Abstract
CRISPR effector proteins introduce double-stranded breaks into the mammalian genome, facilitating gene editing by non-homologous end-joining or homology-directed repair. Unlike the more commonly studied Cas9, the CRISPR effector protein Cas12a/Cpf1 recognizes a T-rich protospacer adjacent motif (PAM) and can process its own CRISPR RNA (crRNA) array, simplifying the use of multiple guide RNAs. We observed that the Cas12a ortholog of Lachnospiraceae bacterium MA2020 (Lb2Cas12a) edited mammalian genes with efficiencies comparable to those of AsCas12a and LbCas12a. Compared to these well-characterized Cas12a orthologs, Lb2Cas12a is smaller and recognizes a narrow set of PAM TTTV. We introduced two mutations into Lb2Cas12a, Q571K and C1003Y, that increased its cleavage efficiency for a range of target sequences beyond those of the commonly used Cas12a orthologs AsCas12a and LbCas12a. In addition to the canonical TTTV PAM, this variant, Lb2-KY, also efficiently cleaved target regions with CTTN PAMs. Finally, we demonstrated that Lb2-KY ribonucleoprotein (RNP) complexes edited two hemoglobin target regions useful for correcting common forms of sickle-cell anemia more efficiently than commercial AsCas12a RNP complexes. Thus, Lb2-KY has distinctive properties useful for modifying a range of clinically relevant targets in the human genome.
Keywords: CRISPR, Cas12a, genome editing, protein engineering, Lb2Cas12a, hemophilia, homology-directed repair, non-homologous end-joining
Graphical Abstract

CRISPR-Cas12a effector proteins edit mammalian genomes using a T-rich PAM with few off-targets. Tran et al. engineered a Cas12a nuclease from Lachnospiraceae bacterium MA2020. They demonstrated that this variant recognized a broadened PAM and edited mammalian genomes with higher efficiencies than commonly used orthologs, expanding Cas12a utility in genome engineering.
Introduction
CRISPR-Cas systems evolved as a component of bacterial and archaeal adaptive immune systems, protecting them against viruses and other mobile genetic elements.1, 2, 3 Cas effector proteins are guided by small CRISPR RNAs (crRNAs) that recognize complementary DNA or RNA sequences next to a protospacer adjacent motif (PAM). After recognition, the effector protein mediates a double-stranded break in the target nucleotide sequence. With their ease of programmability and high efficiency, CRISPR-Cas proteins have been used in a range of gene engineering applications in various organisms.4 Given the range of potential applications, there is a need for additional high-efficiency effector proteins with diverse PAM preferences, low off-target activities, and low immunogenicity.
Two subtypes of CRISPR-Cas effector proteins are commonly used to engineer mammalian genomes. Subtype II-A includes Cas9 proteins, and subtype V-A includes Cas12a proteins, previously known as Cpf1.5 Although less studied than Cas9, Cas12a has useful properties for many genome-editing applications. Cas12a recognizes T-rich PAMs, enabling targeting of genomic regions that lack the G-rich PAM sequences recognized by Cas9.6 Cas12a mediates more efficient homology-directed repair (HDR), in part because it generates staggered DNA ends and because it rapidly releases the PAM-distal end after cleavage.7, 8, 9 Unlike Cas9, Cas12a has an RNase domain that can excise its crRNA from a larger CRISPR array, simplifying editing or gene regulation with multiple crRNAs.10,11 Cas12a may also be safer than Cas9 because it is less permissive to mismatches between crRNA and target sequences, limiting off-target activities.12,13 In addition, commonly used Cas12a proteins derive from non-pathogenic bacteria, and therefore they are less likely to be recognized by pre-existing host immune responses.14 In contrast, major Cas9 variants derive from Streptococcus pyogenes (SpCas9) and Staphylococcus aureus (SaCas9), two major human pathogens.
To date, the most efficient and widely used Cas12a effector proteins derive from Acidaminococcus sp. (AsCas12a) and Lachnospiraceae sp. (LbCas12a). In addition to its canonical TTTV PAM motif, AsCas12a has been engineered to recognize TACV, TYCV, VTTV, and TTCN while maintaining its target specificity.15,16 However, the AsCas12a gene is relatively large, approximately 3.9 kb, precluding its delivery by adeno-associated virus (AAV).17 LbCas12a is smaller and has been reported to have higher editing efficiency than that of AsCas12, but it also has been shown to have higher off-target activity.7,18, 19, 20
Here we characterize another Cas12a ortholog, that of Lachnospiraceae bacterium MA2020 (Lb2Cas12a). Despite the similarity of its name and origin to LbCas12a, it is more closely related to Butyrivibrio sp. NC3005 Cas12a (Figures 1A and 1B; Figure S1). We show that Lb2Cas12a edits mammalian genomes with efficiencies comparable to AsCas12a and LbCas12a at lentivirally integrated targets. We further engineered Lb2Cas12a, markedly enhancing its editing activity and broadening its PAM recognition to include CTTV as well as its cognate TTTV PAM. We show that the improved Lb2Cas12a, namely Lb2-KY, outperformed AsCas12a and LbCas12a at targets with both TTTN and CTTV PAMs when delivered as plasmids or as ribonucleoproteins (RNPs). In addition, Lb2-KY RNP efficiently edited a key target site of the hemoglobin subunit beta gene, as well as the promoter region of hemoglobin subunit gamma 1 and 2 genes, and the Fc region of human IgG1 gene. Thus, this Lb2Cas12a variant, Lb2-KY, increases the potency and expands the range of Cas12a-mediated gene editing.
Figure 1.
Lb2Cas12a efficiently cleaved mammalian genomes at both integrated and endogenous targets
(A) A phylogenetic tree generated by Phylo.io based on an alignment of Cas12a orthologs of the indicated species.21,22 (B) A representation of the indicated Cas12a orthologs with domains indicated. WED: crRNA binding and processing domains, split by REC and PI domains into WED-I, II, and III (green). REC: DNA binding domains REC1 and REC2 (gray). PI: PAM-interacting domain (orange). RuvC: DNA cleavage domains, split by BH and NUC domains into RuvC-I, II, and III (blue). BH: bridge helix (light green). Nuc: DNA processing domain (red). Number at right indicates the number of amino acids of each ortholog. (C) A diagram illustrating the double-stranded-break-induced gain-of-expression assay used in subsequent figures. A construct with an in-frame (+1) crRNA target sequence (orange) preceding an out-of-frame (+3) luciferase gene (light green) is stably integrated into the genomes of HEK293T cells. Upon cleavage by a Cas12a/crRNA complex, non-homologous end-joining (NHEJ) repair places approximately one-third of the downstream luciferase genes in frame (dark green), enabling their expression.11 Luciferase activity reflects the efficiency of Cas12a-mediated cleavage. (D) A list of target sequences used in subsequent panels with their preceding PAMs (blue). (E) The editing efficiency of AsCas12a, LbCas12a, and Lb2Cas12a, measured with the assay shown in panel (C), were compared using the six indicated lentivirally integrated targets. The means of three independent replicates are shown, with error bars indicating ± standard error of the mean (SEM). The significance of differences with Lb2Cas12a are indicated above bars representing AsCas12a and LbCas12 (∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; values of p > 0.05 are indicated in the graph), as determined by two-way ANOVA, followed by Tukey’s multiple comparison tests. (F) Lb2Cas12a edited an endogenous locus of the DNMT1 gene (DNMT1-3) with high efficiency. A T7E1 assay was used to compare the editing efficiencies of the Cas12a orthologs. Percent edited, indicated beneath the figure, is calculated as in Guschin et al.23 and indicates the average of three independent biological replicates. Neg: negative-control cells transfected with vector alone. (G) Expression of AsCas12a, LbCas12a, and Lb2Cas12a in HEK293T cells used in (F). A western blot, representative of three independent biological replicates with similar results, is shown.
Results
Lb2Cas12a efficiently edits mammalian genomes and has a TTTN PAM preference
We investigated the nuclease activity and PAM usage of a range of Cas12a orthologs identified by Zetsche et al.6 In contrast to Zetsche et al.,6 who used the enzyme-mismatch cleavage Surveyor assay, we observed that Lb2Cas12a had significant nuclease activity using a double-stranded-break gain-of-expression assay11 (Figure 1C). This assay uses five 23-nt-target sequences from the EGFP, DNMT, FANCF, or EMX1 genes, each inserted upstream of an out-of-frame luciferase gene (Figure 1D). Cleavage of the target sequence and subsequent non-homologous end-joining (NHEJ) repair puts approximately one-third of the luciferase gene in frame, allowing its correct translation. We observed that Lb2Cas12a-mediated editing efficiency was similar to or better than those of AsCas12a and LbCas12a when targeting these five sequences (Figure 1E). To determine whether the difference in Lb2Cas12a activity between this observation and Zetsche et al. was due to use of different assays, we repeated these studies using the T7E1 nuclease assay and the DNMT1-3 target used in Zetsche et al.6,24 We found that Lb2Cas12a was efficient at cleaving this endogenous target (Figure 1F), again with efficiency comparable to AsCas12a or LbCas12a. Despite its efficiency, we noted that Lb2Cas12a expressed less efficiently than LbCas12a and much less efficiently than AsCas12a (Figure 1G), raising the possibility that modification of this Cas12a ortholog could increase its expression and activity.
Rational engineering of Lb2Cas12a to increase editing efficiency
We reasoned that Lb2Cas12a may contain divergent residues that impair its expression or editing efficiency. Therefore, we aligned Lb2Cas12a peptide sequence with the eight nearest available Cas12a orthologs and converted residues unique to Lb2Cas12a to the consensus residues among these orthologs (Figure S2). These Lb2Cas12a variants, each containing one consensus mutation, were characterized using the gain-of-expression assay with EGFP-1 as a target sequence. Most of these variants were approximately as efficient as wild-type Lb2Cas12a. However, replacement of glutamine 571 with a lysine (Q571K) resulted in a 50% increase in nuclease activity (Figure 2A). We also characterized a subset of these Lb2Cas12a variants and observed that the C1003Y variant consistently expressed more efficiently than wild-type Lb2Cas12a by approximately 40% (Figure 2B). Perhaps due to this higher expression, the C1003Y variant edited more efficiently than wild-type Lb2Cas12a, although this difference was not statistically significant in this initial screening assay (Figure 2A). Alternative mutations at C1003 position did not improve Lb2 editing efficiencies (Figure S3), suggesting that the gain of tyrosine rather than the loss of cysteine contributed to the improved expression of C1003Y mutant. A model of Lb2Cas12a based on the structure of LbCas12a (PDB: 5XUS) localizes Q571 to the loop-lysine-loop complex that interacts with PAM and promotes melting of the target DNA duplex (Figures 2C and 2D).25,26 C1003 is located near a helix bundle of Nuc domain, at the interface between DNA substrate and the nuclease (Figures 2C and 2E). We speculate that the aromatic ring of Y1018 in LbCas12a, equivalent to Lb2Cas12a C1003, associates through π-stacking with single- or double-stranded substrate DNA, stabilizing the DNA.26,27 C1003Y may also form a cation-π interaction with Lb2Cas12a R1111 in the opposite helix, thus stabilizing this domain.28,29
Figure 2.
Improved editing efficiency and expression of Lb2Cas12a variants
(A) A panel of Lb2Cas12a variants were characterized using the gain-of-function assay represented in Figure 1C. Each variant alters an Lb2Cas12a-unique residue to a consensus residue from closely related Cas12a orthologs (see Figures S1 and S2). Brackets indicate the significance of differences between wild-type Lb2Cas12a (WT-Lb2) and each of two variants (Q571K and C1003Y), as determined by one-way ANOVA, followed by Dunnett’s multiple comparisons test (∗∗∗∗p < 0.0001). The means of three independent replicates are shown, with error bars indicating ± SEM. Q571K and C1003Y are described below, respectively, as Lb2-K and Lb2-Y. (B) The expression of selected Lb2Cas12a variants was characterized by western blot using an antibody recognizing a C-terminal hemagglutinin (HA) tag. Expression, quantified by ImageJ densitometry, is indicated underneath the figure as a percentage of wild-type Lb2Cas12a (Lb2), an average of two independent replicates. (C) A model of Lb2Cas12a based on the structure of LbCas12a (PDB: 5XUS). LbCas12a and Lb2Cas12a are shown in light and dark blue, respectively; crRNA is indicated in orange and target DNA is shown in red. Boxes indicate regions shown in detail in (D) and (E). (D) Q571K of Lb2-K and Lb2-KY, like the analogous K579 of LbCas12a, may interact through cation-π interactions with the final nucleotide of the PAM, dA(−1∗), with asterisk indicating the non-target strand. (E) C1003Y of Lb2-Y and Lb2-KY, like the analogous Y1018 of LbCas12a, may stabilize the substrate DNA or the association between two Nuc-domain helices. Note that this figure is rotated 90 degrees about the horizontal axis relative to that in (C).
Lb2-KY is more efficient than wild-type Lb2Cas12a
The Q571K and C1003Y mutations were combined in Lb2Cas12a-KY (Lb2-KY) to determine if they synergized to make a more efficient Cas12a (Figure 3A). Lb2-KY, and variants with either individual mutation (Lb2-K, Lb2-Y), more efficiently cleaved an EGPF-1 target sequence than did wild-type Lb2Cas12a (Lb2). Lb2-KY outperformed both Lb2-K and Lb2-Y, although only the comparison with Lb2-Y was significant. Variants bearing C1003Y mutation (Lb2-Y, Lb2-KY) expressed more efficiently than did wild-type Lb2Cas12 (Figures 3B and 3C). We then compared the activities of Lb2-Y and Lb2-KY with the other commonly studied Cas12a orthologs, namely AsCas12a and LbCas12a, at the integrated target sequence EGFP-3 and the endogenous target sequences DNMT1-3, EMX1, and FANCF. Sanger sequencing and Tracking of Indels by Decomposition assay (TIDE) were used to quantify the editing efficiency in this and the following experiments, because the TIDE assay has been shown to be more reliable and reproducible than endonuclease mismatch cleavage assay in detecting homing endonuclease activity.30,31 Both Lb2-Y and Lb2-KY were more efficient than AsCas12a and Lb2Cas12a with three of these four targets, while AsCas12a and Lb2-KY modified the DNMT1-3 target with comparable efficiency (Figure 3D). Thus, Lb2-KY in mammalian cells is in general more efficient than LbCas12a or AsCas12a.
Figure 3.
Activity of Lb2Cas12a Q571K/C1003Y
(A) The editing efficiency of Lb2Cas12a variants was measured using the gain-of-expression reporter system with an EFGP-1 target, as represented in Figure 1C. Wild-type Lb2Cas12a (Lb2), and Lb2Cas12a variants Lb2Cas12a-Q571K (Lb2-K), Lb2Cas12a-C1003Y (Lb2-Y), and Lb2Cas12a-Q571K/C1003Y (Lb2-KY) were compared. The means of three independent replicates are shown, with error bars indicating ± SEM. ∗p < 0.05; ∗∗p < 0.01; values of p > 0.05 are indicated in the graph; two-tailed paired t test. (B) Expression of Lb2 variants in HEK293T cells from the experiment in (A). The western blot shown is representative of three biological replicates with similar results. (C) Quantification of the three western blots described in (B). Bars indicate mean values ± SEM. ∗p < 0.05, otherwise indicated in the graph; two-tailed paired t test. (D) Indel levels induced by As, Lb, Lb2, and Lb2 variants Lb2-Y and Lb2-KY. Both integrated (EGFP-3) and endogenous (DNMT1, EMX1, FANCF) targets were assayed. Editing efficiency was quantified using TIDE analysis.30 Bars show mean values of three independent replicates ± SEM. Each Cas12a variant was compared with Lb2-KY; ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001; values of p > 0.05 are shown in the graph; two-way ANOVA, followed by Dunnett’s multiple comparisons test.
Lb2-KY utilizes a wider range of PAMs than Lb2Cas12a and has higher editing efficiencies than AsCas12a and LbCas12a
To determine if Lb2-KY utilizes different PAMs than Lb2Cas12a, we established a novel screen, represented in Figure 4A, that assesses PAM usage in mammalian cells. As shown, the EMX1 target preceded by six randomized nucleotides was retrovirally integrated along with a puromycin selection marker into HEK293T cells. Cells were selected with puromycin and transfected with plasmids encoding mCherry and the EMX1-targeting crRNA together with plasmids encoding active Cas12a variants, or with an empty vector. mCherry-positive cells were selected by fluorescent-activated cell sorting, and the sites of integration were analyzed by next-generation sequencing. As shown in Figure 4B, in which preferences for the first three nucleotides of PAM sequence (i.e., positions −4 to −2) were described, wild-type Lb2Cas12a efficiently modified TTTN PAM sequences, but not most other PAMs. In contrast, Lb2-KY more efficiently modified a wider range of PAM sequences including all NTTN PAMs. LbCas12a also showed a broad PAM preference, corroborating previous research, while control mCherry-positive cells transfected with an empty vector had a low level of background variation. Of note, Lb2-KY showed consistent preference for all four CTTN PAMs, as shown in Figure 4C, where the fourth PAM nucleotide was included. We did not observe any significant contribution of the nucleotides at position −5 and −6 to the PAM preferences of the studied Cas12a nucleases.
Figure 4.
PAM usage of Lb, Lb2, and Lb2-KY
(A) A diagram outlining the mammalian-cell PAM identification assay used in subsequent panels. A library of randomized 5′-NNNNNN-3′ PAMs preceding a crRNA target was stably expressed in HEK293T cells. Plasmids encoding Cas12a proteins, crRNAs, and the fluorescent marker mCherry were transfected to these cells. Cells were sorted for mCherry expression, and a region containing the randomized PAM and the target sequence was deep-sequenced with paired-end reads of 300 base pairs. Indel frequencies were then calculated for each four-nucleotide PAM. Note that the first two nucleotides of the 6-mer randomized PAM sequence had no detectable impact on the activities of any Cas12a variant. (B) Heatmaps showing the result of mammalian-cell PAM screen for Lb2Cas12a, Lb2-KY, LbCas12a, and cells lacking a Cas12a ortholog (control). PAMs and the related editing efficiencies were pooled by the first three nucleotide positions. Each heatmap indicates an average of two or three independent replicates. The range of replicates for each 3-nucleotide PAM sequence is less than 10% of the signal at each position. (C) An expansion of the data generated in (B), except that all 64 four-nucleotide PAMs are plotted. The range of replicates for each 4-nucleotide sequence is less than 25% of the average signal for each four-nucleotide PAM.
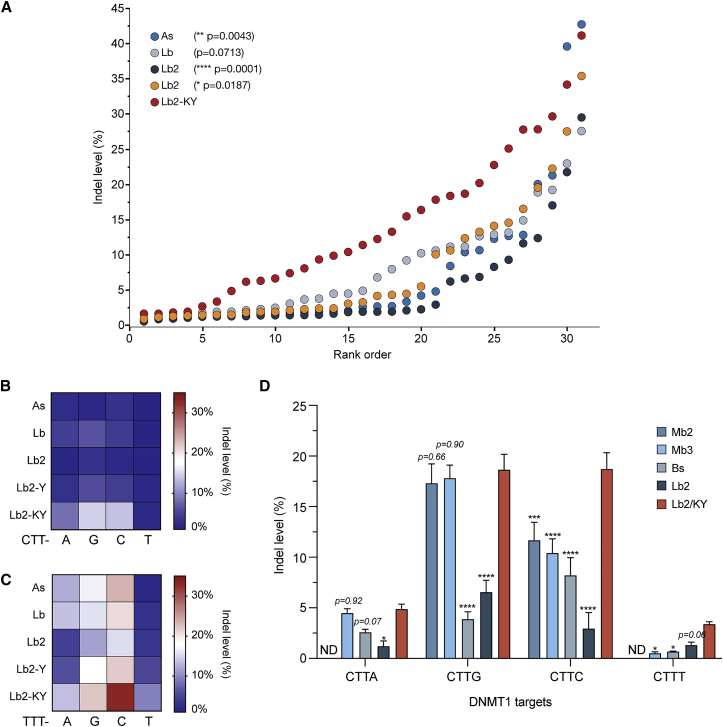
The screen shown in Figure 4 uses a single integrated EMX1 target sequence. To confirm and extend these observations, we used TIDE analysis to compare AsCas12a, LbCas12a, Lb2Cas12a, Lb2-Y, and Lb2-KY for their ability to modify 31 distinct endogenous target sites in four genes. The complete target list is provided in Table S1. As shown in Figure 5A, the editing efficiencies of Lb2-KY markedly exceeded those of AsCas12a and LbCas12a, whereas Lb2-Y activity was similar to those of LbCas12a and AsCas12a for most target regions. Both Lb2-KY and Lb2-Y showed improvements over the wild-type Lb2Cas12a. In accordance with the PAM screening assay, Lb2-KY showed enhanced activity at non-canonical TTTT and CTTV PAMs (Figures 5B and 5C; Figure S4). We conclude that Lb2-KY is more efficient than AsCas12a and LbCas12a at modifying targets with TTTN PAMs and consistently performed better than these latter Cas12a orthologs with CTTV and TTTT PAMs. Finally, we compared Lb2 and Lb2-KY with additional Cas12a orthologs reported recently to be efficient in mammalian cells32 (Figure 5D). Lb2-KY, but not Lb2Cas12a, edited CTTV PAMs similarly to, or more efficiently than, Mb2Cas12a, Mb3Cas12a, and BsCas12a.
Figure 5.
Efficiencies of Cas12a variants at TTTN and CTTN PAMs
(A) As, Lb, Lb2, Lb2-Y, and Lb2-KY Cas12a variants were characterized for their ability to modify 31 endogenous target sequences preceded by TTTN or CTTN PAMs within four genes—DNMT1, EMX1, FANCF, and GRIN2B—as determined by TIDE analysis.30 Indel levels were ranked and compared with Lb2-KY using nonparametric tests with Steel’s corrections for multiple comparisons. p values are indicated in the legends. (B and C) Heatmaps summarizing data from (A) for target regions with CTTN (B) and TTTN (C) PAMs. (D) Editing efficiency of additional Cas12a orthologs at endogenous CTTN loci in the DNMT1 gene. Lb2-KY was compared with Mb2Cas12a (Mb2), Mb3Cas12a (Mb3), and BsCas12a (Bs). The means of three independent replicates are shown, with error bars indicating ± SEM. Significance of comparisons with Lb2-KY was determined by two-way ANOVA, followed by Dunnett’s multiple comparisons tests (∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001; values of p > 0.05 are shown in the graph).
Lb2-KY RNP complexes can precisely edit therapeutically relevant targets bearing CTTV PAMs
RNP complexes composed of a CRISPR effector protein and a crRNA more efficiently edit their targets than plasmid-expressed protein and RNA.33,34 Accordingly, we produced Lb2-KY RNP complexes with an optimized nuclear localization signal (NLS7; Figures S5A, S5B, and S6A) and assessed the editing efficiency of these RNPs using physiologically relevant target sites with CTTV PAM regions. Using the HDR-dependent editing strategy previously described for Cas12a proteins,7,35 we delivered Lb2-KY RNP with single-stranded oligodeoxynucleotide (ssODN) repair templates complementary to the target strand, with either 77/37- or 37/77-nucleotide homology arms. Lb2-KY RNP complexes efficiently used a CTTC PAM to introduce two mutations, M428L and N434S, into the human IgG1 Fc region (IGHG1 gene), which lacks TTTN PAM sequences (Figures 6A and 6B). These mutations have been shown to markedly increase the half-life of antibodies in the serum.37 Note that ssODN enhanced NHEJ-mediated indel formation as well as precise HDR-mediated editing, as previously reported.38
Figure 6.
Precise editing of therapeutically relevant targets bearing CTTV PAMs with Lb2-KY RNP complexes
(A–F) The levels of indels and homology-directed repair (HDR) induced by commercially produced As-V3 (IDT) or Lb2-KY RNP complexes with or without repair templates, as quantified by TIDER assay.36 (A, C, and E) As-V3 or Lb2-KY RNPs were used to introduce the M428L/N434S mutations in IGHG1 gene in JeKo1 cells (A) and the sickle cell mutation E6V in HBB gene in K562 cells (C), and to delete the BCL11A-binding motif preceding the fetal hemoglobin genes HBG1/2 in K562 cells (E). For clarity, only selected statistical significances are shown. The means of three replicates are shown, with error bars indicating ± SEM. ∗p < 0.05; ∗∗∗∗p < 0.0001; values of p > 0.05 are shown in the graph; two-way ANOVA followed by Tukey’s or Sidak’s multiple comparisons tests. (B, D, and F) To the right of each figure is an illustration of the editing strategies used to compare Lb2-KY and As-V3 RNP complexes. Single-stranded oligodeoxynucleotide repair templates (green) with 77/37-base (B and F) or 37/77-base (D) homology arms complimentary to the target strands were used. Red letters denote the designed mutations, blue letters PAM regions, gray box the crRNA hybridization site on the target strand, and red triangles expected Cas12a cut sites. Note that in (B), in addition to the M428L/N434S mutations, silent point mutations were also added on the repair templates at the PAM and seed regions to prevent re-cutting of the repaired gene. (G and H) Indel levels induced by various Cas12a RNP complexes, namely As, commercially produced As-Ultra (IDT), and Lb (NEB), Lb2, and Lb2-KY at endogenous targets as quantified by TIDE assay. PAM sequences are indicated beneath the graph. The means of three independent replicates are shown, with error bars indicating ± SEM. Each RNP was compared with Lb2-KY RNP; ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001; two-way ANOVA followed by Holm-Sidak’s multiple comparisons tests.
We further directed Lb2-KY RNPs to two chromosomal targets relevant to sickle-cell and beta-thalassemia diseases, using a cell line, K562, that resembles early-stage erythrocytes. We compared Lb2-KY RNP with a commercially produced Cas12a RNP, namely Alt-R AsCas12a V3 (denoted here as As-V3; Integrated DNA Technologies [IDT]). We first targeted a region of the hemoglobin subunit beta (HBB) gene proximal to the codon for glutamic acid 6. This glutamic acid is a valine in a common form of sickle cell disease. Although this region lacks a canonical TTTV PAM site, Lb2-KY RNP was still able to mutate this codon to encode a valine by targeting an adjacent CTTG PAM (Figures 6C and 6D), again consistent with the efficiency with which Lb2-KY recognizes CTTV PAM sites. Lb2-KY RNPs were even more efficient at introducing an engineered deletion into the distal BCL11A binding motif preceding the hemoglobin subunit gamma 1 and 2 (HBG1/2) genes (Figures 6E and 6F). This deletion was previously reported to augment expression of fetal hemoglobin in adult reticulocytes.39 Thus, Lb2-KY RNP can be used to efficiently edit CTTV PAM sites in physiologically relevant genomic regions.
During the preparation of our manuscript, a more enhanced commercial version of AsCas12a, namely Alt-R AsCas12a Ultra, was introduced (denoted here as As-Ultra; IDT). As-Ultra was engineered through directed evolution to achieve higher, more robust editing efficiencies compared to the earlier version As-V3. In order to thoroughly compare the efficiencies of available Cas12a RNPs, we produced wild-type Cas12a orthologs, As and Lb2 RNPs (Figures S6B–S6E) and compared the efficiencies of Lb2-KY RNP with As and Lb2 RNPs and the commercially available As-Ultra and EnGen Lba Cas12a (New England BioLabs; denoted here as Lb-NEB; note that neither commercial product is marketed to cleave CTTN PAM motifs). All homemade RNP variants include the optimized nuclear-localization signal NLS7. Lb2-KY RNP outperformed all the tested variants at all but one target from different genes with the canonical TTTN PAMs and the non-canonical CTTN PAMs (Figures 6G and 6H). At the DNMT1-3 target, Lb2-KY activity was slightly less than that of As-Ultra.
The trans-cleavage and off-target effects of Lb2-KY
Cas12a nucleases have been shown to indiscriminately cleave ssDNA in vitro after being activated by a bona fide double-stranded DNA target.40,41 We used ssDNA substrates with fluorophore-quencher to study the trans-cleavage activity of Lb2-KY, as previously described.40,42 Lb2-KY showed comparable trans-cleavage activity to LbCas12a (i.e., EnGen Lba Cas12a, NEB); both were activated by targets containing TTTV and CTTC PAMs, but not AGCG or ACTG targets (Figures S7A–S7D). Note that it is not clear that this trans-cleavage activity is deleterious in physiological settings, because ssDNA in replication bubbles and homologous recombination complexes are protected by DNA-binding proteins,43, 44, 45 and because LbCas12a-mediated HDR with ssDNA repair templates is as efficient as that mediated by AsCas12a.7
We also characterized the target specificity of Lb2-KY RNP compared to those of AsCas12a and As-Ultra RNPs using iGUIDE assay.46 When directed at the DNMT1-3 target, Lb2-KY showed a moderate off-target profile as compared to the other nucleases, with one prominent off-target (Figures S7E–S7H; Table S2). The on-target frequency of Lb2-KY RNP was slightly less than that of AsCas12a RNP but higher than As-Ultra. No novel off-targets containing CTTN PAM were detected with this target. When delivered as plasmids, Lb2-KY on-target frequency was somewhat reduced as expected (Figure S7I; Table S2). Finally, we detected no significant loss of cell viability when Lb2-KY, as well as other Cas12a variants, were expressed from plasmids in HEK293T cells (Figure S7J).
Discussion
In this study, we show that Lb2Cas12a can edit mammalian genomes with efficiencies similar to other, better characterized Cas12a orthologs. By converting unique residues in Lb2Cas12a to residues common among its closely related Cas12a orthologs, we created an Lb2Cas12a variant, Lb2-KY, with enhanced activity and expression in mammalian cells and a broadened PAM preference. Lb2-KY has markedly greater cleavage activity than AsCas12a and LbCas12a at both the canonical PAM TTTN and the non-canonical PAM CTTV. The difference is especially pronounced with sequences bearing CTTV PAM, expanding the range of possible PAMs available for gene editing. In addition to its greater efficiency, Lb2-KY has an additional advantage: at 1,206 residues, it is the smallest of the Cas12a variants that function efficiently in mammalian cells. It thus can be readily introduced into AAV vector along with a crRNA array, perhaps encoded as a single transcript.
We have also developed a mammalian PAM assay that tracked the PAM preference of Cas12a proteins in the mammalian genomes (Figure 4). This assay may better reflect conditions in the eukaryotic nucleus than in vitro assay or those in E. coli.15,47 However, this assay is limited to only a single target sequence per library, and therefore its results should be confirmed with multiple endogenous targets, as shown in Figure 5. Nonetheless, it may be useful as a rapid screen for evaluating Cas12a variants engineered for novel PAM preferences in mammalian cells.
To generate Lb2-KY, we introduce two mutations into wild-type Lb2Cas12a. Noting that wild-type Lb2Cas12a expressed less efficiently than either LbCas12a or AsCas12a (Figure 1G), we initially sought mutations that increased expression in mammalian cells. We observed that replacement of cysteine 1003 with a tyrosine markedly enhanced Lb2Cas12a expression. Although the effect of this mutation on editing was modest, we retained it for two reasons. First, the greater expression may suggest that it is better folded or more stable in mammalian cells. Second, this expression may be more relevant in contexts where Cas12a levels are limiting, for example, in AAV-transduced cells in vivo.
The utility of the Q571K mutation is more obvious. This mutation restores a critical PAM binding lysine present in every Cas12a ortholog that functions efficiently in mammalian cells, including AsCas12a, LbCas12a, FnCas12a, and MbCas12a (Figure S1). Analysis of FnCas12a crystal structure (PDB: 5NFV) has shown that FnCas12a K667, homologous to Lb2Cas12 Q571K, participates in a cation-π interaction with final PAM nucleotide, disrupting base-pairing in the seed region of the target DNA.25,26 Presumably a glutamine at this position does not interact as strongly with this nucleotide.
Finally, we showed that Lb2-KY RNP complexes efficiently edited therapeutically important targets using CTTV PAMs in mammalian cells. At two loci critical to sickle cell and β-thalassemia diseases, namely glutamic acid 6 in HBB gene and BCL11A-binding motif near HBG1/2 genes, Lb2-KY RNPs outperformed the commercially produced As-V3 RNPs. In addition, Lb2-KY RNP showed moderate off-target activity compared other AsCas12a and As-Ultra RNPs. Although SpCas9 RNPs have been shown to efficiently edit these therapeutically relevant targets as well,48, 49, 50 this CRISPR effector protein in general has greater off-target activities, and it presents epitopes from a common human pathogen. Thus, cells expressing SpCas9 might be more efficiently cleared by the adaptive immune system.12,14,51,52 During the preparation of our manuscript, a few novel Cas12a nucleases have been described to utilize non-canonical PAMs53, 54, 55, 56 or utilize more stringent T-rich PAMs with reduced off-target effects compared to AsCas12a and LbCas12a.57 Although the cleavage activities of these nucleases have not been extensively compared with the commonly used Cas12a orthologs, it will be interesting to directly test Lb2-KY activity with these novel variants at both TTTN and CTTN PAMs. In summary, the inherent editing efficiency of Lb2-KY, its smaller size, the ease with which efficient Lb2-KY RNP can be produced and used, and its markedly greater ability to edit regions with CTTV PAM sites suggest that Lb2-KY will be a useful contribution to the expanding toolbox of CRISPR effector proteins.
Materials and methods
Plasmids
Wild-type AsCas12a (pcDNA3.1-hAsCpf1), LbCas12a (pcDNA3.1-hLbCpf1), and Lb2Cas12a (pcDNA3.1-hLb2Cpf1) and pX330 (pX330-U6-chimeric_BB-CBh-hSpCas9) plasmids were gifts from Dr. Feng Zhang (Addgene plasmid numbers 69982, 69988, 69983, and 42230, respectively). pMAL-his-LbCpf1-EC was a gift from Dr. Jin-Soo Kim (Addgene plasmid number 79008) and was used to express Cas12a variants in E. coli for protein production.
Lb2Cas12a point mutations were introduced by PCR using the Phusion site-directed mutagenesis kit (Thermo Fisher Scientific). Changes to the N and C termini of Cas12a plasmids were made by ligation of gBlock gene fragments (IDT) into the original plasmids linearized by appropriate restriction enzymes. pcDNA-3.1 plasmids expressing Cas12a direct repeats and double BsmBI recognition sites under U6 promoters (pcDNA3.1-U6-promoter) were created by PCR amplification of the U6 promoter region from pX330 with a U6-promoter forward primer and a reverse primer carrying the Cas12-direct-repeat-BsmBI sequence in the 5′ end. crRNA plasmids were generated by cloning annealed spacer-sequence between BsmBI-double-digested restriction sites in pcDNA3.1-U6-promoter plasmids. For Cas12a protein production, each Cas12a gene was codon-optimized for E. coli using OptimumGene Codon Optimization (GenScript), synthesized, and cloned into pMAL-his-LbCpf1-EC between EcoRI and NotI restriction sites.
Cell culture
Human embryonic kidney 293T (HEK293T) cells (ATCC) and HEK293T-derived reporter cell lines for gain-of-expression experiments11 were maintained in Dulbecco’s modified Eagle’s medium (DMEM, Life Technologies) supplemented with 10% fetal bovine serum (FBS) (Sigma-Aldrich), 2 mM Glutamax-I (Life Technologies), and 100 μM nonessential amino acids (Life Technologies). JeKo-1 cells (ATCC) were maintained in RPMI-1640 Medium (Life Technologies) supplemented with 20% FBS. K-562 cells (ATCC) were maintained in Iscove’s modified Dulbecco’s medium (IMDM, Life Technologies) supplied with 10% FBS. All growth media were supplemented with 100 U/mL penicillin and 100 μg/mL streptomycin (Life Technologies), except during transfection and electroporation. Cells were confirmed mycoplasma-free by the provider.
Cas12a on-target DNA cleavage assays
HEK293T cells or HEK293T-derived reporter cells were seeded in 48-well plates with antibiotic-free medium 16 h prior to transfection. Cells were transfected with 100 ng of Cas12a plasmids and 100 ng of crRNA plasmids using 0.75 μL of Lipofectamine 2000 (Thermo Fisher Scientific). Eight hours later, transfection medium was completely removed and replaced with regular growth medium containing 2% or 10% FBS. About 72 h post-transfection, cells and culture medium were harvested for reporter gene expression in luminescence assays, T7E1 assay, Sanger sequencing and subsequent TIDE analysis, or western blotting. Luminescence assays measuring Gaussia luciferase, firefly luciferase, and Cypridina luciferase and T7E1 mismatch cleavage assay were conducted as previously described.11
TIDE and TIDER analysis
Genomic DNA from transfected cells was isolated using NucleoSpin Tissue kits (MACHEREY-NAGEL). About 100 ng of the purified DNA was used as templates for PCR amplification with primer pairs flanking a 1.5–2 kbp region inclusive of the target site. PCR products were purified using Nucleospin Gel and PCR Clean Up kit (MACHEREY-NAGEL), then sent to Genewiz for Sanger sequencing using primers binding about 200–400 bp up- or downstream of the Cas12a cleavage site. Table S3 contains a list of primers used. The chromatograms were decomposed and indel levels or precise-editing efficiencies predicted using TIDE package available at http://shinyapps.datacurators.nl/tide/ or TIDER package at http://shinyapps.datacurators.nl/tider/. For TIDER analysis, reference sequences were generated by sequencing gBlocks resembling the target region and carrying the designed mutations.
Western blots
Transfected cells were washed with cold PBS and incubated in PBS on ice for 5 min to detach from the plate, then collected and spun down at 1,600 × g, 4°C, for 5 min. The cell pellet was resuspended in RIPA (Sigma Aldrich) and incubated on a tube rotator at 4°C for 15 min, then spun down at 13,000 × g, 4°C, for 15 min. Supernatant was collected and total protein was quantified with a Pierce BCA Protein Assay kit (Thermo Fisher Scientific). About 10 μg of total protein was mixed with 2× Laemmli buffer (Sigma Aldrich), boiled at 95°C for 5 min, and then resolved on a Novex 8% Tris-glycine mini gel (Invitrogen) in Tris-glycine SDS running buffer. Proteins on the gel were transferred to a 0.45 μm PVDF membrane (Invitrogen) in 20% methanol tris-glycine buffer using a semi-dry transfer module. The membrane was blotted with anti-HA and anti-β-actin antibodies (Sigma Aldrich) in 5% milk blocking buffer overnight at 4°C, then with anti-mouse Igκ light chain antibodies conjugated with horseradish peroxidase (Santa Cruz Biotechnology). Chemiluminescent signal was detected using a SuperSignal West Femto Substrate (Thermo Scientific) and imaged with ImageQuant LAS4000 Mini gel imaging system (GE Healthcare Life Sciences).
PAM screening assay
A randomized PAM library was constructed by amplifying the EMX1 target sequence TCATCTGTGCCCCTCCCTCCCTG with forward primers that contained a randomized 5′-NNNNNN-3′ region adjacent to the EMX1 sequence and a reverse primer (Table S3). The PCR products were assembled into pQCXIP vector between NotI and BamHI sites using In-Fusion cloning (Takara Bio). The ligation products were used to transform Stellar competent cells (Takara Bio), and all the colonies on the plate were collected. The cell suspension was cultured for 6 h at 37°C in a shaking incubator, and all the bacteria were lysed for plasmid library preparation using a Maxi-prep kit (MACHEREY-NAGEL).
A HEK293T cell line integrated with PAM library composed of all 4,096 possible 6-nucleotide sequences preceding a common EMX1 target sequence (TCATCTGTGCCCCTCCCTCCCTG) was created by transducing the parental cells with produced vesicular stomatitis virus (VSV) G protein-pseudotyped murine leukemia viruses (MLVs). Specifically, HEK293T cells were co-transfected with three plasmids, pMLV-gag-pol, pCAGGS-VSV-G, and the library plasmids, and the medium was refreshed after 6-h incubation of transfection mix. The supernatant from producer cells was harvested 72 h post transfection and passed through 0.45 μm filter. Parental HEK293T cells were transduced with generated MLV virus and selected with medium containing 5 μg/mL puromycin (Sigma). After selection, HEK293T-library cells were maintained with medium containing 3 μg/mL puromycin. The integration of the PAM library was confirmed by deep sequencing of the amplified genomic DNA of the target region using a MiSeq (Illumina). To identify the PAM usage of different Cas12a proteins, the polyclonal HEK293T-library cells seeded in 6-well plates were transfected with 1.8 μg of Cas12a-expressing plasmids, 1.8 μg crRNA11 plasmids, and 400 ng of mCherry plasmids using PEI (Polysciences). 48 h post transfection, transfected cells were detached and stained with eBioscience Fixable viability dye eFluor 780 (Invitrogen) and subsequently sorted for viable, mCherry-positive cells using BD FACSAria Fusion cell sorter. Collected cells were washed once and lysed for genomic DNA extraction using a genomic DNA extraction kit (MACHEREY-NAGEL). The region flanking the target site was amplified using nested PCR to add Illumina P5 adapters as well as unique sample-specific barcodes to the target amplicons. PCR products were run on 1% agarose gel (Invitrogen), and gel extraction was performed using a NucleoSpin Gel and PCR Clean-up kit (MACHEREY-NAGEL). Samples were pooled and quantified by Qubit 2.0 Fluorometer (Life Technologies). The prepared samples were sequenced on a MiSeq with a paired-end 300 cycle kit (Illumina).
Approximately 2 million quality-controlled reads were each obtained from Lb2Cas12a- and Lb2Cas12a-KY-transfected cells and from cells transfected without a Cas12a ortholog. Sequences were binned according to the 6-nucleotide randomized region immediately upstream of the EMX1 target. The percentage of each bin containing indels in the 40 nucleotides downstream of this randomized region, averaged across two to four experiments, was determined using the inhouse utility “MakeRGS.” Note that more than 98% of indels did not approach the PAM region, largely eliminating this source of error. Heatmaps representing the percentage of indels observed for the C-terminal 4-nucleotide PAM regions were generated using the inhouse utility “PAMheatmap.” Various 3- and 2-nucleotide combinations were also calculated and represented in Figures 4B–4D.
Cas12a protein production
pMAL-his-Cas12a-EC was expressed in E. coli Rosetta 2 (DE3) (Sigma-Aldrich). Transformed E. coli were grown in 50 mL of Luria-Bertani (LB) broth with 34 μm/mL chloramphenicol and 100 ug/mL carbenicillin in a shaking incubator at 37°C overnight. For a large-scale production of Cas12a protein, this overnight culture was subsequently added to 5 L of LB broth and incubated in the shaking incubator at 37°C for 3–4 h, until its optical density 600 (OD600) reached 0.5–0.6. The culture was put on ice for 15 min, then supplemented with 0.5 mM isopropyl β- d-1-thiogalactopyranoside (IPTG). Cas12a expression was induced overnight in the shaking incubator at 16°C. Bacterial cells were subsequently pelleted by centrifuging at 6,000 × g for 15 min. Before Cas12a purification, cell lysates from the culture before and after induction with IPTG were resolved on PAGE gel to ensure proper Cas12a expression.
Protein purification
Cas12a protein was purified by passage through a Ni affinity column followed by a cation exchange column and a size exclusion column in an Akta explorer fast protein liquid chromatography (FPLC) system (GE Healthcare). In brief, cells were harvested by centrifugation and resuspended in sonication buffer (50 mM NaH2PO4, 500 mM NaCl, 10 mM imidazole [pH 8.0], and 10% glycerol), sonicated on an ice-water bath for 20 min at 18 W output, and centrifuged for 25 min at 50,000 × g. Cas12a was isolated from the sonicated supernatant by adsorption to a 16 mL HisTrap FF column (GE Healthcare) and eluted with linear gradient from 10 mM to 300 mM imidazole in sonication buffer. The elution fraction containing the protein was pooled and concentrated to 20 mL using a 50 kDa cutoff ultrafiltration unit (Millipore). A ratio of 1 mg of TEV protease to 50 mg of Cas12a was added to the concentrate and dialyzed against TEV protease buffer (250 mM NaCl, 20 mM HEPES 7.4, 0.5 mM EDTA, and 1 mM DTT) for 48 h at 4°C.
The digested protein was diluted 2-fold with 20 mM HEPES (pH 7.0) and loaded on a 5 mL HiTrap SP HP column (GE Healthcare) equilibrated with 100 mM NaCl, 20 mM HEPES (pH 7.0). Adsorbed protein was eluted with a linear gradient from 100 mM to 2 M NaCl. Cas12a was purified further by gel filtration through a Superdex 200 26/60 column (GE Healthcare) equilibrated with gel filtration buffer (500 mM NaCl, 20 mM HEPES [pH 7.5], 0.1 mM EDTA, 1 mM DTT, and 10% glycerol). Fractions containing Cas12a were pooled and concentrated to 4 mL with a 50 kDa cutoff ultrafiltration unit (Millipore). Finally, the concentrate was divided and loaded to 1 mL endotoxin removal columns (Pierce) for overnight incubation. The next morning, each column was washed with 3 mL of sample buffer (400 mM NaCl, 20 mM HEPES, pH 7.5, 0.1 mM EDTA, 1 mM DTT, and 10% glycerol). The eluted protein was pooled and concentrated to 40 mg/mL using a 50 kDa cutoff ultrafiltration unit (Millipore). Purity of Cas12a protein was confirmed by SDS-PAGE. A 260/280 ratio of ∼0.5 was measured using NanoDrop (Thermo Scientific), indicating no nucleic acid contamination.
RNP formation and electroporation
crRNAs for Cas12a variants were ordered from IDT. crRNAs were resuspended in IDTE (10 mM Tris, 0.1 mM EDTA; IDT) and annealed by incubation at 95°C for 5 min and cooling down on bench top for 1 h. For each electroporation sample, RNP complexes were formed by mixing 240 pmol of each Cas12a variant with 480 pmol of crRNA and PBS. The RNP mixture was incubated at room temperature for 30 min to 1 h, then added with 400 pmol of single-stranded DNA repair templates and immediately mixed with cell suspension in electroporation solution. HEK293T, K562, and JeKo-1 cells (ATCC) were electroporated using Lonza 2B or 4D modules according to Lonza’s protocols. After electroporation, the cells were resuspended in antibiotics-free media and incubated for 48 h before genomic DNA extraction and sequencing.
Fluorophore-quencher (F-Q) reporter-based trans-cleavage assay.
Fluorophore-quencher DNaseAlert substrate was purchased from IDT. DNA target sequence and experiment procedure were designed according to Swarts and Jinek.42 Each reaction mix contains 150 nM of RNP complex, 50 nM of dsDNA target sequence containing PAM, and 50 nM of DNaseAlert substrate in SEC reaction buffer (500 mM KCl, 20 mM HEPES, 1 mM DTT) supplemented with 5 mM of MgCl2. Fluorescence signal was read every 30 s for 70 min in Tecan Safire 2 plate reader (Tecan Group) with excitation wavelength at 535 nm and emission at 595 nm.
iGUIDE assay
For each sample, approximately one million HEK293T cells were electroporated with RNP complexes and 250 pmol of iGUIDE double-stranded oligodeoxynucleotide (dsODN) using Lonza 2B system. Genomic DNA was extracted and processed as described in Nobles et al.46 and Tsai et al.58 MiSeq data were analyzed using iGUIDE package and are available at https://github.com/cnobles/iGUIDE. Queried PAM sequence was specified as NTTN with allowance for one mismatch. Search window for Cas12a-specific activity was set at 100 bp upstream and 30 bp downstream of the dsODN insertion site, and predicted off-targets can have up to six mismatches from the queried target sequence.
Cell viability assay
HEK293T cells were transfected with varying amounts of Cas12a and crRNA plasmids. 36 h after transfection, glycyl-phenylalanyl-aminofluorocoumarin (GF-AFC) substrate was added to each well as indicated by manufacturer’s protocol (CellTiter-Fluor Cell Viability Assay, Promega) and incubated for 30 min. Digitonin was added to positive control wells minutes before substrate addition. Fluorescence was detected in a SpectraMax M5 plate reader with excitation at 380 nm and emission at 505 nm.
Protein sequence and structure analyses
Alignments of Lb2Cas12a with its orthologs were performed using MUSCLE, available at https://www.ebi.ac.uk/Tools/msa/muscle/ and were visualized using ESPript 3.0, available at http://espript.ibcp.fr/ESPript/ESPript/.
A model of Lb2Cas12a was generated from the LbCas12a structure (42% sequence similarity; PDB: 5XUS) using Chimera Modeler. The model score GA341 equals 1.00, indicating a probability of having the correct fold that is >95%, and the normalized discrete optimized protein energy (zDOPE) score is −0.47.
Statistics
Student’s t test, one-way ANOVA, and two-way ANOVA with Tukey’s, Dunnett’s, or Sidak’s multiple comparison tests were performed in GraphPad Prism 8.0 (GraphPad Software, San Diego, CA, USA). Rank ordered indel % values (Figure 5A) were compared using nonparametric comparisons with control using the Steel method, where control was Lb2-KY. The analysis was done in JMP Pro 15.1.0 (SAS Institute, Cary, NC, USA). In all tests, differences were considered significant at p <0.05.
Code availability
The in-house mammalian PAM screening MakeRGS and PAMheatmap utilities and their C++ source code are available upon request.
Data availability
The datasets generated in this study are available upon reasonable request.
Acknowledgments
This work is supported by NIH grants R37 AI091476 and DP1 DA043912 (PI: M.F.). The authors would like to thank Brian Quinlan, PhD, Shruti Choudhary, PhD, and Matthew Costales, PhD, for experimental advice, and Matthew Gardner, PhD, and Meredith Davis Gardner, PhD, for experimental advice and careful critique of the manuscript.
Author contributions
M.H.T., G.Z., H.M., and M.F. conceived of this study. C.L.N. and M.F. wrote programming codes and provided guidance for experimental design. M.H.T., H.P., P.K., L.P., H.W., W.H., T.O., K.S., I.S., and H.M. performed all experiments. G.C. assisted with statistical analyses. M.H.T., H.M., and M.F. wrote the manuscript.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtn.2021.02.012.
Contributor Information
Huihui Mou, Email: hmu@scripps.edu.
Michael Farzan, Email: mfarzan@scripps.edu.
Supplemental information
References
- 1.Barrangou R., Fremaux C., Deveau H., Richards M., Boyaval P., Moineau S., Romero D.A., Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 2.Marraffini L.A. CRISPR-Cas immunity in prokaryotes. Nature. 2015;526:55–61. doi: 10.1038/nature15386. [DOI] [PubMed] [Google Scholar]
- 3.Mohanraju P., Makarova K.S., Zetsche B., Zhang F., Koonin E.V., van der Oost J. Diverse evolutionary roots and mechanistic variations of the CRISPR-Cas systems. Science. 2016;353:aad5147. doi: 10.1126/science.aad5147. [DOI] [PubMed] [Google Scholar]
- 4.Pickar-Oliver A., Gersbach C.A. The next generation of CRISPR-Cas technologies and applications. Nat. Rev. Mol. Cell Biol. 2019;20:490–507. doi: 10.1038/s41580-019-0131-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Makarova K.S., Wolf Y.I., Iranzo J., Shmakov S.A., Alkhnbashi O.S., Brouns S.J.J., Charpentier E., Cheng D., Haft D.H., Horvath P. Evolutionary classification of CRISPR-Cas systems: a burst of class 2 and derived variants. Nat. Rev. Microbiol. 2020;18:67–83.. doi: 10.1038/s41579-019-0299-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zetsche B., Gootenberg J.S., Abudayyeh O.O., Slaymaker I.M., Makarova K.S., Essletzbichler P., Volz S.E., Joung J., van der Oost J., Regev A. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell. 2015;163:759–771. doi: 10.1016/j.cell.2015.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Y., Liu K.I., Sutrisnoh N.B., Srinivasan H., Zhang J., Li J., Zhang F., Lalith C.R.J., Xing H., Shanmugam R. Systematic evaluation of CRISPR-Cas systems reveals design principles for genome editing in human cells. Genome Biol. 2018;19:62. doi: 10.1186/s13059-018-1445-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Singh D., Mallon J., Poddar A., Wang Y., Tippana R., Yang O., Bailey S., Ha T. Real-time observation of DNA target interrogation and product release by the RNA-guided endonuclease CRISPR Cpf1 (Cas12a) Proc. Natl. Acad. Sci. USA. 2018;115:5444–5449. doi: 10.1073/pnas.1718686115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hewes A.M., Sansbury B.M., Kmiec E.B. The Diversity of Genetic Outcomes from CRISPR/Cas Gene Editing is Regulated by the Length of the Symmetrical Donor DNA Template. Genes (Basel) 2020;11:1160. doi: 10.3390/genes11101160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zetsche B., Heidenreich M., Mohanraju P., Fedorova I., Kneppers J., DeGennaro E.M., Winblad N., Choudhury S.R., Abudayyeh O.O., Gootenberg J.S. Multiplex gene editing by CRISPR-Cpf1 using a single crRNA array. Nat. Biotechnol. 2017;35:31–34. doi: 10.1038/nbt.3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhong G., Wang H., Li Y., Tran M.H., Farzan M. Cpf1 proteins excise CRISPR RNAs from mRNA transcripts in mammalian cells. Nat. Chem. Biol. 2017;13:839–841. doi: 10.1038/nchembio.2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yan W.X., Mirzazadeh R., Garnerone S., Scott D., Schneider M.W., Kallas T., Custodio J., Wernersson E., Li Y., Gao L. BLISS is a versatile and quantitative method for genome-wide profiling of DNA double-strand breaks. Nat. Commun. 2017;8:15058. doi: 10.1038/ncomms15058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Strohkendl I., Saifuddin F.A., Rybarski J.R., Finkelstein I.J., Russell R. Kinetic Basis for DNA Target Specificity of CRISPR-Cas12a. Mol. Cell. 2018;71:816–824.e3. doi: 10.1016/j.molcel.2018.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Charlesworth C.T., Deshpande P.S., Dever D.P., Camarena J., Lemgart V.T., Cromer M.K., Vakulskas C.A., Collingwood M.A., Zhang L., Bode N.M. Identification of preexisting adaptive immunity to Cas9 proteins in humans. Nat. Med. 2019;25:249–254. doi: 10.1038/s41591-018-0326-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gao L., Cox D.B.T., Yan W.X., Manteiga J.C., Schneider M.W., Yamano T., Nishimasu H., Nureki O., Crosetto N., Zhang F. Engineered Cpf1 variants with altered PAM specificities. Nat. Biotechnol. 2017;35:789–792. doi: 10.1038/nbt.3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kleinstiver B.P., Sousa A.A., Walton R.T., Tak Y.E., Hsu J.Y., Clement K., Welch M.M., Horng J.E., Malagon-Lopez J., Scarfò I. Engineered CRISPR-Cas12a variants with increased activities and improved targeting ranges for gene, epigenetic and base editing. Nat. Biotechnol. 2019;37:276–282. doi: 10.1038/s41587-018-0011-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grieger J.C., Samulski R.J. Packaging capacity of adeno-associated virus serotypes: impact of larger genomes on infectivity and postentry steps. J. Virol. 2005;79:9933–9944. doi: 10.1128/JVI.79.15.9933-9944.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim H.K., Song M., Lee J., Menon A.V., Jung S., Kang Y.M., Choi J.W., Woo E., Koh H.C., Nam J.W., Kim H. In vivo high-throughput profiling of CRISPR-Cpf1 activity. Nat. Methods. 2017;14:153–159. doi: 10.1038/nmeth.4104. [DOI] [PubMed] [Google Scholar]
- 19.Tóth E., Weinhardt N., Bencsura P., Huszár K., Kulcsár P.I., Tálas A., Fodor E., Welker E. Cpf1 nucleases demonstrate robust activity to induce DNA modification by exploiting homology directed repair pathways in mammalian cells. Biol. Direct. 2016;11:46. doi: 10.1186/s13062-016-0147-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kleinstiver B.P., Tsai S.Q., Prew M.S., Nguyen N.T., Welch M.M., Lopez J.M., McCaw Z.R., Aryee M.J., Joung J.K. Genome-wide specificities of CRISPR-Cas Cpf1 nucleases in human cells. Nat. Biotechnol. 2016;34:869–874. doi: 10.1038/nbt.3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Robinson O., Dylus D., Dessimoz C. Phylo.io: Interactive Viewing and Comparison of Large Phylogenetic Trees on the Web. Mol. Biol. Evol. 2016;33:2163–2166. doi: 10.1093/molbev/msw080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guschin D.Y., Waite A.J., Katibah G.E., Miller J.C., Holmes M.C., Rebar E.J. A rapid and general assay for monitoring endogenous gene modification. Methods Mol. Biol. 2010;649:247–256. doi: 10.1007/978-1-60761-753-2_15. [DOI] [PubMed] [Google Scholar]
- 24.Vouillot L., Thélie A., Pollet N. Comparison of T7E1 and surveyor mismatch cleavage assays to detect mutations triggered by engineered nucleases. G3 (Bethesda) 2015;5:407–415. doi: 10.1534/g3.114.015834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stella S., Alcón P., Montoya G. Structure of the Cpf1 endonuclease R-loop complex after target DNA cleavage. Nature. 2017;546:559–563. doi: 10.1038/nature22398. [DOI] [PubMed] [Google Scholar]
- 26.Swarts D.C., van der Oost J., Jinek M. Structural Basis for Guide RNA Processing and Seed-Dependent DNA Targeting by CRISPR-Cas12a. Mol. Cell. 2017;66:221–233.e4. doi: 10.1016/j.molcel.2017.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cofsky J.C., Karandur D., Huang C.J., Witte I.P., Kuriyan J., Doudna J.A. CRISPR-Cas12a exploits R-loop asymmetry to form double-strand breaks. eLife. 2020;9:e55143. doi: 10.7554/eLife.55143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wilson K.A., Kellie J.L., Wetmore S.D. DNA-protein π-interactions in nature: abundance, structure, composition and strength of contacts between aromatic amino acids and DNA nucleobases or deoxyribose sugar. Nucleic Acids Res. 2014;42:6726–6741. doi: 10.1093/nar/gku269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gallivan J.P., Dougherty D.A. Cation-pi interactions in structural biology. Proc. Natl. Acad. Sci. USA. 1999;96:9459–9464. doi: 10.1073/pnas.96.17.9459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brinkman E.K., Chen T., Amendola M., van Steensel B. Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res. 2014;42:e168. doi: 10.1093/nar/gku936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sentmanat M.F., Peters S.T., Florian C.P., Connelly J.P., Pruett-Miller S.M. A Survey of Validation Strategies for CRISPR-Cas9 Editing. Sci. Rep. 2018;8:888. doi: 10.1038/s41598-018-19441-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zetsche B., Strecker J., Abudayyeh O.O., Gootenberg J.S., Scott D.A., Zhang F. A Survey of Genome Editing Activity for 16 Cas12a Orthologs. Keio J. Med. 2020;69:59–65. doi: 10.2302/kjm.2019-0009-OA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liang X., Potter J., Kumar S., Zou Y., Quintanilla R., Sridharan M., Carte J., Chen W., Roark N., Ranganathan S. Rapid and highly efficient mammalian cell engineering via Cas9 protein transfection. J. Biotechnol. 2015;208:44–53. doi: 10.1016/j.jbiotec.2015.04.024. [DOI] [PubMed] [Google Scholar]
- 34.Kim S., Kim D., Cho S.W., Kim J., Kim J.S. Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res. 2014;24:1012–1019. doi: 10.1101/gr.171322.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fernandez J.P., Vejnar C.E., Giraldez A.J., Rouet R., Moreno-Mateos M.A. Optimized CRISPR-Cpf1 system for genome editing in zebrafish. Methods. 2018;150:11–18. doi: 10.1016/j.ymeth.2018.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brinkman E.K., Kousholt A.N., Harmsen T., Leemans C., Chen T., Jonkers J., van Steensel B. Easy quantification of template-directed CRISPR/Cas9 editing. Nucleic Acids Res. 2018;46:e58. doi: 10.1093/nar/gky164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zalevsky J., Chamberlain A.K., Horton H.M., Karki S., Leung I.W., Sproule T.J., Lazar G.A., Roopenian D.C., Desjarlais J.R. Enhanced antibody half-life improves in vivo activity. Nat. Biotechnol. 2010;28:157–159. doi: 10.1038/nbt.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Park H.M., Liu H., Wu J., Chong A., Mackley V., Fellmann C., Rao A., Jiang F., Chu H., Murthy N., Lee K. Extension of the crRNA enhances Cpf1 gene editing in vitro and in vivo. Nat. Commun. 2018;9:3313. doi: 10.1038/s41467-018-05641-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu N., Hargreaves V.V., Zhu Q., Kurland J.V., Hong J., Kim W., Sher F., Macias-Trevino C., Rogers J.M., Kurita R. Direct Promoter Repression by BCL11A Controls the Fetal to Adult Hemoglobin Switch. Cell. 2018;173:430–442.e17. doi: 10.1016/j.cell.2018.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen J.S., Ma E., Harrington L.B., Da Costa M., Tian X., Palefsky J.M., Doudna J.A. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science. 2018;360:436–439. doi: 10.1126/science.aar6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li S.Y., Cheng Q.X., Liu J.K., Nie X.Q., Zhao G.P., Wang J. CRISPR-Cas12a has both cis- and trans-cleavage activities on single-stranded DNA. Cell Res. 2018;28:491–493. doi: 10.1038/s41422-018-0022-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Swarts D.C., Jinek M. Mechanistic Insights into the cis- and trans-Acting DNase Activities of Cas12a. Mol. Cell. 2019;73:589–600.e4. doi: 10.1016/j.molcel.2018.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stauffer M.E., Chazin W.J. Physical interaction between replication protein A and Rad51 promotes exchange on single-stranded DNA. J. Biol. Chem. 2004;279:25638–25645. doi: 10.1074/jbc.M400029200. [DOI] [PubMed] [Google Scholar]
- 44.Amunugama R., Fishel R. Homologous recombination in eukaryotes. Prog. Mol. Biol. Transl. Sci. 2012;110:155–206. doi: 10.1016/B978-0-12-387665-2.00007-9. [DOI] [PubMed] [Google Scholar]
- 45.Mondal A., Bhattacherjee A. Mechanism of Dynamic Binding of Replication Protein A to ssDNA. J. Chem. Inf. Model. 2020;60:5057–5069. doi: 10.1021/acs.jcim.0c00564. [DOI] [PubMed] [Google Scholar]
- 46.Nobles C.L., Reddy S., Salas-McKee J., Liu X., June C.H., Melenhorst J.J., Davis M.M., Zhao Y., Bushman F.D. iGUIDE: an improved pipeline for analyzing CRISPR cleavage specificity. Genome Biol. 2019;20:14. doi: 10.1186/s13059-019-1625-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kleinstiver B.P., Prew M.S., Tsai S.Q., Topkar V.V., Nguyen N.T., Zheng Z., Gonzales A.P., Li Z., Peterson R.T., Yeh J.R. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature. 2015;523:481–485. doi: 10.1038/nature14592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.DeWitt M.A., Magis W., Bray N.L., Wang T., Berman J.R., Urbinati F., Heo S.J., Mitros T., Muñoz D.P., Boffelli D. Selection-free genome editing of the sickle mutation in human adult hematopoietic stem/progenitor cells. Sci. Transl. Med. 2016;8:360ra134. doi: 10.1126/scitranslmed.aaf9336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Humbert O., Radtke S., Samuelson C., Carrillo R.R., Perez A.M., Reddy S.S., Lux C., Pattabhi S., Schefter L.E., Negre O. Therapeutically relevant engraftment of a CRISPR-Cas9-edited HSC-enriched population with HbF reactivation in nonhuman primates. Sci. Transl. Med. 2019;11:eaaw3768. doi: 10.1126/scitranslmed.aaw3768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lomova A., Clark D.N., Campo-Fernandez B., Flores-Bjurström C., Kaufman M.L., Fitz-Gibbon S., Wang X., Miyahira E.Y., Brown D., DeWitt M.A. Improving Gene Editing Outcomes in Human Hematopoietic Stem and Progenitor Cells by Temporal Control of DNA Repair. Stem Cells. 2019;37:284–294. doi: 10.1002/stem.2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kosicki M., Tomberg K., Bradley A. Repair of double-strand breaks induced by CRISPR-Cas9 leads to large deletions and complex rearrangements. Nat. Biotechnol. 2018;36:765–771. doi: 10.1038/nbt.4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wienert B., Wyman S.K., Richardson C.D., Yeh C.D., Akcakaya P., Porritt M.J., Morlock M., Vu J.T., Kazane K.R., Watry H.L. Unbiased detection of CRISPR off-targets in vivo using DISCOVER-Seq. Science. 2019;364:286–289. doi: 10.1126/science.aav9023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jacobsen T., Ttofali F., Liao C., Manchalu S., Gray B.N., Beisel C.L. Characterization of Cas12a nucleases reveals diverse PAM profiles between closely-related orthologs. Nucleic Acids Res. 2020;48:5624–5638. doi: 10.1093/nar/gkaa272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Teng F., Li J., Cui T., Xu K., Guo L., Gao Q., Feng G., Chen C., Han D., Zhou Q., Li W. Enhanced mammalian genome editing by new Cas12a orthologs with optimized crRNA scaffolds. Genome Biol. 2019;20:15. doi: 10.1186/s13059-019-1620-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu R.M., Liang L.L., Freed E., Chang H., Oh E., Liu Z.Y., Garst A., Eckert C.A., Gill R.T. Synthetic chimeric nucleases function for efficient genome editing. Nat. Commun. 2019;10:5524. doi: 10.1038/s41467-019-13500-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Aliaga Goltsman D.S., Alexander L.M., Devoto A.E., Albers J.B., Liu J., Butterfield C.N., Brown C.T., Thomas B.C. Novel Type V-A CRISPR Effectors Are Active Nucleases with Expanded Targeting Capabilities. CRISPR J. 2020;3:454–461. doi: 10.1089/crispr.2020.0043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen P., Zhou J., Wan Y., Liu H., Li Y., Liu Z., Wang H., Lei J., Zhao K., Zhang Y. A Cas12a ortholog with stringent PAM recognition followed by low off-target editing rates for genome editing. Genome Biol. 2020;21:78. doi: 10.1186/s13059-020-01989-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tsai S.Q., Zheng Z., Nguyen N.T., Liebers M., Topkar V.V., Thapar V., Wyvekens N., Khayter C., Iafrate A.J., Le L.P. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat. Biotechnol. 2015;33:187–197. doi: 10.1038/nbt.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated in this study are available upon reasonable request.