Abstract
Pneumocystis jirovecii, the fungal agent of human Pneumocystis pneumonia, is closely related to macaque Pneumocystis. Little is known about other Pneumocystis species in distantly related mammals, none of which are capable of establishing infection in humans. The molecular basis of host specificity in Pneumocystis remains unknown as experiments are limited due to an inability to culture any species in vitro. To explore Pneumocystis evolutionary adaptations, we have sequenced the genomes of species infecting macaques, rabbits, dogs and rats and compared them to available genomes of species infecting humans, mice and rats. Complete whole genome sequence data enables analysis and robust phylogeny, identification of important genetic features of the host adaptation, and estimation of speciation timing relative to the rise of their mammalian hosts. Our data reveals insights into the evolution of P. jirovecii, the sole member of the genus able to infect humans.
Subject terms: Coevolution, Fungal genomics
Cissé, Ma et al. utilize genomic data from Pneumocystis species infecting macaques, rabbit, dogs and rats to investigate the molecular basis of host specificity in Pneumocystis. Their analyses provide insight to the specific adaptations enabling the infection of humans by P. jirovecii.
Introduction
The evolutionary history of Pneumocystis jirovecii, a fungus that causes life-threatening pneumonia in immunosuppressed patients such as those with HIV infection, has been poorly defined. P. jirovecii is derived from a much broader group of host-specific parasites that infect all mammals studied to date. Until recently, P. carinii and P. murina (which infect rats and mice, respectively) were the only other species in this genus for which biological specimens suitable for whole-genome sequencing were readily available. Cross-species inoculation studies of P. jirovecii and P. carinii have found that they can only infect humans and rats, respectively1,2. Further, rats are the only mammals known to be coinfected by at least two distinct Pneumocystis species (P. carinii and P. wakefieldiae)3. Within the Pneumocystis genus, P. jirovecii is the only species able to infect and reproduce in humans, although the molecular mechanisms of its host adaptation remain elusive.
Previous efforts to reconstruct the evolutionary history of Pneumocystis have estimated the origins of the genus at a minimum of 100 million years ago (mya)4. Using a partial transcriptome of P. sp. macacae (hereafter referred to as P. macacae), the Pneumocystis species that infects macaques, we recently estimated that P. jirovecii diverged from the common ancestor of P. macacae around ~62 mya5, which substantially precedes the human-macaque split of ~20 mya6. Population bottlenecks in P. jirovecii and P. carinii at 400,000 and 16,000 years ago, respectively5, are also not concordant with population expansions in modern humans (~200,000 years ago7) and rats (~10,000 years ago8), which suggests a decoupled coevolution between Pneumocystis and their hosts. Thus, Pneumocystis species may not be strictly coevolving with their mammalian hosts as suggested by ribosomal RNA-based maximum phylogenies9. A molecular clock has not been tested in any of these phylogenies. A strict coevolution hypothesis has been further challenged by evidence suggesting a relaxation of the host specificity in Pneumocystis infecting rodents10,11. However, the accuracy of speciation times is limited without the complete genomes of additional species including that of P. macacae, the closest living sister species to P. jirovecii identified to date.
The absence of long-term in vitro culture methods or animal models for most Pneumocystis species has precluded obtaining sufficient DNA for full genome sequencing and has hindered investigation of the Pneumocystis genus. To date, only the genomes of human P. jirovecii12,13, rat P. carinii13,14, and mouse P. murina13, are available. These data have provided important insights into the evolution of this genus, including a substantial genome reduction12,13, the presence of intron-rich genes possibly contributing to transcriptome complexity, and an expansion of a highly polymorphic major surface glycoprotein (msg) gene superfamily13, some of which are important for immune evasion. However, the lack of whole-genome sequences for many species of this genus (particularly the closely related P. macacae) has severely constrained the understanding of the implications of these genome features in Pneumocystis evolution and adaptation to hosts.
To explore the evolutionary history of the Pneumocystis genus, and investigate P. jirovecii genetic factors that support its adaptation to humans, we sequenced 2–6 specimens of four additional species: those that infect macaques (P. macacae), rabbits (P. oryctolagi), dogs (P. canis), and rats (P. wakefieldiae).
Results
Direct sequencing of Pneumocystis-host mixed samples
We sequenced the genomes of Pneumocystis species from infected macaques, rabbits, dogs, and rats (see Methods and Supplementary Methods). Specimens originated from immunosuppressed animals as a consequence of simian immunodeficiency virus infection in macaques, corticosteroid treatment (rabbits and rats), immunodeficient knockout (rabbits) or possible congenital immunodeficiencies (dogs). For each species, we sequenced multiple samples from 2–6 animals (Supplementary Tables 1 and 2). These data were used to assemble one nearly full-length genome assembly for each species except P. canis for which we recovered two nearly full-length assemblies and an additional partial assembly from two separate samples (denoted as A, Ck1, and Ck2). Post assembly mapping revealed a negligible amount of genetic variability among samples, for example the average genome-wide single-nucleotide polymorphism (SNP) diversity among six P. macacae isolates excluding highly polymorphic regions such as Msg genes is ~0.1%. The genome of P. macacae was sequenced using Oxford Nanopore long reads and Illumina short read sequences, whereas the other Pneumocystis were sequenced only with Illumina (Supplementary Tables 2 and 3). The genome assemblies range from 7.3 Mb in P. wakefieldiae to 8.2 Mb in P. macacae. The P. macacae and P. wakefieldiae genome assemblies consist of 16 and 17 scaffolds, respectively, both of which are highly contiguous and approach the chromosomal level based on similarities with published karyotypes3,15 and/or the presence of Pneumocystis telomere repeats16 at the scaffold ends (Supplementary Table 3). The genome assemblies of P. oryctolagi and P. canis (assemblies A, Ck1, and Ck2) are less contiguous with 38, 33, 78, and 315 scaffolds, respectively. All these assemblies except for the partial assembly of P. canis Ck2 have very similar total sizes (7.3–8.2 Mb) comparable to previously sequenced genomes of P. jirovecii, P. carinii, and P. murina, all of which are at or near chromosomal level with a size of 7.4–8.3 Mb (Supplementary Table 3). The genome assemblies are all AT-rich (~71%) and ~3% encode DNA transposons and retrotransposons (Supplementary Table 3). We also assembled complete mitochondrial genomes from all species in this study, which are similar in size (21.2–24.5 kilobases) to published rodent Pneumocystis mitogenomes (24.6–26.1 kb)17 but smaller than that of P. jirovecii (~35 kb)17 (Supplementary Table 3). P. macacae has a circular mitogenome similar to P. jirovecii17 whereas all other sequenced species have linear mitogenomes.
Genomic differences among Pneumocystis species
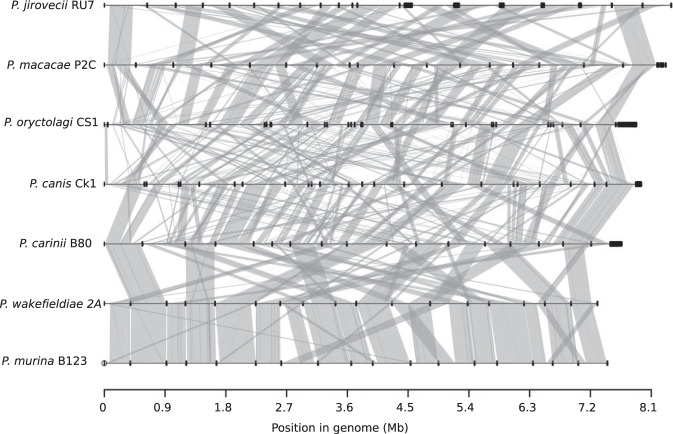
To assess the extent of genome structure variations among species, we generated whole-genome alignment of all representative genome assemblies. We found high levels of interspecies rearrangements ranging from 10 breakpoints between P. wakefieldiae and P. murina to 142 between P. jirovecii and P. oryctolagi (Fig. 1; Supplementary Table 4). The vast majority of chromosomal rearrangements were inversions, which, for example accounted for 23 out of 29 breakpoints between P. jirovecii and P. macacae (Supplementary Table 4). Analysis of aligned raw Nanopore and/or Illumina reads back to the assemblies show no evidence of incorrect contig joins around rearrangement breakpoints. There are clearly fewer rearrangements among rodent Pneumocystis species (P. wakefieldiae, P. carinii, and P. murina) than among all other species (Fig. 1; Supplementary Table 4), which is likely due to the younger evolutionary ages of rodent Pneumocystis (Fig. 2a and c). These rearrangements could have caused incompatibilities between species, thus preventing gene flow for species that infect the same host.
Fig. 1. Whole-genome structure and synteny among Pneumocystis species.
Species names and their genome assembly identifiers are shown on the left. Horizontal black lines on the right represent sequences of all scaffolds for each genome laid end-to-end, with their nucleotide positions indicated at the bottom. Dark thick squares represent short scaffolds. Syntenic regions between genomes are linked with vertical gray lines.
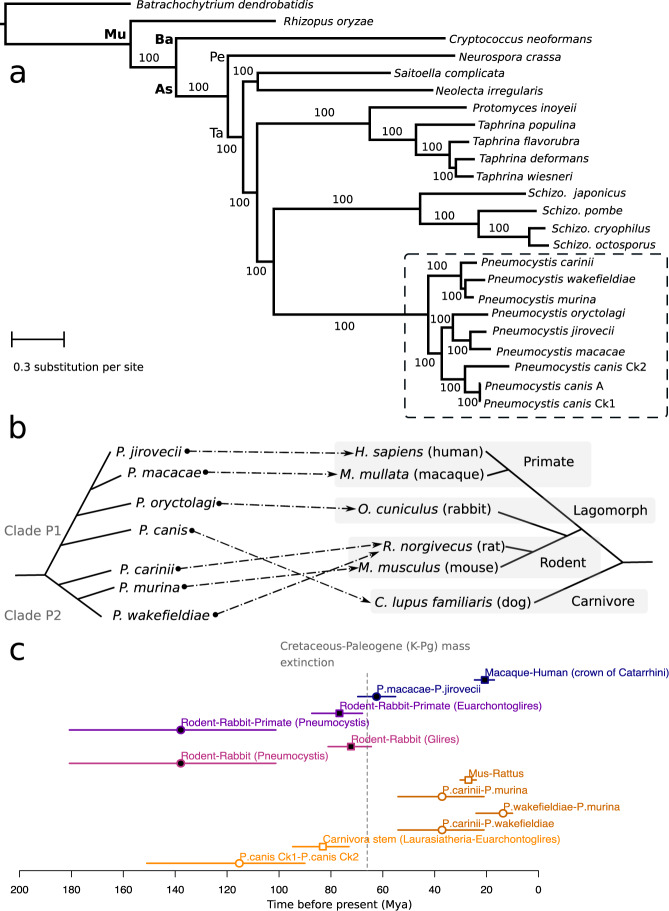
Fig. 2. Phylogeny and divergence times of Pneumocystis species.
a Maximum likelihood phylogeny constructed using 106 single-copy genes based on 1000 replicates from 24 annotated fungal genome assemblies including nine from Pneumocystis (highlighted with a dashed box). Only one assembly is shown for each species except there are three for P. canis (assemblies Ck1, Ck2, and A). Bootstrap support (%) is presented on the branches. The fungal major phylogenetic phyla and subphyla are represented by their initials: As (Ascomycota), Ba (Basidiomycota), Pe (Pezizomycotina), Mu (Mucoromycota), and Ta (Taphrinomycotina). b Schematic representation of species phylogeny and association between Pneumocystis species and their respective mammalian hosts. The dashed arrows represent the specific parasite-host relationships. c Divergence times of Pneumocystis species and mammals (n = 12 taxonomic clades analyzed). Divergence time medians are represented as squares for hosts and as circles for Pneumocystis, and the horizontal lines represent the 95% confidence interval (CI) error bars, which are color-coded the same for each Pneumocystis and its host. Closed elements represent nodes that are different in term of divergence times (nonoverlapping confidence intervals) whereas open elements represent nodes with overlapping confidence intervals. Catarrhini, taxonomic category (parvorder) including humans, great apes, gibbons, and Old-World monkeys. Euarchontoglires, superorder of mammals including rodents, lagomorphs, treeshrews, colugos, and primates. Glires, taxonomic clade consisting of rodents and lagomorphs. Laurasiatheria, taxonomic clade of placental mammals that includes shrews, whales, bats, and carnivorans. Mya, million years ago. K-Pg, Cretaceous-Paleogene. The dotted vertical line representing the K-Pg mass extinction event at 66 mya is included for context only.
Comparison of pairwise whole-genome alignment identities between species indicates a substantial nucleotide divergence: 14% dissimilarity in aligned regions between P. jirovecii and P. macacae; 21% between P. jirovecii and P. oryctolagi; 22% between P. jirovecii and P. canis Ck1; 15% between P. wakefieldiae and P. carinii; and 12% between P. wakefieldiae and P. murina (Supplementary Table 5).
To understand the relationship between the intraspecies and interspecies genetic diversity of Pneumocystis, we generated additional four P. jirovecii and five P. macacae genome assemblies from low sequence coverage samples. All data are expressed as the mean ± standard deviation. The pairwise intraspecies genome divergences among P. jirovecii genome assemblies (0.3 ± 0.2%, n = 8) are significantly lower than those obtained when comparing them to P. macacae assemblies (16.1 ± 0.2%, n = 5) (two sample t-test, p-value = 1.4 × 10−14) or to other Pneumocystis species (21.6 ± 0.9%, n = 7) (p = 2.9 × 10−10). Similarly, mean divergence among P. macacae genome assemblies (0.8 ± 0.3%, n = 5) is lower than divergence when they are compared to P. jirovecii assemblies (15.6 ± 0.2%, n = 8) (p = 1.3 × 10−10) or other Pneumocystis species genome assemblies (21.8 ± 0.9%) (p = 7.2 × 10−11). The results indicate that interspecies divergence exceeds intraspecies divergence, which is consistent with a complete species separation.
Speciation history of the Pneumocystis genus
These new complete genome data enabled us to examine the relationships between different Pneumocystis species and to estimate the timing of speciation events that led to the extant species. We inferred a strongly supported phylogeny of Pneumocystis species rooted with outgroups from distantly related fungal subphyla. Our phylogenomic analysis of 106 single-copy orthologs inferred from all assemblies including the fragmented Ck2 strongly supports monophyly of Pneumocystis species (100% Maximum likelihood bootstrap values; Fig. 2a), Bayesian posterior probabilities (>0.95; Supplementary Fig. 1), and highly significant support from the Shimodaira–Hasegawa test18 (p < 0.001; see Methods). An identical phylogeny was recovered using mitochondrial genome data from 33 specimens representing 7 Pneumocystis (Supplementary Fig. 2). However, we identified unexpected placements of P. wakefieldiae, P. oryctolagi, and P. canis. First, P. wakefieldiae appears as a sister species of P. murina instead of P. carinii (which also infects rats) (Fig. 2b). This observation is supported by the higher similarity in genome size (Supplementary Table 3), lower sequence divergence (Supplementary Table 4), higher genome synteny (Fig. 1; Supplementary Table 5) and higher frequencies of supporting genes (0.64 in 1,718 nuclear gene trees examined; Methods) between P. wakefieldiae and P. murina than between P. wakefieldiae and P. carinii. These relationships contradict the previous phylogenetic placement of P. wakefieldiae as an outgroup of the P. carinii/P. murina clade9 or a sister species of P. carinii19 based on analysis of mitochondrial large and small subunit rRNA genes (mtLSU and mtSSU). Our phylogeny also opposes the prevailing hypothesis for dynamics of host specificity and coevolution within the Pneumocystis genus, that is, P. wakefieldiae shares with P. carinii the same host species (Rattus norvegicus) and thus is expected to be more related to P. carinii than to P. murina.
Similarly, P. oryctolagi would be expected to be phylogenetically closer to rodent Pneumocystis than to primate Pneumocystis, consistent with the closer phylogenetic relationships of rabbits and rodents to each other than to primates20 (Fig. 2a, b). In contrast, P. oryctolagi and P. canis are more closely related to primate Pneumocystis (P. jirovecii and P. macacae) than rodent Pneumocystis (Fig. 2a; Supplementary Figs. 1 and 2; 100% of tree level support in 1718 nuclear genes). The phylogenetic discrepancy between P. oryctolagi and its host (rabbit) suggests that host switching may have occurred in their distant history.
From whole-genome Bayesian phylogenetic estimates (see Methods), the common ancestor of all extant species of the genus emerged around 140 mya (confidence intervals: 180–101 mya; Fig. 2c; Supplementary Fig. 1), with a separation of Pneumocystis and Schizosaccharomyces genera around 512 mya (CI: 822–203 mya), which is consistent with independent estimates of the origin of Taphrinomycota crown group at 530 mya21. The Pneumocystis genus thereafter divided into two main clades, P1 consisting of P. jirovecii, P. macacae, P. oryctolagi, and P. canis, and P2 consisting of species infecting rodents (P. carinii, P. wakefieldiae, and P. murina) (Fig. 2b). Subsequent to the divergence of P1/P2, the clade P1 diversified through a series of speciation events leading either to primate or carnivore species whereas P2 remained localized in rodents. We also found that the divergence time of Pneumocystis in the clade P1 predates that of their hosts, that is, the crown of rodent-rabbit-primate Pneumocystis is clearly more ancient than the corresponding superorder of mammals (Euarchontoglires) (Fig. 2c). The pattern in clade P2 is different as the divergence time estimates overlap with those of their hosts (Fig. 2c). On the basis of coalescent estimates, P. jirovecii began to split from P. macacae at ~62 mya (CI: 69–55 mya), which extended through the Cretaceous-Paleogene mass extinction event at 66 mya, but substantially predates the crown Catarrhini (human-macaque ancestor) of ~20 mya (CI: 24–17 mya; Fig. 2c; Supplementary Fig. 1).
High levels of population differentiation among Pneumocystis genomes support reproductive isolation
To understand the genomic divergence landscape of Pneumocystis populations, we performed genome-wide differentiation tests (FST, relative population divergence) and nucleotide diversity (π). These analyses used 32 genomic datasets, including 26 publicly available datasets in GenBank for P. jirovecii, P. carinii and P. murina and six datasets generated in this study for other four Pneumocystis species (Supplementary Note 1; Supplementary Table 2). We used a trained version LAST22 to account for interspecies divergence during read mapping and ANGSD23 to derive genotype likelihoods instead of genotypes. Since ANGSD’s FST requires outgroups, we analyzed interspecies divergence between P. jirovecii, P. macacae, and P. oryctolagi populations using a sliding window approach of 5-kb and P. carinii as an outgroup species (n samples = 59). P. murina genomic divergence relative to P. carinii and P. wakefieldiae populations was estimated similarly using P. jirovecii as an outgroup species (n = 47). We found high levels of population differentiation among Pneumocystis specimens; 71.9% of the P. jirovecii genome had a Fixation index (FST) > 0.8 compared to the closest species, P. macacae, while 90.2% of the genome had a FST > 0.8 compared to the extant species P. oryctolagi (Supplementary Fig. 3). Similarly, 86.3% and 93.7% of the P. murina genome had a FST > 0.8 compared to P. carinii and P. wakefieldiae, respectively (Supplementary Note 1).
Analyzing historical hybridization in Pneumocystis genus
Topology-based maximum likelihood analysis of 1718 gene trees using PhyloNet24 found no evidence of gene flow among species of clade P1 (P. jirovecii, P. macacae, P. oryctolagi, and P. canis) (Supplementary Fig. 4), which indicates that these lineages were reproductively isolated throughout their evolutionary history, consistent with their isoenzyme diversity25. In contrast, we found strong evidence of ancient hybridization in clade P2, possibly between P. carinii and P. wakefieldiae (Supplementary Fig. 4), which may then have contributed to the formation of the P. murina lineage. We hypothesize that P. murina might have originated as a hybrid between ancestors of P. carinii and P. wakefieldiae in rats, and subsequently shifted to mice, possibly owing to the geographic proximity of ancestral rodent populations (for example in Southern Asia26), which is consistent with the fact that ecological fitting is a major determinant of host switch27. The presumed physiological, cellular and/or immunological similarities among closely related rodent species might also have helped the same Pneumocystis species colonizing multiple closely related rodent species10,27.
Gene families and metabolic pathways linked to host specificity
The predicted protein-coding gene numbers are similar across Pneumocystis genomes and range from 3211 in P. wakefieldiae to 3476 in P. canis strain Ck1 (Supplementary Table 3). Nearly all predicted protein-coding genes in P. macacae (96% of 3471) and P. wakefieldiae (99% of 3221) genomes have RNA-Seq support. Gene models present a complex architecture with ~6 exons per gene on average. High representation of core eukaryotic genes in P. macacae, P. oryctolagi, P. canis and P. wakefieldiae provides evidence that these genomes are nearly complete and comparable in completeness to P. jirovecii, P. murina, and P. carinii genomes: 86.2–93.4% of conserved genes are detectable in all annotated genome assemblies (Supplementary Table 3).
Examination of orthologous genes reveals that ~3100 orthologous clusters had representative genes from all nine analyzed genome assemblies from seven Pneumocystis species (Supplementary Table 3). We found a small number of unique genes in each Pneumocystis species ranging from 25 in P. wakefieldiae to 204 in P. oryctolagi (Supplementary Table 3). Unique genes in most species encode for phylogenetically unrelated proteins with unknown function. A striking exception is observed in P. macacae in which nearly all unique proteins are part of an undescribed large protein family (n = 190). The members of this family are enriched in arginine and glycine amino-acid residues (denoted RG proteins) (Supplementary Fig. 5a) and have no similarities with transposable elements. While RG motifs are often found in eukaryotic RNA-binding proteins28, P. macacae RGs do not possess an RNA-binding domain (Pfam domains PF00076, PF08675, PF05670, PF00035), suggesting a different role. In addition, P. macacae RGs lack functional annotation except for two proteins that encode a Dolichol-phosphate mannosyltransferase domain (PF08285) and a leucine zipper domain (PF10259), respectively. Of the 190 RGs, 134 have RNA-Seq based gene expression support, including five among the top highly expressed genes (Supplementary Fig. 5b). Nearly half of RGs are located at subtelomeric regions, often found in close proximity to msg genes (Supplementary Data 1). RG proteins can be grouped in three main clusters (based on OrthoFinder clustering; Methods), have a reticulate phylogeny (Supplementary Fig. 5c) and a mosaic gene structure (Supplementary Fig. 5d), which suggest frequent gene conversion events.
To investigate the gene loss patterns in sequenced genomes, we compared Pneumocystis gene catalogs to those of related Taphrinomycotina fungi. We found that all sequenced Pneumocystis species have lost ~40% of gene families present in other Taphrinomycotina (Supplementary Fig. 6), and that the metabolic pathways are also very similar among Pneumocystis species with a few minor (possibly stochastic) differences (Supplementary Note 2). This strongly suggests that Pneumocystis ancestry experienced massive gene losses that occurred before the genus diversification.
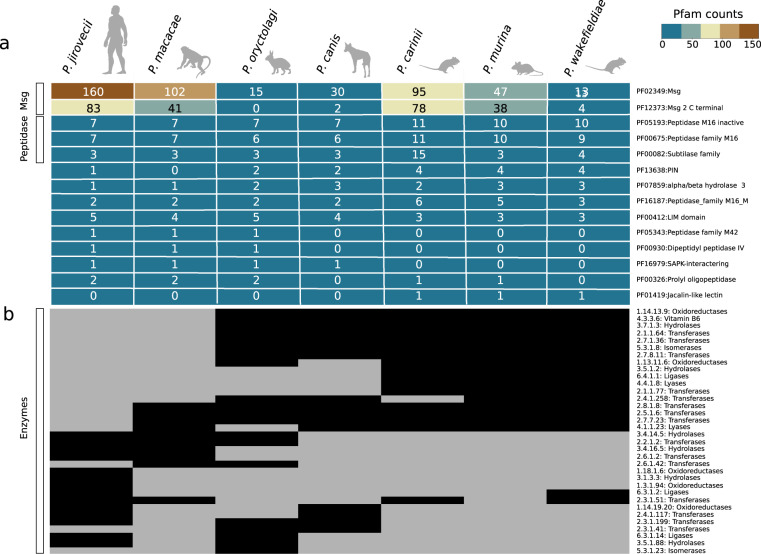
To investigate changes in gene content that might explain interspecies differences among the seven Pneumocystis species, we searched for expansions or contractions in functionally classified gene sets. We identified Pfam domains with significantly uneven distribution among genomes (Wilcoxon signed-rank test p < 0.05). Domains associated with Msg proteins are enriched in P. jirovecii and, to a lesser extent in P. macacae compared to other species (Fig. 3a). Domains associated with peptidases (M16) are enriched in P. carinii, P. murina, and P. wakefieldiae. S8 peptidase family (kexin) is expanded in P. carinii with 13 copies13 whereas all other species have one or three copies (Fig. 3a; Supplementary Fig. 7). Although kexin is localized in other fungi to the Golgi apparatus, and in Pneumocystis is believed to be involved in the processing of Msg proteins, the expanded copies are predicted to be GPI-anchored proteins, and appear to localize to the cell surface; their function is unknown29. We found that P. carinii kexin genes evolved under strong positive selection (p = 0.008) whereas P. wakefieldiae kexin genes did not (p = 0.159).
Fig. 3. Distribution of protein families among Pneumocystis species.
a Heatmap of Pfam protein domains with significant differences (Wilcoxon signed-rank test, p < 0.05) are included if the domain appears at least once in the following comparisons: primate Pneumocystis (P. jirovecii and P. macacae) versus other Pneumocystis, clade P1 (P. jirovecii, P. macacae, P. oryctolagi, and P. canis Ck1) versus clade P2 (P. carinii, P. murina, and P. wakefieldiae), primate Pneumocystis versus clade P2. The number of proteins containing each domain is indicated within each cell for each species. The heatmap is colored according to a score, as indicated by the key at the upper right corner. b Heatmap of distribution of enzymes (represented by Enzyme Commission numbers and their KEGG functional categories), with their presence and absence indicated by black and grey colored cells, respectively. Animal icons were obtained from http://phylopic.org under creative commons licenses https://creativecommons.org/licenses/by/3.0/: mouse (Anthony Caravaggi; license CC BY-NC-SA 3.0); dog (Sam Fraser-Smith and vectorized by T. Michael Keesey; license CC BY 3.0), rabbit (by Anthony Caravaggi; license CC BY-NC-SA 3.0), and rat (by Rebecca Groom; license CC BY-NC-SA 3.0). Icons original black color background were modified to light gray.
Phylogenetic analysis of CFEM (common in fungal extracellular membrane) protein domains, which are important for the acquisition of vital compounds in fungi30, suggest that these domains were likely already present in the last common ancestor of Pneumocystis and were vertically transmitted (Supplementary Fig. 8).
To investigate changes in enzyme gene content that might account for interspecies differences among Pneumocystis species, we searched for enzymes that show clear differences among species, which are represented by Enzyme Commission numbers (ECs) (Fig. 3b). We found 34 ECs, which include 14 that are highly conserved in P. jirovecii but have a patchy distribution in other members of clade P1 (P. macacae, P. canis, and P. oryctolagi) and are lost in clade P2 (P. carinii, P. murina, and P. wakefieldiae). Most of these 14 ECs are assigned to the biosynthesis of antibiotics or secondary metabolites and vitamin B6 metabolism according to KEGG pathways. The latter pathway seems only functional in P2 clade (Supplementary Note 2).
Intron evolution
We analyzed 1211 one-to-one gene orthologs shared by all sequenced Pneumocystis and other Taphrinomycotina fungi (Supplementary Fig. 9a). A total of 9080 homologous sites within 1211 alignments were identified (Supplementary Fig. 9b). While intron densities are similar among Pneumocystis species (ranging from 4842 in P. macacae to 5289 in P. murina), they are markedly more elevated compared to related Taphrinomycotina, including Neolecta irregularis (n = 4202 introns), Schizosaccharomyces pombe (n = 862), and Taphrina deformans (n = 639) (Supplementary Fig. 11b). Predictions of ancestral intron densities show that the Pneumocystis common ancestor had at least 5341 introns, of which 37% were not found in other Taphrinomycotina (Supplementary Fig. 9c). This is in contrast to other fungi; ~26% of Neolecta introns were independently acquired whereas S. pombe and T. deformans genomes have experienced intron losses, which is consistent with published studies31,32. These results suggest that the emergence of Pneumocystis genus was preceded by gain of introns.
Positive selection footprints in P. jirovecii genes
We tested the hypothesis that P. jirovecii has adapted specifically to humans after its separation with P. macacae, and that there will be footprints of directional selection in the genome that point to the molecular mechanisms of this adaptation. To infer P. jirovecii-specific adaptive changes, we compared the P. jirovecii one-to-one orthologs to those of P. macacae and P. oryctolagi using the branch-site likelihood ratio test33. Positive selection was identified as an accelerated nonsynonymous substitution rate. The test identified 244 genes (out of 2466) with a signature of positive selection in the human pathogen P. jirovecii alone (Bonferroni corrected p-value < 0.05; Supplementary Data 2). Gene Ontology enrichment analysis of these genes with accelerate rates identified significant enrichment for the biological process “cellular response to stress” (adjusted using Benjamini–Hochberg p-value = 1.9 × 10−6) and the molecular function “potassium channel regulator activity” (p = 2.8 × 10−10). Among the 244 genes, 197 are conserved in all Pneumocystis genomes available whereas 47 are absent in clade P2 only (P. carinii, P. murina, and P. wakefieldiae; Fig. 2b). While the latter set of genes encode proteins of unknown function, analysis of Pfam domains shows a significant enrichment in the biological process “nucleoside phosphate biosynthetic” process (p = 9.9 × 10−5) and the molecular function “carbon–nitrogen lyase activity” (p = 2.8 × 10−10). Further investigations will be required to determine the precise functions of these genes.
Subtelomeric gene families
Until recently, the only in-depth data on the subtelomeric gene families in Pneumocystis have come from the P. jirovecii, P. carinii, and P. murina genomes13,34. These genes, including msg and kexin, are believed to be important for antigenic variation, and are well represented in the assemblies of P. macacae, P. oryctolagi, P. canis, and P. wakefieldiae.
P. macacae subtelomeres encode numerous arrays of Msg and RG proteins (Supplementary data 1). Phylogenetic analysis of adjacent genes revealed only a few instances of recent paralogs, which suggests that most of the duplications and subsequent positional gene arrangements are ancient. Three P. macacae subtelomeric regions have a nearly perfect synteny in P. jirovecii with the only difference being the absence of RG proteins in P. jirovecii (Supplementary Data 1). P. oryctolagi subtelomeres tend to be enriched in orphan genes that are not members of the Msg superfamily, and are of unknown function. P. canis subtelomeres are enriched in Msg-C family (see Msg section below). P. wakefieldiae subtelomeres are rich in msg genes, though their types are distinct from those of P. carinii and P. murina.
Evolution of msg genes
Up to 6% of the Pneumocystis genomes are comprised of copies of the msg superfamily, which are believed to be crucial mediators of pathogenesis through antigenic variation and interaction with the host cells. The superfamily is classified into five families A, B, C, D, and E based on protein domain architecture, phylogeny and expression mode13,34,35. The A family is the largest of the five, has been subdivided into three subfamilies (A1, A2, and A3) and is generally thought to contribute to antigenic variation. Their protein products contain cysteine-rich domain classified as N1 and M1 to M5.
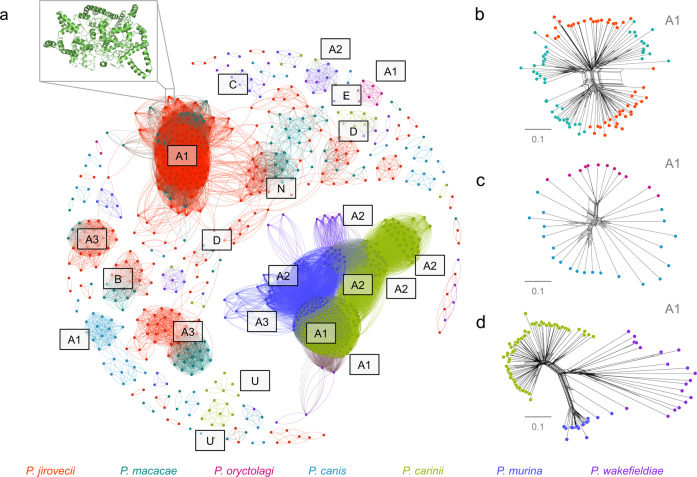
To investigate the origin of msg genes, we used previously developed Hidden Markov Models13 to search for corresponding gene models in the assemblies of P. macacae, P. oryctolagi, P. canis, and P. wakefieldiae and combined these data with published msg sequences annotated in P. jirovecii, P. carinii, and P. murina genomes13,35. Of note, in this study only a subset of msg genes were assembled for P. oryctolagi, P. canis, and P. wakefieldiae due to difficulties in assembling highly similar short reads from Illumina sequencing exclusively while a potentially complete set of msg genes were assembled for P. macacae using Illumina and Nanopore reads (Supplementary Table 3). The number of full-length msg genes available ranges from 9 in P. oryctolagi to 161 in P. jirovecii. Sequence-based clustering and phylogenetic analyses of all msg genes (n = 482) revealed that: (i) there is no evidence of interspecies transfer among Pneumocystis species (Fig. 4b to d; Supplementary Fig. 10), (ii) msg genes may have a polyphyletic origin, i.e., distinct families were present in most recent ancestors of Pneumocystis (Supplementary Fig. 10a), although convergent evolution of msg cannot be ruled out; (iii) msg genes experienced recombination early in their history as estimated by phylogenetic network analysis (Supplementary Fig. 10b and c).
Fig. 4. Clustering of Pneumocystis major surface glycoproteins (Msg).
a Graphical representation of similarity between 482 Msg proteins from seven Pneumocystis species generated using the Fruchterman Reingold algorithm. A 3-D model of a representative member of Msg-A1 protein family (NCBI locus tag T551_00910) generated using DESTINI is presented in the upper left insert. Individual protein sequences are shown as dots and color-coded by species as shown at the bottom of the figure. The edge between two dots indicates a global pairwise identity equal or greater than 45%. The letters represent Msg families (A to E) and subfamilies (A1 to A3). N and U letters represent potentially novel Msg sequences (relative to our prior study35) and unclassified sequences, respectively. For sake of clarity only the major clusters were annotated. b Phylogenetic network of a subset of Msg-A1 family (n = 97) in primate Pneumocystis including P. jirovecii (red) and P. macacae (dark cyan) suggesting recombination events at the root of the network. Nodes with more than two parents represent reticulate events. Bars represent the number of amino acid substitutions per site. c Phylogenetic network of Msg family A1 (n = 33) in P. oryctolagi (red violet) and P. canis (light blue). d Phylogenetic network of Msg-A1 family (n = 113) in rodent Pneumocystis including P. carinii (green), P. murina (dark blue), and P. wakefieldiae (blue violet). The network data are available at 10.5281/zenodo.4450766.
While some gene expansions are relatively recent (for example, msg families A, C, and D) other expansions (msg families E and B) occurred before the emergence of Pneumocystis genus itself (Supplementary Fig. 11). Subsets of msg genes show strong host-specific sequence diversification (Fig. 4a), such as the current A family has emerged relatively recently at 43 mya (CI: 58–28 mya) compared to the emergence of the genus at 140 mya (see Methods; Supplementary Fig. 11). The A1 subfamily displays a substantial expansion in all species (Fig. 4a) and is subject to intraspecies recombination (Fig. 4b to d), which suggest that the most recent Pneumocystis ancestor may have developed a pre-Msg-A family, which then evolved through duplication and recombination after the species separation.
The A3 subfamily has expanded only in clade P1 (especially in P. jirovecii) whereas A2 has expanded only in clade P2 (P. carinii, P. murina and to a lesser extent in P. wakefieldiae) (Fig. 4a). Although all members of the A family might have a shared deep ancestry, we found no evidence suggesting that the A1, A2, A3 subfamilies are directly derived from one another (Supplementary Fig. 10).
The msg-B family underwent a net independent expansion in P. macacae (n = 10) and P. jirovecii (n = 12), while being reduced to only one copy in P. oryctolagi and P. canis, and being completely absent in P. wakefieldiae, P. carinii and P. murina (Fig. 4a). Using Bayesian estimates, we estimated the origin of the B family to be older than the Pneumocystis genus itself (~211 vs. 140 mya; Supplementary Fig. 11).
The msg-D family is expanded only in P. macacae and P. jirovecii. The D family emerged at ~69 mya (CI: 109–40 mya) before the split of these two species (Supplementary Fig. 11), thus suggesting a role in adaptation to primates similar to the A3 subfamily. In contrast, the E family, which is conserved in all species, is much more ancient at ~311 mya (CI: 541–158 mya), again preceding the emergence of the genus (Supplementary Fig. 11).
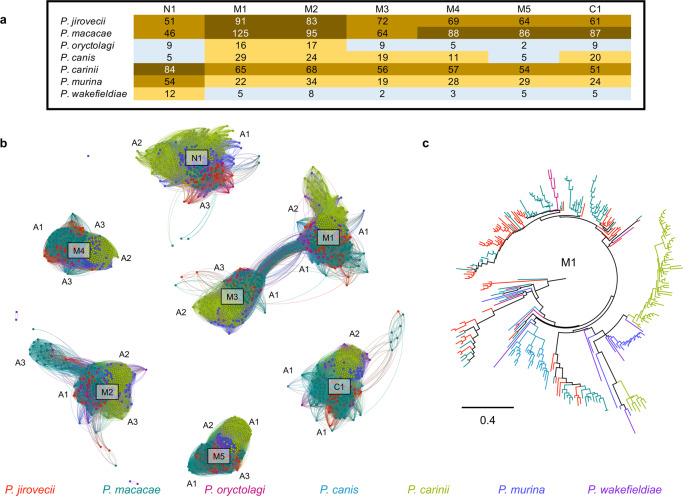
P. jirovecii and P. macacae not only have a larger number of msg-associated cysteine-rich domains than other Pneumocystis species (Fig. 5a) but also a much greater sequence diversity per domain than other Pneumocystis species (Fig. 5c). Domain sequences cluster independently, with each cluster containing sequences from all Pneumocystis species (Fig. 5b). Domains M1 and M3 are more closely related to each other than other domains, which suggests a relatively recent duplication.
Fig. 5. Evolution of Msg cysteine-rich protein domains in Pneumocystis.
a Heatmap showing the distribution of Msg domains in each Pneumocystis species. The color change from blue-orange-brown indicates an increase in the number of domains. b Graphical representation of protein similarity between domains, which highlights that the domains were present in the most recent common ancester and were maintained other than perhaps domains M1 and M3. Domains are clustered by a minimum BLASTp cutoff of 70% protein identity. c Maximum likelihood tree of the M1 domain. In both panels b and c, domains are color-coded by species as shown at the bottom.
Discussion
Surprisingly, analysis of core genomic regions of the nuclear genomes did not identify clear differences that are suggestive of mechanisms for host-specific adaptation; instead it is the highly polymorphic multicopy gene families (msgs) that appear to account for this adaptation. Msgs, which provide Pneumocystis with a mechanism for antigenic variation and consequent immune evasion, may have been important in allowing Pneumocystis organisms to infect mammals successfully, given that an adaptive immune system, which is critical to protection of mammals from exogenous pathogens, arose ~500 mya in the first jawed vertebrates36. Msgs likely played a dual role in avoiding the adapative immunity and in cell adherence.
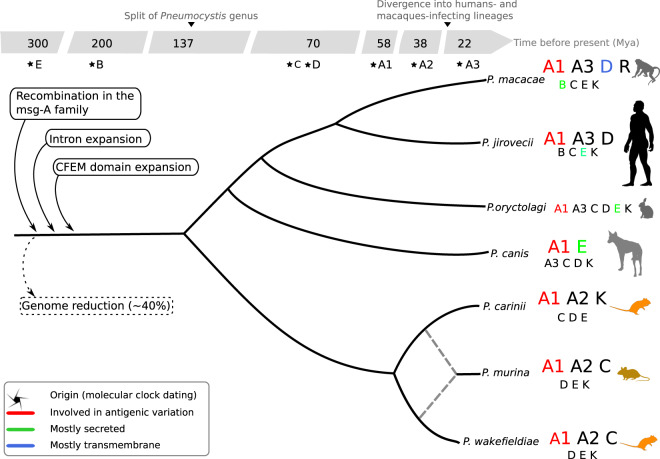
Based on our analysis, we propose the following series of events for the emergence and adaptation of P. jirovecii as a major human opportunistic pathogen (Fig. 6). First, there was a major shift of a pre-Pneumocystis lineage (possibly a soil- or plant-adapted organism) to mammals, which led to a genome reduction but with a proliferation of introns and expansions of cysteine-rich domain-containing proteins involved in immune escape and nutrient scavenging from hosts. Pneumocystis genomes encode multiple gene families that have experienced a rapid accumulation of mutations favoring fungal replication in mammals. Each Pneumocystis species has employed different strategies to adapt to their host including lineage-specific expansions of shared gene families such as msg A1, A3, and D in P. jirovecii or gain and expansion of RG proteins in P. macacae. In addition, some shared gene families also have acquired different properties (e.g., transmembrane domain and secreted signals) potentially contributing to host specificity. The absence of a reliable culture method and the inability to genetically manipulate Pneumocystis prevents directly testing our model. Moreover, for the genes that we have now implicated in the process of host adaptation, only a few have been functionally characterized. Future studies on the role of these genes will be important to elucidate the molecular basis of host-specific adaptation by Pneumocystis pathogens.
Fig. 6. Overview of the genomic evolution of the Pneumocystis genus.
Gene families are represented by letters: A to E for the five families of major surface glycoproteins (Msg) with the A family being further subdivided into three subfamilies A1, A2, and A3; K and R for kexins and arginine-glycine rich proteins, respectively. Larger fonts indicate expansions as inferred by maximum likelihood phylogenetic trees and networks. Dashed lines represent ancient hybridization between P. carinii and P. wakefieldiae. Detailed analysis also reveals distinct phylogenetic clusters within subfamilies. Introns and CFEM (common in fungal extracellular membrane) domains are enriched in Pneumocystis genes which indicate that these elements were likely present in the most recent common ancestor of Pneumocystis species. Animal icons were obtained from http://phylopic.org under creative commons licenses https://creativecommons.org/licenses/by/3.0/: mouse (Anthony Caravaggi; license CC BY-NC-SA 3.0); dog (Sam Fraser-Smith and vectorized by T. Michael Keesey; license CC BY 3.0), rabbit (by Anthony Caravaggi; license CC BY-NC-SA 3.0), and rat (by Rebecca Groom; license CC BY-NC-SA 3.0). Icons original black color background were modified to gray and orange colors.
Methods
Experimental design and Pneumocystis sample sources
Animal and human subject experimentation guidelines of the National Institutes of Health (NIH) were followed in the conduct of this study. Studies of human and mouse Pneumocystis infection were approved by NIH Institutional Review Board (IRB) protocols 99-I-0084 and CCM 19-05, respectively. The collection and processing of a single human P. jirovecii sample from China (Pj55) was approved by the IRB of the First Affiliated Hospital of Chongqing Medical University, China (protocol no. 20172901). Written informed consent was obtained from the patient for participation in this study. The authors confirm that personal information was unidentifiable from this report. The National Institute of Allergy and Infectious Diseases (NIAID) Division of Intramural Research Animal Care and Use Program, as part of the NIH Intramural Research Program, approved all experimental procedures pertaining to the macaques (protocol LVD 26). Nonhuman primate study protocols were approved by the Institutional Animal Care and Use Committee of the University of California, Davis (protocol no. 7092), the Tulane National Primate Research Center (TNPRC), and the Institutional Animal Care and Use Committee (IACUC) (protocol no. P0351R). Studies of rabbit Pneumocystis infection were reviewed and approved by the Institutional Animal Care and Use Committee of the University of Michigan (protocol no. RO00008218). For rabbit samples obtained from France, the conditions for care of laboratory animals stipulated in European guidelines were followed (See: Council directives on the protection of animals for experimental and other scientific purposes, and J. Off. Communautés Européennes, 86/609/EEC, 18 December 1986, L358). Samples from Pneumocystis-infected dog were collected as diagnostic samples and approved only for research purposes. The owner’s consents for using samples and data were obtained on admission of the case and no further ethics permission was required because it was a routine diagnostic case and did not qualify as an animal experiment. Studies of rat Pneumocystis infection were approved by the Veteran Affairs animal protocol (VA ACORP #17-12-05-01). Clinical information and demographic data of the groups of individuals are presented in Supplementary Table 1. Three P. jirovecii samples were obtained as bronchoalveolar lavage from patients at the NIH Clinical Center in Bethesda, MD, USA and Chongqing Medical University in Chongqing, China. Six P. macacae samples were obtained as frozen lung tissues or formalin fixed paraffin embedded (FFPE) tissue sections prepared from SIV-infected rhesus macaques at the NIH Animal Center, Bethesda, Maryland (n = 2), the Tulane National Primate Research Center, Covington, Louisiana (n = 3), and the UC Davis California National Primate Research Center, Davis, California, USA (n = 1).
Four P. oryctolagi samples were obtained as frozen lung tissues from one rabbit with severe combined immunodeficiency at the University of Michigan, Ann Arbor, Michigan, USA, or as DNA from two corticosteroid treated rabbits and one rabbit with spontaneous Pneumocystis infection at the Institut Pasteur de Lille and the Institut National de la Recherche Agronomique de Tours Pathologie Aviaire et Parasitologie, Tours, France. P. canis samples were obtained as DNA from one Cavalier King Charles Spaniel dog at the University of Helsinki, Finland and one Whippet mixed-breed at the University of Veterinary Medicine, Vienna, Austria. The dogs were not laboratory animals. One P. murina sample was obtained from a heavily infected CD40L-KO mouse following a short-term in vitro culture. Genomic data obtained from P. murina isolates were combined with previously sequenced public data (Supplementary Table 2) and used for population genomics analysis (section “Speciation history of the Pneumocystis genus” and Supplementary Note 1). One frozen cell pellet and 4 agarose gel blocks containing P. wakefieldiae and P. carinii were obtained from immunosuppressed rats (one gel block per rat) housed at the Cincinnati VA Medical Center, Veterinary Medicine Unit, Cincinnati, Ohio. Of note, all samples with low sequence coverage were used either in combination with other samples to generate consensus genome assemblies or for population genetic analyses in which variation in sequencing coverage are explicitly accounted for.
Genome sequencing, assembly, and annotation
Genomic DNA in agarose gel blocks was extracted using the Zymoclean Gel DNA Recovery Kit (Zymo Research). Genomic DNA in FFPE sections was extracted using the AllPrep DNA/RNA FFPE Kit (Qiagen). Genomic DNA in frozen lung tissues from two P. macacae-infected macaques and a single P. oryctolagi-infected rabbit was treated with a sequential collagenase type I and DNase I digestion13 to deplete host DNA and extracted using the MasterPure Yeast DNA purification kit (Epicentre Biotechnologies, Madison, WI, USA). Genomic DNA in bronchoalveolar lavage samples from P. jirovecii-infected patients was extracted using the MasterPure Yeast DNA purification kit. Total RNAs for P. macacae, P. wakefieldiae and P. murina were isolated using RNeasy Mini kit (Qiagen). For DNA samples with small quantity, including three P. oryctolagi samples (RABF, RAB1, and RAB2B) and one P. jirovecii sample (RU817), we performed whole-genome amplification prior to Illumina sequencing. Five microliters of each DNA sample were amplified in a 50-ul reaction using an Illustra GenomiPhi DNA V3 DNA amplification kit (GE Healthcare, United Kingdom). Genomic DNA samples were quantified using Qubit dsDNA HS assay kit (Invitrogen) and NanoDrop (ThermoFisher). RNA integrity and quality were assessed using Bioanalyzer RNA 6000 picoassay (Agilent). The identities of Pneumocystis organisms were verified by PCR and Sanger sequencing of mtLSU before high throughput sequencing. No quantitative PCR methods were used. For most of the DNA samples, at least one microgram of each DNA or RNA (depleted of ribosomal RNA using Illumina Ribo-Zero rRNA Removal Kit) sample was sequenced commercially using the Illumina HiSeq2500 platform with 150 or 250-base paired-end libraries (Novogene Inc, USA) or for one DNA sample of P. jirovecii from a Chinese patient using a single-read SE50 library using the MGISeq 2000 platform (MGI Tec, China).
Adapters and low-quality reads were discarded using trimmomatic v0.3637 with the parameters “-phred33 LEADING:3 TRAILING:3 SLIDINGWINDOW:4:15 MINLEN:36”. Host DNA and other contaminating sequences were removed by mapping against host genomes using Bowtie2 v2.4.138. Filtered Illumina reads were assembled de novo using Spades v3.11.139. Details for host DNA sequences removal, Pneumocystis reads recovery and de novo assembly protocols are presented in Supplementary Methods. Completeness of assemblies was estimated using BUSCO v940, FGMP v1.0.141 and CEGMA v2.542. For P. macacae, in addition to Illumina sequencing, Nanopore sequencing was performed on P. macacae DNA samples prepared from a single heavily infected macaque (P2C) with ~68% Pneumocystis DNA based on prior Illumina sequencing (Supplementary Table 2). High molecular weight genomic DNA fragments were isolated using the BluePippin (Sage Science) with the high-pass filtering protocol. A DNA library was prepared using the rapid Sequencing kit (SQK-RAD0004) from Oxford Nanopore Technologies (Oxford, UK) and loaded in the MinION sequencing device. Host reads were removed by mapping to the Rhesus macaque genome (NCBI accession number GCF_000772875.2_Mmul_8.0.1) using Minimap2 v2.1043. Unmapped reads were aligned to the draft version of P. macacae assembly built previously using Illumina data (Supplementary Methods) with ngmlr v0.2.743. A total of 1,633,376 Nanopore reads were obtained, of which ~5% were attributed to Pneumocystis (27-fold coverage), which is much less than the 68% based on Illumina data (Supplementary Table 2). This suggests that many P. macacae genomic DNA fragments were too short to pass the size selection filter. Pneumocystis Nanopore reads were assembled using Canu v.1.8.044, overlapped with the assembly using Racon v.1.3.345 and polished with Pilon v1.2246 using the Illumina reads aligned with BWA MEM v0.7.1747.
Illumina RNA-Seq of the P. macacae sample P2C yielded 22 million reads, of which ~92% were attributed to Pneumocystis (Supplementary Table 2). Filtered reads were mapped to the P. macacae assembly using hisat2 v2.2.048, sorted with SAMtools v1.1049 and filtered with PICARD v2.1.1 (http://broadinstitute.github.io/picard). De novo transcriptome assembly of filtered reads was performed with Trinity50. Quantification of transcript abundance was performed using Kallisto v0.46.151. P. wakefieldiae (2A) and P. murina RNA-Seq data were processed similarly (Supplementary Table 2). DNA transposons, retrotransposons and low complexity repeats were identified using RepeatMasker52, RepBase53 and TransposonPSI (http://transposonpsi.sourceforge.net). Pneumocystis telomere motif “TTAGGG”16 was searched using “FindTelomere” (available at https://github.com/JanaSperschneider/FindTelomeres). The genomes of P. carinii strain Ccin14 and strain SE612 were scaffolded with Satsuma54 using the P. carinii strain B80 as reference genome13. P. macacae, P. oryctolagi, P. canis Ck1, P. canis A, P. wakefieldiae, and P. carinii (strains Ccin and SE6) genome assemblies were annotated using Funannotate v1.5.3 (10.5281/zenodo.1134477). The homology evidence consists of fungal proteins from UniProt55 and BUSCO v9 fungal proteins40. For P. macacae and P. wakefieldiae, RNA-Seq mapping files (BAM) and de novo transcriptome assemblies were used as hints for AUGUSTUS56. Ab initio predictions were performed using GeneMark-ES57. All evidences were merged using EvidenceModeler58. Taphrina genomes (T. deformans, T. wiesneri, T. flavoruba, and T. populina32,59) and P. canis Ck2 genome were annotated using MAKER260 because predicted gene models showed a better quality than those obtained from Funannotate. MAKER2 integrates ab initio prediction from SNAP61, AUGUSTUS built-in Pneumocystis gene models62 and GeneMark-ES as well as BLAST- based homology evidences from a custom fungal proteins database. GPI prediction was performed using PredGPI63, big-PI64 and KohGPI65. Signal peptide leader sequences and transmembrane helices were predicted using Signal-P version 566 and TMpred67, respectively. Protein domains were inferred using Pfam database version 3.168 with PfamScan (https://bio.tools/pfamscan_api). PRIAM69 release JAN2018 was used to predict ECs using the following options: minimum probability > 0.5, profile coverage > 70%, check catalytic—TRUE and e-value < 10−3. Pneumocystis mitochondrial genome assembly and annotations are presented in Supplementary Methods. Three dimensional (3D) protein structure prediction of Msg proteins was performed using DESTINI70 and visualized with PyMol (www.pymol.org).
Comparative genomics
All genomes were pairwise aligned to the P. jirovecii strain RU7 genome NCBI accession GCF_001477535.113 using LAST version 92122 with the MAM4 seeding scheme71. One-to-one pairwise alignments were created using maf-swap utility of LAST package and merged into a single multispecies whole-genome alignment using LAST’s maf-join utility. Pairwise rearrangement distances in terms of minimum number of rearrangements were inferred using GRIMM72 and Mauve73. Breakpoints of genomic rearrangements were refined with Cassis74 and annotated using BEDtools75 ‘annotate’ command. Average pairwise genome-wide nucleotide divergences were computed with Minimap276. Synteny visualization was carried out using Synima77. Msg protein similarity networks were based on global pairwise identity obtained from pairwise alignments of full-length proteins using Needle78 or BLASTp79 identity scores for individual protein domains. The networks presented in Figs. 4 and 5 were generated using the Fruchterman Reingold algorithm as implemented in Gephi v0.9.280. To generate genome sequences for low coverage samples, raw Illumina reads were aligned to reference genomes with LAST. Resulting alignments were filtered and sorted with SAMtools. SNPs and indels were identified, normalized, filtered and used to generate consensus genomes using bcftools81. Pairwise divergence scores were computed using minimap2. Sequence motifs were visualized using WebLogo82. Multi-panel figures were assembled in Inkscape (https://inkscape.org).
To investigate the evolution of introns in Pneumocystis species, we identified unambiguous one-to-one orthologous clusters using reciprocal best Blast hit (e-value of 10−10 as cutoff) in seven Pneumocystis species as well as in three other Taphrinomycotina fungi: S. pombe, T. deformans and N. irregularis. Intron position coordinates were extracted from annotated genomes using Replicer83 and projected onto protein multiple alignments using custom scripts. Homologous splice sites in annotated protein sequence alignments were identified using MALIN84. We required at least 11 unambiguous splicing sites and five minimal nongapped positions. A potential splice was considered unambiguous if the site has at least five nongap positions in the aligned sequences in both the left and right sides. MALIN uses a rates-across sites markov model with branch specific gain and loss rates to infer evolution of introns. Gain and loss rates were optimized through numerical optimizations. Fungi have a strong tendency to intron loss with few exceptions (e.g., Cryptococcus) whereas gain of intron is relatively rare. Thus, we penalized intron gain and set the variation rate to 4/3 for loss and gain levels. Intron evolutionary history was inferred using a posterior probabilistic estimation with 100 bootstrap support values.
Phylogenomics
Orthologous gene families were inferred using OrthoFinder v.2.3.1185. In addition to Pneumocystis and Taphrina species, the predicted proteins for the following species were downloaded from NCBI: Neolecta irregularis (accession no. GCA_001929475.1), S. pombe (GCF_000002945.1), S. cryophilus (GCF_000004155.1), S. octosporus (GCF_000150505.1), S. japonicus (GCF_000149845.2), Saitoella complicata (GCF_001661265.1), Neurospora crassa (GCF_000182925.2), Cryptococcus neoformans (GCF_000149245.1), Rhizopus oryzae (GCA_000697725.1) and Batrachochytrium dendrobatidis (GCF_000203795.1). Single-copy genes were extracted from OrthoFinder output (n = 106) and concatenated into a protein alignment containing 458,948 distinct alignment patterns (i.e., unique columns in the alignment) with a gap proportion of 12.2%. Maximum likelihood tree analysis was performed using RAxML v 8.2.586 with 1000 bootstraps as support values. The LG model87 was selected as the best amino-acid model based on the likelihood PROTGAMMAAUTO in RAxML. One hundred and six gene trees were estimated from each of the single-copy genes. The Shimodaira–Hasegawa test18 was performed on the tree topology for each of the gene trees and the concatenated alignment using IQ-Tree88 with 1000 RELL bootstrap replicates. IQ-Tree was run as follows: “iqtree -s {input.phy} -z {input.t} -n 0 -zb {params.n}”, where {input.phy} represents the concatenated alignment of 106 genes in phylip format (24 species; 543,202 amino-acid sites with 14.2076% of constant sites), {input.t} represents a file containing individual maximum likelihood phylogenetic trees for each of the 106 genes, -n 0 parameter avoid tree search and estimate model parameters based on an initial parsimony tree (the best-fit model according to BIC was LG + F + R7) and the -zb option specifies the number of bootstrap replicates for the resampling estimated log-likelihood method (RELL).
To infer the species phylogeny using mitochondrial genomes, protein-coding genes were extracted, aligned using Clustal Omega89, and concatenated. The resulting alignment was used to infer phylogeny using IQ-Tree v.1.690 with TVM + F + I + G4 as the Best-fit substitution model and 1000 ultrafast bootstraps and SH-aLRT test. A total of 33 mitogenomes from seven Pneumocystis species were used: P. jirovecii [n = 18 including 3 sequences from this study and 4 from previous studies5,12,17], P. macacae (n = 4), P. oryctolagi (n = 4), P. canis [n = 4, refs. 17,91], P. carinii [n = 2, refs. 17,91], P. murina [n = 1, ref. 17], and P. wakefieldiae (n = 1). Phylogenetic reconciliations of species tree and gene trees were performed using Notung92. Ancestral reconstruction of gene family’s history was performed using Count93. Phylogenetic network for Msg protein families was inferred using SplitTree94. The detection of putative mosaic genes was performed using TOPALi v2.595.
Phylodating
Single-copy orthogroup nucleotide sequences were aligned using MACS v0.9b196. Highly polymorphic msg sequences were excluded using BLASTn79 with an e-value of 10−5 as cutoff against 479 published msg sequences13. We inferred the divergence timing using two datasets: (1) 24 single-copy nuclear gene orthologs shared by all Pneumocystis and S. pombe; and (2) 568 nuclear genes found in all Pneumocystis species. BEAST inputs were prepared using BEAUTi v2.5.197. Unlinked relaxed lognormal molecular clock models98,99 and calibrated birth-death tree priors100 were used to estimate the divergence times and the credibility intervals. The substitution site model HKY was applied101. Three secondary calibration priors were used: (i) P. jirovecii/P. macacae divergence with a median time of 65 mya as 95% highest posterior density (HPD)5), (ii) the emergence of the Pneumocystis genus at a minimum age of 100 mya4, and (iii) the Schizosaccharomyces—Pneumocystis split at ~467 mya102. We sampled from various priors and found a minor difference between the marginal posterior distribution on rate and the marginal prior distribution. This indicates that the posterior simply reflects the prior. For the dataset 2, the 568 gene alignments were concatenated in a super alignment with 568 partitions, with each partition defined by one gene. Gene partitions were collapsed using PartitionFinder v2.1.1103 with the “greedy” search to find optimal partitioning scheme. The alignment was split in three partitions in BEAST. Three independent runs for each dataset were performed separately for 60 million generations using random seeds. Run convergence was assessed with Tracer v1.7.1 (minimum effective sampling size of 200 with a burn-in of 10%). Trees were summarized using TreeAnnotator v.2.5.1 (http://beast.bio.ed.ac.uk/treeannotator) and visualized using FigTree v.1.4.4 (http://tree.bio.ed.ac.uk/software/figtree) to obtain the means and 95% HPD. Host divergences were obtained from the most recent mammal tree of life6, available at http://vertlife.org/data/mammals. The dating of fungal gene families was performed similarly. Phylogenetic trees with geological time scale were visualized using strap version 1.4104.
Population genomics
Sequence data sources and primary statistics are presented in Supplementary Table 2. Adapter sequences and low-quality headers of base sequences were removed using Trimmomatic37. Interspecies read alignment was performed using LAST22 with the MAM4 seeding scheme71. Alignments were processed by last-split utility to allow interspecies rearrangements, sorted using SAMtools v1.1049. Duplicates were removed using PICARD v2.1.1. To compute the FST and nucleotide diversity (Watterson, pairwise, FuLi, fayH, L), we calculated the unfolded site frequency spectra for each population using the Analysis of Next Generation Sequencing Data (ANGSD)23. Site frequency spectra was estimated per base site allele frequencies using ANGSD23,105. Hierarchical clustering was performed using ngsCovar106. All data were formatted to fit a sliding windows of 1-10 kb using BEDTools107. For each window, an average value of the statistics was calculated using custom scripts.
Gene flow inference
To infer a phylogenetic network, we used 1718 one-to-one orthologs from gene catalogs of seven Pneumocystis species using reciprocal best BLASTp hit with an e-value of 10−10 as cutoff. Sequences from each orthologous group were aligned using Muscle108. Alignments with evidence of intragenic recombination were filtered out using PhiPack109 with a p-value of 0.05 as cutoff. For each aligned group a maximum likelihood (ML) tree was inferred using RAxML-ng110 with GTR + G model and 100 bootstrap replicates, and Bayesian tree was generated using BEAST297. ML trees were filtered using the following criteria: 0.9 as the maximum proportion of missing data, 100 as the minimum number of parsimony-informative sites, 50 as the minimum bootstrap node-support value and 0.05 as the minimum p-value for rejecting the null hypothesis of no recombination within the alignment. BEAST trees with an effective sampling size <200 were removed. Filtered trees were summarized using Treannotator (https://www.beast2.org/treeannotator/). Summary trees with an average posterior support inferior to 0.8 were discarded. Species network was inferred using PhyloNet option “InferNetwork_MPL”24 with prior reticulation events ranging from 1 to 4. Phylogenetic networks were visualized using Dendroscope 3111.
The highest probability network inferred a hybridization between P. carinii and P. wakefieldiae leading to P. murina followed by a backcrossing between P. murina with P. wakefieldiae (log probability = −12759.4). Analysis of tree topology frequencies revealed that 64% of the trees were consistent with the topology of (P. carinii, (P. murina, P. wakefieldiae)), 28% with the topology of (P. wakefieldiae, (P. carinii, P. murina)) and 8% with the topology of (P. murina, (P. carinii, P. wakefieldiae)).
Detection of positive selection
To search for genes that have been subjected to positive selection in P. jirovecii alone after the divergence from P. macacae, we used the branch-site test33 as implemented in PAML112, which detects sites that have undergone positive selection in a specific branch of the phylogenetic tree (foreground branch). A set of 2466 orthologous groups between P. jirovecii, P. macacae and P. oryctolagi was used for the test. dN/dS ratio estimates per branch per gene were obtained using Codeml (PAML v4.4c) with a free ratio model of evolution. This process identified 244 genes with a signal of positive selection only in P. jirovecii (dN/dS > 1).
Statistics and reproducibility
All analyses were conducted in R version 3.3.2113. Statistical details and experimental design for different data analyses are presented in the respective results and methods sections. No sample-size calculation was performed for genome sequencing. The assessment of protein domains associated Gene Ontologies (GO) term enrichment was performed using hypergeometric test as implemented in dcGOR version 1.0.6114 (p-values adjusted by Benjamini–Hochberg method). The statistical significance of differences among groups was determined using Wilcoxon signed-rank test.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Acknowledgements
This work has been funded in whole or in part with federal funds from the Intramural Research Program of the US National Institutes of Health (NIH) Clinical Center and the National Institute of Allergy and Infectious Diseases (NIAID). This study used the Office of Cyber Infrastructure and Computational Biology (OCICB) High Performance Computing (HPC) cluster at the National Institute of Allergy and Infectious Diseases (NIAID), Bethesda, MD. This study also utilized the high-performance computational capabilities of the Biowulf Linux cluster at the NIH, Bethesda, MD (http://biowulf.nih.gov). The content of this publication neither necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. M.T.C. is a VA Senior Research Career Scientist supported by 5IK6BX005232. Her lab is funded by support from the NIH R01HL146266 and VA Grant I01BX004441. J.E.S. and C.A.C. are CIFAR Fellows in the program Fungal Kingdom: Threats and Opportunities. Animal icons used in Figs. 3 and 6 were obtained from http://phylopic.org under creative commons licenses https://creativecommons.org/licenses/by/3.0/.
Author contributions
O.H.C., L.M. and J.A.K. conceived the project and designed all the experiments. L.M., O.H.C., C.W.L., J.B., J.X., J.S., R.B., B.P., K.V.R., R.K., A.S., M.C., V.H., J.C., L.P., M.T.C., G.K., Y.L. and J.A.K. performed the laboratory work to obtain samples for sequencing. O.H.C., L.M., J.P.D., P.P.K. and J.L. developed and implemented methods for sample processing, library preparation and sequencing. O.H.C., L.M., J.E.S., C.A.C. and N.S.U. analyzed the data. O.H.C., L.M. and J.A.K. drafted the manuscript, which was revised by all authors.
Funding
Open Access funding provided by the National Institutes of Health (NIH).
Data availability
The datasets generated during and/or analyzed during the current study are available at NCBI BioProject portal (https://www.ncbi.nlm.nih.gov/bioproject/): Pneumocystis macacae strain P2C (no. PRJNA632025); P. oryctolagi strain CS1 (PRJNA632560); P. canis strain Ck1 (PRJNA632556); P. canis strain Ck2 (PRJNA632878); P. canis strain A (PRJNA636786);P. wakefieldiae strain 2A (PRJNA632570); P. jirovecii strain 55 (PRJNA647920), P. jirovecii strain 54c (PRJNA648092), P. jirovecii strain 46 (PRJNA648096), P. macacae strain CJ36 (PRJNA648103), P. macacae strain ER17 (PRJNA648108), P. macacae strain UC86 (PRJNA648112), and P. macacae strain GL92 (PRJNA648115).
Code availability
All custom bioinformatic analyses were conducted using Perl v5.26.0 (http://www.perl.org/) or Python v.3.6 (http://www.python.org) scripts. Pipelines were written with Snakemake v5.11.2115. Custom codes generated for this project are available at GitHub: https://github.com/ocisse/pneumocystis_evolution and a stable released version is available at 10.5281/zenodo.4450766.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Ousmane H. Cissé, Liang Ma.
Contributor Information
Ousmane H. Cissé, Email: ousmane.cisse@nih.gov
Liang Ma, Email: mal3@nih.gov.
Joseph A. Kovacs, Email: jkovacs@nih.gov
Supplementary information
The online version contains supplementary material available at 10.1038/s42003-021-01799-7.
References
- 1.Durand-Joly I, et al. Pneumocystis carinii f. sp. hominis is not infectious for SCID mice. J. Clin. Microbiol. 2002;40:1862–1865. doi: 10.1128/JCM.40.5.1862-1865.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gigliotti F, Harmsen AG, Haidaris CG, Haidaris PJ. Pneumocystis carinii is not universally transmissible between mammalian species. Infect. Immun. 1993;61:2886–2890. doi: 10.1128/IAI.61.7.2886-2890.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cushion MT, Keely SP, Stringer JR. Molecular and phenotypic description of Pneumocystis wakefieldiae sp. nov., a new species in rats. Mycologia. 2004;96:429–438. doi: 10.1080/15572536.2005.11832942. [DOI] [PubMed] [Google Scholar]
- 4.Keely SP, Fischer JM, Stringer JR. Evolution and speciation of Pneumocystis. J. Eukaryot. Microbiol. 2003;50:624–626. doi: 10.1111/j.1550-7408.2003.tb00655.x. [DOI] [PubMed] [Google Scholar]
- 5.Cisse OH, et al. Comparative population genomics analysis of the mammalian fungal pathogen Pneumocystis. mBio. 2018;9:e00381–00318. doi: 10.1128/mBio.00381-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Upham NS, Esselstyn JA, Jetz W. Inferring the mammal tree: species-level sets of phylogenies for questions in ecology, evolution, and conservation. PLoS Biol. 2019;17:e3000494. doi: 10.1371/journal.pbio.3000494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McDougall I, Brown FH, Fleagle JG. Stratigraphic placement and age of modern humans from Kibish, Ethiopia. Nature. 2005;433:733–736. doi: 10.1038/nature03258. [DOI] [PubMed] [Google Scholar]
- 8.Suzuki Y, Tomozawa M, Koizumi Y, Tsuchiya K, Suzuki H. Estimating the molecular evolutionary rates of mitochondrial genes referring to Quaternary ice age events with inferred population expansions and dispersals in Japanese Apodemus. BMC Evol. Biol. 2015;15:187. doi: 10.1186/s12862-015-0463-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guillot, J. et al. Parallel phylogenies of Pneumocystis species and their mammalian hosts. J. Eukaryot. Microbiol. Suppl:113S–115S 10.1111/j.1550-7408.2001.tb00475.x (2001). [DOI] [PubMed]
- 10.Latinne A, et al. Genetic diversity and evolution of Pneumocystis fungi infecting wild Southeast Asian murid rodents. Parasitology. 2018;145:885–900. doi: 10.1017/S0031182017001883. [DOI] [PubMed] [Google Scholar]
- 11.Petruzela J, et al. Evolutionary history of Pneumocystis fungi in their African rodent hosts. Infect. Genet. Evol. 2019;75:103934. doi: 10.1016/j.meegid.2019.103934. [DOI] [PubMed] [Google Scholar]
- 12.Cisse OH, Pagni M, Hauser PM. De novo assembly of the Pneumocystis jirovecii genome from a single bronchoalveolar lavage fluid specimen from a patient. mBio. 2012;4:e00428–00412. doi: 10.1128/mBio.00428-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ma L, et al. Genome analysis of three Pneumocystis species reveals adaptation mechanisms to life exclusively in mammalian hosts. Nat. Commun. 2016;7:10740. doi: 10.1038/ncomms10740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Slaven BE, et al. Draft assembly and annotation of the Pneumocystis carinii genome. J. Eukaryot. Microbiol. 2006;53:S89–S91. doi: 10.1111/j.1550-7408.2006.00184.x. [DOI] [PubMed] [Google Scholar]
- 15.Lundgren B, Cotton R, Lundgren JD, Edman JC, Kovacs JA. Identification of Pneumocystis carinii chromosomes and mapping of five genes. Infect. Immun. 1990;58:1705–1710. doi: 10.1128/IAI.58.6.1705-1710.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Underwood AP, Louis EJ, Borts RH, Stringer JR, Wakefield AE. Pneumocystis carinii telomere repeats are composed of TTAGGG and the subtelomeric sequence contains a gene encoding the major surface glycoprotein. Mol. Microbiol. 1996;19:273–281. doi: 10.1046/j.1365-2958.1996.374904.x. [DOI] [PubMed] [Google Scholar]
- 17.Ma L, et al. Sequencing and characterization of the complete mitochondrial genomes of three Pneumocystis species provide new insights into divergence between human and rodent Pneumocystis. FASEB J. 2013;27:1962–1972. doi: 10.1096/fj.12-224444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shimodaira H, Hasegawa M. Multiple comparisons of log-likelihoods with applications to phylogenetic inference. Mol. Biol. Evol. 1999;16:1114–1116. doi: 10.1093/oxfordjournals.molbev.a026201. [DOI] [Google Scholar]
- 19.Aliouat-Denis CM, et al. Pneumocystis species, co-evolution and pathogenic power. Infect. Genet. Evol. 2008;8:708–726. doi: 10.1016/j.meegid.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 20.Kitazoe Y, et al. Robust time estimation reconciles views of the antiquity of placental mammals. PLoS ONE. 2007;2:e384. doi: 10.1371/journal.pone.0000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shen, X. X. et al. Genome-scale phylogeny and contrasting modes of genome evolution in the fungal phylum Ascomycota. Sci. Adv. 10.1126/sciadv.abd0079 (2020). [DOI] [PMC free article] [PubMed]
- 22.Kielbasa SM, Wan R, Sato K, Horton P, Frith MC. Adaptive seeds tame genomic sequence comparison. Genome Res. 2011;21:487–493. doi: 10.1101/gr.113985.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Korneliussen TS, Albrechtsen A, Nielsen R. ANGSD: analysis of next generation sequencing data. BMC Bioinformatics. 2014;15:356. doi: 10.1186/s12859-014-0356-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu Y, Nakhleh L. A maximum pseudo-likelihood approach for phylogenetic networks. BMC Genomics. 2015;16:S10. doi: 10.1186/1471-2164-16-S10-S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mazars E, et al. Isoenzyme diversity in Pneumocystis carinii from rats, mice, and rabbits. J. Infect. Dis. 1997;175:655–660. doi: 10.1093/infdis/175.3.655. [DOI] [PubMed] [Google Scholar]
- 26.Aghova T, et al. Fossils know it best: Using a new set of fossil calibrations to improve the temporal phylogenetic framework of murid rodents (Rodentia: Muridae) Mol. Phylogenet. Evol. 2018;128:98–111. doi: 10.1016/j.ympev.2018.07.017. [DOI] [PubMed] [Google Scholar]
- 27.Araujo SB, et al. Understanding host-switching by ecological fitting. PLoS ONE. 2015;10:e0139225. doi: 10.1371/journal.pone.0139225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McBride AE, Conboy AK, Brown SP, Ariyachet C, Rutledge KL. Specific sequences within arginine-glycine-rich domains affect mRNA-binding protein function. Nucleic Acids Res. 2009;37:4322–4330. doi: 10.1093/nar/gkp349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Russian DA, et al. Characterization of a multicopy family of genes encoding a surface-expressed serine endoprotease in rat Pneumocystis carinii. Proc. Assoc. Am. Physicians. 1999;111:347–356. doi: 10.1046/j.1525-1381.1999.99118.x. [DOI] [PubMed] [Google Scholar]
- 30.Bairwa G, Hee Jung W, Kronstad JW. Iron acquisition in fungal pathogens of humans. Metallomics. 2017;9:215–227. doi: 10.1039/C6MT00301J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stajich JE, Dietrich FS, Roy SW. Comparative genomic analysis of fungal genomes reveals intron-rich ancestors. Genome Biol. 2007;8:R223. doi: 10.1186/gb-2007-8-10-r223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cisse OH, et al. Genome sequencing of the plant pathogen Taphrina deformans, the causal agent of peach leaf curl. mBio. 2013;4:e00055–00013. doi: 10.1128/mBio.00055-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang J, Nielsen R, Yang Z. Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Mol. Biol. Evol. 2005;22:2472–2479. doi: 10.1093/molbev/msi237. [DOI] [PubMed] [Google Scholar]
- 34.Schmid-Siegert E, et al. Mechanisms of surface antigenic variation in the human pathogenic fungus Pneumocystis jirovecii. mBio. 2017;8:e01470–01417. doi: 10.1128/mBio.01470-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ma, L. et al. Diversity and complexity of the large surface protein family in the compacted genomes of multiple Pneumocystis species. mBio10.1128/mBio.02878-19 (2020). [DOI] [PMC free article] [PubMed]
- 36.Flajnik MF, Kasahara M. Origin and evolution of the adaptive immune system: genetic events and selective pressures. Nat. Rev. Genet. 2010;11:47–59. doi: 10.1038/nrg2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bankevich A, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Simao FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31:3210–3212. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- 41.Cisse OH, Stajich JE. FGMP: assessing fungal genome completeness. BMC Bioinformatics. 2019;20:184. doi: 10.1186/s12859-019-2782-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Parra G, Bradnam K, Korf I. CEGMA: a pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics. 2007;23:1061–1067. doi: 10.1093/bioinformatics/btm071. [DOI] [PubMed] [Google Scholar]
- 43.Sedlazeck FJ, et al. Accurate detection of complex structural variations using single-molecule sequencing. Nat. Methods. 2018;15:461–468. doi: 10.1038/s41592-018-0001-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Koren S, et al. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 2017;27:722–736. doi: 10.1101/gr.215087.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vaser R, Sovic I, Nagarajan N, Sikic M. Fast and accurate de novo genome assembly from long uncorrected reads. Genome Res. 2017;27:737–746. doi: 10.1101/gr.214270.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Walker BJ, et al. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE. 2014;9:e112963. doi: 10.1371/journal.pone.0112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv. 2013;1303:3997. [Google Scholar]
- 48.Pertea M, Kim D, Pertea GM, Leek JT, Salzberg SL. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016;11:1650–1667. doi: 10.1038/nprot.2016.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li H, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Grabherr MG, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bray NL, Pimentel H, Melsted P, Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 2016;34:525–527. doi: 10.1038/nbt.3519. [DOI] [PubMed] [Google Scholar]
- 52.Smit, A. F. A., Hubley, R. & Green, P. RepeatMasker Open-4.0. (2013–2015). http://www.repeatmasker.org/asmitpapers.html.
- 53.Bao W, Kojima KK, Kohany O. Repbase Update, a database of repetitive elements in eukaryotic genomes. Mob. DNA. 2015;6:11. doi: 10.1186/s13100-015-0041-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Grabherr MG, et al. Genome-wide synteny through highly sensitive sequence alignment: Satsuma. Bioinformatics. 2010;26:1145–1151. doi: 10.1093/bioinformatics/btq102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.UniProt Consortium, T. UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2018;46:2699. doi: 10.1093/nar/gky092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stanke M, Waack S. Gene prediction with a hidden Markov model and a new intron submodel. Bioinformatics. 2003;19:ii215–ii225. doi: 10.1093/bioinformatics/btg1080. [DOI] [PubMed] [Google Scholar]
- 57.Ter-Hovhannisyan V, Lomsadze A, Chernoff YO, Borodovsky M. Gene prediction in novel fungal genomes using an ab initio algorithm with unsupervised training. Genome Res. 2008;18:1979–1990. doi: 10.1101/gr.081612.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Haas BJ, et al. Automated eukaryotic gene structure annotation using EVidenceModeler and the Program to Assemble Spliced Alignments. Genome Biol. 2008;9:R7. doi: 10.1186/gb-2008-9-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tsai IJ, et al. Comparative genomics of Taphrina fungi causing varying degrees of tumorous deformity in plants. Genome Biol. Evol. 2014;6:861–872. doi: 10.1093/gbe/evu067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Holt C, Yandell M. MAKER2: an annotation pipeline and genome-database management tool for second-generation genome projects. BMC Bioinformatics. 2011;12:491. doi: 10.1186/1471-2105-12-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Korf I. Gene finding in novel genomes. BMC Bioinformatics. 2004;5:59. doi: 10.1186/1471-2105-5-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hauser, P. M. et al. Comparative genomics suggests that the fungal pathogen Pneumocystis is an obligate parasite scavenging amino acids from its host’s lungs. PLoS ONE10.1371/journal.pone.0015152 (2010). [DOI] [PMC free article] [PubMed]
- 63.Pierleoni A, Martelli PL, Casadio R. PredGPI: a GPI-anchor predictor. BMC Bioinformatics. 2008;9:392. doi: 10.1186/1471-2105-9-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Eisenhaber B, Schneider G, Wildpaner M, Eisenhaber F. A sensitive predictor for potential GPI lipid modification sites in fungal protein sequences and its application to genome-wide studies for Aspergillus nidulans, Candida albicans, Neurospora crassa, Saccharomyces cerevisiae and Schizosaccharomyces pombe. J. Mol. Biol. 2004;337:243–253. doi: 10.1016/j.jmb.2004.01.025. [DOI] [PubMed] [Google Scholar]
- 65.Fankhauser N, Maser P. Identification of GPI anchor attachment signals by a Kohonen self-organizing map. Bioinformatics. 2005;21:1846–1852. doi: 10.1093/bioinformatics/bti299. [DOI] [PubMed] [Google Scholar]
- 66.Almagro Armenteros JJ, et al. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 2019;37:420–423. doi: 10.1038/s41587-019-0036-z. [DOI] [PubMed] [Google Scholar]
- 67.Stoffel KHAW. TMbase—a database of membrane spanning proteins segments. Biol. Chem. Hoppe Seyler. 1993;374:166. [Google Scholar]
- 68.El-Gebali S, et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2019;47:D427–D432. doi: 10.1093/nar/gky995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Claudel-Renard C, Chevalet C, Faraut T, Kahn D. Enzyme-specific profiles for genome annotation: PRIAM. Nucleic Acids Res. 2003;31:6633–6639. doi: 10.1093/nar/gkg847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gao M, Zhou H, Skolnick J. DESTINI: a deep-learning approach to contact-driven protein structure prediction. Sci. Rep. 2019;9:3514. doi: 10.1038/s41598-019-40314-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Frith MC, Noe L. Improved search heuristics find 20,000 new alignments between human and mouse genomes. Nucleic Acids Res. 2014;42:e59. doi: 10.1093/nar/gku104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tesler G. GRIMM: genome rearrangements web server. Bioinformatics. 2002;18:492–493. doi: 10.1093/bioinformatics/18.3.492. [DOI] [PubMed] [Google Scholar]
- 73.Darling AE, Mau B, Perna NT. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS ONE. 2010;5:e11147. doi: 10.1371/journal.pone.0011147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Baudet C, et al. Cassis: detection of genomic rearrangement breakpoints. Bioinformatics. 2010;26:1897–1898. doi: 10.1093/bioinformatics/btq301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Quinlan AR. BEDTools: the Swiss-Army tool for genome feature analysis. Curr. Protoc. Bioinformatics. 2014;47:11 12 11–11 12 34. doi: 10.1002/0471250953.bi1112s47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Li H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. 2018;34:3094–3100. doi: 10.1093/bioinformatics/bty191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Farrer RA. Synima: a Synteny imaging tool for annotated genome assemblies. BMC Bioinformatics. 2017;18:507. doi: 10.1186/s12859-017-1939-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rice P, Longden I, Bleasby A. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 2000;16:276–277. doi: 10.1016/S0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 79.Altschul SF, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bastian, M., Heymann, S. & Jacomy, M. In International AAAI Conference on Weblogs and Social Media (2009).
- 81.Li H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics. 2011;27:2987–2993. doi: 10.1093/bioinformatics/btr509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Seton Bocco S, Csuros M. Splice sites seldom slide: intron evolution in oomycetes. Genome Biol. Evol. 2016;8:2340–2350. doi: 10.1093/gbe/evw157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Csuros M. Malin: maximum likelihood analysis of intron evolution in eukaryotes. Bioinformatics. 2008;24:1538–1539. doi: 10.1093/bioinformatics/btn226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Emms DM, Kelly S. OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 2015;16:157. doi: 10.1186/s13059-015-0721-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Le SQ, Gascuel O. An improved general amino acid replacement matrix. Mol. Biol. Evol. 2008;25:1307–1320. doi: 10.1093/molbev/msn067. [DOI] [PubMed] [Google Scholar]
- 88.Minh BQ, et al. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 2020;37:1530–1534. doi: 10.1093/molbev/msaa015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sievers F, Higgins DG. Clustal Omega, accurate alignment of very large numbers of sequences. Methods Mol. Biol. 2014;1079:105–116. doi: 10.1007/978-1-62703-646-7_6. [DOI] [PubMed] [Google Scholar]
- 90.Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sesterhenn TM, et al. Sequence and structure of the linear mitochondrial genome of Pneumocystis carinii. Mol. Genet. Genomics. 2010;283:63–72. doi: 10.1007/s00438-009-0498-7. [DOI] [PubMed] [Google Scholar]
- 92.Stolzer M, et al. Inferring duplications, losses, transfers and incomplete lineage sorting with nonbinary species trees. Bioinformatics. 2012;28:i409–i415. doi: 10.1093/bioinformatics/bts386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Csuros M. Count: evolutionary analysis of phylogenetic profiles with parsimony and likelihood. Bioinformatics. 2010;26:1910–1912. doi: 10.1093/bioinformatics/btq315. [DOI] [PubMed] [Google Scholar]
- 94.Huson DH, Bryant D. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 2006;23:254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- 95.McGuire G, Wright F. TOPAL 2.0: improved detection of mosaic sequences within multiple alignments. Bioinformatics. 2000;16:130–134. doi: 10.1093/bioinformatics/16.2.130. [DOI] [PubMed] [Google Scholar]
- 96.Ranwez V, Douzery EJP, Cambon C, Chantret N, Delsuc F. MACSE v2: toolkit for the alignment of coding sequences accounting for frameshifts and stop codons. Mol. Biol. Evol. 2018;35:2582–2584. doi: 10.1093/molbev/msy159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bouckaert R, et al. BEAST 2: a software platform for Bayesian evolutionary analysis. PLoS Comput. Biol. 2014;10:e1003537. doi: 10.1371/journal.pcbi.1003537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Drummond AJ, Ho SY, Phillips MJ, Rambaut A. Relaxed phylogenetics and dating with confidence. PLoS Biol. 2006;4:e88. doi: 10.1371/journal.pbio.0040088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gernhard T. The conditioned reconstructed process. J. Theor. Biol. 2008;253:769–778. doi: 10.1016/j.jtbi.2008.04.005. [DOI] [PubMed] [Google Scholar]
- 100.Heled J, Drummond AJ. Calibrated birth-death phylogenetic time-tree priors for bayesian inference. Syst. Biol. 2015;64:369–383. doi: 10.1093/sysbio/syu089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hasegawa M, Kishino H, Yano T. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J. Mol. Evol. 1985;22:160–174. doi: 10.1007/BF02101694. [DOI] [PubMed] [Google Scholar]
- 102.Beimforde C, et al. Estimating the Phanerozoic history of the Ascomycota lineages: combining fossil and molecular data. Mol. Phylogenet. Evol. 2014;78:386–398. doi: 10.1016/j.ympev.2014.04.024. [DOI] [PubMed] [Google Scholar]
- 103.Lanfear R, Frandsen PB, Wright AM, Senfeld T, Calcott B. PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol. Biol. Evol. 2017;34:772–773. doi: 10.1093/molbev/msw260. [DOI] [PubMed] [Google Scholar]
- 104.Bell, M. A. & Graeme, T. L. strap: an R package for plotting phylogenies against stratigraphy and assessing their stratigraphic congruence. Palaeontology58, 379–389 (2015).
- 105.Korneliussen TS, Moltke I, Albrechtsen A, Nielsen R. Calculation of Tajima’s D and other neutrality test statistics from low depth next-generation sequencing data. BMC Bioinformatics. 2013;14:289. doi: 10.1186/1471-2105-14-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Fumagalli M, et al. Quantifying population genetic differentiation from next-generation sequencing data. Genetics. 2013;195:979–992. doi: 10.1534/genetics.113.154740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bruen TC, Philippe H, Bryant D. A simple and robust statistical test for detecting the presence of recombination. Genetics. 2006;172:2665–2681. doi: 10.1534/genetics.105.048975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kozlov, A. M., Darriba, D., Flouri, T., Morel, B. & Stamatakis, A. RAxML-NG: A fast, scalable, and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics10.1093/bioinformatics/btz305 (2019). [DOI] [PMC free article] [PubMed]
- 111.Huson DH, Scornavacca C. Dendroscope 3: an interactive tool for rooted phylogenetic trees and networks. Syst. Biol. 2012;61:1061–1067. doi: 10.1093/sysbio/sys062. [DOI] [PubMed] [Google Scholar]
- 112.Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 113.R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org/ (2019).
- 114.Fang H. dcGOR: an R package for analysing ontologies and protein domain annotations. PLoS Comput. Biol. 2014;10:e1003929. doi: 10.1371/journal.pcbi.1003929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Koster J, Rahmann S. Snakemake-a scalable bioinformatics workflow engine. Bioinformatics. 2012;28:2520–2522. doi: 10.1093/bioinformatics/bts480. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
The datasets generated during and/or analyzed during the current study are available at NCBI BioProject portal (https://www.ncbi.nlm.nih.gov/bioproject/): Pneumocystis macacae strain P2C (no. PRJNA632025); P. oryctolagi strain CS1 (PRJNA632560); P. canis strain Ck1 (PRJNA632556); P. canis strain Ck2 (PRJNA632878); P. canis strain A (PRJNA636786);P. wakefieldiae strain 2A (PRJNA632570); P. jirovecii strain 55 (PRJNA647920), P. jirovecii strain 54c (PRJNA648092), P. jirovecii strain 46 (PRJNA648096), P. macacae strain CJ36 (PRJNA648103), P. macacae strain ER17 (PRJNA648108), P. macacae strain UC86 (PRJNA648112), and P. macacae strain GL92 (PRJNA648115).
All custom bioinformatic analyses were conducted using Perl v5.26.0 (http://www.perl.org/) or Python v.3.6 (http://www.python.org) scripts. Pipelines were written with Snakemake v5.11.2115. Custom codes generated for this project are available at GitHub: https://github.com/ocisse/pneumocystis_evolution and a stable released version is available at 10.5281/zenodo.4450766.