Abstract
Objective
To evaluate the accuracy of a multi-stage convolutional neural network (CNN) model-based automated identification system for posteroanterior (PA) cephalometric landmarks.
Methods
The multi-stage CNN model was implemented with a personal computer. A total of 430 PA-cephalograms synthesized from cone-beam computed tomography scans (CBCT-PA) were selected as samples. Twenty-three landmarks used for Tweemac analysis were manually identified on all CBCT-PA images by a single examiner. Intra-examiner reproducibility was confirmed by repeating the identification on 85 randomly selected images, which were subsequently set as test data, with a two-week interval before training. For initial learning stage of the multi-stage CNN model, the data from 345 of 430 CBCT-PA images were used, after which the multi-stage CNN model was tested with previous 85 images. The first manual identification on these 85 images was set as a truth ground. The mean radial error (MRE) and successful detection rate (SDR) were calculated to evaluate the errors in manual identification and artificial intelligence (AI) prediction.
Results
The AI showed an average MRE of 2.23 ± 2.02 mm with an SDR of 60.88% for errors of 2 mm or lower. However, in a comparison of the repetitive task, the AI predicted landmarks at the same position, while the MRE for the repeated manual identification was 1.31 ± 0.94 mm.
Conclusions
Automated identification for CBCT-synthesized PA cephalometric landmarks did not sufficiently achieve the clinically favorable error range of less than 2 mm. However, AI landmark identification on PA cephalograms showed better consistency than manual identification.
Keywords: Artificial intelligence, Convolutional neural networks, Posteroanterior cephalometrics, Cone-beam computed tomography
INTRODUCTION
Posteroanterior (PA) cephalometric analysis is a useful tool for evaluating the cranial-dentofacial structure and growth pattern in the transverse plane.1-3 PA cephalometric analysis in combination with lateral cephalometric analysis provides a substantial amount of diagnostic data for a comprehensive three-dimensional assessment in daily practice.4-6 Therefore, PA cephalograms have proven to be indispensable diagnostic and evaluation tools in planning comprehensive orthodontic treatment.
Nevertheless, some studies have debated the reliability and reproducibility of landmark identification on PA cephalograms. In general, inter-examiner error was reported to be significantly higher than intra-examiner error in assessments based on PA cephalograms.1,3-5 The effect of head positioning on the accuracy of landmark identification with PA cephalometric analysis is greater than with lateral cephalometric analysis.6 Therefore, landmark identification errors in the form of empirical differences that generate major errors in cephalometric analysis are inevitable.7,8 Moreover, conventional two-dimensional radiograms translate a stereoscopic anatomic structure to a planar structure, generating layered images that obscure the landmarks,9,10 and rotation of the head position can generate distortion to induce interference in landmark identification.11
The application of artificial intelligence (AI) techniques in automated landmark identification is a new approach for cephalometric analysis that aims to facilitate landmark identification, since this identification can be performed rapidly with high consistency. Although various machine learning-based automatic landmark identification systems using lateral cephalograms have been proposed to date, we found no corresponding studies that used PA cephalograms.
This study is the first trial to evaluate the validity of a multi-stage convolutional neural network (CNN)-based automatic landmark prediction system using cone-beam computed tomography (CBCT)-synthesized PA cephalograms. The null hypothesis is that there are no differences in the reproducibility of landmark identification between AI prediction and manual identification.
MATERIALS AND METHODS
This retrospective study was performed under approval from the Institutional Review Board of Kyung Hee University Dental Hospital (IRB number: IRB-KH DT19013).
A total of 430 CBCT scans were selected from patients who met following the inclusion criteria: 1) patients who had visited Kyung Hee University Dental Hospital; 2) patients with growth potential, orthodontic appliances or/and dental prostheses, surgical screws or/and plates and patients with or without skeletal asymmetry; 3) no missing upper or lower permanent incisors, missing permanent upper or lower first molars, craniofacial syndromes, or dentofacial traumas.
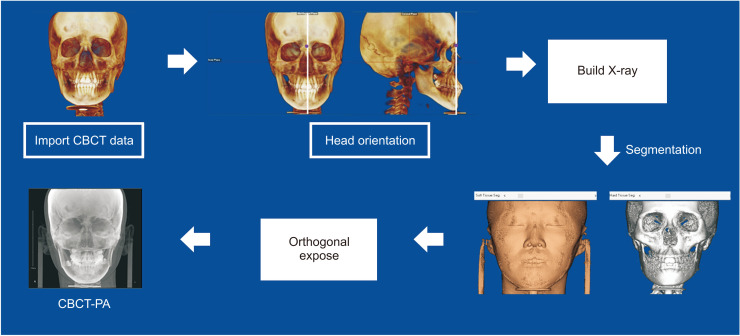
The CBCT scans were taken at the 0.39-mm3 voxel size level, with a 16 × 13-cm field of view and a 30-seconds scan time at 10 mA and 80 kV (Alphard Vega; Asahi Roentgen, Kyoto, Japan). The obtained data were imported as Digital Imaging and Communications in Medicine (DICOM) files to Dolphin software 11.95 Premium (Dolphin Imaging & Management Solutions, Chatsworth, CA, USA). All CBCT images were reoriented according to the anatomic structures of reference. The horizontal plane was established with reference to the right porion, left orbitale, and right orbitale. The sagittal plane was perpendicular to the frontozygomatic suture line and the horizontal plane passing through the nasion. The coronal plane passing through the nasion was made simultaneously perpendicular to the horizontal plane. The PA cephalogram was synthesized from the reoriented CBCT (CBCT-PA) images and saved in the JPG format with a range of pixel sizes, with a width of 2,048 pixels and heights of 1,755–1,860 pix (Figure 1).
Figure 1.
Flow diagram showing the processing of the cone-beam computed tomography (CBCT)-synthesized posteroanterior (PA) cephalograms. The raw CBCT data were imported using Dolphin software 11.95 Premium (Dolphin Imaging & Management Solutions, Chatsworth, CA, USA). The head position was adjusted to reduce layered bilateral structures. The ‘Build X-ray’ button in the software was used to synthesize the CBCT-PA with orthogonal X-ray exposure while eliminating virtual magnification, which causes image distortion.
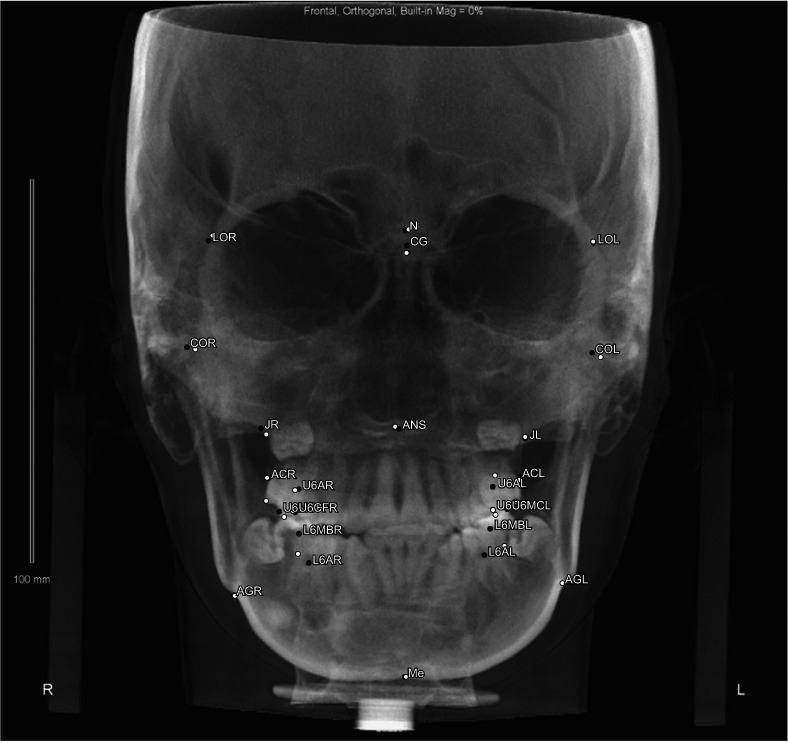
Twenty-three skeletal landmarks used for Tweemac analysis were selected and manually identified on the 345 images used for model training. The landmarks on the remaining 85 images were manually identified two times in a two-week period for testing the model and validating intra-examiner consistency. For comparison with the examiner’s consistency, the AI assessments were also performed twice. All manual identification procedures were completed by a single examiner with more than five years’ orthodontic experience. A detailed description of the PA landmark definitions is given in Table 1. The See-through ceph (See-through Tech Inc., Seoul, Korea) software was used to accomplish landmark identification, with the nasion as the origin point of the coordinate system. The coordinates of each landmark were reported in Microsoft Excel (version 2010; Microsoft, Redmond, WA, USA). Two numeric values were recorded for all landmarks with reference to (X0, Y0).
Table 1.
Landmark definitions
| Landmark | Definition |
|---|---|
| Bilateral skeletal landmarks | |
| Lateral orbit right (LOR) | The most anterior point at the intersection of the frontozygomatic suture on the right inner rim of the orbit |
| Lateral orbit left (LOL) | The most anterior point at the intersection of the frontozygomatic suture on the left inner rim of the orbit |
| Condyle point right (COR) | The most superior (sagittal perspective) and the middle (frontal perspective) point on the contour of the right condyle head |
| Condyle point left (COL) | The most superior (sagittal perspective) and the middle (frontal perspective) point on the contour of the left condyle head |
| Jugal point right (JR) | The intersection of the outline of the right maxillary tuberosity and the zygomatic buttress |
| Jugal point left (JL) | The intersection of the outline of the left maxillary tuberosity and the zygomatic buttress |
| Right antegonial notch (AGR) | Right deepest point on the curvature of the antegonial notch |
| Left antegonial notch (AGL) | Left deepest point on the curvature of the antegonial notch |
| Midline skeletal landmarks | |
| Crista galli (CG) | The most superior and anterior points on the median ridge of bone that projects upward from the cribriform plate of the ethmoid bone |
| Anterior nasal spine (ANS) | Center of the intersection of the nasal septum and the palate |
| Menton (Me) | Midpoint on the inferior border of the mental protuberance |
| Bilateral dentoalveolar landmarks | |
| Upper first molar axis right (U6AR) | Furcation of the upper right first molar |
| Upper first molar axis left (U6AL) | Furcation of the upper left first molar |
| Alveolar crest right (ACR) | The right side of the most cervical rim of the alveolar bone proper |
| Alveolar crest left (ACL) | The left side of the most cervical rim of the alveolar bone proper |
| Upper first molar cup right (U6MCR) | The upper right first molar mesiobuccal cup tip |
| Upper first molar cup left (U6MCL) | The upper left first molar mesiobuccal cup tip |
| Upper first molar central fossa right (U6CFR) | The upper right first molar central fossa |
| Upper first molar central fossa left (U6CFL) | The upper left first molar central fossa |
| Lower first molar mesiobuccal cusp tip right (L6MBR) | The lower right first molar mesiobuccal cup tip |
| Lower first molar mesiobuccal cusp tip left (L6MBL) | The lower left first molar mesiobuccal cup tip |
| Lower first molar axis right (L6AR) | Furcation of the lower right first molar |
| Lower first molar axis left (L6AL) | Furcation of the lower left first molar |
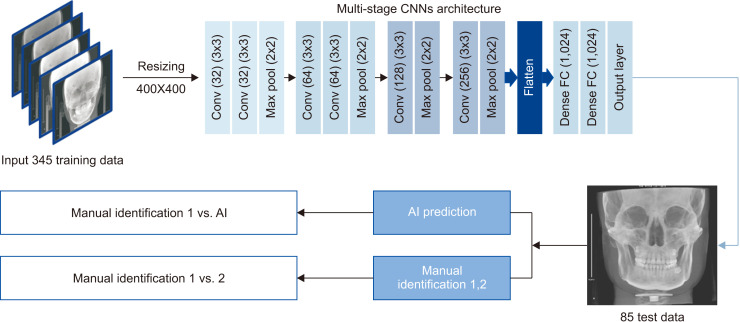
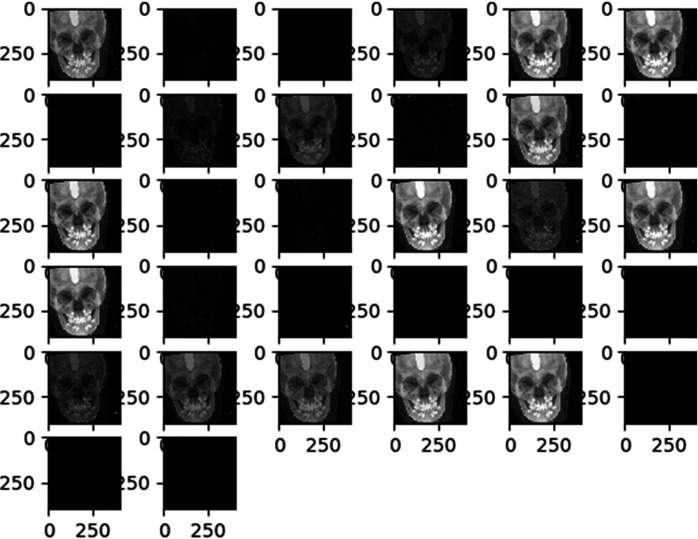
The multi-stage CNN model used in this study was developed on a personal computer with Keras (https://keras.io/, Python deep learning application programming interface). The model consisted of six convolution layers, followed by two dense layers (Figure 2). Deep learning was performed using a GeForce GTX 1080ti GPU (Nvidia Co., Santa Clara, CA, USA) on the Ubuntu 14.04 platform (https://releases.ubuntu.com/14.04/). All images were preprocessed for training by the examiner. The landmarks were identified manually on the row size of the images. To optimize the data for model training, we resized all images to 400 × 400 pixels in the first step. Subsequently, we trained the entire target dataset with the corresponding landmarks for each image in training phase 1. In phase 2, we continued training the model with cropped images in local areas that included the landmarks. The model was trained for each landmark individually with five different image crop sizes: 250, 200, 150, 100, and 50. Briefly, the proposed model was constructed in five stages using the deep CNN model. In the training stage, the learning rate was 0.01, batch size was 100, and the image was composed of a total of 200 epochs. One millimeter corresponding to 10 pixels was used for unit translation. The trained model automatically identified each landmark on the 85 test images twice. Figure 2 illustrates the summarized experimental flow. A visualized effect of each convolutional layer during the first stage training is illustrated in Figure 3.
Figure 2.
Schematic experimental design summary of the multi-stage convolutional neural network (CNN) model.
Conv, convolution; FC, fully connected; AI, artificial intelligence.
Figure 3.
The visualized effect of each convolutional layer during the first stage training.
The absolute value of mean distance difference was calculated in millimeters by the following formula:
Microsoft Excel was used for all calculations.
RESULTS
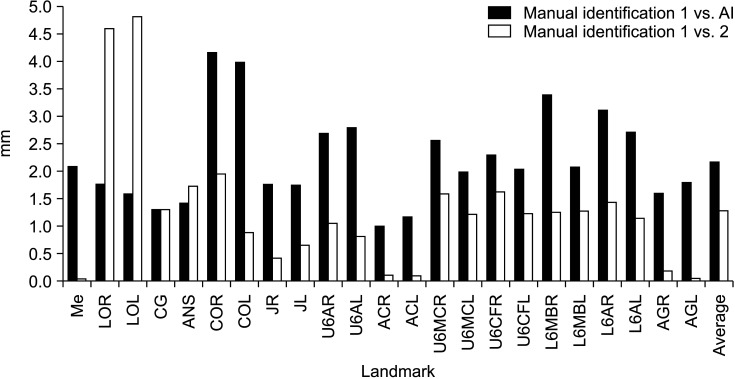
The multi-stage CNN-based AI achieved an MRE of 2.23 ± 2.02 mm. The highest accuracy was obtained for the alveolar crest left (error, 1.2 mm), while the lowest accuracy was obtained for the condyle point left (COL) (error, 4.24 mm). The AI showed errors of less than 2 mm for 10 of the 23 landmarks. The mean intra-examiner error in the repeated manual identification on the 85 images was 1.31 ± 0.94 mm. The repeated measurements of the menton and left antegonial notch showed an average error of 0.05 mm, indicating the highest consistency. In contrast, the average error for the lateral orbit left was 4.91 mm, representing the lowest consistency in repeated measurements (Figure 4 and Table 2). The prediction outcomes are illustrated in Figure 5.
Figure 4.
Mean radial error (MRE) for each landmark, and the average MRE between manual identification 1 and artificial intelligence (AI) (black) and manual identifications 1 and 2 (white).
See Table 1 for definitions of the other landmarks.
Table 2.
The MRE and SD for intra-examiner and AI identification
| Landmark | Manual identification 1 vs. 2 (Intra-examiner) |
Manual identification 1 vs. AI (AI prediction) |
|||
|---|---|---|---|---|---|
| MRE | SD | MRE | SD | ||
| Bilateral | |||||
| LOR | 4.71 | 1.07 | 1.81 | 2.01 | |
| LOL | 4.92 | 1.08 | 1.62 | 2.22 | |
| COR | 1.97 | 0.99 | 4.24 | 2.21 | |
| COL | 0.91 | 1.02 | 4.05 | 2.44 | |
| JR | 0.42 | 0.96 | 1.81 | 2.32 | |
| JL | 0.67 | 0.98 | 1.79 | 1.61 | |
| AGR | 0.19 | 0.79 | 1.65 | 1.91 | |
| AGL | 0.05 | 0.85 | 1.84 | 2.42 | |
| Midline | |||||
| CG | 1.33 | 1.10 | 1.33 | 1.59 | |
| ANS | 1.77 | 0.99 | 1.45 | 2.08 | |
| Me | 0.05 | 0.83 | 2.14 | 1.83 | |
| Dentoalveolar | |||||
| U6AR | 1.08 | 0.90 | 2.75 | 2.48 | |
| U6AL | 0.84 | 0.92 | 2.86 | 2.11 | |
| ACR | 0.12 | 0.93 | 1.03 | 1.68 | |
| ACL | 0.09 | 0.96 | 1.20 | 2.82 | |
| U6MCR | 1.63 | 0.90 | 2.63 | 2.09 | |
| U6MCL | 1.25 | 0.92 | 2.04 | 1.86 | |
| U6CFR | 1.67 | 0.89 | 2.36 | 2.09 | |
| U6CFL | 1.27 | 0.91 | 2.09 | 1.74 | |
| L6MBR | 1.28 | 0.89 | 3.48 | 3.23 | |
| L6MBL | 1.31 | 0.90 | 2.13 | 1.55 | |
| L6AR | 1.48 | 0.85 | 3.19 | 2.46 | |
| L6AL | 1.17 | 0.88 | 2.78 | 2.60 | |
| Average | 1.31 | 0.94 | 2.23 | 2.02 | |
Unit of measurement: millimeter.
MRE, mean radial error; SD, standard deviation; AI, artificial intelligence.
See Table 1 for definitions of the other landmarks.
Figure 5.
Accuracy of the convolutional neural network based on the automatic landmark identification system for cone-beam computed tomography using the synthesized posteroanterior cephalograms. The black dot represents the manually identified landmark and the white dot indicates the automatically identified landmark.
N, nasion.
See Table 1 for definitions of the other landmarks.
DISCUSSION
Although PA cephalometric analysis provides typical valuable information for comprehensive cranial-dentofacial evaluation, it generates more superimposed and layered anatomical structure images than lateral cephalograms. These additional superimposed structures affect the accuracy of landmark identification. It is difficult to identify landmarks with poor reproducibility, so accurate identification strongly depends on the examiner’s experience and skill levels. This may be the reason why PA cephalometric analysis is not routinely performed in orthodontic practice.1,2,4,12,13 This study examined the accuracy of an automated identification system based on a multi-stage CNN model for identification of cephalometric landmarks on CBCT-PA images.
Major et al.1 indicated that the intra-examiner errors ranged from 0.28 mm to 2.23 mm when identification was performed five times. The inter-examiner range of errors for identifying landmarks on PA cephalograms was 0.31 to 4.79 mm, which represents a significantly wide variation. Ulkur et al.2 assessed the intra- and inter-examiner consistencies in identifying landmarks on PA cephalograms and noted higher consistency among trained examiners. On the basis of their findings, in this study, only one examiner was used to minimize the inter-examiner error in landmark identification. Consistent with the findings of previous study, the intra-examiner assessments in the present study showed a large range of errors from 0.05 to 4.91 mm. The fact that the examiner has relatively less experience in PA landmark identification could be the reason for this large range of errors in intra-examiner measurements.
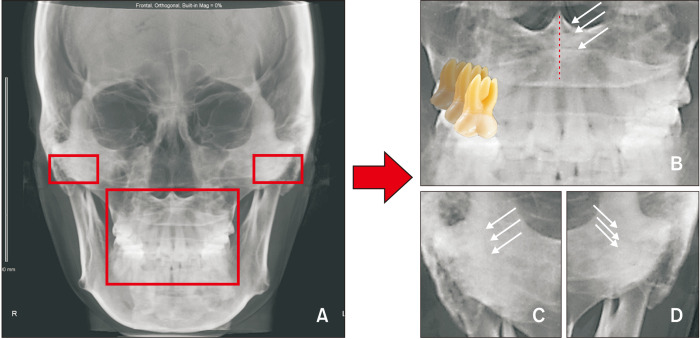
Shokri et al.14 assessed the errors for identification of landmarks in seven different positions on PA cephalograms and observed significant differences in assessments of the antegonion, condyle, and the zygomaticofrontal suture, which are farther from the midline and readily generate identification errors. Thus, we used the CBCT-synthesized PA cephalograms with a consistent head orientation protocol that was more reliable than the conventional protocols for landmark identification. Pirttiniemi et al.5 described the inter-examiner reproducibility of landmark identification on PA cephalograms and found that the mean distance error was higher for the frontozygomatic suture point, pogonion, condyle, and upper and lower incisal midpoints. Some previous studies also mentioned that the condyle, gonion, and zygomatic process points were difficult to locate consistently.1,4,12,15 In this study, we also found relatively higher errors for the condyle and frontozygomatic areas (Figure 6).
Figure 6.
Many structures layered in vertical dimension. The condyle and dentoalveolar areas of the posteroanterior (PA) cephalograms interfere with the artificial intelligence image recognition ability. A, Cone-beam computed tomography-synthesized PA cephalogram. B, Vertical layered images at the intersection of the nasal septum and palatal area (white arrow) and vertical layered and horizontal superimposed images in the dental area. C, D, Vertical layered images in the condyle area.
The emergence of AI in cephalometry has allowed orthodontists to reduce working time and identify landmarks more consistently. Many previous studies have introduced various approaches to improve the accuracy of automated identification. Arık et al.16 applied a deep CNN architecture-based fully automated landmark identification system for the first time. They trained the system with 400 conventional lateral cephalograms including 19 landmarks. The result showed an successful detection rate (SDR) of 75.58% for a range of 2 mm. Park et al.17 trained You-Only-Look-Once version 3 (YOLOv3, https://pjreddie.com/darknet/yolo/) to construct an automated identification system and compared it with a previously introduced model using 1,311 conventional lateral cephalograms with 80 landmarks. They observed an SDR of 80.4% for range of 2 mm, which is approximately 5% higher than the SDR ranges reported in previous studies. Unfortunately, no previous study has used PA cephalograms with an automated identification system. In comparison with similar studies that used lateral cephalograms, our model may have a relatively lower accuracy rate, but it still showed promising feasibility and potential.
Hwang et al.18 used their automatic landmark identification system to compare the stability of manual and AI-assisted landmark identification, and they clearly observed that while AI demonstrated consistency in repetitive identification, the intra-examiner manual landmark identification demonstrated an error of 0.97 ± 1.03 mm. In this study, AI prediction for PA cephalograms also showed consistent results, while the mean intra-examiner error was 1.31 ± 0.94 mm. Thus, AI-based assessments are not affected by subjectivity or external conditions, unlike human performance. Therefore, the AI prediction system for PA cephalograms outlined in this study might offer some advantage because of its ability to identify landmarks consistently in comparison with manual identification.
Deep learning approaches have been recently recommended as a superior technology for automatic location of anatomical landmarks in radiographs. The CNN model shows outstanding ability in image recognition with the application of AI.16-23 This study presented a fully automatic landmark identification system for PA cephalometric analysis based on the multi-stage CNN model. This deep CNN learning model emulates the human examiner’s landmark identification pattern and performs prediction. Thus, AI prediction is affected by the human examiner’s identification pattern. If the examiner shows difficulties in some areas, the AI predictions will reflect these difficulties. In our present study, AI prediction showed the lowest accuracy in the condyle area while the repeated manual identifications showed the lowest consistency for the frontozygomatic suture area. The intra-examiner assessments might show large variability in repetitive tasks. Manual identification in the first measurement yields more variables for the condyle area, but not in the second measurement. However, the human examiner and AI show differences in decision-making. For example, the human examiner can make exclusive decisions based on clinical knowledge, i.e., when the bilateral anatomic structure shows layered images. Thus, the experienced human examiner might show better ability than AI for complicated PA cephalograms requiring comprehensive consideration with subjectivity.
A limitation of this study was that we could not conclusively compare the prediction accuracy of a model trained by a more experienced clinician (who might have shown smaller variations). Additional information on this aspect should be obtained through further investigation. In this study, we developed a feasible automated landmark identification system using PA cephalograms based on a multi-stage CNN model with a personal computer. AI might offer the advantage of consistency in repetitive tasks in comparison with a trained human examiner.
CONCLUSION
We used CBCT-synthesized PA cephalograms to reduce intra-examiner errors and to enhance the ability of Al image recognition.
The null hypothesis was rejected. Our multi-stage CNN model for CBCT-synthesized PA cephalograms did not adequately achieve the clinically acceptable error range of less than the 2 mm, but it showed better consistency than manual identification for repetitive landmark identification on PA cephalograms.
The skeletal landmarks condyle point right and left and most dentoalveolar landmarks showed significant differences between AI prediction and manual identification.
ACKNOWLEDGEMENTS
This article is partly from the PhD thesis of M.J.K. The final form of the machine-learning system was developed by computer engineers of See-through Tech incorporation (Seoul, Korea), which is expected to own the patent in the future.
Footnotes
CONFLICTS OF INTEREST
No potential conflict of interest relevant to this article was reported.
REFERENCES
- 1.Major PW, Johnson DE, Hesse KL, Glover KE. Landmark identification error in posterior anterior cephalometrics. Angle Orthod. 1994;64:447–54. doi: 10.1043/0003-3219(1994)064<0447:LIEIPA>2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 2.Ulkur F, Ozdemir F, Germec-Cakan D, Kaspar EC. Landmark errors on posteroanterior cephalograms. Am J Orthod Dentofacial Orthop. 2016;150:324–31. doi: 10.1016/j.ajodo.2016.01.016. [DOI] [PubMed] [Google Scholar]
- 3.Na ER, Aljawad H, Lee KM, Hwang HS. A comparative study of the reproducibility of landmark identification on posteroanterior and anteroposterior cephalograms generated from cone-beam computed tomography scans. Korean J Orthod. 2019;49:41–8. doi: 10.4041/kjod.2019.49.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sicurezza E, Greco M, Giordano D, Maiorana F, Leonardi R. Accuracy of landmark identification on postero-anterior cephalograms. Prog Orthod. 2012;13:132–40. doi: 10.1016/j.pio.2011.10.003. [DOI] [PubMed] [Google Scholar]
- 5.Pirttiniemi P, Miettinen J, Kantomaa T. Combined effects of errors in frontal-view asymmetry diagnosis. Eur J Orthod. 1996;18:629–36. doi: 10.1093/ejo/18.6.629. [DOI] [PubMed] [Google Scholar]
- 6.Major PW, Johnson DE, Hesse KL, Glover KE. Effect of head orientation on posterior anterior cephalometric landmark identification. Angle Orthod. 1996;66:51–60. doi: 10.1043/0003-3219(1996)066<0051:EOHOOP>2.3.CO;2. [DOI] [PubMed] [Google Scholar]
- 7.Smektała T, Jędrzejewski M, Szyndel J, Sporniak-Tutak K, Olszewski R. Experimental and clinical assessment of three-dimensional cephalometry: a systematic review. J Craniomaxillofac Surg. 2014;42:1795–801. doi: 10.1016/j.jcms.2014.06.017. [DOI] [PubMed] [Google Scholar]
- 8.Kamoen A, Dermaut L, Verbeeck R. The clinical significance of error measurement in the interpretation of treatment results. Eur J Orthod. 2001;23:569–78. doi: 10.1093/ejo/23.5.569. [DOI] [PubMed] [Google Scholar]
- 9.Gribel BF, Gribel MN, Frazäo DC, McNamara JA, Jr, Manzi FR. Accuracy and reliability of craniometric measurements on lateral cephalometry and 3D measurements on CBCT scans. Angle Orthod. 2011;81:26–35. doi: 10.2319/032210-166.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gribel BF, Gribel MN, Manzi FR, Brooks SL, McNamara JA., Jr From 2D to 3D: an algorithm to derive normal values for 3-dimensional computerized assessment. Angle Orthod. 2011;81:3–10. doi: 10.2319/032910-173.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meiyappan N, Tamizharasi S, Senthilkumar KP, Janardhanan K. Natural head position: an overview. J Pharm Bioallied Sci. 2015;7(Suppl 2):S424–7. doi: 10.4103/0975-7406.163488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.El-Mangoury NH, Shaheen SI, Mostafa YA. Landmark identification in computerized posteroanterior cephalometrics. Am J Orthod Dentofacial Orthop. 1987;91:57–61. doi: 10.1016/0889-5406(87)90209-5. [DOI] [PubMed] [Google Scholar]
- 13.Leonardi R, Annunziata A, Caltabiano M. Landmark identification error in posteroanterior cephalometric radiography. A systematic review. Angle Orthod. 2008;784:761–5. doi: 10.2319/0003-3219(2008)078[0761:LIEIPC]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 14.Shokri A, Miresmaeili A, Farhadian N, Falah-Kooshki S, Amini P, Mollaie N. Effect of changing the head position on accuracy of transverse measurements of the maxillofacial region made on cone beam computed tomography and conventional posterior-anterior cephalograms. Dentomaxillofac Radiol. 2017;46:20160180. doi: 10.1259/dmfr.20160180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oshagh M, Shahidi SH, Danaei SH. Effects of image enhancement on reliability of landmark identification in digital cephalometry. Indian J Dent Res. 2013;24:98–103. doi: 10.4103/0970-9290.114958. [DOI] [PubMed] [Google Scholar]
- 16.Arık SÖ, Ibragimov B, Xing L. Fully automated quantitative cephalometry using convolutional neural networks. J Med Imaging (Bellingham) 2017;4:014501. doi: 10.1117/1.JMI.4.1.014501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Park JH, Hwang HW, Moon JH, Yu Y, Kim H, Her SB, et al. Automated identification of cephalometric landmarks: part 1-comparisons between the latest deep-learning methods YOLOV3 and SSD. Angle Orthod. 2019;89:903–9. doi: 10.2319/022019-127.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hwang HW, Park JH, Moon JH, Yu Y, Kim H, Her SB, et al. Automated identification of cephalometric landmarks: part 2-might it be better than human? Angle Orthod. 2019;90:69–76. doi: 10.2319/022019-129.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Takahashi R, Matsubara T, Uehara K. Multi-stage convolutional neural networks for robustness to scale transformation. Paper presented at: 2017 International Symposium on Nonlinear Theory and Its Applications; 2017 Dec 4-7; Cancun, Mexico. Cancun: NOLTA; 2017. pp. 692–5. [Google Scholar]
- 20.Anwar SM, Majid M, Qayyum A, Awais M, Alnowami M, Khan MK. Medical image analysis using convolutional neural networks: a review. J Med Syst. 2018;42:226. doi: 10.1007/s10916-018-1088-1. [DOI] [PubMed] [Google Scholar]
- 21.Yadav SS, Jadhav SM. Deep convolutional neural network based medical image classification for disease diagnosis. J Big Data. 2019;6:113. doi: 10.1186/s40537-019-0276-2. [DOI] [Google Scholar]
- 22.Schwendicke F, Golla T, Dreher M, Krois J. Convolutional neural networks for dental image diagnostics: a scoping review. J Dent. 2019;91:103226. doi: 10.1016/j.jdent.2019.103226. [DOI] [PubMed] [Google Scholar]
- 23.Park JJ, Kim KA, Nam Y, Choi MH, Choi SY, Rhie J. Convolutional-neural-network-based diagnosis of appendicitis via CT scans in patients with acute abdominal pain presenting in the emergency department. Sci Rep. 2020;10:9556. doi: 10.1038/s41598-020-66674-7. [DOI] [PMC free article] [PubMed] [Google Scholar]