Abstract
BACKGROUND:
Pancreatic ductal adenocarcinoma (PDAC) is one of the most lethal malignancies. While the extracellular matrix (ECM) components plays an integral role in PDAC pathogenesis and mediating chemoresistance, its role in predicting response to chemotherapy in PDAC patients remains unclear.
METHODS:
We performed a systematic biomarker discovery by analyzing genomewide transcriptomic profiling data from 423 patients (GSE71729, GSE21501 and TCGA) for predicting overall survival (OS). This was subsequently validated in two independent clinical cohorts of 270 PDAC patients (training cohort; n=121 and validation cohort; n=149). In addition, we investigated EUS-FNA biopsy specimens from 51 PDAC patients with an unresectable cancer for predicting therapeutic response to gemcitabine-based therapy.
RESULTS:
Following rigorous bioinformatic analysis, we identified LAMC2 to be a significant prognostic factor in all three PDAC datasets (GSE71729, HR=2.04, P=0.002; GSE21501, HR=2.17, P=0.031; TCGA, HR=2.57, P<0.001). High LAMC2 expression in PDAC patients associated with a significantly poor OS and relapse-free survival (RFS) in both training (P<0.001, P<0.001) and validation cohorts (P=0.001, P=0.003). More importantly, LAMC2 expression robustly identified PDAC patients with unresectable disease and those who responded to gemcitabine-based therapy (AUC= 0.79; 95%CI, 0.65–0.89). The univariate logistic regression analysis revealed that high LAMC2 expression was the only factor that predicted poor response to gemcitabine in PDAC patients (Odds Ratio [OR]=4.90; 95% CI, 1.45–16.6; P=0.011).
CONCLUSION:
We conclude that LAMC2 is a novel prognostic and predictive biomarker for gemcitabine-based therapy in both adjuvant and palliative setting; which could have significant impact in precision and individualized treatment of PDAC patients.
Keywords: Extracellular matrix, LAMC2, pancreatic ductal adenocarcinoma, gemcitabine, predictive biomarker
INTRODUCTION
Pancreatic ductal adenocarcinoma (PDAC) is one of the most challenging diseases because of the late diagnosis, high rates of disease recurrence, poor survival rates, and availability of limited therapeutic regimens [1, 2]. This issue is compounded further due to the continued rise in PDAC incidence, projecting it to become the second leading cause of cancer-related deaths by 2030 [3].
Gemcitabine based therapy remains the backbone and treatment of choice in PDAC patients – whether it be in a neoadjuvant, adjuvant or palliative treatment setting. Recent developments in gemcitabine-based combination therapies have shown to significantly improve the median and 5-year overall survival (OS) rates in both the resectable and unresectable PDAC patients [4–6]. Nonetheless, the overall prognosis for this malignancy still remains quite poor [7]. In the recent years, the FOLFIRINOX treatment (a combination of 5-fluorouracil, leucovorin, irinotecan, and oxaliplatin) has led to an improvement of OS [8, 9]. However, currently there are no available validated biomarkers that can predict treatment response and facilitate selection of appropriate PDAC patient populations for such treatment regimens. Presently, such therapeutic decision-making and selection of patients with both local and metastatic PDAC primarily relies on patient’s overall health and individual opinion of an oncologist. Several other molecular biomarkers have been proposed for their prognostic potential in PDAC [10–13]; however, their translation into the clinic has been challenging. Collectively, these data highlight the unmet clinical need for developing improved prognostic and predictive biomarkers that can help identify patients who have the highest likelihood of receiving therapeutic benefit from such chemotherapies and spare others from the toxicity and expense associated with these drugs.
The extracellular matrix (ECM) provides the critical scaffold for the tumor microenvironment, and is intimately involved in regulating PDAC progression [14, 15]. In addition, the ECM plays a pivotal role in mediating chemoresistance in cancer [16–19]. While accumulating evidence suggests that the ECM components may serve as potential diagnostic or prognostic biomarkers in PDAC [20–22], their role as biomarkers for predicting response to chemotherapy in PDAC patients have thus far not been explored.
We therefore performed a genome-wide systematic and comprehensive transcriptomic analysis to identify ECM-related molecular biomarkers involved in predicting prognosis and resistance to gemcitabine. We followed this initial discovery effort by validation of our findings in two independent clinical cohorts of surgical resected PDAC patients, as well as another independent cohort of patients with an unresectable disease who were treated with gemcitabine + nab-paclitaxel regimen. Through these comprehensive biomarker discovery and validation efforts, we successfully identified Laminin γ2 (LAMC2) as a novel biomarker for tumor prognosis and predicting response to gemcitabine-based therapy in PDAC patients.
MATERIALS AND METHODS
Study design and patient cohorts
For the systematic biomarker discovery phase, three publicly-available datasets (GSE21501, GSE71729 and the Cancer Genome Atlas [TCGA]) were analyzed to validate the expression of ECM-related genes in PDAC patients. The ECM associated genes were listed and defined as per the Gene Ontology (GO) database [23]. During the biomarker discovery phase, GSE21501 (n=102) and GSE71729 (n=145) datasets were downloaded from the GEO database directly (https://www.ncbi.nlm.nih.gov, accessed on July 17, 2019). In addition, normalized transcriptomic profiling data from the TCGA dataset for 178 PDAC patients was downloaded from the UCSC Xena Browser (https://xenabrowser.net, accessed on July 17, 2019), and used for an independent validation of the discovery cohort.
In the subsequent in-house validation phase, a total of 321 PDAC patients were analyzed. This included a training cohort of 121 patients enrolled at the Kumamoto University, a validation cohort of 149 patients seen at the Nara Medical University, Japan, and a cohort of 51 patients treated with chemotherapy and enrolled at the Tokushima University, Japan. None of the patients with surgical treatment received pre-operative cancer treatment, and all tumors were diagnosed as PDAC. The specimens from the patients with chemotherapy treatment were obtained by endoscopic ultrasound-fine needle aspiration (EUS-FNA), prior to initiation of treatment. All specimens were formalin-fixed paraffin-embedded (FFPE) tissues. The study workflow is summarized in Supplementary Fig. S1. The study was conducted in accordance with the Declaration of Helsinki. A written informed consent was obtained from all patients, and the study was approved by the institutional review boards of all participating institutions.
Total RNA extraction and quantitative reverse transcription polymerase chain reaction (qRT-PCR)
Total RNA was extracted using AllPrep DNA/RNA FFPE Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. The quantitative reverse-transcriptase polymerase chain reaction (qRT-PCR) assays were performed using the QuantStudio 6 Flex RT-PCR System (Applied Biosystems, Foster City, CA). The relative abundance of target transcripts was evaluated and normalized to the expression levels of beta-actin as an internal control using the 2-ΔCt method [24].
Statistical analysis
Statistical analyses were performed using Medcalc statistical software V.16.2.0 (Medcalc Software bvba, Ostend, Belgium), and GraphPad Prism V8.0 (GraphPad Software, San Diego, CA). Continuous variables were expressed as medians and were compared using a t-test or Mann Whitney U test. Categorical variables were compared using χ2 or Fisher’s exact test. All P values were calculated using a two-sided test, and a P<0.05 was considered statistically significant. For time-to-event analyses, survival estimates were calculated using the Kaplan–Meier analysis, and the survival differences between groups were compared using the log-rank test.
RESULTS
Genomewide transcriptional profiling identified LAMC2 as a prognostic biomarker in patients with PDAC
Using the GO search engine and the search for ‘extracellular matrix’ keyword, we identified 852 genes associated with this biological process (Supplementary Table S1). We next analyzed the expression profiles of these genes in the GSE21501 and GSE71729 datasets, specifically in terms of their association with survival outcomes in PDAC patients. Following bioinformatic and biostatistical analysis, we identified a panel of 10 genes that were significantly associated with OS in PDAC patients. We next validated the performance of these genes in an independent cohort of patients within the TCGA dataset and LAMC2 was the only gene that emerged with a robust prognostic potential in PDAC. We observed that high expression of LAMC2 was the singular and significant prognostic factor in all three datasets (GSE71729; Hazard ratio [HR]=2.04; 95%CI, 1.30–3.19; P=0.002, GSE21501; HR=2.17; 95%CI, 1.08–4.38; P=0.031, TCGA; HR=2.57; 95%CI, 1.62–4.07; P<0.001, Supplementary Fig. S2A-C). Furthermore, LAMC2 expression was significantly higher in PDAC tissues compared to the normal mucosa (P<0.0001, Supplementary Fig. S2D).
High LAMC2 expression significantly associates with poorer outcome
Next, we assessed the clinical significance of LAMC2 expression in two independent PDAC patient cohorts (training cohort; n=121 and validation cohort; n=149). All patients were categorized into low- and high-risk groups based on the LAMC2 expression and by utilizing Youden’s index-derived cutoff thresholds in the training cohort (Fig. 1A). To ensure clinical robustness of our findings, we used the same cut-off thresholds in the validation cohort. As illustrated in Table 1, other than the tumor status in the training cohort (P=0.04), no significant differences were observed in the distribution of various clinicopathological variables between the LAMC2-high and low expression groups.
Figure 1.
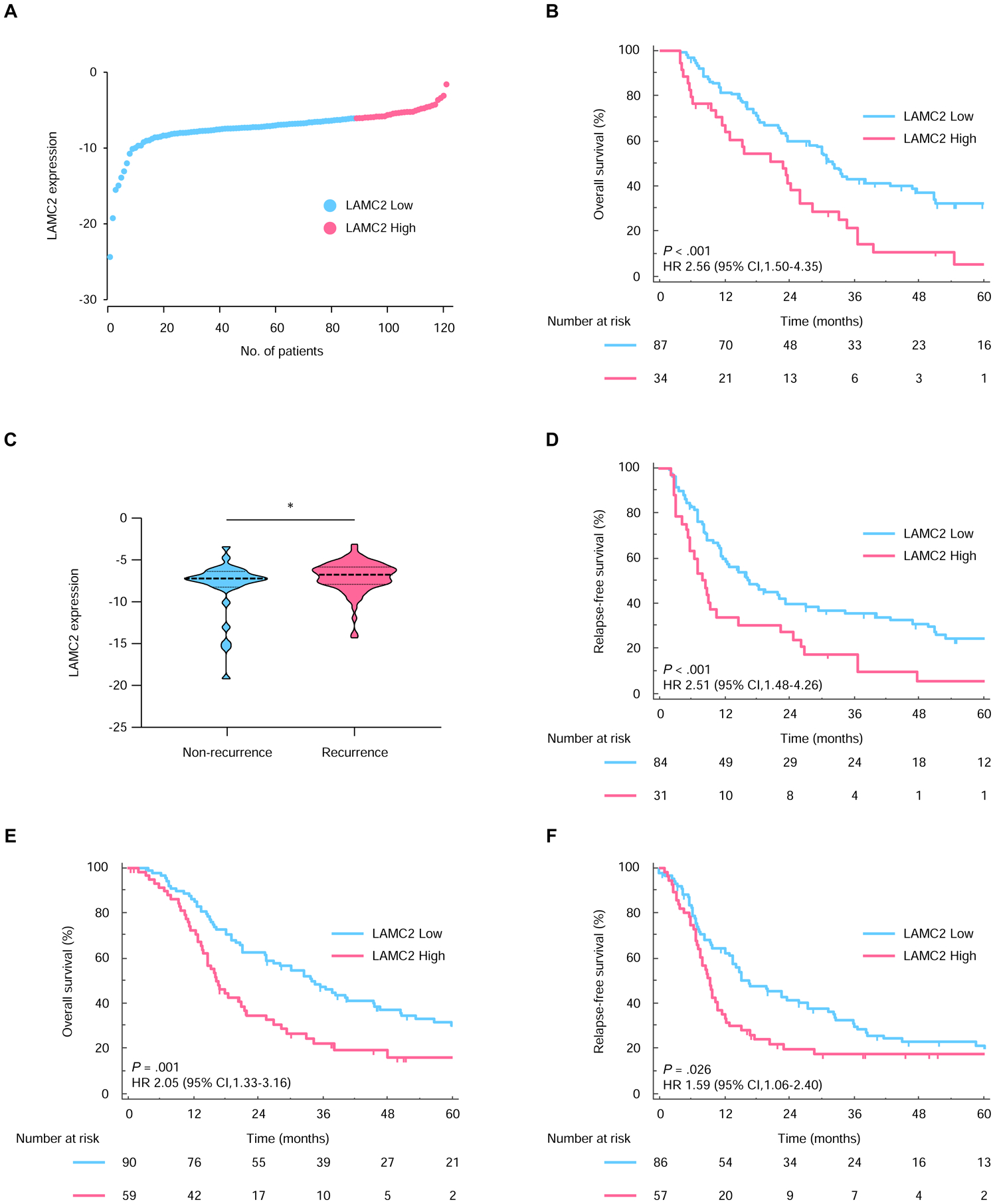
High LAMC2 expression associates with worse prognosis in PDAC patients in the training and validation cohort. (A) The distribution of LAMC2 expression in PDAC patients. (B) Kaplan-Meier curves for OS between PDAC patients with high (pink) and low (blue) LAMC2 expression in the training cohort. (C) Comparison of LAMC2 expression levels in PDAC patients with or without recurrence. (D) Kaplan-Meier curves for RFS between PDAC patients with high (pink) and low (blue) LAMC2 expression in the training cohort. Kaplan-Meier curves for (E) OS and (F) RFS in the validation cohort. *, P<0.05.
Table 1:
Patient characteristics in the resectable cohort of PDAC patients within the training and validation cohorts
| Characteristics | Training cohort | Validation cohort | ||||||
|---|---|---|---|---|---|---|---|---|
| Total | LAMC2 expression | Total | LAMC2 expression | |||||
| n = 121 | Low (n = 87) | High (n = 34) | P-valuea | n = 149 | Low (n = 90) | High (n = 59) | P-valuea | |
| Age, years | 0.27 | 0.52 | ||||||
| < 65, n (%) | 44 | 29 (33.3) | 15 (44.1) | 37 | 24 (26.7) | 13 (22.0) | ||
| ≥ 65, n (%) | 77 | 58 (66.7) | 19 (55.9) | 112 | 66 (73.3) | 46 (78.0) | ||
| Gender | 0.30 | 0.55 | ||||||
| Male, n (%) | 62 | 42 (48.3) | 20 (58.8) | 89 | 52 (57.8) | 37 (62.7) | ||
| Female, n (%) | 59 | 45 (51.7) | 14 (41.2) | 60 | 38 (42.2) | 22 (37.3) | ||
| Tumor status | 0.04b | 0.41b | ||||||
| T1–2 | 16 | 15 (17.2) | 1 (2.9) | 15 | 11 (12.2) | 4 (6.8) | ||
| T3–4 | 105 | 72 (82.8) | 33 (97.1) | 134 | 79 (87.8) | 55 (93.2) | ||
| Nodal status | 0.09b | 0.86 | ||||||
| N0 | 40 | 33 (37.9) | 7 (20.6) | 67 | 41 (45.6) | 26 (44.1) | ||
| N1 | 81 | 54 (62.1) | 27 (79.4) | 82 | 49 (54.4) | 33 (55.9) | ||
| UICC stage (ver. 7) | 0.13 | 0.49 | ||||||
| IA, IB | 14 | 13 (14.9) | 1 (2.9) | 11 | 9 (10.0) | 2 (3.4) | ||
| IIA | 25 | 20 (23.0) | 5 (14.7) | 54 | 31 (34.4) | 23 (39.0) | ||
| IIB | 67 | 45 (51.7) | 22 (64.7) | 68 | 40 (44.4) | 28 (47.5) | ||
| III, IV | 15 | 9 (10.4) | 6 (17.7) | 16 | 10 (11.2) | 6 (10.1) | ||
| CA19-9 (U/mL) | 0.60 | 0.49 | ||||||
| < 37, n (%) | 40 | 30 (34.5) | 10 (29.4) | 40 | 26 (28.9) | 14 (23.7) | ||
| ≥ 37, n (%) | 81 | 57 (65.5) | 24 (70.6) | 109 | 64 (71.1) | 45 (76.3) | ||
| Tumor size (mm) | 0.92 | 0.59 | ||||||
| < 40, n (%) | 93 | 69 (79.3) | 24 (70.6) | 120 | 73 (81.1) | 47 (79.7) | ||
| ≥ 40, n (%) | 27 | 18 (20.7) | 9 (26.5) | 16 | 11 (12.2) | 5 (8.5) | ||
| N/A | 1 | 1 (2.9) | 13 | 6 (6.7) | 7 (11.8) | |||
| Adjuvant therapy | 0.59 | 0.25 | ||||||
| Gemcitabine based | 80 | 58 (66.7) | 22 (64.7) | 92 | 59 (65.6) | 33 (55.9) | ||
| Other | 22 | 17 (19.5) | 5 (14.7) | 26 | 12 (13.3) | 14 (23.7) | ||
| none | 19 | 12 (13.8) | 7 (20.6) | 28 | 17 (18.9) | 11 (18.7) | ||
| Unknown | 0 | 0 | 0 | 3 | 2 (2.2) | 1 (1.7) | ||
Chi-square test
Fisher’s exact test
UICC, International Union Against Cancer; N/A, Not available
For evaluating the performance of LAMC2 expression into a clinically translatable prognostic assay, we first analyzed its relationship with the OS in patients within the training cohort. Interestingly, the median OS in LAMC2-high expression subgroup was 23.0 months vs. 32.1 months in PDAC patients with lower expression of this ECM-related gene (P<0.001; Fig. 1B). To further the prognostic potential of LAMC2, we next interrogated its relationship and cancer recurrence in the training cohort patients. In support of our earlier findings, the LAMC2 expression in patients with recurrence was significantly higher than those without recurrence (P=0.031; Fig. 1C). Moreover, Kaplan-Meier analysis for relapse-free survival (RFS) revealed that high LAMC2 levels in PDAC patients associated with a significantly poor RFS (P<0.001; Fig. 1D).
In accordance with our observations in the training cohort, high tumor LAMC2 expression was associated with poorer OS and RFS compared to the patients with low LAMC2 expression (P=0.001 and P=0.026, respectively; Fig. 1E and F).
High expression of LAMC2 is an independent prognostic risk factor in PDAC patients
When challenged on multivariate analysis in training cohorts (Fig.2A), patients with high LAMC2 expression (HR=2.02; 95% CI, 1.28–3.20; P=0.003), higher levels of CA19-9 (HR=1.68; 95% CI, 1.02–2.78; P=0.043), and those with LNM were associated with poor OS (HR=2.68; 95% CI, 1.59–4.50; P<0.001). Consistent with the training cohort results, in the multivariate analysis, high LAMC2 expression (HR=1.84; 95% CI, 1.24–2.74; P=0.003), CA19-9 status (HR=2.25; 95% CI, 1.36–3.70; P=0.002), and LNM status (HR=2.02; 95% CI, 1.36–2.99; P<0.001) were the only clinicopathological factors that significantly associated with worse OS (Fig.2B).
Figure 2.
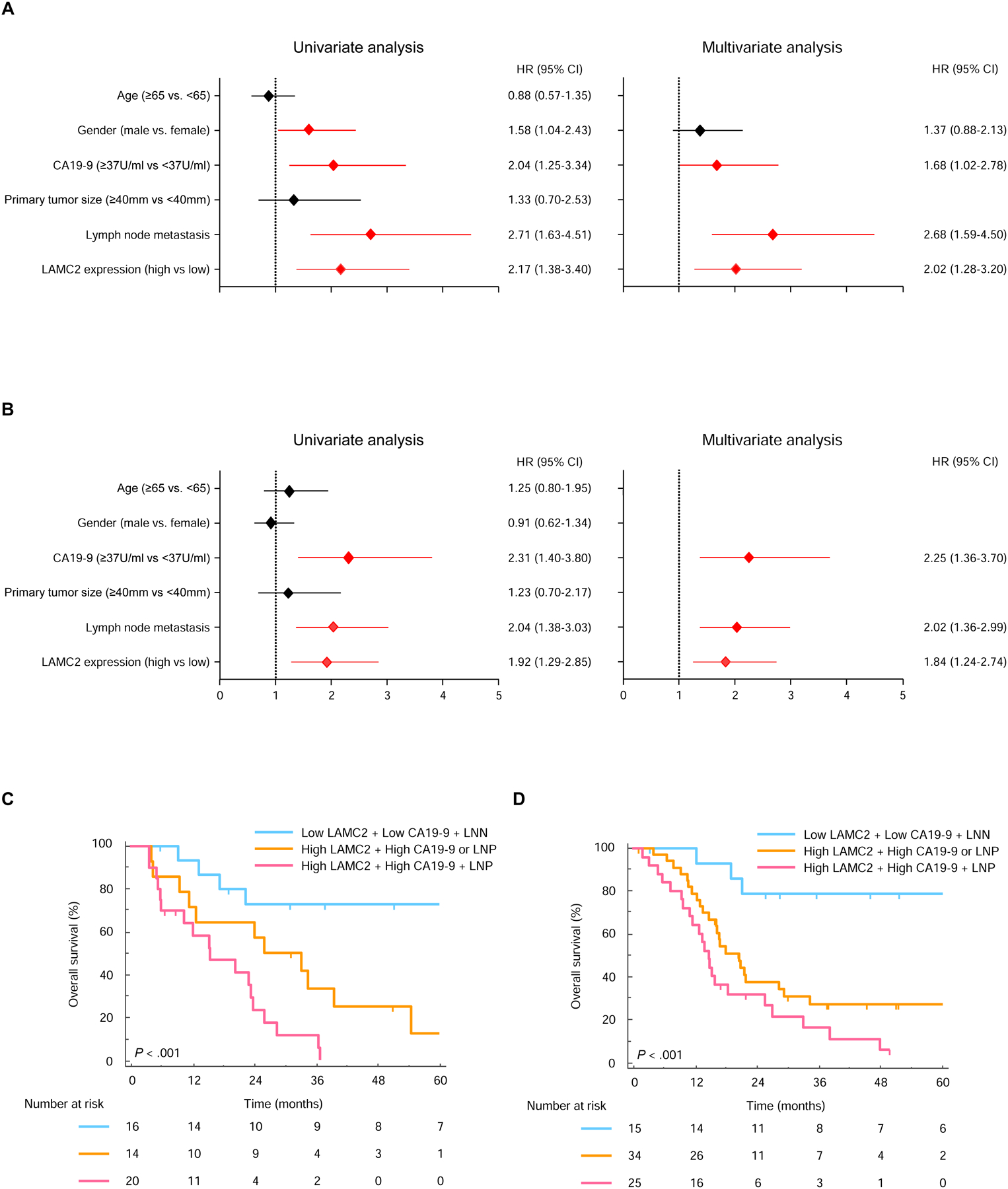
Validation of high LAMC2 expression for predicting poor prognosis in PDAC patients. Univariate and multivariate analysis in the (A) training and (B) validation cohort calculated by Cox regression model. Kaplan-Meier curves for OS among 3 Groups in the (C) training and (D) validation cohort. Lymph node metastasis, LNN; LNM positivity, LNP.
To address a combination of LAMC2 expression together with other clinical factors, we stratified patients into three different groups: Group 1 included patients with low LAMC2 expression, low CA19-9 levels (<37U/ml), and absence of LNM, Group 2 patients included high LAMC2 expression and/or either high CA19-9 levels (≥37U/mL) nor LNM positivity and Group 3 patients were those who exhibited all three risk factors. The median OS was 95.0 months in group 1, 26.0 months in group 2 and 15.5 months in group 3 (P<0.001, Fig. 2C). Similarly, the median RFS was 68.4 months in group 1, 22.5 months in group 2 and 5.93 months in group 3 (P<0.001, Supplementary Fig. S3A). Likewise, we observed that patients in group 3 still exhibited significantly worse OS and RFS (P<0.001 and P=0.001, respectively) in the validation cohort (Fig. 2D and Supplementary Fig. S3B).
High LAMC2 expression predicts therapeutic response to gemcitabine-based therapy
We next analyzed the LAMC2 expression levels in the context of adjuvant chemotherapy. Intriguingly, high LAMC2 expression levels significantly associated with shorter median OS (P=0.018 and P=0.003, respectively) and RFS (P=0.023 and P=0.025, respectively) in the patients who were treated with gemcitabine based therapy in both cohorts (Supplementary Fig. S4A-D). On the other hand, LAMC2 expression in PDAC patients who received 5FU based adjuvant therapy did not associate significantly with OS and RFS (Supplementary Fig. S4E-H).
We next investigated an independent cohort of 51 PDAC patients with an unresectable cancer, who received gemcitabine and nab-paclitaxel regimen as an initial therapy. Patients were classified as either responders (confirmed complete response [CR], partial response [PR], or stable disease [SD]) or non-responders (progressive disease [PD]) based on the best response evaluated by the Response Evaluation Criteria in Solid Tumors (RECIST) version 1.1 and were included in the waterfall plot (Fig. 3A). Among 51 patients, 24 patients exhibited the high LAMC2 expression, while 27 patients had the low expression (Table 2). Of note, the LAMC2 expression within the responder group was significantly lower than patients within the non-responder group (P<0.001, Fig. 3B). Within the responder group, 11 patients exhibited high LAMC2 expression (11/24; 45.8%) and 21 with low LAMC2 expression (21/27; 77.8%; P=0.023, Fig. 3C). More importantly, LAMC2 expression demonstrated robust identification of response in this cohort (AUC= 0.79; 95%CI, 0.65–0.89, Fig. 3D). When we analyzed the OS and progression free survival (PFS) of the patients, patients with high tumor LAMC2 expression had poorer OS and PFS vs. patients with low LAMC2 expression (P=0.031 and 0.040, respectively; Fig. 3E and F).
Figure 3.
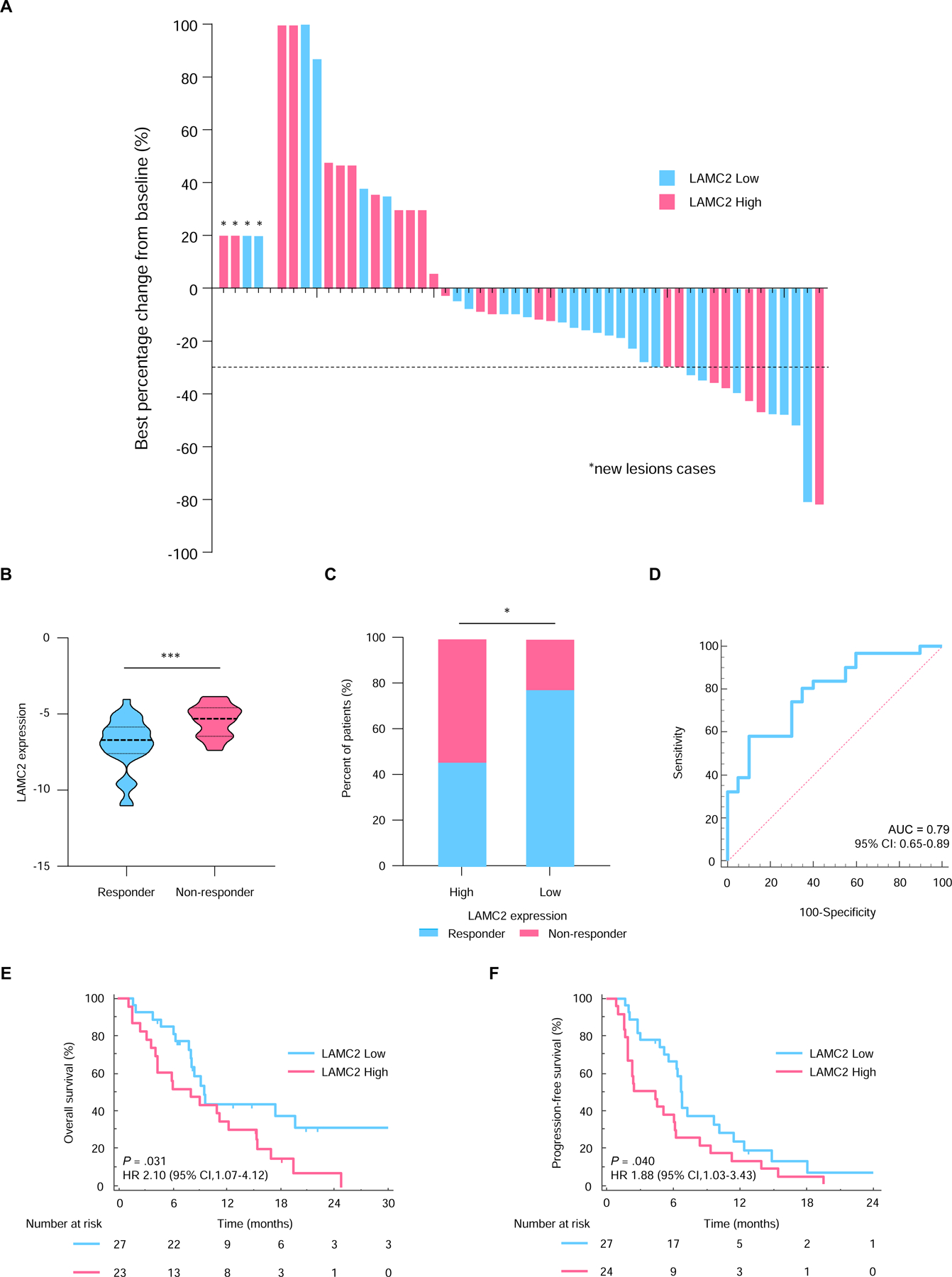
LAMC2 expression predicts therapeutic response to gemcitabine-based therapy. (A) Waterfall plots for predicting best tumor response in PDAC patients treated with gemcitabine and nab-paclitaxel as a primary treatment. (B) Comparison of LAMC2 expression levels in responders and non-responders in the primary chemotherapy cohort. (C) The proportion of responders and non-responders in the LAMC2-high and low patients. (D) ROC curves for the predicting therapeutic response to gemcitabine. Kaplan-Meier curves for (E) OS and (F) PFS in PDAC patients with high (pink) or low (blue) LAMC2 expression in the primary chemotherapy cohort. *, P<0.05; **P<0.001.
Table 2:
Patients characteristics in PDAC patients with an unresectable disease
| Characteristics | Total | LAMC2 expression | |||
|---|---|---|---|---|---|
| n = 51 | Low (n = 27) | High (n = 24) | P-valuea | ||
| Age, years | 0.55 | ||||
| < 65, n (%) | 17 | 10 (37.0) | 7 (29.2) | ||
| ≥ 65, n (%) | 34 | 17 (63.0) | 17 (70.8) | ||
| Gender | 0.20 | ||||
| Male, n (%) | 24 | 15 (55.6) | 9 (37.5) | ||
| Female, n (%) | 27 | 12 (44.4) | 15 (62.5) | ||
| CA19-9 (U/mL) | 0.43b | ||||
| < 37, n (%) | 7 | 5 (18.5) | 2 (8.3) | ||
| ≥ 37, n (%) | 44 | 22 (81.5) | 22 (91.7) | ||
| Tumor size (mm) | 0.44 | ||||
| < 40, n (%) | 22 | 13 (48.1) | 9 (37.5) | ||
| ≥ 40, n (%) | 29 | 14 (51.9) | 15 (62.5) | ||
| Locally or Metastasis | 0.86 | ||||
| Locally advanced | 9 | 5 (18.5) | 4 (16.7) | ||
| Distal metastasis | 42 | 22 (81.5) | 20 (83.3) | ||
| Location | 0.66 | ||||
| Head | 16 | 8 (29.6) | 8 (33.3) | ||
| Body | 26 | 13 (48.1) | 13 (54.2) | ||
| Tail | 9 | 6 (22.3) | 3 (12.5) | ||
| site of metastasis | 0.34 | ||||
| Liver | 28 | 16 (66.7) | 12 (50.0) | ||
| Lung | 8 | 5 (20.9) | 3 (12.5) | ||
| Peritoneum | 5 | 1 (4.1) | 4 (16.7) | ||
| Lymph node | 6 | 2 (8.3) | 4 (16.7) | ||
| Other | 1 | 0 (0.0) | 1 (4.1) | ||
| No. of metastatic sites | 0.46 | ||||
| 0 | 10 | 5 (18.5) | 5 (20.9) | ||
| 1 | 36 | 20 (74.1) | 16 (66.7) | ||
| 2 | 3 | 2 (7.4) | 1 (4.1) | ||
| 3 | 2 | 0 (0.0) | 2 (8.3) | ||
Chi-square test
Fisher’s exact test
Next, the univariate logistic regression analysis revealed that higher levels of LAMC2 expression were the only factor that associated with a poor response to gemcitabine in the cohort (Odds ratio [OR]=4.90; 95% CI, 1.45–16.6; P=0.011; Table 3).
Table 3:
Univariate logistic regression analysis for LAMC2 as a predictive biomarker for therapeutic response in PDAC patients
| Characteristics | OR | 95% CI | P-value |
|---|---|---|---|
| Age (≥65 vs. <65) | 1.89 | 0.55 – 6.57 | 0.31 |
| Gender (Female vs. Male) | 2.26 | 0.71 – 7.19 | 0.17 |
| Primary tumor location (Head vs. Other) | 2.81 | 0.83 – 9.49 | 0.10 |
| Locally Advanced vs. Metastatic | 1.36 | 0.30 – 6.20 | 0.69 |
| CA19-9 (≥37U/mL vs. <37U/mL) | 4.56 | 0.51 – 41.1 | 0.18 |
| LAMC2 status (High vs. Low) | 4.90 | 1.45 – 16.6 | 0.01 |
OR, odds ratio; CI, confidence interval
DISCUSSION
In the present study, using a comprehensive biomarker discovery approach, we initially identified LAMC2 that were significantly associated with poor OS in PDAC patients. Following rigorous training and validation, we validated LAMC2 to be the only gene that consistently exhibited prognostic significance across PDAC patient cohorts. Moreover, we noted that a risk-assessment model that combined high LAMC2 expression, high CA19-9 levels and presence of LNM was significantly superior in predicting the OS and RFS in PDAC patients. Finally, given its biological role as a ECM-related gene, we successfully identified that high expression of LAMC2 are predictive of therapeutic response to gemcitabine-based therapy in adjuvant and palliative settings.
We observed that high LAMC2 expression was significantly associated with poor OS and RFS in PDAC patients. While our results are in line with some of the previous reports [25, 26], the prior studies had several limitations, including inadequate sample size, lack of systematic and comprehensive biomarker discovery approach and lack of independent validation cohorts – all of which were addressed in our current article. Furthermore, we for the first time developed a PCR-based cut-off threshold to assess LAMC2 expression levels in a training cohort, which were successfully applied to an independent validation cohort. More importantly, the multivariate analysis revealed that high LAMC2 expression was an independent prognostic factor in PDAC patients – in large, independent, clinical cohorts.
Following a potentially curative surgery, approximately 80% of PDAC patients often develop metastasis mostly within the first 2 years after surgery [27]. Although adjuvant chemotherapy provides significant survival benefit in PDAC patients [4, 8], there is lack of availability of predictive biomarkers that can guide therapeutic decision-making in individual PDAC patients. Several retrospective studies have investigated whether some nucleoside transporters involved in the uptake of gemcitabine could predict the response [12, 28–30], however, none of these studies have reached clinical significance. In our study, we deliberately focused on ECM-associated pathway and demonstrated that LAMC2 was significantly associated with poor prognosis – both in terms of OS and RFS, in patients who received gemcitabine based adjuvant therapy, while such an effect was not evident for 5FU based adjuvant therapy. Although further studies are required, the results of our study collectively highlight that LAMC2 expression might serve as a potentially attractive biomarker for predicting therapeutic response to gemcitabine chemotherapy in an adjuvant setting.
Thus far, no other biomarkers have reported predictive potential for gemcitabine and nab-paclitaxel therapy in unresectable PDAC patients. Von Hoff et al. demonstrated that secreted protein acidic and rich in cysteine (SPARC) was associated with improved OS in PDAC patients who received gemcitabine and nab-paclitaxel regimen [31]. However, a subsequent study failed to observe any significant associations between stromal SPARC levels and predictive efficacy [32]. Herein, we successfully demonstrated that LAMC2 expression is a robust predictive biomarker against gemcitabine therapy in a palliative setting.
We would like to acknowledge potential limitations of our work. First, this was a retrospective study with the potential inadvertent risk of bias. Hence, a prospective randomized clinical study in future could confirm our analysis before the translation of this biomarker into the clinic. Second, we did not analyze LAMC2 expression in unresectable PDAC patients who received FOLFIRINOX treatments; since such a patient cohort was not available to us at this time. Third, although our cohorts are independent and included reasonably large sample sizes, future studies are required to confirm the utility of LAMC2 in PDAC. To overcome these limitations, a prospective randomized controlled study is required prior to any further consideration regarding the clinical translation of our data.
In conclusion, high LAMC2 expression emerged as a robust prognostic biomarker as it significantly correlated with poor OS and RFS in two large, independent cohorts of PDAC patients. More importantly, our results indicate that LAMC2 expression is a predictor of therapeutic response to gemcitabine resistance in PDAC patients. Collectively, our findings have important implications for the further prospective validation and development of LAMC2 as a prognostic and predictive biomarker for gemcitabine-based treatment in both adjuvant and palliative setting; hence, making a significant advance in precision and individualized treatment of patients suffering from this fatal malignancy.
Supplementary Material
Highlights.
Pancreatic ductal adenocarcinoma (PDAC) is one of the most challenging cancers.
There are no available biomarkers to predict response to gemcitabine in PDAC.
Patients with high Laminin γ2 (LAMC2) expression had poor prognosis.
High LAMC2 expression may not benefit for gemcitabine-based therapy.
LAMC2 can be used for decision-making for the selection of chemotherapy regimens.
ACKNOWLEDGEMENTS
We would like to thank Jinsei Miyoshi and Akira Fukuya for collecting clinical data, and Tatsuhiko Kakisaka, Jasjit K Banwait, Yuma Wada, In Seob Lee, Priyanka Sharma, Souvik Ghatak, Tong Liu, and Huanlin Wang for helping with their important insights into various experiments and data analysis advice.
Funding: The present work was supported by the CA72851, CA187956, CA202797 and CA214254 grants from the National Cancer Institute, National Institute of Health; In addition, this work was also supported by a pilot research award from the City of Hope Ludwig Cancer Research-Hilton Foundation Partnership award.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest: None of the authors has any potential conflicts to disclose.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
REFERENCES
- [1].Hidalgo M, Cascinu S, Kleeff J, Labianca R, Lohr JM, Neoptolemos J, et al. Addressing the challenges of pancreatic cancer: future directions for improving outcomes. Pancreatology : official journal of the International Association of Pancreatology (IAP) [et al]. 2015;15:8–18. [DOI] [PubMed] [Google Scholar]
- [2].Groot VP, Gemenetzis G, Blair AB, Rivero-Soto RJ, Yu J, Javed AA, et al. Defining and Predicting Early Recurrence in 957 Patients With Resected Pancreatic Ductal Adenocarcinoma. Annals of surgery. 2019;269:1154–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Rahib L, Smith BD, Aizenberg R, Rosenzweig AB, Fleshman JM, Matrisian LM. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer research. 2014;74:2913–21. [DOI] [PubMed] [Google Scholar]
- [4].Neoptolemos JP, Palmer DH, Ghaneh P, Psarelli EE, Valle JW, Halloran CM, et al. Comparison of adjuvant gemcitabine and capecitabine with gemcitabine monotherapy in patients with resected pancreatic cancer (ESPAC-4): a multicentre, open-label, randomised, phase 3 trial. Lancet (London, England). 2017;389:1011–24. [DOI] [PubMed] [Google Scholar]
- [5].Jang JY, Han Y, Lee H, Kim SW, Kwon W, Lee KH, et al. Oncological Benefits of Neoadjuvant Chemoradiation With Gemcitabine Versus Upfront Surgery in Patients With Borderline Resectable Pancreatic Cancer: A Prospective, Randomized, Open-label, Multicenter Phase 2/3 Trial. Annals of surgery. 2018;268:215–22. [DOI] [PubMed] [Google Scholar]
- [6].Von Hoff DD, Ervin T, Arena FP, Chiorean EG, Infante J, Moore M, et al. Increased survival in pancreatic cancer with nab-paclitaxel plus gemcitabine. The New England journal of medicine. 2013;369:1691–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Neoptolemos JP, Kleeff J, Michl P, Costello E, Greenhalf W, Palmer DH. Therapeutic developments in pancreatic cancer: current and future perspectives. Nature reviews Gastroenterology & hepatology. 2018;15:333–48. [DOI] [PubMed] [Google Scholar]
- [8].Conroy T, Hammel P, Hebbar M, Ben Abdelghani M, Wei AC, Raoul JL, et al. FOLFIRINOX or Gemcitabine as Adjuvant Therapy for Pancreatic Cancer. The New England journal of medicine. 2018;379:2395–406. [DOI] [PubMed] [Google Scholar]
- [9].Conroy T, Desseigne F, Ychou M, Bouche O, Guimbaud R, Becouarn Y, et al. FOLFIRINOX versus gemcitabine for metastatic pancreatic cancer. The New England journal of medicine. 2011;364:1817–25. [DOI] [PubMed] [Google Scholar]
- [10].Muckenhuber A, Berger AK, Schlitter AM, Steiger K, Konukiewitz B, Trumpp A, et al. Pancreatic Ductal Adenocarcinoma Subtyping Using the Biomarkers Hepatocyte Nuclear Factor-1A and Cytokeratin-81 Correlates with Outcome and Treatment Response. Clinical cancer research : an official journal of the American Association for Cancer Research. 2018;24:351–9. [DOI] [PubMed] [Google Scholar]
- [11].Tiriac H, Belleau P, Engle DD, Plenker D, Deschenes A, Somerville TDD, et al. Organoid Profiling Identifies Common Responders to Chemotherapy in Pancreatic Cancer. Cancer discovery. 2018;8:1112–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Spratlin J, Sangha R, Glubrecht D, Dabbagh L, Young JD, Dumontet C, et al. The absence of human equilibrative nucleoside transporter 1 is associated with reduced survival in patients with gemcitabine-treated pancreas adenocarcinoma. Clinical cancer research : an official journal of the American Association for Cancer Research. 2004;10:6956–61. [DOI] [PubMed] [Google Scholar]
- [13].Pacheco-Barcia V, Mondejar Solis R, France T, Asselah J, Donnay O, Zogopoulos G, et al. A systemic inflammation response index (SIRI) correlates with survival and predicts oncological outcome for mFOLFIRINOX therapy in metastatic pancreatic cancer. Pancreatology : official journal of the International Association of Pancreatology (IAP) [et al]. 2019. [DOI] [PubMed] [Google Scholar]
- [14].Buchheit CL, Weigel KJ, Schafer ZT. Cancer cell survival during detachment from the ECM: multiple barriers to tumour progression. Nature reviews Cancer. 2014;14:632–41. [DOI] [PubMed] [Google Scholar]
- [15].Walker C, Mojares E, Del Río Hernández A. Role of Extracellular Matrix in Development and Cancer Progression. International journal of molecular sciences. 2018;19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Yeldag G, Rice A, Del Rio Hernandez A. Chemoresistance and the Self-Maintaining Tumor Microenvironment. Cancers. 2018;10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Provenzano PP, Cuevas C, Chang AE, Goel VK, Von Hoff DD, Hingorani SR. Enzymatic targeting of the stroma ablates physical barriers to treatment of pancreatic ductal adenocarcinoma. Cancer cell. 2012;21:418–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Li Y, Hong J, Jung BK, Oh E, Yun CO. Oncolytic Ad co-expressing decorin and Wnt decoy receptor overcomes chemoresistance of desmoplastic tumor through degradation of ECM and inhibition of EMT. Cancer letters. 2019;459:15–29. [DOI] [PubMed] [Google Scholar]
- [19].Weaver VM, Lelievre S, Lakins JN, Chrenek MA, Jones JC, Giancotti F, et al. beta4 integrin-dependent formation of polarized three-dimensional architecture confers resistance to apoptosis in normal and malignant mammary epithelium. Cancer cell. 2002;2:205–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Ohlund D, Lundin C, Ardnor B, Oman M, Naredi P, Sund M. Type IV collagen is a tumour stroma-derived biomarker for pancreas cancer. British journal of cancer. 2009;101:91–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Resovi A, Bani MR, Porcu L, Anastasia A, Minoli L, Allavena P, et al. Soluble stroma-related biomarkers of pancreatic cancer. EMBO molecular medicine. 2018;10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Willumsen N, Bager CL, Leeming DJ, Smith V, Karsdal MA, Dornan D, et al. Extracellular matrix specific protein fingerprints measured in serum can separate pancreatic cancer patients from healthy controls. BMC cancer. 2013;13:554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Harris MA, Clark J, Ireland A, Lomax J, Ashburner M, Foulger R, et al. The Gene Ontology (GO) database and informatics resource. Nucleic acids research. 2004;32:D258–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Kandimalla R, Ozawa T, Gao F, Wang X, Goel A. Gene Expression Signature in Surgical Tissues and Endoscopic Biopsies Identifies High-Risk T1 Colorectal Cancers. Gastroenterology. 2019;156:2338–41.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Yang C, Liu Z, Zeng X, Wu Q, Liao X, Wang X, et al. Evaluation of the diagnostic ability of laminin gene family for pancreatic ductal adenocarcinoma. Aging. 2019;11:3679–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Kosanam H, Prassas I, Chrystoja CC, Soleas I, Chan A, Dimitromanolakis A, et al. Laminin, gamma 2 (LAMC2): a promising new putative pancreatic cancer biomarker identified by proteomic analysis of pancreatic adenocarcinoma tissues. Molecular & cellular proteomics : MCP. 2013;12:2820–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Groot VP, Rezaee N, Wu W, Cameron JL, Fishman EK, Hruban RH, et al. Patterns, Timing, and Predictors of Recurrence Following Pancreatectomy for Pancreatic Ductal Adenocarcinoma. Annals of surgery. 2018;267:936–45. [DOI] [PubMed] [Google Scholar]
- [28].Marechal R, Bachet JB, Mackey JR, Dalban C, Demetter P, Graham K, et al. Levels of gemcitabine transport and metabolism proteins predict survival times of patients treated with gemcitabine for pancreatic adenocarcinoma. Gastroenterology. 2012;143:664–74.e6. [DOI] [PubMed] [Google Scholar]
- [29].Farrell JJ, Elsaleh H, Garcia M, Lai R, Ammar A, Regine WF, et al. Human equilibrative nucleoside transporter 1 levels predict response to gemcitabine in patients with pancreatic cancer. Gastroenterology. 2009;136:187–95. [DOI] [PubMed] [Google Scholar]
- [30].Marechal R, Mackey JR, Lai R, Demetter P, Peeters M, Polus M, et al. Human equilibrative nucleoside transporter 1 and human concentrative nucleoside transporter 3 predict survival after adjuvant gemcitabine therapy in resected pancreatic adenocarcinoma. Clinical cancer research : an official journal of the American Association for Cancer Research. 2009;15:2913–9. [DOI] [PubMed] [Google Scholar]
- [31].Von Hoff DD, Ramanathan RK, Borad MJ, Laheru DA, Smith LS, Wood TE, et al. Gemcitabine plus nab-paclitaxel is an active regimen in patients with advanced pancreatic cancer: a phase I/II trial. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2011;29:4548–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Hidalgo M, Plaza C, Musteanu M, Illei P, Brachmann CB, Heise C, et al. SPARC Expression Did Not Predict Efficacy of nab-Paclitaxel plus Gemcitabine or Gemcitabine Alone for Metastatic Pancreatic Cancer in an Exploratory Analysis of the Phase III MPACT Trial. Clinical cancer research : an official journal of the American Association for Cancer Research. 2015;21:4811–8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


