Abstract
Neoadjuvant chemo-radiotherapy (CRT) followed by total mesorectal excision (TME) represents the standard treatment for patients with locally advanced (≥ T3 or N+) rectal cancer (LARC). Approximately 15% of patients with LARC shows a complete response after CRT. The use of pre-treatment MRI as predictive biomarker could help to increase the chance of organ preservation by tailoring the neoadjuvant treatment. We present a novel machine learning model combining pre-treatment MRI-based clinical and radiomic features for the early prediction of treatment response in LARC patients. MRI scans (3.0 T, T2-weighted) of 72 patients with LARC were included. Two readers independently segmented each tumor. Radiomic features were extracted from both the “tumor core” (TC) and the “tumor border” (TB). Partial least square (PLS) regression was used as the multivariate, machine learning, algorithm of choice and leave-one-out nested cross-validation was used to optimize hyperparameters of the PLS. The MRI-Based “clinical-radiomic” machine learning model properly predicted the treatment response (AUC = 0.793, p = 5.6 × 10–5). Importantly, the prediction improved when combining MRI-based clinical features and radiomic features, the latter extracted from both TC and TB. Prospective validation studies in randomized clinical trials are warranted to better define the role of radiomics in the development of rectal cancer precision medicine.
Subject terms: Cancer imaging, Cancer therapy, Tumour biomarkers
Introduction
In recent years, neoadjuvant chemo-radiotherapy (CRT) followed by total mesorectal excision (TME) became the standard treatment for patients with locally advanced (≥ T3 or N+) rectal cancer (LARC)1,2. This approach increased the chance of a significant downstaging and reduced the risk of recurrence3–6. In this context, Magnetic Resonance Imaging (MRI), thanks to its diagnostic and prognostic relevance, is the gold standard imaging approach for local staging and treatment response assessment7. At staging, MRI provides information on the site, the extension and the relation with surrounding organs, thus establishing landmarks for following treatment. Moreover, it reveals prognostic signs such as the mesorectal fascia involvement, the extramural vascular invasion and the distance to the anal sphincter complex8. During the treatment and, mostly, at the end of the CRT right before surgery, MRI plays a crucial role for the response assessment9–15. In fact, approximately 15% of patients with LARC shows a complete response to CRT (complete responders) and it has been suggested that surgery could be omitted in selected patients3. Moreover, an international multicenter registry study on watch and wait (W&W) strategy revealed a 5-year disease specific survival of 94%16,17. In this context, the use of MR as predictive biomarker in oncology quickly became a hot topic in literature, with the primary aim to select the most appropriate treatment thus pursuing precision medicine18. Moreover, the development of organ preservation policies further encouraged the researchers to investigate new MRI-based biomarkers for treatment response3. This approach could be extremely beneficial for patients considered good responder, since the neoadjuvant treatment could be intensified to increase the chance of organ preservation. Moreover, for these patients, a longer interval between CRT and surgery (from 8 to 11 weeks) may increase the pathological complete response rates as well. On the other hand, in poor responders, the treatment could be tailored with an additional boost19. Methods extracting mineable data from clinical images, such as those based on automatic extraction of features from radiological images (radiomic features) and machine learning approaches, revealed promising results when combined to visual morphologic and clinical assessment20–28. Moreover, in other tumor types, prognostic information has been obtained not only from the tumor but also in the tumor surrounding tissue (TST)29–31. We hypothesized that a combination of MRI-based data, including clinical and computational ones, could be useful for treatment response prediction at an early stage.
We here present a novel machine learning model combining MRI-based clinical and radiomic features, the latter extracted from both the “tumor core” (TC) and the “tumor border” (TB) from baseline T2-weighted (T2w) images, for the early prediction of treatment response in patients with LARC.
Methods
Subjects
The study received formal approval from the Ethical Committee of the University G. D’Annunzio of Chieti-Pescara, Italy; informed consent was waived by the same ethics committee that approved the study (Comitato Etico per la Ricerca Biomedica delle Province di Chieti e Pescara e dell’Università degli Studi “G.d’Annunzio” di Chieti e Pescara). The study was conducted according to ethical principles laid down by the latest version of the Declaration of Helsinki. A total of 164 patients who underwent clinically indicated rectal MRI for primary staging between January 2011 and February 2019 at our institution were retrospectively included (Fig. 1). Inclusion criteria were 1) non-mucinous locally advanced rectal cancer confirmed via biopsy, 2) MRI performed using a 3.0 T scanner, 3) availability of final clinical outcome (major pathological response or non-major pathological response), 4) long-course CRT. Ninety-two patients were excluded: 12 were mucinous cancers, 24 because the MRI exam was performed on a 1.5 T scanner, 33 were transferred in other institutes and we had no information regarding the final clinical outcome. Moreover, 18 patients were considered unfit for long-course CRT because of their clinical fragility and 5 patients had severe susceptibility artifacts in the pelvis (hip replacement). The final study population was composed of 72 patients.
Figure 1.
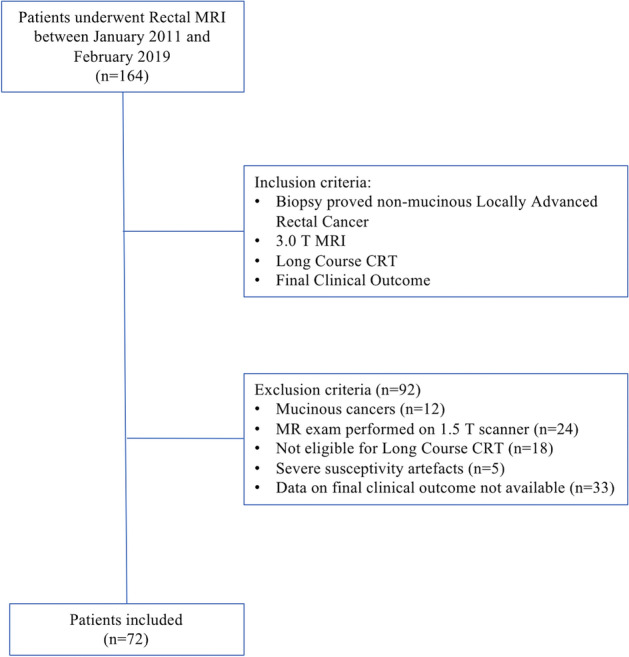
Study flowchart.
Chemoradiotherapy
Radiotherapy was performed by 3D conformal technique, with a total dose of 45 Gy (1.8 Gy/day) to the pelvic nodes, followed by a sequential boost of 5.4 Gy (1.8 Gy/day; total dose 50.40 Gy), or a concomitant boost of 10 Gy (1 Gy/day for 2 times/week; total dose 55 Gy) initially with a 3D-CRT technique and then with simultaneous integrated boost with intensity-modulated radiotherapy (2.2 Gy/day, total dose 55 Gy). Different schedules of drugs including 5-Fluorouracil and leucovorin or capecitabine were administered as concomitant chemotherapy.
MRI protocol
All patients in this cohort underwent clinically indicated rectal MRI consisting of a standard T2w and a diffusion weighted imaging (DWI) acquisition performed using a state-of-the-art 3 T MR scanner (Achieva, Philips Medical System, Best, the Netherlands) equipped with a phased array surface coil. Both T2w and DWI sequences were axially oriented perpendicular to the tumor major axis defined on a sagittal scan. Patients without contraindications received 20 mg of scopolamine butylbromide (Buscopan, Boehringer Ingelheim, Ingelheim am Rhein, Germany) intravenously to reduce bowel motility7. Detailed information regarding the parameters of the MRI protocol are described in Table 1.
Table 1.
MRI protocol parameters.
| T2-weighted Turbo spin echo* |
|
|---|---|
| Repetition time (msec) | 3000–5000 |
| Echo time (msec) | 80 |
| Section thickness (mm) | 3 |
| Section gap (mm) | 0 |
| Acquisition Matrix Size | 188 × 167 |
| No. of signals acquired | 2 |
| Field of view (mm) | 150 × 150 |
| Sensitivity Encoding (SENSE) | Yes |
| Acquisition Time (min) | 2.39 |
| No. of sections | 30 |
*MRI parameters are referred to axial T2-weighted images.
Imaging analysis
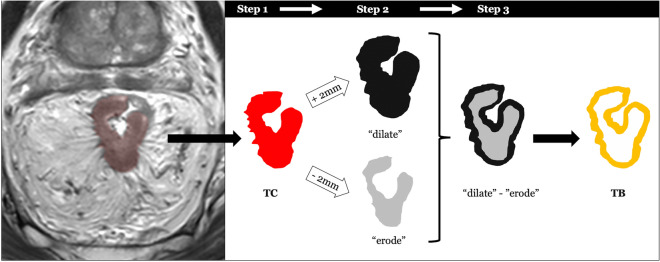
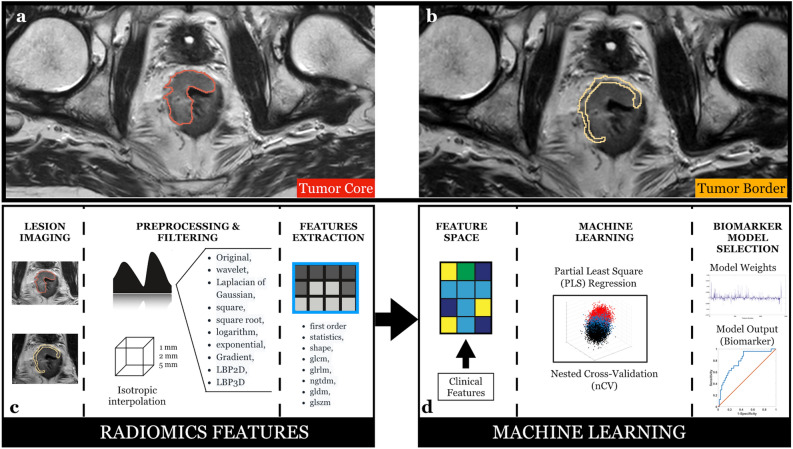
Whole-volume tumor manual segmentation representing the tumor core (TC) was performed on T2w images for each patient by two independent readers with different degrees of experience in abdominal radiology (a radiologist with 5 years of expertise in rectal MRI and a senior resident). All the segmentations were performed on T2w images of the staging MRI and were used as masks for following analysis. The software used for the segmentation was an open-source medical image computing platform, 3DSlicer Version 4.8 (www.3dslicer.org). To create the “border” segmentations (tumor border, TB), a “3dmask_tool” (AFNI) was used32. Firstly, both a 2 mm dilatation (“dilate”) and a 2 mm erosion (“erode”) were obtained from the TC of each patient. Secondly, the two masks were subtracted (“dilate”–“erode”) in order to obtain the TB which was 4 mm thick (Fig. 2). All the “border” masks were then checked by the two readers. If necessary, the segmentation was manually corrected in order to include only the outer border of the tumor and adjacent perivisceral fat. TC and TB were shown in Fig. 3a,b.
Figure 2.
The segmentation process was as follows: in the first step, the whole tumor was manually segmented on axial T2w images and labeled as tumor core (TC). In the second step, the edge of TC was dilated (“dilate”) and eroded (“erode”) by 2 mm, respectively. In the final step, we overlapped the dilated and eroded masks, and subsequently subtracted them to obtain the tumor border (TB). Thus, the TB obtained included the most peripheral portion of the TC and the surrounding tissues.
Figure 3.
(a) Tumor core and (b) tumor border on baseline T2-weighted images. (c) Schematic representation of the radiomic features extraction process and (d) the machine learning approach.
Clinical features
Four weeks after the end of the manual segmentation, the two readers in consensus evaluated, for each patient, nine MRI-based clinical features, namely, tumor location (high = 1, middle = 2, low rectum = 3), whole tumor volume (mm3), cranio-caudal extension (mm), distance from the internal anal sphincter (mm), mesorectal fascia infiltration (absent = 0, present = 1), extramural vascular invasion (absent = 0, present = 1), extramural depth of invasion (mm), T-stage (1–4) and N-stage (1–2). These MRI-based features were defined “clinical features” and were used in the machine learning model.
Radiomic features extraction
The extraction of the radiomic features from the masked (TB and TC) T2w images was performed using PyRadiomics, a flexible open-source platform capable of extracting a large panel of engineered features from medical images; this radiomic quantification platform enables the standardization of both feature definitions and image processing33. The reproducibility assessment of the features extracted by the two readers from the segmentations of all patients was performed. To avoid data heterogeneity bias and to promote reproducibility, the T2w images and masks were resampled using 3 isotropic voxel dimensions (1 × 1 × 1 mm, 2 × 2 × 2 mm, 5 × 5 × 5 mm). For each segmentation and for each image resolution (1 mm, 2 mm and 5 mm) ten built-in filters (Original, wavelet, Laplacian of Gaussian (LoG), square, square root, logarithm, exponential, Gradient, LBP2D, LBP3D) were applied and seven feature classes (first order statistics, shape descriptors, glcm, glrlm, ngtdm, gldm, glszm) were calculated, which resulted in a total of 1688 radiomic features (Fig. 3c).
Ethical statement
This study was approved by the local ethics committee. The study used only pre-existing medical data, therefore patient consent was waived.
Machine learning: partial least square (PLS) regression
Machine learning approaches (also defined multivariate approaches) exploits data multidimensionality to extract useful information. The added value of such approaches is that they identify statistical dependences among variables that are not visible to standard univariate analysis.
However, many radiomic features tend to identify similar image characteristics, generally providing highly redundant information34. This means that, when trying to predict an output based on these features, this information redundancy, coupled with the large numerosity of features with respect to samples (e.g. subjects), corrupts the results making the prediction unstable to noise and prone to overfitting and poor generalization35. To address this problem, in this work, two main approaches were implemented. The first approach to dampen this effect was to reduce the number of used features by selecting only those that were highly repeatable between the two masks (delineated by the two radiologists) used (r > 0.95). The second approach was to implement a machine learning framework based on a linear regression analysis that employed a space dimension reduction procedure, namely the partial least square (PLS) regression36,37. The PLS was used to predict the treatment response in patients with LARC at an early stage from the set of clinical and radiomic features. PLS has been extensively proven to be highly effective in reducing overfitting in the presence of collinearity. The underlying assumption of PLS is that the observed data is generated by a system or process which is driven by a small number of latent variables.
PLS allows the construction of regression equations reducing the predictors to a smaller set of uncorrelated components, i.e. a linear combination of the original predictors, and performs regression on these components36,38. The goal of PLS is to identify components that capture most of the information in the independent variables (e.g. linear combinations of all radiomic features) that is useful for predicting the dependent variable (e.g. treatment response), while reducing the dimensionality of the regression problem by using fewer components than the original number of independent variables. PLS can be conceived as a supervised learning version of the Principal Component Analysis (PCA)39,40. Of note, the learning process (fitting) of the PLS algorithm provides regression loadings that can be used to retrieve the weights (β-weights) linking the original independent variables with the dependent variable, depicting the importance and sign of the original variables in the prediction process. The PLS has one hyperparameter to be optimized, namely the number of uncorrelated components (linear combinations of the original independent variables) to be used in the regression.
In order to perform hyperparameter optimization and evaluate the generalizable performance of the procedure three sets are required, a trained set, where the model is trained as a function of different hyperparameter values, a validation set, where the best hyperparameter is chosen, and a test set, where the final model is tested. However, when the number of samples is small, this separation of data can greatly reduce the training sample, impairing appropriate fitting of the data-driven model.
An extension of this approach that allows to minimize the effect of the loss of samples in the different sets is the nested cross-validation (nCV)41–43. In nCV, data are divided in folds and the model is trained on all data except one-fold in an iterative, nested manner. Whereas the outer loop estimates the performances of the model among iterations (test), the inner loop evaluates the optimal hyperparameter (validation). If the number of folds equals the number of samples (one-fold per sample) the procedure is defined leave-one-out nCV44,45. This approach is highly suited for medical applications where each sample represents one subject. In this work, a leave-one-out nCV was implemented to optimize the PLS number of components and to assess the PLS generalization performance.
The leave-one-out nCV PLS analysis was repeated multiple times considering standalone clinical features, standalone radiomic features (with an inter-radiologist repeatability of r > 0.95) and combined clinical and radiomic features. MRI-based clinical and radiomic features were also analyzed multiple times considering TC only, TB only, as well as both TC and TB radiomic features (Fig. 3d). The machine learning analyses were implemented in Matlab.
Reference standard
The major pathologic response, assessed for 69 patients on surgical specimens, was considered to be Tumor Regression Grade (TRG) 1 or 2 scores according to Mandard’s classification46–48. Alternatively, for 3 patients, a sustained complete clinical response with repeated negative MRI examinations and endoscopy with or without biopsy was considered surrogate for a major pathological response for patients enrolled in watch-and-wait protocols49. More in detail, a sustained complete response was defined as a long-term recurrence-free clinical follow-up (minimum follow-up period of 1 year)50.
Statistical analysis
The classification performances were assessed through Receiver Operating Characteristic (ROC) analyses considering the inferred (out-of-training-sample) treatment response to therapy. Patients who responded to therapy (major pathological response) were attributed to the “negative” group, whereas patient showing a non-major pathological response were attributed to the “positive” group. The ROC analyses were also performed on random shuffled outcomes to simulate the null hypothesis and evaluate its confidence interval (repeated 106 times). The ROC analysis delivered an Area Under the Curve (AUC) which, using the random shuffled outcomes, could be transformed into a z-score for assessing its statistical significance. The Statistical Analysis was performed in Matlab.
Results
Out of the 72 patients included in the study, 48 were male (67%), and the mean age was 65 (IQR: 57.5–73.8) years. 48 (67%) patients showed a major pathological response and 24 (33%) a non-major pathological response (Table 2).
Table 2.
Descriptive baseline characteristics of included patients (n = 72).
| Variable | Value | |
|---|---|---|
| Gender | ||
| Male | 48 (67%) | |
| Female | 24 (33%) | |
| Mean age (IQR*) | 65 (57.5–73.8) | |
| MRI exam | 72 | |
| Clinical MRI assessment | ||
| Mean cancer volume (Mean ± SD) | 21,885 ± 20,539 mm2 | |
| Location | ||
| High | 7 (10%) | |
| Middle | 33 (46%) | |
| Low | 32 (44%) | |
| Craniocaudal extension (Mean ± SD) | 55 ± 22 mm | |
| Distance from IAS (Mean ± SD) | 31 ± 27 mm | |
| Depth of extramural invasion (Mean ± SD) | 7 ± 7 mm | |
| Presence of mesorectal fascia infiltration | 44 (61%) | |
| Presence of EMVI | 50 (69%) | |
| Primary cT stage** | ||
| T1-T2 | 14 (19.4%) | |
| T3 | 55 (76.4%) | |
| T4 | 3 (4.2%) | |
| Primary cN stage** | ||
| N0 | 2 (2.7%) | |
| N1 | 22 (30.6%) | |
| N2 | 48 (66.7%) | |
| Treatment response*** | ||
| MR | 48 (67%) | 34 TRG1 (71%) |
| 14 TRG2 (29%) | ||
| nMR | 24 (33%) | 17 TRG3 (71%) |
| 7 TRG4 (29%) | ||
*IQR Inter-Quartile Range; SD Standard Deviation; IAS Internal Anal Sphincter; EMVI Extramural Vascular Invasion; MR Major Pathological Response; nMR non-Major Pathological Response.
**Assessed with MRI and derived from clinical MRI reports in the hospital’s patient database.
***Assessed according to Mandard Tumor Regression Grade (TRG) system on surgical specimen after neoadjuvant treatment in 69/72 patients. In three patients, a sustained complete clinical response (with repeated negative MRI examinations and endoscopy with or without biopsy) was considered surrogate for a complete response; the follow-up (mean ± SD) was 47 ± 11 months.
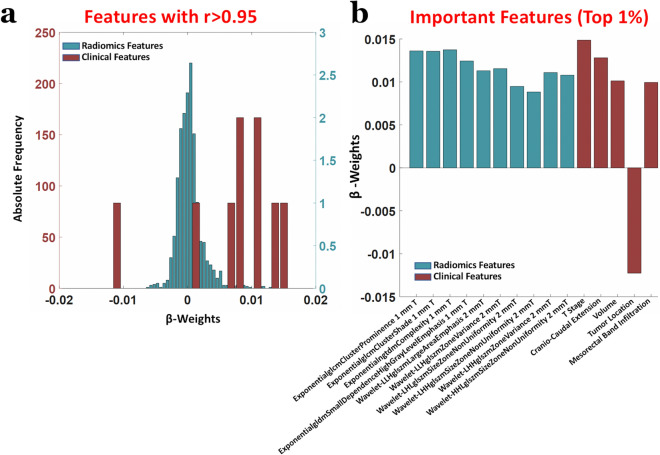
Nine clinical features were available and all of them were considered in the machine learning analysis. Moreover, 1688 radiomic features were extracted for the three image resolutions employed (1 mm, 2 mm and 5 mm) and for TC and TB, leading to a total of 10,128 (1688 × 3 × 2) features. 1405 of these features showed an inter-reader correlation of r > 0.95 and were used for further analysis.
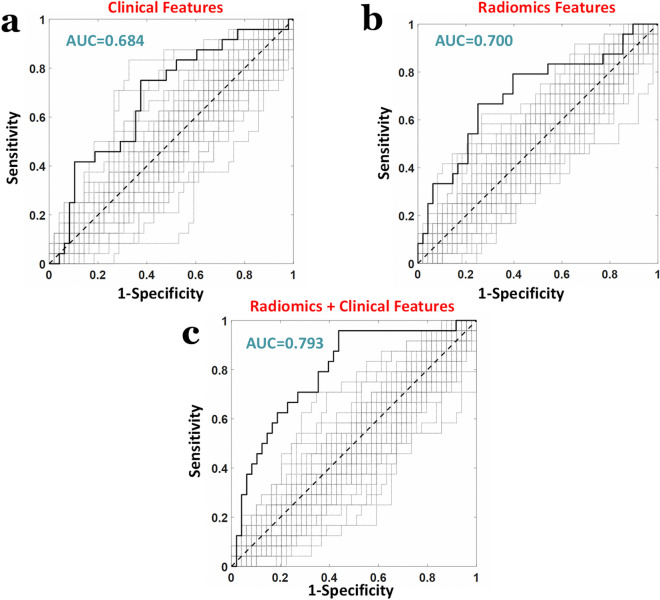
When considering the 9 standalone clinical features an AUC = 0.684 was obtained (z = 2.53, p = 11 × 10−3, Fig. 4a). When employing standalone radiomic features with an r > 0.95, i.e. 1405 radiomic features, an AUC = 0.700 was obtained (z = 2.75, p = 5.9 × 10−3, Fig. 4b). Importantly, the highest AUC was obtained when combining the 9 clinical features with the 1405 radiomic features, obtaining an AUC = 0.793 (z = 4.00, p = 5.6 × 10−5, Fig. 4c).
Figure 4.
ROC analyses of the machine learning (PLS) classification performance predicting response to therapy (major pathological response as “positive”, non-major pathological response labelled as “negative”) when considering (a) standalone clinical features (b) standalone radiomic features and (c) combined clinical and radiomic features.
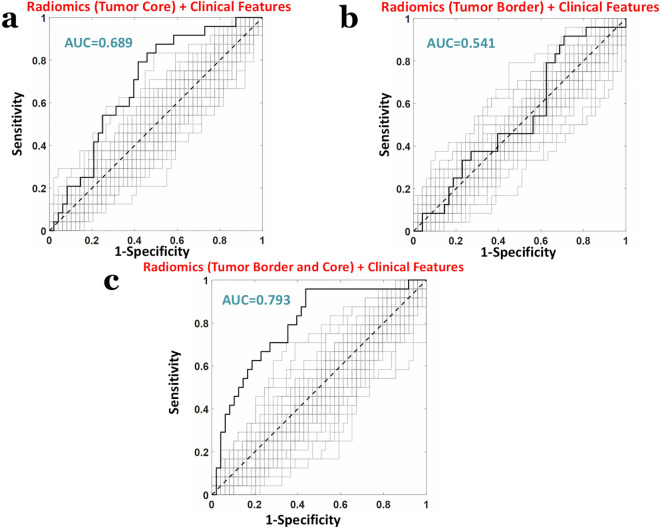
The weights of the PLS (β-weights), when the machinery was trained on both clinical and radiomic features, are shown in Fig. 5. Figure 5a reports the distributions of the β-weights for radiomic and clinical features, whereas Fig. 5b depicts the β-weights associated to the top 1% of features with the largest β-weights in magnitude, i.e. those most impacting the prediction. Importantly, for all except one (tumor location), of the top 1% of features, the weights were positive, that considering the value of “0” attributed to patients with major pathologic response and the value of “1” to the others during the multivariate regression, depicted a worse response to treatment with increasing feature value. Of note, the only important feature with negative weight was tumor location that, considering the labelling value attributed as a function of location (1 = High, 2 = Middle, 3 = Low), delineated a better response to treatment for tumors in the low rectum. Finally, the impact of TC and TB on the results was assessed. When using the 9 clinical features and 790 radiomic features (with an inter-radiologist repeatability of r > 0.95) extracted only from the TC, an AUC = 0.689 was obtained (z = 2.60, p = 9.3 × 10−3, Fig. 6a). When using the 9 clinical features and 626 radiomic features (with an inter-radiologist repeatability of r > 0.95) extracted only from the TB, an AUC = 0.541 was obtained (z = 0.56, p = 0.57, Fig. 6b). Indeed, a highly synergistic effects was obtained when combining TB and TC features, replicating the results previously found with an AUC = 0.793 (z = 4.00, p = 5.6 × 10−5, Fig. 6c).
Figure 5.
(a) Distributions of the PLS β-weights of radiomic and clinical features when the machine learning analysis was performed on combined data. (b) β-weights associated to the top 1% of features with the largest β-weights in magnitude, i.e. those most impacting the prediction.
Figure 6.
ROC analyses of the machine learning (PLS) classification performance predicting response to therapy (major pathological response labelled as “positive”, non-major pathological response labelled as “negative”) when considering (a) clinical and TC radiomic features (b) clinical and TB radiomic features and (c) clinical and TC + TB radiomic features.
Discussion
Our results demonstrated that an MRI-based machine learning “clinical-radiomic” approach was an accurate method to predict, at an early stage, the treatment response of patient with LARC.
Importantly, the addition of novel radiomic features to standard T2w-based clinical features was indeed useful in the improvement of the prognostic model. Moreover, the prediction further improved when both TC and TB radiomic features were included. These results confirm the promising role of baseline T2w-imaging for the prediction of clinical-outcome21,22,51,52. Recent studies have demonstrated the prognostic role of tumor morphology and the adjacent perirectal tissue assessed on T2w-imaging10,53. Furthermore, the importance of the perirectal tissue status for the final clinical outcome was also underlined by the significant reduction of the local recurrences with the introduction of TME thanks to the complete excision of microtumors around the cancer54. To the best of our knowledge our study, apart for the added value of radiomic features to standard MRI-based clinical metrics, is the first demonstrating the synergic role between feature extracted from the tumor core and the tumor border in the early prediction of response to therapy. Since the tumor border naturally include a certain degree of mesorectal compartment, our results confirm that also perirectal tissues contain useful information for the prediction of treatment response, as recently showed by Shaish et al.55.
Other studies based on radiomics were recently proposed in literature. However, different radiomics approaches were used: for example, a delta-radiomics approach was used to analyze the variation of quantitative features over time. In this way, Jeon et al. analyzed the difference in radiomic features before and after CRT. The authors demonstrated that delta radiomics signatures were successful predictors of treatment response and independent prognostic factors56. In other studies, the predictivity of more extensive MRI protocols was explored57,58. For example, Cui et al. investigated the predictive role of a multiparametric MRI approach including T2w, contrast-enhanced T1w and ADC images57. They demonstrated that a radiomics-based nomogram could predict pathological complete response in patients with LARC. In the same way, Liu et al. developed a radiomics model based on T2w images and DWI before and after chemoradiotherapy resulting in 30 selected features showing good discrimination performance59. Additionally, Wan et al., recently published a retrospective study including pre and post neoadjuvant MRI examination from which radiomics features were extracted. Their delta-radiomics signature based on T2w images and DWI showed an AUC of 0.91 for the pathologic complete response prediction60. Furthermore, Liu et al. recently developed and validated a multiparametric MRI-based radiomics signature including T2w images and ADC for the prediction of distant metastasis in LARC61.
Differently from these approaches, our study was focused only on the primary staging and included only T2w images. In fact, our primary aim was to investigate biomarkers of treatment response at an early stage using the most widely used and recommended MRI imaging7. Focusing of the important features deemed as such by the machine learning algorithm, in our study the principal predictive features were mainly focused on tumor heterogeneity and clinical, visually assessed, characteristics. For example, tumors with high degree of internal heterogeneity, N+ and with a high clinical T staging were more likely associated to a poor treatment response. Furthermore, cancers located in low rectum were found to have a more favorable outcome thus confirming the crucial role of neoadjuvant chemo-radiation therapy in this category of patients62. Since most of the predictive features were extracted by 1–2 mm resolution voxels instead of 5 mm, our results support the recommendations of the last European Society of Gastrointestinal and Abdominal Imaging (ESGAR) consensus meeting7. In fact, according to these guidelines, the slice thickness of axial T2-weighted images should be equal or inferior to 3 mm7. Similarly to other studies, the accuracy of the prediction improved when a qualitative MRI assessment is added to a quantitative radiomic-based analysis21,51. In this way we are confident that the development of other “omics” disciplines may further improve the overall accuracy of the treatment response prediction63,64.
Our study has some limitation. First of all, the sample size is limited and our study lacks of a validation cohort. However, the nCV implemented in our study minimized the effect of reduced number of samples and overfitting42. Moreover, the limited sample size was largely due to the exclusion of all patients who underwent rectal MRI on a different magnet (1.5 T), patients who underwent a different radiation therapy than long-course, and patients without known final clinical outcome known. Indeed, future studies on larger cohorts are warranted to confirm our findings, and to further improve the classification outcome.
Secondly, ours is a retrospective single-center study. For this reason, further studies, possibly prospective and multicentric need to be warranted to define a potential standardization of our approach.
Third, some patients did not receive scopolamine butylbromide intravenously due to contraindications. In addition, our 3 T MR scanner is routinely scheduled for a software upgrade which might have improved the image quality over time. Nonetheless, these changes did not affect nor modify the main protocol parameters and reflect the daily clinical practice. Fourth, we only focused our prediction on T2-weighted imaging, without considering other MRI techniques, such as DWI and Dynamic Contrast Enhanced Imaging (DCE). However, since our objective was the prediction at the primary staging, the potential role of DWI and DCE at this time point is controversial. In fact, there is no clear added benefit for DWI regarding T, N, MRF and EMVI assessment based on current evidence. On the other hand, DCE-MRI should be considered as a research tool only and not yet ready to be adopted routinely7.
Conclusion
A pre-treatment, MRI-based, “clinical-radiomic” machine-learning model accurately predict the treatment response in patients with locally advanced rectal cancer. The prediction improved when combining MRI-based clinical features and radiomic features, the latter extracted from both tumor core and the tumor border. Exploiting the method, patients with locally advanced rectal cancer could benefit from a tailored approach including conservative strategies. Prospective validation studies in randomized clinical trials are warranted to better define the role of radiomics in the development of rectal cancer precision medicine.
Acknowledgements
The authors thank Daniele Petrucci and Darien Calvo Garcia for their insightful contribution on the MRI protocol settings and the acquisition of data.
Author contributions
A.D.P. conceived and developed the research idea with the assistance of S.T., D.M.J.L. and R.C. The study design was performed by A.D.P, D.M., R.B., D.G, A.F., R.G.H.B.T. and M.C. M.d’A. and A.D.P. performed the segmentation task. C.R., S.C., D.C., G.C., F.L.S. and P.D.S. performed data collection. P.CR., P.CH. and A.M.C. performed the computational experiments with guidance from R.G.W. and A.F. The paper was written by A.D.P. with technical content from A.M.C. and P.CH, and extensive editorial input from all authors. All authors have read and approved the final version submitted.
Data availability
The datasets generated during and/or analyzed during the current study are not publicly available due to the clinical and confidential nature of the material but can be made available from the corresponding author on reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Nagtegaal I, et al. Morphological changes in tumour type after radiotherapy are accompanied by changes in gene expression profile but not in clinical behaviour. J. Pathol. 2004;204:183–192. doi: 10.1002/path.1621. [DOI] [PubMed] [Google Scholar]
- 2.Heald RJ, Ryall RD. Recurrence and survival after total mesorectal excision for rectal cancer. Lancet. 1986;1:1479–1482. doi: 10.1016/S0140-6736(86)91510-2. [DOI] [PubMed] [Google Scholar]
- 3.Maas M, et al. Long-term outcome in patients with a pathological complete response after chemoradiation for rectal cancer: a pooled analysis of individual patient data. Lancet Oncol. 2010;11:835–844. doi: 10.1016/S1470-2045(10)70172-8. [DOI] [PubMed] [Google Scholar]
- 4.Glimelius B, Tiret E, Cervantes A, Arnold D, Group EGW. Rectal cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2013;24(Suppl 6):vi81–vi88. doi: 10.1093/annonc/mdt240. [DOI] [PubMed] [Google Scholar]
- 5.Krook JE, et al. Effective surgical adjuvant therapy for high-risk rectal carcinoma. N. Engl. J. Med. 1991;324:709–715. doi: 10.1056/NEJM199103143241101. [DOI] [PubMed] [Google Scholar]
- 6.Sauer R, et al. Preoperative versus postoperative chemoradiotherapy for rectal cancer. N. Engl. J. Med. 2004;351:1731–1740. doi: 10.1056/NEJMoa040694. [DOI] [PubMed] [Google Scholar]
- 7.Beets-Tan RGH, et al. Magnetic resonance imaging for clinical management of rectal cancer: updated recommendations from the 2016 European Society of Gastrointestinal and Abdominal Radiology (ESGAR) consensus meeting. Eur. Radiol. 2018;28:1465–1475. doi: 10.1007/s00330-017-5026-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Delli Pizzi A, et al. Rectal cancer MRI: protocols, signs and future perspectives radiologists should consider in everyday clinical practice. Insights Imaging. 2018;9:405–412. doi: 10.1007/s13244-018-0606-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Delli Pizzi A, et al. Performance of diffusion-weighted magnetic resonance imaging at 3.0T for early assessment of tumor response in locally advanced rectal cancer treated with preoperative chemoradiation therapy. Abdom. Radiol. (NY) 2018;43:2221–2230. doi: 10.1007/s00261-018-1457-8. [DOI] [PubMed] [Google Scholar]
- 10.Lambregts DMJ, et al. A pattern-based approach combining tumor morphology on MRI with distinct signal patterns on diffusion-weighted imaging to assess response of rectal tumors after chemoradiotherapy. Dis. Colon Rectum. 2018;61:328–337. doi: 10.1097/DCR.0000000000000915. [DOI] [PubMed] [Google Scholar]
- 11.Beets-Tan RG, Beets GL. MRI for assessing and predicting response to neoadjuvant treatment in rectal cancer. Nat. Rev. Gastroenterol. Hepatol. 2014;11:480–488. doi: 10.1038/nrgastro.2014.41. [DOI] [PubMed] [Google Scholar]
- 12.Lambregts DM, et al. Diffusion-weighted MRI to assess response to chemoradiotherapy in rectal cancer: main interpretation pitfalls and their use for teaching. Eur. Radiol. 2017 doi: 10.1007/s00330-017-4830-z. [DOI] [PubMed] [Google Scholar]
- 13.Delli Pizzi A, et al. Tumor detectability and conspicuity comparison of standard b1000 and ultrahigh b2000 diffusion-weighted imaging in rectal cancer. Abdom. Radiol. (NY) 2019;44:3595–3605. doi: 10.1007/s00261-019-02177-y. [DOI] [PubMed] [Google Scholar]
- 14.Lambregts DMJ, et al. Long-term imaging characteristics of clinical complete responders during watch-and-wait for rectal cancer-an evaluation of over 1500 MRIs. Eur. Radiol. 2020;30:272–280. doi: 10.1007/s00330-019-06396-1. [DOI] [PubMed] [Google Scholar]
- 15.Rosa C, et al. Reproducibility of rectal tumor volume delineation using diffusion-weighted MRI: Agreement on volumes between observers. Cancer Radiother. 2019;23:216–221. doi: 10.1016/j.canrad.2018.10.004. [DOI] [PubMed] [Google Scholar]
- 16.Maas M, et al. Wait-and-see policy for clinical complete responders after chemoradiation for rectal cancer. J. Clin. Oncol. 2011;29:4633–4640. doi: 10.1200/JCO.2011.37.7176. [DOI] [PubMed] [Google Scholar]
- 17.van der Valk MJM, et al. Long-term outcomes of clinical complete responders after neoadjuvant treatment for rectal cancer in the International Watch & Wait Database (IWWD): an international multicentre registry study. Lancet. 2018;391:2537–2545. doi: 10.1016/S0140-6736(18)31078-X. [DOI] [PubMed] [Google Scholar]
- 18.Haak HE, et al. Selection of patients for organ preservation after chemoradiotherapy: MRI identifies poor responders who can go straight to surgery. Ann. Surg. Oncol. 2020;27:2732–2739. doi: 10.1245/s10434-020-08334-8. [DOI] [PubMed] [Google Scholar]
- 19.Burbach JPM, et al. Impact of radiotherapy boost on pathological complete response in patients with locally advanced rectal cancer: A systematic review and meta-analysis. Radiother. Oncol. 2014;113:1–9. doi: 10.1016/j.radonc.2014.08.035. [DOI] [PubMed] [Google Scholar]
- 20.Horvat N, et al. MR Imaging of rectal cancer: radiomics analysis to assess treatment response after neoadjuvant therapy. Radiology. 2018;287:833–843. doi: 10.1148/radiol.2018172300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Petkovska I, et al. Clinical utility of radiomics at baseline rectal MRI to predict complete response of rectal cancer after chemoradiation therapy. Abdom. Radiol. (NY) 2020 doi: 10.1007/s00261-020-02502-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van Griethuysen JJM, et al. Radiomics performs comparable to morphologic assessment by expert radiologists for prediction of response to neoadjuvant chemoradiotherapy on baseline staging MRI in rectal cancer. Abdom. Radiol. (NY) 2020;45:632–643. doi: 10.1007/s00261-019-02321-8. [DOI] [PubMed] [Google Scholar]
- 23.Trebeschi S, et al. Deep learning for fully-automated localization and segmentation of rectal cancer on multiparametric MR. Sci. Rep. 2017;7:5301. doi: 10.1038/s41598-017-05728-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chiacchiaretta P, Cerritelli F, Bubbico G, Perrucci MG, Ferretti A. Reduced dynamic coupling between spontaneous BOLD-CBF fluctuations in older adults: a dual-echo pCASL study. Front. Aging. Neurosci. 2018;10:115. doi: 10.3389/fnagi.2018.00115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chiacchiaretta P, Romani GL, Ferretti A. Sensitivity of BOLD response to increasing visual contrast: spin echo versus gradient echo EPI. Neuroimage. 2013;82:35–43. doi: 10.1016/j.neuroimage.2013.05.069. [DOI] [PubMed] [Google Scholar]
- 26.Cirillo S, Caulo M, Pieri V, Falini A, Castellano A. Role of functional imaging techniques to assess motor and language cortical plasticity in glioma patients: a systematic review. Neural. Plast. 2019;2019:4056436. doi: 10.1155/2019/4056436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Metwali H, Raemaekers M, Ibrahim T, Samii A. The fluctuations of blood oxygen level-dependent signals as a method of brain tumor characterization: a preliminary report. World Neurosurg. 2020;142:e10–e17. doi: 10.1016/j.wneu.2020.04.134. [DOI] [PubMed] [Google Scholar]
- 28.Ren Y, et al. Noninvasive prediction of IDH1 mutation and ATRX expression loss in low-grade gliomas using multiparametric MR radiomic features. J. Magn. Reson. Imaging. 2019;49:808–817. doi: 10.1002/jmri.26240. [DOI] [PubMed] [Google Scholar]
- 29.Caulo M, et al. Data-driven grading of brain gliomas: a multiparametric MR imaging study. Radiology. 2014;272:494–503. doi: 10.1148/radiol.14132040. [DOI] [PubMed] [Google Scholar]
- 30.Akinci D'Antonoli T, et al. CT radiomics signature of tumor and peritumoral lung parenchyma to predict nonsmall cell lung cancer postsurgical recurrence risk. Acad. Radiol. 2020;27:497–507. doi: 10.1016/j.acra.2019.05.019. [DOI] [PubMed] [Google Scholar]
- 31.Mazurowski MA, Zhang J, Grimm LJ, Yoon SC, Silber JI. Radiogenomic analysis of breast cancer: luminal B molecular subtype is associated with enhancement dynamics at MR imaging. Radiology. 2014;273:365–372. doi: 10.1148/radiol.14132641. [DOI] [PubMed] [Google Scholar]
- 32.Cox RW. AFNI: software for analysis and visualization of functional magnetic resonance neuroimages. Comput. Biomed. Res. 1996;29:162–173. doi: 10.1006/cbmr.1996.0014. [DOI] [PubMed] [Google Scholar]
- 33.van Griethuysen JJM, et al. Computational radiomics system to decode the radiographic phenotype. Cancer Res. 2017;77:e104–e107. doi: 10.1158/0008-5472.CAN-17-0339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Traverso A, Wee L, Dekker A, Gillies R. Repeatability and reproducibility of radiomic features: a systematic review. Int. J. Radiat. Oncol. Biol. Phys. 2018;102:1143–1158. doi: 10.1016/j.ijrobp.2018.05.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Magidson, J. in New perspectives in partial least squares and related methods. Springer Proceedings in Mathematics & Statistics Ch. Chapter 3, pp. 65–78 (2013).
- 36.Abdi H, Williams LJ. in Computational Toxicology Methods in Molecular Biology. Ch. Chapter. 2013;23:549–579. doi: 10.1007/978-1-62703-059-5_23. [DOI] [PubMed] [Google Scholar]
- 37.Wold S, Ruhe A, Wold H, Dunn IWJ. The collinearity problem in linear regression. The partial least squares (PLS) approach to generalized inverses. SIAM J. Sci. Stat. Comput. 1984;5:735–743. doi: 10.1137/0905052. [DOI] [Google Scholar]
- 38.Chiarelli AM, Romani GL, Merla A. Fast optical signals in the sensorimotor cortex: general linear convolution model applied to multiple source-detector distance-based data. Neuroimage. 2014;85(Pt 1):245–254. doi: 10.1016/j.neuroimage.2013.07.021. [DOI] [PubMed] [Google Scholar]
- 39.Barlow HB. Unsupervised learning. Neural Comput. 1989;1:295–311. doi: 10.1162/neco.1989.1.3.295. [DOI] [Google Scholar]
- 40.Jolliffe IT, Cadima J. Principal component analysis: a review and recent developments. Philos. Trans. A Math. Phys. Eng. Sci. 2016;374:20150202. doi: 10.1098/rsta.2015.0202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu R, Gillies DF. Overfitting in linear feature extraction for classification of high-dimensional image data. Pattern Recogn. 2016;53:73–86. doi: 10.1016/j.patcog.2015.11.015. [DOI] [Google Scholar]
- 42.Filzmoser P, Liebmann B, Varmuza K. Repeated double cross validation. J. Chemometr. 2009;23:160–171. doi: 10.1002/cem.1225. [DOI] [Google Scholar]
- 43.Krstajic D, Buturovic LJ, Leahy DE, Thomas S. Cross-validation pitfalls when selecting and assessing regression and classification models. J. Cheminf. 2014 doi: 10.1186/1758-2946-6-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chiarelli AM, et al. Electroencephalography-derived prognosis of functional recovery in acute stroke through machine learning approaches. Int. J. Neural Syst. 2020 doi: 10.1142/s0129065720500677. [DOI] [PubMed] [Google Scholar]
- 45.Kearns M, Ron D. Algorithmic stability and sanity-check bounds for leave-one-out cross-validation. Neural Comput. 1999;11:1427–1453. doi: 10.1162/089976699300016304. [DOI] [PubMed] [Google Scholar]
- 46.Mandard, A. M. et al. Pathologic assessment of tumor regression after preoperative chemoradiotherapy of esophageal carcinoma. Clinicopathologic correlations. Cancer73, 2680–2686, doi:10.1002/1097-0142(19940601)73:11<2680::aid-cncr2820731105>3.0.co;2-c (1994). [DOI] [PubMed]
- 47.Valentini V, et al. The INTERACT trial: long-term results of a randomised trial on preoperative capecitabine-based radiochemotherapy intensified by concomitant boost or oxaliplatin, for cT2 (distal)-cT3 rectal cancer. Radiother. Oncol. 2019;134:110–118. doi: 10.1016/j.radonc.2018.11.023. [DOI] [PubMed] [Google Scholar]
- 48.Lupattelli M, et al. Preoperative intensity-modulated radiotherapy with a simultaneous integrated boost combined with Capecitabine in locally advanced rectal cancer: short-term results of a multicentric study. Radiat. Oncol. 2017;12:139. doi: 10.1186/s13014-017-0870-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lambregts DM, et al. Long-term follow-up features on rectal MRI during a wait-and-see approach after a clinical complete response in patients with rectal cancer treated with chemoradiotherapy. Dis. Colon Rectum. 2011;54:1521–1528. doi: 10.1097/DCR.0b013e318232da89. [DOI] [PubMed] [Google Scholar]
- 50.Santiago I, et al. The split scar sign as an indicator of sustained complete response after neoadjuvant therapy in rectal cancer. Eur. Radiol. 2020;30:224–238. doi: 10.1007/s00330-019-06348-9. [DOI] [PubMed] [Google Scholar]
- 51.Bibault JE, et al. Deep learning and radiomics predict complete response after neo-adjuvant chemoradiation for locally advanced rectal cancer. Sci. Rep. 2018;8:12611. doi: 10.1038/s41598-018-30657-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ma X, et al. MRI-based radiomics of rectal cancer: preoperative assessment of the pathological features. BMC Med. Imaging. 2019;19:86. doi: 10.1186/s12880-019-0392-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yoon J, Chung YE, Lim JS, Kim MJ. Quantitative assessment of mesorectal fat: new prognostic biomarker in patients with mid-to-lower rectal cancer. Eur. Radiol. 2019;29:1240–1247. doi: 10.1007/s00330-018-5723-5. [DOI] [PubMed] [Google Scholar]
- 54.Visser O, et al. The influence of total mesorectal excision on local recurrence and survival in rectal cancer patients: a population-based study in Greater Amsterdam. J. Surg. Oncol. 2007;95:447–454. doi: 10.1002/jso.20713. [DOI] [PubMed] [Google Scholar]
- 55.Shaish H, et al. Radiomics of MRI for pretreatment prediction of pathologic complete response, tumor regression grade, and neoadjuvant rectal score in patients with locally advanced rectal cancer undergoing neoadjuvant chemoradiation: an international multicenter study. Eur. Radiol. 2020 doi: 10.1007/s00330-020-06968-6. [DOI] [PubMed] [Google Scholar]
- 56.Jeon SH, et al. Delta-radiomics signature predicts treatment outcomes after preoperative chemoradiotherapy and surgery in rectal cancer. Radiat. Oncol. 2019;14:43. doi: 10.1186/s13014-019-1246-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cui Y, et al. Radiomics analysis of multiparametric MRI for prediction of pathological complete response to neoadjuvant chemoradiotherapy in locally advanced rectal cancer. Eur. Radiol. 2019;29:1211–1220. doi: 10.1007/s00330-018-5683-9. [DOI] [PubMed] [Google Scholar]
- 58.Li ZY, et al. Multi-modal radiomics model to predict treatment response to neoadjuvant chemotherapy for locally advanced rectal cancer. World J. Gastroenterol. 2020;26:2388–2402. doi: 10.3748/wjg.v26.i19.2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu Z, et al. Radiomics analysis for evaluation of pathological complete response to neoadjuvant chemoradiotherapy in locally advanced rectal cancer. Clin. Cancer Res. 2017;23:7253–7262. doi: 10.1158/1078-0432.CCR-17-1038. [DOI] [PubMed] [Google Scholar]
- 60.Wan L, et al. MRI-based delta-radiomics are predictive of pathological complete response after neoadjuvant chemoradiotherapy in locally advanced rectal cancer. Acad. Radiol. 2020 doi: 10.1016/j.acra.2020.10.026. [DOI] [PubMed] [Google Scholar]
- 61.Liu Z, et al. Predicting distant metastasis and chemotherapy benefit in locally advanced rectal cancer. Nat. Commun. 2020;11:4308. doi: 10.1038/s41467-020-18162-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Glynne-Jones R, et al. Rectal cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2017;28(Suppl 4):iv22–iv40. doi: 10.1093/annonc/mdx224. [DOI] [PubMed] [Google Scholar]
- 63.Frydrych LM, et al. Rectal cancer sub-clones respond differentially to neoadjuvant therapy. Neoplasia. 2019;21:1051–1062. doi: 10.1016/j.neo.2019.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Canto LMD, et al. Locally advanced rectal cancer transcriptomic-based secretome analysis reveals novel biomarkers useful to identify patients according to neoadjuvant chemoradiotherapy response. Sci. Rep. 2019;9:8702. doi: 10.1038/s41598-019-45151-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated during and/or analyzed during the current study are not publicly available due to the clinical and confidential nature of the material but can be made available from the corresponding author on reasonable request.