Abstract
Background
The monkey is a primary species used in toxicological research. However, the failures of preclinical studies to predict a life-threatening “cytokine storm”, which, for instance, rapidly occurred in six healthy volunteers with the CD28 superagonist monoclonal antibody (mAb) TGN1412 in the first-in-human phase I clinical trial, have emphasized a need to clarify the differences between human and monkey immune systems.
Methods
In the present study, we analyzed and compared the lymphocyte proliferation, cytokine secretion, and gene expression profiles after phytohemagglutinin (PHA) and lipopolysaccharide (LPS) stimulation of peripheral blood mononuclear cells (PBMCs) from three healthy humans and cynomolgus monkeys (Macaca fascicularis).
Results
The results derived from comparison with the corresponding control groups showed that PHA in humans induced a stronger proliferation and wider range of cytokine secretion, along with a greater number of differently expressed genes (DEGs), than when PHA was applied in cynomolgus monkeys. The significant upregulation of genes involved in the mitotic cell cycle, including cyclin B2, TOP2A, TYMS, and CEP55, was observed in human PBMCs with PHA stimulation, while only infrequent or slight upregulation occurred in cynomolgus monkey PBMCs, which may be one of the reasons for a stronger response to PHA in humans. In contrast to PHA, LPS in both species induced a similar proliferation ratio, cytokine profile, and DEG count, suggesting that human and cynomolgus monkeys have a similar response intensity for innate immune responses. Furthermore, 38 and 20 overlapped genes under PHA and LPS stimulation, respectively, were found in both species. These overlapped DEGs were associated with the same biological functions, including DNA replication, mitosis, immune response, chemotaxis, and inflammatory response. Thus, these results might reflect the highly conserved signatures of immune responses to PHA/LPS stimulation across the primates. Moreover, there were some differences in antigen processing and presentation, and the interferon gamma (INF-γ)–mediated signaling pathway in these species detected by gene expression profile study.
Conclusions
In conclusion, this is the first study to compare data on the responses of PBMCs to PHA and LPS in humans versus cynomolgus monkeys, and these findings may provide crucial insights into translating non-human primate (NHP) studies into human trials.
Keywords: Non-human primates (NHPs), immunotoxicity, toxicogenomics, lipopolysaccharide (LPS), phytohemagglutinin (PHA)
Introduction
Toxicological research requires animal models that are physiologically closely related to one another. There is good evidence that non-human primates (NHPs), including rhesus and cynomolgus monkeys (Macaca fascicularis), closely meet these requirements at both the molecular and developmental levels (1). However, the recently failed first-in-human clinical trial of TGN1412 (theralizumab) raises concerns about whether the immune system of NHPs can be used to predict how TGN1412 affects in humans. In that trial, six healthy volunteers suffered severe adverse events soon after TGN1412 antibody administration, while preclinical safety testing in cynomolgus monkeys had suggested that TGN1412 could be tolerated without toxicity (2,3). It has been suggested that the immune response to exogenous substances differs between these two species. Therefore, comparative studies of the immune system, especially the functional differences of immune cells, in humans and NHPs, could provide valuable information for better extrapolating animal toxicity data in preclinical drug safety studies using NHPs.
The immune system is highly complex and capable of lifelong renewal. The normal physiological range of variation in the immune system is affected by age, sex, environment, and other factors; this diversity presents a major challenge in assessing drug-induced immunotoxicity (4,5). However, research on the anatomical and functional differences between the immune systems of monkeys and humans has been limited. Currently, the morphological characteristics and development of the cynomolgus monkey immune system, including the spleen, lymph nodes, and Peyer’s patches have been reported on (6). Additionally, differences have been found in gene expression of Th1/Th2 cytokines and their receptors in the peripheral blood mononuclear cells (PBMCs) of primates. For example, one study demonstrated a higher gene expression of interleukin 4 (IL-4) in humans and chimpanzees compared with that of baboon/macaques, whereas the expression of interferon gamma (IFN-γ) and IL-12 genes in humans was shown to lower than that in baboon/macaques (7). However, this study was limited to the steady-state gene expression in the resting state of immune cells, while the functions of stimulated immune cells, including cell proliferation, cytokine secretion, pathogen clearance, and others, can better reflect the body’s response to exogenous substances.
As an alternative, ex vivo/in vitro approaches of immune cell function trials using PBMCs can be used to assess immunotoxicity of a given agent. Furthermore, the information obtained from human and NHPs can facilitate the comparative study of different species under the same treatment conditions (8,9). In addition to classical immune cells function trials, microarrays can be a valuable tool for translating data from animals to humans (10-13). Comparing differences between the expression profiles of genes associated with toxicity among various animal and human tissues to identify the significant key genes expressed in each type of host would provide a better translation of animal toxicity data to human scenarios (14,15).
Thus, our aim was to compare the differences in cell proliferation, cytokine secretion, and gene profiles of human and cynomolgus monkey PBMCs under various stimulations in vitro, to provide new specific primate data for comparative immunology. We chose two different stimulations, lipopolysaccharide (LPS) and phytohemagglutinin (PHA), to induce an innate immune response (LPS stimulation) or lymphocyte proliferation (PHA stimulation) respectively. LPS is a part of the outermost layer of Gram-negative bacteria, and is used to study the innate immune response after bacterial infection in vitro by pathogen-associated molecular pattern (PAMP). PHA can activate small lymphocytes to transform into lymphoblasts, then divide and proliferate, release cytokines, and improve the phagocytic ability of macrophages. PHA binds to specific cell surface carbohydrates on T-cells and is often used in immunology studies as a T-lymphocyte proliferation inducer. Both stimulations were chosen as they are typically used to induce distinct types of immune responses for further characterization. We observed the distinct features of cell proliferation, cytokine secretion, and gene expression changes in response to both stimulations with concentration, and here report the molecular mechanisms of the two species in different immune responses based on the information of gene expression microarrays. To our knowledge, this is the first study to report the differences and similarities between the immune responses of PBMCs to LPS and PHA stimulation in humans versus cynomolgus monkeys.
We present the following article in accordance with the Animal Research: Reporting of In Vivo Experiments (ARRIVE) reporting checklist (available at http://dx.doi.org/10.21037/atm-20-4548).
Methods
Isolation of cynomolgus and human PBMCs
Blood from three healthy adult male volunteers who had donated blood to the National Institutes for Food and Drug Control (NIFDC) and who had provided written informed consent, was obtained. Cynomolgus monkey blood was obtained from three apparently healthy 2.5-year-old male cynomolgus monkeys. All procedures in the animal experimental studies were performed under a project license (IACUC-2013-041) granted by the ethics board of the National Center for Safety Evaluation of Drugs (NCSED), in compliance with the Guide for the Care and Use of Laboratory Animals of the NCSED (China).
In all cases, whole blood PBMCs were isolated from the heparinized samples by Ficoll-Hypaque density centrifugation (Histopaque: ρ=1.119 or 1.077; Sigma, St. Louis, MO, USA) at 2,000 rpm for 30 min. Isolated PBMCs had >90% purity based on flow cytometric analyses using CD3, CD20, and CD14 staining (BD Biosciences, San Diego, CA, USA). The major PBMC cell types isolated were T-lymphocytes, B-lymphocytes, and monocytes. The isolated cells were washed twice in Hank’s Balanced Salt Solution (HBSS) and re-suspended to a final concentration of 106 cells/mL in Roswell Park Memorial Institute (RPMI) 1640 medium supplemented with 10% heat-inactivated fetal calf serum (Gibco, Grand Island, NY, USA). All experiments using cultured cells were performed in triplicate.
PBMC lymphocyte proliferation induced by PHA and LPS
The PHA (phytohemagglutinin, Sigma) and LPS (lipopolysaccharide, Type O127:B8 from Escherichia coli, Sigma) were used to induce lymphocyte proliferation among cultured PBMCs. In brief, PBMCs obtained from three cynomolgus monkeys and three human donors were used. For this, 106 cells/well/host were placed into dedicated wells of 96-well plates and then received medium containing PHA (2, 5, or 10 µg/mL final concentration in well) or LPS (2, 5, or 10 µg/mL final concentration in well) and then incubated for 3 days at 37 °C. Cells that received medium only served as controls. Lymphocyte proliferation responses were then measured using a Cell Counting Kit-8 (CCK-8) assay (Dojindo, Tokyo, Japan) according to the manufacturer’s instructions. In brief, 10 µL of CCK-8 solution was added to each well and the plates were incubated for 4 hours at 37 °C. Then, absorbance at 450 nm in each well was measured using a SpectraMax Plus microplate reader (Molecular Devices, Sunnyvale, CA, USA). The proliferation ratio of PBMCs (%) was the ratio of optical density (OD) value of test well to that of the control well. All samples at each stimulant dose were evaluated in triplicate.
Cytokine expression after exposures to PHA and LPS
To detect and compare the expression of cytokines (IFN-γ, tumor necrosis factor alpha [TNF-α, IL-2, IL-4, IL-5, IL-6, IL-8, IL-10, IL-12p70, IL-17A, and IL-23) in human and monkey PBMCs after exposure to PHA/LPS, a multiplex Luminex-based assay (eBioscience Inc., San Diego, CA, USA) was used. In brief, similar to in the proliferation study noted above, the supernatants from cultures after PHA/LPS treatments (2, 5, or 10 µg/mL) were collected and immediately stored at −80 °C until analyzed. Quantification of each cytokine was performed according to manufacturer protocols using a Luminex 200 system (Luminex Corp., Austin, TX, USA). Minimum detectable concentrations of IFN-γ, TNF-α, IL-2, IL-4, IL-5, IL-6, IL-8, IL-10, IL-12p70, IL-17A, and IL-23 were, respectively, 12.2, 8.5, 4.9, 12.2, 7.3, 9.8, 2.4, 2.4, 6.8, 2.4, and 14.6 pg/mL for human cytokines and 1.0, 16.6, 0.5, 8.8, 10.35, 4.03, 3.2, 0.8, 1.5, 7.0, and 6.7 pg/mL for NHP cytokines. All samples at each stimulant dose were evaluated in triplicate.
PBMC gene expression profiles after exposures to PHA and LPS
To compare gene expression profiles of human and cynomolgus monkey PBMCs in response to PHA and LPS, the PBMCs (106 cells/mL) obtained from cynomolgus or human donors were placed in 6-well plates (106 cells/well), treated with PHA (2 or 10 µg/mL) or LPS (2 or 10 µg/mL), and incubated at 37 °C for 72 hours. Cells cultured under similar conditions without stimulation served as controls. Total RNA was extracted from PBMCs using Trizol reagent (Invitrogen, Waltham, MA, USA) according to the manufacturer’s instructions. The RNA purity in each sample was assessed using a NanoDrop system (NanoDrop Technologies, Wilmington, NC, USA); RNA integrity was assessed using standard agarose gel electrophoresis. A total of three RNA samples for each group were selected for subsequent microarray analyses. Purified total RNA was amplified and labeled using Agilent Low Input Quick Amp Labeling Kit (Agilent, Santa Clara, CA, USA). Then, fluorescence dye-labeled complement RNA (cRNA) was hybridized to Agilent 4×44K Human and 4×44K Monkey Whole Genome GeneChips (Agilent, Palo Alto, CA, USA). Hybridization, scanning, and washing were performed on Agilent’s Microarray Platform according to their standard protocols, and the array data were analyzed using Agilent Feature Extraction software (version 10.7). After normalization of the raw data, probes with intensities <400 were removed from the subsequent analysis using significance analysis of microarray (SAM) software. Differentially expressed genes (DEGs) were identified as those having an adjusted q-value <0.05 and an average fold-change (FC) difference of >1.5. Gene Ontology (GO) analysis was performed using hypergeometric distribution in the R-language package software (R Foundation for Statistical Computing, Vienna, Austria).
Validation of the gene expression data using real-time quantitative polymerase chain reaction (qRT-PCR)
A total of 10 genes that exhibited significant differential expression profiles based on the microarray analysis were chosen for validation by qRT-PCR. Here, 1 µg of DNase-treated total RNA (the same RNA as was used in microarrays) was reverse transcribed using an oligo-dT primer and Moloney murine leukemia virus (M-MLV) reverse transcriptase (Life Technologies, San Diego, CA, USA). Real-time PCR amplification was then performed using FastStart Universal SYBR Green Master and a LightCycler 480 system (Roche Diagnostics, Shanghai, China). Running conditions were 3 min at 95 °C, followed by 45 cycles of 30 sec at 94 °C (denaturation), 30 sec at 57 °C (annealing), and 30 sec at 72 °C (extension). Relative mRNA expression levels were analyzed using the 2-ΔΔCt method. A total of 18 primers were used in this study (Table 1). Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as a housekeeping control and to normalize data.
Table 1. Primer sequences for real-time PCR.
| Target gene | Forward Primer (5'---3') | Reverse Primer (3'---5') |
|---|---|---|
| Human IL-8 | ACATACTCCAAACCTTTCCACC | TCAAAAACTTCTCCACAACCC |
| Human PCNA | TTGAAGCACCAAACCAGGA | AGAAGGCATCTTTACTACACAGC |
| Human MCM6 | TCAGAGCAGCGATGGAGAAAT | CCCGACACAGGTAAGGGTAAA |
| Human PLK1 | AATGACGAGTTCTTTACTTCTGGC | TGAGGACTGTGAGGGGCTTC |
| Human CDKN2C | CAGGGGGGACCTAGAGCAA | ATGAATGACAGCGAAACCAGTT |
| Human CCL2 | TCAATGCCCCAGTCACCT | CCACAATGGTCTTGAAGATCA |
| Human CD74 | GCTGACCCCCTGAAGGTGTA | CAAGGAGTGCCTGCTCATTTC |
| Human IL1A | AAGAAGACAGTTCCTCCATTGA | GCTTGGATGTTTTAGAGGTTTC |
| Human CCL3 | AGCCCGGTGTCATCTTCCT | CACACGCATGTTCCCAAGG |
| Monkey IL8 | CAAGAGCCAGGAACAAACCA | GCAAAACTGCACCTTCACACA |
| Monkey PCNA | TCGATAAAGAGGAGGAAGCTG | ACCGTTGAAGAGAGTGGAGTG |
| Monkey MCM6 | CATTGGTTGTGAGTTTTGTGG | GTCTGGTAGGCAGGTCTTGG |
| Monkey PLK1 | AGGAAGAAGACCCTGTGCG | GCAAGAAGTCTCAAAAGGTGGT |
| Monkey CDKN2C | GACCCTAAAGAATGGCCGA | GGGATTTCCAAGTTTCATAACC |
| Monkey CCL2 | TGGTGATTCTTCTATAGCTCGCC | GGTGATTCTTCTATAGCTCGCC |
| Monkey CD74 | CACATCCTGCCCCTCAAAA | AACCCCTGGCTCACCTCTT |
| Monkey IL1A | AATGACACCCTCAATCAAACTA | TCATCTTGGGCACTCACATA |
| Monkey CCL3 | ACTGCTGCCCTTGCTGTCCT | TGGCTGTTGGTCTCAAAGTAGTCA |
Statistical analysis
For the lymphocyte proliferation, F-tests of two regression line equations were used to compare increasing trends for the lymphocytes in the human and cynomolgus monkey groups. For the Luminex data, a Bartlett test for variance homogeneity was performed first; if a result showed no significance (i.e., P>0.05), a one-way analysis of variance (ANOVA) was used. If a result was significant, a Dunnett’s test (parameter method) for multiple comparisons was performed. A P value <0.05 was considered significant in the ANOVA/Dunnett’s evaluations. For GeneChip microarray data, SAM software and R language package software were used to identify the DEG and relevant GO terms. All results, except for the gene expression data, are expressed as mean ± SD.
Results
PBMC lymphocyte proliferation induced by PHA and LPS
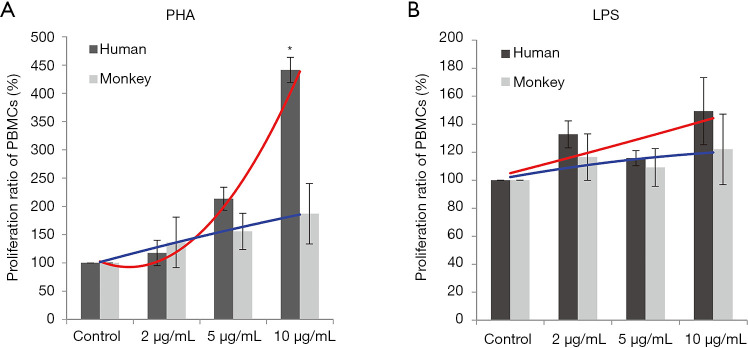
Proliferation responses of PBMC lymphocytes were measured after 72 hours of stimulation with PHA or LPS at 2, 5, and 10 µg/mL. The results showed that with the increasing PHA levels, the proliferation ratio of human and cynomolgus monkey PBMCs increased in a dose-dependent manner. At a high dose, the mean proliferation ratio of human and cynomolgus monkey PBMCs was 441.3% and 187% respectively, and the proliferation of human PBMCs was significantly higher than that of cynomolgus monkeys (Figure 1). The proliferation trend significantly differed between human and monkey cells, which suggested that the proliferation ability of human PBMCs induced by PHA was stronger than that of cynomolgus monkeys (F-test of two regression line equations; P<0.05).
Figure 1.
Lymphocyte proliferation in response to PHA or LPS. Proliferation by human and cynomolgus PBMCs were measured via CCK-8 assay. An F-test of two regression line equations was used to compare trends for proliferation rates between human and monkey cells. Lines in red and blue indicate trend curves for proliferation of the human and monkey PBMCs, respectively. (A) The trend for the proliferation of PBMC was significantly different between humans and cynomolgus monkeys after PHA stimulation (*, P<0.05). (B) No significant difference in trends for proliferation was noted between human and cynomolgus monkey cells after LPS stimulation. PHA, phytohemagglutinin; LPS, lipopolysaccharide; PBMC, peripheral blood mononuclear cells; CCK-8, Cell Counting Kit-8.
After LPS stimulation, compared with negative control groups, the PBMCs of human and cynomolgus monkeys were increased. At a high LPS level, the mean proliferation ratio of human and cynomolgus monkey PBMCs were 149.3% and 122%, respectively, and the proliferation curves of human and cynomolgus monkey PBMCs were similar (P>0.05). Therefore, the differential analysis in these two species showed that the lymphocyte proliferation of PBMCs varied considerably after PHA treatment in comparison to the LPS treatment.
Cytokine formation by human and cynomolgus PBMC induced by PHA and LPS
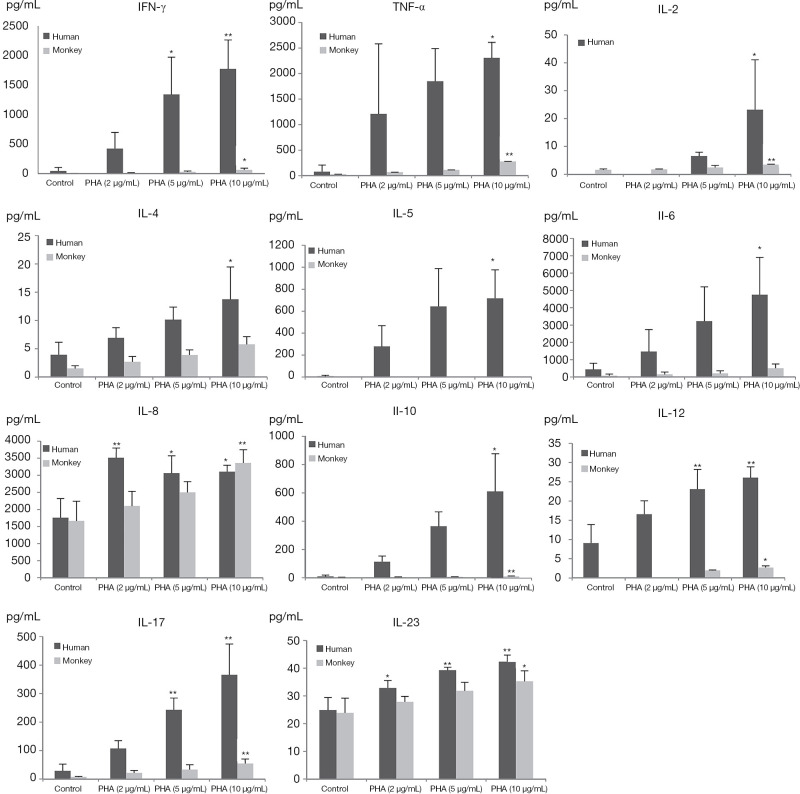
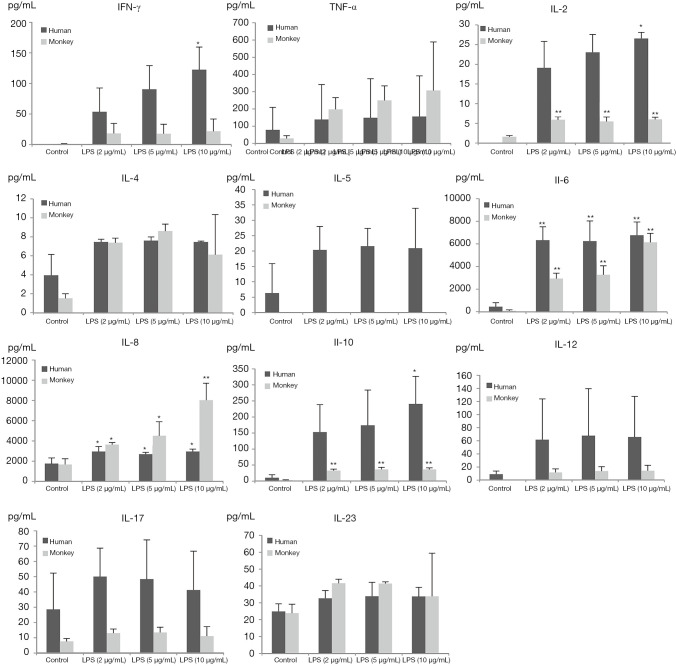
Production of INF-γ, TNF-α, and IL-2, -4, -5, -6, -8, -10, -12p70, -17A, and -23 induced by PHA and LPS in isolated human and cynomolgus PBMCs are shown in Figures 2,3 (summarized in Table 2).
Figure 2.
Cytokine profiles of human and monkey PBMCs treated with PHA or control (n=3). Data are presented as the mean ± SD. Statistically significant differences compared with values of the control group are highlighted by asterisks (*, P<0.05, **, P<0.01). PBMC, peripheral blood mononuclear cells; PHA, phytohemagglutinin; SD standard deviation.
Figure 3.
Cytokine profiles of human and monkey PBMCs treated with LPS or control (n=3). Data are presented as the mean ± SD. Statistically significant differences compared with values of control group are highlighted by asterisks (*, P<0.05, **, P<0.01). PBMC, peripheral blood mononuclear cells; LPS, lipopolysaccharide; SD, standard deviation.
Table 2. Cytokines induced by PHA and LPS in human and monkey.
| Cytokines | PHA | LPS | |||
|---|---|---|---|---|---|
| Human | Monkey | Human | Monkey | ||
| IFN-γ | X | X | X | O | |
| TNF-α | X | X | O | O | |
| IL-2 | X | X | X | X | |
| IL-4 | X | O | O | O | |
| IL-5 | X | O | O | O | |
| IL-6 | X | O | X | X | |
| IL-8 | X | X | X | X | |
| IL-10 | X | X | X | X | |
| IL-12 | X | X | O | O | |
| IL-17 | X | X | O | O | |
| IL-23 | X | X | O | O | |
With PHA/LPS stimulation, statistically significant differences comparing with values of control group are present as “X”. No significant differences comparing with values of control group are present as “O”. Value significantly different from control group at P<0.05 or P<0.01.
Compared with the control group, 10 µg/mL PHA could induce a significant increase in the secretion of all cytokines in human PBMCs, and the secretion of INF-γ, TNFα, IL-2, -8, -10, -12p70, -17A, and -23 in cynomolgus monkey PBMCs was significantly increased. Interestingly, at 10 µg/mL PHA, the secretion of INFγ, TNFα, and IL-5, -6, and -10 in human PBMCs were approximately 10–200 times higher than those in cynomolgus monkey PBMCs, with the contents close to 600–5,000 pg/mL, indicating that human PBMCs secreted a much higher level of cytokines than cynomolgus monkeys under the same conditions of PHA treatment. However, unlike other cytokines, IL-8 and IL-23 secreted by human and cynomolgus monkey PBMCs showed a similarly elevated level after PHA stimulation, with no significant difference between them.
Overall, the secretion of cytokines by human and cynomolgus monkey PBMCs after LPS stimulation, regardless of the types of cytokines or secretion levels, were lower than that of PHA treatment in both species. After 10 µg/mL of LPS stimulation, compared with the control group, the secretion of INF-γ, IL-2, -6, -8, and -10 cytokines in human PBMCs increased significantly, and the secretion of IL-2, -6, -8, and -10 cytokines in cynomolgus monkey PBMCs increased significantly. Except for INF-γ, the secretion levels of cytokines were similar between the two species. Among them, IL-6 and IL-8 secreted by human and cynomolgus monkey PBMCs were significantly increased under three different doses of LPS, and their secretion contents were close to 3000–10,000 pg/mL, which were much higher than those of the other cytokines. These results suggested that LPS induces similar cytokine secretion responses in human and cynomolgus monkey PBMCs and that IL-6 and IL-8 are relatively more pronounced cytokines. However, most notably, IL-5 was not detected (value below standard curve limits) in monkey cultures that received either PHA or LPS.
Comparing numbers of DEGs in human and cynomolgus PBMCs after PHA/LPS stimulation
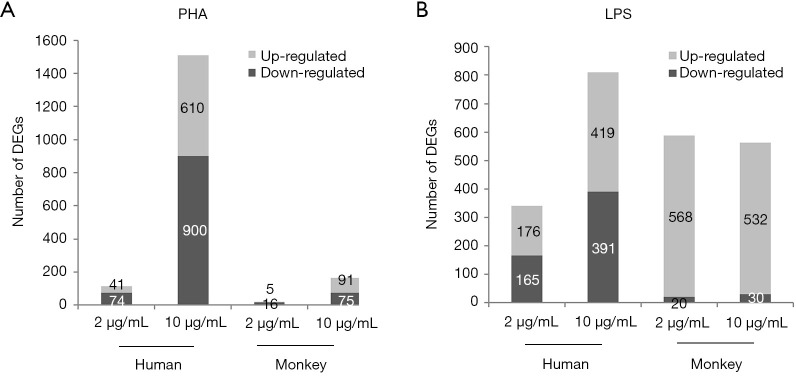
The resulting microarray platform included 11,914 probes in humans and 10,273 probes in monkeys. According to the ratio value of “treatment/control, the DEGs were identified by SAM (two-class unpaired analysis) software. The number of DEGs obtained after PHA/LPS treatments are listed in Figure 4. Compared with the control group, after 2 or 10 µg/mL of PHA stimulation, the number of DEGs in human PBMCs were 115 (41 upregulated genes, 74 downregulated genes) and 1,510 (610 upregulated genes, 900 downregulated genes), respectively; under the same conditions, DEGs in cynomolgus monkey PBMCs were 21 (5 upregulated genes, 16 downregulated genes) and 166 (91 upregulated genes, 75 downregulated genes), respectively (Figure 4A). The results showed that the number of DEGs increased with the increase in dose in both species. Notably, the number of DEGs in human PBMCs was approximately nine times higher than that in cynomolgus monkeys at 10 µg/mL PHA.
Figure 4.
Up- and down-regulated genes in human and cynomolgus PBMCs treated with PHA (2 or 10 µg/mL) or LPS (2 or 10 µg/mL) compared to control. Bar charts in gray and dark represent the number of up- and downregulated genes, respectively. PBMC, peripheral blood mononuclear cells; PHA, phytohemagglutinin; LPS, lipopolysaccharide.
With regard to LPS (Figure 4B), the total DEGs in relation to monkey cells in human cells stimulated with 2 and 10 µg LPS/mL was 341 and 810, and 588 and 562, respectively. In contrast with PHA treatment, there was no apparent difference in the number of DEGs between human and cynomolgus monkey PBMCs after LPS stimulation. The striking differences in the number of DEGs between human and cynomolgus monkeys after PHA treatment might explain the considerable diversities of lymphocyte proliferation results across these two species. For the reader, the raw microarray data have been deposited in the Gene Expression Omnibus (GEO) database under accession number GSE90723.
Comparing functional groups of DEGs by GO enrichment analysis after PHA stimulus
GO enrichment analysis was performed to identify significant functional groups of genes that reflected changes in human and monkey PBMCs after mitogen stimulation. The significant GO terms describing major biological processes (BPs) impacted by PHA/LPS are noted in Table 3.
Table 3. Top-ten biological functions identified by significant enrichment GO analysis in monkey and human PBMCs after PHA stimulation.
| GO term (biological process) | P value# | DEGs* (2 μg/mL PHA) | DEGs (10 μg/mL PHA) |
|---|---|---|---|
| Human | |||
| Immune responseф | −2.22×10–16 | 12 | 71 |
| Mitotic cell cycle | 3.33×10–16 | 11 | 70 |
| Cytokine-mediated signaling pathway | 3.33×10–16 | 2 | 43 |
| DNA strand elongation involved in DNA replication | 5.55×10–16 | 1 | 21 |
| Interferon-gamma-mediated signaling pathway | 2.00×10–15 | 0 | 26 |
| DNA replicationф | 3.79×10–14 | 4 | 37 |
| Cell division | 4.07×10–14 | 10 | 52 |
| Inflammatory response | 5.35×10–14 | 11 | 46 |
| Antigen processing and presentation of peptide or polysaccharide antigen via MHC class II | 1.57×10–13 | 0 | 12 |
| M phase of mitotic cell cycle | 4.08×10–13 | 6 | 28 |
| △Phosphatidylinositol-mediated signaling | 4.74×10–5 | 6 | 14 |
| △Chemotaxisф | △1.53×10–6 | 7 | 25 |
| △Mitosis | △4.75×10–6 | 8 | 31 |
| △Negative regulation of caspase activity | △7.21×10–5 | 4 | 9 |
| Monkey | |||
| DNA-dependent DNA replication initiation | 3.30×10–8 | 0 | 4 |
| Immune responseф | 6.29×10–7 | 6 | 9 |
| DNA replicationф | 3.02×10–5 | 0 | 5 |
| Anti-apoptosis | 7.39×10–5 | 2 | 5 |
| Chromosome organization | 0.00016 | 0 | 3 |
| Regulation of cell shape | 0.00017 | 1 | 4 |
| Astrocyte cell migration | 0.00019 | 1 | 2 |
| Positive regulation of cholesterol efflux | 0.00032 | 1 | 2 |
| Chemotaxisф | 0.00070 | 1 | 3 |
| Apoptotic cell clearance | 0.00087 | 1 | 2 |
| △Antigen processing and presentation of peptide antigen via MHC class I | △3.47×10–13 | 5 | 1 |
| △Antigen processing and presentation | △4.07×10–10 | 5 | 1 |
*, DEGs: number of differently expression genes identified by microarray in 2 or 10 μg/mL PHA groups; #, P value for enrichment; △, that means these top-ten GO terms are identified in 2 μg/mL PHA group; ф, the same GO terms used in both human and monkey groups.
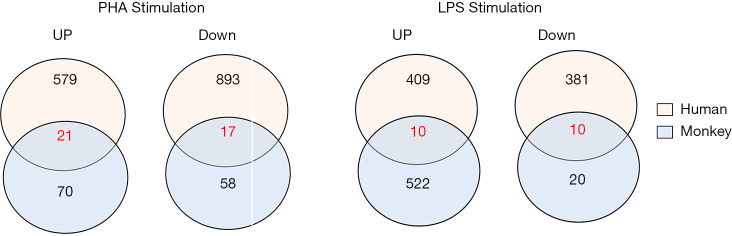
Venn diagrams of the DEGs were generated to compare gene expression profiles between human and monkey PBMCs stimulated with PHA or LPS (Figure 5).
Figure 5.
Venn diagrams of DEGs in human and monkey PBMCs after PHA or LPS stimulation (10 µg/mL). The number of genes that showed expression changes at a two-fold level are shown. DEGs, differentially expressed genes; PBMC, peripheral blood mononuclear cell; PHA, phytohemagglutinin; LPS, lipopolysaccharide.
The results of the GO analysis indicated that the DEGs in human PBMCs under the PHA stimulation were significantly enriched in BPs, which included immune response, mitotic cell cycle, and cytokine-mediated signaling pathway (P value =−2.22×10-16, 3.33×10-16, 3.33×10-16, respectively), in addition to other biological groups related to DNA replication, cell division, etc. (see Table 3). With the increase of PHA concentration, the number of genes involved in the top 10 biological function groups, including immune response and mitotic cell cycle, increased significantly, at a much higher rate than that of monkeys.
Under the PHA stimulation, the biological function groups involved by most DEGs in cynomolgus monkey PBMCs were related to DNA-dependent DNA replication initiation, ommune response, DNA replication (P value =3.30×10-8, 6.29×10-7, and 3.02×10-5, respectively), anti-apoptosis, and other BPs such as chemotaxis and antigen processing and presentation. Similarly, in addition to antigen processing and presentation, there was an increase in PHA concentration in the number of DEGs, but the magnitude of increase was much lower. For example, immune response was the biological function with the highest number of genes involved in cynomolgus monkey PBMCs, but this included only 9 DEGs, which was far from the 71 DEGs for this biological function group in human PBMCs. Through a comparison of the top 10 GO terms in humans versus monkeys, it was found that the similar biological function groups in the two species were mostly associated with immune response, DNA replication, and chemotaxis, suggesting that these three major biological functions may be the most basic and critical biological responses induced by PHA.
The cyclin B2 (CCNB2) gene was by far the most upregulated gene with a positive Fc of 16.7 at 10 µg/mL PHA in humans (see Table 4); it was found to be an important regulator of the G2–M phase. The GO enrichment analysis also showed that CCNB2 participated in 18 GO function groups, mainly involving G2/M transition of mitotic cell cycle, mitotic cell cycle, cell division, and other functions. The other top 10 genes, such as TOP2A, TYMS, CEP55, CENPF, and KIF23 (FC =16.5–11.3) are also involved in mitosis, mitotic cell cycle, G1/S phase of mitotic cell cycle, regulation of G2/M transition of mitotic cell cycle, and DNA replication, among others. The CCL3 [chemokine (C-C motif) ligand 13, FC =0.01] had the most significantly downregulated expression in human PBMCs. Also known as monocyte chemoattractant protein 4, CCL3 is a member of the CC chemokine family and is considered to promote the recruitment of monocytes and lymphocytes and participate in the inflammatory response.
Table 4. Top-ten 10 DE genes in human PBMCs after PHA stimulation.
| Gene symbol | Gene name | FC | Q-value |
|---|---|---|---|
| Up-regulation | |||
| CCNB2 | Cyclin B2 | 16.9 | 0 |
| TOP2A | DNA topoisomerase II alpha | 16.6 | 0 |
| TYMS | Thymidylate synthetase | 13.6 | 0 |
| CEP55 | Centrosomal protein 55 | 13.5 | 0 |
| KIAA0101 | PCNA Clamp Associated Factor | 12.8 | 0 |
| KIF14 | Kinesin family member 14 | 12.2 | 0 |
| CENPF | Centromere protein F | 11.6 | 0 |
| KIF23 | Kinesin family member 23 | 11.3 | 0 |
| HMMR | Hyaluronan-mediated motility receptor | 11.3 | 0 |
| STAG3 | Stromal antigen 3 | 10.9 | 0 |
| Down-regulation | |||
| CCL13 | C-C motif chemokine ligand 13 | 0.01 | 2.840 |
| CLC | Charcot-Leyden crystal galectin | 0.02 | 2.840 |
| FCER1A | Fc fragment of IgE receptor Ia | 0.02 | 2.840 |
| MS4A6A | Membrane spanning 4-domains A 6A | 0.03 | 2.840 |
| CPVL | Carboxypeptidase vitellogenic-like | 0.03 | 2.840 |
| FUCA1 | Alpha-L-fucosidase 1 | 0.03 | 2.840 |
| STAB1 | Stabilin 1 | 0.04 | 2.840 |
| ALDH1A1 | Aldehyde dehydrogenase 1 family member A1" | 0.05 | 2.840 |
| GPNMB | Glycoprotein nmb | 0.05 | 2.840 |
FC, fold change.
For monkeys, the regulator of G-protein signaling 8 (RGS8) gene was the most obvious DEG with a positive FC of 5.8 at 10 µg/mL PHA (see Table 5). The GO enrichment analysis showed that the RGS8 gene was mainly involved in termination of G-protein coupled receptor signaling pathway (GO:0038032). The RGS8 gene can inhibit signaling by increasing the guanosine triphosphate (GTP)ase activity of G protein α-subunit, which then transforms into an inactive guanosine diphosphate (GDP) —bound form. In addition, the top 20 genes, such as MCM4, MCM6, and MCM7 acting as MCM complexes, were found to be upregulated (FC =2.5–3.5). The MCM complex is the putative replicative helicase essential for once-per-cell-cycle DNA replication initiation and elongation in eukaryotic cells, so it participates in DNA replication and promotes cell proliferation. The downregulated gene in cynomolgus monkey PBMCs, CCL2 [chemokine (C-C motif) ligand 2, FC =0.2], is also a CC chemokine family member, and is involved in inflammatory response, immune response, chemotaxis, cytokine activity, and cytokine-mediated signaling pathway, as confirmed by GO enrichment analysis.
Table 5. Top-ten 10 DE genes in monkey PBMCs after PHA stimulation.
| Gene symbol | Gene name | FC | Q-value |
|---|---|---|---|
| Up-regulation | |||
| RGS8 | Regulator of G-protein signaling 8 | 5.8 | 0 |
| XCL1 | Chemokine (C motif) ligand 1 | 4.9 | 0 |
| TOP2A | DNA topoisomerase II alpha | 4.7 | 1.962 |
| CDC20 | Cell division cycle 20 | 4.2 | 0 |
| CENPW | Centromere protein W | 4.1 | 0 |
| MCM4 | Minichromosome maintenance complex component 4 | 3.5 | 0 |
| PTTG1 | PTTG1 regulator of sister chromatid separation, securin | 3.4 | 0 |
| CENPM | Centromere protein M | 3.3 | 0 |
| LOC701198 | Cytochrome c oxidase assembly factor 1 homology | 3.1 | 0 |
| LOC712754 | regulator of G protein signaling 1 | 3.1 | 0 |
| Down-regulation | |||
| MS4A4A | Membrane spanning 4-domains A4A | 0.1 | 0 |
| C1QA | Complement C1q A chain | 0.1 | 0 |
| CCL2 | C-C motif chemokine ligand 2 | 0.2 | 0 |
| HMOX1 | Heme oxygenase 1 | 0.2 | 0 |
| CTSL | Cathepsin L | 0.2 | 0 |
| SLC40A1 | Solute carrier family 40 member 1 | 0.3 | 0 |
| LOC715159 | Cathepsin L-1 pseudogene | 0.3 | 0 |
| SELENOP | Selenoprotein P | 0.3 | 0 |
| SLCO2B1 | Solute carrier organic anion transporter family member 2B1 | 0.3 | 0 |
FC, fold change.
In order to establish bridging biomarkers shared by human and monkey PBMCs, 21 upregulated genes and 17 downregulated genes were obtained by comparing the same DEGs in PBMCs of different species after PHA treatment (see Figure 5). By correlating the same genes in the overlapped biological functional groups of human and cynomolgus monkeys (see Table 6), it was found that the genes related to immune response were C1QC, IL8, and XCL1, and the genes related to inflammatory response and chemotaxis were CCL2 and IL8. In addition, the genes related to DNA replication and DNA-dependent DNA replication initiation were MCM6, MCM3, MCM7, MCM4, and PCNA, while those related to mitosis and cell division were polo-like kinase 1 (PLK1) and CDC20, respectively. The PLK1 variation was most obvious among these overlapping genes, with a positive FC of 6.9 in humans and 2.5 in monkeys. One of the members of the polo kinase family (PLKs), PLK1, is a cell cycle regulatory factor, which plays important roles in mitosis such as spindle assembly, centrosome maturation, chromosome separation, and cytokinesis. It can promote cell mitosis and accelerate cell proliferation in the mitotic M phase.
Table 6. Overlap DE genes in human and monkey PBMCs after PHA stimulation.
| GO term (P value) | Overlap DEGs (FC: h/m) | UniqH DEGs | UniqM DEGs |
|---|---|---|---|
| Immune response: (Ph =−2.22×10–16); (Pm =6.29×10–7) |
C1QC (0.1/0.4); IL8 (0.4/0.5); XCL1 (2.9/4.9) |
1Mar; C3; C5AR1; CBLB; CCL13; CCL8; CCR1; CCR2; CD274; CD28; CD7; CD74; CD8A; CD8B; CFP; CIITA; CMKLR1; CTLA4; CTSC; CTSS; CXCL1; CXCL10; CXCL2; CXCL3; ENPP2; FCGR2B; FCGR2C; FCGR3A; FCGRT; FTH1; HLA-DMA; HLA-DMB; HLA-DPA1; HLA-DPB1; HLA-DQA1; HLA-DQA2; HLA-DQB1; HLA-DRA; HLA-DRB1; HLA-DRB3; HLA-DRB4; HLA-DRB5; ICOS; IGF1R; IGJ; IGLL1; IGSF6; IL10; IL1A; IL1RAP; IL2RA; IL6; IL7R; LILRB2; LST1; LTB; NCF4; PDCD1LG2; PPBP; PTAFR; RGS1; SAMHD1; SECTM1; TGFBR3; TLR1; TLR2; TLR4; TNFRSF4 | CCL11; CCL18; CCL2; CD40LG; LOC714964; SERPINB9 |
| Inflammatory response: (Ph =5.35×10–14); (Pm =0.00095) |
CCL2 (0.2/0.2); IL8 (0.4/0.5) |
AIF1; AOAH; APOL3; C3; C3AR1; CCL13; CCL8; CCR1; CCR2; CCR3; CD14; CD163; CLEC7A; CXCL1; CXCL10; CXCL2; CXCL3; CYBA; CYBB; GGT5; IL10; IL1A; IL1RAP; IL6; LY86; LY96; MGLL; MMP25; MS4A2; NFAM1; NLRP3; OLR1; PTAFR; RXRA; S100A12; S100A8; S100A9; SERPINA1; SIGLEC1; STAB1; TLR1; TLR8; TNFAIP6 | CCL11; CD40LG |
| Chemotaxis: (Ph =1.64×10–8); (Pm =0.00070) |
CCL2 (0.2/0.2); IL8 (0.4/0.5) |
C3AR1; C5AR1; CCL13; CCL8; CCR1; CCR2; CCR3; CMKLR1; CX3CR1; CXCL1; CXCL10; CXCL2; CXCL3; CXCR3; ENPP2; FPR1; FPR3; PLAU; PPBP; PTAFR; S100A8; S100A9 | CCL11 |
| DNA replication: (Ph =3.79×10–14); (Pm =3.02×10–5) |
MCM6 (4.6/2.9); MCM3 (2.8/3.0); MCM7 (3.1/2.5); MCM4 (4.0/3.5); PCNA (4.4/2.8) |
BLM; BRCA1; CDK2; CENPF; CHAF1A; DBF4; DUT; GINS2; GINS3; GINS4; LIG1; MCM2; MCM5; MCM8; NASP; POLA1; POLA2; POLD1; PRIM1; PRIM2; RBBP7; RFC2; RFC3; RFC4; RFC5; RMI1; RPA3; RRM1; RRM2; TK1; TOP2A | |
| DNA–dependent DNA replication initiation: (Ph =1.25×10–8); (Pm =3.30×10–8) |
MCM6 (4.6/2.9); MCM3 (2.8/3.0); MCM7 (3.1/2.5); MCM4 (4.0/3.5) |
CCNE1; MCM2; MCM5; POLA1; POLA2; PRIM1 | |
| Cell division: (Ph =4.07×10–14); (Pm =0.13773) |
CDC20 (3.4/4.2) | 10Sep; ANXA11; AURKA; BIRC5; C13orf34; CCDC99; CCNA2; CCNB1; CCNB2; CCNE1; CCNE2; CDCA8; CDK1; CDK2; CDK6; CENPF; CENPH; CEP55; CKS1B; CKS2; DSN1; FBXO5; HAUS1; HAUS4; HAUS8; KIF11; KIF20B; KNTC1; LATS2; LIG1; MAD2L1; MASTL; MIS18BP1; NCAPD2; NCAPD3; NCAPG2; NUP37; NUSAP1; OIP5; PTTG1; SKA2; SMC4; TIMELESS; TIPIN; TPX2; TUBA1B; TUBB; UBE2C; UBE2S; ZWILCH | |
| Mitosis: (Ph =1.13×10–7); (Pm =0.10771) |
PLK1 (6.9/2.5) | AURKA; BIRC5; CCNA2; CCNB2; CDK1; CDK2; CEP55; HAUS1; HAUS4; HAUS8; KIF11; KIF22; KIF23; LATS2; MAD2L1; MASTL; MIS18BP1; NCAPD2; NCAPD3; NCAPG2; OIP5; PKMYT1; PTTG1; SKA2; TIMELESS; TIPIN; TPX2; TUBB; UBE2C |
FC, fold change; Ph, P value of human; Pm, P value of monkey; h/m, human/monkey.
Comparing biological functions of DEGs by GO enrichment analysis after LPS stimulus
Under LPS stimulation, biological function groups involved by most DEGs in human PBMCs referred to antigen processing and presentation of peptide or polysaccharide antigen via major histocompatibility complex (MHC) class II, IFN-γ mediated signaling pathway, ‘immune response (P=0, 1.11×10-16, 3.33×10-16, respectively), and other BPs related to innate immune response, T cell costimulation, and others (see Table 7). In contrast to PHA, the results showed that the number of genes involved in the top 10 functional groups increased slightly with the increase of LPS concentration.
Table 7. Top-ten biological functions identified by significant enrichment GO analysis in monkey and human PBMCs after LPS stimulation.
| GO term (biological process) | P value# | DEGs* (2 μg/mL LPS) | DEGs (10 μg/mL LPS) |
|---|---|---|---|
| Human | |||
| Antigen processing and presentation of peptide or polysaccharide antigen via MHC class IIф | 0 | 11 | 13 |
| Interferon-gamma-mediated signaling pathway | 1.11×10–16 | 14 | 23 |
| Immune responseф | 3.33×10–16 | 32 | 53 |
| Cytokine-mediated signaling pathway | 1.11×10–16 | 19 | 33 |
| Chemotaxis | 1.11×10–11 | 13 | 21 |
| T cell co-stimulation | 5.18×10–11 | 11 | 16 |
| Inflammatory response | 1.58×10–10 | 17 | 2 |
| T cell receptor signaling pathway | 2.89×10–9 | 11 | 15 |
| Innate immune response | 9.11×10–8 | 13 | 23 |
| Platelet degranulation | 3.13×10–7 | 6 | 13 |
| △Phagocytosis, recognition | △2.60×10–6 | 3 | 3 |
| △Positive regulation vascular endothelial growth factor production | △1.08×10–5 | 4 | 5 |
| Monkey | |||
| Antigen processing and presentation of peptide antigen via MHC Class I | −2.22×10–16 | 13 | 13 |
| Antigen processing and presentation | 0 | 22 | 23 |
| Immune responseф | 8.88×10–16 | 30 | 33 |
| Antigen processing and presentation of peptide or polysaccharide antigen via MHC class IIф | 2.83×10–9 | 6 | 6 |
| Chaperone-mediated protein folding requiring cofactor | 0.00015 | 3 | 3 |
| Sarcomere organization | 0.00028 | 3 | 3 |
| Positive chemotaxis | 0.00061 | 2 | 3 |
| Cellular iron ion homeostasis | 0.00092 | 0 | 3 |
| Aerobic respiration | 0.00148 | 1 | 2 |
| Antigen processing and presentation of exogenous peptide antigen via MHC Class II | 0.00148 | 2 | 2 |
| △Electron transport chain | △0.00017 | 4 | 3 |
| △Translation | △0.00125 | 9 | 8 |
| △ATP synthesis coupled electron transport | △0.00150 | 2 | 2 |
*, DEGs: number of differently expression genes identified by microarray in 2 or 10 μg/mL LPS groups; #, P value for enrichment; △, that means these top-ten GO terms are identified in 2 μg/mL LPS group; ф, the same GO terms used in both human and monkey groups.
For monkeys under LPS stimulation, there was a significant enrichment of DEGs associated with biological functions related to immune response processes such as antigen processing and presentation of peptide antigen via MHC class I, antigen processing and presentation, and immune response (P=−2.22×10-16, 0, 8.88×10-16, respectively). By comparing the top 10 GO biological functional groups of humans and cynomolgus monkeys after 10 µg/mL LPS treatment, we found that antigen processing and presentation of peptide or polysaccharide antigen via the MHC II and immune response were the same functional groups in the two species. However, it was interesting that although antigen processing and presentation via MHC II are common to both human and cynomolgus monkeys, antigen processing and presentation via MHC I (P value =−2.22×10-16) only occur in cynomolgus monkeys.
In the 10 µg/mL LPS treatment group, the most obvious upregulated gene in human PBMCs was metallothionein 1H (MT1H), with a positive FC of 28.6 (see Table 8). The MT1H gene is one of the metallothionein function genes, and is mainly involved in the metabolism of trace metals, detoxification of heavy metals, elimination of free radicals, stress response, DNA replication and transcription, affecting protein and energy metabolism, and promoting cell proliferation. Among the top 10 upregulated genes, the expressions of IL-1β and IL-8 genes were significantly upregulated (FC value =15.6 and 6.4, respectively). Also known as lymphocyte stimulating factor, IL-1 is mainly produced by activated monocyte macrophages, which can synergistically stimulate the activation of antigen-presenting cells (APC) and T cells, promote the proliferation of B cells, secrete antibodies, and contribute to immune regulation. Many different cell types can produce IL-8, including macrophages, neutrophils, and epithelial cells, and its main biological activity is to induce and activate neutrophil effector functions, including the release of reactive oxygen species (ROS), proteinases, and protective antimicrobial peptides. In addition, members of the CXC chemokine family, CXCL1, CXCL5, and CXCL2, were also upregulated. In the top 10 downregulated genes, the change of CXCL1 was also obvious.
Table 8. Top-ten 10 DE genes in human PBMCs after LPS stimulation.
| Gene symbol | Gene name | FC | Q-value |
|---|---|---|---|
| Up-regulation | |||
| MT1H | Metallothionein 1H | 28.6 | 0 |
| CXCL1 | C-X-C motif chemokine ligand 1 | 24.2 | 0 |
| MT1G | Metallothionein 1G | 19.3 | 0 |
| CXCL1 | C-X-C motif chemokine ligand 1 | 16.8 | 0 |
| IL1B | Interleukin 1 beta | 15.6 | 0 |
| IL8 | Interleukin 8 | 6.4 | 0 |
| CXCL5 | C-X-C motif chemokine ligand 5 | 14.0 | 0 |
| CXCL2 | C-X-C motif chemokine ligand 2 | 8.1 | 0 |
| RETN | Resistin | 7.8 | 11.047 |
| MT1M | Metallothionein 1M | 7.6 | 0 |
| Down-regulation | |||
| LTF | Lactotransferrin | 0.04 | 100 |
| ALDH1A1 | Aldehyde dehydrogenase 1 family member A1 | 0.05 | 100 |
| MS4A6A | Membrane spanning 4-domains A6A | 0.05 | 100 |
| C1QA | Complement C1q A chain | 0.05 | 100 |
| IFI27 | Interferon alpha inducible protein 27 | 0.07 | 100 |
| CXCL9 | C-X-C motif chemokine ligand 9 | 0.09 | 100 |
| APOBEC3B | Apolipoprotein B mRNA editing enzyme catalytic subunit 3B | 0.09 | 100 |
| APOC1 | Apolipoprotein C1 | 0.1 | 100 |
| C1QC | Complement C1q C chain | 0.1 | 100 |
FC, fold change.
For monkeys after 10 µg/mL LPS stimulation, the most obvious upregulated DEGs were MHC family members (see Table 9), such as MAMU-A (MHC class I, A), MAMU-B18 (MHC class I, B), MAMU-I (MHC class I, I), MAMU-E (MHC class I, E), and MAMU-F (MHC class I, F), with a FC value of 11.3–8.4. Indeed, this also confirmed that not only were more DEGs involved in antigen processing and presentation via MCH I in cynomolgus monkey PBMCs, but the increase of these DEGs expression was very marked.
Table 9. Top-ten 10 DE genes in monkey PBMCs after LPS stimulation.
| Gene symbol | Gene name | FC | Q-value |
|---|---|---|---|
| Up-regulation | |||
| MAMU-A | Major histocompatibility complex, class I, A | 11.3 | 0 |
| MAMU-B18 | Major histocompatibility complex, class I, B | 11.3 | 0 |
| MAMU-A | Major histocompatibility complex, class I, A | 11.3 | 0 |
| MAMU-B18 | Major histocompatibility complex, class I, B | 10.7 | 0 |
| MAMU-I | Major histocompatibility complex, class I, I | 10.2 | 0 |
| MAMU-E | Major histocompatibility complex, class I, E | 10.2 | 0 |
| LOC714964 | HLA class I histocompatibility antigen, B-67 alpha chain-like | 9.6 | 0 |
| MAMU-E | Major histocompatibility complex, class I, E | 9.2 | 0 |
| MAMU-F | Major histocompatibility complex, class I, F | 8.4 | 0 |
| ACTB | actin beta | 7.7 | 19.539 |
There were 562 DEG in 10 μg/mL LPS groups after LPS stimulation, but only 30 of them were down regulate with Q-value (%) of 103.376 and FC of 0.269 to 0.499. Because of the lack of significance, they were not listed in the table. FC, fold change.
Similarly, we gathered 10 upregulated genes and 10 downregulated genes common to both humans and monkeys after LPS stimulation (see Figure 5). By correlating the same genes in the overlapped GO functional groups of humans and cynomolgus monkeys, we found that the same DEGs involved in immune response were C1QC, CCL3, CD74, CXCL3, IL-1A, and IL-8; those involved in cytokine-mediated signaling pathway were CD74 and IL-1A; those involved in chemotaxis were CCL3 and IL-8; and those involved in inflammatory response were CCL3, IL-1A, and IL-8 (see Table 10). Among these genes, IL-8 gene expression was most the upregulated, with a FC of 6.4 in humans and 2.0 in monkeys.
Table 10. Overlap DE genes in human and monkey PBMCs after LPS stimulation.
| GO term (P value) | Overlap DEGs (FC: h/m) | UniqH DEGs | UniqM DEGs |
|---|---|---|---|
| Immune response: (Ph =3.33×10–16); (Pm =8.88×10–7) | C1QC (0.1/0.4); CCL3 (2.5/2.2); CD74 (0.4/5.2); CXCL3 (7.0/2.7); IL1A (5.4/2.5); IL8 (6.4/2.0) | AQP9, C3, CCL13, CCL3L3, CCR2, CD276, CIITA, CMKLR1, CTSC, CTSS, CTSW, CXCL1, CXCL10, CXCL2, CXCL5, CXCL9, ENPP2, FCAR, FCGRT, HLA-DMA, HLA-DMB, HLA-DOA, HLA-DPA1, HLA-DPB1, HLA-DQA1, HLA-DQA2, HLA-DQB1, HLA-DRA, HLA-DRB1, HLA-DRB3, HLA-DRB4, HLA-DRB5, IFI6, IGSF6, IL18, IL1B, IL6, LST1, NCF4, OAS1, OAS3, P2RY14, PDCD1LG2, PGLYRP1, PPBP, SECTM1, TLR4 | B2M, CCL18, CCL4L1, CCL5, LOC100424348, LOC100427798, LOC694372, LOC714964, LOC720309, LOC720330, LOC720365, LOC722711, MAMU-A, MAMU-AG, MAMU-B, MAMU-B18, MAMU-DPA, MAMU-DPB, MAMU-DQA1, MAMU-DQB1, MAMU-DRA, MAMU-E, MAMU-F, MAMU-I, MAMU_DMA |
| Cytokine–mediated signaling pathway: (Ph =1.11×10–11); (Pm =0.003845) | CD74 (0.4/5.2); IL1A (5.4/2.5) | CCL2, CCR2, CEBPA, CIITA, HLA-DMA, HLA-DMB, HLA-DOA, HLA-DPA1, HLA-DPB1, HLA-DQA1, HLA-DQA2, HLA-DQB1, HLA-DRA, HLA-DRB1, HLA-DRB3, HLA-DRB4, HLA-DRB5, IFI27, IFI30, IFI6, IFNG, IL1B, IL6, IRF5, JAK2, MT2A, MX1, OAS1, OAS3, SOCS3 | IL2RG, JAK1 |
| Chemotaxis: (Ph =1.11×10–11); (Pm =0.00647) | CCL3 (2.5/2.2); IL8 (6.4/2.0) | C3AR1; CCL13; CCL2; CCL3L3; CCR2; CMKLR1; CXCL1; CXCL10; CXCL16; CXCL2; CXCL3; CXCL5; CXCL9; ENPP2; FPR2; FPR3; PLAU; PLAUR | LOC721048 |
| Inflammatory response: (Ph =1.58×10–10); (Pm =0.002622) | CCL3 (2.5/2.2); IL1A (5.4/2.5); IL8 (6.4/2.0) | C3, C3AR1, CCL13, CCL2, CCL3L3, CCR2, CD163, CLEC7A, CXCL1, CXCL10, CXCL2, CXCL3, CXCL9, FPR2, GGT5, IL1B, IL6, IRAK2, MMP25, NFKBIZ, SIGLEC1, SPP1, STAB1, TNFAIP6 | CCL5 |
FC, fold change; Ph, P value of human; Pm, P value of monkey; h/m, human/monkey.
Comparing KEGG pathways of DEGs by enrichment analysis after PHA/LPS stimulus
We analyzed the pathway function enrichment of the selected differential genes, and listed the top 10 pathway names according to P value (see Tables S1,S2).
In the PHA treatment groups, the two most important pathways in both human and cynomolgus monkey PBMCs were DNA replication [P value =1.11×10-16 (human), 7.57×10-7 (monkey)] and cell cycle [P value =1.44×10-15 (human), 1.39×10-7 (monkey)]. In the DNA replication pathway, there were 18 DEGs in human PBMCs, among which MCM2-7, PCNA, RFC2-5, CDK2, and RPA were significantly upregulated; meanwhile, there were 6 DEGs in cynomolgus monkey PBMCs, with MCM2-7, PCNA, and RPA being similarly upregulated. The overlapped cell cycle pathway contained 27 DEGs in human PBMCs, but only 10 DEGs were present in cynomolgus monkey PBMCs after PHA stimulation. The results showed that the numbers of upregulated human genes were higher than those of cynomolgus monkeys despite these species having the same pathways.
In the LPS treatment groups, only graft-versus-host disease [P value: 0.00215 (human), 4.44×10-16 (monkey)] was the same signaling pathway in the two species. Interestingly, there were 16 genes involved in cynomolgus monkey PBMCs, and only 4 involved in human PBMCs. The obvious upregulated genes in cynomolgus monkey PBMCs were mainly MAMU family members, which participate in antigen processing and presentation, and this was consistent with the results of GO analysis. In contrast, IFNγ, IL-1α, IL-1β, and IL-6 were the upregulated genes in the human PBMC pathway. These genes were also involved in the cytokine-cytokine receptor interaction signaling pathway and other parasitic infectious diseases such as trypanosomiasis, amoebiasis, malaria, and leishmaniasis.
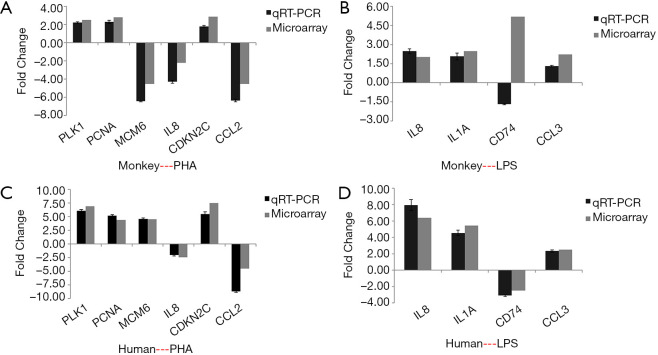
Validation of DEGs by qRT-PCR
A set of 10 genes was selected for microarray data validation by qRT-PCR. The genes were chosen according to their similar biological functions in both human and monkey PBMCs, and included CCL2, IL8, PCNA, MCM6, PLK1, and CDKN2C after PHA stimulation, and CD74, IL1A, IL8, and CCL3 after LPS stimulation (see Figure 6). The direction of change in the two methods showed a good correlation in spite of there being small discrepancies in the magnitude of change.
Figure 6.
Effects of PHA and LPS on select gene expression in PBMCs. Impact of 10 µg/mL of PHA on the expression of PLK1, PCNA, MCM6, IL8, CDKN2C, and CCL2, and that of 10 µg/mL of LPS on IL-8, IL-1A, CD74, and CCL3, were validated by qRT-PCR. Fold-changes in expression levels between the PHA group and control are indicated in A (monkey) and B (human). Fold-changes in the expression levels between the LPS and control groups are indicated in C (monkey) and D (human). The qRT-PCR data are presented as means ± SD of three independent samples. Array data were calculated using SAM software. PHA, phytohemagglutinin; LPS, lipopolysaccharide; qRT-PCR, quantitative reverse transcription polymerase chain reaction; SD, standard deviation; SAM, significance analysis of microarray.
Discussion
Comparative studies of the physiological or pathological status of the immune system in monkeys and humans can give us invaluable information for performing immune-related biomedical studies using NHPs. In vitro approaches can evaluate immunotoxicity using human PBMSs to accommodate its complexity, with several cell components present in a relatively intact environment (16,17). Also, such information from humans can be likewise compared with animals (8). In general, primary immunotoxicologic functional observations can be monitored in vitro by a proliferative response and cytokine releases of lymphocytes provoked with different stimulations (18). Therefore, in the current study, the different stimulations expected to induce either T cell-specific proliferation (PHA) or classic innate immune response (LPS) were used for the first time to evaluate the similarities and differences in immune responses of PBMCs of humans and cynomolgus monkeys, including the aspects of lymphocyte proliferation, cytokine secretions, and gene expression profile, which provided primate-specific data for comparative immunology, and also provided valuable information for extrapolating animal data to human in biomedical research using cynomolgus monkeys.
PBMC proliferation induced by PHA/LPS
Here, the proliferation ratio of PHA-stimulated PBMCs of human and cynomolgus monkeys was significantly increased with an increase in concentration. In particular, human PBMCs showed a higher proliferative capacity, for which the proliferation ratio was ~2.5 fold higher compared with cynomolgus monkey PBMCs to 10 µg/mL PHA. This result was also consistent with the gene expression profile, in which the number of DEGs in human PBMCs after PHA stimulation was found to be almost nine times higher than that of cynomolgus monkeys. In contrast to PHA, no significant difference in the proliferation of PBMCs was detected between human and cynomolgus monkey PBMCs stimulated with LPS. These results showed a distinct difference of immune responses to the various activators in primates, suggesting humans were more susceptible to T cell-specifical mitogen PHA than cynomolgus monkeys.
Cytokine profile induced by PHA/LPS
In the cytokine profiles study, the levels of most cytokines secreted by PHA-stimulated human PBMCs were more pronounced than those of cynomolgus monkeys; in particular, the secretion level of INFγ and IL-10 were almost 30 and 50 times that of the cynomolgus monkeys. With consideration to previous proliferation assay, the increased secretion of INFγ might have been due to the increased number of activated T lymphocytes by PHA in humans. There was additional evidence in our gene expression profile study showing that an enrichment of DEGs in PHA-stimulated human PBMCs was principally related to IFNγ-mediated signaling pathway (GO:0060333, P value =2.00×10-15). Of note, the significant functional group of genes under both PHA and LPS stimulation were also mostly associated with IFNγ-mediated signaling pathway (GO:0060333, P value: 1.11×10-11) in human PBMCs, which is consistent with the significant secretion of IFN-γ in human PBMCs by LPS stimulation. Therefore, the pronounced secretion and upregulated genes associated with IFN-γ in human PBMCs may reflect a functional difference between the two species after PHA or LPS stimulation.
IFN-γ, one of the most important cytokines in innate and adaptive immune responses, is mainly secreted by T cells and natural killer (NK) cells. It plays an important role in eliminating pathogens, increasing macrophage function, promoting the expression of MHC molecules and antigen presenting molecules, and preventing allergic inflammation (19). Although previous studies have confirmed that PHA can induce human PBMCs to secrete a large amount of IFN-γ (20,21), we found for the first time that the secretion level in humans was much higher than that in cynomolgus monkeys. Unequal cytokine formation by dendritic cell (DC)/other cell types was also shown by Gujer et al. (22), in which human plasmacytoid (p)DC displayed rapid IFN-γ secretion after TLR7/8-L activation, and that secretion was more pronounced in humans than in rhesus cells after CpG C stimulation. Similar research results also exist in comparative studies of some zoonotic diseases [e.g., human immunodeficiency virus (HIV) or yellow fever (YF)-17D]; for example, YF-17D stimulation of sooty mangabey (SM) PBMCs resulted in a significantly reduced IFNα production compared with the PBMCs of rhesus macaques and humans, which could possibly be explained by the diminished NKs and effector CD8+T cells in the SMs (23). The extent of early CD8+T cell proliferation has been found to vary between different animals with various stimulations (24,25). Another explanation may be the genetic variation across primates. Obvious sequence differences in the promoter or the proximal region of cytokine genes (e.g., IL-4, IL-10, IL-12β, TNF-α, and IFN-γ) among human, mangabey, and macaque monkeys, which affect regulation of cytokine synthesis have been reported (26). Although the IFN data here are just general examples, these could potentially reflect strikingly different mechanisms in the innate and adaptive immune responses across the primates.
The current study also examined the cytokine changes by LPS stimulation in human and cynomolgus monkeys. The results showed that the significantly increased secretion of IL-2, IL-6, IL-8, and IL-10 was observed in both species. The levels of IL-6 and IL-8 in the supernatant of PBMCs with LPS were much higher than those other cytokines in the two species. Indeed, IL-8 is a chemokine known to act as one of the major mediators of inflammatory response and has been reported to be upregulated after LPS stimulation in many mammals including cattle, mice, and pigs (27-30). Recent studies have shown that bacterial and host interactions can affect the histone acetylation, phosphorylation, and methylation status of the TLR4 and IL-8 promoters in host cells (31). The similar changes in IL-8 in different species after LPS stimulation suggested that IL-8 could be a bridge biomarker to reflect the activation state of innate immune response in mammals.
Most notably, in the cytokine profile study, IL-5 was not detected (value below standard curve limits) in monkey cultures that received either PHA or LPS. It has been reported that Th2-type cytokines like IL-5 (also IL-4 and IL-13) were associated with promotion of immunoglobulin E (IgE) and eosinophilic responses (32). Recently, it was found that IL-5 was produced by memory CD4+ TH2 cell subsets that play an important role in allergic inflammation in humans and mice (33). Although we have no direct evidence to account for these differences in IL-5 secretion between humans and cynomolgus monkeys, it has been previously reported that a marked difference in memory CD4+ TH2 cells was found between the two species (24,34). Previous studies have shown that human memory T cells can be divided into central T cells (CD28+CCR7+) and effector T cells (CD28+CCR7-), while cynomolgus monkey memory T cells can be divided into central T cells (CD28+CCR7+), transitional T cells (CD28+CCR7+), and effector T cells (CD28-CCR7-) (35,36). Thus, one cannot rule out that the lack of cell surface marker CD28 in effector T cells of monkeys reflects a functional difference between humans and monkeys. Eastwood et al. found that the failure of the monoclonal antibody TGN1412 trial was due to species differences in CD28 expression on CD4+ effector memory T cells (34). In addition, age-related phenotypic and functional changes were reported in cynomolgus monkey T cells; for example, only adult monkey T cells could be activated and induced to released high concentrations of IFN-γ after stimulation with anti-CD28 and/or anti-CD3 antibodies, while young monkey T cells showed very low responses to these stimulations (37). Therefore, the differences in memory T cells in humans and cynomolgus monkeys, especially in juvenile monkeys, might have contributed to the differential secretion of IL-5 in PBMCs stimulated with PHA or LPS in this study, and further experiments will be required to specifically address this question.
Biologic functional features of DEGs in human and monkey PBMCs with PHA stimulation
Comparative study of gene expression profiles can be used to better understand the mechanisms of similar and different immune responses to PHA or LPS between the two species. Therefore, the GO enrichment analysis of DEGs was performed to identify significant functional groups of genes in PHA/LPS-stimulated PBMCs. According to the results from GO analysis, the DEGs in human PBMCs with PHA treatment were significantly enriched in BPs, including immune response, mitotic cell cycle, cytokine-mediated signaling pathway, DNA replication, and inflammatory response. Although the DEGs in cynomolgus monkey PBMCs were also mainly involved in the same biological functions, including immune response and DNA replication, the number of DEGs in the same BP was far fewer than that of humans. The most upregulated gene in human PBMCs after 10 µg/mL PHA stimulation was cyclin B2 (FC =16.7), an important regulator of the G2/M phase of the cell cycle that initiates the G2/M phase transition by activation of CDK1 kinase in the cell cycle. Moreover, several of top 10 DEGs, including TOP2A, TYMS, CEP55, CENPE, and KIF23, also participated in the BPs of mitosis, mitotic cell cycle, and the G1/S phase of mitotic cell cycle, with upregulated expression >10-fold in PHA-stimulated human PBMCs compared to control PBMCs. A large number of upregulated genes related to mitosis seemed to suggest that a stronger response to PHA in humans may be implicated in rapidly increased mitosis, leading to proliferation of PBMCs, which may be one of the important molecular mechanisms responsible for the difference in immune responses to PHA between the two species in question.
By comparing the top 10 gene functional groups between humans and cynomolgus monkeys, we found similar characteristics in the gene expression profiles of the two species after PHA treatment, including in immune response, DNA replication, and chemotaxis. Also, the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis results revealed that the DEGs were involved in DNA replication and the cell cycle in both species. For identification of conserved signatures of immune responses to PHA in human and cynomolgus monkey PBMCs, we found an overlap of 21 upregulated and 17 downregulated genes between human and monkey DEG lists, notably the MCM complexes (MCM3, MCM4, MCM6, and MCM7) and proliferating cell nuclear antigen (PCNA) involved in DNA replication pathway. The MCM complex, listed in the top 20 upregulated genes in cynomolgus monkeys, is a hexameric protein complex required for the initiation and regulation of DNA replication. PCNA is an auxiliary protein of DNA polymerase delta and is involved in DNA replication/repair. Previous studies have shown that PCNA was necessary for entering S phase, and appeared in active lymphocytes during late G1, increased until the beginning of S phase, and reached its peak 72 hours later (38,39). In our study, the PLK1 gene, a highly conserved serine/threonine kinase widely found in eukaryotes, was the most significant upregulated gene shared by humans and cynomolgus monkeys under PHA stimulation. Indeed, PLK1 performs several important functions throughout the M phase of the cell cycle, such as the regulation of centrosome maturation and spindle assembly, the removal of cohesin from chromosome arms, the regulation of mitotic exit, and cytokinesis (40). Thus, our results are in agreement with these studies, suggesting that these overlapped DEGs associated with DNA replication and cell cycle reflect the highly conserved signatures of immune responses to PHA stimulation across the primates.
Biologic functional features of DEGs in human and monkey PBMCs with LPS stimulation
Our results have confirmed a wide range of genes which were affected by LPS stimulation in human and cynomolgus PBMCs. For human PBMCs, the DEGs with LPS treatment were significantly enriched in antigen processing and presentation of peptide or polysaccharide antigen via MHC class II, interferon-gamma-mediated signaling pathway, immune response, cytokine-mediated signaling pathway, chemotaxis, T cell costimulation, and inflammatory response. Also, the DEGs in cynomolgus monkey PBMCs were mainly involved in the same biological functions, including antigen processing and presentation of peptide antigen via MHC Class II, antigen processing and presentation, and immune response. In contrast to PHA, the count of DEGs in the same BP was similar in the two species under LPS stimulation, suggesting humans and cynomolgus monkeys have similar characteristics in immune pathways and response intensity in the innate immune response to LPS.
In human PBMCs, the genes MT1H, CXCL1, MT1G, IL1B, and IL-8 were the top five genes found over-expressed with 10 µg/mL LPS treatment. IL-1β is a key cytokine that modulates the expression of other secondary cytokines including IL-8 and TNFAIP6 (41). IL-8 is a chemokine known to act as one of the major mediators of inflammatory response for mobilization and activation of neutrophils (42). We also observed increased IL-8 cytokine expression after LPS stimulation in both human and monkey PBMCs by Luminex-based analysis. In addition, upregulation of a wider range of inflammatory cytokines, including IL-6, IL-1A, IL-1B, and TNF, was also observed in other species after LPS stimulation (43). In the 10 µg/mL LPS groups, we found an overlap of the 10 upregulated and top 10 downregulated genes across the human and monkey DEG lists; notably, IL-8 was the most upregulated gene in humans and cynomolgus monkeys. These data indicated that IL-8 was highly conserved among the primates and might play an important role in the innate immune response to LPS.
In monkey PBMCs, the genes MAMU-A, MAMU-B18, MAMU-I, MAMU-E, and LOC714964 were the top five genes found to be strongly upregulated with 10 µg/mL LPS treatment. It could be seen that, unlike the overexpression of cytokines and chemokines genes in the top genes lists in humans, the most upregulated genes in monkeys belonged to the MHC I antigen. These results, confirmed by the GO analysis, suggest that the upregulated DEGs in monkeys were primarily associated with antigen processing and presentation of peptide antigen via MHC class I (P value =−2.22×10-16). Also, we found that the DEGs were enriched in graft-versus-host disease during the KEGG enrichment analysis in both humans and monkeys by LPS stimulation. Although they shared the same pathway, the 16 genes involved in this pathway are MAMU family members in monkeys, while the 4 genes in humans were IFNG, IL-1A, IL1-B, and IL-6, which suggests that antigen processing and presentation is the most characteristic response to LPS in cynomolgus monkeys.
Interestingly, the antigen processing and presentation was found mainly via MHC class II rather than MHC class I in humans after LPS stimulation. Molecules of MHC class I are found on almost every nucleated cell of the body and generally interact with CD8+ T cells. Benefiting from the development of molecular genetics, the study of Mafa MHC polymorphism demonstrated that the MHC of cynomolgus macaques (Mafa MHC) was organized in the same way as that of humans, but it differed from the human type by its high degree of classical class I gene duplication (44). In other words, the MHC-I locus in macaques was astoundingly complex, with each individual haplotype expressing multiple A alleles and potentially >10 B alleles, while a single A, B, and C allele were expressed on a haplotype in human MHC-I locus. Several published studies are in agreement with regards to the important role that the MHC class IB region plays in the mechanism of controlling simian immunodeficiency virus (SIV) infection in a Mauritian macaque model (45). Additionally, the magnitude and specificity of vaccine-elicited T-cell responses are variable in currently utilized nonhuman primate populations, owing to the heterogeneity in MHC class I genetics (46). The genetic difference in the two species might by accounted for by the distinct findings in the gene expression profile after LPS stimulation. As the MHC class I and II regions influence the ability to mount an immune response against infectious pathogens and vaccines, the genetic and functional differences between humans and monkeys should be considered in experimental medicine in macaques.
Conclusions
In the present study, we have characterized the cytokine secretion and gene expression profiles of human and cynomolgus monkey PBMCs upon in vitro stimulation by either PHA or LPS, and thus have identified the similarities and differences of immune responses between these species. These differences suggest the species-specific nature implicated in the T cell–specific mitogen or innate immune responses, including lymphocyte proliferation, cytokine secretion (e.g., IFN-γ and IL-5), antigen processing and presentation via MHC I, and so on, which should be carefully considered in biomedical research using NHPs. Additionally, the evaluation of genes conserved between the species under the same stimulation conditions could provide a new way to discern biomarkers and to transfer animal data to humans. Accordingly, future studies exploring the effects of other stimulations on human and monkey immune cells are needed.
Supplementary
The article’s supplementary files as
Acknowledgments
Funding: This work was supported by Project of the 13th National Five-Year Plan for Significant New Drug Creation ‘Key Technologies for Nonclinical Safety Evaluation of Innovative Drugs’ (Project No. 2018ZX09201-017).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures of animal experimental studies were performed under a project license (No. IACUC-2013-041) granted by the ethics board of National Center for Safety Evaluation of Drugs (NCSED), in compliance with the Guide for the Care and Use of Laboratory Animals of the NCSED [China].
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at http://dx.doi.org/10.21037/atm-20-4548
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-20-4548
Peer Review File: Available at http://dx.doi.org/10.21037/atm-20-4548
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-4548). The authors have no conflicts of interest to declare.
(English Language Editors: J. Jones and J. Gray)
References
- 1.Buse E. Development of the immune system in the cynomolgus monkey: Appropriate model in human targeted toxicology? J Immunotoxicol 2005;2:211-6. 10.1080/15476910500362937 [DOI] [PubMed] [Google Scholar]
- 2.Suntharalingam G, Perry M, Ward S, et al. Cytokine storm in a Phase 1 trial of the anti-CD28 monoclonal antibody TGN1412. N Engl J Med 2006;355:1018-28. 10.1056/NEJMoa063842 [DOI] [PubMed] [Google Scholar]
- 3.Wadman M. London's disastrous drug trial has serious side effects for research. Nature 2006;440:388-9. 10.1038/440388a [DOI] [PubMed] [Google Scholar]
- 4.Wanda M. Haschek, Colin G, Rousseaux T. Handbook of Toxicologic Pathology, Second Edition. New York: Academic Press. 2002; pp.585-646. [Google Scholar]
- 5.Elmore SA. Enhanced histopathology of the immune system: A review and update. Toxicol Pathol 2012;40:148-56. 10.1177/0192623311427571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moriyama A, Fujishima J, Furukawa T, et al. Quantitative analyses of lymphoid tissue in spleen, lymph nodes, and Peyer's patches in cynomolgus monkeys. J Vet Med Sci 2011;73:1459-64. 10.1292/jvms.11-0207 [DOI] [PubMed] [Google Scholar]
- 7.Jeong AR, Nakamura S, Mitsunaga F. Gene expression profile of Th1 and Th2 cytokines and their receptors in human and nonhuman primates. J Med Primatol 2008;37:290-6. 10.1111/j.1600-0684.2008.00289.x [DOI] [PubMed] [Google Scholar]
- 8.Galbiati V, Mitjans M, Corsini E. Present and future of in vitro immunotoxicology in drug development. J Immunotoxicol 2010;7:255-67. 10.3109/1547691X.2010.509848 [DOI] [PubMed] [Google Scholar]
- 9.Grote-Wessels S, Frings W, Smith C, et al. Immunotoxicity testing in non-human primates. Methods Mol Biol 2010;598:341-59. 10.1007/978-1-60761-401-2_23 [DOI] [PubMed] [Google Scholar]
- 10.Luebke RW, Holsapple MP, Ladics GS, et al. Immunotoxicogenomics: Potential of genomics technology in immuno-toxicity risk assessment process. Toxicol Sci 2006;94:22-7. 10.1093/toxsci/kfl074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mei N, Fuscoe J, Lobenhofer E, et al. Application of microarray-based analysis of gene expression in the field of toxicogenomics. Methods Mol Biol 2010;597:227-41. 10.1007/978-1-60327-389-3_16 [DOI] [PubMed] [Google Scholar]
- 12.Currie RA. Toxicogenomics: Challenges and opportunities to identify biomarkers, signa-tures, and thresholds to support mode-of-action. Mutat Res 2012;746:97-103. 10.1016/j.mrgentox.2012.03.002 [DOI] [PubMed] [Google Scholar]
- 13.Wilson VS, Keshava N, Hester S, et al. Utilizing toxicogenomic data to understand chemical mechanism of action in risk assessment. Toxicol Appl Pharmacol 2013;271:299-308. 10.1016/j.taap.2011.01.017 [DOI] [PubMed] [Google Scholar]
- 14.Bartosiewicz M, Trounstine M, Barker D, et al. Development of a toxicological gene array and quantitative assessment of this technology. Arch Biochem Biophys 2000;376:66-73. 10.1006/abbi.2000.1700 [DOI] [PubMed] [Google Scholar]
- 15.Bender AT, Wu Y, Cao Q, et al. Assessment of the translational value of mouse lupus models using clinically-relevant biomarkers. Transl Res 2014;163:515-32. 10.1016/j.trsl.2014.01.003 [DOI] [PubMed] [Google Scholar]
- 16.Fu K, Cheng Q, Liu Z, et al. Immunotoxicity assessment of rice-derived recombinant human serum albumin using human peripheral blood mononuclear cells. PLoS One 2014;9:e104426. 10.1371/journal.pone.0104426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rosas LE, Elgamal OA, Mo X, et al. In vitro immunotoxicity assessment of culture-derived extracellular vesicles in human monocytes. J Immunotoxicol 2016;13:652-65. 10.3109/1547691X.2016.1148089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.House RV. An overview of in vitro/ex vivo assays for preclinical evaluation of immunomodulation. Hum Exp Toxicol 2000;19:246-50. 10.1191/096032700678815800 [DOI] [PubMed] [Google Scholar]
- 19.Carneiro MW, Fukutani KF, Andrade BB, et al. Gene expression profile of high IFN-γ producers stimulated with Leishmania braziliensis identifies genes associated with Cutaneous Leishmaniasis. PLoS Negl Trop Dis 2016;10:e0005116. 10.1371/journal.pntd.0005116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang H, Sun J, Li Y, et al. Human umbilical cord-derived mesenchymal stem cells suppress proliferation of PHA-activated lymphocytes in vitro by inducing CD4(+)CD25(high)CD45RA(+) regulatory T cell production and modulating cytokine secretion. Cell Immunol 2016;302:26-31. 10.1016/j.cellimm.2016.01.002 [DOI] [PubMed] [Google Scholar]
- 21.Mohammadi AM, Javadi A, Firooz A, et al. Comparison of cytokine profile of IFN-γ, IL-5 and IL-10 in cutaneous leishmaniasis using PBMC vs. whole blood. Iran J Microbiol 2019;11:431-9. 10.18502/ijm.v11i5.1962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gujer C, Sundling C, Seder R, et al. Human and rhesus plasmacytoid dendritic cell and B-cell responses to Toll-like receptor stimulation. Immunology 2011;134:257-69. 10.1111/j.1365-2567.2011.03484.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mandl JN, Akondy R, Lawson B, et al. Distinctive TLR7 signaling, type I IFN production, and attenuated innate and adaptive immune responses to yellow fever virus in a primate reservoir host. J Immunol 2011;186:6406-16. 10.4049/jimmunol.1001191 [DOI] [PubMed] [Google Scholar]
- 24.Medina M, Couturier J, Feske M, et al. Granzyme B- and Fas ligand-mediated cytotoxic function induced by mitogenic CD28 stimulation of human memory CD4+ T-cells. J Leukocyte Biol 2012;91:759-71. 10.1189/jlb.0511264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kolumam GA, Thomas S, Thompson LJ, et al. Type I interferons act directly on CD8 T cells to allow clonal expansion and memory formation in response to viral infection. J Exp Med 2005;202:637-50. 10.1084/jem.20050821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bostik P, Watkins M, Villinger F, et al. Ansari AA: Genetic analysis of cytokine promoters in nonhuman primates: implications for Th1/Th2 profile characteristics and SIV disease pathogenesis. Clin Dev Immunol 2004;11:35-44. 10.1080/10446670410001670472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Doherty R, O'Farrelly C, Meade KG. Epigenetic regulation of the innate immune response to LPS in bovine peripheral blood mononuclear cells (PBMC). Vet Immunol Immunopathol 2013;154:102-10. 10.1016/j.vetimm.2013.05.004 [DOI] [PubMed] [Google Scholar]
- 28.Gao Y, Flori L, Lecardonnel J, et al. Transcriptome analysis of porcine PBMCs after in vitro stimulation by LPS or PMA-ionomycin using an expression array targeting the pig immune response. BMC Genomics 2010;11:292. 10.1186/1471-2164-11-292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Uddin MJ, Nuro-Gyina PK, Islam MA, et al. Expression dynamics of Toll-like receptors mRNA and cytokines in porcine peripheral blood mononuclear cells stimulated by bacterial lipopolysaccharide. Vet Immunol Immunopathol 2012;147:211-22. 10.1016/j.vetimm.2012.04.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ding Q, Liu G, Zeng Y, et al. Glycogen synthase kinase 3β inhibitor reduces LPS induced acute lung injury in mice. Mol Med Rep 2017;16:6715-21. 10.3892/mmr.2017.7469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Raymond B, Batsche E, Boutillon F, et al. Anthrax lethal toxin impairs IL-8 expression in epithelial cells through inhibition of histone H3 modification. PLoS Pathog 2009;5:e1000359. 10.1371/journal.ppat.1000359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gereda JE, Leung DY, Thatayatikom A, et al. Relation between house-dust endotoxin exposure, Type 1 T-cell development, and allergen sensitiza-tion in infants at high risk of asthma. Lancet 2000;355:1680-3. 10.1016/S0140-6736(00)02239-X [DOI] [PubMed] [Google Scholar]
- 33.Shinoda K, Hirahara K, Nakayama T. Maintenance of pathogenic TH2 cells in allergic disorders. Allergol Int 2017;66:369-76. 10.1016/j.alit.2017.03.005 [DOI] [PubMed] [Google Scholar]
- 34.Eastwood D, Findlay L, Poole S, et al. Monoclonal antibody TGN1412 trial failure explained by species differences in CD28 expression on CD4+ effector memory T-cells. Br J Pharmacol 2010;161:512-26. 10.1111/j.1476-5381.2010.00922.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nam KH, Illés Z, Terao K, et al. Characterization of expanded T-cell clones in healthy macaques: Ontogeny, distribution, and stability. Dev Comp Immunol 2000;24:703-15. 10.1016/S0145-305X(00)00004-5 [DOI] [PubMed] [Google Scholar]
- 36.Lee WW, Nam KH, Terao K, et al. Age-related increase of peripheral CD4+ CD8+ double-positive T lymphocytes in cynomolgus monkeys: longitudinal study in relation to thymic involution. Immunology 2003;109:217-25. 10.1046/j.1365-2567.2003.01646.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Terao K. Essentials for starting a pediatric clinical study (3): Dynamic changes in early development of immune system in macaque monkeys‐‐the significance from standpoint of preclinical toxicity test using nonhuman primates. J Toxicol Sci 2009;34 Suppl 2:P321-5. 10.2131/jts.34.SP321 [DOI] [PubMed] [Google Scholar]
- 38.Ohta S, Shiomi Y, Sugimoto K, et al. A proteomics approach to identify proliferating cell nuclear antigen (PCNA)-binding proteins in human cell lysates. J Biol Chem 2002;277:40362-7. 10.1074/jbc.M206194200 [DOI] [PubMed] [Google Scholar]
- 39.Giacomelli L, Nicolini C. Gene expression of human T-lymphocyte cell cycle: Experimental and bioinformatic analysis. J Cell Biochem 2006;99:1326-33. 10.1002/jcb.20991 [DOI] [PubMed] [Google Scholar]
- 40.Kothe M, Kohls D, Low S, et al. Structure of the catalytic domain of human polo-like kinase 1. Biochemistry 2007;46:5960-71. 10.1021/bi602474j [DOI] [PubMed] [Google Scholar]
- 41.Brochu C, Cabrita MA, Melanson BD, et al. NF-κB-dependent role for cold-inducible RNA binding protein in regulating interleukin 1β. PLoS One 2013;8:e57426. 10.1371/journal.pone.0057426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Caswell JL, Middleton DM, Gordon JR. Production and functional characterization of recombi-nant bovine IL-8 as a specific neutrophil activator and chemoattractant. Vet Immunol Immunopathol 1999;67:327-40. 10.1016/S0165-2427(99)00007-0 [DOI] [PubMed] [Google Scholar]
- 43.Jacquier V, Estellé J, Schmaltz-Panneau B, et al. Genome-wide immunity studies in the rabbit: transcriptome variations in peripheral blood mononuclear cells after in vitro stimulation by LPS or PMA-Ionomycin. BMC Genomics 2015;16:26. 10.1186/s12864-015-1218-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shiina T, Blancher A. The Cynomolgus Macaque MHC Polymorphism in Experimental Medicine. Cells 2019;8:978. 10.3390/cells8090978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Antony JM, MacDonald KS. A critical analysis of the cynomolgus macaque, Macaca fascicularis, as a model to test HIV-1/SIV vaccine efficacy. Vaccine 2015;33:3073-83. 10.1016/j.vaccine.2014.12.004 [DOI] [PubMed] [Google Scholar]
- 46.Florese RH, Wiseman RW, Venzon D, et al. Comparative study of Tat vaccine regimens in Mauritian cynomolgus and Indian rhesus macaques: influence of Mauritian MHC haplotypes on susceptibility/resistance to SHIV(89.6P) infection. Vaccine 2008;26:3312-21. 10.1016/j.vaccine.2008.03.100 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as