Abstract
Pathogenic variants in TP53 have been classically thought to cause Li-Fraumeni syndrome (LFS), a cancer predisposition with high risks for various childhood- and adult-onset malignancies. However, increased genetic testing has lately revealed, that pathogenic variant carriers exhibit a broader range of phenotypes and that penetrance may be dependent both on variant type and modifiers. Using next generation sequencing and short tandem repeat analysis, we identified germline pathogenic variants in TP53 and RAD51C located in cis on chromosome 17 in a 43-year-old male, who has developed a rare sebaceous gland carcinoma (SGC) but so far no tumors of the LFS spectrum. This course mirrors a Trp53-Rad51c-double-mutant cis mouse-model, which similarly develops SGC, while the characteristic Trp53-associated tumor spectrum occurs with significantly lower frequency. Therefore, we propose that co-occurent pathogenic variants in RAD51C and TP53 may predispose to SGC, reminiscent of Muir-Torre syndrome. Further, this report supports the diversity of clinical presentations associated with germline TP53 alterations, and thus, the proposed expansion of LFS to heritable TP53-related cancer syndrome.
Subject terms: Genetics research, Cancer genetics
Introduction
Li-Fraumeni syndrome (LFS) is an autosomal dominantly inherited multicancer predisposition covering a spectrum of five core malignancies: breast cancer, soft tissue or bone sarcoma, brain tumors, and adrenocortical carcinoma [1, 2]. The genetic basis of LFS are germline pathogenic variants in TP53 [1, 3]. Carriers have a 58% risk of developing cancer before age 40 and about 80% before age 70 [4]. Penetrance varies according to age, sex, and variant type [4]. Thus, penetrance is higher in males than in females during childhood and adolescence, but by age 35 the initial male bias is offset by the burden of breast cancer in women [4]. Lifetime risk of developing cancer has been estimated to 70% or higher for men, while women’s risk is close to 100% [4–7]. The predominance of familial cases included in these studies likely leads to an overestimation of the disease penetrance [1]. Thus, estimating the cancer risk for TP53 variant carriers remains a great challenge [1]. However, the fact that about 20% of carriers detected in a familial context do not develop cancer until the age of 70 years, suggests that additional genetic or nongenetic factors may create an environment that is either promoting, or restricting LFS development. The presence of modulators may also explain the higher than expected frequency of pathogenic TP53 variants in the general population [8].
In support of genetic modulators of TP53, a mouse model harboring a Trp53-null-allele and a Rad51c-null-allele displayed different phenotypes depending on the location of the mutant alleles (on the same (cis) or on the alternate mouse chromosome 11 (trans)). Trans mice developed tumors with latency and spectrum similar to LFS, while cis mice had sebaceous glands carcinoma (SGC) and developed fewer tumors characteristic of Trp53-null-allele [9] (for a detailed description of the mouse model and its limitations see Supplementary Material).
Germline pathogenic variants in RAD51C predispose to hereditary breast and ovarian cancer [10, 11], and, to our knowledge, SGC does not belong to the tumor spectrum. SGC is frequently observed in the context of Muir-Torre syndrome (MTS) due to mismatch repair (MMR) deficiency related to hereditary non-polyposis colorectal carcinoma syndrome [12], but do not belong to the typical LFS-related tumor spectrum. To our knowledge SGC has been previously described in only one patient with a pathogenic TP53 variant (c.818G>A, p.(Arg273His)) [13]; in this case, it remains unclear whether pathogenic variants affecting other genes co-occurred.
We present a patient that harbors a pathogenic missense variant in TP53 located in cis with a deletion within RAD51C on human chromosome 17. Based on the IARC TP53 [14] database about 71% of the pathogenic variations are missense. Our patient is a 43-year-old male, who was diagnosed with a mid-occipital basal cell carcinoma (BCC) at age 38. At the age of 41, a right upper lid SGC was diagnosed. Both tumors had been fully excised shortly after diagnosis and the further clinical course was uneventful without any additional treatment necessary. Apart from colon cancer of the maternal grandmother at >80 years, family history is negative for any tumor diseases. This is not unexpected since 7–20% of the TP53 variants occur de novo [15, 16], and hence tumor family history is absent. The proband has two healthy 4 year-old twin children of different gender (Fig. 1C).
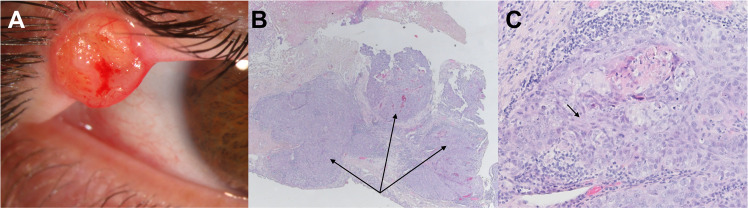
Fig. 1. Presentation of the sebaceous gland carcinoma.
A Clinical appearance of the sebaceous gland carcinoma as a right upper lid mass. B Hematoxylin eosin staining of the resected mass (10×). The arrow indicates irregular lobules and sheets of atypical sebaceous cells. C Hematoxylin eosin staining of the resected mass (20×). The arrows indicate undifferentiated, atypical cells with considerable nuclear pleomorphism and eosinophilic cytoplasm.
Although our observations are limited given the LFS known penetrance of around 58% at the age of 40 [4], the proband appears to recapitulate the mouse phenotype with no LFS typical tumors, but SGC reminiscent of MTS.
Methods
Histologic examination
An initial biopsy of the tumor followed by hematoxylin and eosin staining showed a malignant epithelial tumor containing basophilic sebaceous tumor cells with a nodular and in part trabecular order and no peripheral palisading.
Molecular and bioinformatic analyses
Genomic DNA was extracted from peripheral blood of the proband and his relatives, and, to determine whether the variant is of germline-origin or mosaicism, in the proband also from finger nails. We performed panel sequencing (Illumina TruSight Cancer Panel) targeting 94 cancer-related genes (Table S1 in the Supplementary Material) on a NextSeq500/550 High Output platform. Coverage of at least 20-fold was obtained for 99.4% of the target sequences. We evaluated data using Varvis software (Limbus, Rostock). Copy number variations (CNV) were analyzed using the coverage information of the panel with the current version of the Varfeed CNV software. The deletion of exons 5–9 in RAD51C was validated by Multiplex Ligation-dependent Probe Amplification (MLPA) (Kit P260, MRC-Holland) and the TP53 variant was validated by Sanger sequencing.
To investigate possible recombination events we performed a short tandem repeat (STR) analysis in the proband, his parents, and his children using loci located on chromosome 17 (Table S2 in the Supplementary Material).
To assess a potential loss of heterozygosity (LOH) in the tumor, we performed the same panel sequencing. The proportion of normal tissue within the tumor section was approx. 30%. Of the target sequences 99.9% were covered at least 20 times. Variant calling was performed with Strelka2 [17], to increase sensitivity. We used samtools [18] to determine the read depth. For a detailed description of the methods, see the Supplementary Material.
Results
Diagnostic evaluation
Histological examination of the resected tumor established the diagnosis of an SGC (Fig. 1A–C).
In line with the normal MMR protein expression patterns (Fig. S1 in the Supplementary Material), we did not detect any sequence alteration in MMR genes by panel sequencing of DNA from blood and tumor. However, we detected the heterozygous missense germline variant chr17:7,578,536; NM_000546.5, c.394A>G, p.(Lys132Glu) in TP53, which was classified as pathogenic [19] (Supplementary Material). The variant is located in the DNA-binding domain of p53 and functional examination in yeast revealed a dominant negative (DN) effect of the altered allele with a mean residual activity of 1% or less compared to wild-type [20, 21]. The variant has been described in the context of Li-Fraumeni [21, 22] in one pedigree and has been recently classified as a true recurrent pathogenic LFS variant [8] (for phenotype description and variant classification see Supplementary Material). CNV analysis of the proband further revealed a deletion of exons 5–9 of RAD51C (NM_058216.2, c.(235 + 1_236-1)_(*120_?)del), located also on chromosome 17. This deletion has already been described as pathogenic in the context of familial breast and ovarian cancer [23–25] (for phenotype description and variant classification see Supplementary Material). Both variants were validated using a second detection method (Fig. 2A, B) and in an additional tissue (Figs. S2A and S3A in the Supplementary Material). No further clinically relevant variants could be identified in cancer-relevant genes using the above panel.
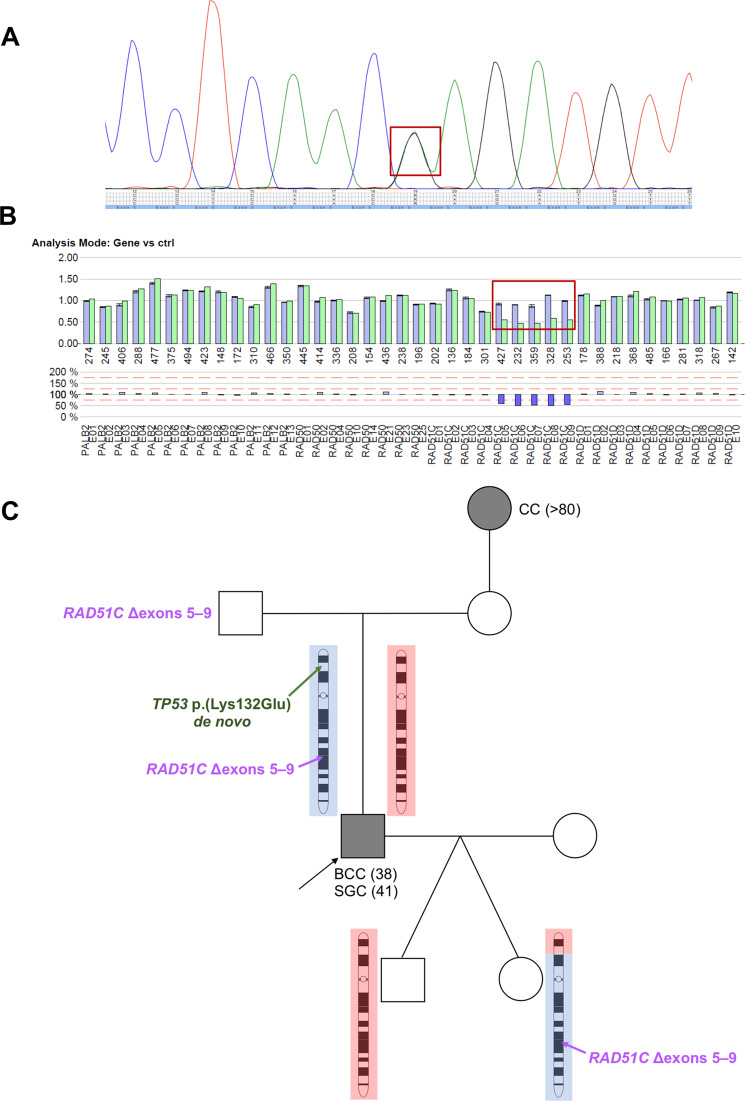
Fig. 2. Identified pathogenic variants.
A Electropherogram of the Sanger sequencing and the heterozygous NM_000546.5, c.394A>G TP53 variant (marked in the red quadrant). B MLPA validation of the exons 5–9 deletion, NM_058216.2 in RAD51C (marked in the red quadrant). C Pedigree chart of the family: the proband is indicated with an arrow. The origin of the paternal (blue) and maternal (red) chromosome has been established based on STR markers length (Fig. S4 in Supplementary Material). The son inherited the chromosome originating from his paternal grandmother, without recombination, and with wild type alleles of TP53 and RAD51C. The daughter inherited a recombined chromosome 17 with the beginning of p arm originating from the paternal grandmother, with a wild type TP53 allele, and the rest originating from the paternal grandfather, harboring the deletion within RAD51C. Location of the de novo mutated TP53 allele (in green) on the paternal originating chromosome of the proband, thus situated in cis with the deletion within RAD51C. CC colon carcinoma, BCC basal cell carcinoma, SGC sebaceous gland carcinoma, age at diagnosis in years is given between brackets.
Chromosomal localization of the variants
Segregation analysis in the proband’s family revealed that the deletion within RAD51C is paternally inherited, while the TP53 variant occurred de novo (Pedigree in Figs. 2C and S2 in the Supplementary Material). Phasing of the two variants was done using STR analysis (Fig. S4 in the Supplementary Material). We initially excluded the TP53 variant in both children of the proband (Figs. 2C and S2D–E in the Supplementary Material). Using STR analysis we show that the daughter inherited a recombined chromosome 17, while there was no evidence for a crossing-over event in the son (Figs. 2C and S4, Table S3 in the Supplementary Appendix). In accordance with the STR analysis, we confirmed the deletion within RAD51C in the daughter (Figs. 2C and S3 in the Supplementary Material). We evaluated the probability of recombination for the TP53 and RAD51C variants to be 31.6%, using , where d represents the distance in centimorgans (the two genes are at ca. 51 × 106 bp apart corresponding roughly to 51 centimorgans [26]). Thus, the TP53 variant occurred de novo on the paternal chromosome of the proband, and thereafter in cis with the deletion within RAD51C (Fig. S4 in the Supplementary Material).
Discussion
Here, we describe a proband with a de novo pathogenic TP53 variant associated with an in cis inherited pathogenic deletion within RAD51C (Fig. 2C). Since the TP53 variant occurred de novo the family history for LFS was absent. It is remarkable that, although the proband harbors a variant of severe deficiency with a dominant negative effect, known to significantly increase cancer risk [21], no LFS-typical tumors occurred until his current age of 43 years. This is in high accordance with a Trp53-Rad51c-double-mutant mouse model, which carries the mutant alleles in cis [9]. The Rad51c loss promoted SGCs and skin malignancies, but reduced tumors characteristic of Trp53-mutant mice [9]. The human proband also developed SGC and a BCC, recapitulating the mouse phenotype.
To date, MTS due to dysfunctional MMR is the only known autosomal dominant mendelian condition predisposing to SGC. In both our proband and the mouse model, [9] MMR was intact (Fig. 1D–F), suggesting an alternative mechanism for these tumors. Indeed, p53 dysfunction was previously suggested to be a divergent pathway in the molecular pathogenesis of SGC that show strong nuclear p53 staining and intact MMR [27].
To our knowledge, SGC has been described in only one patient with a pathogenic TP53 variant [13]. It remains unclear whether this individual had either typical LFS as well as an additional predisposition to SGC, or potentially an underlying genetic disorder similar to the mechanism described in this manuscript (no information is available on sequence alterations of RAD51C in this published individual). Although further studies are required to validate and fully elucidate the molecular mechanism, our observations point towards two major clinical implications: (1) SGC could be related to co-occurrence of pathogenic TP53 and RAD51C variants, and cause a phenotype reminiscent of MTS independent of MMR deficiency. Moreover, TP53 was found to harbor the highest number of pathogenic variants in a set of SGCs [28], suggesting that germline pathogenic variants in TP53 potentially associated with other modifiers may be more frequent than expected. (2) In line with the observations in Trp53-Rad51c-double mutant cis mice [9], also human in cis co-occurrence of pathogenic TP53 and RAD51C variants may substantially transform the Li-Fraumeni phenotype to a predisposition to SGC; however, this may not preclude the development of LFS typical tumors.
In order to establish a clear genotype–phenotype correlation future patients with co-occurrence of TP53 and RAD51C pathogenic variants, additional to the mouse model are needed. Thus, our recommendation for the present patient was to undergo regular LFS screening [1] (for ethical considerations and patient management see Supplementary Material). Lately, multiple studies have demonstrated the phenotypic variability of TP53 pathogenic variants carriers [29, 30]. This has been related to both variant type and potential modifiers [1, 29, 30]. Bougeard and colleagues suggested that a clinical gradient can be identified in TP53 pathogenic variants carriers, depending on the variant type. Hence, they suggested that future studies should characterize genotype–phenotype correlations and modifiers of the phenotype, such that patients could benefit from a stratified clinical management [29]. Our brief report adds to the heterogeneity of the heritable TP53-related cancers and aims to raise awareness on potential modifiers. If this is confirmed by other studies a clinical management stratification could be implemented to the benefit of such patients.
Database submissions
Variants have been submitted to ClinVar (SUB8180034): https://www.ncbi.nlm.nih.gov/clinvar/
TP53: NM_000546.5:c.394A>G, SCV001429318;
RAD51C: NM_058216.2:c.(235 + 1_236-1)_(*120_?)del, SCV001438810.
Supplementary information
Acknowledgements
DLD is funded through the “Clinician Scientist Programm, Medizinische Fakultät der Universität Leipzig”. We are grateful that our patient and his family agreed in participating in this study. We thank Sandra Schinkel, Kathleen Lehmann, and Sophie Behrendt for their great technical assistance. Discussions with Torsten Schöneberg were very helpful in elucidating this case.
Author contributions
DLD designed the study, performed the genetic analyses, and contributed to the writing of the manuscript. JH performed genetic analyses and contributed to the writing of the manuscript. SN contacted the family, contributed to diagnosis, and contributed to the writing of the manuscript. MS performed genetic analyses. CM performed genetic analyses and contributed to the writing of the manuscript. EJ performed genetic analyses. RAJ performed genetic analyses and contributed to the writing of the manuscript. KM performed genetic analyses and contributed to the writing of the manuscript. AM performed histology and contributed to the writing of the manuscript. MZ diagnosed the sebaceous gland carcinoma and contributed to the writing of the manuscript. AlM diagnosed the sebaceous gland carcinoma and contributed to the writing of the manuscript. HB coordinated the histology analyses and contributed to histologic diagnosis. JRL coordinated the contact to patients, contributed to genetic diagnosis, and contributed to the writing of the manuscript.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This study was approved by the ethics committee of the University of Leipzig, Germany (224/16-ek and 402/16-ek). Informed consent was obtained from each tested individual prior to genetic testing (for thorough description and discussion of ethical and management aspects see Supplementary Material).
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Diana Le Duc, Email: gabriela-diana.leduc@medizin.uni-leipzig.de.
Johannes R. Lemke, Email: johannes.lemke@medizin.uni-leipzig.de
Supplementary information
The online version of this article (10.1038/s41431-020-00781-x) contains supplementary material, which is available to authorized users.
References
- 1.Frebourg T, Bajalica Lagercrantz S, Oliveira C, Magenheim R, Evans DG. Guidelines for the Li-Fraumeni and heritable TP53-related cancer syndromes. Eur J Hum Genet. 2020;26:1–8. [DOI] [PMC free article] [PubMed]
- 2.Li FP, Fraumeni JF., Jr Soft-tissue sarcomas, breast cancer, and other neoplasms. A familial syndrome? Ann Intern Med. 1969;71:747–52. doi: 10.7326/0003-4819-71-4-747. [DOI] [PubMed] [Google Scholar]
- 3.Malkin D, Li FP, Strong LC, Fraumeni JF, Jr, Nelson CE, Kim DH, et al. Germ line p53 mutations in a familial syndrome of breast cancer, sarcomas, and other neoplasms. Science. 1990;250:1233–8. doi: 10.1126/science.1978757. [DOI] [PubMed] [Google Scholar]
- 4.Amadou A, Achatz MIW, Hainaut P. Revisiting tumor patterns and penetrance in germline TP53 mutation carriers: temporal phases of Li-Fraumeni syndrome. Curr Opin Oncol. 2018;30:23–9. doi: 10.1097/CCO.0000000000000423. [DOI] [PubMed] [Google Scholar]
- 5.Guha T, Malkin D. Inherited TP53 mutations and the Li-Fraumeni syndrome. Cold Spring Harbor Perspect Med. 2017;7:a026187. [DOI] [PMC free article] [PubMed]
- 6.Mai PL, Best AF, Peters JA, DeCastro RM, Khincha PP, Loud JT, et al. Risks of first and subsequent cancers among TP53 mutation carriers in the National Cancer Institute Li-Fraumeni syndrome cohort. Cancer. 2016;122:3673–81. doi: 10.1002/cncr.30248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schneider K, Zelley K, Nichols KE, Garber J. Li-Fraumeni syndrome: GeneReviews®[Internet]. Seattle: University of Washington; 2019.
- 8.Soussi T, Leroy B, Devir M, Rosenberg S. High prevalence of cancer-associated TP53 variants in the gnomAD database: a word of caution concerning the use of variant filtering. Hum Mutat. 2019;40:516–24. doi: 10.1002/humu.23717. [DOI] [PubMed] [Google Scholar]
- 9.Kuznetsov SG, Haines DC, Martin BK, Sharan SK. Loss of Rad51c leads to embryonic lethality and modulation of Trp53-dependent tumorigenesis in mice. Cancer Res. 2009;69:863–72. doi: 10.1158/0008-5472.CAN-08-3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meindl A, Hellebrand H, Wiek C, Erven V, Wappenschmidt B, Niederacher D, et al. Germline mutations in breast and ovarian cancer pedigrees establish RAD51C as a human cancer susceptibility gene. Nat Genet. 2010;42:410–4. doi: 10.1038/ng.569. [DOI] [PubMed] [Google Scholar]
- 11.Osorio A, Endt D, Fernández F, Eirich K, de la Hoya M, Schmutzler R, et al. Predominance of pathogenic missense variants in the RAD51C gene occurring in breast and ovarian cancer families. Hum Mol Genet. 2012;21:2889–98. doi: 10.1093/hmg/dds115. [DOI] [PubMed] [Google Scholar]
- 12.Fusaro RM, Lemon SJ, Lynch HT. Muir-Torre syndrome and defective DNA mismatch repair genes. J Am Acad Dermatol. 1996;35:493–4. doi: 10.1016/S0190-9622(96)90644-1. [DOI] [PubMed] [Google Scholar]
- 13.Baumuller S, Herwig MC, Mangold E, Holz FC, Loeffler KU. Sebaceous gland carcinoma of the eyelid masquerading as a cutaneous horn in Li-Fraumeni syndrome. Br J Ophthalmol. 2011;95:1470–8. doi: 10.1136/bjo.2009.175158. [DOI] [PubMed] [Google Scholar]
- 14.Bouaoun L, Sonkin D, Ardin M, Hollstein M, Byrnes G, Zavadil J, et al. TP53 variations in human cancers: new lessons from the IARC TP53 database and genomics data. Hum Mutat. 2016;37:865–76. doi: 10.1002/humu.23035. [DOI] [PubMed] [Google Scholar]
- 15.Gonzalez KD, Buzin CH, Noltner KA, Gu D, Li W, Malkin D, et al. High frequency of de novo mutations in Li-Fraumeni syndrome. J Med Genet. 2009;46:689–93. doi: 10.1136/jmg.2008.058958. [DOI] [PubMed] [Google Scholar]
- 16.Renaux-Petel M, Charbonnier F, Théry JC, Fermey P, Lienard G, Bou J, et al. Contribution of de novo and mosaic TP53 mutations to Li-Fraumeni syndrome. J Med Genet. 2018;55:173–80. doi: 10.1136/jmedgenet-2017-104976. [DOI] [PubMed] [Google Scholar]
- 17.Kim S, Scheffler K, Halpern AL, Bekritsky MA, Noh E, Kallberg M, et al. Strelka2: fast and accurate calling of germline and somatic variants. Nat Methods. 2018;15:591–4. doi: 10.1038/s41592-018-0051-x. [DOI] [PubMed] [Google Scholar]
- 18.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–9. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–24.. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kato S, Han SY, Liu W, Otsuka K, Shibata H, Kanamaru R, et al. Understanding the function-structure and function-mutation relationships of p53 tumor suppressor protein by high-resolution missense mutation analysis. Proc Natl Acad Sci USA. 2003;100:8424–9. doi: 10.1073/pnas.1431692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Monti P, Perfumo C, Bisio A, Ciribilli Y, Menichini P, Russo D, et al. Dominant-negative features of mutant TP53 in germline carriers have limited impact on cancer outcomes. Mol Cancer Res. 2011;9:271–9. doi: 10.1158/1541-7786.MCR-10-0496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goi K, Takagi M, Iwata S, Delia D, Asada M, Donghi R, et al. DNA damage-associated dysregulation of the cell cycle and apoptosis control in cells with germ-line p53 mutation. Cancer Res. 1997;57:1895–902. [PubMed] [Google Scholar]
- 23.Carter NJ, Marshall ML, Susswein LR, Zorn KK, Hiraki S, Arvai KJ, et al. Germline pathogenic variants identified in women with ovarian tumors. Gynecol Oncol. 2018;151:481–8. doi: 10.1016/j.ygyno.2018.09.030. [DOI] [PubMed] [Google Scholar]
- 24.Schnurbein G, Hauke J, Wappenschmidt B, Weber-Lassalle N, Engert S, Hellebrand H, et al. RAD51C deletion screening identifies a recurrent gross deletion in breast cancer and ovarian cancer families. Breast Cancer Res. 2013;15:R120. doi: 10.1186/bcr3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schubert S, van Luttikhuizen JL, Auber B, Schmidt G, Hofmann W, Penkert J, et al. The identification of pathogenic variants in BRCA1/2 negative, high risk, hereditary breast and/or ovarian cancer patients: high frequency of FANCM pathogenic variants. Int J Cancer. 2019;144:2683–94. doi: 10.1002/ijc.31992. [DOI] [PubMed] [Google Scholar]
- 26.Lodish H, Berk A, Zipursky SL, Matsudaira P, Baltimore D, Darnell J. Molecular cell biology 4th edition. 4th ed. New York: National Center for Biotechnology Information, Bookshelf; 2000.
- 27.Shalin SC, Sakharpe A, Lyle S, Lev D, Calonje E, Lazar AJ. p53 staining correlates with tumor type and location in sebaceous neoplasms. Am J Dermatopathol. 2012;34:129–35. doi: 10.1097/DAD.0b013e3181ed39f9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tetzlaff MT, Singh RR, Seviour EG, Curry JL, Hudgens CW, Bell D, et al. Next-generation sequencing identifies high frequency of mutations in potentially clinically actionable genes in sebaceous carcinoma. J Pathol. 2016;240:84–95. doi: 10.1002/path.4759. [DOI] [PubMed] [Google Scholar]
- 29.Bougeard G, Renaux-Petel M, Flaman JM, Charbonnier C, Fermey P, Belotti M, et al. Revisiting Li-Fraumeni syndrome from TP53 mutation carriers. J Clin Oncol. 2015;33:2345–52. doi: 10.1200/JCO.2014.59.5728. [DOI] [PubMed] [Google Scholar]
- 30.Rana HQ, Clifford J, Hoang L, LaDuca H, Black MH, Li S, et al. Genotype-phenotype associations among panel-based TP53+ subjects. Genet Med. 2019;21:2478–84. doi: 10.1038/s41436-019-0541-y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.