Abstract
Kabuki syndrome (KS) is a rare cause of intellectual disability primarily caused by loss-of-function mutations in lysine-specific methyltransferase 2D (KMT2D), which normally adds methyl marks to lysine 4 on histone 3. Previous studies have shown that a mouse model of KS (Kmt2d+/βGeo) demonstrates disruption of adult neurogenesis and hippocampal memory. Proof-of-principle studies have shown postnatal rescue of neurological dysfunction following treatments that promote chromatin opening; however, these strategies are non-specific and do not directly address the primary defect of histone methylation. Since lysine-specific demethylase 1A (LSD1/KDM1A) normally removes the H3K4 methyl marks added by KMT2D, we hypothesized that inhibition of KDM1A demethylase activity may ameliorate molecular and phenotypic defects stemming from KMT2D loss. To test this hypothesis, we evaluated a recently developed KDM1A inhibitor (TAK-418) in Kmt2d+/βGeo mice. We found that orally administered TAK-418 increases the numbers of newly born doublecortin (DCX)+ cells and processes in the hippocampus in a dose-dependent manner. We also observed TAK-418-dependent rescue of histone modification defects in hippocampus both by western blot and chromatin immunoprecipitation sequencing (ChIP-seq). Treatment rescues gene expression abnormalities including those of immediate early genes such as FBJ osteosarcoma oncogene (Fos) and FBJ osteosarcoma oncogene homolog B (Fosb). After 2 weeks of TAK-418, Kmt2d+/βGeo mice demonstrated normalization of hippocampal memory defects. In summary, our data suggest that KDM1A inhibition is a plausible treatment strategy for KS and support the hypothesis that the epigenetic dysregulation secondary to KMT2D dysfunction plays a major role in the postnatal neurological disease phenotype in KS.
Keywords: epigenetics, adult neurogenesis, histone modification, therapeutics, H3K4me1, H3K4me3, LSD1, ERK, splenomegaly, chromatin
Graphical Abstract

KMT2D and KDM1A have opposing effects on histone 3 lysine 4 methylation. In this study, we show that TAK-418, an inhibitor of KDM1A, rescues abnormal gene expression and chromatin levels as well as defects of adult neurogenesis and hippocampal memory in a mouse model of Kabuki syndrome carrying a mutation in Kmt2d.
Introduction
Kabuki syndrome (KS) is a rare Mendelian disorder that affects multiple systems, including neuro, immune, auditory, and cardiac systems.1 It is characterized by distinctive facial features, growth retardation, and mild to moderate intellectual disability.2 Human genetics studies have revealed that this autosomal-dominant/X-linked condition is caused by heterozygous/hemizygous loss of function in either of two genes with complementary function, that is, KMT2D on human chromosome 12 or lysine-specific demethylase 6A (KDM6A) on human X chromosome.3,4 Both of these disease genes encode histone modifiers that contribute to the opening of chromatin. Most (>70%) molecularly confirmed cases of KS have loss-of-function variants in KMT2D. KMT2D catalyzes the addition of methyl groups to lysine 4 of histone 3 (H3K4me1 and H3K4me3),5,6 which are marks associated with open chromatin. KDM6A also participates in chromatin opening by removing H3K27me3, a closed chromatin mark.7 It is therefore likely that the observed gene dosage sensitivity in KS results from an imbalance between open and closed chromatin states at or around critical target genes. We hypothesize that it may be possible to restore this balance with drugs that promote open chromatin states.8
Our laboratory characterized a mouse model of KS1 (Kmt2d+/βGeo) and found that these mice have many features that overlap with KS patient phenotypes, including craniofacial abnormalities, growth retardation, and immune dysregulation.9, 10, 11 These mice also demonstrate an ongoing deficiency of adult neurogenesis in the subgranular zone (SGZ) of the dentate gyrus (DG) in the hippocampus and hippocampal memory defects. We have previously shown that these deficiencies improve after a short-term (2–3 weeks) oral treatment with a histone deacetylase inhibitor (HDACi), AR-42.9 Modulation of epigenetic modifications through dietary elevation of beta-hydroxybutyrate, an endogenous HDACi, may also rescue these same phenotypes.10 However, both of these strategies are indirect, affecting chromatin opening through modulation of histone acetylation rather than histone methylation. Furthermore, HDACis have been poorly tolerated in clinical trials,12 and a ketogenic diet is very stringent and hard to implement in children who often have baseline feeding problems and growth retardation, as is the case in KS. Recently, genetic disruption of KDM1A, the factor that normally removes H3K4me1/2, was shown to rescue disrupted chromatin states in KMT2D-deficient embryonic stem cells.13 In this study, we used a specific inhibitor of KDM1A activity, TAK-418, to demonstrate in vivo rescue of adult DG neurogenesis, improvement of hippocampal memory deficits, chromatin remodeling, and gene expression abnormalities in Kmt2d+/βGeo mice. TAK-418, also called 5-{(1R,2R)-2-[(cyclopropylmethyl)-amino]cyclopropyl}-N-(tetrahydro-2H-pyran-4-yl)thiophene-3-carboxamide monohydrochloride, was successfully advanced in preclinical development and was recently given orphan designation by the European Medicines Agency (EMA) and the US Food and Drug Administration (FDA) for the treatment of KS. TAK-418 has already undergone phase 1 studies on healthy volunteers to establish safety and tolerability (ClinicalTrials.gov: NTC03228433 and NCT03501069). Therefore, the TAK-418-dependent rescue of disease phenotypes in Kmt2d+/βGeo mice provides support for a possible therapeutic role in KS.
Results
Kmt2d+/βGeo mice demonstrate a global decrease of histone H3K4 methylation in the hippocampus
Kmt2d+/βGeo mice contain an expression cassette encoding a β-galactosidase neomycin resistance fusion protein (β-Geo) inserted into intron 50 of the Kmt2d gene locus on mouse chromosome 15. This cassette contains a 5′ end splice acceptor sequence and a 3′ end cleavage and polyadenylation signal. The mutated allele, therefore, results in a truncated KMT2D protein with the peptide encoded by the first 50 exons of KMT2D fused to β-Geo at the C terminus.9 The truncated KMT2D fusion protein, therefore, lacks the C-terminal catalytic SET domain of KMT2D, which is responsible for its H3K4 methyltransferase activity. Because KMT2D is a prominent mammalian H3K4 methyltransferase and Kmt2d+/βGeo mice have disrupted hippocampal neurogenesis, we hypothesized that we would see diminished levels of mono- and di- methylated H3K4 (H3K4me1/2) in the granule cell layer of the DG in Kmt2d+/βGeo mice. To test this, we performed western blots of histone extracts from mouse hippocampi and observed significantly (p < 0.0001) reduced levels of H3K4me1 and H3K4me2 in Kmt2d+/βGeo mice compared to Kmt2d+/+ littermates when normalized to total histone 3 (H3) (Figures 1A and 1B; Figures S1A and S1B). Therefore, although KMT2D and lysine methyltransferase 2C (KMT2C) have some overlapping functions, our results indicate that the latter is unable to compensate for the heterozygous loss of Kmt2d in the hippocampus.
Figure 1.
TAK-418 rescues histone H3K4 methylation abnormalities and neurogenesis defects in hippocampus of Kmt2d+/βGeo mice
(A–C) Quantification of H3K4me1, H3K4me2, and H3K4me3 levels from western blots of hippocampal lysates from both genotypes with and without 2 weeks of TAK-418 treatment. (D and E) A deficiency of DCX+ cells in the granule cell layer of the hippocampus seen in Kmt2d+/βGeo mice compared to littermates normalized after 2 weeks of TAK-418. (F) A defect of the number of DCX+ processes is also rescued after 2 weeks on TAK-418. All sample values were normalized to the average of the untreated wild-type animals. ANOVA: ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.005, ∗∗∗∗p < 0.001.
Previously it has been reported that KMT2D is required for H3K4 trimethylation (H3K4me3) in bivalent promoters.6,14 Indeed, we found that the level of H3K4me3 was also reduced (Figure 1C; Figure S1C), and that the ratio of H3K4me3 to total histone 3 was significantly reduced in Kmt2d+/βGeo mice compared to Kmt2d+/+ littermates (p < 0.0016), although to a lesser extent than the observed deficiencies of H3K4me1/2.
TAK-418 treatment rescues the deficiency of mono- and di-methylated, but not tri-methylated, H3K4 in the hippocampus of Kmt2d+/βGeo mice
TAK-418 is a selective inhibitor of the lysine-specific demethylase 1 (KDM1A/LSD1) recently developed by Takeda Pharmaceutical Company. KDM1A, in association with the corepressor of REST1 (CoREST) complex, removes methylation at H3K4me1/2 sites.15, 16, 17, 18 Previously, genetic disruption of KDM1A was found to rescue differentiation defects in KMT2D-deficient embryonic stem cells.13 To test whether TAK-418 is able to functionally modify KDM1A activity and thereby rescue epigenetic abnormalities in vivo in the hippocampus, mice were given TAK-418 at 1 mg/kg/day by oral administration for 2 weeks and H3K4 methylation levels were characterized. Histones were then extracted from dissected hippocampus for analysis. We observed significant rescue (p < 0.05) by TAK-418 treatment of the deficient H3K4me1 levels in Kmt2d+/βGeo mice compared to Kmt2d+/+ littermates (Figure 1A; Figure S1A). Similarly, we observed a significant rescue (p < 0.05) of H3K4me2 levels in Kmt2d+/βGeo mice with TAK-418 compared to littermates on vehicle (Figure 1B; Figure S1B). H3K4me3 levels were also increased by TAK-418, albeit not to statistical significance (Figure 1C; Figure S1C). Thus, in summary, we observed biochemical rescue of H3K4 methylation in the hippocampus, a disease-relevant tissue, although we note that this was not a full rescue to wild-type (WT) levels in the treated Kmt2d+/βGeo mice (Figures 1A–1C). As expected, the effects of TAK-418 on all three H3K4 methylation marks were largely correlated. We did not observe any changes in H3K4 methylation levels in the WT animals on TAK-418.
TAK-418 rescues defects of adult neurogenesis in Kmt2d+/βGeo mice
Adult hippocampal neurogenesis is an ongoing process that persists throughout life19,20 in which adult-born neural progenitors in the SGZ give rise to excitatory granule cell neurons residing in the hippocampal DG.20 The neuroblast migration protein DCX is highly expressed in immature neurons and sharply decreases with maturation of neurons,21,22 providing a cell stage/type-specific marker to quantify adult neurogenesis in vivo. We tested the effects of TAK-418 on adult neurogenesis as measured by DCX+ cells per mm2 in the DG and SGZ. As before,9,10 we found decreased numbers of DCX+ cells in Kmt2d+/βGeo mice compared to Kmt2d+/+ littermates. On TAK-418 treatment, this phenotype showed dose-dependent normalization (Figure S2) with full rescue at the medium dose (1 mg/kg/day, Figures 1D and 1E). Dendrites can be visualized in some DCX+ cells, as DCX is a cytoplasmic protein that is associated with microtubules present in dendrites.23,24 We counted the DCX+ cells with dendrites and found that Kmt2d+/βGeo mice have fewer DCX+ cells with dendrites compared to Kmt2d+/+ littermates (Figure 1F), although this likely, to some extent, reflects absolute cell numbers. Kmt2d+/βGeo mice treated with TAK-418 have significantly increased numbers of DCX+ cells with dendrites compared to Kmt2d+/βGeo mice treated with vehicle (Figure 1F). The dendrites in the Kmt2d+/βGeo mice also appeared shorter than the dendrites in Kmt2d+/+ littermates, which reached farther into the granule cell layer and even into the molecular cell layer. TAK-418 treatment restored the dendrite length and characteristics in Kmt2d+/βGeo mice, resulting in longer dendrites extending throughout the granule cell layer and into the molecular cell layer (Figure S3).
TAK-418 treatment rescues the genome-wide deficiency of H3K4 methylation in Kmt2d+/βGeo mice
Given the global H3K4 methylation deficiency and rescue by TAK-418 in the hippocampus of Kmt2d+/βGeo mice, we next interrogated the genome-wide histone profiles in hippocampus by chromatin immunoprecipitation high-throughput sequencing (ChIP-seq). We first performed H3K4me1-ChIP-seq on hippocampi harvested from Kmt2d+/βGeo and Kmt2d+/+ mice treated with either vehicle or TAK-418. Most H3K4me1 peaks (∼57.7%) appeared in intergenic regions, which is consistent with known enhancer-associated roles of this mark.5 A third of peaks (∼31.2%) appeared to be in introns, and fewer were at the promoter/transcription start site (∼2.7%) or transcription start site (∼1.8%). There was no obvious difference between the two genotypes although there were a number of sites that significantly changed in Kmt2d+/+ littermates with TAK-418 treatment (Figure S4A). In untreated hippocampi, we observed a total of 621 differentially bound peaks of H3K4me1 in Kmt2d+/βGeo mice relative to Kmt2d+/+ mice (Figure 2A; Table S1). Of these, 396 peaks were decreased and 225 were increased in vehicle-treated Kmt2d+/βGeo mice compared to Kmt2d+/+ littermates (Figure 2A). The 28 most statistically significant peaks are shown in Table S1. In general, Kmt2d+/+ mice had more differentially H3K4me1-bound peaks compared to Kmt2d+/βGeo mice when either genotype was on vehicle (396/225, 1.76/1, Figure 2A). Upon treatment, we observed 622 differentially bound H3K4me1 peaks, with 322 increased and 300 decreased in Kmt2d+/βGeo on TAK-418 compared to Kmt2d+/+ mice on vehicle (Figure 2B; Table S2). The ratio of bound H3K4me1 in Kmt2d+/βGeo mice on TAK-418 compared to Kmt2d+/+ mice on vehicle was 1 to 0.93 (Figure 2B). An inverse linear regression revealed a correlation coefficient of −0.632 (Figure 2C) of log2 fold change of the common bound loci between the comparison of Kmt2d+/βGeo/Kmt2d+/+ mice on vehicle and Kmt2d+/βGeo on TAK-418/Kmt2d+/βGeo mice on vehicle. These results indicate that TAK-418 treatment leads to generalized, albeit partial, rescue of the H3K4me1 deficiency.
Figure 2.
Hippocampal genome-wide levels of H3K4me1 and H3K4me3 demonstrate TAK-418-dependent rescue
(A–C) Comparison of hippocampal H3K4me1 levels of Kmt2d+/βGeo mice on vehicle or TAK-418 compared to Kmt2d+/+ mice on vehicle. (D–F) Comparison of hippocampal H3K4me3 levels of Kmt2d+/βGeo mice on vehicle or TAK-418 compared to Kmt2d+/+ mice on vehicle. Each point corresponds to a genomic location of a peak with a statistically significant difference between the two genotypes. Log2 fold change >0.2 or <0.2 are in red, and others are in dark gray. To allow visualization of changes upon TAK-418 treatment, the green dots indicate H3K4me1/3-bound locations that were significantly increased in Kmt2d+/βGeo mice on vehicle (B and E), and blue dots indicate H3K4me1/3-bound peaks that were decreased in Kmt2d+/βGeo mice on vehicle (B and E).
We next performed H3K4me3 ChIP-seq. As expected, the most H3K4me3 peaks are intragenic, with ∼18%–19% in the promoter region, ∼16%–17% peaks in exons, ∼33%–35% peaks in CpGs in introns, and ∼7% intergenic. There was no obvious difference in overall distribution between the two genotypes, although again we did observe a number of significant sites when comparing Kmt2d+/+ littermates on and off TAK-418 (Figure S4B). We observed 262 loci that were increased and 1,650 loci that were decreased in the Kmt2d+/βGeo mice compared to the Kmt2d+/+ mice when vehicle treated (Figure 2D; Table S3). On average, there is much more H3K4me3 bound in the Kmt2d+/+ mice compared to Kmt2d+/βGeo mice with a ratio of 6.63/1 (Figure 2D, both genotypes on vehicle). In contrast, when we compared Kmt2d+/βGeo mice treated with TAK-418 to Kmt2d+/+ mice treated with vehicle control, we observed 612 significantly increased loci and 1,972 significantly decreased loci in Kmt2d+/βGeo mice compared to Kmt2d+/+ littermates. As before, we observed more H3K4me3 binding upon treatment (3.22/1, Kmt2d+/βGeo mice on TAK-418/Kmt2d+/+ mice on vehicle, Figure 2E; Table S4). An inverse linear regression revealed a correlation coefficient (r) of −0.62636 upon plotting log2 fold change of the common bound loci between the comparison of Kmt2d+/βGeo/Kmt2d+/+ mice on vehicle and Kmt2d+/βGeoon TAK-418/vehicle mice on TAK-418, suggesting that regions with low levels in Kmt2d+/βGeo mice on vehicle increased with TAK-418 and vice versa, indicating a partial rescue of the genome-wide deficiency of H3K4me3 by TAK-418 (Figure 2F).
Global gene expression changes are rescued in Kmt2d+/βGeo mice on TAK-418
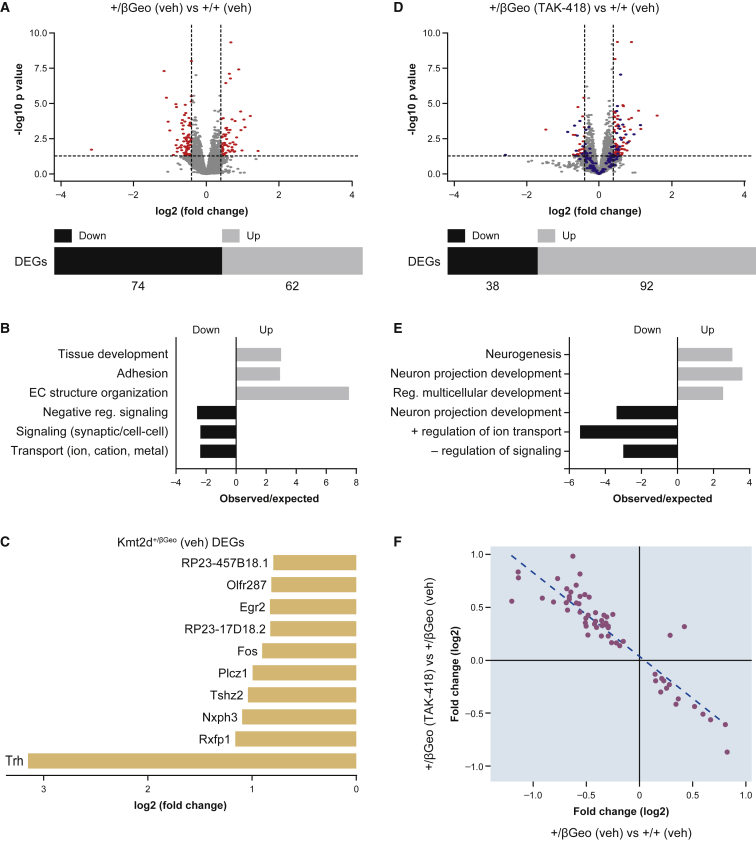
We next interrogated functional effects of the observed differential histone modifications, as measured by changes in gene expression, in order to define a list of potential KMT2D target genes in the hippocampus. We performed RNA sequencing (RNA-seq) on samples from whole hippocampi harvested from Kmt2d+/βGeo mice and Kmt2d+/+ littermates treated with or without TAK-418. Differential gene expression analysis between Kmt2d+/βGeo mice and Kmt2d+/+ littermates treated with vehicle control revealed a total of 136 differentially expressed genes (DEGs) (absolute log2 fold change >0.5, Figure 3A; Table S5). Among the 136 DEGs, 74 genes were downregulated and 62 genes were upregulated in hippocampus from Kmt2d+/βGeo mice compared to Kmt2d+/+ littermates (Figure 3A). Genes that were downregulated in Kmt2d+/βGeo mice compared to Kmt2d+/+ littermates were enriched for networks of ionic transport and negative regulation of synaptic signaling (Figure 3B). The pathways affected by genes upregulated in Kmt2d+/βGeo mice compared to Kmt2d+/+ littermates included tissue development, adhesion, and extracellular structure organization (Figure 3B). After treatment with TAK-418, however, we observed a reversed pattern of expression effects. Among 130 DEGs, only 38 genes were downregulated and 92 genes were upregulated in Kmt2d+/βGeo mice on TAK-418 compared to Kmt2d+/+ littermates treated with vehicle control (Figure 3D; Table S6). Pathways enriched among genes upregulated in Kmt2d+/βGeo mice treated with TAK-418 were involved in neurogenesis and neuronal projection development (Figure 3E). The majority of DEGs (absolute log2 fold change <0.5) in Kmt2d+/βGeo mice compared to Kmt2d+/+ littermates were normalized with TAK-418 treatment (blue dots, Figure 3D). Among the genes that were highly differentially expressed in untreated Kmt2d+/βGeo animals and rescued with TAK-418 were the immediate early genes (IEGs) Fos and Fosb, in addition to neurexophilin-3 (Nxph3), relaxin/insulin-like family peptide receptor 1 (Rxfp1), downstream targets of extracellular signal-regulated kinase (ERK) signaling, and ribosomal protein genes (Figure 3C; Figure S5A). When we plotted the log2 fold change of the common genes found in the comparison of Kmt2d+/βGeo/Kmt2d+/+ mice on vehicle and Kmt2d+/βGeo mice on TAK-418/Kmt2d+/βGeo mice on vehicle, we observed a highly significant inverse correlation (R = −0.9), indicating that TAK-418 treatment of Kmt2d+/βGeo mice rescued the disrupted expression levels of genes in untreated Kmt2d+/βGeo mice (low genes became more highly expressed and vice versa) to approximately what is normally seen in Kmt2d+/+ littermates (Figure 3F). Finally, these three genes appeared to show some favorable responses when H3K4me1 and H3K4me3 traces in treated animals were examined (Figures S5B and S5C).
Figure 3.
TAK-418 rescues gene expression abnormalities in Kmt2d+/βGeo mice
(A and B) An overview of gene expression abnormalities (A, volcano plot) and a summary of major gene categories (B) among differentially expressed genes (DEGs) in hippocampus when comparing Kmt2d+/βGeo and Kmt2d+/+ mice on vehicle. (C–E) Many of the DEGs from vehicle treatment (A) were normalized (D, blue dots) upon a comparison of hippocampal gene expression levels between Kmt2d+/βGeo mice on TAK-418 and Kmt2d+/+ mice on vehicle. Red dots are DEGs with absolute log2 fold change ≥0.5 and p value <0.05. (C) A view of representative (top 10, all downregulated) DEGs between Kmt2d+/βGeo and Kmt2d+/+ mice on vehicle. (F) A correlation of DEGs in Kmt2d+/βGeo (TAK-418/vehicle) and the two genotypes (Kmt2d+/βGeo/Kmt2d+/+ mice on vehicle) reveals TAK-418-dependent rescue.
The IEG Fos shows decreased expression in Kmt2d+/βGeo mice that is rescued with TAK-418
We validated gene expression of some of the most interesting candidate genes by qRT-PCR. NXPH3 is a specific ligand of synaptic alpha-neurexins and is essential for efficient neurotransmitter release.25 Nxph3 expression is significantly downregulated (p < 0.009) in Kmt2d+/βGeo mice compared to Kmt2d+/+ littermates (Figure 4A). Upon treatment with TAK-418, we saw a modest increase of Nxph3 levels (p < 0.09, Figure 4A). Fos and FosB are immediately early genes that respond to extracellular stimuli. Fos, a marker of neuronal activity, has been associated with a number of neural and behavioral responses to acute stimuli expression. Fos expression is downregulated in Kmt2d+/βGeo mice (p < 0.05); however, upon treatment with TAK-418, the expression level of Fos is upregulated significantly (p < 0.03) (Figure 4B). FosB expression trends mirrored those of Fos, but to a lesser degree (Figure 4C). At the protein level, these gene expression abnormalities did not appear to translate to significantly decreased levels in total hippocampus lysate from Kmt2d+/βGeo mice compared with Kmt2d+/+ littermate controls (Figures 4D and 4E). We hypothesize that the lack of differences between the genotypes could be related to the differential cell type composition in samples from brain tissue. In support of this hypothesis, when we stained the mouse brain slices for FOS and counted FOS+ cells and calculated the average number of FOS+ cells per mm2 from 9 to 10 slices per mouse with three to five mice in each examination group (Figure S6), we noticed significantly fewer FOS+ cells per mm2 of DG in Kmt2d+/βGeo mice compared with Kmt2d+/+ littermate controls (Figures 4F and 4G). After 2 weeks of treatment with TAK-418 in Kmt2d+/βGeo mice, the average FOS+ cells per mm2 increased, although not to a significant level (Figures 4F and 4G; Figure S6). Fos mediates a signaling cascade involving phosphorylation by ERKs. Activated (phosphorylated) ERK (phospho-ERK) is known to be one of the regulators of Fos expression and activity. The decrease in Fos RNA and protein levels in Kmt2d+/βGeo mice prompted us to look at the phospho-ERK level in total hippocampal lysate. The ratio of phospho-ERK to total ERK was decreased significantly in Kmt2d+/βGeo mice compared with Kmt2d+/+littermates (Figure S7), supporting the hypothesis that the ERK-based activation of the Fos signaling cascade is disrupted in Kmt2d+/βGeo mice.
Figure 4.
Gene expression abnormalities of disease-relevant candidate genes in Kmt2d+/βGeo mice are reflected in abnormal qRT-PCR, western blot, and immunofluorescence staining
Nxph3, fos, and fosb are disease-relevant DEGs and all demonstrate consistent decreased gene expression in Kmt2d+/βGeo mice compared to littermates. (A–C) For fos, the gene expression change shows significant rescues with TAK-418. (D and E) Western blotting of FOS did not reveal a significant defect at the protein level in Kmt2d+/βGeo mice compared to littermates. (F and G) Immunofluorescence staining of tissue slices from Kmt2d+/βGeo mice and littermates demonstrates a significant defect that rescues upon TAK-418 treatment. (F) is a representative immunofluorescence image of FOS from Kmt2d+/βGeo mice treated with TAK-418. ANOVA: ∗p < 0.05, ∗∗p < 0.01.
TAK-418 rescues the visuospatial learning and memory defect in Kmt2d+/βGeo mice
Adult-born hippocampal neurons become integrated in the DG circuitry where they mediate neuronal plasticity in support of visuospatial learning and pattern discrimination. Previously we have shown that Kmt2d+/βGeo mice have defects in spatial learning and memory formation as evaluated by a Morris water maze test.9 Herein, we confirm9,10 that vehicle-treated Kmt2d+/βGeo mice persistently crossed the platform less frequently (p < 0.0007, Figure 5A) than did vehicle-treated Kmt2d+/+ littermates in the Morris water maze. We also found that Kmt2d+/βGeo mice demonstrated a longer latency to reach the platform (p < 0.0031, Figure 5B) and spend less time on the platform (Figure 5C) and in the quadrant with the platform (p < 0.0026, data not shown) compared to Kmt2d+/+ littermates. The TAK-418 treatment considerably ameliorated the defect observed in Kmt2d+/βGeo mice in the Morris water maze test (Figure 5A). In the final probe trial, the Kmt2d+/βGeo mice treated with TAK-418 had increased frequency of crossing the platform compared to untreated Kmt2d+/βGeo mice (p < 0.0206, Figure 5A). Similarly, Kmt2d+/βGeo mice treated with vehicle consistently had increased latency to find the platform (p < 0.0088, Figure 5B) and spent less time on the platform (p < 0.0131, Figure 5C), both of which were rescued with TAK-418. There did not appear to be any obvious confounders of either drug or genotype on vision or muscle strength based on the results collected from the Morris water maze. Specifically, there were no major side effects on muscles or vision after treatment with TAK-418 for 2–3 weeks, as the average swimming speed was very similar in both genotypes on and off TAK-418 (Figure 5D) and there was no difference in the 3-day visual trial with regard to the latency to find the platform (Figure 5E). During the 5 days of training, the mice improved over time, demonstrating appropriate learning; however, the Kmt2d+/βGeo mice appeared to perform worse (Figure S8) and were significantly worse on day 4 (p < 0.05) but appeared to demonstrate some rescue on TAK-418, although it was not significant (repeated-measures ANOVA).
Figure 5.
TAK-418 rescues the visual spatial learning and memory defect in Kmt2d+/βGeo mice
(A–C) Kmt2d+/βGeo mice have significant abnormalities in the number of times crossing the platform area during the probe trial (A), latency to find the platform (B), and time spent on the platform (C); all defects are rescued on TAK-418. (D and E) These results are not confounded by muscle strength or vision, as Kmt2d+/βGeo mice have similar average speed (D) and no significant defects in a visual flag-finding regimen compared to Kmt2d+/+ mice (E). ANOVA (A–D), repeated-measures ANOVA (E): ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.005.
TAK-418 is well tolerated in mice at therapeutic levels and leads to rescue of splenomegaly, another disease-relevant phenotype
We observed no obvious side effects in the mice that were on TAK-418, including no effect on weight or general well-being (data not shown). We have previously observed splenomegaly at or around 4 months of age (data not shown). Given this finding, we kept a cohort of mice on TAK-418 for 8 weeks (starting at 2 months of age) and again observed no obvious side effect or impact on weight (no weight loss or weight gain). We did, however, observe splenomegaly in the Kmt2d+/βGeo mice treated with vehicle, but correction of the splenomegaly in Kmt2d+/βGeo mice on TAK-418 (Figure S9), indicating that this phenotype may also be malleable by TAK-418 treatment.
Discussion
Post-translational modifications of histone proteins, such as acetylation, methylation, phosphorylation, and ubiquitination, are thought to serve as crucial regulatory signals to control gene expression in eukaryotic cells and are important for maintaining genomic integrity.26 Emerging evidence suggests that dysregulation of epigenetic modifications is mechanistically linked to both cancer and developmental defects, including neurodevelopmental disorders.27 Histone methylation, in particular, confers active or repressive chromatin states in a locus-specific manner. The histone methylation state is meticulously regulated by the balance between two opposing enzyme systems: KMTs and lysine demethylases (KDMs). Disease-causing variants in KMTs and KDMs have been identified in multiple disorders in patients with intellectual disability, indicating that altered regulation of histone methylation can lead to intellectual disability.28 Pharmaceutical inhibition of epigenetic targets counteracting the epigenetic effects of these loss-of-function variants has been investigated as a possible therapeutic option in several epigenetic conditions, including KS.9,10,29
In recent years, data have emerged that support the notion that KMT2D dysfunction affects cellular function in the hippocampus. These include several studies that report more severe visuospatial disruption (linked to hippocampus) in molecularly confirmed patients with KS compared to other individuals with non-KS intellectual disability.30,31 MRI images also suggest a grossly smaller hippocampus in individuals with molecularly confirmed KS.32 These studies help set the stage for a potential clinical trial, as do recently developed international diagnostic criteria for KS.2 These data also support the strategy to focus on hippocampus and DG to estimate the effect of treatment, although they in no way exclude that other brain regions or cell types play a role in the neurological disease phenotype in KS.
Recently, genetic targeting of KDM1A was found to rescue a cellular phenotype observed with loss of function of KMT2D in embryonic stem cells.13 KDM1A, also known as LSD1 or AOF2, is the first identified FAD-dependent histone demethylase capable of specifically demethylating mono- and di-methylated lysine 4 of histone H3 (H3K4me1 and H3K4me2), the very marks placed by KMT2D.15 This suggests the feasibility of a more targeted therapeutic strategy for KS, namely KDM1A inhibition. KDM1A associates with HDAC1/2, CoREST, BHC80, and BRAF35.16 BRAF35, with its HMG DNA-binding domain, is thought to recruit the KDM1A/CoREST complex to the target sites. Subsequent deacetylation of the histone tail by the HDAC1/2 at the target sites then enables KDM1A to demethylate H3K4.17 It has been demonstrated that hyperacetylated nucleosomes are less susceptible to CoREST/LSD1-mediated demethylation,33 suggesting that hypoacetylated nucleosomes may be the preferred physiological substrates. We have previously shown that AR-42, an HDACi, can rescue the learning and memory defect in Kmt2d+/βGeo mice9 after a short term (2–4 weeks) of treatment. However, AR-42, a pan-histone deacetylase inhibitor, likely impacts deacetylase activities indiscriminately across a range of distinct HDAC-containing multiprotein complexes. Such broad cellular effects may result in a narrow therapeutic window between disease efficacy and toxicity. Indeed, when we treated Kmt2d+/βGeo mice with AR-42, we began to observe the effect at 5 mg/kg/day, with the full effect obtained at 10 mg/kg/day. At 25 mg/kg/day, however, we started to observe a cytotoxicity effect, reflected by fewer DCX+ cells compared with Kmt2d+/βGeo mice treated with vehicle.9 We had postulated that the treatment effect was likely indirect, primarily affecting histone acetylation, with a secondary effect on histone methylation.9 An alternative hypothesis is that AR-42 exerts its effect through inhibition of HDAC1/2 activity in the KDM1A-CoREST complex, leading to hyperacetylation of the target sites, which are more resistant to KDM1A-mediated demethylation, thus retaining the H3K4 methylation and thus over time retaining more H3K4 methylation. This may mean that low-dose combined treatment with deacetylase inhibitor and demethylase inhibitor or dual histone deacetylase and demethylase inhibitors targeting the CoREST complex may have synergistic effect and be a particularly effective treatment for KS. In contrast to AR-42, TAK-418 achieved the full effect at 1 mg/kg/day with regard to DCX+ cells, and no adverse effects have been observed with a higher dose of TAK-418, and H3K4 methylation levels did not change in WT animals on TAK-418. This may suggest that Kmt2d+/βGeo mice may be more susceptible to such changes than WT littermates and perhaps through ongoing epigenetic compensation through the many layers of epigenetic modification.34 Thus, our data suggest that TAK-418, as a specific inhibitor to KDM1A, may have the potential to be a single-agent treatment for KS through its effects on H3K4 methylation; however, it is noteworthy that this phenotypic rescue occurred in the context of incomplete rescue of H3K4 methylation. This suggests that perhaps the histone methylation needs to reach some threshold that lies under the normal levels of H3K4 methylation.
In addition to the expected global increase of H3K4me1 and H3K4me2 after treatment with TAK-418 in Kmt2d+/βGeo mice, we also observed a global increase of H3K4me3, likely due to the accumulation of H3K4me2 and subsequent conversion into H3K4me3 by H3K4 methyltransferases. Alternatively, the KDM1A complex has been shown to be unstable while binding to methylated targets sites. iBRAF, the paralog of BRAF35,35 may compete with BRAF35 for the same target sites and recruit the KMT2A complex, which subsequently may enhance trimethylation of H3K4, however; this will need to be explored in future studies. Although KDM1A-inhibiting treatment is promising for KS, the dose will need to be optimized. This is clear because missense mutations in KDM1A have been identified in three individuals with developmental delay,36, 37, 38 indicating that too much KDM1A inhibition can be damaging. The developmental symptoms in these individuals are similar to those of KS (MIM: 147920), characterized by distinct craniofacial features, including widely spaced teeth and palatal abnormalities, indicating that too much disruption of KDM1A activity could also be detrimental to intellectual function. Although this protein also has non-epigenetic function, it is likely that the cause of the phenotypes relates to its epigenetic function because biochemical studies have demonstrated that these mutant proteins exhibit reduced stability and demethylase activity,38 indicating a loss-of-function mechanism.
Our RNA and ChIP-seq data indicate that the IEGs such as Fos and FosB may be one class of genes that are affected in KS and show rescue on TAK-418. Rusconi et al.39,40 have shown that the KDM1A complex interacts with serum response factor (SRF) under resting conditions, and an enrichment of the KDM1A complex with SRF has been detected in the c-fos promoter, which contains the serum response element (SRE). SRF is known to be constitutively bound to the DNA of its target genes, and the interaction of the KDM1A complex with SRF modulates the H3K4 methylation level at the fos promoter. We also noted gene expression changes that would be consistent with decreased ERK signaling and, in fact, ERK activation appears deficient in Kmt2d+/βGeo hippocampi compared to those in littermates. Previous data from fibroblasts from KS individuals and zebrafish models of KS have implicated decreased activation of this pathway,41,42 which is concordant with our observations. Thus, both studies reveal abnormalities of ERK signaling, and future studies should further elucidate whether ERK abnormalities play a mechanistic role in the pathogenesis of KS.
In summary, in this study we show that oral administration of a KDM1A specific inhibitor, TAK-418, can ameliorate neurological problems at the cellular, molecular, gene expression, and functional levels in a mouse model of KS (Kmt2d+/βGeo mice). TAK-418 treatment increases adult neurogenesis in adult mice, as indicated by increased DCX+ cells in the granule cell layer of the hippocampus. At the molecular level, TAK-418 increases the global level of mono-, di-, and tri-methylated H3K4 (H3K4me1/2/3) in Kmt2d+/βGeo mice as assessed by both western blot and ChIP-seq. TAK-418 treatment also corrects the differential gene expression profile abnormalities found in Kmt2d+/βGeo mice compared to Kmt2d+/+ littermates. Finally, and most importantly, we show that TAK-418 can correct the functional deficits by improving the learning and memory behavior of Kmt2d+/βGeo mice. Currently, we do not know whether these effects will translate in humans. However, the present data are informative in respect to the TAK-418 dose range and exposure to be achieved to produce pharmacologically relevant effects in Kmt2d+/βGeo mice. In summary, our data support the hypothesis that KS is a treatable cause of intellectual disability and that KDM1A inhibition may be a novel and effective mechanism of action for the treatment of KS.
Materials and methods
Animals
Our mouse model, Kmt2d+/βGeo, also named Mll2Gt(RRt024)Byg, was originally acquired from Bay Genomics (University of California) but backcrossed in the Bjornsson laboratory. All experimental mice were on a fully backcrossed C57BL/6J background (99% verified using a mouse SNP chip). For treatment with TAK-418, mice were orally gavaged daily with drug (TAK-418, Takeda) solubilized in vehicle (methylcellulose) or with vehicle alone. Both drug and vehicle (methylcellulose) were shipped from Takeda. Drug was administered for 14 days for adult neurogenesis studies, after which mice were sacrificed on day 15. For behavioral studies, Morris water maze testing was initiated at day 15 after treatment start at ∼8 weeks of age, and the drug was continued throughout the behavioral studies (until at least day 23). A dose curve was initially performed with three doses (0.5, 1, and 2 mg/kg/day). However, after that initial study, all other experiments were done with a dose of 1 mg/kg/day. For quantification of spleen size, evaluation was performed after 8 weeks of TAK-418 started at ∼8 weeks of age, since splenomegaly is not observed until 12–16 weeks. For this reason, this cohort was kept on TAK-418 for ∼8 weeks and to evaluate for side effects. Mouse numbers for individual studies were as follows: ChIP-seq/RNA-seq, 3–4 per group; immunohistofluorescence, 11–12 per group; qRT-PCR, 5–6 per group; and behavioral testing, 24–30 per group. Genotyping was performed using the following primers: Mll2_exon50F (forward), 5′-CTGTGTGGAACCGCATCATTG-3′; Mll2_exon51R (reverse), 5′-CGGTTCTGATCTGGCACAGCC-3′; and β-GeoR1, 5′-CTCAGTGCAGTGCAGTCAGG-3′. The Mll2_exon50F and Mll2_exon51R pair amplifies sequences from the wild allele of KMT2D, and the Mll2_exon50F and β-GeoR1 pair amplifies sequences specific for the targeted allele. All experiments were performed using mouse protocols approved by the Animal Care and Use Committee of Johns Hopkins University School of Medicine. The mouse protocols used for this study are in accordance with the guidelines used by the National Institutes of Health (NIH) for mouse care and handling.
Perfusion and cryosectioning
Mice were sacrificed with a lethal dose of xylazine/ketamine combination, after which they were transcardially flushed with PBS (1×) with heparin and then perfused with 4% paraformaldehyde (PFA)/PBS. Brains were removed from the skulls and cryopreserved in 30% sucrose 0.1 M phosphate solution overnight at 4°C. Brains were frozen and sectioned using a Microm HM550 cryostat (Thermo Scientific). Sectioning was performed at 30-μm intervals, and every section of the brain was collected onto the slide in a series of six slides with 12 slices on each slide and stored in a −80°C freezer prior to use.
Immunofluorescence staining
Pre-mounted slides were thawed and washed once in Tris-buffered saline (TBS) (1×). Slices were briefly permeabilized in TBS (1×) with 0.4% Triton X-100 (30 min), followed by blocking with TBS with 3% donkey serum and 0.05% Triton X-100 for 1 h at room temperature. Next, each slide was incubated with primary antibody (in TBS with 3% donkey serum and 0.05% Triton X-100) overnight (O/N) at 4°C. After three to five washes in TBS with 0.05% Triton X-100, Alexa Fluor-conjugated secondary antibodies with DAPI counterstain were added to the slide for 1 h at room temperature. Slides were mounted with ProLong antifade (Thermo Fisher Scientific, catalog #P36930) after several washes in TBS with 0.05% Triton X-100 and a final wash of TBS. Images were taken with an LSM 780 confocal microscope. Antibodies used included anti-DCX (Santa Cruz, catalog #sc-8066) and anti-c-fos (Abcam, catalog #190289).
Granule cell layer and quantification of DCX+ cells
The area of the granule cell layer and DCX+ cells was measured using ImageJ as previously described.9 Briefly, we traced the granule cell layer by hand and counted DCX+ cells within this area in all slides. We performed an independent count of all DCX+ processes. Counting was performed by an investigator who was blinded to genotype and/or drug exposure.
Western blotting
Hippocampus was dissected, snap-frozen, and kept at −80°C. Histones were extracted following a published acid extraction protocol.43 Briefly, 250 μL of TED buffer (0.5% Triton X-100, 2 mM PMSF, and 0.02% NaN3 in PBS) was added and hippocampal tissue was disaggregated by 50–60 strokes with a pellet pestle. Nuclear pellets were collected at 10,000 rpm for 1 min at 4°C and resuspended in 50 μL of acid extraction buffer (0.5 N HCl in 10% glycerol). After O/N incubation in acid extraction buffer, the supernatant was collected and histones were precipitated with acetone O/N at −20°C. Histone pellets were subsequently dissolved in water. For whole-cell lysate, we disaggregated hippocampal tissue in radioimmunoprecipitation assay (RIPA) buffer (150 mM NaCl, 1.0% Nonidet P-40 [NP-40], 0.5% sodium deoxycholate, 0.1% SDS, 50 mM Tris [pH 8.0]) by 50–60 strokes with a pellet pestle. After incubation for 2 h at 4°C, supernatant was collected. Amount of protein was quantified using a BSA protein assay. Proteins were fractionated in 4%–12% NuPAGE Bis-Tris gel. Antibodies used included the following: anti-histone H3 (Cell Signaling Technology, catalog #3638), anti-H3K4me3 (Cell Signaling Technology, catalog # 9727), anti-H3K4me2 (Cell Signaling Technology, catalog #9725), anti-H3K4me1(Abcam, catalog #ab8895), anti-c-fos (Abcam, catalog #190289), anti-phospho-p44/42 mitogen-activated protein kinase (MAPK) (Erk1/2) (Cell Signaling Technology, catalog #9106), anti-p44/42 MAPK (Erk1/2) (Cell Signaling Technology, catalog #9102), anti-β-actin (Cell Signaling Technology, catalog#3700).
Behavioral testing
No exclusion criteria were used other than decreased visual acuity as evaluated by visible platform training (see below). The investigator was blinded to genotype and drug exposure status during testing. For Morris water maze testing, mice were placed in a 1.1-m diameter tank filled with room temperature water dyed with nontoxic white paint. For analytical purposes, the tank was divided into four quadrants, with one quadrant containing a small platform submerged 1.5 cm beneath the water. On each day of training, mice were placed in the tank in a random quadrant facing away from the center and allowed to swim until they found the platform. Once they reached the platform, they were left there for 30 s. If they did not reach the platform after 60 s, they were placed on it for 30 s. Each mouse was given four trials per day (for 5 days) with an inter-trial interval of 5–20 min and subsequently returned to its home cage. Latency to reach the platform was measured during each trial. The day after the final day of training, the platform was removed for a probe trial where mice were placed in the tank for 90 s. The average number of crossings of the platform’s previous location was recorded. Visible/flagged platform training was also performed for 3 days right before the hidden platform to ensure no problems with sight. During visible training, a visible flag was placed on the submerged platform, and the time for each mouse to reach the platform was measured during each 60-s trial, four of which were run in the same way as the hidden platform training. For all training and probe testing, data were recorded both manually and electronically with ANY-maze software (San Diego Instruments) when applicable. Differences in the number of platform crossings as well as the latency of the first crossing of the platform during the probe trial were compared between groups with an ANOVA with the significance value set at p < 0.05.
RNA-Seq/ChIP-Seq
For RNA-Seq and ChIP-Seq, 2-week-old mice were given TAK-418 at a dose of 1 mg/kg body weight for a period of 2 weeks. Hippocampi were harvested at the end of the 2-week treatment. RNA was extracted with a Direct-zol RNA microprep kit (Zymo Research), and RNA-seq libraries were constructed with an NEBNext Ultra RNA library prep kit for Illumina. Pooled RNA libraries were sequenced on a HiSeq 2500 using 150-bp paired-end sequencing. For ChIP-seq library construction, nuclear lysate preparation and ChIP followed the ENCODE protocol from the Bing Ren laboratory with chip graded anti-H3K4me3 (Abcam, catalog #ab8580) and anti-H3K4me1 (Abcam, catalog #ab8895). After reverse cross-linking and purifying the DNA fragments, library construction was performed with the NEBNext Ultra II DNA library prep kit for Illumina based on the manufacturer’s recommendations. Pooled ChIP DNA libraries were sequenced on a HiSeq 2500 using a flow cell for 150-bp paired-end size.
RNA-Seq/ChIP-Seq analysis
RNA-Seq
Transcriptomic data collected by RNA-seq were analyzed to determine the genes that were present in each sample and condition, their expression levels, and the differences between expression levels among different experimental conditions. Following quality checking with the software FastQC, reads were mapped to the mouse genome version mm10 with the alignment tool TopHat2 v2.1.0,44 which allows for large “gaps” in the alignment, representing introns. The aligned reads were assembled with CLASS2 v2.1.745 to create partial genes and transcript models (transfrags). Transfrags from all samples were further merged with cuffmerge and mapped to the GENCODE M17 gene models46 to create a unified set of gene annotations for differential analyses. Lastly, gene (transcript) expression levels were computed, and DEGs (transcripts) were determined separately with the tools Cuffdiff2 v2.2.147 and DESeq.48 DEGs were further analyzed and graphed with R, a public free resource for statistical computing and graphing.
ChIP-Seq
The reads collected by H3K4me1 and H3K4me3 ChIP sequencing were first trimmed 5 bp at 5′ end with the software seqtk (https://github.com/lh3/seqtk), followed by mapping with the short sequence alignment tool Bowtie 2.49 The aligned sequences were indexed with software SAMtools for easy viewing in IGV (Integrated Genome Viewer). The THOR software50 was then used to detect and analyze differential peaks in two sets of ChIP-seq data from distinct biological conditions with replicates. HOMER51 and Gene Ontology (GO) analysis (http://geneontology.org/) was used to annotate the peaks, and custom scripts were written to filter THOR output, gather statistics, and reformat files. Differential peaks were further analyzed and graphed with R. All data have been posted to GEO (GEO: GSE146727, GSE146728, and GSE146729).
Acknowledgments
We are thankful for statistical analysis by Dr. Liliana Florea and Corina Antonescu, through the Computational Biology Consulting Core, and support by Dr. Pletnikov in the JHMI behavioral core. H.T.B. is funded by the following sources: NIH (DP5OD017877), USA, the Louma G. Foundation, USA the Walter Zaitzeff Fund, USA the Icelandic Research Fund (195835-051 and 206806-051), Iceland and, for this project, with a grant from Takeda Pharmaceutical Company, Japan.
Author contributions
E.M.P, J.D., A.N., and H.T.B. came up with idea; L.Z. and G.P. performed all experiments; L.Z., G.P., and H.T.B. wrote the manuscript and analyzed all data; and R.B., S.M., M.D., Y.H., M.I., and H.K. developed and provided TAK-418.
Declaration of interest
This work was partially supported with a grant from Takeda Pharmaceutical Company, who owns rights to TAK-418. E.M.P., J.D., A.N., R.B., S.M., M.D., Y.H., S.M., M.I., and H.K. are employees of Takeda Pharmaceutical Company.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtm.2021.02.011.
Supplemental information
References
- 1.Adam M.P., Hudgins L. Kabuki syndrome: a review. Clin. Genet. 2005;67:209–219. doi: 10.1111/j.1399-0004.2004.00348.x. [DOI] [PubMed] [Google Scholar]
- 2.Adam M.P., Banka S., Bjornsson H.T., Bodamer O., Chudley A.E., Harris J., Kawame H., Lanpher B.C., Lindsley A.W., Merla G., Kabuki Syndrome Medical Advisory Board Kabuki syndrome: international consensus diagnostic criteria. J. Med. Genet. 2019;56:89–95. doi: 10.1136/jmedgenet-2018-105625. [DOI] [PubMed] [Google Scholar]
- 3.Ng S.B., Bigham A.W., Buckingham K.J., Hannibal M.C., McMillin M.J., Gildersleeve H.I., Beck A.E., Tabor H.K., Cooper G.M., Mefford H.C. Exome sequencing identifies MLL2 mutations as a cause of Kabuki syndrome. Nat. Genet. 2010;42:790–793. doi: 10.1038/ng.646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lederer D., Grisart B., Digilio M.C., Benoit V., Crespin M., Ghariani S.C., Maystadt I., Dallapiccola B., Verellen-Dumoulin C. Deletion of KDM6A, a histone demethylase interacting with MLL2, in three patients with Kabuki syndrome. Am. J. Hum. Genet. 2012;90:119–124. doi: 10.1016/j.ajhg.2011.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guo C., Chen L.H., Huang Y., Chang C.C., Wang P., Pirozzi C.J., Qin X., Bao X., Greer P.K., McLendon R.E. KMT2D maintains neoplastic cell proliferation and global histone H3 lysine 4 monomethylation. Oncotarget. 2013;4:2144–2153. doi: 10.18632/oncotarget.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim J.H., Sharma A., Dhar S.S., Lee S.H., Gu B., Chan C.H., Lin H.K., Lee M.G. UTX and MLL4 coordinately regulate transcriptional programs for cell proliferation and invasiveness in breast cancer cells. Cancer Res. 2014;74:1705–1717. doi: 10.1158/0008-5472.CAN-13-1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Agger K., Cloos P.A., Christensen J., Pasini D., Rose S., Rappsilber J., Issaeva I., Canaani E., Salcini A.E., Helin K. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature. 2007;449:731–734. doi: 10.1038/nature06145. [DOI] [PubMed] [Google Scholar]
- 8.Fahrner J.A., Bjornsson H.T. Mendelian disorders of the epigenetic machinery: tipping the balance of chromatin states. Annu. Rev. Genomics Hum. Genet. 2014;15:269–293. doi: 10.1146/annurev-genom-090613-094245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bjornsson H.T., Benjamin J.S., Zhang L., Weissman J., Gerber E.E., Chen Y.C., Vaurio R.G., Potter M.C., Hansen K.D., Dietz H.C. Histone deacetylase inhibition rescues structural and functional brain deficits in a mouse model of Kabuki syndrome. Sci. Transl. Med. 2014;6:256ra135. doi: 10.1126/scitranslmed.3009278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Benjamin J.S., Pilarowski G.O., Carosso G.A., Zhang L., Huso D.L., Goff L.A., Vernon H.J., Hansen K.D., Bjornsson H.T. A ketogenic diet rescues hippocampal memory defects in a mouse model of Kabuki syndrome. Proc. Natl. Acad. Sci. USA. 2017;114:125–130. doi: 10.1073/pnas.1611431114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pilarowski G.O., Cazares T., Zhang L., Benjamin J.S., Liu K., Jagannathan S., Mousa N., Kasten J., Barski A., Lindsley A.W., Bjornsson H.T. Abnormal Peyer patch development and B-cell gut homing drive IgA deficiency in Kabuki syndrome. J. Allergy Clin. Immunol. 2020;145:982–992. doi: 10.1016/j.jaci.2019.11.034. [DOI] [PubMed] [Google Scholar]
- 12.Subramanian S., Bates S.E., Wright J.J., Espinoza-Delgado I., Piekarz R.L. Clinical toxicities of histone deacetylase inhibitors. Pharmaceuticals (Basel) 2010;3:2751–2767. doi: 10.3390/ph3092751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cao K., Collings C.K., Morgan M.A., Marshall S.A., Rendleman E.J., Ozark P.A., Smith E.R., Shilatifard A. An Mll4/COMPASS-Lsd1 epigenetic axis governs enhancer function and pluripotency transition in embryonic stem cells. Sci. Adv. 2018;4:eaap8747. doi: 10.1126/sciadv.aap8747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dhar S.S., Zhao D., Lin T., Gu B., Pal K., Wu S.J., Alam H., Lv J., Yun K., Gopalakrishnan V. MLL4 is required to maintain broad H3K4me3 peaks and super-enhancers at tumor suppressor genes. Mol. Cell. 2018;70:825–841.e6. doi: 10.1016/j.molcel.2018.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shi Y., Lan F., Matson C., Mulligan P., Whetstine J.R., Cole P.A., Casero R.A., Shi Y. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 16.Shi Y.J., Matson C., Lan F., Iwase S., Baba T., Shi Y. Regulation of LSD1 histone demethylase activity by its associated factors. Mol. Cell. 2005;19:857–864. doi: 10.1016/j.molcel.2005.08.027. [DOI] [PubMed] [Google Scholar]
- 17.Hou H., Yu H. Structural insights into histone lysine demethylation. Curr. Opin. Struct. Biol. 2010;20:739–748. doi: 10.1016/j.sbi.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kooistra S.M., Helin K. Molecular mechanisms and potential functions of histone demethylases. Nat. Rev. Mol. Cell Biol. 2012;13:297–311. doi: 10.1038/nrm3327. [DOI] [PubMed] [Google Scholar]
- 19.van Praag H., Schinder A.F., Christie B.R., Toni N., Palmer T.D., Gage F.H. Functional neurogenesis in the adult hippocampus. Nature. 2002;415:1030–1034. doi: 10.1038/4151030a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao C., Deng W., Gage F.H. Mechanisms and functional implications of adult neurogenesis. Cell. 2008;132:645–660. doi: 10.1016/j.cell.2008.01.033. [DOI] [PubMed] [Google Scholar]
- 21.Couillard-Despres S., Winner B., Schaubeck S., Aigner R., Vroemen M., Weidner N., Bogdahn U., Winkler J., Kuhn H.G., Aigner L. Doublecortin expression levels in adult brain reflect neurogenesis. Eur. J. Neurosci. 2005;21:1–14. doi: 10.1111/j.1460-9568.2004.03813.x. [DOI] [PubMed] [Google Scholar]
- 22.von Bohlen Und Halbach O. Immunohistological markers for staging neurogenesis in adult hippocampus. Cell Tissue Res. 2007;329:409–420. doi: 10.1007/s00441-007-0432-4. [DOI] [PubMed] [Google Scholar]
- 23.Francis F., Koulakoff A., Boucher D., Chafey P., Schaar B., Vinet M.C., Friocourt G., McDonnell N., Reiner O., Kahn A. Doublecortin is a developmentally regulated, microtubule-associated protein expressed in migrating and differentiating neurons. Neuron. 1999;23:247–256. doi: 10.1016/s0896-6273(00)80777-1. [DOI] [PubMed] [Google Scholar]
- 24.Gleeson J.G., Lin P.T., Flanagan L.A., Walsh C.A. Doublecortin is a microtubule-associated protein and is expressed widely by migrating neurons. Neuron. 1999;23:257–271. doi: 10.1016/s0896-6273(00)80778-3. [DOI] [PubMed] [Google Scholar]
- 25.Beglopoulos V., Montag-Sallaz M., Rohlmann A., Piechotta K., Ahmad M., Montag D., Missler M. Neurexophilin 3 is highly localized in cortical and cerebellar regions and is functionally important for sensorimotor gating and motor coordination. Mol. Cell. Biol. 2005;25:7278–7288. doi: 10.1128/MCB.25.16.7278-7288.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tessarz P., Kouzarides T. Histone core modifications regulating nucleosome structure and dynamics. Nat. Rev. Mol. Cell Biol. 2014;15:703–708. doi: 10.1038/nrm3890. [DOI] [PubMed] [Google Scholar]
- 27.Bjornsson H.T. The Mendelian disorders of the epigenetic machinery. Genome Res. 2015;25:1473–1481. doi: 10.1101/gr.190629.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Faundes V., Newman W.G., Bernardini L., Canham N., Clayton-Smith J., Dallapiccola B., Davies S.J., Demos M.K., Goldman A., Gill H., Clinical Assessment of the Utility of Sequencing and Evaluation as a Service (CAUSES) Study. Deciphering Developmental Disorders (DDD) Study Histone lysine methylases and demethylases in the landscape of human developmental disorders. Am. J. Hum. Genet. 2018;102:175–187. doi: 10.1016/j.ajhg.2017.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alarcón J.M., Malleret G., Touzani K., Vronskaya S., Ishii S., Kandel E.R., Barco A. Chromatin acetylation, memory, and LTP are impaired in CBP+/− mice: a model for the cognitive deficit in Rubinstein-Taybi syndrome and its amelioration. Neuron. 2004;42:947–959. doi: 10.1016/j.neuron.2004.05.021. [DOI] [PubMed] [Google Scholar]
- 30.Harris J., Mahone E.M., Bjornsson H.T. Molecularly confirmed Kabuki (Niikawa-Kuroki) syndrome patients demonstrate a specific cognitive profile with extensive visuospatial abnormalities. J. Intellect. Disabil. Res. 2019;63:489–497. doi: 10.1111/jir.12596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van Dongen L.C.M., Wingbermühle P.A.M., van der Veld W.M., Stumpel C., Kleefstra T., Egger J.I.M. Exploring the cognitive phenotype of Kabuki (Niikawa-Kuroki) syndrome. J. Intellect. Disabil. Res. 2019;63:498–506. doi: 10.1111/jir.12597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boisgontier J., Tacchella J.M., Lemaître H., Lehman N., Saitovitch A., Gatinois V., Boursier G., Sanchez E., Rechtman E., Fillon L. Anatomical and functional abnormalities on MRI in Kabuki syndrome. Neuroimage Clin. 2019;21:101610. doi: 10.1016/j.nicl.2018.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee M.G., Wynder C., Bochar D.A., Hakimi M.A., Cooch N., Shiekhattar R. Functional interplay between histone demethylase and deacetylase enzymes. Mol. Cell. Biol. 2006;26:6395–6402. doi: 10.1128/MCB.00723-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.LaSalle J.M., Powell W.T., Yasui D.H. Epigenetic layers and players underlying neurodevelopment. Trends Neurosci. 2013;36:460–470. doi: 10.1016/j.tins.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wynder C., Hakimi M.A., Epstein J.A., Shilatifard A., Shiekhattar R. Recruitment of MLL by HMG-domain protein iBRAF promotes neural differentiation. Nat. Cell Biol. 2005;7:1113–1117. doi: 10.1038/ncb1312. [DOI] [PubMed] [Google Scholar]
- 36.Tunovic S., Barkovich J., Sherr E.H., Slavotinek A.M. De novo ANKRD11 and KDM1A gene mutations in a male with features of KBG syndrome and Kabuki syndrome. Am. J. Med. Genet. A. 2014;164A:1744–1749. doi: 10.1002/ajmg.a.36450. [DOI] [PubMed] [Google Scholar]
- 37.Chong J.X., Yu J.H., Lorentzen P., Park K.M., Jamal S.M., Tabor H.K., Rauch A., Saenz M.S., Boltshauser E., Patterson K.E. Gene discovery for Mendelian conditions via social networking: de novo variants in KDM1A cause developmental delay and distinctive facial features. Genet. Med. 2016;18:788–795. doi: 10.1038/gim.2015.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pilotto S., Speranzini V., Marabelli C., Rusconi F., Toffolo E., Grillo B., Battaglioli E., Mattevi A. LSD1/KDM1A mutations associated to a newly described form of intellectual disability impair demethylase activity and binding to transcription factors. Hum. Mol. Genet. 2016;25:2578–2587. doi: 10.1093/hmg/ddw120. [DOI] [PubMed] [Google Scholar]
- 39.Rusconi F., Grillo B., Ponzoni L., Bassani S., Toffolo E., Paganini L., Mallei A., Braida D., Passafaro M., Popoli M. LSD1 modulates stress-evoked transcription of immediate early genes and emotional behavior. Proc. Natl. Acad. Sci. USA. 2016;113:3651–3656. doi: 10.1073/pnas.1511974113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rusconi F., Grillo B., Toffolo E., Mattevi A., Battaglioli E. NeuroLSD1: splicing-generated epigenetic enhancer of neuroplasticity. Trends Neurosci. 2017;40:28–38. doi: 10.1016/j.tins.2016.11.002. [DOI] [PubMed] [Google Scholar]
- 41.Bögershausen N., Tsai I.C., Pohl E., Kiper P.Ö., Beleggia F., Percin E.F., Keupp K., Matchan A., Milz E., Alanay Y. RAP1-mediated MEK/ERK pathway defects in Kabuki syndrome. J. Clin. Invest. 2015;125:3585–3599. doi: 10.1172/JCI80102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tsai I.C., McKnight K., McKinstry S.U., Maynard A.T., Tan P.L., Golzio C., White C.T., Price D.J., Davis E.E., Amrine-Madsen H., Katsanis N. Small molecule inhibition of RAS/MAPK signaling ameliorates developmental pathologies of Kabuki Syndrome. Sci. Rep. 2018;8:10779. doi: 10.1038/s41598-018-28709-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shechter D., Dormann H.L., Allis C.D., Hake S.B. Extraction, purification and analysis of histones. Nat. Protoc. 2007;2:1445–1457. doi: 10.1038/nprot.2007.202. [DOI] [PubMed] [Google Scholar]
- 44.Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S.L. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Song L., Sabunciyan S., Florea L. CLASS2: accurate and efficient splice variant annotation from RNA-seq reads. Nucleic Acids Res. 2016;44:e98. doi: 10.1093/nar/gkw158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Frankish A., Diekhans M., Ferreira A.M., Johnson R., Jungreis I., Loveland J., Mudge J.M., Sisu C., Wright J., Armstrong J. GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res. 2019;47(D1):D766–D773. doi: 10.1093/nar/gky955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Trapnell C., Hendrickson D.G., Sauvageau M., Goff L., Rinn J.L., Pachter L. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol. 2013;31:46–53. doi: 10.1038/nbt.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Anders S., Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Allhoff M., Seré K., F Pires J., Zenke M., G Costa I. Differential peak calling of ChIP-seq signals with replicates with THOR. Nucleic Acids Res. 2016;44:e153. doi: 10.1093/nar/gkw680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., Glass C.K. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.