Abstract
Diphtheria is a respiratory disease caused by the bacterium Corynebacterium diphtheriae. Although the development of a toxin-based vaccine in the 1930s has allowed a high level of control over the disease, cases have increased in recent years. Here, we describe the genomic variation of 502 C. diphtheriae isolates across 16 countries and territories over 122 years. We generate a core gene phylogeny and determine the presence of antimicrobial resistance genes and variation within the tox gene of 291 tox+ isolates. Numerous, highly diverse clusters of C. diphtheriae are observed across the phylogeny, each containing isolates from multiple countries, regions and time of isolation. The number of antimicrobial resistance genes, as well as the breadth of antibiotic resistance, is substantially greater in the last decade than ever before. We identified and analysed 18 tox gene variants, with mutations estimated to be of medium to high structural impact.
Subject terms: Bacterial evolution, Bacterial genomics, Bacterial toxins, Pathogens
Cases of diphtheria have increased in recent years. Here, the authors analyse the genomes of 502 Corynebacterium diphtheriae isolates across 16 countries and territories over 122 years, describing an increase in antimicrobial resistance genes and identifying toxin variants.
Introduction
Diphtheria, traditionally regarded as a toxin-driven disease of the respiratory system, usually begins with angina or tonsillitis symptoms, sore throat and mild fever. However, the clinical picture can quickly escalate leading to death if the disease is not treated1. A white-grey pseudomembrane over the pharynx, larynx and tonsils is considered stereotypical to the disease, as is a swollen bull neck, although these symptoms are not displayed in all the infected cases2,3. Once a common cause of infection, diphtheria has been vaccine preventable for decades and is now rarely observed in high income countries2,4. In low and middle income countries, however, diphtheria is still of concern as it can cause sporadic infections or outbreaks in unvaccinated and partially vaccinated communities3,5. Any drop in the levels of diphtheria vaccine coverage could potentially lead to an opportunistic return of this communicable disease, as already being reported for other vaccine-preventable diseases like measles and pertussis6–9. The number of diphtheria cases reported globally has followed a gradually increasing trend in recent years, with the cases in 2018 (16,651) being over double the 1996–2017 average (8105)10.
Diphtheria is primarily caused by toxigenic Corynebacterium diphtheriae, that colonise the upper respiratory tract11. The route of transmission between humans is likely through droplets contaminated with C. diphtheriae12. The diphtheria toxin is encoded on a tox+ corynephage in C. diphtheriae. Related species including C. ulcerans and C. pseudotuberculosis can also be lysogenised by this phage type, and such strains are also associated with a clinical disease almost identical to diphtheria13–16. In addition, non-toxigenic C. diphtheriae can cause disease, often in the form of systemic infections17,18. Non-toxigenic but toxin gene-bearing (NTTB) C. diphtheriae have also been recorded19,20. Since the universal diphtheria vaccine is a partially pure toxoid formulation, it may not be as effective for preventing NTTB led infections. Acute diphtheria is usually treated with anti-diphtheria toxin serum, alongside a course of antibiotics21. While C. diphtheriae resistant to antibiotics have been reported, the extent of such resistance in this pathogen remains largely unknown2,22–24. A complete reference genome for C. diphtheriae has been available for almost two decades25. Subsequent genomic analysis has predominantly focused on small, geographically clustered isolates, alongside reference genomes to provide a context26–29.
In this work we seek to understand the genomic dynamics of C. diphtheriae more widely with a detailed focus on India, where over 50% of globally reported cases occurred in 201810. We interrogate the genomes of a large collection of C. diphtheriae isolates to determine their phylogenetic structure, assess the presence of antimicrobial resistance (AMR) genes and assess toxin variation, which is the key target of the current diphtheria vaccine.
Results
Genomic insight into 122 years (1896–2018) of C. diphtheria
A collection of 502 C. diphtheriae genomes was established by sequencing 61 novel Indian isolates and combining these with 441 publicly available genomes. The isolates in our study cover 16 countries and territories and they were collected across a period of 122 years. As India has reported the highest number of cases in the last few years, a separate Indian subset totalling 122 genomes with two isolates from 1973 and the rest collected between 2015 and 2018 (including the 61 novel isolates), was created and analysed.
An initial analysis of the total 502 C. diphtheriae genomes showed that, as previously reported, it is a genetically diverse species. An initial mapping analysis revealed evidence for extensive recombination across the whole species, not conducive to an accurate phylogeny (Supplementary Fig. 1). 100 Multilocus sequence types (MLST) were determined, with 73 isolates designated as ‘novel STs’. The core gene list was calculated to be 1035 genes with an extended pan gene list accumulating to 23,447 genes. The Indian subset had 1367 core genes but a lower number of 7436 genes in the pan gene list across all 122 genomes (Supplementary Fig. 2).
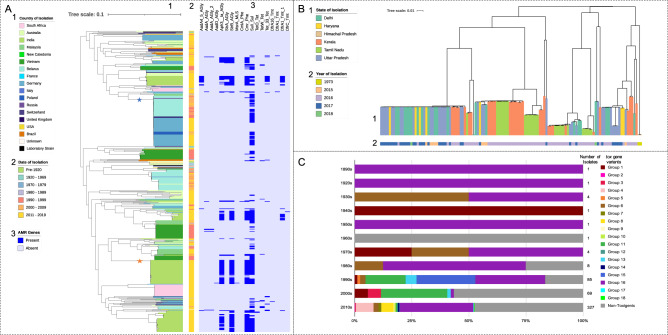
Figure 1A displays the outcome of the phylogenetic analysis of all 502 genomes included in our study. Built from a 49,454 nucleotide long core gene single nucleotide polymorphism (SNP) alignment, the phylogenetic tree in Fig. 1A indicated the presence of several clusters across the collection, each containing isolates from wide temporal and spatial ranges. Within these clusters, single monophyletic groups were readily identifiable, and these predominantly showed strong geographic and temporal association. Isolates within a particular monophyletic group shared a single ST, although some individual isolates within these groups were also ‘novel STs’. One subclade spread extensively to cover multiple neighbouring European countries—predominantly Belarus and Germany—and persisted over a large period of time (marked by a blue star). Isolates within this lineage were isolated as early as 1996 and persisted until at least 2017 when isolates were found in Germany. Clusters contained groups isolated from multiple continents, most commonly Asia and Europe. This clearly indicates that C. diphtheriae has been established in the human population for at least over a century and has spread, potentially by population movements, across time and space. This also shows that there are multiple distinct clonal populations circulating in the same geographical setting1,29.
Fig. 1. Global and Indian core gene phylogenies of Corynebacterium diphtheriae and tox gene variants by decade.
A Maximum likelihood phylogenetic tree based on the core gene single nucleotide polymorphisms from the 502 global Corynebacterium diphtheriae genome collection. The country of isolation (1), decade of isolation (2), and AMR gene presence/absence heatmap made using ARIBA50 (3), are all annotated. Most monophyletic groups within the tree represent only one country and decade, with the major exception of the large Belarussian/German dominated group, marked with a blue star. Both the blue and orange stars highlight groups used for BEAST analysis. B The core gene maximum likelihood phylogeny of only the 122 Indian isolates, coloured by state (1) and year of isolation (2). C The proportion of the 18 tox gene variants found across 291 tox+ and 211 non-toxigenic isolates per decade, with the number of isolates per decade shown.
Within India, isolates from individual monophyletic groups were from multiple states, spanning both Northern and Southern India (Fig. 1B). The closely related monophyletic groups of similar origins suggest clonal outbreaks occurring opportunistically, perhaps emerging from commensalism when favourable factors occurred.
tox gene diversity
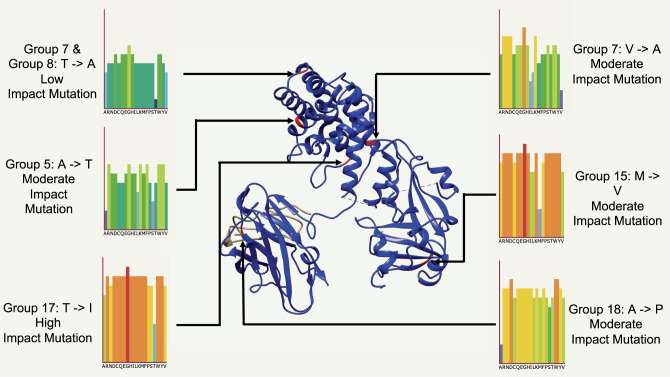
Diphtheria toxin is the main virulence-associated factor in the disease. In total, 18 allelic variants of the tox gene were found across the 291 tox+ isolates, which make 58% of the total collection of 502. Of these 18 allelic types, eight were found to contain non-synonymous SNP changes with types 5, 8, 14, 15, 17 and 18 each containing one amino acid substitution. Toxin type 7 contained two amino acid substitutions, one of which was shared with type 8. Group 13 instead harboured a deletion, previously reported to result in the strain being NTTB. The impact of these non-synonymous mutations on the protein structure was estimated using PHYRE2 and SuSPect, and were plotted onto the diphtheria toxin protein model 1XDT (https://www.rcsb.org/structure/1xdt) from the Protein Data bank using UCSF ChimeraX (Fig. 2)30–34. Groups 13 deletion and group 14 mutations were both present in the signal sequence of the gene, and thus could not be mapped to the protein structure. Groups 7 and 8 shared mutation was estimated to have low impact, the second mutation in group 7 as well as groups 5, 15, 18 were calculated as moderate impact mutation, while group 17 mutation was calculated as a high impact mutation. Notably, the site of groups 15, 17 and 18 mutations had a much higher average mutation impact, with many of the potential amino acid substitutions showing a high risk of mutations impacting on the toxin structure. The proportions of the tox allelic variants found in each decade is shown in Fig. 1C. The most common toxin variant was type 16 (49.5% of toxigenic isolates), with 4, 6, 8, 11 and 15 being the next most common alleles. Type 16 also includes the tox variant found in the vaccine strain, PW835. Three C. diphtheriae isolates carried two identical copies of the tox gene, two type 7 isolated in Germany and Switzerland, and one type 16 isolated in Germany. tox variant diversity significantly increases by decade (r (9) = 0.70, p = 0.02), although this may be because of unavoidable bias of sampling. Supplementary Figs. 3, 4 show the breakdown of tox types by country of origin and year of isolation. Non-toxigenic isolates were much more common in mainland Europe, Brazil and Australia than in India and Southeast Asia, as well as being more common in more recently isolated genomes (non-toxigenic isolates per decade (r (9) = 0.60, p = 0.05). The difference between the number of non-toxigenic and toxigenic isolates in HICs and LMICs was significantly different (r (1) = 202.67, p < 0.001). Group 16 was the predominant variant across South Asia, while being much less common outside the region. Group 16 was also the predominant variant in recent years, despite the rise in non-toxigenic isolates. Some tox variants were present only within a single monophyletic group and location e.g. type 8, only present in 18 South African isolates, and groups 17 and 18, only present in one isolate each from Australia. Others including types 4 and 16 were present throughout the phylogeny, across numerous clusters and groups.
Fig. 2. Six non-synonymous mutations plotted onto diphtheria toxin model 1XDT (https://www.rcsb.org/structure/1xdt) from the Protein Data Bank using PHYRE2.
The impact of these mutations is estimated by SuSPect, with a gradient per mutation of low (dark blue) to high (orange/red).
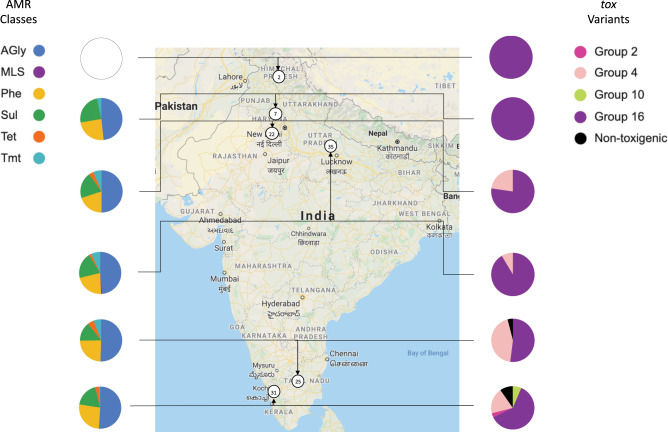
Figure 3 shows the proportion of each tox type per state across India. Four of the eighteen toxin types were represented among the Indian isolates, with Group 16 still being the dominant variant. Types 2 and 11 were only present in Kerala. The only non-toxigenic Indian isolates were found in the South, in Tamil Nadu and Kerala.
Fig. 3. The antimicrobial resistance (AMR) gene and tox gene variant proportions represented in the 122 Corynebacterium diphtheriae genomes from India at state level.
The number of isolates from the six states sampled are shown in circles on the map. The AMR genes are coloured by the classes of antibiotic the genes offer resistance to. AGly (blue) aminoglycosides, MLS (purple) macrolide–lincosamide–streptogramin, Phe (yellow) phenicols, Sul (green) sulfonamides, Tet (orange) tetracyclines, Tmt (light blue) trimethoprim. The map was taken from Google Maps47.
Antimicrobial resistance in C. diphtheria
The AMR gene portfolio (Fig. 1A) and the average number of AMR genes per decade (Supplementary Fig. 5) were evaluated. More recently isolated genomes (including those from India) showed a disproportionately high number of AMR genes, with a significant positive correlation between the average number of AMR genes per genome and the decade of isolation (r (9) = 0.68, p = 0.02). Similar resistance profiles were found across a small number of recent Swiss and German isolates. The sulphonamide resistance gene (sulI) was the most common, being consistently found in Indian and sporadically in other Asian and European isolates. Aminoglycoside (aadA4, aph3_Ia, strA and strB), chloramphenicol (cmr) and trimethoprim (dfrA1) resistance genes were also present in most recent Indian isolates. Only one isolate (from India) was found to contain a macrolide resistance-encoding gene (msrA), while no isolates harboured β-lactam resistance genes. Isolates from the decade of 2010 to 2019 shows the highest average number of AMR genes per genome, almost four times as many genes on average than in the next highest decade; the 1990s (Supplementary Fig. 5). The 2010s also showed more variation in the classes resisted (six), compared to other decades.
Within the Indian isolates, C. diphtheriae from Himachal Pradesh (isolated in 1973) had no detectable AMR genes (Fig. 3). Isolates from Haryana harboured genes encoding resistance to four classes of antibiotic, whereas isolates from Delhi, Kerala and Tamil Nadu had genes for resistance to five. Isolates from the most populous state of India, Uttar Pradesh, showed resistance to six classes, including the only isolate of our study with macrolide resistance. Five isolates from Kerala (all isolated in 2016) and one from Uttar Pradesh (isolated in 2017) showed no AMR genes present in their genomes. Phenotypic AMR testing was carried out for the 61 novel Indian isolates and these results align well with the genes found in silico (Supplementary Data 1).
Time scaled phylogenetic analysis
The European and Indian groups (marked with blue and orange stars respectively on Fig. 1A) are shown in Supplementary Figs. 6, 7. BEAST estimates the European group’s most recent shared ancestor was in September 1983. Three clades are present within the main European clade with the first spanning March 1987–2008. The second and third clades are estimated to have diverged in July 1985 with the former present from September 1988 to 2010 and the latter estimated to being in circulation between April 1990 and 2017. The ratio of pre- and post-recombination removal SNPs included in the alignment was 1.1 (1944:1783).
The Indian group is estimated to have shared an ancestor with the reference isolate NCTC11397 in October 1955, before diverging into two clades in February 2009. The first clade is estimated to have run from April 2012 to 2017, with the second from June 2013 to 2018. The ratio of pre- and post-recombination removal SNPs included in the alignment was 1.2 (26879:22717).
Corynephage variation
The maximum likelihood phylogeny of 11 toxigenic corynephages from complete C. diphtheriae genomes can be seen in Supplementary Fig. 8, based on a mapped alignment of 36,570 nucleotide bases. There were no major phylogenetic correlations with the country of isolation and the decade of isolation as closely related corynephages being isolated over large temporal and geographical distances. The tox gene variant carried by the corynephage also does not correspond with the phylogenetic structure, with groups 6 and 16 found in distantly related isolates across the phylogeny.
Discussion
By analysing a large collection of C. diphtheriae genomes we identified numerous clades distributed across the globe. Individual clades contained monophyletic groups isolated from multiple countries and regions across our sampling time frame. In India, where the majority of recently reported diphtheria cases originate, numerous monophyletic groups were found across the phylogeny as several independent lineages. These sub-lineages of C. diphtheriae thus represent a persisting and apparently successful diverse population. This is in contrast to other bacterial pathogens such as Mycobacterium abscessus, which are represented by largely clonal lineages that have spread across the globe. The high level of diversity and recombination once again highlights the major challenges of creating high confidence phylogenies of C. diphtheriae. Core gene approaches remain the most robust methodologies for building phylogenies of large C. diphtheriae collections to cope with the large diversity and recombination across major clades. Mapping approaches and recombination removal have been demonstrated as possible for closely related groups, highlighting the need for more high-quality reference genomes representing these currently circulating lineages. We were able to use BEAST to create time-scaled phylogenies of individual closely related monophyletic groups, reaffirming the importance of these high-quality reference genomes. This demonstrates that time-scaled phylogenies could be used to accurately analyse individual outbreaks and support public health measures, provided a single group of C. diphtheriae is responsible, rather than numerous clusters concurrently as seems to be the case in India and other parts of the world.
Although our study is based on a comprehensive clinical strain collection, our study lacks data on carriage isolates from non-clinical cases, as C. diphtheriae is not routinely screened for in healthy individuals. Nevertheless, the spatio-temporal structure of the phylogeny suggests that carriage could be playing a significant role in the overall persistence and evolution of the C. diphtheriae species. Therefore, in future studies, it is important to monitor the human population for asymptomatic carriage in both vaccinated and unvaccinated individuals, especially in at-risk communities and known epidemiological hot spots.
While our data does not, at present, highlight any efficacy concern in the currently used tox variant type 16 based diphtheria toxoid vaccine, the continually increasing toxin diversity and prevalence of non-toxigenic strains do however forecast a real possibility of vaccine escape and anti-toxin treatment failure in future. While the current vaccine against diphtheria continues to remain largely effective, it is vital to conduct further in vitro and in vivo studies to investigate the benefit, if any, that the six variants with non-synonymous allelic mutations that impact protein structure may provide to C. diphtheriae, especially those estimated to have moderate and high impacts on structure. Further sequencing of C. diphtheriae isolates may reveal additional variant types and show the full extent of the spread of the variants catalogued by us. This evolutionary tracking is of critical importance as it will forewarn the public health agencies of any possible vaccine escape or anti-toxin treatment failure, allowing the planning of early alternate intervention. Our analysis of the corynephage genome, while preliminary, suggests the diversity in the tox gene is not merely a product of wider phage diversity. Due to being based on only the limited number of complete genomes that carried the corynephage, this conclusion requires further investigation as the number of public completed C. diphtheriae genomes increases. With a steady global increase in vaccination rates for diphtheria, the selection pressure on the toxin as the main antigen is bound to increase, and this may be the reason behind the increase in non-toxigenic isolates recorded, as well as the significantly lower proportion of the 18 tox variants found among higher income countries. Among the isolates from Europe, where variant 16 now makes up only a small fraction of tox variants found, and where non-toxigenic isolates have been more commonly reported, further research utilising this toxin diversity information is imperative, as is more detailed investigations into the mechanisms of non-toxigenic infection. Using advanced phylogenomics, our data guides public health preparedness and suggests that a plan B in hybrid toxin-based vaccine candidates and antitoxins must be pre-emptively prepared for action if a non-synonymous non-group 16 toxin carrying C. diphtheriae were to abruptly become prevalent.
AMR has rarely been considered as a major problem in the treatment of C. diphtheriae. Here, we demonstrate that recently in some parts of the world, genomes are acquiring resistance to numerous classes of antibiotics, likely driven through the overall exposure to antimicrobials and the common co-occurrence of different resistance elements on the same mobile elements. The resistance genotypes are not limited to a single country or region but present in both Asia and Europe. Despite erythromycin and penicillin being the traditionally recommended antibiotics of choice for treating confirmed cases of early-stage diphtheria, only one macrolide resistance gene was found, and no resistance to β-lactamases. This indicates that AMR acquisition in C. diphtheriae could be happening during its purported carriage state and in response to the exposure to antibiotics in their environment or during patient treatment against other infections. The independent acquisition of highly similar AMR genes in C. diphtheriae indicates recombination between these lineages and acquisition of mobile elements from other species. It could also be due to collateral selection, where pressure to acquire resistance to one antimicrobial agent may drive the advancement of resistance to other agents by its very development36.
Although the results from our study are based on a comprehensive clinical strain collection of isolates and genomes, our study lacks data on carriage isolates, which could be playing a highly significant role in the overall evolution of the C. diphtheriae species. Our phylogenomic findings strengthen the hypothesis that while the diphtheria vaccine may be effective in preventing the symptomatic infection, carriage in vaccinated individuals or asymptomatically infected individuals could be continuing to provide a suitable ecosystem for C. diphtheriae sustenance for prolonged periods. COVID-19 has negatively impacted childhood vaccination schedules worldwide, with diphtheria doses scheduled at 6, 10 and 14 weeks. This coupled with reported case numbers rising over the past decade, and the year 2018 showing the highest incidence in 22 years, it is more important than ever to understand this historically important disease, to prevent it from becoming a major global threat ever again in its original or a modified, better adapted, form.
Methods
Data collection
Sixty-one novel isolates were identified from 3 Northern Indian states. Of these, 22 isolates were from patients in Delhi, 7 from Haryana, and 32 from Uttar Pradesh. Nine were isolated in 2015, 16 in 2016, 34 in 2017, and 2 in 2018. Genomes of all these isolates have been made publicly available, with accession numbers found in Supplementary Data 1. Novel Indian isolates were grown, and DNA extracted for sequencing. Whole Genome Sequencing was carried out using an Illumina HiSeq v4 platform at Wellcome Sanger Institute, producing short read whole genome sequences that were assembled and annotated using SPAdes assembly (v3.13.0) and Prokka annotation (v1.5)37,38. Publicly available genomic data and metadata were collated following a literature review, to frame a global representation. We also analysed our novel isolates within an Indian context, to allow comparison between the global picture and the country with the highest reported case numbers to the World Health Organisation. Microreact (v5.93.0) plots have been produced for both the global (https://microreact.org/project/CZaifLUuW) and the Indian collections (https://microreact.org/project/TuIdKrIfc) to aid in data sharing39.
Core gene phylogeny
Due to the extremely high recombinogenic variation between the genomes of C. diphtheriae identified using Gubbins (v2.4.1) and Phandango (v0.9)40,41, which is not conducive to an accurate phylogeny, we chose Roary (v3.13.0)42 instead to investigate the phylogenetic relationships between isolates. Using annotated genomes produced by Prokka (v1.5)38, Roary extracted the genes present in 99% of the isolates, concatenating them to produce a core gene alignment. The number of genes across the pan-genome categories was determined. SNPs were identified using SNP-sites (v2.5.1)43 and used to produce a core gene maximum likelihood phylogenetic tree, using IQ-TREE (v1.6.10) and the inbuilt ModelFinder over 1000 pseudo-bootstrap replicates44,45.
Genomic analysis
Results, alongside the spatial and temporal metadata, were annotated to the phylogenetic trees using the Interactive Tree of Life (iTOL) (v5.5.1)46, to determine any epidemiological relatedness between the different phylogenetic and phylogeographic clades. To display the toxin variants and AMR genes per Indian state, a map was taken from Google Maps47. The MLST of all isolates was determined using MLSTcheck (v2.1.1706216)48.
tox Gene analysis
Variation in the tox gene was investigated by obtaining the gene sequence from 291 toxigenic isolates using in silico PCR49. Primers are available in Supplementary Table 1. The non-synonymous mutations found were plotted onto diphtheria toxin model 1XDT from the Protein Data Bank using UCSF ChimeraX (v1.1), and the risk of those mutations impacting on protein structure was calculated using PHYRE2 (v2.0) and the inbuilt SuSPect30–34.
Antimicrobial resistance testing
ARIBA (v2.14.6) was used to interrogate the genome for the presence of antimicrobial resistance (AMR) genes, to investigate if resistance was now becoming a concern in C. diphtheriae globally50. Antimicrobial susceptibility test was performed using MICs gradient test (E-test, BioMèrieux, Marcy, l’Etoile, France). The E-test strips of ampicillin (AM), amoxicillin/clavulanic acid (XL), azithromycin (AZ), ciprofloxacin (CI), chloramphenicol (CL), clindamycin (CM), cefotaxime (CT), doxycycline (DC), erythromycin (EM), gentamycin (GM), imipenem (IP), linezolid (LZ), moxyfloxacin (MX), penicillin (PG), rifampicin (RI), tetracycline (TC, ceftriaxone (TX), vancomycin (VA), trimethoprime–sulphamethoxazole (SXT) on Mueller-Hinton agar plates with 5% horse blood. Corynebacterium spp. MIC interpretative standard was based on the Clinical and Laboratory Standards Institute51.
Statistics
RStudio (v4.6.1) was used to carry out Pearson’s product-moment correlation to determine the significance between decade of isolation and tox gene variety, the average number of AMR genes per genome, and the number of non-toxigenic isolates per decade, while a chi-squared test of non-toxigenic and toxigenic isolates from HICs and LMICs52,53.
Time scaled phylogenetic analysis
Two closely related clades were chosen for time-scaled phylogenetic analysis. Recombination was removed from the mapped alignments using Gubbins (v2.4.1) before using the BEAST (v1.10) package to carry out the Bayesian time-scaled phylogenetic analysis40,54. The Hasegawa, Kishino and Yano model (HKY) substitution model with different demographic models (Bayesian skyline, exponential and constant) was investigated. Markov chain Monte Carlo runs of 100 million generations were carried out with sampling of 20,000 generations55. The convergence of each run was manually inspected using Tracer (v1.7)56. A burn in of 20% was discarded from the runs and maximum clade credibility tree was finally generated using Treeannotator (v1.8.2)57. The annotated phylogenetic tree was visualised using FigTree (v1.4)58.
Corynephage analysis
Corynephage diversity was investigated by mapping the 11 toxigenic complete genomes publicly available from NCBI Genbank to the corynephage sequence annotated in C. diphtheriae isolate NCTC 13129 using BWA (v0.7.17-r1188)59,60. This mapped alignment was then used to produce a maximum likelihood phylogenetic tree with IQ-TREE (v1.6.10) and the inbuilt ModelFinder over 1000 pseudo-bootstrap replicates, and annotated using iTOL (v5.5.1)44–46.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Descriptions of Additional Supplementary Files
Acknowledgements
The authors thank Prof. P.A. Hoskisson, and Z.A. Dyson for their expert knowledge and advice. This work primarily received funding from the Medical Research Council under the University of Cambridge Medical Research Council Doctoral Training Programme. Additionally, the Horizon 2020-MSCA-IF-2018; 843405-DIFTERIA. This research was supported by the NIHR Cambridge Biomedical Research Centre and AMR Research Capital Funding Scheme [NIHR200640]. The views expressed are those of the author(s) and not necessarily those of the NIHR or the Department of Health and Social Care.
Source data
Author contributions
Conceptualisation: R.C.W., T.R.M., N.C.S., G.D. and A.M. Data curation: R.C.W. Formal analysis: R.C.W., A.K.P., K.V., and D.K. Funding acquisition: R.C.W., T.R.M., N.C.S., B.V., V.M., G.D., and A.M. Investigation: T.R.M. and N.C.S., Methodology: R.C.W., E.H., V.S., G.D., A.M. Supervision: B.V., L.S., P.H., B.D., E.H., V.M., S.B., V.S., G.D. and A.M. Validation: R.C.W. Visualisation: R.C.W. and K.V. Writing—original draft: R.C.W. Writing—review and editing: all authors.
Data availability
The novel genome sequences generated during and analysed during the current study are available in the NCBI Genbank repository, under the study ID PRJEB20897. Source data are provided with this paper.
Competing interests
The authors declare no competing interests.
Footnotes
Peer review information Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. Peer review reports are available.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Robert C. Will, Thandavarayan Ramamurthy, Naresh Chand Sharma.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-021-21870-5.
References
- 1.Sangal V, Hoskisson PA. Evolution, epidemiology and diversity of Corynebacterium diphtheriae: new perspectives on an old foe. Infect. Genet. Evol. 2016;43:364–370. doi: 10.1016/j.meegid.2016.06.024. [DOI] [PubMed] [Google Scholar]
- 2.Edwards D, et al. Transmission of toxigenic Corynebacterium diphtheriae by a fully immunised resident returning from a visit to West Africa, United Kingdom, 2017. Eur. Surveill. 2018;23:1700681. doi: 10.2807/1560-7917.ES.2018.23.39.1700681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jain A, Samdani S, Meena V, Sharma MP. Diphtheria: It is still prevalent!!! Int. J. Pediatr. Otorhinolaryngol. 2016;86:68–71. doi: 10.1016/j.ijporl.2016.04.024. [DOI] [PubMed] [Google Scholar]
- 4.Dangel A, Berger A, Konrad R, Bischoff H, Sing A. Geographically diverse clusters of nontoxigenic corynebacterium diphtheriae infection, Germany, 2016-2017. Emerg. Infect. Dis. 2018;24:1239–1245. doi: 10.3201/eid2407.172026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.World Health Organisation. WHO | Data, statistics and graphics. WHO (2019).
- 6.Kubin L. Is there a resurgence of vaccine preventable diseases in the U.S.? J. Pediatr. Nurs. 2019;44:115–118. doi: 10.1016/j.pedn.2018.11.011. [DOI] [PubMed] [Google Scholar]
- 7.Nelson R. US measles outbreak concentrated among unvaccinated children. Lancet Infect. Dis. 2019;19:248. doi: 10.1016/S1473-3099(19)30074-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hopkins J, York TN. Measles: two US outbreaks are blamed on low vaccination rates. BMJ. 2019;364:l312. doi: 10.1136/bmj.l312. [DOI] [PubMed] [Google Scholar]
- 9.National Statistics. Childhood vaccination coverage statistics—England 2018–19—NHS digital. Childhood Vaccination Coverage Statisticshttps://digital.nhs.uk/data-and-information/publications/statistical/nhs-immunisation-statistics/england-2018-19 (2019).
- 10.World Health Organisation. Diphtheria reported cases. WHO vaccine-preventable diseases: monitoring system 2018 global summaryhttp://apps.who.int/immunization_monitoring/globalsummary/timeseries/tsincidencediphtheria.html (2019).
- 11.Sharma NC, et al. Diphtheria. Nat. Rev. Dis. Prim. 2019;5:81. doi: 10.1038/s41572-019-0131-y. [DOI] [PubMed] [Google Scholar]
- 12.Hoskisson PA. Microbe profile: corynebacterium diphtheriae-an old foe always ready to seize opportunity. Microbiology. 2018;164:865–867. doi: 10.1099/mic.0.000627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wagner KS, et al. Diphtheria in the postepidemic period, Europe, 2000–2009. Emerg. Infect. Dis. 2012;18:217–225. doi: 10.3201/eid1802.110987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guaraldi, A. L. de M., Hirata, R. & Azevedo, V. A. de C. Corynebacterium diphtheriae, corynebacterium ulcerans and corynebacterium pseudotuberculosis—general aspects. in Corynebacterium diphtheriae and Related Toxigenic Species 15–37 (Springer Netherlands, 2014).
- 15.Wagner, K. S. et al. Diphtheria surveillance. in Corynebacterium diphtheriae and Related Toxigenic Species 207–224 (Springer Netherlands, 2014).
- 16.Moore LSP, et al. Corynebacterium ulcerans cutaneous diphtheria. Lancet Infect. Dis. 2015;15:1100–1107. doi: 10.1016/S1473-3099(15)00225-X. [DOI] [PubMed] [Google Scholar]
- 17.Benamrouche N, et al. Microbiological and molecular characterization of corynebacterium diphtheriae isolated in Algeria between 1992 and 2015. Clin. Microbiol. Infect. 2016;22:1005.e1–1005.e7. doi: 10.1016/j.cmi.2016.08.013. [DOI] [PubMed] [Google Scholar]
- 18.Sangal V, et al. Adherence and invasive properties of Corynebacterium diphtheriae strains correlates with the predicted membrane-associated and secreted proteome. BMC Genom. 2015;16:765. doi: 10.1186/s12864-015-1980-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zakikhany K, Neal S, Efstratiou A. Emergence and molecular characterisation of non-toxigenic tox gene-bearing Corynebacterium diphtheriae biovar mitis in the United Kingdom, 2003–2012. Eurosurveillance. 2014;19:20819. doi: 10.2807/1560-7917.ES2014.19.22.20819. [DOI] [PubMed] [Google Scholar]
- 20.Billard-Pomares T, et al. Diagnosis of a non-toxigenic tox gene-bearing strain of Corynebacterium diphtheriae in a young male back from Senegal to France. Open Forum Infect. Dis. 2017;4:ofw271. doi: 10.1093/ofid/ofw271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Both L, White J, Mandal S, Efstratiou A. Access to diphtheria antitoxin for therapy and diagnostics. EuroSurveillance. 2014;19:20830. doi: 10.2807/1560-7917.ES2014.19.24.20830. [DOI] [PubMed] [Google Scholar]
- 22.Floros P, Gunaratne DA, Chang A, Coman WB. A rare case of toxigenic diphtheria tonsillitis resistant to penicillin causing sepsis and death. Aust. J. Otolaryngol. 2018;1:29. doi: 10.21037/ajo.2018.10.01. [DOI] [Google Scholar]
- 23.FitzGerald RP, Rosser AJ, Perera DN. Non-toxigenic penicillin-resistant cutaneous C. diphtheriae infection: A case report and review of the literature. J. Infect. Public Health. 2015;8:98–100. doi: 10.1016/j.jiph.2014.05.006. [DOI] [PubMed] [Google Scholar]
- 24.Mina NV, et al. Canada’s first case of a multidrug-resistant corynebacterium diphtheriae strain, isolated from a skin abscess downloaded from. J. Clin. Microbiol. 2011;49:4003–4005. doi: 10.1128/JCM.05296-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cerdeño-Tárraga AM, et al. The complete genome sequence and analysis of Corynebacterium diphtheriae NCTC13129. Nucleic Acids Res. 2003;31:6516–6523. doi: 10.1093/nar/gkg874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lodeiro-Colatosti A, et al. Diphtheria outbreak in Amerindian communities, Wonken, Venezuela, 2016–2017. Emerg. Infect. Dis. 2018;24:1340–1344. doi: 10.3201/eid2407.171712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Santos LS, et al. Diphtheria outbreak in Maranhão, Brazil: microbiological, clinical and epidemiological aspects. Epidemiol. Infect. 2014;143:791–798. doi: 10.1017/S0950268814001241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mahomed S, et al. An isolated outbreak of diphtheria in South Africa, 2015. Epidemiol. Infect. 2017;145:2100–2108. doi: 10.1017/S0950268817000851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grosse-Kock S, et al. Genomic analysis of endemic clones of toxigenic and non-toxigenic Corynebacterium diphtheriae in Belarus during and after the major epidemic in 1990s. BMC Genom. 2017;18:873. doi: 10.1186/s12864-017-4276-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJE. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015;10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yates CM, Filippis I, Kelley LA, Sternberg MJE. SuSPect: Enhanced prediction of single amino acid variant (SAV) phenotype using network features. J. Mol. Biol. 2014;426:2692–2701. doi: 10.1016/j.jmb.2014.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goddard TD, et al. UCSF ChimeraX: meeting modern challenges in visualization and analysis. Protein Sci. 2018;27:14–25. doi: 10.1002/pro.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Berman H, Henrick K, Nakamura H. Announcing the worldwide Protein Data Bank. Nat. Struct. Mol. Biol. 2003;10:980–980. doi: 10.1038/nsb1203-980. [DOI] [PubMed] [Google Scholar]
- 34.Louie GV, Yang W, Bowman ME, Choe S. Crystal structure of the complex of diphtheria toxin with an extracellular fragment of its receptor. Mol. Cell. 1997;1:67–78. doi: 10.1016/S1097-2765(00)80008-8. [DOI] [PubMed] [Google Scholar]
- 35.Trost E, et al. Pangenomic study of corynebacterium diphtheriae that provides insights into the genomic diversity of pathogenic isolates from cases of classical diphtheria, endocarditis, and pneumonia. J. Bacteriol. 2012;194:3199–3215. doi: 10.1128/JB.00183-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Love WJ, Zawack KA, Booth JG, Grӧhn YT, Lanzas C. Markov networks of collateral resistance: national antimicrobial resistance monitoring system surveillance results from escherichia coli isolates, 2004–2012. PLoS Comput. Biol. 2016;12:1005160. doi: 10.1371/journal.pcbi.1005160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bankevich A, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 39.Argimón S, et al. Microreact: visualizing and sharing data for genomic epidemiology and phylogeography. Microb. Genom. 2016;2:e000093. doi: 10.1099/mgen.0.000093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Croucher NJ, et al. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res. 2015;43:e15. doi: 10.1093/nar/gku1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hadfield J, et al. Phandango: an interactive viewer for bacterial population genomics. Bioinformatics. 2018;34:292–293. doi: 10.1093/bioinformatics/btx610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Page AJ, et al. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics. 2015;31:3691–3693. doi: 10.1093/bioinformatics/btv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Page AJ, et al. SNP-sites: rapid efficient extraction of SNPs from multi-FASTA alignments. Microb. Genom. 2016;2:e000056. doi: 10.1099/mgen.0.000056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nguyen L-T, Schmidt HA, Von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kalyaanamoorthy S, Minh BQ, Wong TKF, Von Haeseler A, Jermiin LS. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods. 2017;14:587–589. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Letunic I, Bork P. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016;44:W242–W245. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Google Maps. India—Google Maps. https://www.google.co.uk/maps (2020).
- 48.Page AJ, Taylor B, Keane JA. Multilocus sequence typing by blast from de novo assemblies against PubMLST. J. Open Source Softw. 2016;8:118. doi: 10.21105/joss.00118. [DOI] [Google Scholar]
- 49.Harris, S. R. GitHub—simonrharris/in_silico_pcr: script to run an in silico pcr for a set of primer pairs on an assembly. https://github.com/simonrharris/in_silico_pcr (2016).
- 50.Hunt M, et al. ARIBA: rapid antimicrobial resistance genotyping directly from sequencing reads. Microb. Genom. 2017;3:e000131. doi: 10.1099/mgen.0.000131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Clinical and Laboratory Standards Institute. Methods for Antimicrobial Dilution and Disk Susceptibility Testing of Infrequently Isolated or Fastidious Bacteria, CLSI Guideline M45 (Standards Institute, 2015). [DOI] [PubMed]
- 52.RStudio Team. RStudio: integrated development for R. (RStudio, Inc., rstudio.com, 2015).
- 53.R Core Team. R: a Language and Environment for Statistical Computing, 3–36 (R Foundation for Statistical Computing, 2014).
- 54.Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 2007;7:1–8. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rue Hå, Martino S. Approximate Bayesian inference for hierarchical Gaussian Markov random field models. J. Stat. Plan. Inference. 2007;137:3177–3192. doi: 10.1016/j.jspi.2006.07.016. [DOI] [Google Scholar]
- 56.Rambaut A, Drummond AJ, Xie D, Baele G, Suchard MA. Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Syst. Biol. 2018;67:901–904. doi: 10.1093/sysbio/syy032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Helfrich, P., Rieb, E., Abrami, G., Lücking, A. & Mehler, A. TREEANNOTATOR: versatile visual annotation of hierarchical text relations. in Proceedings of the Eleventh International Conference on Language Resources and Evaluation (LREC 2018) (European Language Resources Association (ELRA), 2018).
- 58.Rambaut, A. Figtree, version 1.4.3. http://tree.bio.ed.ac.uk/software/figtree (2009).
- 59.Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. Preprint at https://arxiv.org/abs/1303.3997v2 (2013).
- 60.Benson DA, et al. GenBank. Nucleic Acids Res. 2012;41:D36–D42. doi: 10.1093/nar/gks1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Descriptions of Additional Supplementary Files
Data Availability Statement
The novel genome sequences generated during and analysed during the current study are available in the NCBI Genbank repository, under the study ID PRJEB20897. Source data are provided with this paper.