Abstract
The human adult liver has a multi-cellular structure consisting of large lobes subdivided into lobules containing portal triads and hepatic cords lined by specialized blood vessels. Vital hepatic functions include filtering blood, metabolizing drugs, and production of bile and blood plasma proteins like albumin, among many other functions, which are generally dependent on the location or zone in which the hepatocyte resides in the liver. Due to the liver’s intricate structure, there are many challenges to design differentiation protocols to generate more mature functional hepatocytes from human stem cells and maintain the long-term viability and functionality of primary hepatocytes. To this end, recent advancements in three-dimensional (3D) stem cell culture have accelerated the generation of a human miniature liver system, also known as liver organoids, with polarized epithelial cells, supportive cell types and extra-cellular matrix deposition by translating knowledge gained in studies of animal organogenesis and regeneration. To facilitate the efforts to study human development and disease using in vitro hepatic models, a thorough understanding of state-of-art protocols and underlying rationales is essential. Here, we review rapidly evolving 3D liver models, mainly focusing on organoid models differentiated from human cells.
Introduction
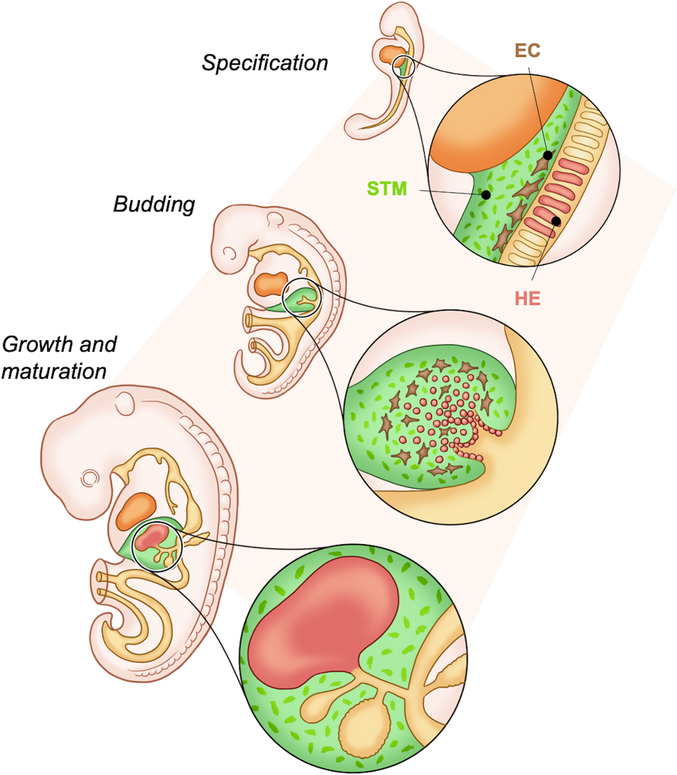
During early development, the liver is first specified from the endoderm-derived ventral foregut at embryonic (E) day 7.5 in the mouse, and the third week of human gestation (Gordillo et al., 2015). It has been shown that three regions of foregut endoderm contribute to hepatic progenitors: the paired lateral domains, the ventral midline of the endoderm lip and other endodermal tissues (Zaret, 2016). This layer of bi-potent hepatoblasts thickens and buds away from the endoderm during days E8.5–10 (Zaret, 2016) (Fig. 1). These hepatoblasts express alpha-fetoprotein (AFP) and markers of hepatocytes, including albumin and hepatocyte nuclear factor (HNF) 4α, as well as markers of cholangiocytes, like cytokeratin (CK) 19, as they delaminate from the foregut and expand into the surrounding septum transversum mesenchyme (STM) (Gordillo et al., 2015; Ober and Lemaigre, 2018) (Fig. 1).
Figure 1. Schematic of liver bud formation.
Liver progenitors are first specified from the ventral foregut endoderm which is surrounded by the STM and endothelial cells that supply necessary factors. The hepatoblast cell layer thickens and begins to delaminate from the epithelium and bud out through the STM to form the liver bud. The liver bud then undergoes growth and maturation throughout development to form the fetal liver. (abbreviations: septum transversum mesenchyme (STM), endothelial cells (EC), hepatic endoderm (HE)).
The liver bud receives critical developmental signals from the surrounding cardiac mesoderm, the STM, and the endothelium that include ligands of the bone morphogenic protein (BMP), Wnt, and fibroblast growth factor (FGF) families (Gordillo et al., 2015; Han et al., 2020; Zaret, 2016; Zorn, 2008). Depending on the signals received and their location in relation to the intrahepatic bile ducts the bi-potent hepatoblasts differentiate into either hepatocytes or cholangiocytes which continue to mature until after birth through intricate transcriptional regulations (reviewed in Huppert and Iwafuchi-Doi, 2019; Zorn, 2008). For example, hepatocyte maturation is dependent on several factors including oncostatin M (OSM), glucocorticoid hormones, hepatocyte growth factor (HGF), and Wnt signaling (Shin and Monga, 2013; Zorn, 2008). Additional liver-resident cells which are derived from the mesoderm include fibroblasts and stellate cells, sinusoidal endothelial cells and immune cells like Kupffer cells (Zorn, 2008) all of which are involved in signaling and cell-cell interactions.
Within the human body the liver is located behind the rib cage, and above the stomach and small intestine. This anatomical location is critical as the liver has functional connections with many organs of the gastrointestinal tract. The liver receives oxygen-rich blood from the hepatic artery and nutrient-rich blood from the gastrointestinal tract, spleen, and pancreas. The liver filters and detoxifies this blood, metabolizes xenobiotics as well as endogenous metabolic byproducts, and is one of the main sites for protein synthesis. The liver is also responsible for producing and excreting bile which drains directly through the biliary tree into the duodenum and aids in lipid digestion.
The human liver is divided into 4 lobes, and these lobes can be further sub-divided into hexagonal shaped lobules, consisting of the central vein at the center and the portal triad, composed of the portal vein, hepatic artery, and intrahepatic bile duct system at the corners (Huppert and Iwafuchi-Doi, 2019). Within each lobule zones named periportal, mid-, or pericentral zones exist based on hepatocyte proximity to the central or portal veins (Shin and Monga, 2013). These different zones are distinguished by variations in amounts of hormones, oxygen and nutrients and the cells in each zone, and hepatocytes in each zone have specialized functions like fatty acid oxidation, urea and cholesterol synthesis and gluconeogenesis in the periportal zone (Huppert and Iwafuchi-Doi, 2019). For example, the signaling molecule β-catenin is active in the pericentral zone and downstream genes such as glutamine synthetase, cytochrome p450 (CYP) 2e1 and 1a2 are expressed at high levels by hepatocytes in this zone (Shin and Monga, 2013).
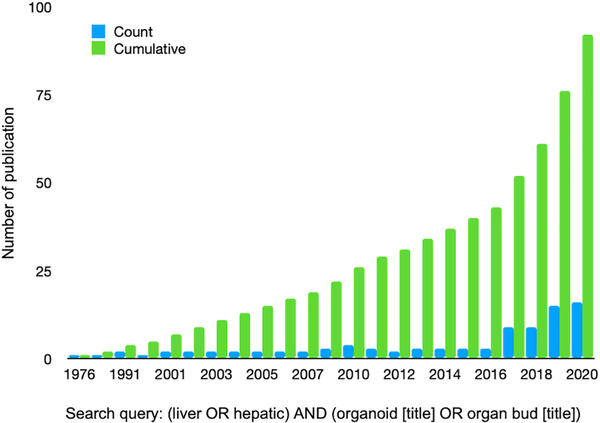
In a dish liver modeling studies with the use of human cells have focused on a variety of important research topics including liver development, drug toxicity and metabolism studies, liver disease modeling, cross population genetic studies, liver regeneration, and therapeutic transplantation. There is a need for more predictive and reproducible in vitro liver models and over the past few years, the amount of publications for liver organoids has increased greatly (Fig. 2). Given the complex 3D structure and functional regionalization of the liver, while 2D monolayer-based approaches have been informative, 3D liver models including organoids are imperative. These include more closely mimicking the cellular heterogeneity, spatial organization, and microenvironment, and recapitulating important cell-cell and cell-extracellular matrix (ECM) contacts that stimulate proliferation, differentiation, expression of relevant hepatic genes and proteins, and responsiveness to exogenous stimuli (Edmondson et al., 2014; Godoy et al., 2013). Due to the limited interactions between hepatocytes in 2D cultures there is a reduction in polarization, reduced bile canaliculi formation, and a decrease in signaling pathways that have been demonstrated to be critical for normal hepatocyte function (Godoy et al., 2013). For example, specific transporter proteins are expressed on the sinusoidal, basolateral and apical membranes of hepatocytes and this expression is lost in 2D cultures in which the hepatocytes are not polarized and have a more flattened morphology (Godoy et al., 2013). Furthermore, many studies utilizing primary human hepatocytes (PHHs)-, or primary stem cell- or pluripotent stem cell (hPSC)-derived hepatocytes cultured in 3D have demonstrated prolonged hepatic viability, gene expression, signaling, and/or function compared to a variety of 2D hepatic cultures (Bell et al., 2017; Berger et al., 2015; Gieseck et al., 2014; Kamei et al., 2019; Kim et al., 2015; Luo et al., 2018; Ma et al., 2016; Meier et al., 2017; Messner et al., 2013; Nagata et al., 2020; Pettinato et al., 2019; Proctor et al., 2017; Ramasamy et al., 2013; Schyschka et al., 2013; Sendi et al., 2018; Takayama et al., 2013; Tasnim et al., 2016; Vorrink et al., 2017; Wang et al., 2016; Wang et al., 2018). Hereafter, we will focus on the emerging 3D model system covering organoids, spheroids, aggregates and scaffold based engineered tissues. The detailed features and protocols of the recent literatures discussed in this review are summarized in Supplementary Table 1.
Figure 2. Research interest and publications for liver organoids has increased greatly over the past 5 years.
A pubmed search using the query (liver OR hepatic) AND (organoid [title] OR organ bud [title]) demonstrates a continued increase in liver organoid papers.
Overview of hPSC derived 3D models
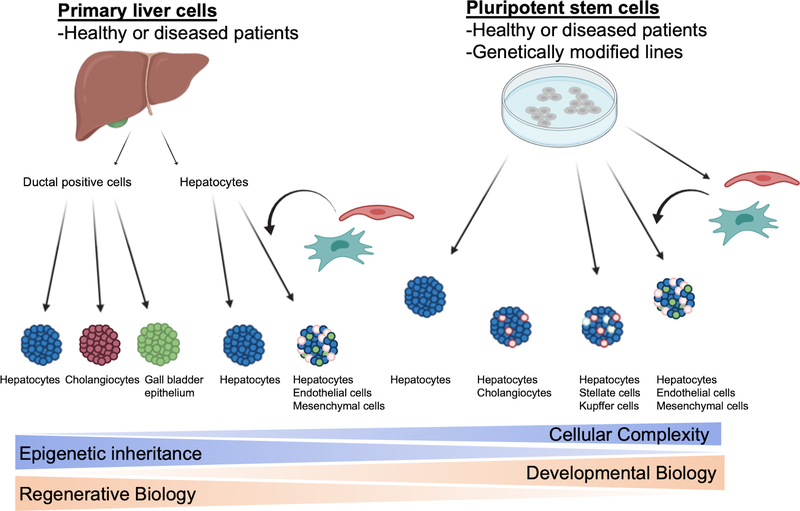
Human induced PSCs (hiPSCs) offer a near unlimited source of genetically diverse pluripotent cell lines that can be generated from healthy and diseased patients (Fig. 4). Furthermore, these cells are amenable to genetic modification using the CRISPR/Cas system to introduce single base changes to generate isogenic pairs of mutant and control iPSCs to facilitate powerful disease modeling. The protocols for the differentiation of human embryonic stem cells (hESCs) and hiPSCs to diverse organoid types is largely informed by studies identifying important developmental stages and signaling pathways in model organisms. However, hESCs and hiPSCs themselves have advantages to developing and refining methods for generating organoids including their experimental tractability and ability to empirically determine the activity of candidate signaling molecules on human development in the absence of the confounding effects of signaling from adjacent tissues encountered in animal models. One important limitation is the relative immaturity of hepatocyte-like cells generated from hPSCs. This is demonstrated by continued alpha-fetoprotein (AFP) and lower albumin expression, and distinctive CYP expression and activities. However, recent protocols have emphasized methods to enhance functional maturity by modifying culture conditions. These include improvements to medium composition (e.g. inclusion of specific growth factors, small molecules, hormones and corticosteroids), co-culture with additional supportive cell types (e.g. mesenchymal cells), and culture in substrates and scaffolds (e.g. laminins and hydrogels).
Figure 4: Two sources of human liver organoids include cells taken directly from the human liver as well as cells differentiated from hPSCs.
Both can use cells derived from healthy or diseased patients, and hPSCs can also be genetically modified before differentiation. Cells taken from the liver include primary hepatocytes as well as ductal cells and primary hepatocytes can be mixed with other supporting cell types. hPSCs can be co-differentiated in a dish to hepatocytes and cholangiocytes, or hepatocytes and stellate and Kupffer cells, and hepatocytes can also be mixed with other supporting cell types generated from hPSCs.
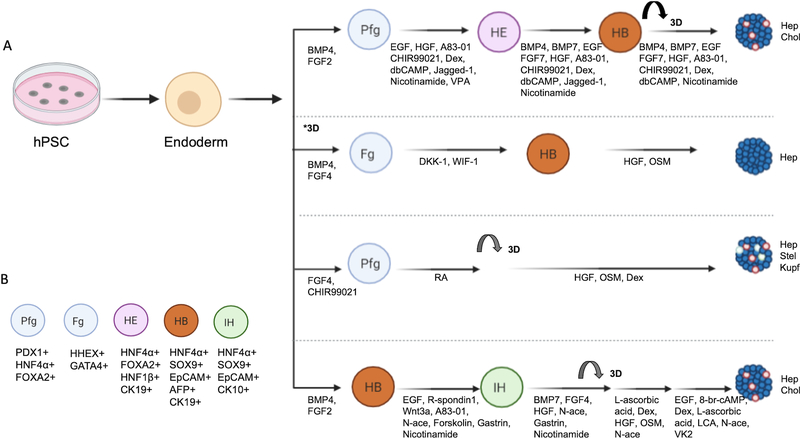
Many of the hPSC-generated hepatic 3D models rely on a stepwise directed differentiation approach that attempts to recapitulate in vivo development (Fig. 3) based on earlier papers (Cai et al., 2007; Si-Tayeb et al., 2010). First, either a monolayer culture grown on a diverse range of substrates including Matrigel or laminin or an embryoid body/spheroid culture is used to differentiate hPSCs into definitive endoderm (DE), the stage of endoderm development corresponding to E6.5–7 in the mouse. Embryoid bodies are 3D aggregates of hPSCs capable of differentiating into all 3 germ layers. Cultures at the DE stage express Forkhead Box A2 (FoxA2), SRY-Box Transcription Factor 17 (Sox17), and (C-X-C motif chemokine receptor 4) CXCR4 and downregulate stemness markers such as Oct 4 and Nanog. Typically, 3 or more days of Activin A, a member of the transforming growth factor β (TGFβ) superfamily, is used to mimic Nodal activity which specifies the endoderm (D’Amour et al., 2005). Often, Activin A is combined with BMP or Wnt cytokines to efficiently induce DE (D’Amour et al., 2006; Teo et al., 2012). Around DE stage, some literatures identified an expandable endodermal progenitors that can give rise to hepatic, pancreatic and intestinal lineages in both 2D and 3D (Cheng et al., 2012; Hannan et al., 2013; Zhang et al., 2018b).
Figure 3. A. Examples of step-wise directed differentiation of 3D hPSC derived hepatic organoids or spheroids.
Nearly all the hPSC derived hepatic 3D models first undergo a DE stage before modifications are made to the standard hepatic endoderm, hepatoblast and hepatocyte-like cell sequence. *3D: this differentiation starts out as embryoid bodies in suspension culture. B. Markers that correspond to each stage of organoid development. (abbreviations: Posterior foregut (Pfg), Hepatic endoderm (HE), Hepatoblast (HB), Foregut (Fg), Intermediate Hepatoblast (IH), Hepatocytes (Hep), Cholangiocytes (Chol), Stellate cells (Stel), Kupffer cells (Kupf), Dexamethasone (Dex), Epidermal Growth Gactor (EGF), N6,2’-ODibutyryladenosine 3’,5’-cyclic 35 monophosphate sodium salt (dbCAMP), Dickkopf-related protein 1 (DKK-1), Wnt inhibitory factor 1 (WIF-1), Retinoic Acid (RA), Lithocholic Acid (LCA), Vitamin K2 (VK2), Valproic Acid (VPA), N-acetylcysteine (N-ace), Pancreatic and Duodenal Homeobox 1 (PDX1), and Hematopoietically-expressed homeobox (HHEX)).
The next stage is differentiation of DE into either a foregut or a hepatic endoderm stage, also sometimes referred to as a hepatocyte precursor. This stage is often when many of the monolayer cultures are transferred to a 3D culture. To generate hepatic endoderm a combination of FGF2 and BMP4 is often used, resulting in cells that are hepatocyte nuclear factor (HNF)4α, HNF1β, and FoxA2 positive. Next, a hepatoblast (HNF4α, AFP, and FOXA2 positive) stage that is capable of differentiating into either hepatocytes or cholangiocytes is specified. To differentiate the cells into hepatocyte-like cells there is often one or more final steps involving OSM, HGF, and steroids to generate hepatocytes that are generally albumin, CYP3A4, E-cadherin, Asialoglycoprotein Receptor 1 (ASGR1), Bile Salt Export Pump (BSEP), Tryptophan 2,3-dioxygenase (TDO2), Transthyretin (TTR), Alpha-1 antitrypsin (A1AT), HNF4α and Zona Occludens 1 (ZO1) positive. Although hPSC hepatocytic models have advanced greatly over the past 10 years, new studies are continually improving hepatocyte functionality compared to PHH.
Organoids are self-organizing mini-organs derived from stem cells that can recapitulate many of the functions and cell-types seen in the original organ (Lancaster and Huch, 2019). In the past few years there have been several methods described for generating hepatic organoids from hPSCs, each incorporating modifications on the basic stepwise differentiation protocol described above (Table 1). For example, Akbari and colleagues separated the epithelial cell adhesion molecule (Epcam)-positive endodermal cells and embedded these cells into Matrigel using media derived from protocols from Huch et al to reproducibly differentiate the cells into hepatic organoids that are capable of being expanded (Akbari et al., 2019; Huch et al., 2015). In addition, several papers have designed organoids to generate multiple cell types by co-differentiating hepatocytes and cholangiocytes (Bin Ramli et al., 2020; Guan et al., 2017; Wang et al., 2019; Wu et al., 2019), or hepatocytes with other supporting cell types like stellate-like and Kupffer-like cells (Ouchi et al., 2019) by modifying the culture conditions (Fig. 4). Bin Ramli et al showed the specification of bipotent hepatoblasts by treating hepatic endoderm with BMP4, BMP7 and FGF7 which can then form organoids in a high-throughput 96-well plate approach and after further differentiation resulted in functional bile canaliculi systems (Bin Ramli et al., 2020). Ouchi et al, demonstrated that differentiation to posterior foregut endoderm resulted in co-development of a mesoderm population that could differentiate further into stellate and Kupffer cells (Ouchi et al., 2019). These multi-cellular liver organoids are capable of modeling fibrosis occurring after non-alcoholic steatohepatitis. Further, Wu et al discovered that by including 25% mTeSR1 in the media for differentiating hPSC to DE both endodermal and mesodermal commitment was induced and subsequently generated hepatobiliary organoids by activation of the NOTCH2 and TGF-β pathway that could survive for more than 8 weeks in immune-deficient mice (Wu et al., 2019).
Table 1: Representative articles of 3D liver organoids and liver buds from the past 5 years.
This table notes the cellular source of hepatocytes, the cell types present in the resulting organoid, which scaffold (if any) is used in the model, and major advantages of each study.
| Reference | Year | Source | Cell composition | Matrix support | Major advantages |
|---|---|---|---|---|---|
| Akbari et al Stem Cell Reports | 2019 | hPSC | hepatocytes, cholangiocytes | Matrigel | reproducible, efficient, long term culture, disease modelling |
| Bin Ramli et al Gastroenterology | 2020 | hPSC | hepatocytes, cholangiocytes | Matrigel, low attachment plate | high throughput approach, bile canaliculi network, disease modelling |
| Guan et al JCI insight | 2017 | hPSC | hepatocytes, cholangiocytes | Matrigel | hepatocytes that surround bile duct like structures, disease modelling |
| Hu et al Cell | 2018 | Primary | hepatocytes | Matrigel | long term culture, disease modelling, transplantable |
| Huch et al Cell | 2015 | Primary | hepatocytes or cholangiocytes | Matrigel | long term culture, genome stable, disease modelling |
|
Mun et al Journal of Hepatology Mun et al International Journal of Stem Cells |
2019, 2020 | hPSC | hepatocytes | Matrigel | long-term culture, exhibited hepatoxicity to drugs, disease modelling |
| Ng et al Biomaterials | 2018 | hPSC | hepatocytes | inverted colloidal crystal with collagen coating | functional, transplantable, bioengineered platform |
| Ouchi et al Cell Metabolism | 2019 | hPSC | hepatocytes, stellate cells, Kupffer cells | Matrigel | co-differentiation with multiple cell types, disease modelling |
| Sgodda et al Stem Cells and Development | 2017 | hPSC | hepatocytes | suspension culture | homogenous population, exhibited hepatoxicity to drugs |
| Takebe et al Nature, Takebe et al Cell Reports | 2013, 2017 | hPSC | hepatocytes, mesenchymal cells, endothelial cells | Matrigel, microwell plate | functional liver bud, vascularized upon transplantation, massive scale |
| Wang et al Cell Research | 2019 | hPSC | hepatocytes, cholangiocytes | Matrigel, low attachment plate | long term culture, disease modelling, transplantable |
| Wu et al Journal of Hepatology | 2019 | hPSC | hepatocytes, cholangiocytes | buds off 2D culture | hepatic and biliary function, transplantable |
| Zabulica et al Stem Cells Dev | 2019 | hPSC | hepatocytes | Matrigel | strong hepatic function |
Self-renewal of the hepatocyte progenitors is critical for generating and expanding large numbers of viable hepatocytes. Wang et al developed a protocol for generating organoids with functional hepatocytes and cholangiocytes that could be expanded for more than 20 passages resulting in 1018 cells after 5 months first by specifying hepatic endoderm/progenitors and then treatment with a chemically defined/ serum free protocol (Wang et al., 2019). Mun et al generated hepatic organoids that were self-renewing and mature, could model steatosis, and could be passaged for 1 year by collecting the 3D spheroids that formed and then further differentiating them in media also adapted from Huch et al (Huch et al., 2015; Mun et al., 2020; Mun et al., 2019).
A key issue is hepatic functionality and maturation of hPSC compared to PHH and human liver. Zabulica et al generated hepatic organoids from an ornithine transcarbamylase deficient hiPSC line and used these organoids to create a catalog of 60 hepatic, pluripotent, and developmental genes from both fetal and adult livers to be used to assess and optimize the maturation status of hepatic models (Zabulica et al., 2019). Other articles have used single cell RNA sequencing to compare the organoid models to fetal or adult primary hepatocytes demonstrating many of the models remain in a more fetal-like state (Camp et al., 2017), and modifications to protocols are ongoing (Ouchi et al., 2019). These diverse liver organoid models all highlight the distinct outcomes possible from harnessing the power of pluripotent cells and following directed differentiation in a dish.
hPSC derived hepatic 3D models are further facilitated by the use of evolving culture platforms: ultra-low attachment plates, microwell plates, or spinner/suspension cultures. These will enable the scalable generation of aggregates/spheroids either before organoid differentiation begins or during one of the later steps in each protocol. The surface on ultra-low attachment plates and microwell plates is a hydrophilic and neutrally charged yet biologically inert hydrogel coating and this prevents cells from binding to the surface forcing them to stay in suspension and promotes one spheroid formation per well. The spheroid can then be kept in these plates for further maturation, transferred to suspension cultures, or embedded in Matrigel or other hydrogels. These methods are especially useful when incorporating several cell types into the same spheroid, can be used completely scaffold-free, and are amenable to automation and high-throughput screening due to their consistently sized spheroids. Lu et al and Pettinato et al start with aggregated spheroids generated in a microwell plate or embryoid bodies before differentiating DE, and while Lu embeds these cells into a hydrogel, Pettinato leaves the spheroids in suspension for the entire differentiation (Luo et al., 2018; Pettinato et al., 2016). Several other studies aggregated hepatic endoderm, hepatoblasts, or hepatocyte progenitors and further differentiated the spheroids to generate hepatic models (Chen et al., 2020; Kim et al., 2015; Ng et al., 2018; Sgodda et al., 2017; Yang et al., 2020; Zhang et al., 2014). In general these methods allow for increased production of hepatic spheroids on larger scales; Chen and group were able to transfer the hepatic spheroids into a suspension culture and used ~1 × 109 cells in a bio-artificial liver to rescue a porcine model of liver failure (Chen et al., 2020). However, the terminology of these models are unclear at times in the literature, as they do not necessarily self-organize and may therefore lack certain cellular spatial organization seen in organoids.
Overview of primary cell derived 3D models
Another source of human hepatocytes for liver research is by isolation of primary cells from the mature liver, typically from surgical or biopsy specimens. Both PHH and bi-potent ductal epithelial cells can be isolated and cultured from these samples, these cells may retain epigenetic memory, and these methods hold great promise for regenerative therapies (Fig. 4). However, costly biopsies are needed to generate enough cells for study and donor availability can limit access to study materials. Furthermore, most mature PHH can only be cultured for a short period of time before rapidly de-differentiating in culture (Godoy et al., 2013). In contrast, culturing PHH as 3D spheroids has been shown to result in retention of some hepatocyte functions, morphology, and gene expression and remain metabolically stable through several weeks in culture (Bell et al., 2016; Vorrink et al., 2017).
Of the recent 3D models using primary cells a main focus has been on modifying media components to maximize hepatocyte viability and continued growth and proliferation (reviewed in Schutgens and Clevers, 2020). Huch et al generated human liver organoids from ductal EpCAM+ cells, that were grown in a defined human liver media that was developed by a systematic approach and contained a TGF-β inhibitor A83–01, forskolin and Wnt signals which were necessary for continued proliferation and extended the ability to culture the cells, that could be passaged for over 6 months and frozen and thawed (Huch et al., 2015). To differentiate these cells into a mature hepatocyte phenotype, the growth stimuli forskolin and R-spo were removed from the media, and a NOTCH inhibitor, FGF19, BMP7, and dexamethasone were added, which resulted in hepatocytes that had CYP3A4 activity similar to freshly isolated hepatocytes, secreted albumin, and could engraft into a murine model of liver damage (Huch et al., 2015). Further, this organoid model has been shown to efficiently model diseases such as α1-antitrypsin (A1AT) deficiency (Huch et al., 2015), liver cancers (Broutier et al., 2017), as well as to study drug-induced phospholipidosis (Lee et al., 2020), among others. Zhang et al used cryopreserved PHH grown in a defined media in which the Wnt3a pathway and hypoxia conditions were found to be important for PHH proliferation, and could passage the cells for more than one month with an over 10,000-fold expansion (Zhang et al., 2018a). These cells could then be used to generate liver organoids in the differentiation medium from Huch et al. Finally, Hu et al established organoids directly from human primary hepatocytes that could be grown and passaged for months and retained key mature hepatocyte functions and gene expression comparable to PHH after developing a media with key ingredients including Wnt/R-spondin signaling and HGF (Hu et al., 2018).
Of note is the method of direct reprogramming of fibroblasts into hepatocyte-like cells that are functionally active (Du et al., 2014; Huang et al., 2014; Sekiya and Suzuki, 2011; Xie et al., 2019). This method holds another promise towards regenerative studies and has been reviewed elsewhere (Ge et al., 2019).
ECM variations/synthetic scaffolds/specialized plates
Many of the 3D hepatic organoid models rely on Matrigel or similar preparations from the Engelbreth-Holm-Swarm mouse sarcoma that mimics the native extracellular matrix (ECM) and consists of laminin, collagen, proteoglycans as well as growth factors. This matrix allows organoids to self-organize and differentiate within a supportive physical ECM for cells to attach to, as well as providing factors and hormones that can affect gene and protein expression. However, disadvantages of animal derived hydrogels include lot-to-lot variability, an incompletely defined matrix, and species differences which limit applications for translational and in vivo use. Recent progress has systematically developed chemically-defined synthetic hydrogels to culture stem cell derived human intestinal and liver organoids that results in organoids that are comparable to those embedded in Matrigel and in some cases can be frozen and thawed (Gjorevski et al., 2016; Kruger et al., 2020; Sorrentino et al., 2020). This new work opens customizable possibilities for clinically relevant studies necessitating defined materials.
Other 3D models have utilized specialized types of biological scaffolds including de-cellularized livers, specialized plates like pillar plates or hanging drop plates, perfused liver on a chip technology, 3D printing, or bioreactors that offer alternatives to standard cultures. Biological scaffolds like de-cellularized livers offer a cell-free 3D support structure that has preserved the native liver form and consists of both liver-specific ECM as well as other biological molecules like growth factors. Several variations exist using whole animal acellular liver scaffolds or slices of the liver to grow and mature hepatocytes with and without non-parenchymal cells. Vyas et demonstrated that de-cellularized ferret livers could provide liver ECM and structure for human fetal liver progenitor cells that allowed for 3D liver organoids to form with both hepatic and biliary structures (Vyas et al., 2018). When hiPSC generated hepatocytes were introduced into an acellular scaffold derived from rat livers they became polarized, formed bile canaliculi-like structures, and expressed higher levels of Cyp2C9, Cyp3A4, and Cyp1A2 than control 2D cells, and albumin levels close to PHH, with a corresponding decrease in AFP (Wang et al., 2016). Collin de l’Hortet et al showed that repopulating a de-cellularized rat liver could be used for modeling fatty liver by continually perfusing the scaffold, and re-populating the liver with genetically modified hPSC derived hepatocytes, mesenchymal cells, fibroblasts and macrophages (Collin de l’Hortet et al., 2019). Recently, Takeishi et al biofabricated a human liver resulting in mature hepatocytes by first re-populating the vasculature and biliary structure of an acellular rat liver using hiPSC generated biliary and vascular endothelial cells, and then the parenchyma with hiPSC generated hepatocytes along with human fibroblasts and mesenchymal stem cells (Takeishi et al., 2020). These approaches offer an exciting opportunity to reconstruct macro-anatomical composition including major blood vessels and establish a basis for future transplant studies.
In addition, several other novel 3D methods have modified ECM environments and demonstrate increased hepatocyte viability, functions and/or maturity. Nagata et al used hiPSC generated hepatocytes in a core-shell gel microfiber that encapsulates the hepatocytes in Matrigel resulting in higher expression of hepatic genes than in spheroid cultures, and demonstrated that this method is amenable to transplantation (Nagata et al., 2020). Using hPSC generated hepatocytes in spheroid form atop pillar plates and overlaid with Matrigel allowed Takayama et al to mature the hepatocytes compared to earlier methods (Takayama et al., 2013). Messner et al cultured PHH with stellate cells and endothelial cells in a hanging drop spheroid culture which enabled the hepatocytes to remain viable and functional for 5 weeks in culture demonstrated by stable ATP content and persistent albumin secretion (Messner et al., 2013). These results parallel a study using 3D printed primary PHH with endothelial cells and stellate cells with maintained ATP and albumin expression over 4 weeks (Nguyen et al., 2016). Finally, liver-on-a-chip 3D models mimics the liver microenvironment within a chip along with perfusion using microfluidic devices. hPSC derived hepatocytes placed in simple perfusable liver-on-a- chip models generated an increase in mature and functional hepatocytes (Kamei et al., 2019; Wang et al., 2018). These studies all highlight the importance of hepatocyte- substrate interactions for maturation and functionality, and the need for new innovations in chemical biology and material science towards the potential for liver regeneration.
Next-generation models with other liver cell types
Developmentally, interactions with non-parenchymal cells are essential and have been shown to aid in maturity and functional ability of hepatocytes. Models with other cell types can more accurately model liver disease where the non-parenchymal cells play a role, such as in fibrotic or inflammatory disease (Sharma et al., 2020). In in vitro 3D models addition of non-parenchymal cells increases heterotypic cell-cell contacts, soluble trophic factors and cytokines are released from each cell type, and the liver microenvironment is better recapitulated as non-parenchymal cells account for about 40% of total liver cells. Stellate cells are myofibroblasts that reside in the subendothelial space of Disse, can store vitamin A, and once activated can produce large amounts of extracellular matrix that can be found after chronic viral infections or nonalcoholic steatohepatitis (Tsuchida and Friedman, 2017). Liver sinusoidal endothelial cells line the hepatic sinusoids and are located where they filter blood coming from both the gut and the systemic circulation, as well as have immunological functions (Shetty et al., 2018).
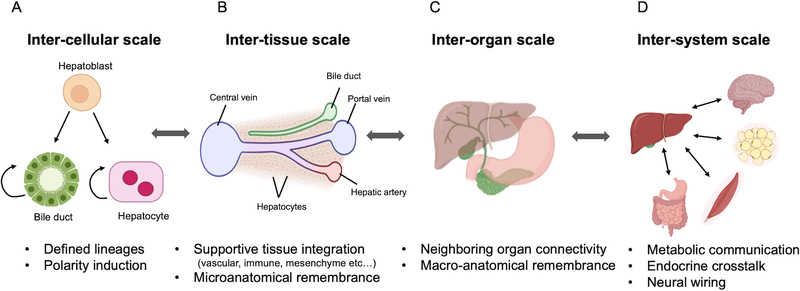
Takebe et al first demonstrated a functional complex, self-organizing liver bud by combining hPSC generated hepatic endoderm, human umbilical vein endothelial cells (HUVECs), and mesenchymal stem cells (MSCs) that once transplanted into the mouse could differentiate into target cells types and vascularize (Takebe et al., 2013). The liver bud method was later modified to use all hPSC derived cells from the same individual, using a high throughput protocol to generate liver buds on a massive scale (Takebe et al., 2017a). Recent studies using hPSC generated hepatic cells co-cultured in 3D models with other supporting cell types such as endothelial cells, stellate, and/or cholangiocyte cells have increased hepatic gene expression, albumin secretion, and CYP activity with a decrease in AFP expression when compared to hepatic only spheroids (Ardalani et al., 2019; Pettinato et al., 2019). Similar results have been seen in hiPSC hepatic co-culture 3D printed and reconstituted de-cellularized liver models in increasing hepatic functions (Goulart et al., 2019; Ma et al., 2016; Takeishi et al., 2020). By incorporating supportive cell types, these models all aim to more closely mimic human physiology by improving the hepatocyte microenvironment and demonstrate the importance of cell-cell cross talk and endogenous secretion of growth factors, signaling molecules and ECM (Fig 5A).
Figure 5: Framing next generation organoid systems.
On an inter-cellular scale there can be mixed cell types in one model generated from defined lineages. On an inter-tissue scale supportive tissues are incorporated including vascular, immune or mesenchymal lineages, and there is microanatomical remembrance. An inter-organ scale will include neighboring organ connectivity and macro-anatomical remembrance, while an inter-system scale can have connectivity between distant organs.
A next step up to build additional complexity in the 3D multi-cellular liver are models that incorporate and exhibit surrounding organ connectivity in a dish (Fig. 5B). Impaired inter-organ connectivity lead to critical and, in most cases, lethal organ failures, as seen in biliary atresia, thus, engineering organoid systems that incorporate connectivity between neighboring organs is a critical unmet challenge. It is vital to understand the mechanisms that drive the intricate inter-organ orchestrations that occur during organogenesis, which can then be leveraged for developing appropriate multi-organ three-dimensional (3-D) stem cell culture systems that allow for seamless connections to neighboring tissues (Song et al., 2019; Xiang et al., 2019; Xiang et al., 2017). Recently, hepatic, biliary, and pancreatic structures were patterned from hiPSCs and stemmed from the boundary region of anterior and posterior gut spheroids, which is a viable model to study complex endoderm organogenesis (Koike et al., 2019). This paper demonstrates that in vitro it is possible to study the development and interactions of multiple organs at one time. These models demonstrate the potential of using tissue-tissue models for predictive toxicity and metabolism studies as well as developmental studies (Fig. 5B–C).
Finally, ultimate goals of 3D tissue based modeling will recapitulate an inter-system scale interaction, combining more distant organs with the liver in a dish, by potentially incorporating metabolic communication, endocrine crosstalk, and neural wiring (Fig. 5D). Recent excitement of microphysiological system (MPS) is equipped to address some of these key challenges. For example, liver and intestine models-on-a-chip have been able to mimic a first pass metabolism system (Esch et al., 2014), liver and skin tissue chips (Wagner et al., 2013), liver-kidney biochips (Choucha-Snouber et al., 2013) or gut, liver and kidney or bone marrow, liver and kidney chips (Herland et al., 2020). The multi-organ on a chip models allow for precise control over the culture conditions (including nutrients, flow conditions, ECM, etc), as well as for the cellular inputs including differentiated state, cellular number when combined, and are amenable to higher-throughput and real-time monitoring (Takebe et al., 2017b). These models will further require unique, customizable systems capable of producing multiple, viable and functional mini-organs in one enclosed MPS, also known as “body-on-a-chip” or “physiome-on-a-chip” (Sharma et al., 2020; Takebe et al., 2017b). These models will allow an in vitro representation of human physiology and pathology on an ultimate scale.
Conclusions
In the past five years remarkable advancements have been made using 3D in vitro hepatic models in terms of functionality and maturity of hepatocytes, long term viability, and more precise representations of the microarchitecture of the in vivo liver. These new models offer scientists the opportunity to more accurately portray human disease in a dish, as well as to study liver development and regeneration, and hepatic toxicity and metabolism studies. By exploring novel scaffolds and substrates, as well as investigating the signaling pathways necessary for hepatic growth and survival much progress has been made in the world of in vitro 3D hepatic models.
Supplementary Material
Supplementary Table 1: Representative articles of 3D human hepatic models from the past 10 years. This spreadsheet is an overview of current (after 2010) articles that discuss human liver/hepatic spheroids/organoids/liver on a chip, liver scaffolds models. The cellular source of hepatocytes, the cell types present in the resulting organoid, which scaffold (if any) is used in the model, the step-by-step protocol, quality control steps, and major advantages of each study are listed, along with complete references.
Acknowledgements
We thank Misaki Ouchida for illustration materials in Figure 1A. We would like to also express sincere gratitude to the other Takebe lab members, Wells-Zorn lab members, PSCF lab members for their support and excellent technical assistance. This work was supported by Cincinnati Children’s Research Foundation grant, NIH Director’s New Innovator Award (DP2 DK128799-01) and CREST (20gm1210012h0001) grant from Japan Agency for Medical Research and Development (AMED) to TT. This work was also supported by an NIH grant UH3 DK119982, Cincinnati Center for Autoimmune Liver Disease Fellowship Award, PHS Grant P30 DK078392 (Integrative Morphology Core and Pluripotent Stem Cell and Organoid Core) of the Digestive Disease Research Core Center in Cincinnati, the Falk Catalyst Research Awards Program, Takeda Science Foundation award, Mitsubishi Foundation award and AMED JP19fk0210037, JP19bm0704025, JP19fk0210060, JP19bm0404045, and JSPS JP18H02800, 19K22416. TT is a New York Stem Cell Foundation – Robertson Investigator.
References
- Akbari S, Sevinc GG, Ersoy N, Basak O, Kaplan K, Sevinc K, Ozel E, Sengun B, Enustun E, Ozcimen B, et al. (2019). Robust, Long-Term Culture of Endoderm-Derived Hepatic Organoids for Disease Modeling. Stem Cell Reports 13, 627–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardalani H, Sengupta S, Harms V, Vickerman V, Thomson JA, and Murphy WL (2019). 3-D Culture and Endothelial Cells Improve Maturity of Human Pluripotent Stem Cell-Derived Hepatocytes. Acta Biomaterialia 95, 371–381. [DOI] [PubMed] [Google Scholar]
- Bell CC, Hendriks DFG, Moro SML, Ellis E, Walsh J, Renblom A, Fredriksson Puigvert L, Dankers ACA, Jacobs F, Snoeys J, et al. (2016). Characterization of primary human hepatocyte spheroids as a model system for drug-induced liver injury, liver function and disease. Scientific Reports 6, 25187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell CC, Lauschke VM, Vorrink SU, Palmgren H, Duffin R, Andersson TB, and Ingelman-Sundberg M (2017). Transcriptional, Functional, and Mechanistic Comparisons of Stem Cell-Derived Hepatocytes, HepaRG Cells, and Three-Dimensional Human Hepatocyte Spheroids as Predictive In Vitro Systems for Drug-Induced Liver Injury. Drug Metab Dispos 45, 419–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger DR, Ware BR, Davidson MD, Allsup SR, and Khetani SR (2015). Enhancing the functional maturity of induced pluripotent stem cell-derived human hepatocytes by controlled presentation of cell-cell interactions in vitro. Hepatology 61, 1370–1381. [DOI] [PubMed] [Google Scholar]
- Bin Ramli MN, Lim YS, Koe CT, Demircioglu D, Tng W, Uy Gonzales KA, Tan CP, Szczerbinska I, Liang H, Soe EL, et al. (2020). Human Pluripotent Stem Cell-Derived Organoids as Models of Liver Disease. Gastroenterology. [DOI] [PubMed] [Google Scholar]
- Broutier L, Mastrogiovanni G, Verstegen MM, Francies HE, Gavarró LM, Bradshaw CR, Allen GE, Arnes-Benito R, Sidorova O, Gaspersz MP, et al. (2017). Human primary liver cancer-derived organoid cultures for disease modeling and drug screening. Nature Medicine 23, 1424–1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai J, Zhao Y, Liu Y, Ye F, Song Z, Qin H, Meng S, Chen Y, Zhou R, Song X, et al. (2007). Directed differentiation of human embryonic stem cells into functional hepatic cells. Hepatology 45, 1229–1239. [DOI] [PubMed] [Google Scholar]
- Camp JG, Sekine K, Gerber T, Loeffler-Wirth H, Binder H, Gac M, Kanton S, Kageyama J, Damm G, Seehofer D, et al. (2017). Multilineage communication regulates human liver bud development from pluripotency. Nature 546, 533–538. [DOI] [PubMed] [Google Scholar]
- Chen S, Wang J, Ren H, Liu Y, Xiang C, Li C, Lu S, Shi Y, Deng H, and Shi X (2020). Hepatic spheroids derived from human induced pluripotent stem cells in bio-artificial liver rescue porcine acute liver failure. Cell Res 30, 95–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X, Ying L, Lu L, Galvao AM, Mills JA, Lin HC, Kotton DN, Shen SS, Nostro MC, Choi JK, et al. (2012). Self-renewing endodermal progenitor lines generated from human pluripotent stem cells. Cell Stem Cell 10, 371–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choucha-Snouber L, Aninat C, Grsicom L, Madalinski G, Brochot C, Poleni PE, Razan F, Guillouzo CG, Legallais C, Corlu A, et al. (2013). Investigation of ifosfamide nephrotoxicity induced in a liver-kidney co-culture biochip. Biotechnology and Bioengineering 110, 597–608. [DOI] [PubMed] [Google Scholar]
- Collin de l’Hortet A, Takeishi K, Guzman-Lepe J, Morita K, Achreja A, Popovic B, Wang Y, Handa K, Mittal A, Meurs N, et al. (2019). Generation of Human Fatty Livers Using Custom-Engineered Induced Pluripotent Stem Cells with Modifiable SIRT1 Metabolism. Cell Metab 30, 385–401 e389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Amour KA, Agulnick AD, Eliazer S, Kelly OG, Kroon E, and Baetge EE (2005). Efficient differentiation of human embryonic stem cells to definitive endoderm. Nat Biotechnol 23, 1534–1541. [DOI] [PubMed] [Google Scholar]
- D’Amour KA, Bang AG, Eliazer S, Kelly OG, Agulnick AD, Smart NG, Moorman MA, Kroon E, Carpenter MK, and Baetge EE (2006). Production of pancreatic hormone-expressing endocrine cells from human embryonic stem cells. Nat Biotechnol 24, 1392–1401. [DOI] [PubMed] [Google Scholar]
- Du Y, Wang J, Jia J, Song N, Xiang C, Xu J, Hou Z, Su X, Liu B, Jiang T, et al. (2014). Human hepatocytes with drug metabolic function induced from fibroblasts by lineage reprogramming. Cell Stem Cell 14, 394–403. [DOI] [PubMed] [Google Scholar]
- Edmondson R, Broglie JJ, Adcock AF, and Yang L (2014). Three-dimensional cell culture systems and their applications in drug discovery and cell-based biosensors. Assay Drug Dev Technol 12, 207–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esch MB, Mahler GJ, Stokor T, and Shuler ML (2014). Body-on-a-chip simulation with gastrointestinal tract and liver tissues suggests that ingested nanoparticles have the potential to cause liver injury. Lab Chip 14, 3081–3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge JY, Zheng YW, Liu LP, Isoda H, and Oda T (2019). Impelling force and current challenges by chemicals in somatic cell reprogramming and expansion beyond hepatocytes. World J Stem Cells 11, 650–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gieseck RL, Hannan NRF, Bort R, Hanley NA, Drake RAL, Cameron GWW, Wynn TA, and Vallier L (2014). Maturation of induced pluripotent stem cell derived hepatocytes by 3D-culture. PLOS ONE 9, e86372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gjorevski N, Sachs N, Manfrin A, Giger S, Bragina ME, Ordonez-Moran P, Clevers H, and Lutolf MP (2016). Designer matrices for intestinal stem cell and organoid culture. Nature 539, 560–564. [DOI] [PubMed] [Google Scholar]
- Godoy P, Hewitt NJ, Albrecht U, Andersen ME, Ansari N, Bhattacharya S, Bode JG, Bolleyn J, Borner C, Bottger J, et al. (2013). Recent advances in 2D and 3D in vitro systems using primary hepatocytes, alternative hepatocyte sources and non-parenchymal liver cells and their use in investigating mechanisms of hepatotoxicity, cell signaling and ADME. Arch Toxicol 87, 1315–1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordillo M, Evans T, and Gouon-Evans V (2015). Orchestrating liver development. Development 142, 2094–2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulart E, de Caires-Junior LC, Telles-Silva KA, Araujo BHS, Rocco SA, Sforca M, de Sousa IL, Kobayashi GS, Musso CM, Assoni AF, et al. (2019). 3D bioprinting of liver spheroids derived from human induced pluripotent stem cells sustain liver function and viability in vitro. Biofabrication 12, 015010. [DOI] [PubMed] [Google Scholar]
- Guan Y, Xu D, Garfin PM, Ehmer U, Hurwitz M, Enns G, Michie S, Wu M, Zheng M, Nishimura T, et al. (2017). Human hepatic organoids for the analysis of human genetic diseases. JCI Insight 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han L, Chaturvedi P, Kishimoto K, Koike H, Nasr T, Iwasawa K, Giesbrecht K, Witcher PC, Eicher A, Haines L, et al. (2020). Single cell transcriptomics identifies a signaling network coordinating endoderm and mesoderm diversification during foregut organogenesis. Nat Commun 11, 4158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannan NR, Fordham RP, Syed YA, Moignard V, Berry A, Bautista R, Hanley NA, Jensen KB, and Vallier L (2013). Generation of multipotent foregut stem cells from human pluripotent stem cells. Stem Cell Reports 1, 293–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herland A, Maoz B, Das D, Somayaji MR, Prantil-Baun R, Novak R, Cronce M, Huffstater T, Jeanty SSF, Ingram M, et al. (2020). Quantitative prediction of human pharmacokinetic responses to drugs via fluidically coupled vascularized organ chips. Nature Biomedical Engineering. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H, Gehart H, Artegiani B, LÖpez-Iglesias C, Dekkers F, Basak O, van Es J, Chuva de Sousa Lopes SM, Begthel H, Korving J, et al. (2018). Long-Term Expansion of Functional Mouse and Human Hepatocytes as 3D Organoids. Cell 175, 1591–1606.e1519. [DOI] [PubMed] [Google Scholar]
- Huang P, Zhang L, Gao Y, He Z, Yao D, Wu Z, Cen J, Chen X, Liu C, Hu Y, et al. (2014). Direct reprogramming of human fibroblasts to functional and expandable hepatocytes. Cell Stem Cell 14, 370–384. [DOI] [PubMed] [Google Scholar]
- Huch M, Gehart H, van Boxtel R, Hamer K, Blokzijl F, Verstegen MMA, Ellis E, van Wenum M, Fuchs SA, de Ligt J, et al. (2015). Long-term culture of genome-stable bipotent stem cells from adult human liver. Cell 160, 299–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huppert SS, and Iwafuchi-Doi M (2019). Molecular regulation of mammalian hepatic architecture. Curr Top Dev Biol 132, 91–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamei KI, Yoshioka M, Terada S, Tokunaga Y, and Chen Y (2019). Three-dimensional cultured liver-on-a-Chip with mature hepatocyte-like cells derived from human pluripotent stem cells. Biomed Microdevices 21, 73. [DOI] [PubMed] [Google Scholar]
- Kim JH, Jang YJ, An SY, Son J, Lee J, Lee G, Park JY, Park HJ, Hwang DY, Kim JH, et al. (2015). Enhanced Metabolizing Activity of Human ES Cell-Derived Hepatocytes Using a 3D Culture System with Repeated Exposures to Xenobiotics. Toxicol Sci 147, 190–206. [DOI] [PubMed] [Google Scholar]
- Koike H, Iwasawa K, Ouchi R, Maezawa M, Giesbrecht K, Saiki N, Ferguson A, Kimura M, Thompson WL, Wells JM, et al. (2019). Modelling human hepato-biliary-pancreatic organogenesis from the foregut-midgut boundary. Nature 574, 112–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruger M, Oosterhoff LA, van Wolferen ME, Schiele SA, Walther A, Geijsen N, De Laporte L, van der Laan LJW, Kock LM, and Spee B (2020). Cellulose Nanofibril Hydrogel Promotes Hepatic Differentiation of Human Liver Organoids. Adv Healthc Mater 9, e1901658. [DOI] [PubMed] [Google Scholar]
- Lancaster MA, and Huch M (2019). Disease modelling in human organoids. Dis Model Mech 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JY, Han HJ, Lee SJ, Cho EH, Lee HB, Seok JH, Lim HS, and Son WC (2020). Use of 3D Human Liver Organoids to Predict Drug-Induced Phospholipidosis. Int J Mol Sci 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y, Lou C, Zhang S, Zhu Z, Xing Q, Wang P, Liu T, Liu H, Li C, Shi W, et al. (2018). Three-dimensional hydrogel culture conditions promote the differentiation of human induced pluripotent stem cells into hepatocytes. Cytotherapy 20, 95–107. [DOI] [PubMed] [Google Scholar]
- Ma X, Qu X, Zhu W, Li YS, Yuan S, Zhang H, Liu J, Wang P, Lai CS, Zanella F, et al. (2016). Deterministically patterned biomimetic human iPSC-derived hepatic model via rapid 3D bioprinting. Proc Natl Acad Sci U S A 113, 2206–2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier F, Freyer N, Brzeszczynska J, Knospel F, Armstrong L, Lako M, Greuel S, Damm G, Ludwig-Schwellinger E, Deschl U, et al. (2017). Hepatic differentiation of human iPSCs in different 3D models: A comparative study. Int J Mol Med 40, 1759–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messner S, Agarkova I, Moritz W, and Kelm JM (2013). Multi-cell type human liver microtissues for hepatotoxicity testing. Arch Toxicol 87, 209–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mun SJ, Hong Y-H, Ahn H-S, Ryu J-S, Chung K-S, and Son MJ (2020). Long-Term Expansion of Functional Human Pluripotent Stem Cell-Derived Hepatic Organoids. Int J Stem Cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mun SJ, Ryu J-S, Lee M-O, Son YS, Oh SJ, Cho H-S, Son M-Y, Kim D-S, Kim SJ, Yoo HJ, et al. (2019). Generation of expandable human pluripotent stem cell-derived hepatocyte-like liver organoids. Journal of Hepatology 71, 970–985. [DOI] [PubMed] [Google Scholar]
- Nagata S, Ozawa F, Nie M, and Takeuchi S (2020). 3D culture of functional human iPSC-derived hepatocytes using a core-shell microfiber. PLOS ONE 15, e0234441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng SS, Saeb-Parsy K, Blackford SJI, Segal JM, Serra MP, Horcas-Lopez M, No DY, Mastoridis S, Jassem W, Frank CW, et al. (2018). Human iPS derived progenitors bioengineered into liver organoids using an inverted colloidal crystal poly (ethylene glycol) scaffold. Biomaterials 182, 299–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ober EA, and Lemaigre FP (2018). Development of the liver: Insights into organ and tissue morphogenesis. J Hepatol 68, 1049–1062. [DOI] [PubMed] [Google Scholar]
- Ouchi R, Togo S, Kimura M, Shinozawa T, Koido M, Koike H, Thompson W, Karns RA, Mayhew CN, McGrath PS, et al. (2019). Modeling Steatohepatitis in Humans with Pluripotent Stem Cell-Derived Organoids. Cell Metabolism 30, 374–384.e376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettinato G, Lehoux S, Ramanathan R, Salem MM, He L-X, Muse O, Flaumenhaft R, Thompson MT, Rouse EA, Cummings RD, et al. (2019). Generation of fully functional hepatocyte-like organoids from human induced pluripotent stem cells mixed with Endothelial Cells. Scientific Reports 9, 8920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettinato G, Ramanathan R, Fisher RA, Mangino MJ, Zhang N, and Wen X (2016). Scalable Differentiation of Human iPSCs in a Multicellular Spheroid-based 3D Culture into Hepatocyte-like Cells through Direct Wnt/β-catenin Pathway Inhibition. Scientific Reports 6, 32888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proctor WR, Foster AJ, Vogt J, Summers C, Middleton B, Pilling MA, Shienson D, Kijanska M, Ströbel S, Kelm JM, et al. (2017). Utility of spherical human liver microtissues for prediction of clinical drug-induced liver injury. Archives of Toxicology 91, 2849–2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramasamy TS, Yu JS, Selden C, Hodgson H, and Cui W (2013). Application of three-dimensional culture conditions to human embryonic stem cell-derived definitive endoderm cells enhances hepatocyte differentiation and functionality. Tissue Eng Part A 19, 360–367. [DOI] [PubMed] [Google Scholar]
- Schutgens F, and Clevers H (2020). Human Organoids: Tools for Understanding Biology and Treating Diseases. Annu Rev Pathol 15, 211–234. [DOI] [PubMed] [Google Scholar]
- Schyschka L, Sánchez JJM, Wang Z, Burkhardt B, Müller-Vieira U, Zeilinger K, Bachmann A, Nadalin S, Damm G, and Nussler AK (2013). Hepatic 3D cultures but not 2D cultures preserve specific transporter activity for acetaminophen-induced hepatotoxicity. Archives of Toxicology 87, 1581–1593. [DOI] [PubMed] [Google Scholar]
- Sekiya S, and Suzuki A (2011). Direct conversion of mouse fibroblasts to hepatocyte-like cells by defined factors. Nature 475, 390–393. [DOI] [PubMed] [Google Scholar]
- Sendi H, Mead I, Wan M, Mehrab-Mohseni M, Koch K, Atala A, Bonkovsky HL, and Bishop CE (2018). miR-122 inhibition in a human liver organoid model leads to liver inflammation, necrosis, steatofibrosis and dysregulated insulin signaling. PLOS ONE 13, e0200847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sgodda M, Dai Z, Zweigerdt R, Sharma AD, Ott M, and Cantz T (2017). A Scalable Approach for the Generation of Human Pluripotent Stem Cell-Derived Hepatic Organoids with Sensitive Hepatotoxicity Features. Stem Cells Dev 26, 1490–1504. [DOI] [PubMed] [Google Scholar]
- Sharma A, Sances S, Workman MJ, and Svendsen CN (2020). Multi-lineage Human iPSC-Derived Platforms for Disease Modeling and Drug Discovery. Cell Stem Cell 26, 309–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shetty S, Lalor PF, and Adams DH (2018). Liver sinusoidal endothelial cells - gatekeepers of hepatic immunity. Nat Rev Gastroenterol Hepatol 15, 555–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin D, and Monga SP (2013). Cellular and molecular basis of liver development. Compr Physiol 3, 799–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si-Tayeb K, Noto FK, Nagaoka M, Li J, Battle MA, Duris C, North PE, Dalton S, and Duncan SA (2010). Highly efficient generation of human hepatocyte-like cells from induced pluripotent stem cells. Hepatology 51, 297–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L, Yuan X, Jones Z, Griffin K, Zhou Y, Ma T, and Li Y (2019). Assembly of Human Stem Cell-Derived Cortical Spheroids and Vascular Spheroids to Model 3-D Brain-like Tissues. Sci Rep 9, 5977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorrentino G, Rezakhani S, Yildiz E, Nuciforo S, Heim MH, Lutolf MP, and Schoonjans K (2020). Mechano-modulatory synthetic niches for liver organoid derivation. Nat Commun 11, 3416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayama K, Kawabata K, Nagamoto Y, Kishimoto K, Tashiro K, Sakurai F, Tachibana M, Kanda K, Hayakawa T, Furue MK, et al. (2013). 3D spheroid culture of hESC/hiPSC-derived hepatocyte-like cells for drug toxicity testing. Biomaterials 34, 1781–1789. [DOI] [PubMed] [Google Scholar]
- Takebe T, Sekine K, Enomura M, Koike H, Kimura M, Ogaeri T, Zhang R-R, Ueno Y, Zheng Y-W, Koike N, et al. (2013). Vascularized and functional human liver from an iPSC-derived organ bud transplant. Nature 499, 481–484. [DOI] [PubMed] [Google Scholar]
- Takebe T, Sekine K, Kimura M, Yoshizawa E, Ayano S, Koido M, Funayama S, Nakanishi N, Hisai T, Kobayashi T, et al. (2017a). Massive and Reproducible Production of Liver Buds Entirely from Human Pluripotent Stem Cells. Cell Reports 21, 2661–2670. [DOI] [PubMed] [Google Scholar]
- Takebe T, Zhang BY, and Radisic M (2017b). Synergistic Engineering: Organoids Meet Organs-on-a-Chip. Cell Stem Cell 21, 297–300. [DOI] [PubMed] [Google Scholar]
- Takeishi K, Collin de l’Hortet A, Wang Y, Handa K, Guzman-Lepe J, Matsubara K, Morita K, Jang S, Haep N, Florentino RM, et al. (2020). Assembly and Function of a Bioengineered Human Liver for Transplantation Generated Solely from Induced Pluripotent Stem Cells. Cell Rep 31, 107711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasnim F, Toh YC, Qu Y, Li H, Phan D, Narmada BC, Ananthanarayanan A, Mittal N, Meng RQ, and Yu H (2016). Functionally Enhanced Human Stem Cell Derived Hepatocytes in Galactosylated Cellulosic Sponges for Hepatotoxicity Testing. Mol Pharm 13, 1947–1957. [DOI] [PubMed] [Google Scholar]
- Teo AK, Ali Y, Wong KY, Chipperfield H, Sadasivam A, Poobalan Y, Tan EK, Wang ST, Abraham S, Tsuneyoshi N, et al. (2012). Activin and BMP4 synergistically promote formation of definitive endoderm in human embryonic stem cells. Stem Cells 30, 631–642. [DOI] [PubMed] [Google Scholar]
- Tsuchida T, and Friedman SL (2017). Mechanisms of hepatic stellate cell activation. Nat Rev Gastroenterol Hepatol 14, 397–411. [DOI] [PubMed] [Google Scholar]
- Vorrink SU, Ullah S, Schmidt S, Nandania J, Velagapudi V, Beck O, Ingelman-Sundberg M, and Lauschke VM (2017). Endogenous and xenobiotic metabolic stability of primary human hepatocytes in long-term 3D spheroid cultures revealed by a combination of targeted and untargeted metabolomics. FASEB J 31, 2696–2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vyas D, Baptista PM, Brovold M, Moran E, Gaston B, Booth C, Samuel M, Atala A, and Soker S (2018). Self-assembled liver organoids recapitulate hepatobiliary organogenesis in vitro. Hepatology 67, 750–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner I, Materne EM, Brincker S, Sussbier U, Fradrich C, Busek M, Sonntag F, Sakharov DA, Trushkin EV, Tonevitsky AG, et al. (2013). A dynamic multi-organ-chip for long-term cultivation and substance testing proven by 3D human liver and skin tissue co-culture. Lab Chip 13, 3538–3547. [DOI] [PubMed] [Google Scholar]
- Wang B, Jakus AE, Baptista PM, Soker S, Soto-Gutierrez A, Abecassis MM, Shah RN, and Wertheim JA (2016). Functional Maturation of Induced Pluripotent Stem Cell Hepatocytes in Extracellular Matrix-A Comparative Analysis of Bioartificial Liver Microenvironments. Stem Cells Translational Medicine 5, 1257–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Wang X, Tan Z, Su Y, Liu J, Chang M, Yan F, Chen J, Chen T, Li C, et al. (2019). Human ESC-derived expandable hepatic organoids enable therapeutic liver repopulation and pathophysiological modeling of alcoholic liver injury. Cell Res 29, 1009–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Wang H, Deng P, Chen W, Guo Y, Tao T, and Qin J (2018). In situ differentiation and generation of functional liver organoids from human iPSCs in a 3D perfusable chip system. Lab Chip 18, 3606–3616. [DOI] [PubMed] [Google Scholar]
- Wu F, Wu D, Ren Y, Huang Y, Feng B, Zhao N, Zhang T, Chen X, Chen S, and Xu A (2019). Generation of hepatobiliary organoids from human induced pluripotent stem cells. Journal of Hepatology 70, 1145–1158. [DOI] [PubMed] [Google Scholar]
- Xiang Y, Tanaka Y, Cakir B, Patterson B, Kim KY, Sun P, Kang YJ, Zhong M, Liu X, Patra P, et al. (2019). hESC-Derived Thalamic Organoids Form Reciprocal Projections When Fused with Cortical Organoids. Cell Stem Cell 24, 487–497.e487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y, Tanaka Y, Patterson B, Kang YJ, Govindaiah G, Roselaar N, Cakir B, Kim KY, Lombroso AP, Hwang SM, et al. (2017). Fusion of Regionally Specified hPSC-Derived Organoids Models Human Brain Development and Interneuron Migration. Cell Stem Cell 21, 383–398 e387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie B, Sun D, Du Y, Jia J, Sun S, Xu J, Liu Y, Xiang C, Chen S, Xie H, et al. (2019). A two-step lineage reprogramming strategy to generate functionally competent human hepatocytes from fibroblasts. Cell Res 29, 696–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Han Y, Nilsson-Payant BE, Gupta V, Wang P, Duan X, Tang X, Zhu J, Zhao Z, Jaffre F, et al. (2020). A Human Pluripotent Stem Cell-based Platform to Study SARS-CoV-2 Tropism and Model Virus Infection in Human Cells and Organoids. Cell Stem Cell 27, 125–136 e127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabulica M, Srinivasan RC, Vosough M, Hammarstedt C, Wu T, Gramignoli R, Ellis E, Kannisto K, Collin de l’Hortet A, Takeishi K, et al. (2019). Guide to the Assessment of Mature Liver Gene Expression in Stem Cell-Derived Hepatocytes. Stem Cells Dev 28, 907–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaret KS (2016). From Endoderm to Liver Bud: Paradigms of Cell Type Specification and Tissue Morphogenesis. Curr Top Dev Biol 117, 647–669. [DOI] [PubMed] [Google Scholar]
- Zhang K, Zhang L, Liu W, Ma X, Cen J, Sun Z, Wang C, Feng S, Zhang Z, Yue L, et al. (2018a). In Vitro Expansion of Primary Human Hepatocytes with Efficient Liver Repopulation Capacity. Cell Stem Cell 23, 806–819 e804. [DOI] [PubMed] [Google Scholar]
- Zhang R-R, Koido M, Tadokoro T, Ouchi R, Matsuno T, Ueno Y, Sekine K, Takebe T, and Taniguchi H (2018b). Human iPSC-Derived Posterior Gut Progenitors Are Expandable and Capable of Forming Gut and Liver Organoids. Stem Cell Reports 10, 780–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R-R, Takebe T, Miyazaki L, Takayama M, Koike H, Kimura M, Enomura M, Zheng Y-W, Sekine K, and Taniguchi H (2014). Efficient hepatic differentiation of human induced pluripotent stem cells in a three-dimensional microscale culture. Methods in Molecular Biology (Clifton, NJ) 1210, 131–141. [DOI] [PubMed] [Google Scholar]
- Zorn AM (2008). Liver development. In StemBook (Cambridge (MA)). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1: Representative articles of 3D human hepatic models from the past 10 years. This spreadsheet is an overview of current (after 2010) articles that discuss human liver/hepatic spheroids/organoids/liver on a chip, liver scaffolds models. The cellular source of hepatocytes, the cell types present in the resulting organoid, which scaffold (if any) is used in the model, the step-by-step protocol, quality control steps, and major advantages of each study are listed, along with complete references.