Summary
Anti-sigma factors play a critical role in regulating the expression of sigma factors in response to environmental stress signals. PG1659 is cotranscribed with an upstream gene PG1660 (rpoE), which encodes for a sigma factor that plays an important role in oxidative stress resistance and the virulence regulatory network of P. gingivalis. PG1659, which is annotated as a hypothetical gene, is evaluated in this study. PG1659, composed of 130 amino acids, is predicted to be transmembrane protein with a single calcium (Ca2+) binding site. In P. gingivalis FLL358 (ΔPG1659::ermF), the rpoE gene was highly upregulated compared to the wild-type W83 strain. RpoE induced genes were also upregulated in the PG1659-defective isogenic mutant. Both protein-protein pull-down and bacterial two-hybrid assays revealed that the PG1659 protein could interact with/bind RpoE. The N-terminal domain of PG1659, representing the cytoplasmic fragment of the protein, is critical for interaction with RpoE. In the presence of PG1659, the initiation of transcription by the RpoE sigma factor was inhibited. Taken together, our data suggest that PG1659 is an anti-sigma factor which plays an important regulatory role in the modulation of the sigma factor RpoE in P. gingivalis.
Keywords: Porphyromonas gingivalis, sigma factor, anti-sigma factor, oxidative stress
1. Introduction
An ability to synchronize a cellular response with environmental stimuli is vital for the survival and adaptation of bacteria in diverse environmental conditions. In general, the response and adaptation mechanisms are known be regulated mainly at the level of transcription initiation (Helmann, 2002). This regulation primarily involves alternative sigma factors that interact with the RNA polymerase to facilitate specific promoter recognition resulting in transcription initiation. Extracytoplasmic function (ECF) sigma (σ) factors, the largest group of alternative sigma factors, represent a diverse group that belongs to the sigma factor subfamily 4 of the σ70 class (Helmann, 2002; Staron et al., 2009). In addition to the key role they play in the adaption to environmental conditions, they are important in the pathogenesis of several microorganisms (Bashyam & Hasnain, 2004; Schneider & Glickman, 2013).
The ECF σ factor is autoregulated at the transcriptional level by binding to its own promoter under certain environmental signals including outer membrane stress. A comparative analysis of the ECF σ factors indicate that they carry conserved domains (σR4 and σR2) which are responsible for the interaction with the RNA polymerase core enzyme and specific promoter sites (−35 and −10 respectively) (Lin et al., 2019; Li, Fang, Zhuang, Wang, & Zhang, 2019). The −35 promoter element recognized by ECF sigma factors includes an “AAC” conserved sequence motif in more than half of the observed examples which is in contrast to the more diverged sequences at the −10 element (Lane & Darst, 2006; Gaballa et al., 2018). Currently, there are few mechanisms known to control both the cellular concentration of the ECF sigma factors and their association with the RNAP core enzyme (Paget 2015). In many cases, the ECF sigma factors are regulated by their cognate anti-sigma factors located on the inner membrane of the bacterial cell (Pinto, Liu, & Mascher, 2019). Unlike σ factors, the anti-σ factors are less conserved at the primary sequence level but are usually cotranscribed with the cognate σ factor genes (Paget 2015).
There are several mechanisms facilitate the release of the ECF σ factors in response to the appropriate signals. One of the well-illustrated examples in Escherichia coli is σE, which has been shown to control the extra-cytoplasmic stress response needed to maintain cell integrity and stationary-phase cell survival (Nicoloff, Gopalkrishnan, & Ades, 2017). In the absence of environmental stress, the membrane-spanning anti-σ factor RseA binds σE through its cytoplasmic anti-sigma domain (ASD). Under stress conditions, the C-termini of unassembled outer membrane proteins in the periplasm trigger the activation of the membrane-associated protease DegS through binding to its PDZ domain. The activated DegS cleaves the periplasmic domain of RseA at site-1 (Wilken, Kitzing, Kurzbauer, Ehrmann, & Clausen, 2004; Chaba et al., 2011). Because of dual requirements for σE induction that will enable the cell to integrate multiple signals, RseB, another periplasmic protein is normally bound to RseA to inhibit the DegS-dependent cleavage. This inhibition is relieved when lipopolysaccharide (LPS) that accumulates in the periplasm (in response to dysfunction of outer-membrane biogenesis) binds directly to RseB (Cezairliyan & Sauer, 2007). With RseB dissociation and RseA cleavage by GegS, RseP then cleaves the transmembrane region of RseA to release the σE/RseA complex into the cytoplasm (Hizukuri & Akiyama, 2012; Li et al., 2009). σE is released when RseA is finally degraded by ClpXP, the ATP-dependent protease (Flynn, Levchenko, Sauer, & Baker, 2004). Release of the ECF σ factor can also occur with the direct sensing of the environmental signal by the anti-sigma factor. In Streptomyces coelicolor, σR, in response to oxidative stress, modulates the expression of the thioredoxin system (Paget, Kang, Roe, & Buttner, 1998). The anti-σ factor RsrA binds and inactivates σR via its zinc binding anti-sigma domain (ZASD) domain (Paget et al., 2001). The ZASD domain, which coordinates zinc ion binding, is composed of three cysteine and one histidine (Zdanowski et al., 2006). Under oxidative stress conditions, the formation of an intramolecular disulfide bond triggers the expulsion of the single metal ion, which causes dramatic structural changes in RsrA that result in its dissociation from σR and an ability to activate the transcription of antioxidant genes (Paget, Molle, Cohen, Aharonowitz, & Buttner, 2001; Kalifidas, Thomas, Doughty, & Paget, 2010). In Bacillus subtilis, σB responds to general environmental stresses and is regulated by the anti-σ factor RsbW (Haldenwang 1995). RsbV is an anti-anti-σ factor, which can be phosphorylated by RsbW and leads to the interaction between σB and RsbW. Under stress conditions, alternative phosphatases (RsbTU or RsbQP) dephosphorylate RsbV-P leading to the association of RsbV with RsbW, triggering the release, activation and initiation of antioxidants related genes by σB (Delumeau, Lewis, & Yudkin, 2002; Hardwick et al., 2007).
Porphyromonas gingivalis is a keystone pathogen that is involved in the development of periodontal disease leading to the loss of tissues supporting the teeth (Lamont & Jenkinson, 1998; Darveau 2010). P. gingivalis is also associated with other important chronic inflammatory disorders including rheumatoid arthritis, cardiovascular disease and Alzheimer’s disease (Perricone et al., 2019; Damgaard et al., 2017; Liccardo et al., 2019; Dominy et al., 2019). A genome analysis of P. gingivalis W83 revealed 6 ECF sigma factors including PG0162, PG0214, PG0985, PG1318, PG1660 (RpoE) and PG1827 (Nelson et al., 2003). The roles of these sigma factors in oxidative stress and virulence are not yet fully understood. PG0162- and RpoE-deficient isogenic mutants showed reduced gingipain activity (Dou, Osbourne, McKenzie, & Fletcher, 2010); a PG1318-deficient mutant showed a mutator phenotype (Kikuchi et al., 2009); RpoE- and PG1827-deficient mutants showed increased sensitivity to oxidative stresses (Dou et al., 2010; Yanamandra, Sarrafee, Anaya-Bergman, Jones, & Lewis, 2012). Regulation of the ECF sigma factor in response to oxidative stress in other bacteria has been reviewed (Donohue 2019). However, the activation mechanism of ECF sigma factor RpoE in P. gingivalis is unknown. Our previous study showed that RpoE played a role in the response to hydrogen peroxide-induced oxidative stress (Dou et al., 2018). The PG1659 gene, which encodes for a hypothetical protein, is located downstream and co-transcribed with rpoE. The PG1659 protein is predicted to contain two transmembrane helices. Here, we have further characterized PG1659. We now demonstrate that this protein has anti-sigma factor properties for RpoE in P. gingivalis.
2. Material and Methods
2.1. Bacterial strains, plasmids, and culture conditions
Strains and plasmids used in this study are as listed in Table 1. Briefly, P. gingivalis was cultured in Brain Heart Infusion (BHI) broth supplemented with yeast extract (0.5%), hemin (5 μg/ml), vitamin K (0.5 μg/ml), and cysteine (0.1%) in an anaerobic chamber in 10% H2, 10% CO2, and 80% N2 at 37°C. For RNA isolation, the cells were grown to OD600 of 0.6~0.8 and collected by centrifugation at 10,000 g for 10 min.
Table 1.
Strains and plasmids used in this study
| Strains and plasmids | Relevant characteristics | References |
|---|---|---|
| Strains | ||
| Porphyromonas gingivalis | ||
| W83 | Wild-type | (Dou et al., 2010) |
| FLL357 | ΔrpoE::ermF, polar | This study |
| FLL358 | ΔPG1659::ermF | This study |
| Escherichia coli | ||
| BL21 | Host cell for protein overexpression | Life Technology |
| BTH101 | Host cell for BACTH | EuroMedex |
| Plasmids | ||
| pVA2198 | Emr | (Fletcher et al., 1995) |
| pET102-TOPO | Apr | Life Technology |
| pUT18 | Apr | EuroMedex |
| pUT18C | Apr | EuroMedex |
| pKT25 | Kmr | EuroMedex |
| pKNT25 | Kmr | EuroMedex |
| pUT18C-zip | pUT18C containing GCN4 leucine zipper | EuroMedex |
| pKT25-zip | pKT25 containing GCN4 leucine zipper | Euromedex |
| pUTrpoE | pUT18 containg rpoE | This study |
| pUTrpoEC | pUT18C containing rpoE | This study |
| pUT1659 | pUT18 containing PG1659 | This study |
| pKNT1659 | pKNT25 containing PG1659 | This study |
| pKT1659 | pKT25 containing PG1659 | This study |
| pKTrpoE | pKT25 containing rpoE | This study |
| pKNT1659N48 | pKNT25 containing N-terminal 48 amino acids coding region of PG1659 | This study |
| pKNT1659N20 | pKNT25 containing N-terminal 20 amino acids coding region of PG1659 | This study |
| pKNTC82 | pKNT25 containing C-terminal 82 amino acids coding region of PG1659 | This study |
| pKNTC70 | pKNT25 containing C-terminal 70 amino acids coding region of PG1659 | This study |
| pKNT0162 | pKNT25 containing PG0162 | This study |
2.2. Bioinformatics analyses
The protein structures were predicted and modeled using the online software package I-Tasser (http://zhanglab.ccmb.med.umich.edu/I-TASSER/) (Yang et al., 2015), and PredictProtein (http://www.predictprotein.org).
2.3. Creation of PG1659-deletion mutant and rpoE-deletion polar mutant
Primers used to create the PG1659-deletion mutant are listed in Supplemental Table-1. One kilobase flanking fragments of both upstream and downstream of PG1659 were PCR amplified from chromosomal DNA of P. gingivalis W83. The promoterless ermF cassette was amplified from the plasmid pVA2198 (Fletcher et al., 1995), by using oligonucleotide primers that contained overlapping 24~25 nucleotides for the upstream and downstream fragments. These three fragments were fused by PCR, and then used to transform P. gingivalis W83 by electroporation as previously described (Dou et al., 2010). The cells were incubated on BHI agar plates containing 10 μg/ml of erythromycin at 37°C for 7 days. To delete the rpoE gene with a polar effect on the expression of PG1659 downstream gene, the promoter containing ermF cassette was PCR amplified from the pVA2198 plasmid. The purified PCR fragment was fused to the 1 kb flanking upstream and downstream region of the rpoE gene. The fused fragments were used to transform P. gingivalis W83 by electroporation as previously described (Dou et al., 2010). The isogenic mutants were confirmed by PCR and DNA sequencing.
2.4. Real-time quantitative PCR and reverse transcription PCR
Briefly, RNA isolation and cDNA synthesis were performed as previously described (Dou et al., 2014). For real-time PCR, amplification was performed by using the SYBR Green Mix kit, and real-time fluorescence was detected using the Applied Biosystems Real Time PCR apparatus (Life Technology, Carlsbad, CA). The primers used for the reactions are listed in Supplemental Table-1. The 16S rRNA was used as an internal control to normalize variation due to differences in reverse transcription efficiency. The 2−ΔΔCT method was used to calculate fold change of the genes been tested (Livak & Schmittgen, 2001). The PCR reaction was performed as follows: 50°C, 2 min; 95°C, 2 min; then 95°C, 15 sec; 55°C, 15 sec; 72°C, 30 sec for 40 cycles. Each amplification reaction was performed in triplicate. The reverse transcription PCR (RT-PCR) amplification was performed as follows: 94°C, 5 min; then 94°C, 30 sec; 54°C, 30 sec; 72°C, 30 sec for 25 cycles. The amplification reaction for each PCR was performed in triplicate. The products were analyzed by agarose gel electrophoresis.
2.5. Growth study and survival assay
Growth and hydrogen peroxide sensitivity studies of P. gingivalis strains were tested as previously described (Dou et al., 2018). For the growth study, overnight cultures of P. gingivalis were used to inoculate pre-warmed BHI broth and then the optical density of the cultures was monitored spectrophotometrically (OD600) at regular intervals. For the survival assay, hydrogen peroxide was added to the testing cultures at OD600≈0.8 to the final concentration of 0.25 mM. After exposure for 1 hr or 2 hrs, the cultures were serially diluted and plated on BHI agar. Bacterial cultures in the absence of hydrogen peroxide were used as control. Colony forming units were enumerated after incubation for 7 days anaerobically at 37°C. At least three independent experiments performed in triplicate in this study.
2.6. Protein expression and purification
Recombinant proteins, rRpoE and rPG0162, were expressed and purified from E. coli as previously described (Dou et al., 2018; Dou et al., 2016). For the overexpression of the PG1659 protein, a 3×FLAG tag was inserted before the stop codon of the PG1659 gene carried on the pET102-TOPO plasmid. The rPG1659 protein was purified using resin containing the anti-FLAG antibody according to manufacturers’ protocols (Sigma-Aldrich, St. Louis, MO). The primers used are listed in Supplemental Table-1. The rRpoE and rPG0162 proteins were purified by using the Ni-NTA agarose resin (Qiagen, Valencia, CA).
2.7. Protein-protein pull-down assay
The pull-down assay was performed as previously reported elsewhere (Hasan et al., 2004; Gerace & Moazed, 2015). The purified His-RpoE or His-PG0162 protein was first immobilized on Ni-NTA agarose beads and washed with wash buffer (50 mM NaH2PO4, 300 mM NaCl, 20 mM imidazole, pH8.0). The immobilized His-RpoE or His-PG0162 protein was then incubated with the purified FLAG-PG1659 protein for 1 hr at 4°C (in PBS buffer). Finally, the immobilized protein samples were washed twice with wash buffer and then collected in the elution buffer (50 mM NaH2PO4, 300 mM NaCl, 250 mM imidazole, pH8.0). Similarly, the purified His-RpoE or His-PG0162 was incubated with immobilized FLAG-PG1659 on anti-FLAG agarose beads for 1 hr at 4°C (in PBS buffer). After washing, the bound proteins were collected in elution buffer (0.1 M glycine HCl, pH 3.5), then analyzed using SDS-PAGE.
2.8. Western-blot
The protein samples were resolved by SDS-PAGE, and then transferred to a Bio-Trace nitrocellulose membranes at 15 V for 30 min using a Semi-Dry Trans-blot apparatus (Bio-Rad, Hercules, CA). The membrane containing protein samples were first probed with primary antibody against the His-tag or FLAG-tag (mouse), and then probed with the horseradish peroxidase conjugated secondary goat anti-mouse antibody (Zymed Laboratories, San Francisco, CA). Immunoreactive proteins were detected using the Western Lightning Chemiluminescence Reagent Plus Kit (Perkin-Elmer Life Sciences, Boston, MA).
2.9. In vitro transcription assay
The in vitro transcription assay was performed using the E. coli RNA polymerase core enzyme (New England Biolabs, Beverly, MA), and the purified rRpoE and/or rPG1659 proteins as previously reported (Dou et al., 2018). The DNA fragment carrying the rpoE promoter region was used as the template. The primers used are as listed in Supplemental Table-1. The reactions were incubated at 37°C for 2 hrs in transcription buffer, with 2.5 mM of NTP mix, RNA polymerase core enzyme, rRpoE or rPG1659, DNA template and an RNase inhibitor (Roche, Indianapolis, IN). The samples were analyzed on a 1.4% agarose gel.
2.10. Bacterial adenylate cyclase two-hybrid (BACTH) assay
The in vivo interaction between PG1659 and RpoE was detected by using Bacterial Adenylate Cyclase Two-Hybrid System (EuroMedex, Souffelweyersheim, France) according to the manufacturer’s protocols. In brief, PG1659 and rpoE were inserted into the multiple cloning sites of the vectors from the kit respectively (Table 1), and the derived plasmids were then co-transformed into E. coli BTH101 strain. The transformants were incubated on LB plates containing 50 μg/ml of ampicillin, 50 μg/ml of kanamycin, 40 μg/ml of X-gal and 0.5 mM of IPTG at 37°C for 24 hrs or 30°C for 48 hrs. Colonies showing a blue color on LB plate were further incubated on M63 minimal medium plates containing 1% maltose, 50 μg/ml of ampicillin, 50 μg/ml of kanamycin, 40 μg/ml of X-gal, and 0.5 mM of IPTG at 30°C for 48 hrs. The β-galactosidase activity assay was performed according to the manufacturer’s protocols (EuroMedex, Souffelweyersheim, France).
2.11. Gingipain activity assays
The presence of Arg-X- and Lys-X-specific cysteine protease activity (Rgp and Kgp, respectively) was determined as previously reported (Dou et al., 2015). In brief, activities of Arg-X and Lys-X gingipains were measured using 1 mM BAPNA (Nα-benzoyl-DL-arginine-p-nitroanilide) and 1 mM ALNA (Ac-Lys-p-nitroanilide HCl) respectively in an activated protease buffer (0.2 M Tris-HCl, 0.1 M NaCl, 5 mM CaCl2, 10 mM L-cysteine, pH 7.6). The O.D. at 405 nm were then measured against a BHI broth sample containing no bacteria.
3. Results
3.1. In silico analysis of PG1659 reveals peptide binding domain and transmembrane domain
PG1659 is 393 bp in length and is predicted to encode for a 130 amino acid protein (www.ncbi.nlm.nih.gov/genome). Based on the sequence of this protein annotated as hypothetical, modeling using web servers for I-TASSER and PredictProtein predicted a structure that carries two helix-coil-helix structures (positions I5-A60 and F78-R121, respectively), and a β-strand (V61-L67), though the final 3D-model generated in I-TASSER presents only a set of four α-helices (1. K2-R15; 2. A22-E42; 3. R44-V61; and 4. I104-T128). Helix 2 (A22-E42) and helix 3 (R44-V61) overlap with the first predicted transmembrane helix, while helix 4 (I104-T128) overlaps with the second transmembrane helix (Figure 1). A putative motif that binds calcium (Ca2+) is identified at positions D77 and D79 (Figure 1). The N-terminal portion of PG1659 (helix 1) is predicted to be essential for binding to sigma factor RpoE. The amino acid sequence blast of PG1659 did not show any identity to other known anti-sigma factors. However, subdomains that carry helix-1 (K2-R15) and the cytoplasmic part of helix-2 showed 36.1% similarity to the N-terminal subdomain of the anti-sigma factor CnrY from Cupriavidus metallidurans CH34 (Supplemental Figure 1A) (Maillard 2014).
Figure 1.
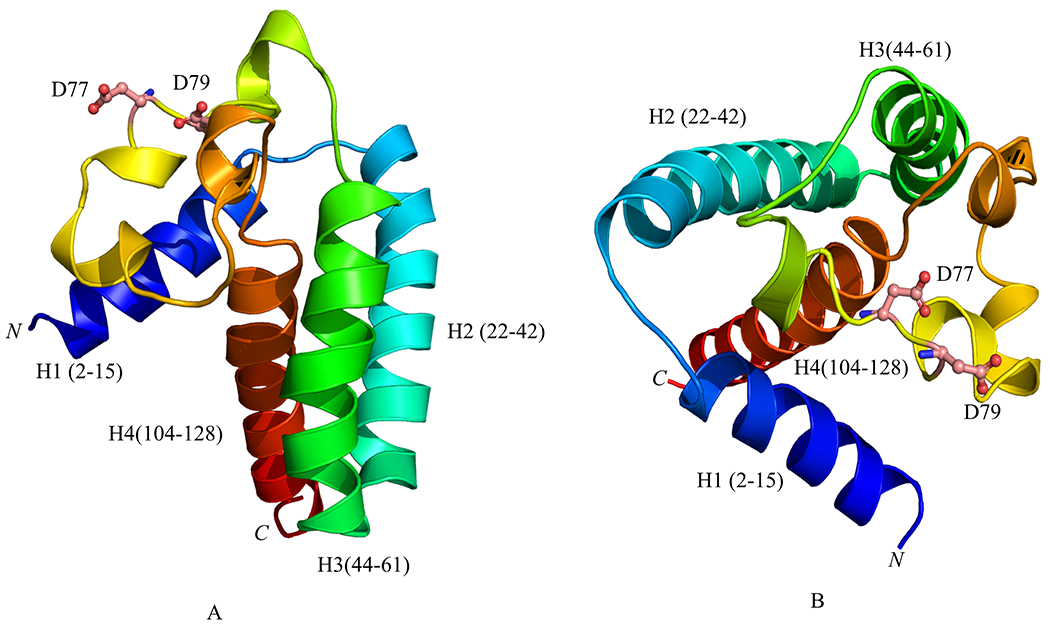
A, I-Tasser model of PG1659 is made up of three transmembrane helices H2 (cyan), H3 (green), and H4 (red). The putative calcium binding site D77 and D79 is shown in ball and stick model (pink). B, switched angle of PG1659 modelling. The C-score=-4.06, TM-score=0.28±0.09 for this model.
3.2. Inactivation of the PG1659 gene can modulate the expression of the upstream gene in the rpoE-PG1659 operon
PG1659 has a 4-bp overlap with the upstream gene rpoE and is part of the same transcriptional unit as shown in Figure 2A (Dou et al., 2018). To further evaluate the polar effects of the genes is this operon, deletion mutants were constructed in the PG1659 and rpoE genes using the ermF cassette as described in the Materials and Methods. After electroporation of P. gingivalis with the purified PCR fusion fragment (see Materials and Methods), several black pigmented erythromycin-resistant colonies were detected after 7 days of incubation. The replacement of the rpoE or PG1659 gene with the ermF cassette in these isogenic mutants was confirmed by colony PCR and DNA sequencing (data not shown). One randomly selected isogenic mutant, designated P. ginivalis FLL357 (ΔrpoE::ermF) or FLL358 (ΔPG1659::ermF) was selected for further studies. The isogenic mutants FLL357 and FLL358 showed similar growth rates compared to the wild-type W83 strain (Figure 2B).
Figure 2.
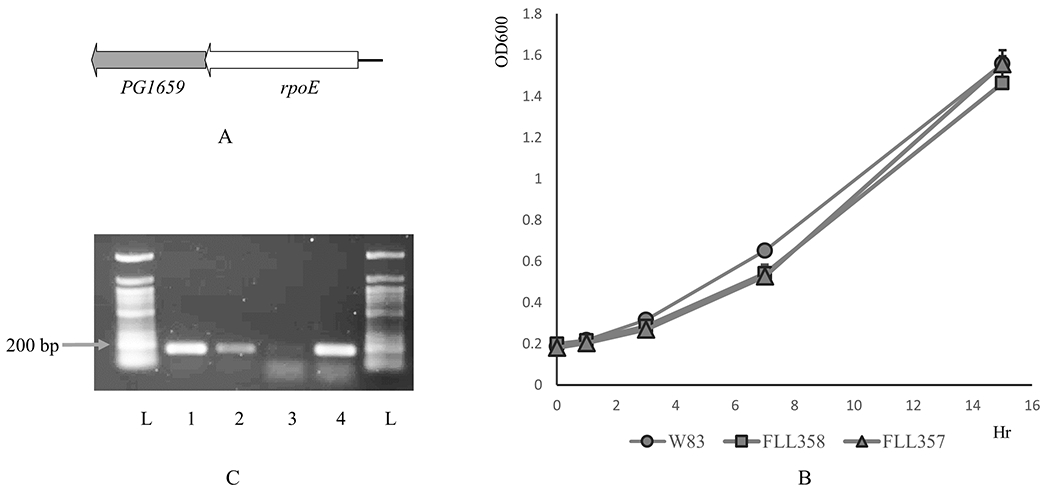
A, Schematic representation of PG1659-rpoE. B, FLL377 (rpoE polar mutant) and FLL358 (PG1659 mutant) show similar growth rate as the wild-type W83. C, RT-PCR results show that rpoE is upregulated in FLL358. Lane 1, W83 cDNA with 16S rRNA primers; Lane 2, FLL358 cDNA with 16S rRNA primers; Lane 3, W83 cDNA with rpoE primers; Lane 4, FLL358 cDNA with rpoE primers; L, 50 bp DNA ladder. The data shows average of three repeats.
To clarify the effect of the PG1659 gene on the expression of the transcriptional unit, quantitative real time-PCR was used to determine the expression of the rpoE gene in the PG1659-deficient mutant using primers as listed in Supplemental Table-1. The results show that rpoE was upregulated 94.2 fold in the P. gingivalis FLL358 (ΔPG1659::ermF) compared to the W83 wild-type strain (Table 2). This is consistent with a RT-PCR analysis (Figure 2C) which showed a more intense band for RpoE in P. gingivalis FLL358 compared to the wild-type W83 strain under normal non-inducing growth conditions. In the FLL357 (ΔrpoE::ermF) isogenic mutant that carried the promoter containing ermF cassette in the reverse orientation to the PG1659 downstream gene, no expression of the PG1659 gene was detected (Supplemental Figure 1B).
Table 2.
Genes dysregulated by RpoE were upregulated in FLL358 (PG1659 mutant)
| Gene | Fold change (FLL358/W83) | Standard deviation |
|---|---|---|
| rpoE | 94.20 | 6.98 |
| PG0844 | 134.24 | 45.51 |
| PG1459 | 151.32 | 8.21 |
| PG1511 | 199.53 | 6.30 |
3.3. PG1659 can dysregulate gene expression
RpoE is autoregulated and is directly associated with the initiation of expression of several genes in response to hydrogen peroxide-induced stress conditions (Dou et al., 2018). In the absence of their cognate anti-σ factors, autoregulated sigma factors can become constitutively active, resulting in increased abundance of the sigma factor and upregulation of other specific genes. If RpoE is modulated by PG1659, the expression of genes in response to hydrogen peroxide-induced stress conditions should be upregulated in the FLL358 (ΔPG1659::ermF) under normal anaerobic conditions. Three RpoE induced genes encoding small hypothetical proteins including PG0844, PG1459 and PG1511 were also upregulated in the PG1659-deficient mutant (Table 2).
3.4. The interaction of PG1659 and RpoE
The recombinant proteins rPG1659 containing a FLAG-tag and rRpoE carrying a His-tag were overexpressed and purified from E. coli. Purified His-RpoE was incubated with the immobilized FLAG-PG1659 protein on anti-FLAG agarose beads. The purified FLAG-PG1659 protein was incubated with the immobilized His-RpoE on Ni-NTA agarose beads. As controls, purified His-RpoE and FLAG-PG1659 were incubated with the anti-FLAG and Ni-NTA agarose beads respectively. The proteins eluted from the agarose beads and analyzed by SDS-PAGE showed an interaction between the RpoE and PG1659 proteins (Figure 3A). The purified His-RpoE protein did not bind to the anti-FLAG agarose beads nor the FLAG-PG1659 proteins to the Ni-NTA agarose beads. Immunoreactive bands in Western-blot analysis using either the His-tag (Figure 3B), or FLAG-tag (Figure 3C) antibodies, showed interaction between the RpoE and PG1659 proteins.
Figure 3.
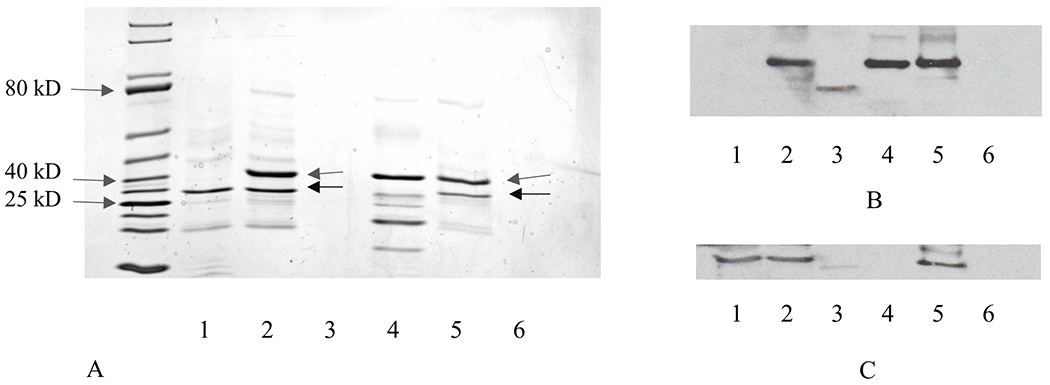
Interaction between RpoE and PG1659 by in vitro protein-protein pull-down assay. A, SDS-PAGE gel analyses of protein-protein pull-down products, and the arrows indicate the rRpoE and rPG1659 respectively. B, Western-blot by using anti-His tag antibody. C, Western-blot by using anti-FLAG antibody. Lanes order as follows: Lane 1, rPG1659 purification; Lane 2, rPG1659 pull down rRpoE; Lane 3, rRpoE elution from anti-FLAG resin; Lane 4, rRpoE purification; Lane 5, rRpoE pull down rPG1659; Lane 6, rPG1659 elution from Ni-NTA resin.
The bacterial adenylate cyclase two-hybrid system (BACTH) was used to further confirm the in vivo interaction between RpoE and PG1659. The rpoE was first cloned into the pUT18 then this recombinant plasmid was transformed into E. coli BTH101 with the pKNT25-derived plasmid carrying the PG1659 gene. The transformant colonies showed a blue color on LB plates containing antibiotics, X-gal and IPTG. Colonies grown on LB plates also grew and showed a similar color on M63 minimal medium containing antibiotics, maltose, X-gal and IPTG. The interaction between RpoE and PG1659 proteins was also demonstrated with E. coli BTH101 transformants carrying pUT1659 and pKTrpoE or pUTrpoEC and pKNT1659. These bacterial colonies appeared in blue color on both LB and M63 plates containing antibiotics, X-gal and IPTG (Figure 4A). The highest β-galactosidase activity was observed in bacterial cells that contained the pUTrpoE and pKNT1659, or the pUT1659 and pKTrpoE constructs (Figure 4B). It is noteworthy that the interaction between RpoE and PG1659 is influenced by their N- or C- terminal fusion to either pUT18 or pKT25.
Figure 4.
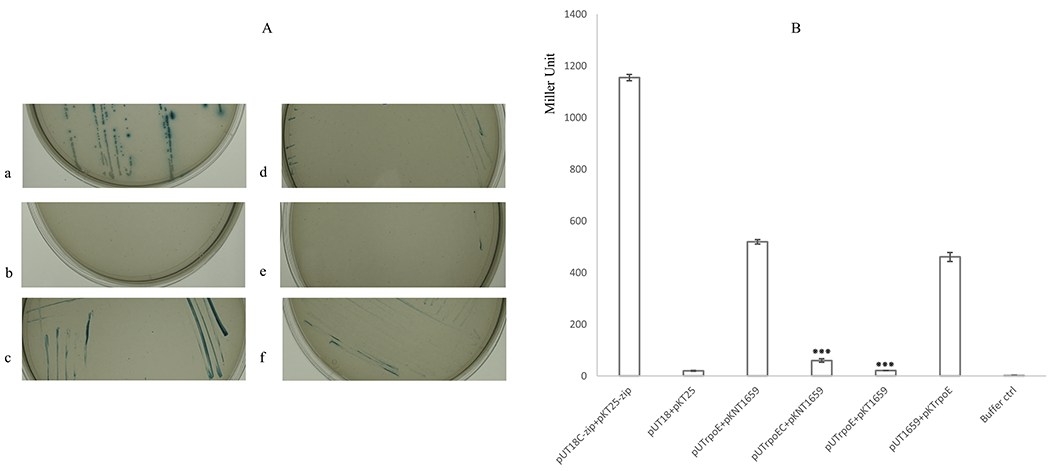
BACTH assay results show interaction of RpoE and PG1659. A, E. coli BTH101 containing PG1659 and rpoE inserted recombinant plasmids can grow on M63 minimal medium. a, BTH101 containing pUT18C-zip + pKT25-zip; b, BTH101 containing pUT18 + pKT25; c, BTH101 containing pUT1660 + pKNT25; d, BTH101 containing pUTrpoEC + pKNT1659; e, BTH101 containing pUTrpoE + pKT1659; f, BTH101 containing pUT1659 + pKTrpoE. B, β-galactosidase assay results show the interaction between RpoE and PG1659. The data shows average of three repeats and “***” indicates the P value of T-test <0.01.
3.5. The N-terminus of PG1659 is critical for the interaction with RpoE
The transformant cells carrying the pUTrpoE and pKNT1659 plasmids were used to evaluate the interaction domain of PG1659 with RpoE. Because the N-terminal 48 amino acids of PG1659 is homologous to the N-terminus of CnrY, which is essential to the interaction of CnrY and sigma factor CnrH in C. metallidurans, the M1~L48 amino acid residues of PG1659 were evaluated in this study. To determine if the N-terminal region of PG1659 was capable of interacting with RpoE, DNA fragments encoding the 48 or 20 amino acids at the N-terminus were cloned into pKNT25, and the recombinant plasmids along with pUTrpoE were used to cotransform BTH101. As shown in Figure 5A and Supplemental Figure 2, blue colonies on M63 medium plates with antibiotics, X-gal, and IPTG were only observed for those transformants that carried the 48 amino acid N-terminal fragment. β-galactosidase activity of the transformant containing pUTrpoE and pKNT1659N48 was similar to the cell that carried the full length PG1659 protein. The cells carrying the pUTrpoE and pKNT1659N20 plasmids showed little or no β-galactosidase activity (Figure 5B). To determine whether the C-terminus of the protein can interact with RpoE, DNA fragments spanning 82 and 70 amino acids from the C-terminal end of PG1659 were cloned onto the pKNT25 vector, then was cotransformed with pUTrpoE into E. coli BTH101. No blue colonies on M63 plates with antibiotics, X-gal, and IPTG were observed for those transformants (Supplemental Figure 2). These results indicate that the N-terminal 48 amino acids of PG1659 are involved in the interaction with RpoE. However, the site-specific mutagenesis of amino acid Q26 or R41, which was predicated to be involved in the protein-protein binding, did not show any influence on the interaction of PG1659 and RpoE (Supplemental Figure 3).
Figure 5.
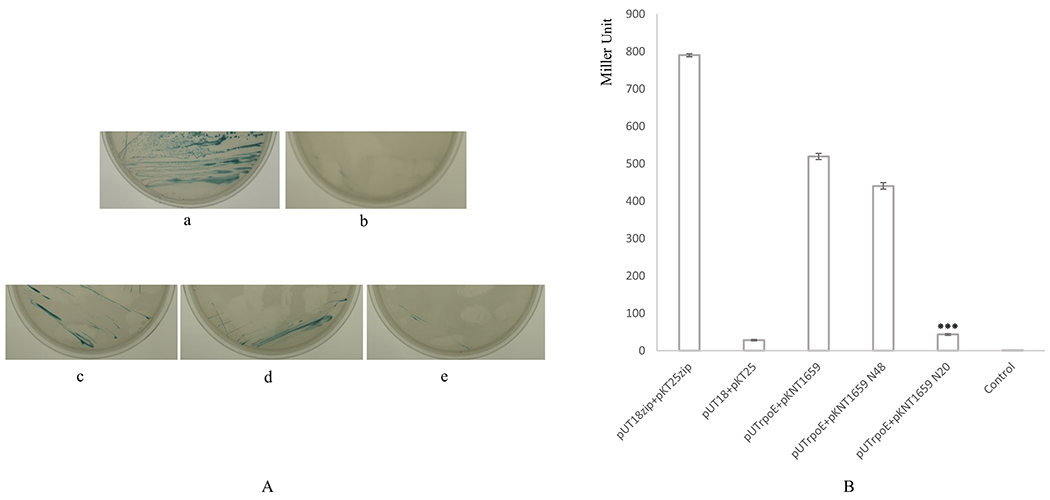
BACTH assay results show N-terminus of PG1659 is essential for the interaction of RpoE and PG1659. A, E. coli BTH101 containing PG1659 and rpoE inserted recombinant plasmids can grow on M63 minimal medium. a, BTH101 containing pUT18-zip + pKT25-zip; b, BTH101 containing pUT18 + pKT25; c, BTH101 containing pUTrpoE + pKNT1659; d, BTH101 containing pUTrpoE + pKNT1659 N48; e, BTH101 containing pUTrpoE + pKNT1659 N20. B, β-galactosidase assay shows that N-terminal 48 amino acids of PG1659 is essential for the interaction between RpoE and PG1659. The data shows average of three repeats and “***” indicates the P value of T-test <0.01.
Another ECF sigma factor PG0162, which is involved in virulence regulation and CTD protein regulation (Kadowaki et al., 2016), shares similarity with RpoE (Dou et al., 2016). Using the BACTH assay and protein-protein pull-down assay, no interaction between PG0162 and PG1659 was detected (Supplemental Figure 4).
3.6. PG1659 can inhibit the ability of RpoE to initiate transcription
The P. gingivalis RpoE sigma factor can initiate transcription from its own promoter (Dou et al., 2018). To test if PG1659 can inhibit the rRpoE initiated transcription, a 636-bp (-347 to +289) DNA fragment predicted to carry the promoter region of the rpoE gene was used as a template in an in vitro run-off transcription reconstitution assays. The rRpoE and rPG1659 proteins, overexpressed and purified from E. coli, were first confirmed by Western-blot using anti-His and anti-FLAG antibodies, respectively (Figure 3), and then used in an in vitro transcription assay. As shown in Figure 6, when the promoter region of rpoE was used as a template, a fragment of approximately 300-nt in length was observed in the presence of the E. coli core RNAP and the rRpoE protein (lane 2, indicated with arrow). There was no transcription product detected with the addition of PG1659 to the reaction (lanes 5&6, Figure 6). In the control experiments, when the rRpoE protein was absent, in the presence of PG1659 or with the presence of RNaseA, no transcription product was detected (lanes 1&3, Figure 6). These results further confirm that RpoE can function as sigma factor, in contrast to PG1659. In conclusion, the interaction of RpoE and PG1659 negatively regulates the function of RpoE and prevent the initiation of the transcription.
Figure 6.
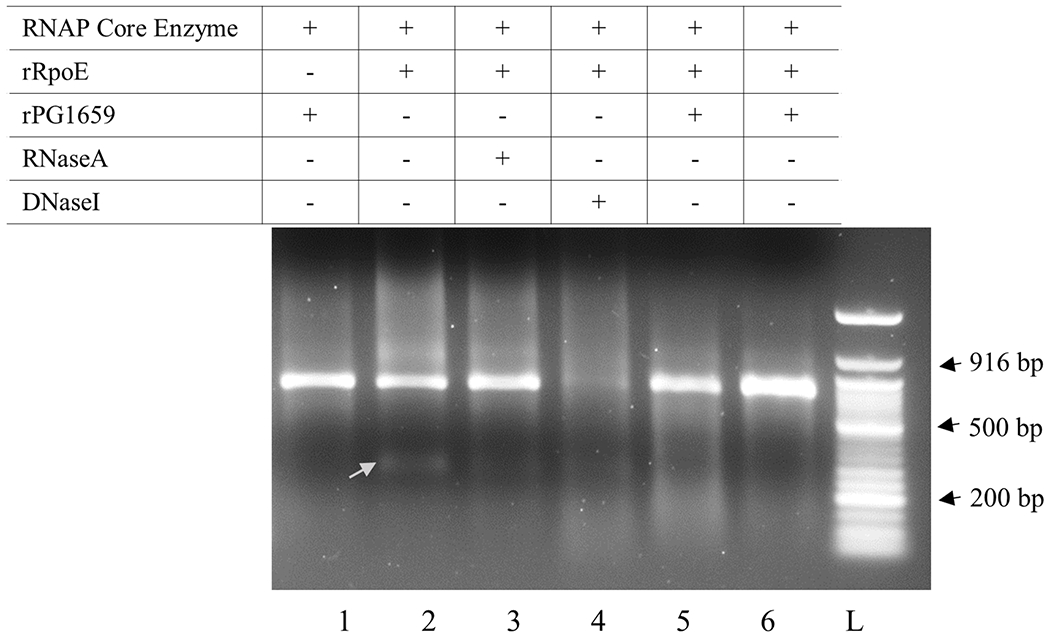
In vitro transcription assay results show that rPG1659 inhibits transcription activity of rRpoE. Lanes, 1, reaction containing only rPG1659; Lane 2, reaction containing only rRpoE; Lane 3, reaction containing only rRpoE and followed by RNase A treatment; Lane 4, reaction containing only rRpoE and followed by DNaseI treatment; Lanes 5&6, reaction containing both rPG1659 and rRpoE. The arrow shows the transcription product.
3.7. Sensitivity of P. gingivalis to H2O2
The rpoE-defective mutant was previously shown to have increased sensitivity to hydrogen peroxide compared to the parent strain (Dou et al., 2010). Moreover, it was demonstrated that several genes associated with oxidative stress resistance in P. gingivalis are modulated by RpoE (Dou et al., 2018). If these genes are upregulated in P. gingivalis FLL358, an increased resistance to hydrogen peroxide-induced stress should be observed. As shown in Figure 7A, FLL358 has a higher survival rate in the presence of 0.25 mM of hydrogen peroxide compared to the parental W83 strain. These results indicate that FLL358 is more resistant to oxidative stress as compared to the wild-type parental strain.
Figure 7.
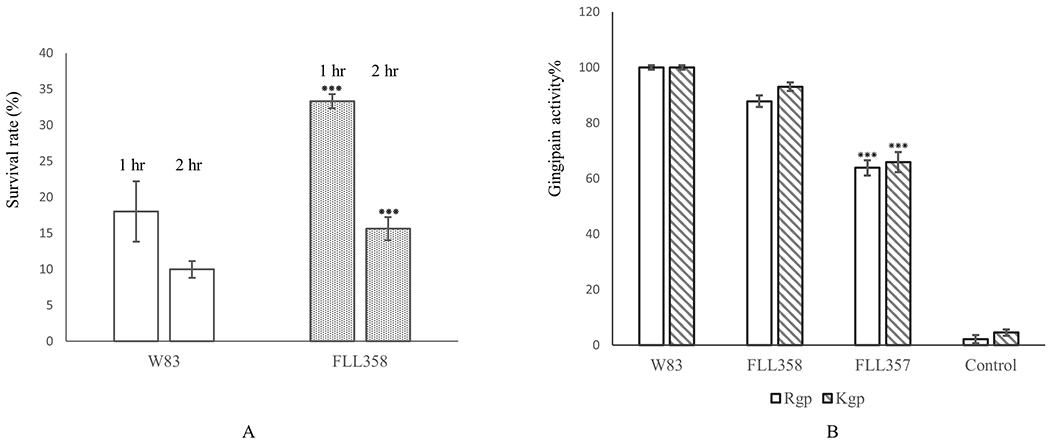
A, FLL358 showed a higher survival rate as compared to the wild-type W83 strain under 0.25 mM of hydrogen peroxide stress. B, PG1659 mutant has no effect on gingipain activity of P. gingivalis. FLL357, RpoE polar mutant; FLL358, PG1659 mutant. The data present shows average of three repeats and “***” indicates the P value of T-test <0.05.
3.8. FLL357 shows gingipain activity reduction
We previously reported that both PG0162-deficient mutant and rpoE-deficient mutant showed gingipain activity reduction (Dou et al., 2010). To further characterize the mutants FLL357 and FLL358, gingipain activity of these two mutants were assayed. The results show that the rpoE polar mutant FLL357 has more than 30% reduction in both Rgp and Kgp activities (Figure 7B), which is similar to the rpoE-deficient mutant FLL354 (Dou et al., 2010). However, the PG1659-deficient mutant does not show any reduction of either Rgp or Kgp activity, which indicates that deficiency of PG1659 in P. gingivalis has no influence on the expression of gingipains.
4. Discussion
Because of the central role that transcription initiation plays in the adaptation and survival of bacteria, regulation of this process has been a major focus in understanding pathogenesis. Bacterial ECF sigma factors are known to function as regulators that will facilitate a response to extracellular signals via a modular design that primarily functions in transmembrane signal transduction (Brooks & Buchanan, 2008). The impact of DNA sequencing with an increased knowledge of more microbial genomes have highlighted the abundance and diversity of ECF sigma factors in bacteria. Regulation of ECF sigma factors, which is well illustrated in bacteria such as E. coli, Pseudomonas aeruginosa, and B. subtilis, typically occurs by an anti-sigma factor that will sequester their cognate ECF sigma factors and prevent interaction with the RNA polymerase core enzyme (Trevino-Quintanilla, Freyre-Gonzale, & Martinez-Flores, 2013; Potvin, Sanschagrin, & Levesque, 2008; Asai 2018). At the sequence level, the poor conservation of the anti-sigma factor proteins compared to the sigma factor family makes this group of proteins diverse. In the present study, we have provided evidence for the anti-sigma factor PG1659 and its role in the modulation the ECF sigma factor RpoE.
In addition to the ability of the RpoE sigma factor to be modulated by hydrogen peroxide-induced stress, and the recognition of a unique promoter in P. gingivalis, there is also emerging evidence that this factor could be involved in the regulation of a yet-to-be defined oxidative stress resistance pathway (Dou et al., 2018). In most cases, the ECF sigma factor is regulated by its cognate anti-sigma factor protein, which is encoded by a downstream gene on the same transcriptional unit. A 4-bp overlap in the coding sequence of rpoE and PG1659 suggests strong transcriptional coupling and they have been demonstrated to be part of the same transcriptional unit (Dou et al., 2018). PG1659 has no homology to any previously characterized protein but has now shown in our in silico analyses to have secondary structure predictions consistent with anti-sigma factor protein function. An isogenic mutant strain of P. gingivalis defective in the PG1659 gene showed a significant upregulation of the upstream rpoE gene under normal non-inducing growth conditions and more resistance to the hydrogen peroxide as compared to the parental strain. Because overexpression of ECF sigma factors usually results in the expression of the sigma factor-dependent genes in the absence of the inducing signal (Beare, For, Martin, & Lamont, 2003; Llamas et al., 2008), our study of several of the RpoE induced genes, has also indicated their upregulation in the PG1659-deficient mutant. Further confirmation of an anti-sigma factor function for PG1659 is its ability to modulate RpoE via transcription initiation. In the presence of PG1659, the activity of RpoE to initiate transcription from its own promoter was inhibited (Figure 6).
Our data have demonstrated the interaction of PG1659 with RpoE. Using the FLAG-tag or His-tag approach co-purification of PG1659 only occurred in the presence of RpoE. This interaction was confirmed using the bacterial adenylate cyclase two-hybrid system (BACTH) which has been widely used to study protein interaction in bacteria. The results not only showed positive interaction between these two proteins but also indicated that protein polarity can impact the interaction. The N-terminal fusion of these two proteins to the T25 or T18 subunit of adenylate cyclase showed the highest β-galactosidase activity as compared to the C-terminal fusion. Consistent with the observation that the N-terminal cytoplasmic region of ECF anti-σ factors can be sufficient for anti-σ factor activity (McGuffle, Vallet-Gely, & Dove, 2016), the N-terminal 48 residues of PG1659 are essential for the interaction between PG1659 and RpoE. The N-terminal region of a similar size was also shown to be vital for the function of the anti-sigma factor CnrY from C. metallidurans (Maillard et al., 2014). In addition to the N-terminal 20 residues of PG1659 that were insufficient for the interaction between PG1659 and RpoE, site-specific mutagenesis of other amino acids (Q26, R41) that were predicated to be involved in protein-protein interaction of the N-terminal 48 residues indicated they were not significant in that process (Supplemental Figure 3). It is unclear what effect the mutations have on protein structure and the minimal structural conservation that is required for interaction. This is under further study in the laboratory.
The amino acids sequence blast from NCBI does not show any known homology to other anti-sigma factors, and the bioinformatics analyses does not show any conserved anti-sigma factor domain neither. The first group of anti-sigma factor contains an anti-sigma domain, which features a conserved “H-X3-C-X2-C” zinc binding motif (Chabert, Lebrun, Lebrun, Latour, & Seneque, 2019). However, PG1659 does not exhibit any putative zinc-binding motif and there is only one cysteine residue located in the transmembrane domain of PG1659. The secondary and tertiary structure of the N-terminal region indicates that PG1659 might belong to class II anti-sigma factor family which containing two helix at the N-terminal (Maillard et al., 2014; Casas-Pastor, Diehl, & Fritz, 2020). The binding of PG1659 to RpoE inhibits the activity of RpoE to start transcription from its own promoter suggesting that PG1659 functions as an anti-sigma factor. The lack of interaction between ECF sigma factor PG0162 which shares similarity with RpoE and PG1659 may indicate that either another anti-sigma factor could be involved in regulation of PG0162, or the putative fused regulatory subdomain of PG0162 functions as an anti-sigma factor as reported (Sineva et al., 2017). This is under further investigation in the laboratory.
In addition to its role in oxidative stress resistance, the ECF sigma factors can also modulate the virulence potential of P. gingivalis (Dou et al., 2010). In contrast to RopE, inactivation of its anti-sigma factor PG1659 had no effect gingipain activity in P. gingivalis. Gingipain activity via its role in heme accumulation on the cell surface can be part of a network of mechanisms associated with oxidative stress resistance in P. gingivalis (Smalley, Birss, Szmigielski, & Potempa, 2006; Henry, McKenzie, Robles, & Fletcher, 2012; Phillips et al., 2018). The oxidative response that may involve binding of oxygen and its toxic derivatives to iron accumulated on the surface of the cell can trigger their catalytic destruction (Lewis 2010). The regulation of gingipain activity can occur at multiple levels (including transcriptional and posttranslational), with an accumulated impact on oxidative stress resistance. Because RopE can likely regulate gingipain activity at the posttranscriptional level via sialidase activity and/or secretion using the type IX secretion system (PorSS), its upregulation in the PG1659-defective mutant would not be expected, as observed in our study, to reduce those activities.
We have demonstrated the functional properties of anti-sigma factor PG1659 in P. gingivalis. It is likely that PG1659 may be involved in a complex regulatory network that may represent a yet-to-be described oxidative stress resistance pathway in P. gingivalis. The protein folding and post-translational modification including acetylation and phosphorylation in E. coli would be different from P. gingivalis. In this case, an efficient expression system for P .gingivalis proteins is necessary to characterize the PG1659 protein. The regulatory mechanism of PG1659 under certain stress conditions, and/or whether any anti-anti-sigma factor is involved, and the detailed information including whether phosphorylation is involved in the interaction of PG1659 and RpoE needs further study.
Supplementary Material
Acknowledgement
This study is supported by Public Health Services Grants R-56-DE13664, DE019730, DE022508, DE022724 from NIDCR (to H.M.F). Thanks to Dr. Charles Wang of Genomic Center of Loma Linda University for generous help and support.
Footnotes
Conflict of interests
The authors declare no conflicts of interest.
Reference list
- Asai K (2008). Anti-sigma factor-mediated cell surface stress response in Bacillus subtilis. Genes & Genetic Systems, 92, 223–234. 10.1266/ggs.17-00046. [DOI] [PubMed] [Google Scholar]
- Bashyam MD, & Hasnain SE (2004). The extracytoplasmic function sigma factors: role in bacterial pathogenesis. Infection, Genetics and Evolution, 4, 301–308. 10.1016/j.meegid.2004.04.003. [DOI] [PubMed] [Google Scholar]
- Beare PA, For RJ, Martin LW, & Lamont IL (2003). Siderophore-mediated cell signalling in Pseudomonas aeruginosa: divergent pathways regulate virulence factor production and siderophore receptor synthesis. Molecular Microbiology, 47, 195–207. 10.1046/j.1365-2958.2003.03288.x. [DOI] [PubMed] [Google Scholar]
- Brooks BE, & Buchanan SK (2008). Signaling mechanisms for activation of extracytoplasmic function (ECF) sigma factors. Biochimica et Biophysica Acta, 1778,1930–1945. 10.1016/j.bbamem.2007.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casas-Pastor D, Diehl A, & Fritz G (2020). Coevolutionary analysis reveals a conserved dual binding interface between extracytoplasmic function σ factors and class I anti-σ factors. mSystems, 5, e00310–20. 10.1128/mSystems.00310-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cezairliyan BO, & Sauer RT (2007). Inhibition of regulated proteolysis by RseB. Proceedings of the National Academy of Sciences of the United States of America, 104, 3771–3776. 10.1073/pnas.0611567104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaba R, Alba BM, Guo MS, Sohn J, Ahuja N, Sauer RT, & Gross CA (2011). Signal integration by DegS and RseB governs the σE-mediated encelope stress response in Escherichia coli. Proceedings of the National Academy of Sciences of the United States of America, 108, 2106–2111. 10.1073/pnas.1019277108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabert V, Lebrun V, Leburn C, Latour JM, & Seneque O (2019). Model peptide for anti-sigma factor domain HHCC zinc fingers: high reactivity toward 1O2 leads to domain unfolding. Chemical Science, 10, 3608–3615. 10.1039/C9SC00341J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damgaard C, Reinholdt J, Enevold C, Fiehn NE, Nielsen CH, & Holmstrup P (2017). Immunoglobulin G antibodies against Porphyromonas gingivalis and Aggregatibacter actinomycetemcomitans in cardiovascular disease and periodontitis. Journal of Oral Microbiology, 9, 1374154. 10.1080/20002297.2017.1374154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darveau RP (2010). Periodontitis: a polymicrobial disruption of host homeostasis. Nature reviews. Microbiology, 8, 481–490. 10.1038/nrmicro2337. [DOI] [PubMed] [Google Scholar]
- Delumeau O, Lewis RJ, & Yudkin MD (2002). Protein-protein interactions that regulate the energy stress activation of σB in Bacillus subtilis. Journal of Bacteriology, 184, 5583–5589. 10.1128/JB.184.20.5583-5589.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominy SS, Lynch C, Ermini F, Benedyk M, Marczyk A, Konradi A, … & Potempa J (2019). Porphyromonas gingivalis in Alzheimer’s disease brains: Evidence for disease causation and treatment with small-molecule inhibitors. Science Advances, 5, eaau3333. 10.1126/sciadv.aau3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donohue TJ (2019). Shedding light on a group IV (ECF11) alternative σ factor. Molecular Microbiology, 112, 374–384. 10.1111/mmi.14280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou Y, Aruni W, Luo T, Roy F, Wang C, & Fletcher HM (2014). Involvement of PG2212 a Zinc-finger protein in the regulation of oxidative stress resistance in Porphyromonas gingivalis W83. Journal of Bacteriology, 196, 4057–4070. 10.1128/JB.01907-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou Y, Aruni W, Muthiah A, Roy F, Wang C, & Fletcher HM (2016). Studies of the extracytoplasmic function sigma factor PG0162 in Porphyromonas gingivalis. Molecular Oral Microbiology, 31, 270–283. 10.1111/omi.12122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou Y, Osbourne D, McKenzie R, & Fletcher HM (2010). Involvement of extracytoplasmic function sigma factors in virulence regulation in Porphyromonas gingivalis W83. FEMS Microbiol Letters, 312, 24–32. 10.1111/j.1574-6968.2010.02093.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou Y, Robles A, Roy F, Aruni AW, Sandberg L, Nothnagel E, & Fletcher HM (2015). The roles of RgpB and Kgp in late onset gingipain activity in the vimA-defective mutant of Porphyromonas gingivalis W83. Molecular oral microbiology, 30, 347–360. 10.1111/omi.12098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou Y, Rutanhira H, Chen X, Mishra A, Wang C, & Fletcher HM (2018). Role of extracytoplasmic function sigma factor PG1660 (RpoE) in the oxidative stress resistance regulatory network of Porphyromonas gingivalis. Molecular Oral Microbiology, 33, 89–104. 10.1111/omi.12204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher HM, Schenkein HA, Morgan RM, Bailey KA, Berry CR, & Macrina FL (1995). Virulence of a Porphyromonas gingivalis W83 mutant defective in the prtH gene. Infection and Immunity, 63, 1521–1528. 10.1128/IAI.63.4.1521-1528.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn JM, Levchenko I, Sauer RT, & Baker TA (2004). Modulating substrate choice: the SspB adaptor delivers a regulator of the extracytoplasmic-stress response to the AAA+ protease ClpXP for degradation. Genes & Development, 18, 2292–2301. 10.1101/gad.1240104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaballa A, Guariglia-Oropeza V, Durr F, Butcher FD, Chen AY, Chandransu P, & Helmann JD (2018). Modulation of extracytoplasmic function (ECF) sigma factor promoter selectivity by spacer region sequence. Nucleic Acids Research, 46, 134–145. 10.1093/nar/gkx953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerace E, & Moazed D (2015). Affinity pull-down of proteins using anti-FLAG M2 agarose beads. Methods in Enzymology, 559, 99–110. 10.1016/bs.mie.2014.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldenwang WG (1995). The sigma factors of Bacillus subtilis. Microbiological Reviews, 59(1), 1–30. 10.1128/mmbr.59.1.1-30.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardwick SW, Pane-Farre J, Delumeau O, Marles-Wright J, Murray JW, Hecker M, & Lewis RJ (2007). Structural and functional characterization of partner switching regulating the environmental stress response in Bacillus subtilis. The Journal of Biological Chemistry, 282, 11562–11572. 10.1074/jbc.M609733200. [DOI] [PubMed] [Google Scholar]
- Hasan MK, Yaguchi T, Minoda Y, Hirano T, Taira K, Wadhwa R, & Kaul SC (2004). Alternative reading frame protein (ARF)-independent function of CARF (collaborator of ARF) involves its interactions with p53: evidence for a novel p53-activation pathway and its negative feedback control. The Biochemical Journal, 380, 605–610. 10.1042/BJ20040337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmann JD (2002). The extracytoplasmic function (ECF) sigma factors. Advances in Microbial Physiology, 46, 47–110. 10.1016/s0065-2911(02)46002-x. [DOI] [PubMed] [Google Scholar]
- Henry LG, McKenzie RM, Robles A, & Fletcher HM (2012). Oxidative stress resistance in Porphyromonas gingivalis. Future microbiology, 7, 497–512. 10.2217/fmb.12.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hizukuri Y, & Akiyama Y (2012). PDZ domains of RseP are not essential for sequential cleavage of RseA or stress-induced σE activation in vivo. Molecular Microbiology, 86, 1232–1245. 10.1111/mmi.12053. [DOI] [PubMed] [Google Scholar]
- Kadowaki T, Yukitake H, Naito M, Sato K, Kikuchi Y, Kondo Y, … & Nakayama K (2016). A two-component system regulates gene expression of the type IX secretion component proteins via an ECF sigma factor. Scientific Reports, 6, 23288. 10.1038/srep23288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallifidas D, Thomas D, Doughty P, & Paget MS (2010). The σR regulon of Streptomyces coelocolor A32 reveals a key role in protein quality control during disulphide stress. Microbiology, 156, 1661–1672. 10.1099/mic.0.037804-0. [DOI] [PubMed] [Google Scholar]
- Kikuchi Y, Ohara N, Ueda O, Hirai K, Shibata Y, Nakayama K, & Fujimura S (2009). Porphyromonas gingivalis mutant defective in a putative extracytoplasmic function sigma factor shows a mutator phenotype. Oral Microbiology and Immunology, 24, 377–383. 10.1111/j.1399-302X.2009.00526.x. [DOI] [PubMed] [Google Scholar]
- Lamont RJ, & Jenkinson HF (1998). Life below the gum line: pathogenic mechanisms of Porphyromonas gingivalis. Microbiology and Molecular Biology Review, 62, 1244–1263. 10.1128/MMBR.62.4.1244-1263.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane WJ, & Darst SA (2006). The structural basis for promoter −35 element recognition by the group IV σ factors. PLOS Biology, 4, e269. 10.1371/journal.pbio.0040269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis JP (2010). Metal uptake in host-pathogen interactions: role of iron in Porphyromonas gingivalis interactions with host organisms. Periodontology, 2000, 52, 94–116. 10.1111/j.1600-0757.2009.00329.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Fang C, Zhuang N, Wang T, & Zhang Y (2019). Structural basis for transcription initiation by bacterial ECF σ factors. Nature Communications, 10, 1153. 10.1038/s41467-019-09096-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Wang B, Feng L, Kang H, Qi Y, Wang J, & Shi Y (2009). Cleavage of RseA by RseP requires a carboxyl-terminal hydrophobic amino acid following DegS cleavage. Proceedings of the National Academy of Sciences of the United States of America, 106, 14837–14842. 10.1073/pnas.0903289106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liccardo D, Cannavo A, Spagnuolo G, Ferrara N, Cittadini A, Rengo C, & Rengo G (2019). Periodontal disease: a risk factor for disbetes and cardiovascular disease. International Journal of Molecular Sciences, 20, 1414. 10.3390/ijms20061414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W, Mandal S, Degen D, Cho MS, Feng Y, Das K, & Ebright RH (2019). Structural basis of ECF-σ-factor-dependent transcription initiation. Nature Communications, 10, 710. 10.1038/s41467-019-08443-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, & Schmittgen TD (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods (San Diego, Calif), 25, 402–408. 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Llamas MA, Mooij MJ, Sparrius M, Vandenbroucke-Grauls CM, Ratledge C, & Bitter W (2008). Characterization of five novel Pseudomonas aeruginosa-surface signalling systems. Molecular Microbiology, 67, 458–472. 10.1111/j.1365-2958.2007.06061.x. [DOI] [PubMed] [Google Scholar]
- Maillard AP, Girard E, Ziani W, Petit-Hartlein I, Kahn R, & Coves J (2014). The crystal structure of the anti-σ factor CnrY in complex with the σ factor CnrH shows a new structure class of anti-σ factors targeting extracytoplasmic function σ factors. Journal of Molecular Biology, 426, 2313–2327. 10.1016/j.jmb.2014.04.003. [DOI] [PubMed] [Google Scholar]
- McGuffle BA, Vallet-Gely I, & Dove SL (2015). σ factor and anti-σ factor that control swarming motility and biofilm formation in Pseudomonas aeruginosa. Journal of Bacteriology, 198, 755–765. 10.1128/JB.00784-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson KE, Fleischmann RD, DeBoy RT, Paulsen IT, Fouts DE, Eisen JA, … & Fraser CM (2003). Complete genome sequence of the oral pathogenic Bacterium Porphyromonas gingivalis strain W83. Journal of Bacteriology, 185, 5591–5601. 10.1128/JB.185.18.5591-5601.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicoloff H, Gopalkrishnan S, & Ades SE (2017). Appropriate regulation of the σE-dependent envelope stress response is necessary to maintain cell envelope integrity and stationary-phase survival in Escherichia coli. Journal of Bacteriology, 199, e00089–17. 10.1128/JB.00089-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paget MS (2015). Bacterial sigma factors and anti-sigma factors: structure, function and distribution. Biomolecules, 5, 1245–1265. 10.3390/biom5031245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paget MS, Bae J, Hahn M, Li W, Kleanthous C, Roe J, & Buttner MJ (2001). Mutational analysis of RsrA, a zinc-binding anti-sigma factor with a thiol-disulphide redox switch. Molecular Microbiology, 39, 1036–1047. 10.1046/j.1365-2958.2001.02298.x. [DOI] [PubMed] [Google Scholar]
- Paget MS, Kang JG, Roe JH, & Buttner MG (1998). σR, an RNA polymerase sigma factor that modulates expression of the thioredoxin system in response to oxidative stress in Streptomyces coelicolor A3(2). The EMBO Journal, 17, 5776–5782. 10.1093/emboj/17.19.5776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paget MS, Molle V, Cohen G, Aharonowitz Y, & Buttner MJ (2001). Defining the disulphide stress response in Streptomyces coelicolor A3(2): identification of the σR regulon. Molecular Microbiology, 42, 1007–1020. 10.1046/j.1365-2958.2001.02675.x. [DOI] [PubMed] [Google Scholar]
- Perricone C, Ceccarelli F, Saccucci M, Di Carlo G, Bogdanos DP, Lucchetti R, Pilloni A, Valesini G, Polimeni A, & Conti F (2019). Porphyromonas gingivalis and rheumatoid arthritis. Current Opinion in Rheumatology, 31, 517–524. 10.1097/BOR.0000000000000638. [DOI] [PubMed] [Google Scholar]
- Phillips PL, Reyes L, Sampson EM, Murrell EA, Whitlock JA, & Progulske-Fox A (2018). Deletion of a conserved transcript PG_RS02100 expressed during logarithmic growth in Porphyromonas gingivalis results in hyperpigmentation and increased tolerance to oxidative stress. PloS one, 13, e0207295. 10.1371/journal.pone.0207295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto D, Liu Q, & Mascher T (2019). ECF σ factors with regulatory extensions: the one-component systems of the σ universe. Molecular Microbiology, 112, 399–409. 10.1111/mmi.14323. [DOI] [PubMed] [Google Scholar]
- Potvin E, Sanschagrin F, & Levesque RC (2008). Sigma factors in Pseudomonas aeruginosa. FEMS Microbiology Review, 32: 38–55. 10.1111/j.1574-6976.2007.00092.x. [DOI] [PubMed] [Google Scholar]
- Schneider JS, & Glickman MS (2013). Function of site-2 proteases in bacteria andbacterial pathogens. Biochimica et Biophysica Acta, 1828, 2808–2814. 10.1016/j.bbamem.2013.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sineva E, Savkina M, & Ades SE (2017). Themes and variations in gene regulation by extracytoplasmic function (ECF) sigma factors. Current Opinion in Microbiology, 36, 128–137. 10.1016/j.mib.2017.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalley JW, Birss AJ, Szmigielski B, & Potempa J (2006). The HA2 haemagglutinin domain of the lysine-specific gingipain (Kgp) of Porphyromonas gingivalis promotes micro-oxo bishaem formation from monomeric iron(III) protoporphyrin IX. Microbiology (Reading, England), 152, 1839–1845. 10.1099/mic.0.28835-0. [DOI] [PubMed] [Google Scholar]
- Staron A, Sofia HJ, Dietrich S, Ulrich LE, Liesegang H, & Mascher T (2009). The third pillar of bacterial signal transduction: classification of the extracytoplasmic function (ECF) sigma factor protein family. Molecular Microbiology, 74, 557–581. 10.1111/j.1365-2958.2009.06870.x. [DOI] [PubMed] [Google Scholar]
- Trevino-Quintanilla LG, Freyre-Gonzale JA, & Martinez-Flores I (2013). Anti-sigma factors in E. coli: common regulatory mechanisms controlling sigma factors availability. Current Genomics, 14, 378–387. 10.2174/1389202911314060007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilken C, Kitzing K, Kurzbauer R, Ehrmann M, & Clausen T (2004). Crystal structure of the DegS stress sensor: how a PDZ domain recognizes misfolded protein and activates a protease. Cell, 117, 483–494. 10.1016/s0092-8674(04)00454-4. [DOI] [PubMed] [Google Scholar]
- Yanamandra SS, Sarrafee SS, Anaya-Bergman C, Jones K, & Lewis JP (2012). Role of the Porphyromonas gingivalis extracytoplasmic function sigma factor, SigH. Molecular Oral Microbiology, 27, 202–219. 10.1111/j.2041-1014.2012.00643.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Yan R, Roy A, Xu D, Poisson J, & Zhang Y (2015). The I-TASSER suite: protein structure and function prediction. Nature Methods, 12, 7–8. 10.1038/nmeth.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zdanowski K, Doughty P, Jakimowicz P, O’Hara L, Buttner MJ, Paget MS, & Kleanthous C (2006). Assignment of the zinc ligands in RsrA, a redox-sensing ZAS protein from Streptomyces coelicolor. Biochemistry, 45, 8294–8300. 10.1021/bi060711v. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


