-ovarian cancer and Lynch syndrome: a systematic review of the literature and implications for clinical practice. Fam Cancer. 2019;18:127–35. doi: 10.1007/s10689-018-0089-z. [DOI] [PubMed] [Google Scholar]
- 41.Hampel H, Bennett RL, Buchanan A, Pearlman R, Wiesner GL. A practice guideline from the American College of Medical Genetics and Genomics and the National Society of Genetic Counselors: referral indications for cancer predisposition assessment. Genet Med. 2015;17:70–87. doi: 10.1038/gim.2014.147. [DOI] [PubMed] [Google Scholar]
- 42.Läll K, Lepamets M, Palover M, Esko T, Metspalu A, Tõnisson N, et al. Polygenic prediction of breast cancer: comparison of genetic predictors and implications for risk stratification. BMC Cancer. 2019;19:557. doi: 10.1186/s12885-019-5783-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.