Summary
Enteroviruses, such as EV-A71 and CVA16, mainly infect the human gastrointestinal tract. Human coronaviruses, including SARS-CoV and SARS-CoV-2, have been variably associated with gastrointestinal symptoms. We aimed to optimize the human intestinal organoids and hypothesize that these optimized intestinal organoids can recapitulate enteric infections of enterovirus and coronavirus. We demonstrate that the optimized human intestinal organoids enable better simulation of the native human intestinal epithelium, and that they are significantly more susceptible to EV-A71 than CVA16. Higher replication of EV-A71 than CVA16 in the intestinal organoids triggers a more vigorous cellular response. However, SARS-CoV and SARS-CoV-2 exhibit distinct dynamics of virus-host interaction; more robust propagation of SARS-CoV triggers minimal cellular response, whereas, SARS-CoV-2 exhibits lower replication capacity but elicits a moderate cellular response. Taken together, the disparate profile of the virus-host interaction of enteroviruses and coronaviruses in human intestinal organoids may unravel the cellular basis of the distinct pathogenicity of these viral pathogens.
Keywords: intestinal organoids, EV-A71, CVA16, SARS-CoV, SARS-CoV-2, pathogenicity
Highlights
-
•
An optimized differentiation protocol improves maturation of intestinal organoids
-
•
SARS-CoV-2 and SARS-CoV infection triggers less robust response than enteroviruses
-
•
Coronaviruses show lower sensitivity to type III IFNs than enteroviruses
-
•
Intestinal organoids recapitulate disparate pathogenicity of CoVs and enteroviruses
Zhao and colleagues modified the composition of differentiation medium and demonstrated that the optimized medium improves the maturation state of human intestinal organoids. Two coronaviruses, SARS-CoV-2 and SARS-CoV, and two enteroviruses, EV-A71 and CVA16, reveal distinct profiles of virus-host interaction in the optimized intestinal organoids. Overall, human intestinal organoids adequately recapitulate disparate pathogenicity of coronaviruses and enteroviruses in the human intestines.
Introduction
Enteroviruses of the Piconaviridae family are important human pathogens causing a broad spectrum of clinical diseases, ranging from diarrhea, respiratory symptoms, and skin rashes, to more severe diseases, such as meningitis and paralysis (Solomon et al., 2010; Zaoutis and Klein, 1998). Enteroviruses mainly infect human gastrointestinal tract and are transmitted via the fecal-oral route. Upon primary infection in the gastrointestinal and respiratory tract, enteroviruses can enter the bloodstream, systematically spread, and lead to complications, such as neurologic disorders (Solomon et al., 2010). Enterovirus 71 (EV-A71) and coxsackievirus A serotype 16 (CVA16), both classified as members of human enteroviruses A, have caused major outbreaks of hand-foot-and-mouth disease, which normally affects children under 5 years (Muehlenbachs et al., 2015; Wong et al., 2010; Yeung et al., 2018). Despite the genetic relatedness of these two viruses, many epidemiological studies demonstrated that EV-A71 is more pathogenic than CVA16 and more likely to associate with severe neurological disorder and fatality (Chen et al., 2007; Xing et al., 2014).
Human enteroviruses are mainly transmitted via the fecal-oral route, and the intestinal epithelium is the primary target of viral invasion (Solomon et al., 2010). The human intestinal tract is lined by a simple columnar epithelium. The LRG5+ stem cells, in close contact with Paneth cells in the base of crypts, divide every day, and eventually produce major cell types of the small intestinal epithelium, including enterocytes, goblet cells, enteroendocrine cells (Sato et al., 2009). Absorptive enterocytes, the most abundant cell type, account for over 80% of all intestinal epithelial cells (Cheng and Leblond, 1974). Paneth cells, which secrete antimicrobial cryptdins or defensins, digestive enzymes, and growth factors, are found throughout the small intestine, yet are absent in the colonic epithelium. The multi-cellular architecture constitutes the structural basis of functional complexity of the intestinal epithelium. Apparently, it is implausible that conventional cell lines, which are composed of homogeneous clonal cells, can recapitulate the morphological and functional complexity of human intestinal epithelium, let alone model virus-host interaction in human intestinal cells.
Advances in understanding of the adult stem cell niche have enabled 3D reconstitution of various tissues as organoids (Clevers, 2016). Sato et al. (2011) reported culture of human intestinal organoids, which allows long-term expansion of human intestinal epithelium in vitro. The expansion medium enables the initial derivation and long-term maintenance of intestinal organoids by directing the organoids toward an undifferentiated state, while the differentiation protocol upon withdrawal of Wnt and mitogens drives differentiation along the crypt-villi axis and generates epithelial domains encompassing four mature cell types of native human intestinal epithelium. These physiologically relevant human intestinal organoids have become the most popular model for studying enteric infections in recent years (Ettayebi et al., 2016; Zhou et al., 2017, 2020a).
A novel human coronavirus, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has caused the global COVID-19 pandemic since December 2019 (Chan et al., 2020b; Zhou et al., 2020b). In fact, apart from respiratory infection, all human coronaviruses have been variably associated with gastrointestinal symptoms (Cimolai, 2020). We and others have reported SARS-CoV-2 and MERS-CoV infection in human intestinal organoids (Lamers et al., 2020; Zhou et al., 2017, 2020a). Enteric involvement was actually more common in SARS patients (Leung et al., 2003). In this study, we optimized the differentiation medium of human intestinal organoids. We examined whether the optimized differentiated intestinal organoids can recapitulate higher pathogenicity of EV-A71 than CVA16. In addition, we examined the infectivity of SARS-CoV in human intestinal organoids, in comparison with phylogenetically related SARS-CoV-2. Our results highlight the competence of these intestinal organoids as an optimal in vitro model to recapitulate human enteric infections of enterovirus and coronavirus.
Results
The Differentiation Medium Was Optimized to Improve Villous Differentiation of Intestinal Organoids
Human intestinal organoids are maintained in the long-term in expansion medium supplemented with various niche factors and mitogens. Based on the protocol established by Sato et al. (2011), withdrawal of Wnt, nicotinamide, and p38 inhibitor from the expansion medium drove villous differentiation, while R-spondin and BMP inhibitor Noggin were retained in the differentiation medium. R-Spondins are a major driver of Wnt-mediated crypt self-renewal and proliferation (de Lau et al., 2014; Yan et al., 2017). BMP signaling acts as a negative regulator of crypts (He et al., 2004; Qi et al., 2017). Thus, R-spondin and BMP inhibitor Noggin are essential components to maintain the stemness and long-term expansion of intestinal organoids. We deduced that further removal of R-spondin and Noggin in the previously refined differentiation medium may promote villous differentiation. To test the hypothesis, we reduced the amount of R-spondin- and Noggin-conditioned medium in the originally refined differentiation medium (H10) in which both were supplemented at 10%, to 5% (H5 medium), or total removal of both (H0 medium). The small intestinal organoids (enteroids) were transferred to three differentiation media or maintained in the expansion (Ex) medium (Table 1), and incubated for 5 days. As expected, the enteroids in the expansion medium gradually enlarged over time, while those in three differentiation media became more compact (Figure S1).
Table 1.
Composition of the Expansion Medium and Differentiation Medium for Intestinal Organoid Culture
| Reagent (Concentration) | Expansion medium | H10 | H5 | H0 |
|---|---|---|---|---|
| Wnt3a-conditioned mediuma | 50% | − | − | − |
| Rspondin1-conditioned mediuma | 20% | 10% | 5% | − |
| Noggin-conditioned mediuma | 10% | 10% | 5% | − |
| B27 supplement (1×) | + | + | + | + |
| Nicotinamide (10 mM) | + | − | − | − |
| A8301 (500 nM) | + | + | + | + |
| SB202190 (10 μM) | + | − | − | − |
| hEGF (50 ng/mL) | + | + | + | + |
| Y-27632 (10 μM) | + | + | + | + |
| hGastrin I (10 nM) | + | + | + | + |
| N-Acetylcysteine (1 mM) | + | + | + | + |
| HEPES 1M (10 mM) | + | + | + | + |
| GlutaMAX (2 mM) | + | + | + | + |
| Primocin (50 μg/mL) | + | + | + | + |
| Penicillin/Streptomycin (100 U/mL) | + | + | + | + |
| Advanced F12/DMEM | + | + | + | + |
Percentage of total volume.
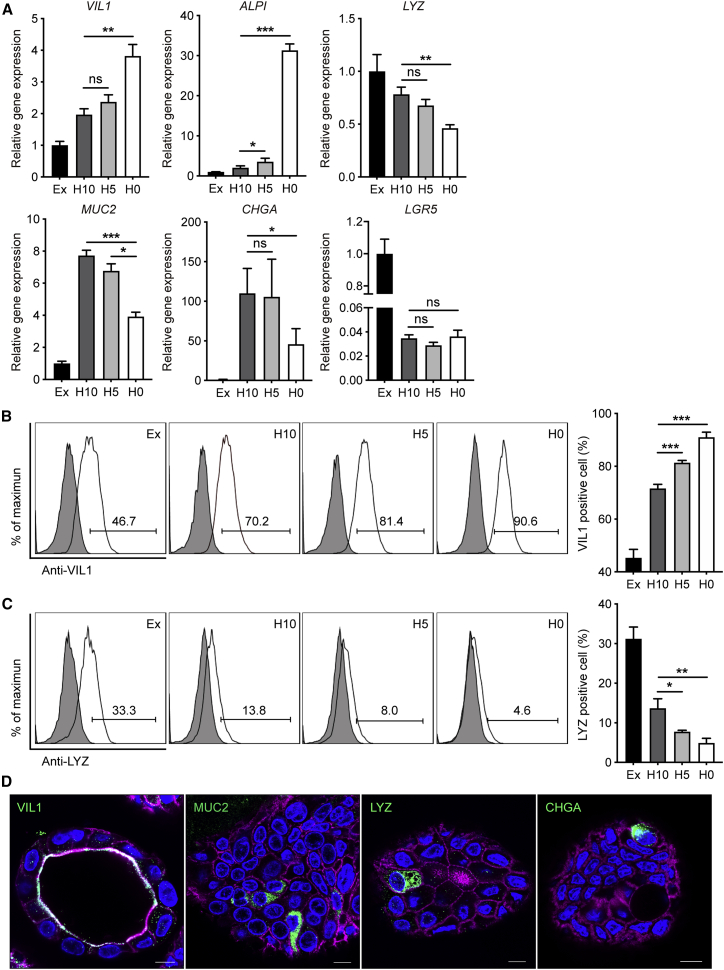
We compared the expression levels of cell-type markers in these organoids. The expression level of enterocyte marker Villin (VIL1) was elevated in the organoids in three differentiation media compared with those in expansion medium (Figure 1A). VIL1 was significantly upregulated in H0 organoids than in H10 organoids. Another enterocyte marker, intestinal alkaline phosphatase (ALPI), was highly elevated upon withdrawal of R-spondin and Noggin. The expression of Paneth cell marker lysozyme (LYZ) was significantly but modestly lower in H0 organoids than H10 organoids. Mucin 2 (MUC2), the marker of goblet cells, decreased moderately in H0 organoids compared with that in H10 and H5 organoids. A decreased CHGA (chromogranin A, enteroendocrine cell marker) expression was discernible in H0 organoids in comparison with H10 organoids. As expected, a dramatic downregulation of stem cell marker LGR5 was observed in the organoids incubated in three differentiation media, while its expression levels in H0, H5, and H10 organoids were comparable.
Figure 1.
Characterization of the Differentiation Status of Human Enteroids
(A–C) (A) Fold change of expression levels of cell-type markers in the organoids cultured in different differentiation media (H10, H5, and H0) versus those cultured in expansion medium (Ex). Data show the mean and SD of three independent experiments. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 analyzed by Student's t test, two tailed. The percentages of enterocytes (B) and Paneth cells (C) in the organoids cultured in different differentiation media and expansion medium. The representative histograms of one experiment (left), and means and SD of three independent experiments (right) are presented.
(D) Confocal images of VIL1+ enterocytes, MUC2+ goblet cells, LYZ+ Paneth cells, and CHGA+ enteroendocrine cells in the organoids cultured in H0 medium. Nuclei and cellular actin filaments are counterstained with DAPI (blue) and Phalloidin-647 (purple). Scale bar, 10 μm.
See also Figure S1.
We then inspected the cellular composition of the enteroids in different media by flow cytometry. The percentage of enterocytes increased from 70% in H10 organoids to 81% in H5 organoids and 90% in H0 organoids (Figure 1B), whereas that of Paneth cell decreased with descending amounts of R-spondin and Noggin (Figure 1C). Despite the downregulation of non-enterocyte markers and reduction of Paneth cells, the differentiated enteroids in H0 medium contain all mature cell types of human intestinal epithelium, i.e., enterocytes, Paneth cells, goblet cells, and enteroendocrine cells, as revealed by immunofluorescence staining (Figure 1D). Consistent with a recent report (Co et al., 2019), intestinal organoids, maintained three-dimensionally in Matrigel, invariably display an apical-in polarity. Accordingly, VIL1, a member protein on the apical surface of enterocytes, is located in the interior lumen of the organoid. Collectively, removal of R-spondin and Noggin in the original differentiation medium improved the differentiation status of intestinal organoids. These optimized intestinal organoids possess more enterocytes with better simulation of the native human intestinal epithelium, in which enterocytes account for over 80% of all epithelial cells (Cheng and Leblond, 1974). Thereafter, H0 medium was used to generate the differentiated intestinal organoids for the subsequent experiments.
EV-A71 More Productively Infected Differentiated Human Enteroids and Colonoids than CVA16
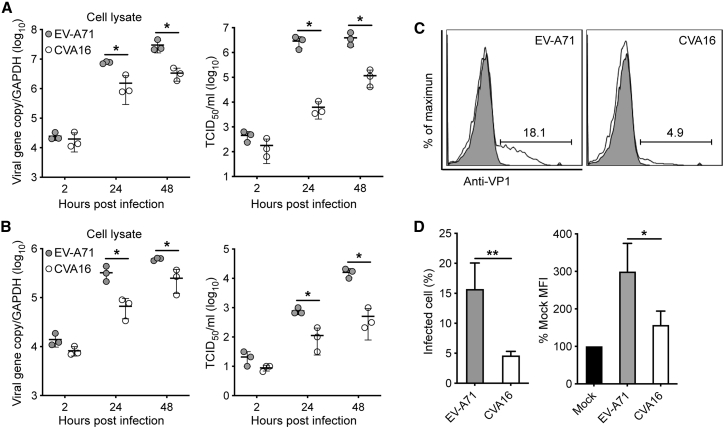
As mentioned above, EV-A71 is more pathogenic than CVA16 in patients. To assess whether the differentiated enteroids can recapitulate the distinct pathogenicity, we compared the replication potential of EV-A71 and CVA16 in human enteroids. Both EV-A71 and CVA16 actively propagated in the enteroids, as shown by the significantly increased viral load and viral titer in cell lysates and culture media, respectively (Figure 2A). Of note, EV-A71 replicated more robustly than CVA16. At 24 h post infection (hpi), viral titer was more than 2 log units higher in EV-A71-infected organoids than in CVA16-infected organoids. Higher replication capacity of EV-A71 than CVA16 was reproduced in the differentiated colonoids. Viral load and viral titer were significantly higher in EV-A71-infected colonoids than in CVA16-infected organoids (Figure 2B).
Figure 2.
More Productive Infection of EV-A71 than CVA16 in the Differentiated Human Enteroids and Colonoids
The differentiated human enteroids (A) and colonoids (B) were inoculated with EV-A71 or CVA16 virus at an MOI of 0.01. Cell lysates and culture media of the infected organoids were harvested at the indicated time points for the quantification of viral gene copy number and viral titration, respectively. The viral gene copy in the cell lysate is normalized with the transcript of GAPDH and presented. The experiment was performed three times independently. ∗p < 0.05 analyzed by Student's t test, two tailed.
(C and D) The differentiated human enteroids were inoculated with EV-A71 or CVA16 virus at an MOI of 5. At 10 hpi, the infected- and mock-infected enteroids were fixed, labeled, and applied to flow cytometry to evaluate the percentage of VP1-positive cells. (C) The histogram shows the results of one representative experiment. (D) Mean and SD of three independent experiments are presented. ∗p < 0.05, ∗∗p < 0.01 analyzed by Student's t test, two tailed.
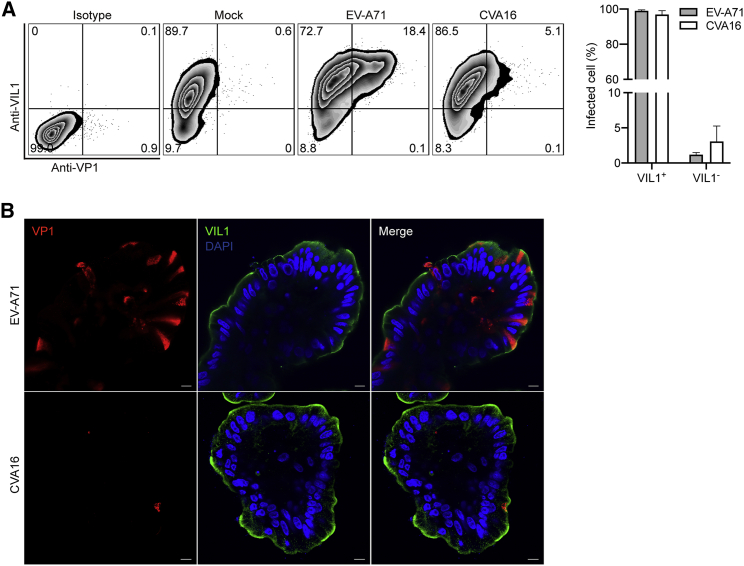
We then examined the infection rate by flow cytometry. At 10 hpi with an MOI of 5, around 20% cells in EV-A71-infected enteroids were viral protein VP1 positive, whereas approximately 5% cells were VP1 positive by CVA16 inoculation (Figure 2C). The abundance of VP1 as shown by mean fluorescence intensity was significantly higher in EV-A71-infected enteroids than those infected by CVA16 (Figure 2D). We also examined the cellular tropism of EV-A71 and CVA16 after double-staining with α-VP1 and α-Villin. As shown in Figure 3A, in both EV-A71- and CVA16-infected enteroids, over 90% VP1-positive cells were VIL1 positive, indicating that enterocytes are the predominant target cells of EV-A71 and CVA16. Immunofluorescence staining of viral protein VP1 consistently displayed more productive infection of EV-A71 than CVA16 (Figure 3B). Enterocyte tropism of the two virus strains was also verified by the colocalization of viral antigens VP1 and VIL1 in the infected organoids (Figure 3B). As mentioned above, intestinal organoids maintained within Matrigel normally display an apical-in polarity (Figure 1D). However, for infection experiments, differentiated organoids had to be mechanically sheared before virus inoculation to render the apical surface of organoids to be fully exposed to the virus inoculum. This shearing procedure may revert the polarity of some organoids and give rise to apical-out organoids, as shown in Figure 3B. Collectively, both human enteroids and colonoids are more susceptible to EV-A71 than CVA16.
Figure 3.
Cellular Tropism of EV-A71 and CVA16 in Human Enteroids
The differentiated human enteroids were inoculated with EV-A71 or CVA16 virus at an MOI of 5 or mock-infected. At 10 hpi, the infected and mock-infected human enteroids were fixed, co-labeled with α-VP1 and α-Villin, and then applied to flow cytometry.
(A) The histogram shows the results of one representative experiment (left). Data in the right panel show the mean and SD of three independent experiments.
(B) At 10 hpi, EV-A71- and CVA16-infected human enteroids, after fixation and co-staining of α-VP1 (red) and α-Villin (green), were applied to confocal imaging. Nuclei are counterstained with DAPI (blue). Scale bar, 10 μm.
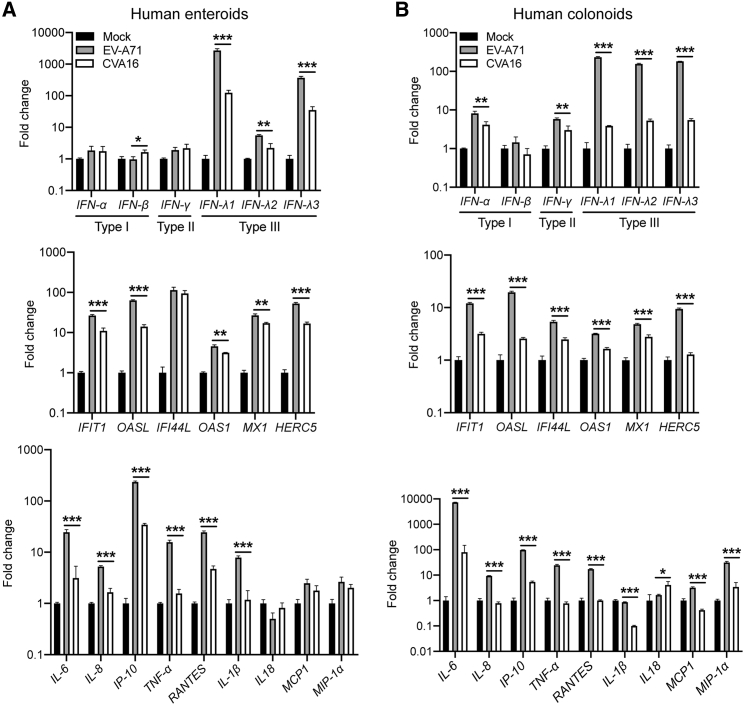
EV-A71 Triggered Higher Interferon and Proinflammatory Response than CVA16
We proceeded to examine the cellular response of human enteroids after infection, including the induction of interferons, interferon-stimulated genes (ISGs), and proinflammatory cytokines. As shown in Figure 4A, type I and type II IFNs were barely induced in both EV-A71-infected and CVA16-infected enteroids, whereas type III IFNs, particularly IFN-λ1 and IFN-λ3, were highly stimulated. EV-A71-infected enteroids produced significantly higher levels of IFN-λ1 and IFN-λ3 than CVA16-infected enteroids. ISGs were upregulated after infection, with significantly higher magnitude in the former than the latter. In addition, most proinflammatory cytokines, including IL-6, IP-10, and TNF-α, were induced to a significantly higher level by EV-A71 than CVA16. Higher cytokines response triggered by EV-A71 than CVA16 was reproduced in colonoids (Figure 4B). Interestingly, the induction of IFNs and ISGs was stronger in enteroids than colonoids, whereas inflammatory response, especially IL-6 induction, was more vigorous in the colonoids. Collectively, EV-A71 stimulated significantly higher induction of type III IFNs, ISGs, and proinflammatory cytokines than CVA16 in the infected enteroids and colonoids.
Figure 4.
EV-A71 Triggers More Vigorous Cellular Response than CVA16 in Human Enteroids and Colonoids
The differentiated human enteroids (A) and colonoids (B) were inoculated with EV-A71 or CVA16 virus at an MOI of 5 or mock-infected. At 10 hpi, cell lysates were harvested to detect mRNA expression levels of IFNs (top), ISGs (middle), and proinflammatory cytokines (bottom). The results present fold change of GAPDH-normalized expression levels in the infected organoids relative to that in the mock-infected organoids. Data show the mean and SD of three independent experiments. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 analyzed by Student's t test, two tailed.
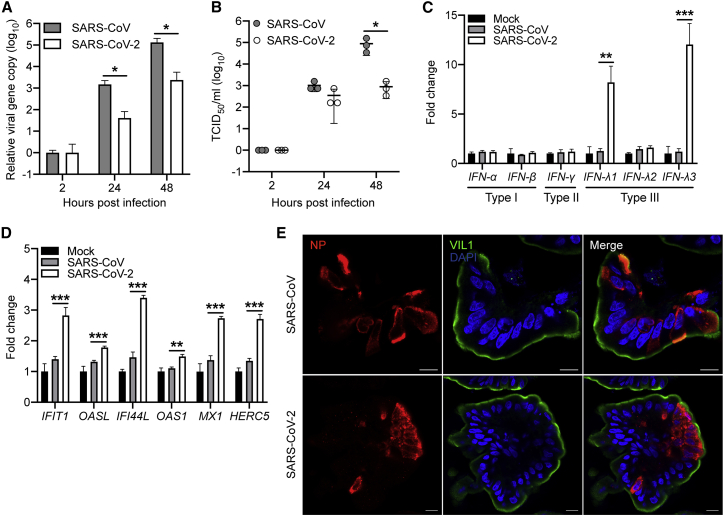
SARS-CoV Replicated More Actively in Differentiated Enteroids than SARS-CoV-2 with Dampened Cellular Response
As enteric involvement was reported in SARS patients (Leung et al., 2003), we tested the susceptibility of human enteroids to SARS-CoV. We inoculated the differentiated enteroids with SARS-CoV and monitored viral growth, and SARS-CoV-2 was inoculated in parallel for comparison. As shown in Figure 5A, SARS-CoV robustly propagated in the enteroids. At 48 hpi, viral loads in the culture media increased by ∼4 log units. A significantly increased viral titer of 4–5 log units was observed (Figure 5B). The replication capacity of SARS-CoV was significantly higher than that of SARS-CoV-2. At 48 hpi, viral titer was approximately 2 log units higher in SARS-CoV than SARS-CoV-2. We also detected the infection rates of two viruses in the differentiated enteroids. At 10 h after an MOI of 2 inoculation, approximately 3% of cells in the organoids were infected by SARS-CoV and SARS-CoV-2; the two viruses displayed similar infection rates at early time points after infection (Figure S2).
Figure 5.
Productive Infection and Cellular Tropism of SARS-CoV and SARS-CoV-2 in the Differentiated Human Enteroids
The differentiated human enteroids were inoculated with SARS-CoV or SARS-CoV-2 at an MOI of 1. Cell-free culture media were harvested at the indicated time points to detect viral loads (A) and viral titer (B). The experiment was performed three times independently. ∗p< 0.05 analyzed by Student's t test, two tailed.
(C and D) At 10 hpi of SARS-CoV or SARS-CoV-2 with an MOI of 2, the differentiated human enteroids were lysed and applied to detect mRNA expression levels of IFNs (C) and ISGs (D). The results present the fold change of GAPDH-normalized expression level in the infected enteroids relative to that in the mock-infected enteroids. Data show the mean and SD of three independent experiments. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 analyzed by Student's t test, two tailed.
(E) At 48 hpi, SARS-CoV- and SARS-CoV-2-infected human enteroids, after fixation and staining of α-NP (red) and α-Villin (green), were applied to confocal imaging. Nuclei are counterstained with DAPI (blue).
Scale bar, 10 μm.
See also Figure S2.
We next examined the cellular response in SARS-CoV- and SARS-CoV-2-infected human enteroids. As shown in Figure 5C, SARS-CoV-2 infection elicited significant production of IFN-λ1 and IFN-λ3, while neither type I nor type II IFNs was induced, consistent with the observation in enterovirus infections. ISGs were also significantly elicited in SARS-CoV-2-infected enteroids (Figure 5D). In contrast to the acute cytokine response in SARS-CoV-2-infected enteroids, three types of IFNs and all the tested ISGs were not stimulated in SARS-CoV-infected organoids, in which viral replication was more prominent. We also examined the cellular tropism of SARS-CoV and SARS-CoV-2 in human enteroids. As shown in Figure 5E, most infected cells in the enteroids were VIL1 positive, indicating that enterocytes are the major target cell of both SARS-CoV and SARS-CoV-2. Collectively, both SARS-CoV and SARS-CoV-2 productively infected human enteroids, in which SARS-CoV replicated more robustly than SARS-CoV-2. In contrast to the acute induction of cellular response in SARS-CoV-2 infection, IFNs and ISGs were barely stimulated in SARS-CoV-infected enteroids.
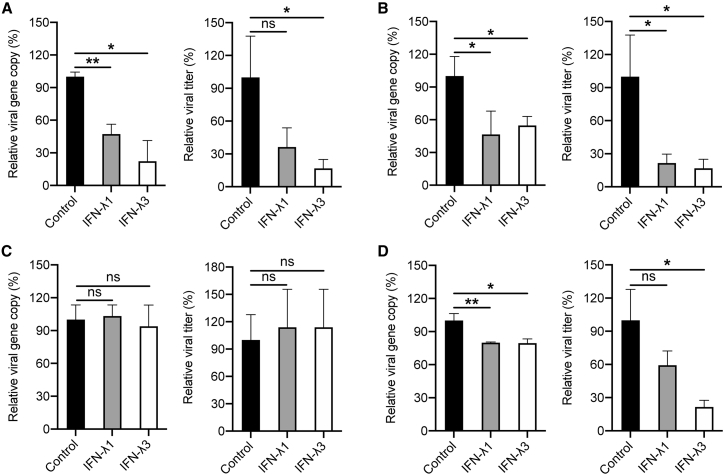
Enteroviruses and Coronaviruses Showed Differential Sensitivity to Type III IFNs
Two enteroviruses and two coronaviruses exhibited disparate profiles of viral replication and IFN response, as demonstrated above. Namely, higher replication of EV-A71 than CVA16 triggered more vigorous IFN response in the former. However, two coronaviruses showed an inverse profile; more robustly replicated SARS-CoV than SARS-CoV-2 was accompanied with weaker cellular response. It is likely that, apart from the distinct induction of IFNs, two enteroviruses and two coronaviruses may show differential susceptibility to the antiviral response triggered in the early phase of infection, particularly the induction of type III IFNs. To this end, we tested the sensitivity of these viruses to IFN-λ1 and IFN-λ3, two IFN molecules that are highly induced during early infections in intestinal organoids. Vero-E6 cells that are defective in IFN production were pre-treated with recombinant IFN-λ1 or IFN-λ3 for 18 h, infected with four viruses, and further incubated with IFNs for 48 h. The propagation of EV-A71 (Figure 6A) and CVA16 (Figure 6B) was significantly and comparably suppressed by the IFN treatment. A significant viral reduction in the culture medium was demonstrated by viral load detection and verified by viral titration. However, SARS-CoV seemed refractory to both IFN-λ1 and IFN-λ3 treatment (Figure 6C). The viral titer of SARS-CoV-2 significantly decreased to 20%–30% by IFN-λ3 treatment (Figure 6D). Therefore, the two enteroviruses are more or less similarly susceptible to IFN-λ treatment. However, two highly pathogenic coronaviruses show disparate sensitivity to IFN-λ treatment. SARS-CoV-2 is susceptible to IFN-λ3, while SARS-CoV seems resistant to both IFN-λ1 and IFN-λ3.
Figure 6.
Differential Sensitivity of Enteroviruses and Coronaviruses to IFN-λ
After 18 h pretreatment with recombinant IFN-λ1 or IFN-λ3, Vero-E6 cells were inoculated with EV-A71 (A) or CVA16 (B) at an MOI of 0.01, or SARS-CoV (C) or SARS-CoV-2 (D) at an MOI of 0.001. IFN-λ1 and IFN-λ3 were supplemented after inoculation. At 48 hpi, cell-free culture media were harvested to detect viral gene copy and viral titer. Results represent the viral load and viral titer in the organoids treated with IFN-λ1 or IFN-λ3 relative to those in mock-treated organoids. Data show the mean and SD of three independent experiments. ∗p < 0.05, ∗∗p < 0.01 analyzed by Student's t test, two tailed.
Discussion
Human intestinal organoids, the first adult stem cell-derived organoid culture, have become a popular and robust in vitro model to study microbe-host interactions in human intestinal cells (Ettayebi et al., 2016; Heo et al., 2018). In this study, we sought to improve the differentiation status of intestinal organoids through optimizing the composition of the differentiation medium. As a result, the differentiated enteroids that are generated upon removal of R-spondin and Noggin possess more enterocytes, meanwhile they harbor the other three mature intestinal cell types. Our results suggest that the modified enteroids enable better simulatation of the native human intestinal epithelium (Figure 1), in which enterocytes account for over 80% of all intestinal epithelial cells (Cheng and Leblond, 1974). In the optimized intestinal organoids, EV-A71 shows higher replication capacity in intestinal organoids than CVA16, although both viruses predominately target enterocytes (Figures 2 and 3). Both viruses trigger a potent induction of type III IFNs, ISGs, and proinflammatory cytokines. Notably, the cellular response is significantly more vigorous in EV-A71 infection than CVA16 infection in enteroids and colonoids (Figure 4). Namely, the higher replication of EV-A71 than CVA16 elicits more intensive cytokine response in intestinal organoids. The profile of viral replication kinetics and cellular response of EV-A71 and CVA16 in human intestinal organoids may unravel the cellular basis of higher pathogenicity of EV-A71 than CVA16 in patients. Clinically, enteric infections of EV-A71 and CVA16 are commonly self-limiting. This can be ascribed to the acute and potent elicitation of IFN-λs and ISGs in the intestinal epithelial cells, which primes the innate and adaptive immune response, and resolves the infection in a timely manner. Moreover, the sensitivity to IFN treatment (Figures 6A and 6B) apparently contributes to favorable clinical manifestations of these enteroviruses.
We and others have previously reported productive infection of MERS-CoV and SARS-CoV-2 in human intestinal organoids (Lamers et al., 2020; Zhou et al., 2017, 2020a). Gastrointestinal symptoms, mainly in the form of diarrhea, were very common in SARS patients. SARS-CoV infection of human intestinal cells was verified by electron microscopy of a patient's endoscopic biopsies (Leung et al., 2003). In this study, we demonstrated, for the first time, active replication of SARS-CoV in human intestinal organoids, and examined the susceptibility of enteroids to SARS-CoV in comparison with SARS-CoV-2. Notably, SARS-CoV replicates to a titer 1–2 log units higher than SARS-CoV-2 within 2 days after infection (Figure 5). The productive infection of both SARS-CoV and SARS-CoV-2 in human intestinal organoids are not unexpected, since the essential host factors for cellular entry of the two viruses, cellular receptor ACE2 (angiotensin-converting enzyme 2), and virus activation protease TMPRSS2, are highly expressed in the differentiated intestinal organoids, as we demonstrated elsewhere (Zhou et al., 2020a). SARS-CoV-2 replication in enteroids elicits a moderate induction of IFN-λ1 and IFN-λ3, and a modest induction of ISGs, which is consistent to our previous observation (Zhou et al., 2020a). Intriguingly, more actively replicating SARS-CoV triggers minimal cytokine response within the same time frame. Given these observations, we believe that coronaviruses, especially SARS-CoV, apparently develop antagonism to evade IFN response in the early phase of infection. Apart from this, we asked whether the sensitivity of these viruses to type III IFNs could contribute to the distinct dynamics of virus-host interaction. We note that EV-A71 and CVA16 are comparably susceptible to IFN-λ treatment (Figure 6). SARS-CoV-2 is also significantly sensitive to IFN-λ3, whereas SARS-CoV is resistant to both IFN-λ3 and IFN-λ1. Namely, in response to replicating enteroviruses, intestinal cells promptly mount a robust type III IFN response, which in turn restricts virus propagation. In contrast, SARS-CoV not only deploys antagonism to evade IFN production in the infected intestinal cells, but also shows an intrinsic resistance to IFN-λ, the major antiviral player in the intestinal tissues. These mechanisms synergistically give SARS-CoV an edge to robustly propagate in intestinal organoids. SARS-CoV is phylogenetically related to SARS-CoV-2 with 79%–82% sequence similarity (Chan et al., 2020a). SARS-CoV emerged in 2002–2003 and infected more than 8,000 individuals with a case-fatality rate of approximately 10% (Perlman and Dandekar, 2005). Apart from the genetic relatedness, SARS-CoV-2 shares with SARS-CoV the same cell receptor, human ACE2. Accordingly, we observed that SARS-CoV and SARS-CoV-2 show a similar infection rate at early infection (Figure S2). Nevertheless, the higher capacity to evade IFN response and stronger resistance to IFN enable SARS-CoV to achieve a higher replication potential than SARS-CoV-2, and this may underlie the higher fatality rate of SARS-CoV than SARS-CoV-2.
Type I IFNs have been considered as key cytokines in the frontline of cellular response against viral infections. However, the plethora of in vivo studies have demonstrated the critical role of IFN-λ in the immune protection of barrier tissues, including the epithelium lining the respiratory and gastrointestinal tract. Intestinal epithelial cells respond exclusively to IFN-λ because these cells show high levels of the IFN-λ receptor and low levels of the IFN-α/β receptor (Wack et al., 2015). In addition, epithelial cells, including intestinal epithelial cells, are potent inducers of IFN-λ. In mouse reovirus and norovirus infections, type I Ifn is indispensable for the restriction of systematic infection, but virus control in the intestinal tract depends on the intact type III Ifn signaling (Nice et al., 2015; Pott et al., 2011). These findings are generally obtained from mouse studies. The advent of adult stem cell-derived organoids provides an abundant and readily available source of human epithelial cells in which the interaction of native intestinal epithelial cells with viruses can be resolved in culture dishes. We and others reported the induction of type III IFNs in virus-infected human intestinal organoids (Good et al., 2019; Zhou et al., 2020a). While verifying the sensitivity of enteroviruses to type III IFNs as described previously (Good et al., 2019), we further demonstrate that SARS-CoV is resistant to IFN-λ, in contrast to the phylogenetic-related SARS-CoV-2, which is both an active stimulator of IFN-λ and an acute responder to IFN-λ.
We modified the differentiation protocol and generated differentiated human intestinal organoids that better simulate human native intestinal epithelium. Our studies unravel the distinct dynamics of the virus-host interaction of enteroviruses and coronaviruses in human intestinal organoids. The competence of human intestinal organoids to recapitulate enteric enterovirus and coronavirus infections underscores the fact that these organoids as a physiologically relevant model to study biology and pathology in human intestinal tract.
Experimental Procedures
Cell Culture and Virus Propagation
RD cell (ATCC, CCL-136) and Vero-E6 (ATCC, CRL-1586) were cultured in Dulbecco’s modified Eagle’s medium (Gibco) supplemented with 10% fetal bovine serum (Gibco) and 100 units/mL penicillin and streptomycin at 37°C with 5% CO2. Clinical isolates of EV-A71 (GenBank accession number DQ341368.1) and CVA16 were propagated and titrated in RD cells with a 50% cell culture infective dose (TCID50) assay as described elsewhere (Zhou et al., 2014). SARS-CoV-2 (GenBank accession number MT230908) and SARS-CoV (GZ50, GenBank accession number AY304495) were propagated in Vero E6 cells and titrated as we described previously (Zhou et al., 2018). All experiments with live viruses were conducted in biosafety level 2 or 3 laboratories upon institutional approval.
Establishment, Maintenance, and Differentiation of Human Intestinal Organoids
After ethics approval by the institutional review board of the University of Hong Kong/Hospital Authority Hong Kong West Cluster (UW13-364), human intestinal organoids were previously established using the human small and large intestinal tissues from patients who underwent surgical resection (Zhou et al., 2020a). Human intestinal organoids are maintained in the expansion medium and passaged every 7 days. To induce differentiation, 4 days after passage, the expansion medium was replaced with three differentiation media, H10, H5, and H0, and incubated for 5 days (Table 1). Photomicrographs of the organoids were acquired using a Nikon Eclipse TS100 inverted routine microscope.
Virus Infection and Detection
Differentiated intestinal organoids were sheared mechanically and incubated with the indicated virus for 2 h at 37°C. After the inoculum was removed, the intestinal organoids were rinsed with PBS, re-embedded in Matrigel and maintained in the differentiation medium. To assess replication kinetics, enteroviruses and coronaviruses were inoculated in organoids at MOIs of 0.01 and 1, respectively. At the indicated hours after inoculation, cell-free culture media were harvested and applied to RNA extraction using the MiniBEST Viral RNA/DNA Extraction Kit (TaKaRa), detection of viral loads by qRT-PCR assay, and viral titration (Zhou et al., 2014). Viral loads of EV-A71 and CVA16 were detected by qRT-PCR assay (primer sequences are listed in Table S1), those of SARS-CoV and SARS-CoV-2 were detected using a one-step qRT-PCR assay (QuantiNova Probe RT-PCR Kit, QIAGEN), as described elsewhere (Zhou et al., 2020a).
Quantification of Cellular mRNA Transcript by qRT-PCR Assay
To assess the cellular response upon infection, the infected or mock-infected human intestinal organoids were harvested at 10 hpi and applied to RNA extraction, followed by reverse transcription and qPCR assay to measure mRNA expression levels of cellular genes as described elsewhere (Zhang et al., 2019; Zhou et al., 2019). In brief, cell lysates were applied to RNA extraction using a MiniBEST Universal RNA Extraction Kit (TaKaRa), followed by reverse transcription using oligo(dT). The resultant cDNAs were used for qPCR assay with SYBR Green I Master (Roche) to measure mRNA expression level of cellular gene, which is normalized with that of GAPDH, and presented. The primer sequences used in qPCR assay are shown in Table S1.
Flow Cytometry Analysis and Immunofluorescence Staining
The infected or mock-infected human intestinal organoids were dissociated into a single-cell suspension with TrypLE (Gibco) and then fixed with 4% paraformaldehyde at 10 hpi. The fixed cells were immune stained, followed by confocal imaging and flow cytometry for the examination of infection rate and cell tropism. In brief, after fixation and permeabilization, the human intestinal organoids were labeled with primary antibodies or antibodies of the isotype control, followed by labeling with secondary antibodies. Rabbit anti-Villin (ab130751), mouse anti-Lysozyme (ab36362), mouse anti-CHGA (ab199014), rabbit (ab172730), and mouse (ab18421) isotypic antibodies were purchased from Abcam. Mouse anti-Mucin 2 (sc-515032) and anti-dsRNA (10010500) were purchased from Santa Cruz and Scicons, respectively. EV-A71- and CVA16-infected cells were detected using in-house made mouse anti-VP1 antibody, followed by fluorescein isothiocyanate (FITC)-conjugated goat anti-mouse IgG (Life Technologies). Viral structural protein VP1 is highly conserved in EV-A71 and CVA16. SARS-CoV and SARS-CoV-2 were detected using in-house made mouse anti-NP antibody (Zhou et al., 2020a), followed by FITC-conjugated goat anti-mouse IgG (Life Technologies). Flow cytometry analysis was performed using a BD FACSCanto II flow cytometer (BD Biosciences), and the data were analyzed using FlowJo v.10 (Tree Star, USA). For confocal imaging, nuclei and actin filaments were counterstained with 4′-6-diamino-2-phenylindole (Thermo Fisher Scientific) and Phalloidin-647 (Sigma Aldrich), respectively. The organoids were whole-mounted with ProLong Gold antifade reagent (Life Technologies) and imaged using a Carl Zeiss LSM 800 confocal microscope.
IFN Treatment
Recombinant IFN-λ1 or IFN-λ3 (300 ng/mL; 1598-IL-025 and 5259-IL-025, R&D Systems) were used to pretreat Vero-E6 cells for 18 h before virus inoculation and maintained in the culture medium throughout after inoculation. At 48 hpi, cell-free culture media were harvested and applied to RNA extraction and detection of viral loads, and viral titration by TCID50 assay.
Statistics
Unpaired t test was performed for data analysis using GraphPad Prism 6. A p value < 0.05 was considered to be statistically significant. Data are presented as mean and standard deviation (SD) of representative experiments.
Author Contributions
J.Z. and K.Y.Y. designed and supervised the study. X.Z., C.L., X.L., M.C.C., D.W., Y.W., and H.C. performed the experiments. J.-P.C. prepared the antibodies. J.Z., X.Z., C.L., X.L., J.F.-W.C., and K.K.-W.T. analyzed the data. I.H.-Y.C. and K.K.-Y.W. provided human tissues. J.Z., X.Z., and K.Y.Y. wrote the manuscript.
Declaration of interests
The authors declare no competing interests.
Acknowledgments
We thank the Center of PanorOmic Sciences and Electron Microscope Unit, Li Ka Shing Faculty of Medicine, University of Hong Kong, for assistance in confocal imaging flow cytometry and electron microscopy.
This work was partly supported by funding from the Health and Medical Research Fund (HMRF, 17161272 and 19180392) of the Food and Health Bureau of the HKSAR government to J.Z.; General Research Fund (GRF, 17105420) of the Research Grants Council of HKSAR government to J.Z.; Theme-based Research Scheme (T11-707/15-R) of the Research Grants Council, the HKSAR government, to K.Y.Y.; the High Level Hospital-Summit Program in Guangdong, The University of Hong Kong-Shenzhen Hospital to K.Y.Y.; and donations of the Shaw Foundation Hong Kong, May Tam Mak Mei Yin, Richard Yu and Carol Yu, Michael Seak-Kan Tong, Respiratory Viral Research Foundation Limited, Hui Ming, Hui Hoy & Chow Sin Lan Charity Fund Limited, and Chan Yin Chuen Memorial Charitable Foundation to K.Y.Y.
Published: March 9, 2021
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.stemcr.2021.02.009.
Supplemental Information
References
- Chan J.F., Kok K.H., Zhu Z., Chu H., To K.K., Yuan S., Yuen K.Y. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg. Microbes Infect. 2020;9:221–236. doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J.F., Yuan S., Kok K.H., To K.K., Chu H., Yang J., Xing F., Liu J., Yip C.C., Poon R.W. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020;395:514–523. doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K.T., Chang H.L., Wang S.T., Cheng Y.T., Yang J.Y. Epidemiologic features of hand-foot-mouth disease and herpangina caused by enterovirus 71 in Taiwan, 1998–2005. Pediatrics. 2007;120:e244–e252. doi: 10.1542/peds.2006-3331. [DOI] [PubMed] [Google Scholar]
- Cheng H., Leblond C.P. Origin, differentiation and renewal of the four main epithelial cell types in the mouse small intestine. I. Columnar cell. Am. J. Anat. 1974;141:461–479. doi: 10.1002/aja.1001410403. [DOI] [PubMed] [Google Scholar]
- Cimolai N. Features of enteric disease from human coronaviruses: implications for COVID-19. J. Med. Virol. 2020;92:1834–1844. doi: 10.1002/jmv.26066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevers H. Modeling development and disease with organoids. Cell. 2016;165:1586–1597. doi: 10.1016/j.cell.2016.05.082. [DOI] [PubMed] [Google Scholar]
- Co J.Y., Margalef-Catala M., Li X., Mah A.T., Kuo C.J., Monack D.M., Amieva M.R. Controlling epithelial polarity: a human enteroid model for host-pathogen interactions. Cell Rep. 2019;26:2509–2520 e2504. doi: 10.1016/j.celrep.2019.01.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lau W., Peng W.C., Gros P., Clevers H. The R-spondin/Lgr5/Rnf43 module: regulator of Wnt signal strength. Genes Dev. 2014;28:305–316. doi: 10.1101/gad.235473.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ettayebi K., Crawford S.E., Murakami K., Broughman J.R., Karandikar U., Tenge V.R., Neill F.H., Blutt S.E., Zeng X.L., Qu L. Replication of human noroviruses in stem cell-derived human enteroids. Science. 2016;353:1387–1393. doi: 10.1126/science.aaf5211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good C., Wells A.I., Coyne C.B. Type III interferon signaling restricts enterovirus 71 infection of goblet cells. Sci. Adv. 2019;5:eaau4255. doi: 10.1126/sciadv.aau4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X.C., Zhang J., Tong W.G., Tawfik O., Ross J., Scoville D.H., Tian Q., Zeng X., He X., Wiedemann L.M. BMP signaling inhibits intestinal stem cell self-renewal through suppression of Wnt-beta-catenin signaling. Nat. Genet. 2004;36:1117–1121. doi: 10.1038/ng1430. [DOI] [PubMed] [Google Scholar]
- Heo I., Dutta D., Schaefer D.A., Iakobachvili N., Artegiani B., Sachs N., Boonekamp K.E., Bowden G., Hendrickx A.P.A., Willems R.J.L. Modelling Cryptosporidium infection in human small intestinal and lung organoids. Nat. Microbiol. 2018;3:814–823. doi: 10.1038/s41564-018-0177-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamers M.M., Beumer J., van der Vaart J., Knoops K., Puschhof J., Breugem T.I., Ravelli R.B.G., Paul van Schayck J., Mykytyn A.Z., Duimel H.Q. SARS-CoV-2 productively infects human gut enterocytes. Science. 2020;369:50–54. doi: 10.1126/science.abc1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung W.K., To K.F., Chan P.K., Chan H.L., Wu A.K., Lee N., Yuen K.Y., Sung J.J. Enteric involvement of severe acute respiratory syndrome-associated coronavirus infection. Gastroenterology. 2003;125:1011–1017. doi: 10.1016/j.gastro.2003.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muehlenbachs A., Bhatnagar J., Zaki S.R. Tissue tropism, pathology and pathogenesis of enterovirus infection. J. Pathol. 2015;235:217–228. doi: 10.1002/path.4438. [DOI] [PubMed] [Google Scholar]
- Nice T.J., Baldridge M.T., McCune B.T., Norman J.M., Lazear H.M., Artyomov M., Diamond M.S., Virgin H.W. Interferon-lambda cures persistent murine norovirus infection in the absence of adaptive immunity. Science. 2015;347:269–273. doi: 10.1126/science.1258100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlman S., Dandekar A.A. Immunopathogenesis of coronavirus infections: implications for SARS. Nat. Rev. Immunol. 2005;5:917–927. doi: 10.1038/nri1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pott J., Mahlakoiv T., Mordstein M., Duerr C.U., Michiels T., Stockinger S., Staeheli P., Hornef M.W. IFN-lambda determines the intestinal epithelial antiviral host defense. Proc. Natl. Acad. Sci. U S A. 2011;108:7944–7949. doi: 10.1073/pnas.1100552108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Z., Li Y., Zhao B., Xu C., Liu Y., Li H., Zhang B., Wang X., Yang X., Xie W. BMP restricts stemness of intestinal Lgr5(+) stem cells by directly suppressing their signature genes. Nat. Commun. 2017;8:13824. doi: 10.1038/ncomms13824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T., Stange D.E., Ferrante M., Vries R.G., Van Es J.H., Van den Brink S., Van Houdt W.J., Pronk A., Van Gorp J., Siersema P.D. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett's epithelium. Gastroenterology. 2011;141:1762–1772. doi: 10.1053/j.gastro.2011.07.050. [DOI] [PubMed] [Google Scholar]
- Sato T., Vries R.G., Snippert H.J., van de Wetering M., Barker N., Stange D.E., van Es J.H., Abo A., Kujala P., Peters P.J. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- Solomon T., Lewthwaite P., Perera D., Cardosa M.J., McMinn P., Ooi M.H. Virology, epidemiology, pathogenesis, and control of enterovirus 71. Lancet Infect. Dis. 2010;10:778–790. doi: 10.1016/S1473-3099(10)70194-8. [DOI] [PubMed] [Google Scholar]
- Wack A., Terczynska-Dyla E., Hartmann R. Guarding the frontiers: the biology of type III interferons. Nat. Immunol. 2015;16:802–809. doi: 10.1038/ni.3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong S.S., Yip C.C., Lau S.K., Yuen K.Y. Human enterovirus 71 and hand, foot and mouth disease. Epidemiol. Infect. 2010;138:1071–1089. doi: 10.1017/S0950268809991555. [DOI] [PubMed] [Google Scholar]
- Xing W., Liao Q., Viboud C., Zhang J., Sun J., Wu J.T., Chang Z., Liu F., Fang V.J., Zheng Y. Hand, foot, and mouth disease in China, 2008–12: an epidemiological study. Lancet Infect. Dis. 2014;14:308–318. doi: 10.1016/S1473-3099(13)70342-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan K.S., Janda C.Y., Chang J., Zheng G.X.Y., Larkin K.A., Luca V.C., Chia L.A., Mah A.T., Han A., Terry J.M. Non-equivalence of Wnt and R-spondin ligands during Lgr5(+) intestinal stem-cell self-renewal. Nature. 2017;545:238–242. doi: 10.1038/nature22313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung M.L., Jia L., Yip C.C.Y., Chan J.F.W., Teng J.L.L., Chan K.H., Cai J.P., Zhang C., Zhang A.J., Wong W.M. Human tryptophanyl-tRNA synthetase is an IFN-gamma-inducible entry factor for enterovirus. J. Clin. Invest. 2018;128:5163–5177. doi: 10.1172/JCI99411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaoutis T., Klein J.D. Enterovirus infections. Pediatr. Rev. 1998;19:183–191. doi: 10.1542/pir.19-6-183. [DOI] [PubMed] [Google Scholar]
- Zhang K., Li C., Luo Y.S., Wen L., Yuan S., Wang D., Ho-Yin Wong B., Zhao X., Chiu M.C., Ye Z.W. Establishment of a lethal aged mouse model of human respiratory syncytial virus infection. Antivir. Res. 2019;161:125–133. doi: 10.1016/j.antiviral.2018.11.015. [DOI] [PubMed] [Google Scholar]
- Zhou J., Chu H., Li C., Wong B.H., Cheng Z.S., Poon V.K., Sun T., Lau C.C., Wong K.K., Chan J.Y. Active replication of Middle East respiratory syndrome coronavirus and aberrant induction of inflammatory cytokines and chemokines in human macrophages: implications for pathogenesis. J. Infect. Dis. 2014;209:1331–1342. doi: 10.1093/infdis/jit504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Li C., Liu X., Chiu M.C., Zhao X., Wang D., Wei Y., Lee A., Zhang A.J., Chu H. Infection of bat and human intestinal organoids by SARS-CoV-2. Nat. Med. 2020;26:1077–1083. doi: 10.1038/s41591-020-0912-6. [DOI] [PubMed] [Google Scholar]
- Zhou J., Li C., Sachs N., Chiu M.C., Wong B.H., Chu H., Poon V.K., Wang D., Zhao X., Wen L. Differentiated human airway organoids to assess infectivity of emerging influenza virus. Proc. Natl. Acad. Sci. U S A. 2018;115:6822–6827. doi: 10.1073/pnas.1806308115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Li C., Zhao G., Chu H., Wang D., Yan H.H., Poon V.K., Wen L., Wong B.H., Zhao X. Human intestinal tract serves as an alternative infection route for Middle East respiratory syndrome coronavirus. Sci. Adv. 2017;3:eaao4966. doi: 10.1126/sciadv.aao4966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Wang D., Wong B.H., Li C., Poon V.K., Wen L., Zhao X., Chiu M.C., Liu X., Ye Z. Identification and characterization of GLDC as host susceptibility gene to severe influenza. EMBO Mol. Med. 2019;11:e9528. doi: 10.15252/emmm.201809528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.