Abstract
Background
Hemorrhoids are a frequently-occurring disease of the anorectal system that is often accompanied by vascular hyperplasia and edema. A METTL14-mediated RNA N-6 methyladenosine (m6A) modification can improve mRNA stability and increase its transcriptional and translational activities, closely related to the occurrence of many diseases.
Methods
Western blot, qPCR, and immunofluorescence staining were used to detect the levels of gene and protein expression. Haematoxylin and eosin staining was used for histopathological examination. RNA immunoprecipitation-PCR and RNA dot blotting were used to detect mRNA m6A modification.
Results
Obvious signs of angiogenesis (CD31+/vWF+) were identified in the hemorrhoids. High levels of METTL14 expression on vascular endothelial cells (CD31+) suggested that angiogenesis was accompanied by differential modification of m6A RNA. It was subsequently found that the level of miR-4729 expression was significantly decreased in hemorrhoid tissues. The luciferase reporter enzyme assay results suggested that miR-4729 silenced its expression by targeting the 3'UTR of METTL14 mRNA. MiR-4729 overexpression in human umbilical vein endothelial cells (HUVECs) inhibited the proliferation and migration of HUVECs in vitro and vascular structure formation in the outer matrix. MiR-4729 overexpression significantly inhibited endogenous METTL14 expression in HUVECs and reduced the entire m6A RNA modification, especially the level of m6A methylation at the specific site of the 3' UTR of TIE1 mRNA. Moreover, miR-4729 overexpression significantly inhibited the molecular loop of the TIE1/VEGFA signaling pathway in HUVECs.
Conclusions
Our findings confirmed that the down-regulation of miR-4729 in hemorrhoid vascular endothelial cells was one of the main reasons for vascular proliferation. The overexpression of miR-4729 in vascular endothelial cells decreased the global mRNA methylation and TIE1 mRNA 3'UTR-specific site methylation by silencing METTL14 expression, reducing TIE1 mRNA stability, down-regulating the TIE1/VEGFA signal molecular loop expression, and weakening angiogenesis ability.
Keywords: Hemorrhoids, vascular hyperplasia, microRNA, TIE1, METTL14
Introduction
Hemorrhoids are a frequently-occurring disease of the anorectal system in Chinese populations (1-7). Based on their location, they can be classified as internal, external, or mixed hemorrhoids. A visible serrated line termed the anal dentate line at the junction of the anal skin and rectal mucosa (3,5,7). External hemorrhoids are located below the dentate line and are covered with anal mucosa. These can be further divided into connective tissue external hemorrhoids, external varicose hemorrhoids, and thrombotic external hemorrhoids. The causes of hemorrhoids are extremely complex, though the following 2th eories are currently accepted: (I) the theory of varicose veins, which holds that hemorrhoids are venous masses formed by congestion or dilatation; and (II) flexion of the venous plexus under the submucous membrane of the rectum and under the skin of the anal canal (1-7). Blood vessel proliferation and dilatation of blood vessels and edema of vascular endothelial cells are commonly observed in the hemorrhoid nuclear tissue of external hemorrhoids (1-7). However, there are few reports on the mechanisms of hemorrhoids and their development from an epigenetic perspective.
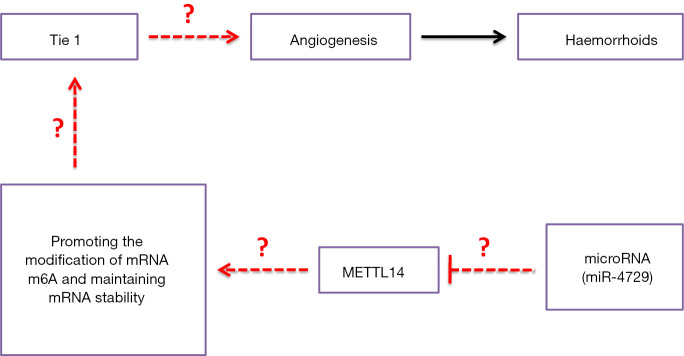
MicroRNAs (miRNAs) are endogenous single-stranded non-coding RNA molecules, approximately 19–23 nucleotides in length, with no open reading frames (8-14). Through complementary pairing with the 3' untranslated region (3'-UTR) of the target gene mRNA molecule, inhibition induces the Dicer enzyme to specifically cleave the target mRNA and silence target gene expression (8-14). Accumulating studies suggest that miRNAs participate in important physiological and biochemical activities, such as cellular proliferation, development, differentiation, migration, and apoptosis. Moreover, miRNAs play an extremely important role in regulating body development and disease occurrence (8-14). Our previous study’s findings revealed that DLK1-DIO3 imprinted cluster miRNA expression differed between the clinical hemorrhoid samples and normal perianal tissues (Figure 1). An in-depth study found that miRNA-412-5p regulates the epigenetic mechanism of exportin1 (Xpo1) and the downstream p53/p66Shc/p16 pathway on vascular endothelial cell proliferation and hemorrhoid formation (15). Therefore, studying miRNAs in angiogenesis in hemorrhoids and nuclear tissue is extremely important.
Figure 1.
Hypothesis of this study. We investigated the miR-4729-targeted regulation of vascular endothelial cell METTL14 expression, m6A methylation of the 3' untranslated region (3'UTR) of TIE1 mRNA, as well as its epigenetic regulation mechanism on vascular endothelial cell proliferation and hemorrhoid formation.
RNA N-6 methyladenosine (m6A) refers to a methylation modification on the sixth nitrogen atom (N) of adenine RNA. Methylation of m6ARNA has been widely identified in most eukaryotic species (e.g., yeast, plants, fruit flies, and mammals) and viral mRNA and plays a key role in post-transcriptional mRNA regulation and metabolism (16-20). The m6A methyltransferases, METTL14 and METTL3, are 2 components of the m6A methyltransferase complex. These 2 proteins can form a stable complex with 1:1 stoichiometry and catalyze RNA m6A modification, creating methylation “writers” (19,21-23). The FTO protein can remove m6A RNA methylation, making it an “eraser” (19,21-23). Therefore, m6A RNA's modification is a dynamic and reversible enzymatic reaction (19,21-23). Some studies have suggested that the m6A RNA modification can improve mRNA stability, increase its transcriptional and translational activity, promote tumorigenesis and invasion, and improve stem cell reprogramming efficiency (20-22,24,25). However, the dynamic modification changes and mechanism of m6A RNA in the process of hemorrhoid neovascularization remains poorly understood.
Based on the above evidence, this study analyzed the differences in the expression of TIE1, CD31, METTL14, and other proteins between clinical hemorrhoid samples and normal perianal tissues (Figure 1). We clarified the details of the miR-4729 targeted regulation of METTL14 expression in vascular endothelial cells, m6A methylation of the 3'UTR of TIE1 mRNA, and its epigenetic regulation mechanism on vascular endothelial cell proliferation and hemorrhoid formation.
We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/atm-20-3399).
Methods
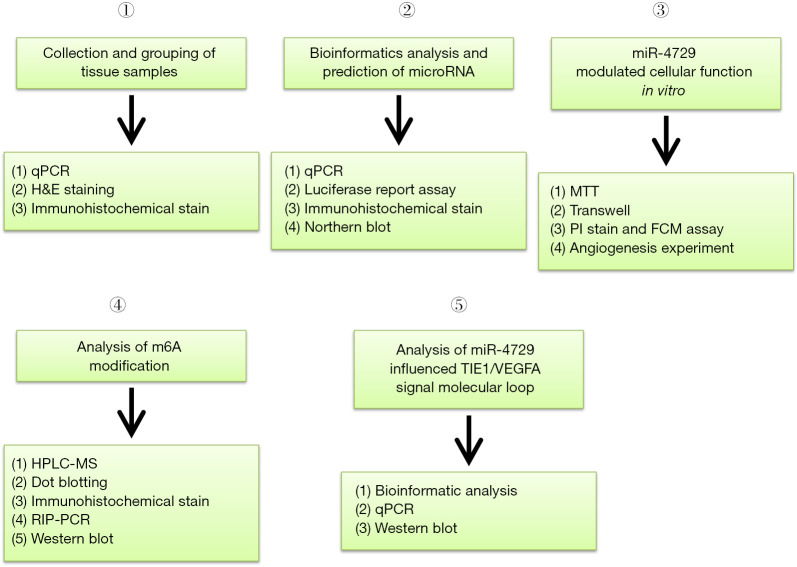
Collection and grouping of tissue samples (Figure 2)
Figure 2.
Experimental and research processes.
The hemorrhoid samples were collected from 20 patients undergoing hemorrhoid surgery at the department of anorectal surgery, Shuguang Hospital affiliated with the Shanghai University of Traditional Chinese Medicine between April 2018 and August 2019. All patients were aged between 28 and 50 years old, with either third or fourth degree hemorrhoids. The Regional Ethics Committee approved the study protocol of Shuguang Hospital, Shanghai University of Traditional Chinese Medicine (Permission No. 201701.4). This study conformed to the provisions of the Declaration of Helsinki (as revised in 2013), and written informed consent was obtained from every participant.
Human umbilical vein endothelial cell (HUVECs) culture
Human umbilical cords of healthy puerperant women were collected in asepsis after parturition. HUVECs were isolated with 1% trypsin. The HUVECs were grown on 1% gelatin-coated culture plates in McCoy’s 5A (Sigma) supplemented with 15% fetal bovine serum (Hyclone), 100 U/mL penicillin, and 100 µg/mL streptomycin (Hyclone), at 37 °C in a humidified atmosphere containing 5% CO2. The cells were used within 2–3 passages and were identified as endothelial cells by their cobblestone monolayer appearance in culture, and by the expression of cell-surface antigens CD34 and CD31 as detected by flow cytometry.
RNA isolation and reverse transcription into cDNA
An RNAprep pure Tissue Kit (TIANGEN Biotech, Beijing Co., Ltd) was used following the manufacturer’s instructions. Approximately 20 mg of human tissue samples were collected, 800 µL of lysate was added, and grinding homogenization was performed. Then 200 µL of chloroform was added to the supernatant, the mixture was inverted, centrifuged at 4 °C at 13,400 ×g for 15 min, and the supernatant was collected. The volume of anhydrous ethanol was added two times, and the mixture was inverted and centrifuged at 4 °C at 13,400 ×g for 30 min. RNA was precipitated by adding 500 µL of 75% ethanol before resuspending and centrifuging for 5 min at 4 °C at 13,400 ×g. The excess liquid was removed, and the precipitate was fully dissolved in 300 µL DEPC water. The OD260/OD280 ratio was detected using 1 µL RNA solution (the ratio should generally range between 1.8 and 2.0) to determine the RNA purity and total concentration. Based on the miRcute miRNA First-strand cDNA (TIANGEN Biotech, Beijing Co., Ltd) manual, 20 µL (100 ng/µL) of total RNA, 25 µL of 2× miRNART Reaction Buffer, 4 µL of 1× miRNART Enzyme Mix, and 6 µL of RNase-free deionized water were combined and mixed well. The reactions were performed in a thermal cycler, followed by a miRNA plus A tail reaction and reverse transcription reaction at 42 °C for 60 min, and an enzyme inactivation reaction at 95 °C for 3 min.
qPCR
According to the miRcute miRNA qPCR Detection (TIANGEN Biotech, Beijing Co., Ltd) instructions, the reagents, samples, and primers were added in the following amounts: 10 µL of 2× miRcute PlusmiRNA Premix (with SYBR), 1 µL each (10 µM) of 1× forward primer and reverse primer, 4 µL of miRNA first strand cDNA, and 4 µL of deionized water. The following reactions were performed using a real-time fluorescence quantitative PCR instrument: 95 °C for 15 min, 94 °C for 20 s, and 60 °C for 34 s before reading the fluorescence values. The above reactions were performed for 40 cycles. The following qPCR primers were used:hsa-miR-3200-5p-F: AATCTGAGAAGGCGCACAAGGT; hsa-miR-548p-F: TAGCAAAAACTGCAGTTACTTT; hsa-miR-4760-5p-F: TTTAGATTGAACATGAAGTTAG; hsa-miR-150-5p-F: TCTCCCAACCCTTGTACCAGTG; hsa-miR-4729-F: TCATTTATCTGTTGGGAAGCTA; hsa-miR-593-3p-F: TGTCTCTGCTGGGGTTTCT; hsa-miR-3168-F: GAGTTCTACAGTCAGAC; hsa-miR-4713-5p-F: TTCTCCCACTACCAGGCTCCCA; hsa-miR-2276-3p-F: TCTGCAAGTGTCAGAGGCGAGG; hsa-miR-3921-F: TCTCTGAGTACCATATGCCTTGT; VEGFR2-F: GTGATCGGAAATGACACTGGAG; VEGFR2-R: CATGTTGGTCACTAACAGAAGCA; IFNG-F: TCGGTAACTGACTTGAATGTCCA; IFNG-R: TCGCTTCCCTGTTTTAGCTGC; VEGFA-F: AGGGCAGAATCATCACGAAGT; VEGFA-R: AGGGTCTCGATTGGATGGCA; vWF-F: CCTTGACCTCGGACCCTTATG; vWF-R: GATGCCCGTTCACACCACT; TIE1-F: AAGCAGACAGACGTGATCTGG; TIE1-R: GCACGATGAGCCGAAAGAAG; CD31-F: AACAGTGTTGACATGAAGAGCC; CD31-R: TGTAAAACAGCACGTCATCCTT; 18SrRNA-F: CAGCCACCCGAGATTGAGCA; 18SrRNA-R: TAGTAGCGACGGGCGGTGTG.
Haematoxylin-eosin staining
Tissue samples were fixed with 4% paraformaldehyde, dehydrated, and embedded in paraffin. Thin slices (4 µm) were cut using the paraffin slicer and affixed to glass slides. Xylene was used for dewaxing, and ethanol gradient dehydration was performed. A hematoxylin staining solution was applied for 5 min at room temperature, followed by 1% hydrochloric acid ethanol to differentiate for 30 s. Light ammonia water was added to return to blue for 1 min, and the sections were rinsed in distilled water for 5 min. Subsequently, the eosin staining solution was added at room temperature for 2 min, followed by rinsing the samples in distilled water for 2 min. Ethanol gradient decolorization was performed. Xylene was allowed to permeate for 2 min. Finally, the slides were sealed with neutral gum.
Northern blot
The northern blot was performed as previously described (26,27). In brief, each group’s cells were treated with aTrizol kit to obtain the total RNA, and 20 µg of high quality total RNA was resolved using 7.5M urea-12% formaldehyde (PAA) denaturing gel electrophoresis. The RNA was transferred to a Hybond N+ nylon membrane (Amersham, Freiburg, Germany). The membrane was crosslinked with 1,200 mjoule/cm2 UV for the 30s. A miR-4729 antisense DNA probe was used for hybridization to detect miR-4729 expression. After hybridization and washing, the membrane was exposed to Kodak XAR-5 film for 20-40 h (Sigma-Aldrich, Chemical). U6 snRNA was used as an internal reference.
Western blot
Western blot was performed as previously reported (26,27). Briefly, total proteins from each group were resolved by 12% SDS-PAGE denaturing gel electrophoresis before being transferred to a PVDF membrane (Millipore). The membrane was sealed and washed before being incubated with primary antibodies for 45 min (Table 1) at 37 °C. After the membrane was washed, secondary antibodies were added and incubated for 45 min (Table 1) at 37 °C. The membrane was washed 4times with TBST at room temperature for 14 min. Finally, ECL enhanced chemiluminescence (ECL kit, Pierce Biotechnology) (Sigma-Aldrich, Chemical) was used to visualize the PVDF membrane’s proteins.
Table 1. List of primary antibodies.
| Antibodies | Companies | Applications |
|---|---|---|
| Mouse anti-TIE1 antibody [8C9] (ab27851) | Abcam, MA, USA | IF(1:300)/WB(1:1,000) |
| Rabbit anti-CD31 antibody (ab28364) | Abcam, MA, USA | IF(1:300)/WB(1:1,000) |
| Mouse anti-VEGF Receptor 2 antibody (ab39256) | Abcam, MA, USA | IF(1:300)/WB(1:1,000) |
| Rabbit anti-VEGFA antibody (ab9570) | Abcam, MA, USA | IF(1:300)/WB(1:1,000) |
| Rabbit anti-Von Willebrand Factor antibody [F8/86] (ab778) | Abcam, MA, USA | IF(1:300)/WB(1:1,000) |
| Rabbit anti-METTL14 antibody [CL4252] (ab220030) | Abcam, MA, USA | IF(1:300)/WB(1:1,000) |
| Rabbit anti-GAPDH antibody [EPR16891] (ab181602) | Abcam, MA, USA | WB (1:1,000) |
| Goat anti-Rabbit IgG H&L (HRP) (ab97051) | Abcam, MA, USA | WB (1:1,000) |
| Goat anti-Mouse IgG H&L (HRP) (ab6789) | Abcam, MA, USA | WB (1:1,000) |
| Goat Anti-Rabbit IgG H&L (Alexa Fluor® 488) (ab150077) | Abcam, MA, USA | IF (1:300) |
| Donkey Anti-Rabbit IgG H&L (Alexa Fluor® 647) (ab150075) | Abcam, MA, USA | IF (1:300) |
| Goat anti-Mouse IgG H&L (Alexa Fluor® 488) (ab150113) | Abcam, MA, USA | IF (1:300) |
| Goat Anti-Mouse IgG H&L (Alexa Fluor® 647) (ab150115) | Abcam, MA, USA | IF (1:300) |
IF, immunofluorescence staining; WB, Western blotting; HRP, horseradish peroxidase; H&L, heavy chain & light chain.
Immunohistochemistry
All fresh tissues were briefly soaked at room temperature and fixed in 4% paraformaldehyde (Sigma-Aldrich, St. Louis, USA) for 30 min. Ethanol gradient dehydration, paraffin embedding, slicing (thickness 6 µm), and dewaxing was performed using xylene. The tissue sections were incubated with an immunohistochemical blocking solution (Beyotime Biotechnology Co., Ltd., Zhejiang, China) at 37 °C for 30 min. The blocking solution was discarded, and an immunohistochemical cleaning solution (Beyotime Biotechnology) was added to rinse the tissue sections at room temperature 3 times for 5 min. The first antibody (Table 1) was added and incubated at 37 °C for 45 min. The antibody was discarded, and an immunohistochemical cleaning solution (Beyotime Biotechnology) was added to rinse the tissue for 5 minutes 3 times at room temperature. The secondary antibody (Table 1) was added and incubated at 37 °C for 45 min. The antibody was discarded before adding an immunohistochemical cleaning solution (Beyotime Biotechnology) to rinse the tissue at room temperature 3 times for 5 min. Finally, an immunofluorescence sealing solution (Sigma-Aldrich) was added to seal the slides.
Propidium iodide (PI) staining and flow cytometry (FCM)
PI staining and an FCM assay were performed based on previously published protocols (26,27). Briefly, 5×105/mL cells were collected and fixed in 1 mL 70% precooled ethanol for 48 h. The cell precipitates were collected after centrifuging at 1,500 r/min at 4 °C for 5 min. The PI staining solution (Sigma, Chemicals) was added in the dark at 4 °C for 30 min. The cell cycle distribution of each group was analyzed by flow cytometry (BD FACSAria), and the data were analyzed using CellQuest software.
Dot blotting
Different concentrations of genomic DNA from each group were spotted on a Hybond-N+ membrane. The spotted DNA was cross-linked to the membrane using a UV crosslinker. The membrane was blocked in 5% BSA and subsequently incubated with an anti-m6A antibody (CST) and horseradish peroxidase (HRP)-conjugated anti-mouse secondary antibody (CST). Finally, the blot was developed with enhanced chemiluminescence reagents and exposed to imaging film.
RNA immunoprecipitation (RIP)-PCR
RIP experiments were performed using the Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore, Bedford, MA). All RIP steps were performed as previously described (28,29). Briefly, the cells from all groups were lysed (500 µL per plate) in a modified cell lysis buffer used for western blotting and IP (20 mM Tris, pH7.5, 150 mM NaCl, 1% Triton X-100, 1 mM EDTA, sodium pyrophosphate, β-glycerophosphate, Na3VO4, and leupeptin; Beyotime Biotechnology). After lysis, each sample was centrifuged to clear the insoluble debris and pre-incubated with 20 µg protein A agarose beads (Beyotime Biotechnology) while rocking for 30 min at 4 °C before centrifuging and transferring to a fresh 1.5 mL tube. A mouse anti-human m6A monoclonal antibody (1:100; Santa Cruz Biotechnology, California, USA) was added and incubated for 90 min before 20 µg of protein A agarose beads were re-added to capture any immune complexes. The agarose beads were washed 3times in an ice-cold homogenization buffer.
Statistical analysis
Each experiment was performed at least 3times, and data are presented as the mean ± SE, where applicable. Differences were evaluated using a Student’s t-test. A threshold of P<0.05 was considered statistically significant.
Results
High expression of vascular markers and METTL14 in hemorrhoids
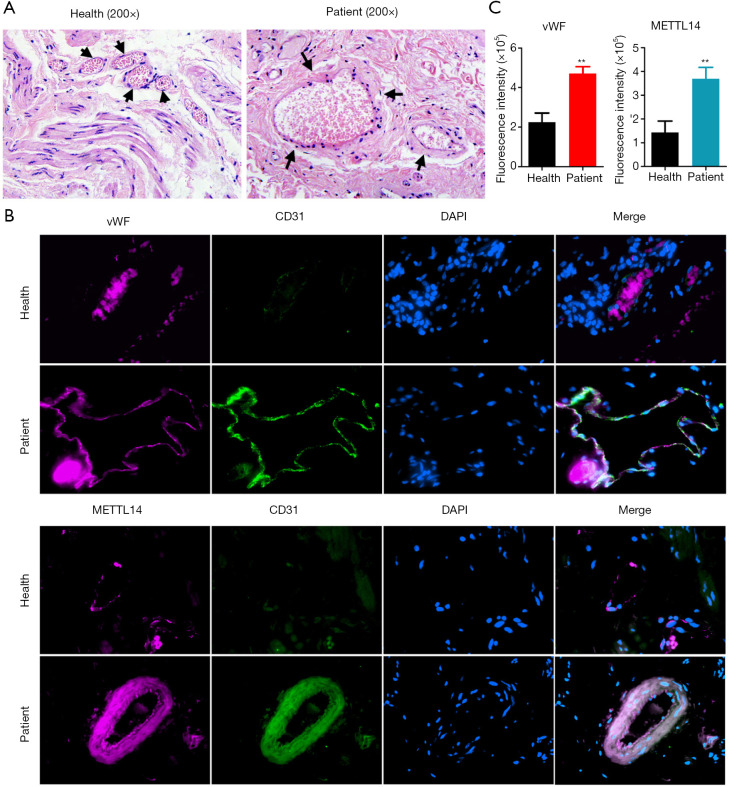
The pathological examination demonstrated marked blood vessel proliferation in hemorrhoids, significant enlargement of the vascular endothelium nucleus, and overall edema in the cells. The interstitial layer connection around the blood vessel was not tight (Figure 3A). The immunofluorescence staining results showed that staining of the vascular endothelial cell markers vWF and CD31 in hemorrhoids was strongly positive compared to normal tissues (Figure 3B). Simultaneously, the proportion of vWF+ cells in hemorrhoids was significantly higher than in the normal tissues (Figure 3C). The immunofluorescence staining results showed that METTL14 expression in CD31+ cells in hemorrhoids was significantly enhanced (Figure 3B and C). These results suggest that vascular proliferation is evident in hemorrhoid tissues, and the hyperplastic vascular endothelial cells express high levels of theRNA m6A “writer” protein, METTL14.
Figure 3.
Vascular markers and METTL14 were highly expressed in hemorrhoids. Pathological examination results using (A) hematoxylin-eosin (HE) staining. The arrows indicate blood vessels. Immunofluorescence staining results in each group (B). Magnification: 200×. The proportion of vWF+ and CD31+/METTL14+ cells in (C) hemorrhoids was significantly higher than that in normal tissues. **P<0.01 vs. healthy tissue; t-test; n=4.
MiR-4729 is expressed at low levels in hemorrhoids and targets METTL14
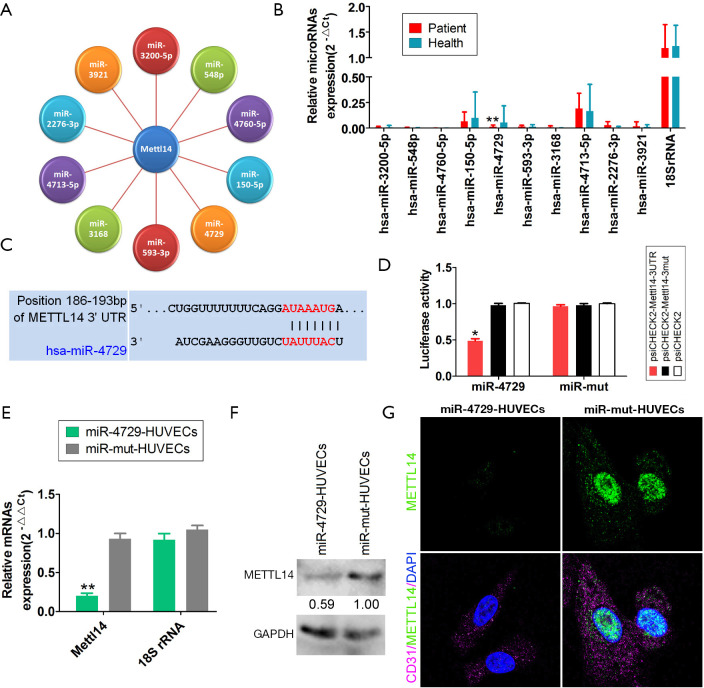
The bioinformatics analysis (Targetscantool: http://www.targetscan.org/vert_72/) showed 10 miRNAs that can target METTL14 expression (Figure 4A). The qPCR results showed that only miR-4729 expression in the hemorrhoid tissues was significantly low (Figure 4B). Further analysis showed that mature miR-4729 could complement 7bases (UAUUUAC) at the specific site (+186bp~+193bp) of METTL14, which suggests that METTL14 may be a target of miR-4729 (Figure 4C). Subsequently, the luciferase reporter assay analysis showed that WT miR-4729 overexpression significantly decreased WT Mettl14-associated luciferase expression, whereas the other combinations did not affect luciferase expression (Figure 4D). Finally, the qPCR results for miR-4729 overexpression showed that Mettl14 mRNA expression was significantly lower in HUVECs than in the control group (transfected miR-mut) (Figure 4E). Western blotting also showed that the METTL14 protein expression level in miR-4729 overexpressing HUVEC cells was significantly lower than that of the miR-mut control cells at 72 h (Figure 4F). The immunofluorescence staining results were consistent with the western blotting results (Figure 4G). The results showed that miR-4729 overexpression could significantly inhibit METTL14 expression in vascular endothelial cells.
Figure 4.
MiR-4729 was expressed at low levels in hemorrhoids, and targets METTL14. (A) The bioinformatics analysis showed that there were 10 microRNAs that could potentially regulate METTL14 expression. (B) miR-4729 expression was significantly decreased in hemorrhoids. **P<0.01 vs. healthy tissue; t-test; n=8. (C) Mature miR-4729 and the 7 bases at specific METTL14 sites can be completely complementary and paired. (D) The luciferase reporter assay analysis showed that wild type (WT) miR-4729 overexpression significantly decreased luciferase expression carrying WT Mettl14. Other combinations did not affect the level of luciferase expression. *P<0.05 vs. psiCHECK2; t-test; n=4. (E) The quantitative real-time PCR (qPCR) results showed that the level of Mettl14 mRNA expression in miR-4729-HUVECs was significantly lower than that of the control group (miR-mut-HUVECs). **P<0.01 vs. miR-mut-HUVECs; t-test; n=3. (F) The western blot results showed that the level of METTL14 protein expression in miR-4729-HUVECs was significantly lower than that of the miR-mut control cells. (G) Immunofluorescence staining showed that the level of METTL14 protein expression in the miR-4729-HUVECs was significantly lower than that in the miR-mut-HUVECs. Magnification: 400×.
MiR-4729 overexpression affects vascular endothelial cell function
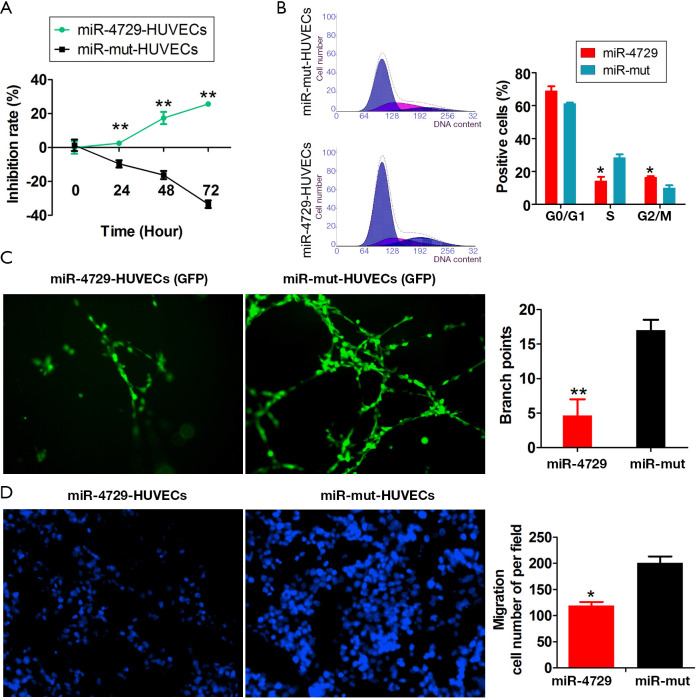
Either miR-4729 or miR-mut was overexpressed in HUVECs (control group) to verify the regulatory effect of miR-4729 on vascular endothelial cell function. The MTT assay results showed that the proliferation inhibition rate in the miR-4729-HUVEC group was significantly higher over time than that of the control group (Figure 5A). The FCM results showed that the proportion of S phase cells decreased significantly after 72 h, whereas the proportion of cells in the G2/M phase significantly increased in the miR-4729-HUVEC cell cycle. This suggested that the miR-4729-HUVEC cell cycle was arrested in the G2/M phase (Figure 5B). The in vitro angiogenesis results revealed that the ability of miR-4729-HUVECs to form official lumen and branches in METROGEL was significantly lower than that of the control cells after 72 h (Figure 5C). Moreover, the transwell chamber experiment results showed that the miR-4729-HUVEC extracellular matrix’s migratory ability was significantly lower than that of the control cells (Figure 5D). Therefore, these experimental results suggest that miR-4729 overexpression weakens vascular endothelial cell function.
Figure 5.
MiR-4729 overexpression affects the function of vascular endothelial cells. The results of (A) an 3-(4,5)-dimethylthiahiazo(-z-y1)-3,5-di-phenytetrazoliumromide (MTT) assay showed that the rate at which cellular proliferation was inhibited in the miR-4729-HUVEC group was significantly higher than the control group over time. **P<0.01 vs. miR-mut-HUVECs; t-test; n=3. (B) The flow cytometry (FCM) results showed that the cell cycle of miR-4729-HUVECs was arrested in the G2/M phase. *P<0.05 vs. miR-mut-HUVECs; t-test; n=3. (C) The angiogenesis in vitro results showed that the ability of miR-4729-HUVECs to form official lumen and branches in METROGEL was significantly lower than that of the control group. Magnification: 200×. **P<0.01 vs. miR-mut-HUVECs; t-test; n=3. (D) The transwell chamber assay results showed that the migratory ability of miR-4729-HUVECs in the extracellular matrix was significantly lower than the control cells. *P<0.05 vs. miR-mut-HUVECs; t-test; n=3. Magnification: 200×.
MiR-4729 overexpression affects TIE1 expression and modification of m6A mRNA methylation
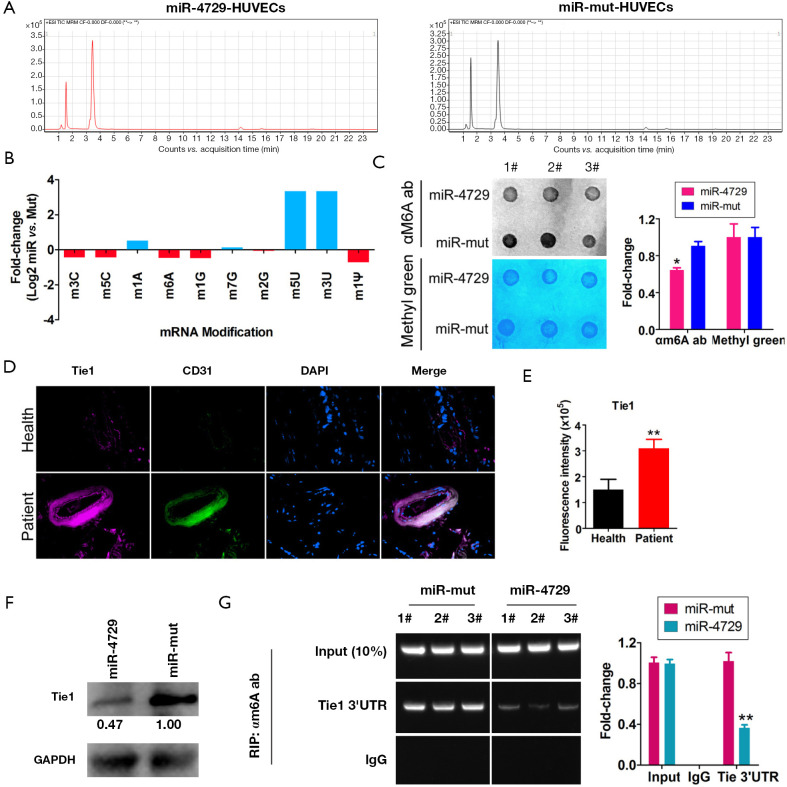
First, the HPLC-MS detection results showed that methylation modifications (e.g., m3C, m5C, m6A, and m1G) were significantly lower compared tothe miR-4729-HUVEC group (Figure 6A and B; Table 2). The dot blot results showed that the overall level of mRNA m6A modification in the miR-4729-HUVEC group was significantly lower than that of the control group (Figure 6C). The immunofluorescence staining results showed that TIE1 expression in CD31+ cells in hemorrhoids was also significantly enhanced (Figure 6D and E). Also, the western blot results showed that TIE1 protein expression was significantly lower in the miR-4729-HUVEC group than the miR-mut-HUVEC group (Figure 6F). Finally, the RIP-PCR results showed that a specific product of 3'UTR of the TIE2 gene mRNA could be amplified using PCR from miR-mut-HUVECs in the complex cross-linked with an anti-m6A antibody (α m6A Ab) (Figure 6G). Few of the above products were amplified in miR-mut-HUVECs using PCR in the complexes cross-linked with an anti-m6A antibody (α m6A Ab) (Figure 6G). These results indicate that miR-4729 overexpression significantly affected the overall level of mRNA methylation in vascular endothelial cells and especially decreased TIE1 mRNA m6A methylation level and expression.
Figure 6.
MiR-4729 overexpression affects TIE1 expression and mRNA m6A methylation. (A) High performance liquid chromatography/mass spectrometry (HPLC/MS) detection of mRNA modified cells in each group. (B) The HPLC-MS results showed that the miR-4729-HUVEC mRNA was significantly lower than that ofthemiR-4729-HUVEC group in 3-methylcytidine (m3C), 5-methylcytidine (m5C), 1-methylguanosine (m1G), N6-methyladenosine (m6A) modifications. (C) The dot blotting results showed that the overall level of m6A mRNA modification in the miR-4729-HUVEC group was significantly lower than that of the control group. *P<0.05 vs. miR-mut-HUVECs; t-test; n=3. (D) Immunofluorescence staining results of CD31 and TIE1. Magnification: 200×. (E) The immunofluorescence results showed that the level of cellular TIE1 expression in CD31+ cells in hemorrhoids also significantly increased. **P<0.01 vs. miR-mut-HUVECs; t-test; n=3. (F) The western blotting results showed that the level of TIE1 protein expression in the miR-4729-HUVEC group was significantly lower than the miR-mut-HUVEC group. (G) The RNA immunoprecipitation-polymerase chain reaction (RIP-PCR) results showed that the specific products of the 3'UTR of the TIE2 gene mRNA in miR-4729-HUVECs were significantly decreased in the complexes cross-linked with anti-m6A antibody (α-m6Aab). **P<0.01 vs. miR-mut-HUVECs; t-test; n=3.
Table 2. Raw and normalized peak information.
| Nucleoside | Symbol | Log2(Fold Change) miR-4729 vs. miR-mut |
|---|---|---|
| 3-methylcytidine | m3C | −0.441458398 |
| 5-methylcytidine | m5C | −0.441458398 |
| 1-methyladenosine | m1A | 0.506928195 |
| N6-methyladenosine | m6A | −0.471422896 |
| 1-methylguanosine | m1G | −0.48635495 |
| 7-methylguanosine | m7G | 0.131056973 |
| N2-methylguanosin | m2G | −0.071694974 |
| 5-methyluridine | m5U | 3.341422273 |
| 3-methyluridine | m3U | 3.341422273 |
| 1-methylpseudouridine | m1Ψ | −0.714714526 |
MiR-4729 overexpression inhibits the TIE1/VEGFA signal molecular loop in vascular endothelial cells
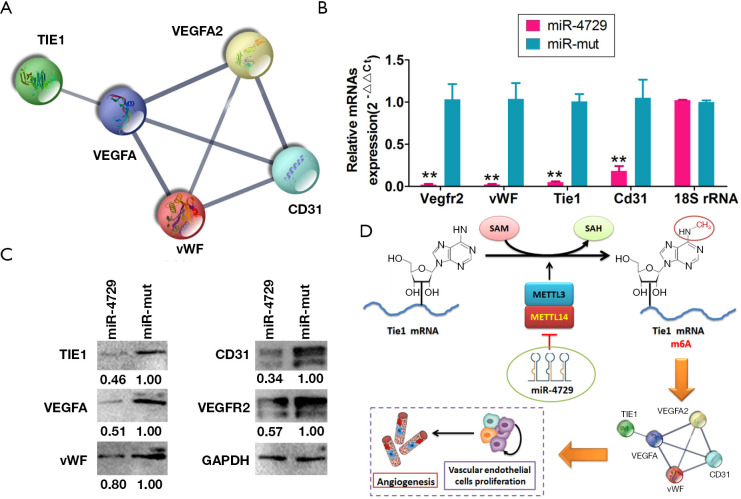
The interaction network associated with TIE1 and angiogenesis-related proteins was identified via bioinformatics analysis (STRING: functional protein association networks; https://string-db.org/). Software predictions showed that TIE1 is intrinsically related to the proteins encoded by the 4 genes associated with VEGFA, VEGFR2, vWF, and CD31, the latter of which can form a completely closed signal loop (Figure 7A). The qPCR and western blotting results showed that the expression of TIE1, VEGFA, VEGFR2, vWF, and CD31 and other genes and proteins in the miR-4729-HUVEC group was significantly lower than that of the miR-mut-HUVEC group (Figure 7B and C). Therefore, these experimental results showed that miR-4729 overexpression inhibits TIE1/VEGFA signaling molecule expression in vascular endothelial cells.
Figure 7.
MiR-4729 overexpression inhibits the molecular loop of the TIE1/VEGFA signaling pathway in vascular endothelial cells. The interaction networks with TIE1 and angiogenesis related proteins were identified using (A) bioinformatics tools. (B) The qPCR results showed that the levels of TIE1, VEGFA, VEGFR2, vWF, and CD31 and other gene expression in the miR-4729-HUVEC group were significantly lower than that of the miR-mut-HUVEC group. **P<0.01 vs. miR-mut-HUVECs; t-test; n=3. (C) The western blotting results showed that the levels of TIE1, VEGFA, VEGFR2, vWF, CD31, and other protein expression in the miR-4729-HUVEC group were significantly lower than that of the miR-mut-HUVEC group. (D) MiR-4729 weakens the m6A modification of the 3'UTR of TIE1mRNA by inhibiting METTL14, inhibiting the expression of TIE1/VEGFA signaling molecules in vascular endothelial cells, the proliferation of vascular endothelial cells, and the vascular proliferation mechanism.
Discussion
The relationship between hemorrhoid occurrence and the regulation of epigenetic abnormalities remains unclear. Several studies have shown that vascular hyperplasia and edema of vascular endothelial cells and the surrounding tissues are particularly obvious in hemorrhoids (1-7); however, the causes of vascular endothelial cell proliferation and edema are poorly understood. TIE1 is a member of the TIE family of receptor tyrosine kinases (30-33). TIE1 is structurally similar to its homolog, TIE2, but differs from TIE2 in that it has no known ligand, making TIE1 an orphan receptor (30-33). One of the key functions of TIE1 is to regulate TIE2 signal transduction by forming heterodimers with TIE2 on the cell surface (30-33). The influence of TIE1-TIE2 interaction depends on the environment. Heterodimerization can promote or inhibit downstream TIE2 signal transduction, depending on the level of local TIE2 expression (30-33). Previous studies have also shown that TIE1 is associated with angiogenesis, vascular maturation, tissue remodeling, and inflammation. Recent studies have found that increased TIE1 expression is also closely related to tumor stem cell “dryness” and atherosclerosis development (32,34-37). However, no inherent relationship between TIE1 and angiogenesis in hemorrhoids has been reported. In this study, focusing on TIE1 expression, we found that the blood vessels in the hemorrhoid nucleus swelled and proliferated.
Furthermore, the proliferated vascular endothelial cells expressed high levels ofTIE1. Subsequently, the bioinformatics predictions found that the proteins encoded by 4 TIE1 related genes (VEGFA, VEGFR2, vWF, and CD31) were intrinsically related to each other, the latter of which can form a complete closed signaling pathway (TIE1/VEGFA signal molecular loop). These findings suggest that there is a close relationship between TIE1 and hemorrhoid nuclear angiogenesis.
The m6A RNA refers to the methylation modification on the sixth nitrogen atom (N) of adenine RNA. Also, m6A RNA methylation has been widely observed in most eukaryotic species (including yeast, plants, fruit flies, and mammals) and viral mRNA and also plays a key role in post-transcriptional mRNA regulation and metabolism (16-20). Meyer et al. and Dominissini et al. used the same m6A-specific combination immunoprecipitation high-throughput sequencing method to identify human and mouse genes, respectively. They also studied m6A RNA distribution in the full transcriptome range. The results showed that m6A RNA was primarily distributed near the mRNA 3'-UTR and the stop codon in the coding region. It was also found that the m6A distribution was highly conserved in humans and mice.
Moreover, it has also been reported that m6A RNA can improve RNA stability (16,17,21,24). Further studies have shown that m6A is mainly concentrated near the mRNA stop codon and on the long exons in them RNA3' UTR. The conserved order of the main distribution was G (m6A) C (70%) or A (m6A) C (30%) (16,18,38). Thus, we first analyzed the level of m6A “writer” METTL14 expression. We found that its expression in hemorrhoid vascular endothelial cells was higher than that of the control group. Subsequently, we speculated that differences in expression might be caused by differential microRNA regulation. Based on bioinformatics predictions and a luciferase reporter assay, miR-4729 was identified as one of the target miRNA molecules that could regulate METTL14.
Moreover, miR-4729 expression was significantly decreased in hemorrhoids. After overexpressing exogenous miR-4729 in vascular endothelial cells, METTL14 and expression of the TIE1/VEGFA signaling molecular loop decreased significantly. At the same time, miR-4729 was also found to weaken the physiological function of vascular endothelial cells. Therefore, based on its function, we speculate that METTL14 may affect its expression and other related molecules by impacting the m6A methylation of the 3'UTR of TIE1 mRNA, resulting in the loss of its stability. Using an analysis of the total mRNA methylation in each group, it was found that miR-4729 overexpression reduced the methylation of nucleic acid mRNA in target cells, especially m6A modifications.
Further analysis showed that miR-4729 overexpression significantly reduced m6A modification at specific TIE1 mRNA sites. Also, inhibition of METTL14 expression caused abnormal writing of multiple genes associated with m6A mRNA in target cells, including TIE1. Since TIE1 is closely related to hemorrhoid vascular hyperplasia, METTL14 expression is also closely related to hemorrhoid vascular proliferation; however, there are currently no reports investigating the relationship between hemorrhoid angiogenesis and RNA methylation. Therefore, this study provides novel insights into the potential RNA epigenetic mechanisms of hemorrhoid nuclear angiogenesis.
The results of this study enrich our understanding of the regulation mechanism of TIE1 on a post-transcriptional level. Since the discovery of TIE1, most studies have focused on the interaction between TIE1 and TIE2, protein regulation, protein activity, and endothelial cell function. However, there are few reports on transcriptional regulation and post-transcriptional regulation of TIE1 levels (30-37). In addition to the TIE1/TIE2/VEGFR2 signaling pathway itself, our study also revealed that TIE1 could induce the expression of vWF and CD31 by activating VEGFA/VEGFR2 receptor ligands and promote angiogenesis. The results provide a theoretical basis for the mechanism of TIE1 in promoting angiogenesis. It is suggested that TIE1 may not activate the VEGFR2 signaling pathway in the presence of TIE2 but can activate the VEGFA and VEGFR2 signaling pathway. Although we did not reveal the regulatory mechanism between TIE1 and VEGFA, the above results suggest a novel regulatory phenomenon of TIE1.
Moreover, this study revealed the epigenetic regulation of TIE1 self-stability. It has previously been reported that the m6A mRNA has been modified in some genes; however, regulation of TIE1 gene mRNA after transcription is rare. We found that the level of TIE1 mRNA and protein was significantly increased during the process of hemorrhoid tissue angiogenesis, suggesting that its activity and stability were greatly enhanced. Quantitative analysis finally revealed that the specific location of TIE1 mRNA was modified by m6A, which led to its abnormal stability in hemorrhoid tissues, providing strong support for enhanced TIE1 protein translation. Therefore, the in-depth mechanism revealed in this study is of great significance regarding angiogenesis and the formation of hemorrhoids, and the post-transcriptional regulation and modification of TIE1.
In summary, the present study's findings showed that miR-4729 down-regulation in vascular endothelial cells of hemorrhoids is one of the main causes of vascular proliferation. MiR-4729 overexpression in vascular endothelial cells can decrease mRNA methylation and 3'UTR-specific site methylation of TIE1mRNA by silencing the expression of the METTL14 target gene. This reducesTIE1 mRNA stability, down-regulates the expression of the TIE1/VEGFA signal molecular loop, and inhibits vascular endothelial cells' physiological activity.
Supplementary
The article’s supplementary files as
Acknowledgments
Funding: This work was supported by grant from the National Natural Science Foundation of China (No. 81673768, 81574033, 81973899). And, grant from the projects sponsored by the development fund for Shanghai talents (2017054). And, grant from Shanghai Natural Science Foundation (No. 16401971600). And, grant from Shanghai Municipal Health Bureau Fund (No. SHDC12014222). And, grant from Shanghai University of TCM GaoFengGaoYuan Foundation (2505). And, grant from Shanghai University of TCM Foundation (2016YSN44). And, grant from the Sixth Batch of national senior TCM experts' academic experience inheritance project of the State Administration of traditional Chinese Medicine ((2017)-29). And, grant from construction project of Shanghai famous and old TCM academic experience research studio (SHGZS-2017028). We declared no potential conflicts of interest.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The Regional Ethics Committee approved the study protocol of Shuguang Hospital, Shanghai University of Traditional Chinese Medicine (Permission No.: 201701.4). This study conformed to the provisions of the Declaration of Helsinki (as revised in 2013), and written informed consent was obtained from every participant.
Footnotes
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/atm-20-3399
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-20-3399
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-3399). The authors have no conflicts of interest to declare.
References
- 1.Fox A, Tietze PH, Ramakrishnan K. Anorectal conditions: hemorrhoids. FP Essent 2014;419:11-9. [PubMed] [Google Scholar]
- 2.Rakinic J, Poola VP. Hemorrhoids and fistulas: new solutions to old problems. Curr Probl Surg 2014;51:98-137. 10.1067/j.cpsurg.2013.11.002 [DOI] [PubMed] [Google Scholar]
- 3.Sugerman DT. JAMA patient page. Hemorrhoids. JAMA 2014;312:2698. 10.1001/jama.2014.281 [DOI] [PubMed] [Google Scholar]
- 4.Lohsiriwat V. Treatment of hemorrhoids: A coloproctologist's view. World J Gastroenterol 2015;21:9245-52. 10.3748/wjg.v21.i31.9245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Qureshi WA. Office management of hemorrhoids. Am J Gastroenterol 2018;113:795-8. 10.1038/s41395-018-0020-0 [DOI] [PubMed] [Google Scholar]
- 6.Mott T, Latimer K, Edwards C. Hemorrhoids: Diagnosis and Treatment Options. Am Fam Physician 2018;97:172-9. [PubMed] [Google Scholar]
- 7.Jacobs D. Clinical practice. Hemorrhoids. N Engl J Med 2014;371:944-51. 10.1056/NEJMcp1204188 [DOI] [PubMed] [Google Scholar]
- 8.van Rooij E. The art of microRNA research. Circ Res 2011;108:219-34. 10.1161/CIRCRESAHA.110.227496 [DOI] [PubMed] [Google Scholar]
- 9.Borghini A, Andreassi MG. Genetic polymorphisms offer insight into the causal role of microRNA in coronary artery disease. Atherosclerosis 2018;269:63-70. 10.1016/j.atherosclerosis.2017.12.022 [DOI] [PubMed] [Google Scholar]
- 10.Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature 2005;435:834-8. 10.1038/nature03702 [DOI] [PubMed] [Google Scholar]
- 11.Leung AK, Sharp PA. MicroRNA functions in stress responses. Mol Cell 2010;40:205-15. 10.1016/j.molcel.2010.09.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mishra P, Singh D, Ganju L, et al. MicroRNA in gastrointestinal cell signalling. Inflammopharmacology 2018;26:1-14. 10.1007/s10787-017-0414-4 [DOI] [PubMed] [Google Scholar]
- 13.Mendell JT, Olson EN. MicroRNAs in stress signaling and human disease. Cell 2012;148:1172-87. 10.1016/j.cell.2012.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Creugny A, Fender A, Pfeffer S. Regulation of primary microRNA processing. FEBS Lett 2018;592:1980-96. 10.1002/1873-3468.13067 [DOI] [PubMed] [Google Scholar]
- 15.Wang C, Lu H, Luo C, et al. miR-412-5p targets Xpo1 to regulate angiogenesis in hemorrhoid tissue. Gene 2019;705:167-76. 10.1016/j.gene.2019.04.058 [DOI] [PubMed] [Google Scholar]
- 16.Meyer KD, Saletore Y, Zumbo P, et al. Comprehensive analysis of mRNA methylation reveals enrichment in 3' UTRs and near stop codons. Cell 2012;149:1635-46. 10.1016/j.cell.2012.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dominissini D, Moshitch-Moshkovitz S, Schwartz S, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 2012;485:201-6. 10.1038/nature11112 [DOI] [PubMed] [Google Scholar]
- 18.Yue Y, Liu J, He C. RNA N6-methyladenosine methylation in post-transcriptional gene expression regulation. Genes Dev 2015;29:1343-55. 10.1101/gad.262766.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jia G, Fu Y, Zhao X, et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol 2011;7:885-7. 10.1038/nchembio.687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen T, Hao YJ, Zhang Y, et al. m(6)A RNA methylation is regulated by microRNAs and promotes reprogramming to pluripotency. Cell Stem Cell 2015;16:289-301. 10.1016/j.stem.2015.01.016 [DOI] [PubMed] [Google Scholar]
- 21.Tang C, Klukovich R, Peng H, et al. ALKBH5-dependent m6A demethylation controls splicing and stability of long 3'-UTR mRNAs in male germ cells. Proc Natl Acad Sci U S A 2018;115:E325-33. 10.1073/pnas.1717794115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen M, Wei L, Law CT, et al. RNA N6-methyladenosine methyltransferase-like 3 promotes liver cancer progression through YTHDF2-dependent posttranscriptional silencing of SOCS2. Hepatology 2018;67:2254-70. 10.1002/hep.29683 [DOI] [PubMed] [Google Scholar]
- 23.Liu J, Yue Y, Han D, et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol 2014;10:93-5. 10.1038/nchembio.1432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang X, Lu Z, Gomez A, et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 2014;505:117-20. 10.1038/nature12730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barbieri I, Tzelepis K, Pandolfini L, et al. Promoter-bound METTL3 maintains myeloid leukaemia by m(6)A-dependent translation control. Nature 2017;552:126-31. 10.1038/nature24678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cheng W, Liu T, Wan X, et al. MicroRNA-199a targets CD44 to suppress the tumorigenicity and multidrug resistance of ovarian cancer-initiating cells. FEBS J 2012;279:2047-59. 10.1111/j.1742-4658.2012.08589.x [DOI] [PubMed] [Google Scholar]
- 27.Gao Y, Liu T, Huang Y. MicroRNA-134 suppresses endometrial cancer stem cells by targeting POGLUT1 and Notch pathway proteins. FEBS Lett 2015;589:207-14. 10.1016/j.febslet.2014.12.002 [DOI] [PubMed] [Google Scholar]
- 28.Liu T, Chi H, Chen J, et al. Curcumin suppresses proliferation and in vitro invasion of human prostate cancer stem cells by ceRNA effect of miR-145 and lncRNA-ROR. Gene 2017;631:29-38. 10.1016/j.gene.2017.08.008 [DOI] [PubMed] [Google Scholar]
- 29.Liu T, Cheng W, Huang Y, et al. Human amniotic epithelial cell feeder layers maintain human iPS cell pluripotency via inhibited endogenous microRNA-145 and increased Sox2 expression. Exp Cell Res 2012;318:424-34. 10.1016/j.yexcr.2011.12.004 [DOI] [PubMed] [Google Scholar]
- 30.Korhonen EA, Lampinen A, Giri H, et al. Tie1 controls angiopoietin function in vascular remodeling and inflammation. J Clin Invest 2016;126:3495-510. 10.1172/JCI84923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim M, Allen B, Korhonen EA, et al. Opposing actions of angiopoietin-2 on Tie2 signaling and FOXO1 activation. J Clin Invest 2016;126:3511-25. 10.1172/JCI84871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Savant S, La Porta S, Budnik A, et al. The Orphan Receptor Tie1 Controls Angiogenesis and Vascular Remodeling by Differentially Regulating Tie2 in Tip and Stalk Cells. Cell Rep 2015;12:1761-73. 10.1016/j.celrep.2015.08.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seegar TC, Eller B, Tzvetkova-Robev D, et al. Tie1-Tie2 interactions mediate functional differences between angiopoietin ligands. Mol Cell 2010;37:643-55. 10.1016/j.molcel.2010.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tiainen L, Korhonen EA, Leppanen VM, et al. High baseline Tie1 level predicts poor survival in metastatic breast cancer. BMC Cancer 2019;19:732. 10.1186/s12885-019-5959-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.La Porta S, Roth L, Singhal M, et al. Endothelial Tie1-mediated angiogenesis and vascular abnormalization promote tumor progression and metastasis. J Clin Invest 2018;128:834-45. 10.1172/JCI94674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Torigata M, Yamakawa D, Takakura N. Elevated expression of Tie1 is accompanied by acquisition of cancer stemness properties in colorectal cancer. Cancer Med 2017;6:1378-88. 10.1002/cam4.1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Garcia J, Sandi MJ, Cordelier P, et al. Tie1 deficiency induces endothelial-mesenchymal transition. EMBO Rep 2012;13:431-9. 10.1038/embor.2012.29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bartosovic M, Molares HC, Gregorova P, et al. N6-methyladenosine demethylase FTO targets pre-mRNAs and regulates alternative splicing and 3'-end processing. Nucleic Acids Res 2017;45:11356-70. 10.1093/nar/gkx778 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as